Abstract
Dermatomyositis (DM) is a rare, systemic autoimmune disease that most frequently affects the skin, muscles, and lungs. The inflammatory infiltrate in skin has not been fully characterized and in this study we took an single cell, unbiased approach by using Imaging mass cytometry (IMC). Substantial monocyte-macrophage diversity was observed with the CD14+ population correlated positively with cutaneous dermatomyositis disease area and severity index (CDASI) scores (p=0.031). The T cell compartment revealed CD4+ T, CD8+ T, and FOXP3+ T cells. Activated (CD69+) circulating memory T cells correlated positively with CDASI scores (p=0.0268). IFNβ protein was highly upregulated in the T cell, macrophage, dendritic cell, and endothelial cell populations of DM skin. Myeloid DCs (mDCs) expressed pPPARγ, pIRF3, IL4, and IL31 and their quantity correlated with itch as measured in the Skindex-29. Plasmacytoid DCs (pDCs) colocalized with IFNγ in addition to the known colocalization with IFNβ. Nuclear pPPARγ expression was found in the DM endothelium. IMC allows us to characterize single cells in the immune cell population and identify upregulated cytokines and inflammatory pathways in DM. These findings have important implications for the development of future targeted therapies for DM.
INTRODUCTION
Dermatomyositis (DM) is a rare, chronic systemic autoimmune disorder that most frequently affects the skin, muscles, and lungs (Krathen et al. 2008). Cutaneous signs of disease are frequently present on sun-exposed areas such as the v of the neck and upper back/shoulders, and are accentuated on extensor stretch surfaces, including elbows, knees, and joints of the hands (Concha et al. 2019).
Histopathologically, the disease is characterized by a lymphocytic infiltrate at the dermo-epidermal junction, of which most identified cells are CD4+ T cells (Caproni et al. 2004; Hausmann et al. 1991; Wenzel et al. 2006). B cells were typically absent from the inflammatory infiltrate (Hausmann et al. 1991). Plasmacytoid dendritic cells (pDCs) have also been implicated in the disease with increased numbers noted in the skin (McNiff and Kaplan 2008; Wenzel et al. 2006).
The pathogenesis of DM is currently linked to the type I interferon (IFN) system. IFNβ, in particular, appears to correlate closely with the type I IFN signature in blood and skin (Liao et al. 2011; Wong et al. 2012). Further identification of the sources and consequences of these type I IFNs are important as this may yield useful therapeutic targets to modulate disease activity (Crow et al. 2019; Lee and Ashkar 2018).
One would typically need a multitude of markers traditionally used on eluted skin cells through flow cytometry to investigate the highly diverse inflammatory profile in skin. IMC, a novel technique, incorporates flow cytometry principles but preserves the histological and architectural components of the skin for analysis. We screened for potential therapeutic targets by identifying the inflammatory infiltrate in DM compared to healthy control (HC) at a single cell level and correlated these findings with two important outcomes used to clinically assess DM severity: the itch question from the Skindex-29 and the cutaneous dermatomyositis disease area and severity index (CDASI).
RESULTS
DM skin lesions are composed of numerous immune cell populations expressing novel pathways
We identified 13 total cell populations based on expression patterns (Figure 1a). The 13 identified and 5 unidentified clusters (UC, negative for panel markers) for 5 DM (Figure 1b) and 5 HC (Figure 1c) were plotted using the t-distributed stochastic neighbor embedding (tSNE). The average relative composition of immune cells in DM skin lesions was as follows: CD14+ macrophages>CD11c+ mDC>CD14+CD16+ macrophages>CD4+ T Cell>MAC387+ macrophages>mast cells>CD8+ T cells>FOXP3+ T cells>pSTING+ macrophages>BDCA2+ pDC>CD56+ cells>B cells (Figure 1d). The most common cells include macrophages and mDCs, followed by T cells (Figure 1d), with individual patient compositions in Figure 1e. UCs consisted of 24 ± 38.6% (median ± IQR) of the population in patients.
Figure 1.
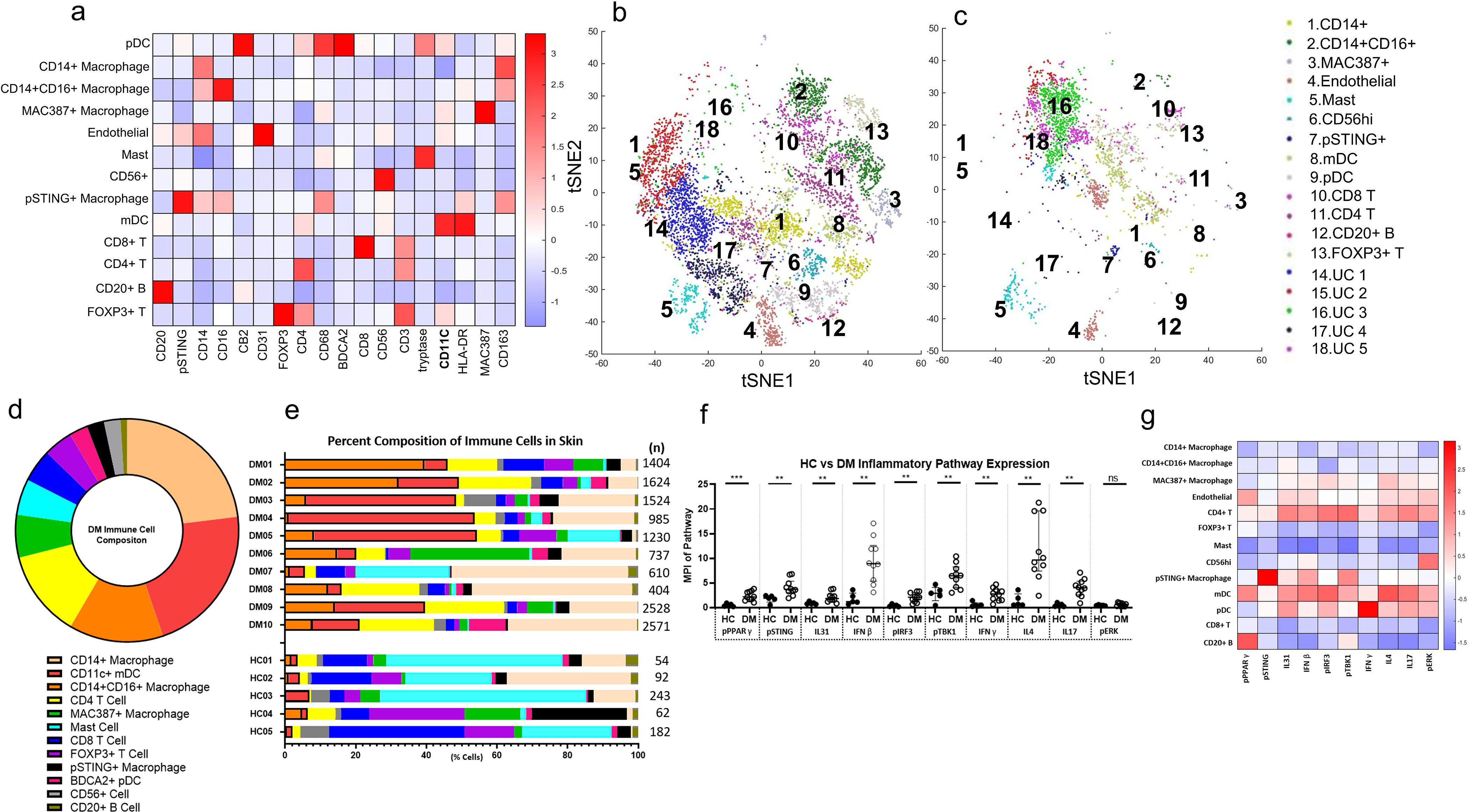
Unbiased Clustering of Immune Cells in DM Skin Lesions using PhenoGraph a) Heatmap of Different Cell Type Markers expressed the 13 cell populations identified: CD14+ macrophages, CD14+CD16+ macrophages, MAC387+ macrophages, CD31+ endothelial cells, CD4+ T cells, FOXP3+ Tregs, tryptase+ mast cells, CD56+ cells, pSTING+ macrophages, CD11c+ mDCs, BDCA2+ pDCs, and CD8+ T cells, and CD20+ B cells. b) tSNE dimensional reduction plot of the different cell clusters identified in lesional DM skin. c) tSNE dimensional reduction plot of the corresponding cell clusters in HC skin. Clusters 14–18 represent unidentified clusters (UC). d) Average percent composition of immune cells identified in DM skin from DM1-DM10 (%): CD14+ macrophage (23.0), CD11C+ mDC (21.7), CD14+CD16+ macrophage (13.7), CD4 T Cell (12.6), MAC387+ macrophage (6.2), mast (5.2), CD8 T Cell (4.8), FOXP3+ T Cell (4.2), pSTING+ macrophage (2.5), BDCA2+ pDC (2.4), CD56+ cells (2.4), and CD20+ B Cell (1.0) Data are represented as mean ± SEM. e) The percent composition of immune cells identified in each DM patient and HC. f) Expression of pPPARγ, pSTING, pIRF3, pTBK1, IFNβ, IFNγ, IL4, IL17, and IL31 is increased in DM skin compared to HC skin. There is no significant difference between DM and HC skin pERK expression. g) Heatmap displaying the relative mean expression of each inflammatory pathway in each cell cluster. Data are represented as median ± IQR. * p<0.05, ** p<0.01, *** p<0.001
The mean pathway marker expression in lesional DM and HC skin was compared for the following: phosphorylated peroxisome proliferator-activated γ (pPPARγ), phosphorylated stimulator of interferon genes (pSTING), phosphorylated interferon factor 3 (pIRF3), phosphorylated TANK binding-kinase 1 (pTBK1), IFNβ, IFNγ, IL4, IL17, IL31, and phosphorylated extracellular signal-related kinase (pERK). All pathway markers except pERK had increased mean pixel intensity (MPI) per cell in DM skin compared to HC skin (Figure 1f, p<0.05). Pathway markers specifically related to the type I IFN system, such as pSTING, pIRF3, pTBK1, and IFNβ were all increased in DM, with the largest difference in expression noted with IFNβ (p=0.0013) and pIRF3 (p=0.0013) (Figure 1f). The pSTING pathway correlated with downstream pTBK1 (Figure S1a, r=0.8788, p=0.0016) and pIRF3 (Figure S1b, r=0.8415, p=0.0035), but did not correlate with overall IFNβ production (Figure S1c, r=0.4134, p=0.2353). Furthermore, IFNβ production did not significantly correlate with any specific cell population; however, a trend was observed with both mDCs (Figures S2a, p=0.1099) and pDCs (Figure S2b, p=0.2337). Of the cytokines examined, IL4 was the most dramatically upregulated in DM (Figure 1f, p=0.0013). It was observed that certain patients expressed very high IL4 compared to others. pPPARγ, a novel pathway marker not previously investigated in the scope of DM, had dramatically increased expression in DM compared to HC skin (Figure 1f, p<0.0007). A heatmap analysis displays relative expression of various pathway markers and intracellular cytokines in the 13 identified cell clusters (Figure 1g). This heatmap gives insight to the pathways and cytokines that are upregulated in each cell cluster, revealing mDC expression of type I IFN pathways and cytokines, suggesting they are highly active in DM.
Monocyte-Macrophage diversity in DM reveals CD14+ macrophages correlating with CDASI
Skin lesions of DM have four distinct monocyte-macrophage populations identified based on the markers we selected, as seen in a multiplexed image (Figure 2a). CD14 single positive macrophages were found to express CD163 but expressed neither CD16 nor MAC87. CD14+CD16+ macrophages also expressed CD163, but had higher HLA-DR expression and were divided into two populations; those that were CD68lo, pSTING− were referred to as CD14+CD16+ macrophages, whereas the CD68hi, pSTING+ cells were considered the unique pSTING+ macrophages (Figure 2c and 2d, respectively). The fourth population of macrophages is MAC387+CD16+CD163− (Figure 2e). Although the pSTING marker exclusively co-localized with the pSTING+ mo-MAC population in the dermis, some pSTING expression was also noted in the lower layers of the epidermis (Figure 2a). Cell counts/region of interest (ROI) were higher for all four populations in DM compared to HC (Figure 2b–e, p<0.05) with the largest difference seen in CD14+CD16+ macrophages (p=0.0007). The CD14 single positive macrophages correlated positively with the Cutaneous Dermatomyositis Disease Area and Severity Index (CDASI), (r=0.697, p=0.031, Figure 2f).
Figure 2.
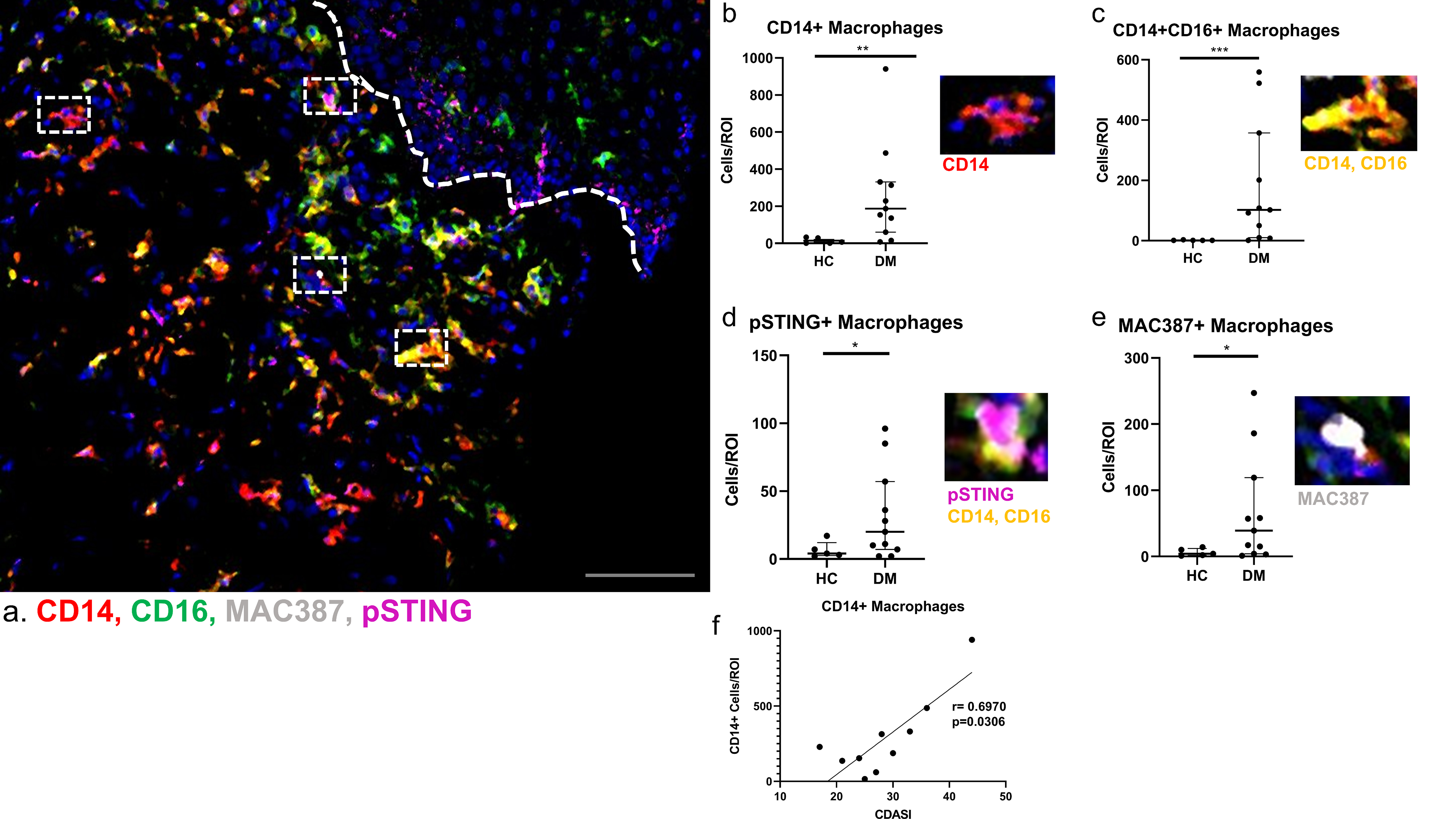
Monocyte-Macrophage diversity in Dermatomyositis Skin
a) Multiplexed image depicting CD14+ (red), CD16+ (green), Nuclei (blue), MAC387+ (white), and pSTING+ (magenta) cells. b-e) In DM skin there are increased numbers of b) CD14+ macrophages (red), c) CD14+CD16+ macrophages (yellow), d) MAC387+ macrophages (white), and e) pSTING+ macrophages (magenta) cells compared to HC skin. Dermal-epidermal junction shown with white dotted line. Representative images from a) shown next to corresponding graphs with nuclei seen with Ir-DNA (blue). f) Correlation of the CDASI scores from cutaneous DM lesions revealed a positive correlation with CD14+ macrophages. Scale bar=120 μm (gray). Data are represented as median ± IQR. * p<0.05, ** p<0.01, *** p<0.001
The T cell compartment in DM skin reveals recently activated CD69+ T cells that express the circulating memory phenotype of CCR7+CD45RA− and correlate with CDASI
There are increased numbers of CD4, CD8, and FOXP3+ T cells in DM compared to HC skin (Figure 3a, p<0.05). The average percent composition of naïve, memory, CD69+ naïve, and CD69+ memory T cells shows that CD4+ and CD8+ memory T cells are the most prevalent in DM (Figure 3a). The CD3+ cells were further evaluated based on their expression of CCR7, CD3, and CD45RA (Figure 3b). We found that CD3+ cells express CCR7; their co-localization is seen in yellow. Most CD3+ cells are also CD45RA− as seen with the lack of co-localization of CD3 (green) and CD45RA (white), supporting a dominant memory T cell infiltrate. Based on this phenotype (CCR7+CD45RA−), we concluded that these are circulating memory T cells. CD69+ T cells were also identified, with CCR7 and CD69 overlapping with CD3 (Figure 3c). Further gating of the T cell population in FlowJo for frequencies of CD69+CD45RA− activated circulating memory cells (TACM) and IL31+ CD4+ T cells revealed positive correlations with CDASI (Figure 3d, TACM: r=0.709, p=0.0268 and Figure 4e, IL31+ CD4+ T: r=0.754, p=0.0151).
Figure 3.
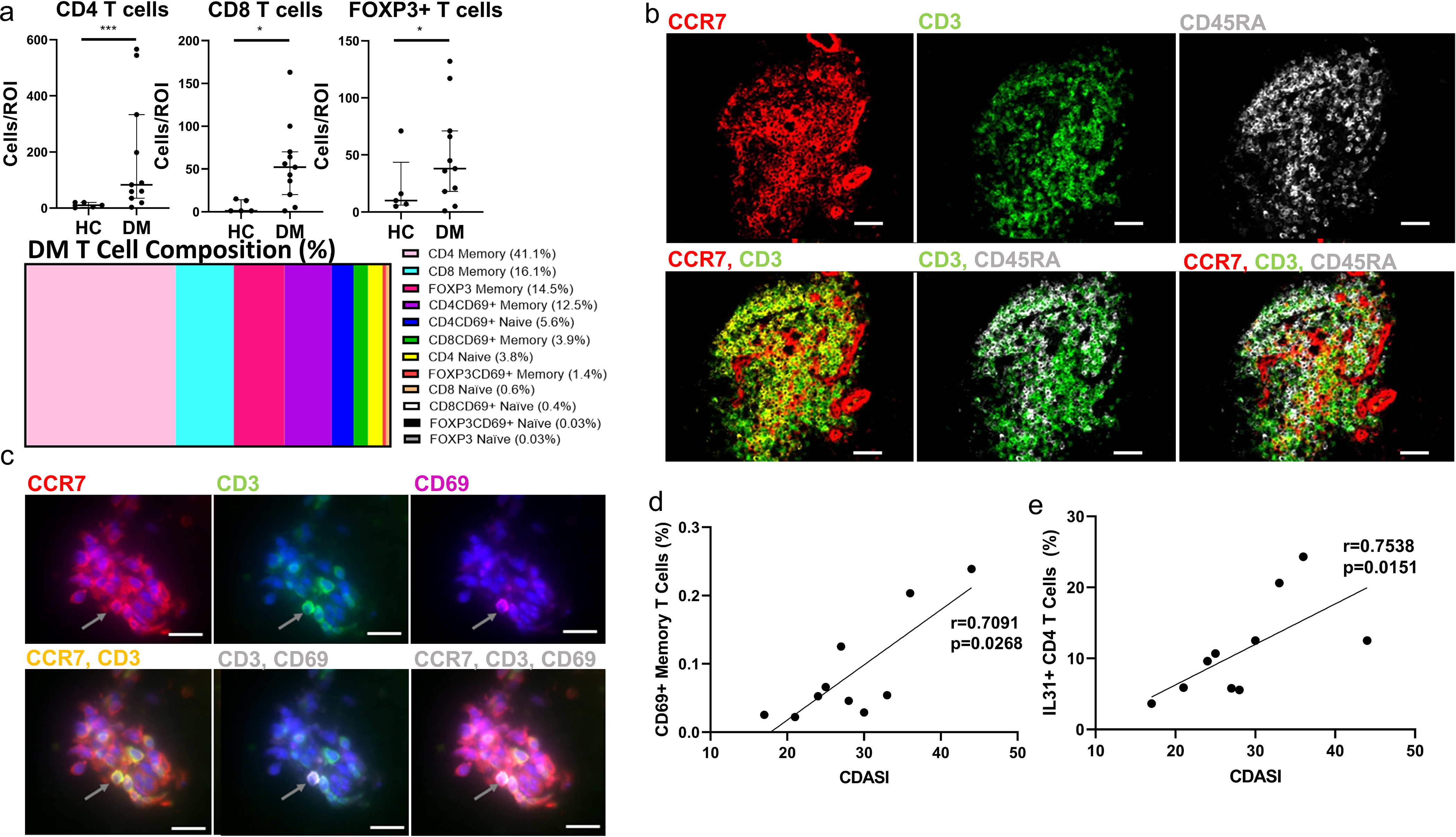
T Cell Distribution in Dermatomyositis skin identifies CD69+ memory T, and IL31+ T cells correlating with CDASI
a) There is an increase in CD4, CD8, and FOXP3+ T cells in DM skin compared to HC, and the average percent composition of T Cells in the colored bar graph identifies predominant memory T cells (Data represented as mean). b) IMC images showing CCR7 (red), CD3 (green), CD45RA (white). Colocalization of CCR7 and CD3 (yellow) reveals circulating T cells and the lack of overlap of CD3 and CD45RA further identifies them as memory T cells. IMC images show most T Cells in DM are CCR7+CD3+CD45RA− and can be characterized as circulating memory T cells. c) Immunofluorescence staining was done to show CD69+ T cells in DM skin CCR7 (red) is seen to colocalize with CD3 (green), and CD69 (magenta) with gray arrows representing the same cell in each panel, and DAPI seen in blue. d-e) Correlation of CDASI scores with different T cell subsets revealed a positive correlation with d) CD69+ Memory T Cells and e) IL31+ CD4 T cells. Scale bars (b)= 60 μm (white), Scale bars (c)= 12 μm (white). Data are represented as median ± IQR. * p<0.05,*** p<0.001
Figure 4.
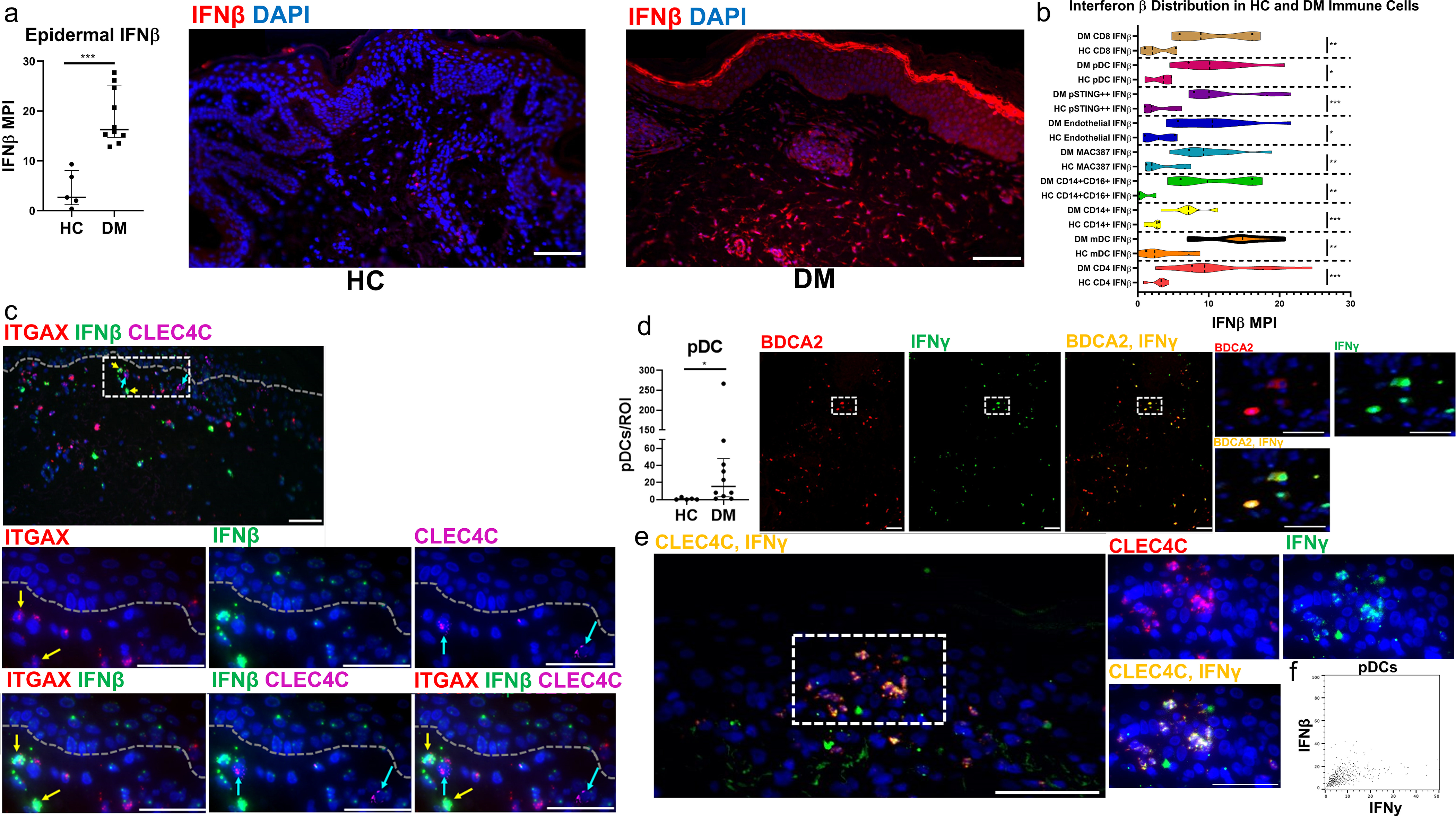
Interferon β and γ expression in immune cells of Dermatomyositis a) Immunofluorescence staining quantification reveals increased Epidermal IFNβ in DM skin compared to HC skin. Immunofluorescence staining reveals little expression of IFNβ in HC skin, and high IFNβ in the epidermis and dermal inflammatory infiltrate of DM skin. b) Violin plot showing the quantification of IFNβ expressed by cells in HC and corresponding cells in DM skin lesions reveals increased IFNβ production by DM CD14+ macrophages, CD8 T cells, pDCs, pSTING+ macrophages, endothelial cells, MAC387+ macrophages, CD14+CD16+ macrophages, mDCs, and CD4 T cells. There is a trend for increased IFNβ by mDCs compared to other inflammatory cells represented. Correlations for IFNβ seen in Figure S2–S3. c) ISH shows mRNA of ITGAX mDCs (CD11C gene, red), IFNβ (green), and CLEC4C pDCs (BDCA2 gene, magenta). ITGAX+ mDCs are seen to colocalize with more IFNβ mRNA compared to CLEC4C+ pDCs. Nuclei depicted by DAPI (a,c). Scale bars (a,c) = 70 μm (white) d) DM skin lesions have increased pDCs compared to HC (p<0.05) IMC of DM skin shows the distribution of BDCA2 (red) and IFNγ (green). Colocalization of BDA2 and IFNγ shows that pDCs are capable of producing IFNγ (yellow). e) ISH shows mRNA colocalization of CLEC4C+ (BDCA2 gene, red) pDCs with IFNγ (green) in yellow further supporting pDC IFNγ production. f) Plot showing pDC expression of high IFNγ but not IFNβ. Nuclei represented with Ir-intercalator (blue, a) and DAPI (blue, b). Data are represented as median ± IQR. Scale bars(d,e) = 100 μm (white, d) and 30 μm (white, e). * p<0.05. ** p<0.01,*** p<0.001
Interferon β is highly upregulated in T cells, macrophages, dendritic cells, and endothelial cells in DM skin while Plasmacytoid dendritic cells express IFNγ
Immunofluorescence (IF) staining of IFNβ in lesional DM skin and HC skin reveals increased epidermal IFNβ in DM (Figure 4a). IF shows minimal IFNβ expression in HC skin (Figure 4a), whereas expression of IFNβ occurs in almost all nucleated DM skin cells (Figure 4a). We stratified IFNβ production by cell population in the dermis and compared production of IFNβ between HC and DM cell populations. Increased IFNβ production was noted in virtually all DM cell populations, including CD8 T cells, pDC, pSTING+ macrophages, endothelial cells, MAC387+ macrophages, CD14+CD16+ macrophages, CD14+ macrophages, mDCs, and CD4 T cells, compared to the corresponding HC populations.(Figure 4b, p<0.05). Of these cell populations, mDCs and pSTING+ macrophages were observed to have the highest expression of IFNβ, followed by CD4 T cells, suggesting mDC importance in the type I IFN system (Figure 4b). The MPI of total IFNβ production correlated with individual cell population IFNβ production for all immune cells, but did not correlate for endothelial cells (Figure S3); the most significant IFNβ MPI correlations occurred with mDCs (Figure S3b, r=0.936, p=0.0002) and CD14+CD16+ macrophages (Figure S3e, r=0.9483, p=0.0001), suggesting a greater contribution to the overall IFNβ signature or regulation by the type I IFN system. In situ hybridization (ISH) for Integrin alpha X (ITGAX), IFNβ, and C-Type lectin domain family 4 member (CLEC4C) mRNA is shown in Figure 4c). Similar to IMC, this revealed increased IFNβ localization with ITGAX+ mDCs compared to CLEC4C+ pDCs (Figure 4c).
There are increased number of pDCs in DM compared to HC skin (p=0.015) (Figure 4d). Based on the heatmap of expressions, we found that pDCs had the highest relative IFNγ production when compared to other cell populations with a median of 44.93 ± 56.67 % IFNγ+ pDCs (Figure 1g). To confirm this, we isolated IMC images and found that BDCA2+ pDCs colocalized with IFNγ as shown in yellow (Figure 4d). ISH of mRNA also supported these results and showed CLEC4C+ pDCs overlapping with IFNγ as shown in yellow (Figure 4e). Figure 4f shows via IMC that pDCs produce high IFNγ but not IFNβ, which is a relatively novel concept and may be unique to DM.
Myeloid dendritic cells in DM skin may produce IL4 and IL31
The distribution of IL4 amongst different cell types shows a trend that mDCs may be the major producers (Figure 5a). We further investigated the contribution of mDCs to the itch pathway in DM by examining their IL4-mediated IL31 production and found colocalization of IL4 and IL31 in CD11c+ mDCs, all co-labeled (Figure 5b). ISH of mRNA again supported our findings, as IL4 and IL31 colocalized with ITGAX+ mDCs (Figure 5c). To quantify IL4+ and IL31+ mDCs and evaluate their role in pruritus, single cell data was imported, gated in FlowJo (Figure S4a–b), and correlated with the Skindex-29 itch question. The Skindex-29 itch question correlated with the percent of IL4+ mDCs (Figure 5d, r=0.6547, p=0.0448) and percent of IL31+ mDCs (Figure 5e, r=0.7310, p=0.0194). Notably, the overall IL31 protein also correlated with IL4 protein (Figure S4c, r=0.8774, p=0.0016). At a single cell level, the overall IL31 protein correlated with mDC IL31 (Figure S4d, r=0.7915, p=0.0091) and mDC IL4 (Figure S4f, r=0.6831, p=0.0388). Myeloid DC production of IL31 and IL4 also correlated positively (Figure S4f, r=0.8075, p=0.0047). Total single cell correlations suggest IL4 and IL31 correlate with each other in numerous cell populations (Figure S4g–h). In addition, IL-31 correlates with IFNβ but not to the same extent with IFNγ (Figure S4g).
Figure 5.
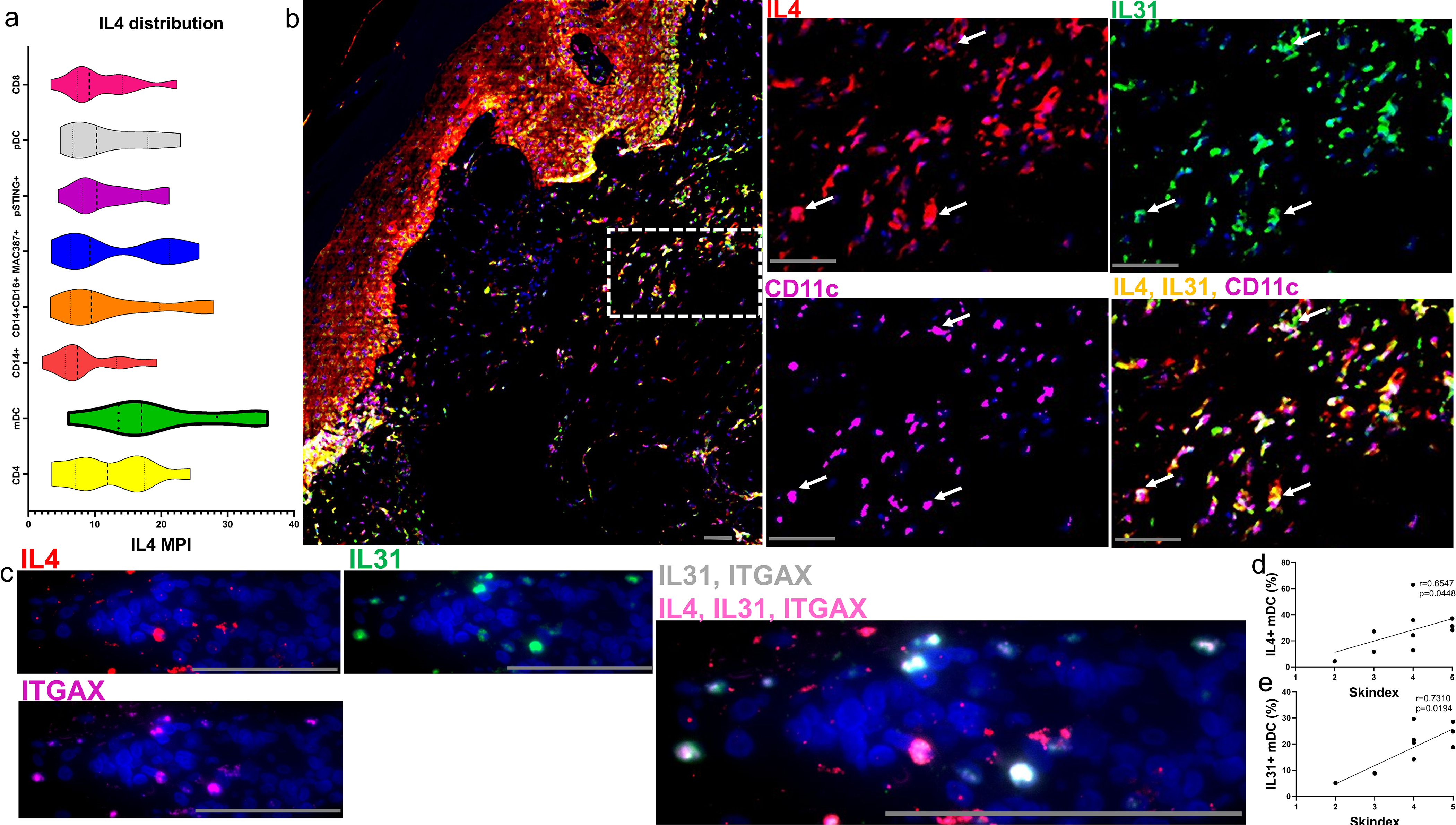
CD11C+ mDCs co-express IL4, IL31 and correlate with the Skindex-29 itch score a) The distribution of IL4 amongst different cell types in DM skin shows a trend for mDCs being the major producers followed by CD4 T Cells. b) IMC of a DM skin lesions reveals an image of IL4(red), IL31(green), and CD11C (magenta). Corresponding dotted region from (b) shown at a higher magnification with IL4(red), IL31(green), and CD11C (magenta). Colocalization of IL4 and IL31 can be seen (yellow) and overlaps with CD11C+ cells (magenta) with white arrows highlighting corresponding mDCs. c) ISH images of mRNA similarly show IL4+ (red), IL31+ (green), and ITGAX+ mDCS (CD11C gene, magenta) with IL4+, IL31+, and IL4+IL31+ mDCs. Single positive IL4 mDCs are seen by red arrows, single positive IL31 mDCs by green arrows and double positive IL4+IL31+ mDCs by yellow arrows. d-e) Correlation with the Skindex-29 itch score showed a positive correlation for the percent of IL4+ mDCs (d) and percent of IL31+ mDCs (e). Nuclei represented with Ir-intercalator (b, blue) and DAPI (c). Scale bars (b-c, gray)= 60 μm. Data are represented as median ± IQR. See Figure S4
Neighborhood Analysis of immune cells identifies positive mDC interactions
Neighborhood analysis was done using histoCAT to identify significant cell interactions in DM (Schapiro et al. 2017). A typical image displays that mDCs interact with CD3+ T cells, whereas pDCs do not interact with T cells based on their relative proximity (Figure 6a). A heatmap for each cell interaction is displayed in Figure 6b with red signifying positive (present) interactions (p<0.05) and blue representing negative (avoidant) interactions (p<0.05). The heatmap shows that mDCs interact with CD4 T cells, CD14+CD16+ macrophages, CD8 T cells, CD56 (NCAM) hi cells, and FOXP3+ T cells. The pDCs did not appear to significantly interact with T cells, although did associate with the endothelium and pSTING+ macrophages.
Figure 6.
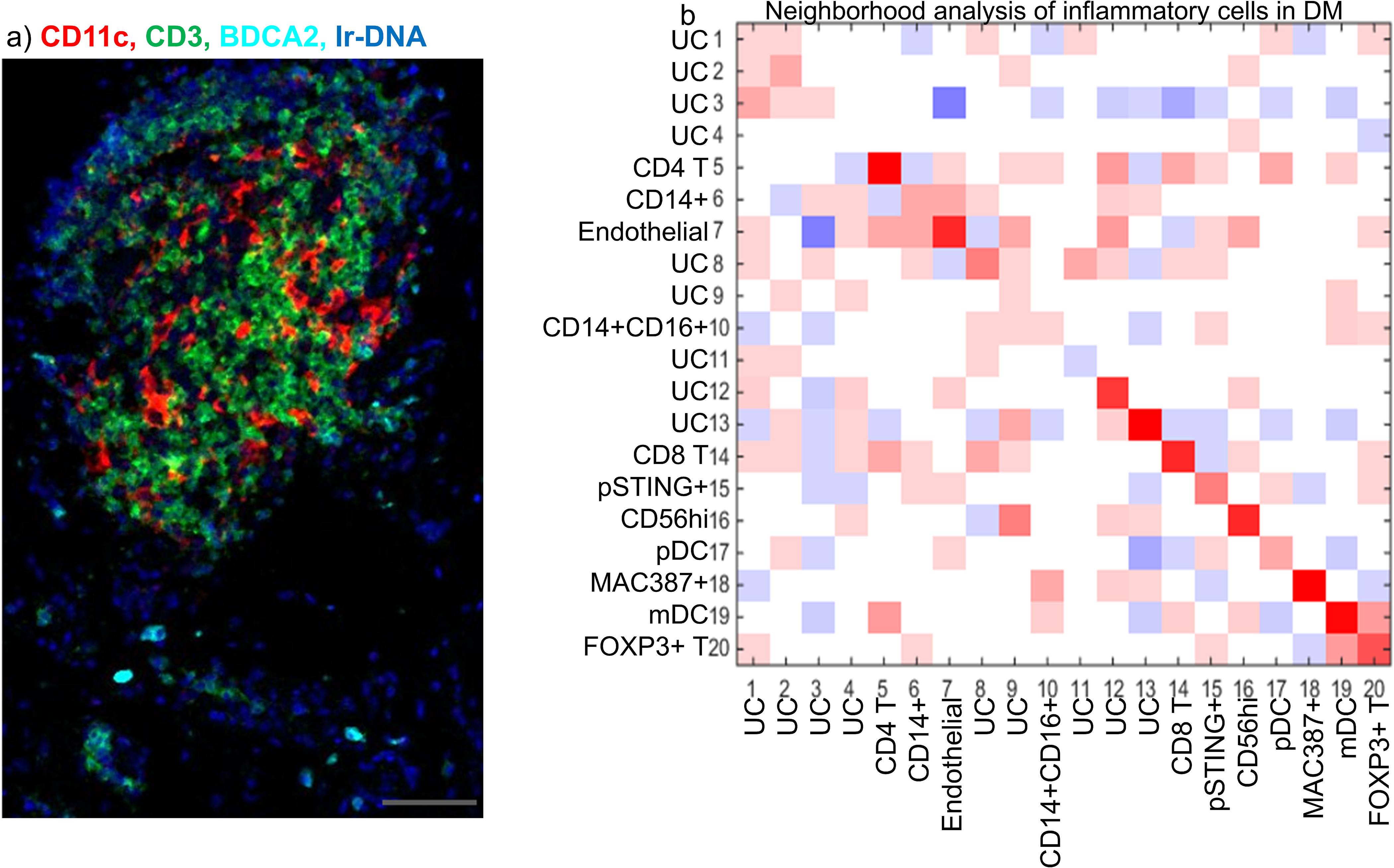
Neighborhood analysis of Immune cells in DM identified mDC interactions with T cells, macrophages, and CD56+ cells
a) IMC allowed for the visualization of different immune cells in DM skin and their spatial relationship to each other. CD11C+ mDCs (red) are seen to be intertwined with CD3+ T cells (green) whereas BDCA2+ pDCs are scattered throughout the tissue (cyan). b) Neighborhood analysis of n=7 DM lesional skin biopsies using histoCAT (b) reveals cell to cell interactions displayed as a heatmap with red representing a positive (neighbored) association (p<0.05), white as an insignificant association (p>0.05), and blue as a negative (avoided) association (p<0.05). Rows signify cells surrounding a cell type of interest. Columns signify the cell type of interest surrounding other cell types. The heatmap shows mDCs may interact with CD4 T, CD14+CD16+ macrophages, CD8 T, CD56(NCAM) hi cells, and FOXP3+ T cells while pDCS may interact more with the endothelial cells and pSTING+ macrophages. Scale bar (a) = 60 μm (gray).
Sources of increased pPPARγ in DM: the endothelium, mDCs, and B cells
The expression of pPPARγ was found to be the most significantly different pathway when comparing HC and DM (Figure 1f). Based on the heatmap analysis, pPPARγ expression is high in endothelial cells, mDCs, and B cells (Figure 1g). We quantified pPPARγ expression using MPI and found greater expression in the DM endothelium than in the HC endothelium (p=0.008) (Figure S5a). The marker for nuclear pPPARγ and endothelial marker CD31 colocalized, showing nuclear pPPARγ expression inside the CD31 surface-stained endothelial cell (Figure S5a). There are increased mDCs in DM skin compared to HC skin (p=0.001) that express increased pPPARγ (p=0.005) (Figure S5b). Colocalization of pPPARγ and CD11c+ mDCs is seen in yellow (Figure S5b). Similarly, B cells were also increased in DM skin compared to HC skin (p=0.003) (Figure S5c) and also expressed significantly increased pPPARγ compared to HC (p=0.017). Colocalization of pPPARγ within CD20+ B cells can be seen in Figure S5c.
DISCUSSION
The current study allows an in-depth analysis of the DM inflammatory infiltrate not previously possible. Contrary to previous findings that the DM infiltrate is mainly composed of CD4+ lymphocytes, we found CD14+ macrophages to be the most populous, followed by CD11c+ mDCs and CD14+CD16+ macrophages. We attribute the previous finding of a primarily CD4+ lymphocytic infiltrate to the expression of CD4 by non T-cells such as macrophages and mDCs. We identified four different monocyte-macrophage populations; this diversity and abundance of monocytes and macrophages in DM may be attributed to the type I IFN response, as monocyte recruitment occurs in a type I IFN-dependent fashion (Ramos et al. 2017; Seo et al. 2011). The potential therapeutic importance of monocyte/macrophage recruitment is further highlighted by our recent findings, as we show that the CD14+ monocyte/macrophage population correlates with disease activity.
The T cell compartment in DM skin reveals a CD4, CD8, and FOXP3+ infiltrate with predominantly memory T cells. IMC images reveal CCR7+CD45RA− circulating memory T cells that may have implications in systemic disease as a migratory pro-inflammatory population. The CD69+ memory cells that correlate with CDASI may be of importance in DM pathogenesis, as CD69 is an early activation marker which programs T cells to a memory phenotype and has been associated with tissue residence and chronic, refractory inflammation (Cibrián and Sánchez-Madrid 2017; Osborn et al. 2019). IL31+ CD4 T cells also correlated positively with CDASI, suggesting that they also contribute to disease severity. Other populations analyzed did not show correlations with CDASI.
CD8 cytotoxic T cells have predominantly been found in the muscles, but we show their presence in skin that may contribute to apoptosis, but requires investigation (Fujiyama et al. 2013). An increase in FOXP3+ cells may be reactive to inflammation, dysfunctional, or possibly be activated T cells themselves (Angerami et al. 2017; Bonelli et al. 2014; Horwitz 2010). Further work is needed to identify the properties of these FOXP3+ cells and to identify their role in DM as well as possible defects in suppressive function.
It is important to note post-translational modifications, such as phosphorylation, when interpreting protein results. With regards to pPPARγ, we focused on deactivated PPARγ that had been phosphorylated at Ser273 (Brunmeir and Xu 2018; Diradourian et al. 2005). Active PPARγ (non-phosphorylated) may promote co-inhibitory molecules such as B7H1, resulting in CD4 anergy (Klotz et al. 2009; Klotz et al. 2007; Nencioni et al. 2002). The increased amounts of inactivated pPPARγ observed in DM, thus, may cause effects opposite to those of activated PPARγ and may assist mDC function (Ishikawa et al. 2007; Mishra 2017; Le Naour et al. 2001; Zaccagnino et al. 2012). Furthermore, the increased pPPARγ in DM endothelial cells compared to the HC endothelium highlights a possible role in regulating the perivascular infiltrate, as several studies have observed decreased PPARγ resulting in the increased recruitment of inflammatory cells via leukocyte-endothelial interactions (Jackson et al. 1999; Marx et al. 2000; Pasceri et al. 2000). For this reason, PPARγ agonists such as Lenabasum may successfully reduce the migration of inflammatory cells seen in DM (Stasiulewicz et al. 2020; Werth et al. 2018).
Recent research has highlighted the role of the STING pathway in stimulating the type I IFN response in systemic lupus erythematosus (SLE), a disease with similar pathological characteristics to DM (Elkon 2018; Kato et al. 2018; Thim-uam et al. 2019). Here we report increased pSTING expression in DM skin compared to HC skin, with the sources being lower epidermal cells and dermal macrophages. The major source of STING production in SLE was found to be peripheral monocytes, and the main source of STING in DM was the closely related monocyte-derived macrophage, highlighting similarities in disease pathogenesis (Murayama et al. 2020). Downstream STING pathways pTBK1 and pIRF3 were also upregulated in DM skin compared to HC skin, suggesting propagation of the pre-type I IFN signal.
IFNβ expression is abundant in all cells present in the epidermis and dermis. Supporting this observation, there are studies showing that almost any nucleated cell can produce this antiviral response (Kalliolias and Ivashkiv 2010). Conventionally pDCs are thought to be the major producers of type I IFN, but similar to others, we show that monocyte/macrophages, mDCs, and T cells can also produce type I IFNs (Azuma et al. 2012; Jelinek et al. 2011; Larkin et al. 2017; Tatematsu et al. 2013). The correlations shown between cellular IFNβ and total IFNβ suggest a possible common stimulus. Type I IFNs can be produced through a variety of pattern recognition, damage recognition, toll-like receptors, and intracellular DNA/RNA receptors. This pathway is unique in that many of the regulatory transcription molecules are regulated by IFN itself creating autocrine amplification loops (Miettinen et al. 2001; Sato et al. 1998). This may explain the correlations as IFNβ production by one cell type may augment another, and itself thereby propagate the signal in regions of clustering cells.
Of the inflammatory cells, mDCs seem to interact the most with other immune cells, are the major producers of IFNβ, and have upstream activation of IRF3 in mDCs. They are a promising target as they may augment the immune response via antigen presentation and type I IFN production.
IFNγ is an important cytokine mainly thought to be produced by Th1 cells, however we show pDC expression. IFNγ+ pDCs have been reported previously in a murine model, but not frequently in humans (Suto et al. 2005). These IFNγ+ pDCs may be alternatively activated or exhausted thereby transitioning to a type II vs type I IFN response, as has been recently suggested in autoimmunity (Psarras et al. 2020). Further research to characterize these pDCs may help identify functional plasticity between diseases.
Increased IL4 mRNA has been previously reported in DM skin and muscle, and may augment the pro-itch cytokine IL31 (Giriş et al. 2017; Kim et al. 2018). We found mDCs to be a major source of IL4, which has been rarely described (Maroof et al. 2006). IL4 in mDCs may relate to pruritus, as IL4+ mDCs also overlapped with IL31, suggesting a common source and potential synergy of both cytokines. Both IL4+mDCs and IL31+ mDCs correlated positively with the Skindex itch question, and spatial information may suggest mDCs interacted positively with CD56+ hi cells which may include NCAM+ neurons capable of transmitting itch signals.
This initial study is limited by a small sample and overlap between cell markers and secreted proteins due to the relatively low resolution of IMC also exists. To best address this, we corroborated our findings using IF and ISH while conducting user-guided data analysis in order to compare findings with actual staining patterns. UC cell clusters were also not included in analysis; however, these may represent cell types for which we did not have adequate markers such as fibroblasts, follicular cells, and glandular cells.
Nonetheless, we provide a comprehensive summary of the adaptive immune system response and phosphorylated pathways in DM skin tissue by taking a true image mass cytometry approach with segmentation, single cell data extraction, and unsupervised clustering from skin, highlighting its use in dermatologic research. In summary, this study identifies previously unknown inflammatory profiles for the cells in DM via IMC and highlights the use of this technology for therapeutic discovery in dermatologic research.
MATERIALS AND METHODS
Patients
All patients and healthy controls were recruited from the Department of Dermatology at the University of Pennsylvania Hospital with appropriate Institutional Review Board approval. All subjects signed written informed consent before participating in this study.
Patients were diagnosed with DM using Bohan and Peter criteria, Sontheimer criteria, or investigator expert experience and subsequently placed in a longitudinal DM database. Lesional skin biopsies were obtained from 10 patients with newly diagnosed moderate-severe DM and 5 healthy controls. The CDASI score was determined for each DM patient at the time of skin biopsy. DM patients were also given the Skindex-29; this study used question 10 from the Skindex to quantify patient reported itch on a scale of 1 (never) to 5 (all the time).
Immunofluorescence staining
RNA In Situ Hybridization
In situ detection of ITGAX, IFNβ, CLEC4C, IL4, IL31, and IFNγ mRNA transcripts was done using the RNAscope kit and probes (Acd Bio, Newark, CA). See supplementary methods.
Image Mass Cytometry
Antibody Staining and Image acquisition
Prior to processing of tissue, antibodies were conjugated to different metal isotopes using the Maxpar Antibody Labeling Kit (Fluidigm, San Francisco, CA). Tissue sections were incubated in a cocktail of 32 metal conjugated antibodies and processed with the Hyperion Imaging System (Fluidigm, San Francisco, CA). Regions of interest (ROI) up to 2mm x 1mm were ablated at a frequency of 200Hz. (see supplementary methods)
Image processing and data analysis
Images were segmented as shown in Figure S5 (see supplementary methods). The resulting mask and image TIFF files were then imported into histoCAT where per object mean pixel intensity (MPI) data was generated. The Phenograph algorithm was used for unsupervised clustering of cell populations using markers that had the best signal to noise ratio: pSTING, CD14, CD16, CD31, FOXP3, CD4, CD68, CD8, CD45RA, CD3, tryptase, CD11C, HLA-DR, MAC387, and CD163 with a second round of clustering to resolve under-clustered populations with merging phenotypes. CD20+ B cells were manually gated in FlowJo (FlowJo Ashland, Oregon). Further subset gating for T cells was also done using CD69 and CD45RA using gate boundaries based on the corresponding image pixel values for the respective channels. Similarly, IL4 and IL31 positive mDC populations were gated in FlowJo (Figure S4). MPI for different cell populations and related heatmaps were gathered using histoCAT. Colocalization images were created using ImageJ and sliding scale pixel threshold values based corresponding marker signal/noise ratio.
Quantification and Statistical Analysis
Statistical analysis was done using GraphPad Prism v8.3 (GraphPad, San Diego, CA). Non-parametric tests were used to report all data as median, except for average percent composition of cell populations in Figure 1d and Figure 3a. The Spearman rank correlation test was used to correlate CDASI and Skindex scores with cell counts and pathways. The Mann-Whitney test was used to compare the HC group with the DM group with p<0.05 being significant.
Supplementary Material
Funding/Support:
R21 AR066286 and R01 AR076766. This work was supported by the United States Department of Veterans Affairs (Veterans Health Administration, Office of Research and Development and Biomedical Laboratory Research and Development).
Abbreviations:
- DM
dermatomyositis
- mDC
myeloid dendritic cell
- pDC
plasmacytoid dendritic cell
- IFN
interferon
- CDASI
cutaneous dermatomyositis disease area and severity index
- UC
unidentified clusters
- tSNE
t-distributed stochastic neighbor embedding
- pPPARγ
phosphorylated peroxisome proliferator-activated γ
- pSTING
phosphorylated stimulator of interferon genes
- pIRF3
phosphorylated interferon factor 3
- pTBK1
phosphorylated TANK binding-kinase 1
- pERK
phosphorylated extracellular signal-related kinase
- MPI
Mean pixel intensity
- TACM
activated circulating memory cells
- ISH
In situ hybridization
- ITGAX
Integrin alpha X
- CLEC4C
C-Type lectin domain family 4 member
Footnotes
Conflict of Interest:
The authors state no conflict of interest
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data availability statement
No datasets were generated for this submission.
REFERENCES
- Angerami MT, Suarez GV., Vecchione MB, Laufer N, Ameri D, Ben G, et al. Expansion of CD25-negative forkhead Box P3-positive T cells during HIV and mycobacterium tuberculosis infection. Front. Immunol 2017;8:528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma M, Ebihara T, Oshiumi H, Matsumoto M, Seya T. Cross-priming for antitumor ctl induced by soluble ag + polyi: C depends on the ticam-1 pathway in mouse cd11c+/CD8α+ dendritic cells. Oncoimmunology 2012;1:581–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonelli M, Göschl L, Blüml S, Karonitsch T, Steiner CW, Steiner G, et al. CD4+CD25-Foxp3+ T cells: A marker for lupus nephritis? Arthritis Res. Ther 2014;16:R104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunmeir R, Xu F. Functional regulation of PPARs through post-translational modifications. Int. J. Mol. Sci 2018;19:1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caproni M, Torchia D, Cardinali C, Volpi W, Del Bianco E, D’Agata A, et al. Infiltrating cells, related cytokines and chemokine receptors in lesional skin of patients with dermatomyositis. Br. J. Dermatol 2004;151:784–91. [DOI] [PubMed] [Google Scholar]
- Cibrián D, Sánchez-Madrid F. CD69: from activation marker to metabolic gatekeeper. Eur. J. Immunol 2017;47:946–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concha JSS, Tarazi M, Kushner CJ, Gaffney RG, Werth VP. The diagnosis and classification of amyopathic dermatomyositis: a historical review and assessment of existing criteria. Br. J. Dermatol 2019;180:1001–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow MK, Olferiev M, Kirou KA. Type i Interferons in Autoimmune Disease. Annu. Rev. Pathol. Mech. Dis 2019;14:369–93. [DOI] [PubMed] [Google Scholar]
- Diradourian C, Girard J, Pégorier JP. Phosphorylation of PPARs: From molecular characterization to physiological relevance. Biochimie 2005;87:33–8. [DOI] [PubMed] [Google Scholar]
- Elkon KB. Cell Death, Nucleic Acids, and Immunity: Inflammation Beyond the Grave. Arthritis Rheumatol 2018;70:805–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiyama T, Ito T, Tokura Y, Hashizume H. Tug-of-war between IFN-γ-producing CD8+T cells and IL-4-producing CD4+T cells destines the severity of muscle injury in dermatomyositis. J. Dermatol. Sci 2013;69:e11 [Google Scholar]
- Giriş M, Durmuş H, Yetimler B, Taşli H, Parman Y, Tüzün E. Elevated IL-4 and IFN-γ levels in muscle tissue of patients with dermatomyositis. In Vivo (Brooklyn) 2017;31:657–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausmann G, Herrero C, Cid MC, Casademont J, Lecha M, Mascaro JM. Immunopathologic study of skin lesions in dermatomyositis. J. Am. Acad. Dermatol 1991;25:225–30. [DOI] [PubMed] [Google Scholar]
- Horwitz DA. Identity of mysterious CD4+CD25-Foxp3+cells in systemic lupus erythematosus. Arthritis Res. Ther 2010;12:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa F, Niiro H, Iino T, Yoshida S, Saito N, Onohara S, et al. The developmental program of human dendritic cells is operated independently of conventional myeloid and lymphoid pathways. Blood 2007;110:3591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SM, Parhami F, Xi XP, Berliner JA, Hsueh WA, Law RE, et al. Peroxisome proliferator-activated receptor activators target human endothelial cells to inhibit leukocyte-endothelial cell interaction. Arterioscler. Thromb. Vasc. Biol 1999;19:2094–104. [DOI] [PubMed] [Google Scholar]
- Jelinek I, Leonard JN, Price GE, Brown KN, Meyer-Manlapat A, Goldsmith PK, et al. TLR3-Specific Double-Stranded RNA Oligonucleotide Adjuvants Induce Dendritic Cell Cross-Presentation, CTL Responses, and Antiviral Protection. J. Immunol 2011;186:2422–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalliolias GD, Ivashkiv LB. Overview of the biology of type I interferons. Arthritis Res. Ther 2010;12:S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Park J, Takamatsu H, Konaka H, Aoki W, Aburaya S, et al. Apoptosis-derived membrane vesicles drive the cGAS-STING pathway and enhance type i IFN production in systemic lupus erythematosus. Ann. Rheum. Dis 2018;77:1507–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Zeidi M, Bonciani D, Pena SM, Tiao J, Sahu S, et al. Itch in dermatomyositis: the role of increased skin interleukin-31. Br. J. Dermatol 2018;179:669–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotz L, Dani I, Edenhofer F, Nolden L, Evert B, Paul B, et al. Peroxisome Proliferator-Activated Receptor γ Control of Dendritic Cell Function Contributes to Development of CD4 + T Cell Anergy. J. Immunol 2007;178:2122–31. [DOI] [PubMed] [Google Scholar]
- Klotz L, Hucke S, Thimm D, Classen S, Gaarz A, Schultze J, et al. Increased Antigen Cross-Presentation but Impaired Cross-Priming after Activation of Peroxisome Proliferator-Activated Receptor γ Is Mediated by Up-Regulation of B7H1. J. Immunol 2009;183:129–36. [DOI] [PubMed] [Google Scholar]
- Krathen M, Fiorentino D, Werth V. Dermatomyositis. Curr. Dir. Autoimmun 2008;10:313–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin B, Ilyukha V, Sorokin M, Buzdin A, Vannier E, Poltorak A. Cutting Edge: Activation of STING in T Cells Induces Type I IFN Responses and Cell Death. J. Immunol 2017;199:397–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AJ, Ashkar AA. The dual nature of type I and type II interferons. Front. Immunol 2018;9:2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao AP, Salajegheh M, Nazareno R, Kagan JC, Jubin RG, Greenberg SA. Interferon β is associated with type 1 interferon-inducible gene expression in dermatomyositis. Ann. Rheum. Dis 2011;70:831–6. [DOI] [PubMed] [Google Scholar]
- Maroof A, Penny M, Kingston R, Murray C, Islam S, Bedford PA, et al. Interleukin-4 can induce interleukin-4 production in dendritic cells. Immunology 2006;117:271–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx N, Mach F, Sauty A, Leung JH, Sarafi MN, Ransohoff RM, et al. Peroxisome Proliferator-Activated Receptor-γ Activators Inhibit IFN-γ-Induced Expression of the T Cell-Active CXC Chemokines IP-10, Mig, and I-TAC in Human Endothelial Cells. J. Immunol 2000;164:6503–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNiff JM, Kaplan DH. Plasmacytoid dendritic cells are present in cutaneous dermatomyositis lesions in a pattern distinct from lupus erythematosus. J. Cutan. Pathol 2008;35:452–6. [DOI] [PubMed] [Google Scholar]
- Miettinen M, Sareneva T, Julkunen I, Matikainen S. IFNs activate toll-like receptor gene expression in viral infections. Genes Immun 2001;2:349–55. [DOI] [PubMed] [Google Scholar]
- Mishra A. Metabolic Plasticity in Dendritic Cell Responses: Implications in Allergic Asthma. J. Immunol. Res 2017;2017:5134760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama G, Chiba A, Kuga T, Makiyama A, Yamaji K, Tamura N, et al. Inhibition of mTOR suppresses IFNα production and the STING pathway in monocytes from systemic lupus erythematosus patients. Rheumatology 2020;59:2992–3002. [DOI] [PubMed] [Google Scholar]
- Le Naour F, Hohenkirk L, Grolleau A, Misek DE, Lescure P, Geiger JD, et al. Profiling Changes in Gene Expression during Differentiation and Maturation of Monocyte-derived Dendritic Cells Using Both Oligonucleotide Microarrays and Proteomics. J. Biol. Chem 2001;276:17920–31. [DOI] [PubMed] [Google Scholar]
- Nencioni A, Grünebach F, Zobywlaski A, Denzlinger C, Brugger W, Brossart P. Dendritic Cell Immunogenicity Is Regulated by Peroxisome Proliferator-Activated Receptor γ. J. Immunol 2002;169:1228–35. [DOI] [PubMed] [Google Scholar]
- Osborn JF, Hobbs SJ, Mooster JL, Khan TN, Kilgore AM, Harbour JC, et al. Central memory CD8+ T cells become CD69+ tissue-residents during viral skin infection independent of CD62l-mediated lymph node surveillance. PLoS Pathog 2019;15:e1007633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psarras A, Alase A, Antanaviciute A, Carr IM, Md Yusof MY, Wittmann M, Emery P, Tsokos GC, Vital EM. Functionally impaired plasmacytoid dendritic cells and non-haematopoietic sources of type I interferon characterize human autoimmunity. Nat Commun 2020;11:6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasceri V, Wu HD, Willerson JT, Yeh ETH. Modulation of vascular inflammation in vitro and in vivo by peroxisome proliferator-activated receptor-γ activators. Circulation 2000;101:235–8. [DOI] [PubMed] [Google Scholar]
- Ramos JMP, Bussi C, Gaviglio EA, Arroyo DS, Baez NS, Rodriguez-Galan MC, et al. Type I IFNs are required to promote central nervous system immune surveillance through the recruitment of inflammatory monocytes upon systemic inflammation. Front. Immunol 2017;8:1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Hata N, Asagiri M, Nakaya T, Taniguchi T, Tanaka N. Positive feedback regulation of type I IFN genes by the IFN-inducible transcription factor IRF-7. FEBS Lett 1998;441:106–10. [DOI] [PubMed] [Google Scholar]
- Schapiro D, Jackson HW, Raghuraman S, Fischer JR, Zanotelli VRT, Schulz D, et al. HistoCAT: Analysis of cell phenotypes and interactions in multiplex image cytometry data. Nat. Methods 2017;14:873–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo SU, Kwon HJ, Ko HJ, Byun YH, Seong BL, Uematsu S, et al. Type I interferon signaling regulates Ly6Chi monocytes and neutrophils during acute viral pneumonia in mice. PLoS Pathog 2011;7:e1001304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasiulewicz A, Znajdek K, Grudzień M, Pawinski T, Sulkowska JI. A guide to targeting the endocannabinoid system in drug design. Int. J. Mol. Sci 2020;21:2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suto A, Nakajima H, Tokumasa N, Takatori H, Kagami S, Suzuki K, et al. Murine Plasmacytoid Dendritic Cells Produce IFN-γ upon IL-4 Stimulation. J. Immunol 2005;175:5681–9. [DOI] [PubMed] [Google Scholar]
- Tatematsu M, Nishikawa F, Seya T, Matsumoto M. Toll-like receptor 3 recognizes incomplete stem structures in single-stranded viral RNA. Nat. Commun 2013;4:1833. [DOI] [PubMed] [Google Scholar]
- Thim-uam A, Prabakaran T, Tansakul M, Makjaroen J, Wongkongkathep P, Chantaravisoot N, et al. Inhibition of Sting rescues lupus disease by the regulation of Lyn-mediated dendritic cell differentiation. bioRxiv 2019;810192. [Google Scholar]
- Wenzel J, Schmidt R, Proelss J, Zahn S, Bieber T, Tüting T. Type I interferon-associated skin recruitment of CXCR3+ lymphocytes in dermatomyositis. Clin. Exp. Dermatol 2006;31:576–82. [DOI] [PubMed] [Google Scholar]
- Werth VP, Hejazi E, Pena S, Haber J, Feng R, Patel B, et al. 605 Study of safety and efficacy of lenabasum, a cannabinoid receptor type 2 agonist, in refractory skin-predominant dermatomyositis. J. Invest. Dermatol 2018;138:S103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong D, Kea B, Pesich R, Higgs BW, Zhu W, Brown P, et al. Interferon and biologic signatures in dermatomyositis skin: Specificity and heterogeneity across diseases. PLoS One 2012;7:e29161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaccagnino P, Saltarella M, Maiorano S, Gaballo A, Santoro G, Nico B, et al. An active mitochondrial biogenesis occurs during dendritic cell differentiation. Int. J. Biochem. Cell Biol 2012;44:1962–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated for this submission.


