Abstract
Kidney fibrosis constitutes the shared final pathway of nearly all chronic nephropathies, but biomarkers for the non-invasive assessment of kidney fibrosis are currently not available. To address this, we characterize five candidate biomarkers of kidney fibrosis: Cadherin-11 (CDH11), Sparc-related modular calcium binding protein-2 (SMOC2), Pigment epithelium-derived factor (PEDF), Matrix-gla protein, and Thrombospondin-2. Gene expression profiles in single-cell and single-nucleus RNA-sequencing (sc/snRNA-seq) datasets from rodent models of fibrosis and human chronic kidney disease (CKD) were explored, and Luminex-based assays for each biomarker were developed. Plasma and urine biomarker levels were measured using independent prospective cohorts of CKD: the Boston Kidney Biopsy Cohort, a cohort of individuals with biopsy-confirmed semiquantitative assessment of kidney fibrosis, and the Seattle Kidney Study, a cohort of patients with common forms of CKD. Ordinal logistic regression and Cox proportional hazards regression models were used to test associations of biomarkers with interstitial fibrosis and tubular atrophy and progression to end-stage kidney disease and death, respectively. Sc/snRNA-seq data confirmed cell-specific expression of biomarker genes in fibroblasts. After multivariable adjustment, higher levels of plasma CDH11, SMOC2, and PEDF and urinary CDH11 and PEDF were significantly associated with increasing severity of interstitial fibrosis and tubular atrophy in the Boston Kidney Biopsy Cohort. In both cohorts, higher levels of plasma and urinary SMOC2 and urinary CDH11 were independently associated with progression to end-stage kidney disease. Higher levels of urinary PEDF associated with end-stage kidney disease in the Seattle Kidney Study, with a similar signal in the Boston Kidney Biopsy Cohort, although the latter narrowly missed statistical significance. Thus, we identified CDH11, SMOC2, and PEDF as promising non-invasive biomarkers of kidney fibrosis.
Keywords: Kidney disease, fibrosis, histopathology, biopsy, biomarker
INTRODUCTION
Fibrosis is a hallmark of chronic kidney disease (CKD) which affects around 10% of the world’s population.1,2 Fibrosis is attributed to the pathologic deposition of extracellular matrix components in response to chronic injury and inflammation,1,3 which can be induced by a variety of stimuli, including autoimmune reactions, infections, or ischemic and toxic insults. In the setting of acute and chronic kidney diseases, sustained injury leads to uncontrolled accumulation of fibrotic matrix, thereby making kidney fibrosis the shared final pathway of almost all progressive and chronic nephropathies.1–4 Interstitial fibrosis / tubular atrophy (IFTA) is among the best histologic predictors for CKD progression irrespective of disease etiology.5–7 Kidney fibrosis is characterized by interstitial expansion through accumulation of proliferating myofibroblasts. Under conditions of chronic injury, myofibroblasts secrete matrix proteins that disrupt kidney architecture and lead to parenchymal loss.8–10 Myofibroblasts are therefore an important therapeutic target in fibrosis and CKD. Antifibrotic agents targeting myofibroblasts are being developed, but a major roadblock to successful translation of these therapies is the current lack of non-invasive measures of kidney fibrosis. The traditional biomarkers of CKD—estimated glomerular filtration rate (eGFR) and albuminuria—are not direct measures of fibrosis and therefore ill-suited for clinical trials of novel antifibrotic agents. New biomarkers that can serve as surrogate markers and capture the degree and evolution of fibrosis are critically required to facilitate trial design in nephrology.
We hypothesized that the ideal kidney fibrosis biomarkers would be expressed specifically in myofibroblasts or would be extracellular matrix proteins, since both accumulate in fibrotic disease. To develop and validate promising biomarkers of fibrosis, we chose five candidate proteins from published preclinical models and clinical studies of kidney fibrosis. Sparc-related modular calcium binding protein-2 (SMOC2), cadherin-11 (CDH11), and matrix-gla protein (MGP) were selected from bulk RNA sequencing findings that demonstrated significantly higher gene expression of these three candidates in kidney fibrosis.11 Pigment epithelium-derived factor (PEDF, also called SERPINF1) and thrombospondin-2 (THBS2) were selected as myofibroblast-specific genes identified in translational profiling experiments of fibrotic mouse kidney.12
To determine cell-specific gene expression profiles and to confirm the initial findings from preclinical experiments, we analyzed a publicly available single-cell RNA sequencing (scRNA-seq) dataset of murine unilateral ureteral obstruction (UUO) kidney,13 a well-established rodent model of renal fibrosis, a single-nucleus RNA sequencing (snRNA-seq) dataset of healthy human kidneys,14 and an snRNA-seq dataset from 8 individuals with chronic kidney disease (CKD). We then developed microbead-based ELISA (Luminex) assays and measured each biomarker in plasma and urine samples of patients enrolled in two independent CKD cohort studies. We tested each biomarker’s association with the degree of kidney fibrosis and the risks of progression to end-stage kidney disease (ESKD) and death.
METHODS
scRNA and snRNA-sequencing and bioinformatic analysis
To map biomarker-associated genes to gene expression data, we explored four different datasets: (1) an scRNA-seq dataset and (2) a bulk-RNA seq dataset derived from mice that underwent sham surgery or UUO with kidneys harvested at day 2 and day 7 post UUO or ureteral reimplantation to reverse obstruction before euthanasia at 7 days, 14 days, or 28 days post UUO (R-UUO) (n=4/timepoint).13 (3) A human snRNA-seq dataset derived from 3 healthy adult kidneys,14 and (4) a human snRNA-seq dataset derived from 8 kidney biopsy specimens of patients with CKD. The mouse UUO datasets were acquired from the Gene Expression Omnibus (GEO) database under accession numbers GSE 145053. In the reversible unilateral-ureteric-obstruction (R-UUO) model, ureteric obstruction was surgically reversed by anastomosis of the previously obstructed ureter to the bladder.15 The subsequent decompression of the kidney and restoration of urinary flow has been shown to induce a regression of established fibrosis, which allows to investigate immune processes and subsequent tissue remodeling following the removal of an injurious stimulus.13,15,16 The healthy human kidney snRNA-seq dataset was obtained from the GEO database accession number GSE131882. Details on these datasets have been previously described.13,14 The human CKD snRNA-seq dataset was derived from 8 individuals with CKD who underwent research biopsies as part of the Kidney Precision Medicine Project (KPMP). Details on the KPMP snRNA-seq protocol are described at kpmp.org. Data were acquired from the KPMP Atlas data repository (https://atlas.kpmp.org/repository).
Unique molecule index (UMI) count matrices were uploaded into the R package Seurat. For normalization, the digital gene expression (DGE) matrix was scaled by total UMI counts, multiplied by 10,000 and transformed to log space. Only genes found to be expressed in >10 cells were retained and cells with a relatively high percentage of UMIs mapped to mitochondrial genes (>=0.5 for the UUO scRNA-seq and >=0.3 for the human snRNA-seq) were discarded. Batch effect was corrected by the integration analysis from Seurat 3.0 (https://satijalab.org/seurat/v3.2/integration.html). Linear dimensional reduction was performed by using principal component (PC) analysis. For data exploration and visualization, the first 20 PCs were selected for two-dimensional uniform manifold approximation and projection (UMAP) implemented by the Seurat software in R Version 3.6.2 (R Foundation for Statistical Computing, Vienna, Austria). Biomarker gene expression was visualized by feature plot, dot plot, or violin plot with the internal plotting functions in Seurat.
Study populations
The BKBC is a prospective, observational cohort study of individuals undergoing native kidney biopsy at three tertiary care hospitals in Boston, MA. Details of the study design have been previously described.5 The study includes adults ≥18 years of age who underwent a clinically indicated native kidney biopsy between September 2006 and October 2018. Exclusion criteria were the inability to provide written consent, severe anemia, pregnancy, or enrollment in competing studies. Participants provided blood and urine samples on the day of kidney biopsy. For this study, we evaluated a subset of 721 participants with available baseline urine samples and 514 participants with available baseline plasma samples. Baseline samples were collected at the day of native kidney biopsy and progression to ESKD and death was defined from that date.
The Seattle Kidney Study (SKS) is a prospective, nephrology clinic-based, observational cohort study of individuals with CKD.17,18 Since 2004, the SKS has enrolled 691 participants >18 years of age with non-dialysis CKD (defined as eGFR <90 ml/min/1.73m2 or UACR ≥30 mg/g) from outpatient nephrology clinics at three University of Washington–affiliated hospitals in Seattle, WA. Exclusion criteria were prior or current kidney transplantation, dementia, non-English speaking or the expectation to start renal replacement therapy within 3 months. For this study, we evaluated a subset of 236 participants with available baseline urine samples and 252 participants with available baseline plasma samples who were enrolled between 2006 and 2013. Baseline samples were collected at the study enrollment date and progression to ESKD was defined from that date.
Biomarker assay development and measurements
Multiplex microbead-based ELISA (Luminex) assays were developed for each biomarker including assay and antibody evaluation for cross-reactivity and non-specificity, pairing, tuning, and determination of lower limits of blank and detection. Recombinant proteins were generated for standard curves using a mammalian expression vector. After collection, plasma and urine samples were aliquoted and immediately stored at −800C until analysis. Biomarkers were measured in triplicates at 1:10 dilution and blind split replicates were included to assess coefficients of variation (CV). CVs across blind split replicates were <20% for each plasma and urine biomarker. Assay development and biomarker measurements were performed at ProtAtOnce Ltd, Athens, Greece as previously described19 and amended according to the EMEA/CHMP/EWP/192217/2009, CLSI EP17-A Vol. 24 No. 34, and CKD BioCon II Quality Control Committee Version 11.0 5/2/16 guidelines. Details on the final selection of antibodies can be found in Supplemental Table 1. Capture antibodies were coupled to Luminex magnetic beads and detection antibodies were biotinylated. 2500 coupled beads per analyte were incubated with the samples using a flat bottom 96-well plate on a shaker at 900 revolutions per minute (rpm) for 90 minutes at room temperature followed by addition of the detection mix and incubation for another 60 minutes on a shaker at 900 rpm at room temperature. For signal detection, samples were then incubated with SAPE solution (Streptavidin, R-Phycoerythrin conjugate, Cat Nr: SAPE-001, Moss Inc.) for 15 minutes and the signal was measured with the Luminex FlexMAP 3D instrument.
Evaluation of histopathology
Kidney biopsy specimens of BKBC participants were adjudicated under light microscopy by two experienced renal pathologists who provided semiquantitative scores for histopathologic lesions. Lesions were jointly graded by the two pathologists for this study without knowledge of the suspected clinical diagnosis or the final pathology report. IFTA was graded as involvement of <10%, 11–25%, 26–50%, or > 50% of total cortical volume. The weighted kappa statistic from 26 randomly selected biopsies for repeat review months after the initial scoring was 0.72 (95% CI, 0.52–0.93) for IFTA scores.5 We limited statistical analyses on histopathologic lesions to cases with adjudicated biopsies (n=602 (84%) for urine biomarkers/cr and n= 438 (85%) for plasma biomarkers).
Clinical outcomes and covariates
The primary outcome was progression to ESKD defined as receiving dialysis or kidney transplantation. The secondary endpoint was all-cause mortality. In the BKBC, eGFR during follow-up was obtained from the electronic medical record (EMR) and ESKD status was confirmed by reviewing the EMR and linkage with the United States Renal Data System (USRDS) database. Mortality status was confirmed with the Social Security Death Index (SSDI). Participants were followed up until the occurrence of death, voluntary study withdrawal, loss to follow-up, or February 1, 2020. Participant information was collected at the biopsy visit, including demographics, medical history, medication lists, and laboratory data. We obtained serum creatinine from the EMR on the day of biopsy. In participants for whom this was unavailable, we measured serum creatinine in blood samples collected on the day of biopsy. We obtained spot urine protein-to-creatinine ratio from the date of kidney biopsy up to 3 months before biopsy from the EMR. We used the creatinine-based Chronic Kidney Disease Epidemiology Collaboration (CKD-EPI) equation to calculate the eGFR.20,21
In the SKS, information on the initiation of dialysis, kidney transplantation, and mortality status were ascertained via in-person examinations, telephone contacts, and medical chart review. Information on procedures and events were verified using the EMR and linkage to the USRDS and the SSDI. Annual, in-person SKS exams included health questionnaires, measurements of physical performance, and collection of blood and urine specimens. Baseline laboratory values including serum creatinine and UACR were measured at the Kidney Research Institute, Seattle, WA using an in-house Beckman Coulter DXC600 chemistry analyzer. We used the CKD-EPI equation to calculate the eGFR.20,21
Statistical analyses
Descriptive statistics were summarized as count with percentages for categorical variables and mean ± standard deviation or median with interquartile range (IQR) for continuous variables. For skewed data distributions, we performed logarithmic transformation as appropriate. Urinary biomarkers were normalized to urinary creatinine (indicated as biomarker/cr). Spearman correlation coefficients were used to determine associations between eGFR and each biomarker and multivariable-adjusted linear regression models were used to assess associations between biomarkers and demographic and clinical characteristics. We used multivariable-adjusted ordinal logistic regression models, including age, sex, race, and baseline eGFR, to evaluate associations of each urine and plasma biomarker with the degree of IFTA. In these models, the ordinal IFTA score (<10%, 11–25%, 26–50%, and > 50% of total cortical volume involved) was used as the dependent variable and each biomarker as the independent variable. Conceptually, ordinal logistic regression can be understood as fitting parallel logistic regression models with a common odds ratio for the parameters at each IFTA threshold while still taking the ordinal scale of IFTA into account.22,23 The Brant test24 was used to confirm the assumption of proportionality of the odds across response categories. We provided the pseudo-R squared25 and the Akaike information criterion (AIC)26 as measures to compare the performance of each ordinal logistic regression model. We first calculated both measures for the base model that included age, sex, race, and eGFR and after the addition of plasma and urine biomarkers. For the outcomes of progression to ESKD and all-cause-mortality, we fit Cox proportional hazards regression models. Models were stratified by site with multivariable adjustment for covariates, including age, sex, race, baseline eGFR/ log-transformed eGFR (SKS), and baseline log-transformed proteinuria. We modeled each biomarker continuously on a log2-scale and in tertiles. For urinary biomarkers, additional analyses were performed to compare urinary creatinine normalization (i.e., expressing urine markers as a ratio to urinary creatinine, as is done with albuminuria) to adjustment for urinary creatinine as a covariate in multivariable models. We used complete case analysis since there was less than 5% missing data. In the SKS, we only modeled biomarkers continuously because of the smaller sample size and smaller number of events. We confirmed no violations of the proportional hazard assumption through assessment of Schoenfeld residuals. All statistical tests were two-sided, and P-values <0.05 were considered significant. Statistical analyses were performed using STATA 15.0 (STATACorp, College Station, TX).
Study approval
The Partners Human Research Committee (the Brigham and Women’s Hospital Institutional Review Board, Boston, MA) approved the BKBC Study protocol for recruitment and plasma and urine sample collection which was performed with written informed consent of the participants. The SKS Study protocol was approved by the Institutional Review Board (IRB) at the University of Washington Medical Center, the Harborview Medical Center, and the Veterans Affairs Puget Sound Health Care Center in Seattle, WA. All SKS participants provided informed consent. All mouse experiments carried out to generate the UUO snRNA-seq dataset (GSE11953) were performed according to the animal experimental guidelines issued by the Animal Care and Use Committee at Washington University. The KPMP study is reviewed and approved through the central IRB process at The Human Research Protection Office and Washington University in St. Louis.
RESULTS
scRNA-seq and snRNA-seq reveals biomarker gene expression in mouse and human kidney fibroblasts
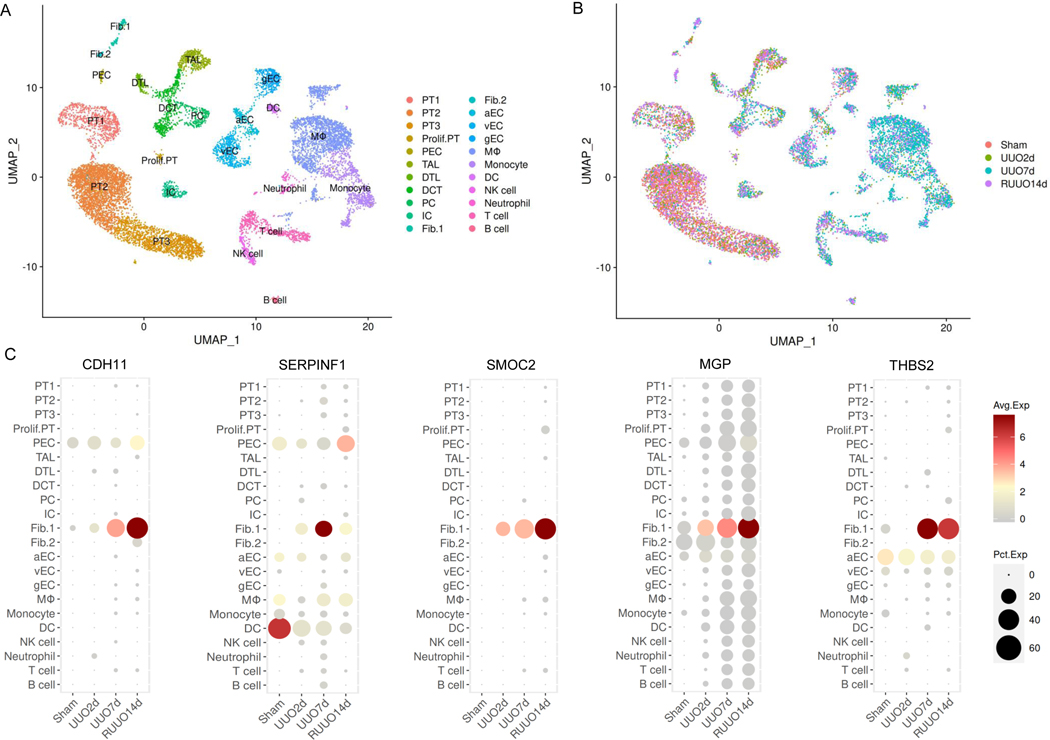
We analyzed an scRNA-seq dataset of murine kidneys at day 2 and day 7 after UUO and at 14 days after ureter reimplantation to reverse obstruction (R-UUO).13 The uniform manifold approximation and projection (UMAP) plot shows 22 separate cell clusters (Figure 1A and B), two of which represent distinct activated fibroblast populations (fibroblast cell clusters type 1 and 2). All five biomarker genes were expressed in fibroblast cluster 1 (Figure 1C). We observed strong expression of CDH11, SMOC2, MGP, and THBS2 7 days post UUO as well as sustained expression after 14 days in the R-UUO model. Similar expression post R-UUO was observed for α-smooth muscle actin (Acta2), an established myofibroblast marker, as well as for the matrix proteins Collagen 12A1 (Col12a1) and Collagen 15A1 (Col 15a1) (Supplemental Figure 1).27 SERPINF1, the corresponding gene for PEDF, was most strongly expressed 7 days post UUO, but expression attenuated after ureter reimplantation. The increase of myofibroblasts at day 14 post R-UUO coincided with decreased expression of Collagen 1A1 (Col1a1) and Collagen 5A2 (Col5a2) and increased expression in matrix degrading genes such as matrix metalloproteinase-14 (MMP-14), consistent with the partial regression of fibrosis seen in the R-UUO model (Supplemental Figure 1).13 Additional analyses using bulk RNA-sequencing data confirmed results from scRNA-seq and showed the strongest expression of all biomarker genes at 7 days post UUO and at 7 days post R-UUO (Supplemental Figure 2).
Figure 1. Single-cell RNA-sequencing shows biomarker gene expression in mouse kidney fibroblasts.
Biomarker gene expression in scRNA-sequencing datasets from animals undergoing sham surgery, or at UUO day 2 (UUO2d), UUO day 7 (UUO7d), or 14 days post R-UUO (RUUO14d) were analyzed. (A) Uniform manifold approximation and projection (UMAP) plot of mouse UUO kidneys reveals 22 separate cell clusters. (B) Biomarker gene expression across cell clusters at different time points. (C) Quantification of differential gene expression at different time points shows that fibrosis biomarkers are primarily expressed in kidney fibroblasts.
UUO, unilateral ureteral obstruction; R-UUO, ureter reimplantation to reverse obstruction; PT1: proximal tubule S1 segment; PT2: proximal tubule S2 segment; PT3: proximal tubule S3 segment; Prolif. PT, proliferating proximal tubule population; PEC, parietal epithelial cells; TAL, thick ascending limb of Loop of Henle; DTL: descending limb of Loop of Henle; DCT, distal convoluted tubule; PC, principal cells; IC, intercalated cells; Fib., fibroblasts type 1; Fib. 2, fibroblasts type 2; aEC, arterial endothelial cells; vEC, venous endothelial cells; gEC, glomerular endothelial cells; Mø: macrophages; DC, dendritic cells; NK cells, natural killer cells. The SERPINF1 (Serine Proteinase Inhibitor-F1) gene encodes PEDF.
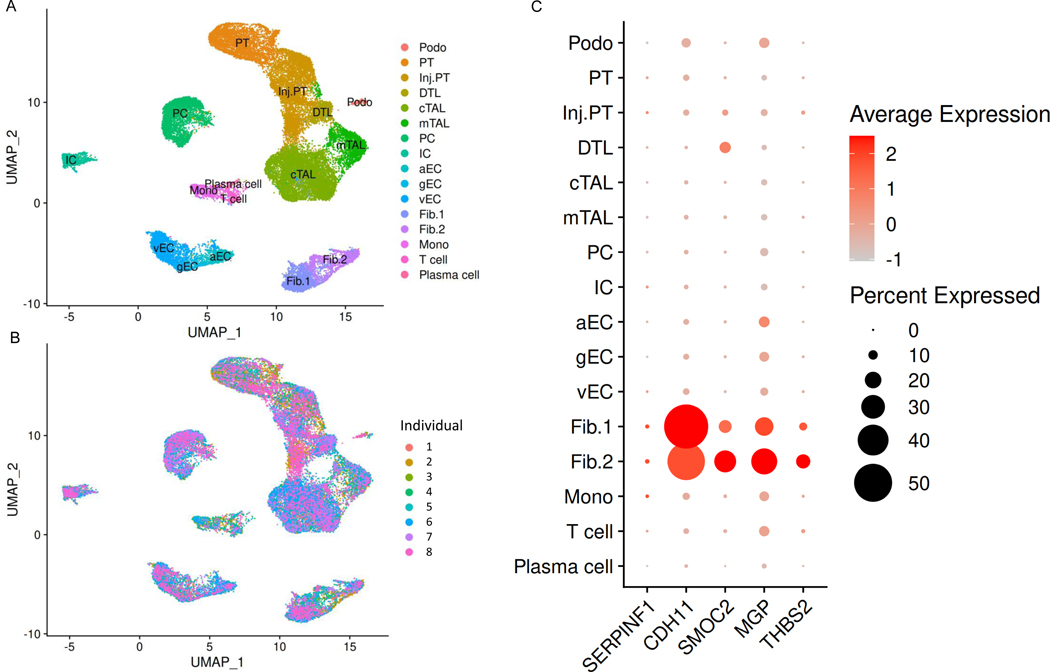
SnRNA-sequencing of three healthy human kidneys revealed expression of all five biomarker genes in human fibroblasts, with highest expression levels for CDH11, MGP, SMOC2, and THBS2 (Supplemental Figure 3). In snRNA-sequencing data analyses from 8 individuals with CKD (Supplemental Table 2), we found cell-specific expression of CDH11, SMOC2, MGP, and THBS2 in type 1 and type 2 fibroblasts. Compared to healthy human kidney, CDH11 and SMOC2 were strongly expressed in CKD kidney. PEDF expression was relatively weak in both CKD and healthy human kidney (Figure 2 and Supplemental Figure 3).
Figure 2. Single-nucleus RNA-sequencing analyses reveal biomarker gene expression in individuals with chronic kidney disease (CKD).
(A) Uniform manifold approximation and projection (UMAP) plot of 8 human kidneys from individuals with CKD reveals 16 separate cell clusters. (B) UMAP plot annotated by cell type and individual. (C) Dot plots shows quantification of gene expression across cell clusters. Podo, podocyte; PT, proximal tubule; Inj. PT, injured proximal tubule; DTL: descending limb of Loop of Henle; cTAL, cortical thick ascending limb of Loop of Henle; mTAL, medullary thick ascending limb of Loop of Henle; PC, principal cell; IC, intercalated cell; aEC, arterial endothelial cells; vEC, venous endothelial cells; gEC, glomerular endothelial cells; Fib., fibroblasts type 1; Fib. 2, fibroblasts type 2; Mono, monocyte. The SERPINF1 (Serine Proteinase Inhibitor-F1) gene encodes PEDF.
Baseline characteristics and correlations with eGFR
Clinical characteristics and median urinary and plasma biomarker levels of participants in both cohorts are shown in Table 1. The mean age was 52.4 (± 16.4) years and 53.4 (± 14.2) years in the BKBC and the SKS, respectively. The mean eGFR was 56.6 (± 35.9) ml/min/1.73m2 in BKBC and 53.4 (± 30.7) ml/min/1.73m2 in SKS participants. Median proteinuria [IQR] was 1.7 [0.4, 4.4] g/g creatinine in the BKBC and median urine albumin-to-creatinine ratio (UACR) was 0.1 [0.01, 0.6] g/g in the SKS. Spearman pairwise correlations between biomarkers, eGFR, and proteinuria are shown in Supplemental Table 4. In the BKBC and the SKS, plasma CDH11, SMOC2, and PEDF, and all urine biomarkers/cr correlated inversely with eGFR (rho= −0.14 to −0.53, all p<0.05 for plasma biomarkers and rho= −0.12, to −0.46, all p<0.01 for urine biomarkers/cr). In both cohorts, plasma SMOC2 (BKBC: rho= 0.12, SKS: rho= 0.18, p<0.01) and urine CDH11, SMOC2, and PEDF correlated positively with proteinuria (rho= 0.16 to 0.41, all p<0.01, respectively). Factors associated with plasma and urine CDH11, SMOC2, and PEDF in the two cohorts are shown in Supplemental Table 5. In multivariable-adjusted linear regression models, lower eGFR was consistently associated with higher levels of each biomarker in both cohorts.
Table 1.
Baseline characteristics of the Boston Kidney Biopsy Cohort (BKBC) and the Seattle
| BKBC (nurine=721, nplasma=514a) | SKS (n=252) | |
|---|---|---|
|
| ||
| Plasma biomarker concentrations | ||
|
| ||
| CDH11, pg/ml | 1818.8 [867.7, 3320.3] | 906.2 [765.5, 1104.4] |
| SMOC2, pg/ml | 5093.9 [3381.8, 7441.8] | 5460.9 [4047.1, 7190.1] |
| PEDF, pg/ml | 24988.5 [18961.8, 32124.8] | 18325.0 [14036.8, 22593.2] |
| MGP, pg/ml | 268.8 [231.2, 424.2] | 229.1 [207.0, 265.5] |
| THBS2, pg/ml | 190.8 [163.0, 249.3] | 178.7 [153.2, 211.9] |
|
| ||
| Urine biomarker/cr concentrations | ||
|
| ||
| CDH11, pg/mg | 4315.2 [1861.1, 10545.8] | 3542.2 [1723.3, 8464.2] |
| SMOC2, pg/mg | 3779.5 [2152.9, 8288.9] | 3649.9 [1554.0, 7980.3] |
| PEDF, pg/mg | 1965.2 [717.4, 13849.9] | 549.8 [291.3, 1992.4] |
| MGP, pg/mg | 370.2 [217.2, 703.7] | 458.2 [284.4, 839.0] |
| THBS2, pg/mg | 145.4 [93.6, 245.2] | 260.4 [158.5, 393.6] |
|
| ||
| Clinical characteristics | ||
|
| ||
| Age, years | 52.4 (± 16.4) | 53.4 (± 14.2) |
| Female | 360 (49.9) | 177 (70.2) |
| Race | ||
| White | 452 (62.7) | 148 (58.7) |
| Black | 143 (19.8) | 55 (21.8) |
| Other | 125 (17.3) | 49 (19.4) |
| eGFR, ml/min/1.73m2 | 56.6 (± 35.9) | 53.4 (± 30.7) |
| Proteinuria, UACRb | 1.7 [0.4 – 4.4] | 0.1 [0.01 – 0.6] |
|
| ||
| Co-morbid conditions | ||
|
| ||
| Diabetes mellitus | 153 (21.2) | 95 (37.7) |
| Hypertension | 359 (49.8) | 214 (84.9) |
| Systemic lupus erythematosus | 91 (12.6) | 22 (8.7) |
| Malignancy | 104 (14.4) | 21 (8.3) |
|
| ||
| Medications | ||
|
| ||
| ACEi | 232 (32.2) | 114 (45.2) |
| ARB | 106 (14.7) | 72 (28.6) |
| Calcium channel blockers | 162 (22.5) | 77 (30.6) |
| Beta-blockers | 213 (29.5) | 88 (34.9) |
| Immunosuppression | 124 (17.2) | 0 (0) |
| Corticosteroids | 125 (17.3) | 26 (10.3) |
|
| ||
| Clinical site | ||
|
| ||
| Site 1 | 441 (61.2) | 252 (100.0) |
| Site 2 | 211 (29.3) | |
| Site 3 | 69 (9.6) | |
Kidney Study (SKS).
Data presented as mean ± standard deviation, median [interquartile range], and count with frequencies (%) for binary and categorical variables. Data on race were missing for one individual.
Baseline characteristics of n=514 individuals with available plasma biomarker measurements are shown in Supplemental Table 3.
BKBC: Proteinuria (g/g creatinine), SKS: UACR (g/g). ACEi, angiotensin converting enzyme inhibitor; ARB, angiotensin II receptor blocker; eGFR, estimated glomerular filtration rate; UACR, urine albumin-to-creatinine ratio; cr, creatinine
Associations of fibrosis biomarkers with interstitial fibrosis and tubular atrophy (IFTA)
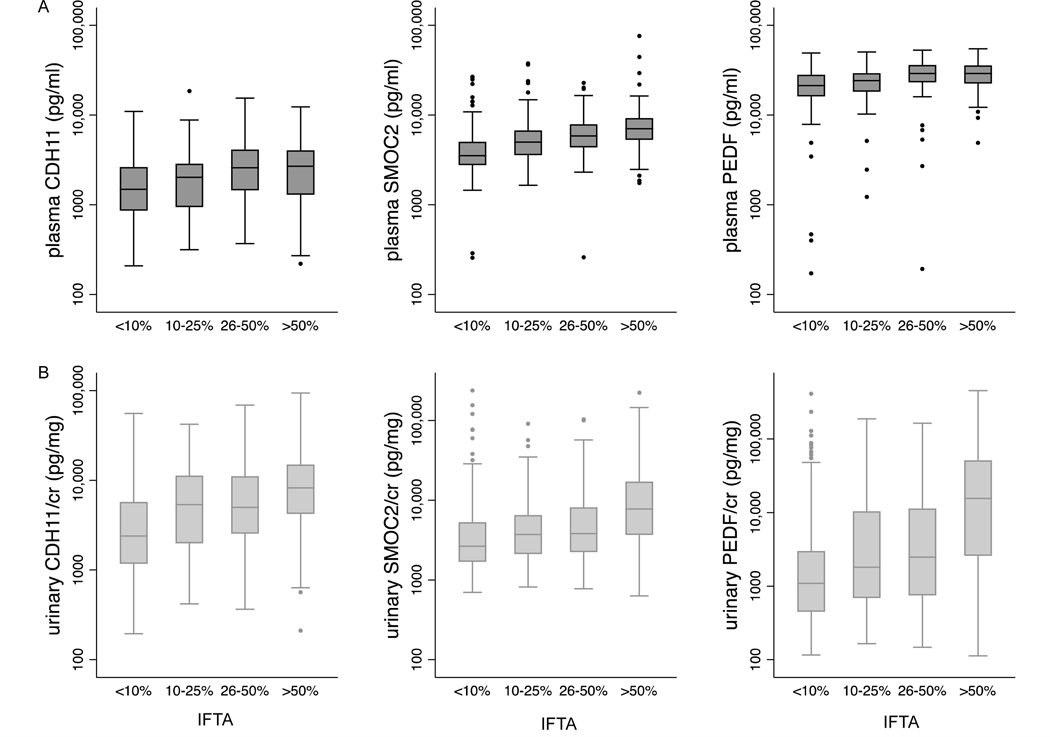
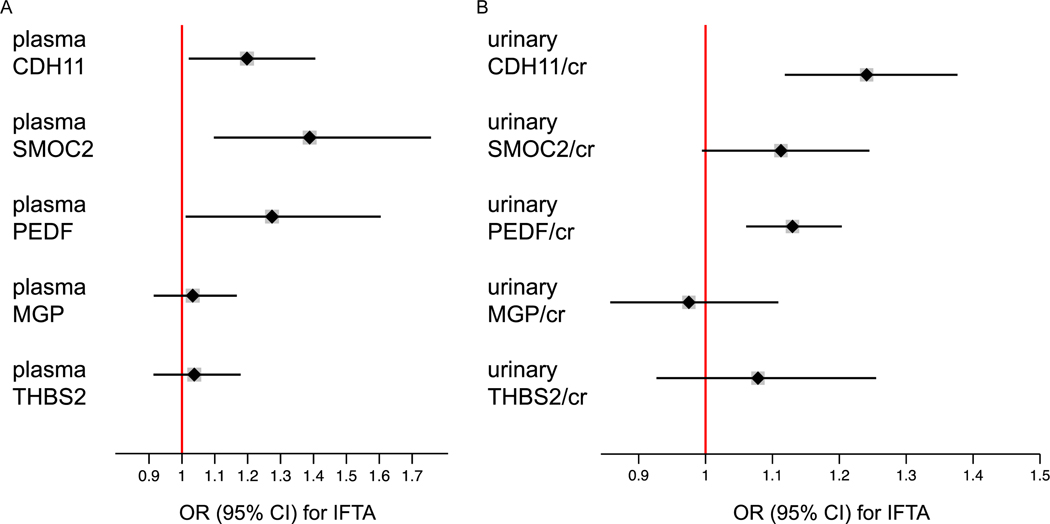
Figure 3andSupplemental Table 6 show differences in plasma and urinary fibrosis biomarker levels across categories of IFTA in the BKBC. Differences in diagnostic sub-groups including glomerulopathies, diabetic nephropathy, and IgA-nephropathy are shown in Supplemental Figures 4–6. In multivariable ordinal logistic regression models adjusted for age, sex, race, and eGFR (Figure 4), each doubling of plasma SMOC2, CDH11, and PEDF associated with increased odds of being in a higher IFTA category (OR=1.39, 95% CI 1.10 to 1.76; OR=1.20, 95% CI 1.02 to 1.41, and OR=1.27, 95% CI 1.01 to 1.60, respectively). Across urinary biomarkers, each doubling of CDH11/cr and PEDF/cr was associated with greater odds of being in a higher IFTA category (OR=1.24; 95% CI 1.12 to 1.38 and OR=1.13; 95% CI 1.06 to 1.20, respectively). The association between urinary SMOC2/cr and IFTA narrowly missed conventional levels of significance (OR=1.11; 95% CI 0.99 to 1.24). The observed associations of urinary biomarkers with IFTA remained qualitatively unchanged when adjusted for urinary creatinine as a covariate, rather than normalized for urinary creatinine in the denominator (Supplemental Table 7). Addition of each biomarker to the baseline model improved the model performance as shown by the increase in pseudo R2 and the decrease in the AIC (Supplemental Table 8).
Figure 3. Differences in plasma (A) and urine (B) CDH11/cr, SMOC2/cr, and PEDF/cr levels by grades of interstitial fibrosis and tubular atrophy (IFTA) in kidney biopsy specimens.
Boxplots show median and interquartile range (IQR) of fibrosis biomarkers on a log scale. Whiskers span data within 1.5 times of the IQR of the lower and upper quartile (25th and 75th percentile, respectively). P value from Kruskal Wallis test <0.001 for each biomarker, n=438 and n=602 for plasma and urinary biomarkers/cr, respectively.
Figure 4. Associations between fibrosis biomarkers and interstitial fibrosis and tubular atrophy (IFTA) in kidney biopsy specimens.
Odds Ratios (OR) are obtained from ordinal logistic regression models using IFTA (graded as involvement of <10%, 11–25%, 26–50%, or > 50% of total cortical volume) as the dependent variable and log2-transformed plasma biomarkers (A) and log2-transformed urinary biomarkers/cr (B) as predictor variables. OR are expressed per change in IFTA score. Models are adjusted for age, sex, race, and eGFR. N=438 and n=602 for plasma and urine biomarkers, respectively.
Associations of fibrosis biomarkers with progression to ESKD
During a median follow-up time of 4.4 and 4.1 years, 182 BKBC participants with available urine biomarker levels and 122 BKBC participants with available plasma biomarker levels progressed to ESKD, respectively (Supplemental Table 9). Table 2 A and Supplemental Figure 7 show multivariable-adjusted associations between each plasma and urinary biomarker with ESKD. In fully adjusted models including age, sex, race, log(proteinuria), and eGFR, two plasma biomarkers associated with an increased risk of developing the outcome: compared to tertile 1, individuals in tertile 3 of plasma SMOC2 and plasma PEDF had an increased risk of progression to ESKD. Across all urine fibrosis markers, three biomarkers associated with higher risk of developing ESKD: compared to tertile 1, individuals in tertile 3 of urinary CDH11/cr, SMOC2/cr, and THBS2/cr were at increased risk of progressing to ESKD. We observed a similar trend for the association of higher levels of urinary PEDF/cr with ESKD, but the latter was confounded by eGFR and narrowly missed conventional levels of statistical significance. The associations of urinary CDH11 and THBS2 with ESKD were sensitive to normalization versus adjustment to urinary creatinine which attenuated the effect (Supplemental Table 10).
Table 2.
Associations of plasma and urinary kidney fibrosis biomarkers with ESKD in the BKBC (A) and the SKS (B).
| (A) BKBC | HR (95% CI)a per doubling | HR (95% CI)a Tertile 1 | HR (95% CI)a Tertile 2 | HR (95% CI)a Tertile 3 |
|---|---|---|---|---|
| Plasma CDH11 | 1.00 (0.86 – 1.16) | Reference | 1.24 (0.75 – 2.03) | 0.94 (0.60 – 1.49) |
| Plasma SMOC2 | 1.17 (0.92 – 1.48) | Reference | 1.40 (0.74 – 2.63) | 1.86 (1.02 – 3.39) |
| Plasma PEDF | 1.04 (0.77 – 1.40) | Reference | 1.63 (0.92 – 2.92) | 1.89 (1.11 – 3.23) |
| Plasma MGP | 0.90 (0.79 – 1.03) | Reference | 0.71 (0.45 – 1.12) | 0.74 (0.47 – 1.17) |
| Plasma THBS2 | 0.97 (0.84 – 1.13) | Reference | 0.87 (0.56 – 1.36) | 0.94 (0.60 – 1.48) |
| Urine CDH11/cr | 1.09 (1.00 – 1.20) | Reference | 1.52 (0.96 – 2.40) | 1.75 (1.12 – 2.72) |
| Urine SMOC2/cr | 1.11 (1.02 – 1.22) | Reference | 1.33 (0.83 – 2.15) | 1.66 (1.03 – 2.66) |
| Urine PEDF/cr | 1.06 (1.00 – 1.13) | Reference | 1.09 (0.68 – 1.76) | 1.46 (0.92 – 2.32) |
| Urine MGP/cr | 1.00 (0.91 – 1.11) | Reference | 1.52 (1.04 – 2.22) | 1.25 (0.84 – 1.88) |
| Urine THBS2/cr | 1.09 (0.96 – 1.24) | Reference | 2.07 (1.38 – 3.12) | 1.64 (1.07 – 2.52) |
| (B) SKS | HR (95% CI)b per doubling |
|---|---|
| Plasma CDH11 | 1.64 (1.02 – 2.63) |
| Plasma SMOC2 | 1.56 (1.02 – 2.39) |
| Plasma PEDF | 0.93 (0.61 – 1.43) |
| Plasma MGP | 1.13 (0.97 – 1.33) |
| Plasma THBS2 | 1.51 (0.96 – 2.36) |
| Urine CDH11/cr | 1.28 (1.05 – 1.57) |
| Urine SMOC2/cr | 1.26 (1.04 – 1.53) |
| Urine PEDF/cr | 1.22 (1.09 – 1.36) |
| Urine MGP/cr | 1.13 (0.88 – 1.45) |
| Urine THBS2/cr | 1.21 (0.87 – 1.69) |
Model is stratified by site and adjusted for age, sex, race, log(proteinuria), and eGFR.
Model is adjusted for age, sex, race, log(UACR), and log(eGFR); Number of participants and ranges of urinary and plasma biomarker concentrations by tertiles are shown in Supplemental Table 9. HR, Hazard Ratio; cr, creatinine.
In the SKS, 59 participants suffered kidney disease progression to ESKD over a median follow-up time of 4.6 years. Results from multivariable-adjusted analyses including age, sex, race, log(UACR), and log(eGFR) are shown in Table 2 B. Higher levels of plasma SMOC2 and CDH11 were independently associated with an increased risk of ESKD. Across all urine biomarkers, higher levels of urinary CDH11/cr, SMOC2/cr, and PEDF/cr were associated with an increased risk of progression to ESKD. In models adjusted for urinary creatinine as a covariate, the association of urinary THBS2 with ESKD also became significant. All other associations after urinary creatinine adjustment were qualitatively unchanged (Supplementary Table 11).
Associations of fibrosis biomarkers with all-cause mortality
During a median follow-up time of 5.7 and 5.1 years, 134 BKBC participants with available urine biomarker levels and 87 BKBC participants with available plasma biomarker levels died, respectively (Supplemental Table 9). Across all plasma biomarkers, higher levels of plasma SMOC2 were associated with all-cause-mortality after multivariable adjustment for age, sex, race, log(proteinuria), and eGFR (Supplemental Table 12 and Supplemental Figure 8). Across all urinary biomarkers, individuals in tertile 3 of CDH11/cr had an increased risk of death compared to individuals in tertile 1 after multivariable adjustment. These associations did not remain statistically significant when adjusted for urinary creatinine as a covariate (Supplemental Table 13).
DISCUSSION
The present study illustrates the sequential evaluation of candidate biomarkers of kidney fibrosis, starting from initial selection based on rodent models, to exploration in scRNA and snRNA-seq datasets, to assay development, and then finally to measurement in prospective cohort studies. Of the five biomarkers we chose to study, the strongest signals were observed with SMOC2, CDH11, and PEDF. Higher levels of plasma SMOC2 and plasma and urinary CDH11 and PEDF were independently associated with increasing severity of IFTA on human kidney biopsies. In two independent cohort studies, plasma and urinary SMOC2 and urinary CDH11 associated with progression to ESKD after multivariable adjustment. Higher levels of urinary PEDF were also associated with ESKD in the SKS with consistent signals in the BKBC, although the latter narrowly missed conventional levels of statistical significance. These findings demonstrate that SMOC2, CDH11, and PEDF may have the potential to serve as biomarkers for the non-invasive assessment of kidney fibrosis.
SMOC2 is a member of the secreted protein acidic and rich in cysteine (SPARC) family of matricellular proteins. We are not aware of prior studies of plasma or urine SMOC2 in prospective cohort studies of individuals with CKD. In preclinical models, SMOC2 is upregulated upon kidney injury and activates fibroblast-to-myofibroblast transition, thereby stimulating extracellular matrix production.4,28,29 Targeting SMOC2 by siRNA attenuated TGF-β1-mediated myofibroblast activation and protected from kidney fibrosis development in mice.4
PEDF belongs to the serine protease inhibitor family and is a regulator of the Wnt-signaling pathway.30 In rodent models of fibrotic kidney disease, decreased kidney PEDF expression has been demonstrated in the diabetic mouse and rat kidney.30,31 Consistent with our findings, studies in humans have shown that higher plasma PEDF is associated with CKD progression in type 2 diabetes, suggesting it may represent a compensatory change in diabetic patients with kidney disease and may have potential to serve as a prognostic biomarker in diabetic nephropathy.32–34 We also found higher urinary and plasma PEDF levels to be associated with higher degrees of IFTA and increased risk of ESKD. No previous studies, to our knowledge, have examined urinary PEDF in prospective cohort studies of individuals with diverse kidney diseases.
CDH11 is a transmembrane glycoprotein that mediates cell-cell adhesion and is involved in the pathogenesis of lung fibrosis through regulation of TGF-β production.35 Studies in mouse models of kidney fibrosis have shown that CDH11-mRNA expression increased with greater severity of kidney fibrosis.11 In cross-sectional analyses of 53 individuals with and without CKD, urinary CDH11 distinguished patients with CKD from healthy controls.11 Our findings are the first, to our knowledge, to investigate CDH11 as a biomarker for kidney fibrosis and disease progression in prospective cohort studies.
The other biomarkers that we studied, THBS2 and MGP, did not demonstrate consistent associations with IFTA and adverse clinical outcomes. THBS2 is a glycoprotein which is synthesized and secreted by a variety of cells including fibroblasts, epithelial cells, and immune cells.36,37 In our study, urinary THBS2 did appear to associate with greater risk of ESKD but did not correlate with IFTA cross-sectionally. Neither plasma nor urinary MGP appeared to be associated with fibrosis or kidney disease progression in this study.
Significant strengths of our study include the availability of biomarker measurements in both urine and plasma in two independent cohort studies of individuals with CKD. Both studies have a relatively long duration of follow-up and low rates of missing outcome data. Specific strengths of the BKBC study include the availability of adjudicated histopathologic scores on lesion severity for IFTA as well as the inclusion of individuals across a diverse spectrum of kidney diseases.
Our study has several limitations that warrant consideration as well. Although we identified statistically significant associations of protein biomarkers with interstitial fibrosis and clinical outcomes, and demonstrated cell-specific expression in published datasets, we cannot determine whether the proteins studied here are kidney-specific fibrosis biomarkers or synthesized by other organs or in other clinical conditions. We also had relatively limited numbers of participants with specific clinical diagnoses, so the comparative performance in different types of kidney diseases will require additional study. Because measured GFR was not available for the two cohorts, we used eGFR for all analyses included in the study. Lastly, whether these biomarkers can be used to monitor and are modulated by anti-fibrotic treatments that are currently in development remains an important area for future investigation. Further investigation in additional validation studies is needed to verify our findings and pave the foundation for preclinical and clinical approval of the most promising candidate biomarkers.
In conclusion, we identified SMOC2, PEDF, and CDH11 as promising biomarkers of kidney fibrosis that carry prognostic value to estimate the risk of kidney disease progression in individuals with CKD. These biomarkers may be used in studies to make non-invasive estimates of the degree of fibrosis and could assist with estimating prognosis and clinical decision-making. Additional studies are warranted to determine if these biomarkers track over time with kidney disease progression, which could lead to their use as biomarkers of therapeutic response, for example facilitating the investigation of anti-fibrotic therapies that are under development for the treatment of commons forms of CKD.38
Supplementary Material
Acknowledgements
The authors thank the participants of the studies for their important contributions and the study staff for their invaluable assistance. This study was supported by National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) grant U01DK104308 (S.S.W. and B.D.H.) and National Institutes of Health (NIH) grant R01DK107931 and R01DK103986 (B.R.K.). The KPMP, UG-DK-114907, is a multiyear project (see Supplemental Acknowledgements for consortium details) and funded by the following grants from the NIDDK: U2C DK114886, UH3DK114861, UH3DK114866, UH3DK114870, UH3DK114908, UH3DK114915, UH3DK114926, UH3DK114907, UH3DK114920, UH3DK114923, UH3DK114933, and UH3DK114937. We are grateful to Kun Zhang (UCSD) and Sanjay Jain (Washington University) for providing unpublished snRNA-seq biopsy data supported by UH3DK114933. I.M.S. is supported by the American Philosophical Society Daland Fellowship in Clinical Investigation. S.S.W. is also supported by NIH grants UH3DK114915, U01DK085660, U01DK104308, R01DK103784, and R21DK119751. The Vaidya laboratory at Brigham and Women’s Hospital is supported by Outstanding New Environmental Sciences (ONES) award from NIH/NIEHS (ES017543). A.S. is supported by NIH grant K23DK120811 and core resources from the George M. O’Brien Kidney Research Center at Northwestern University (NU-GoKIDNEY) P30DK114857. This work was conducted with support from Harvard Catalyst. The Harvard Clinical and Translational Science Center (National Center for Advancing Translational Sciences, National Institutes of Health Award UL1TR001102) and financial contributions from Harvard University and its affiliated academic healthcare centers. The content is solely the responsibility of the authors and does not necessarily represent the official views of Harvard Catalyst, Harvard University and its affiliated academic healthcare centers, or the NIH. Part of this work was presented as a poster presentation at the 2020 American Society of Nephrology Scientific Session on October 22th.
Funding
National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) grant U01DK104308 and National Institutes of Health (NIH) grant R01DK107931 and R01DK103986.
Footnotes
Disclosures
S.S.W. reports personal fees from Public Health Advocacy Institute, CVS, Roth Capital Partners, Kantum Pharma, Mallinckrodt, Wolters Kluewer, GE Health Care, GSK, Allena Pharmaceuticals, Mass Medical International, Barron and Budd (vs. Fresenius), JNJ, Venbio, Strataca, Takeda, Cerus, Pfizer, Bunch and James, Harvard Clinical Research Institute (aka Baim), Oxidien, Sironax, Metro Biotechnology, Biomarin, and Bain. A.S. reports personal fees from Horizon Pharma, PLC, AstraZeneca, CVS Caremark, and Tate & Latham (Medicolegal consulting). V.S.V. is an employee of Pfizer and has a patent US 10,119,168 B2 licensed to Mediar Therapeutics. B.R.K. reports personal fees from Reatta Pharmaceuticals. B.D.H. reports grants and personal fees from Chinook Therapeutics, Janssen, Celgene, Genentech, Chinook Therapeutics, and Indalo Therapeutics. I.M.S., R.P., M.R.C., I.E.S., H.G.R., J.L., L.G.A., and H.W. have nothing to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wynn TA. Cellular and molecular mechanisms of fibrosis. J Pathol. 2008;214(2):199–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Humphreys BD. Mechanisms of Renal Fibrosis. Annu Rev Physiol. 2018;80:309–326. [DOI] [PubMed] [Google Scholar]
- 3.Ferenbach DA, Bonventre JV. Mechanisms of maladaptive repair after AKI leading to accelerated kidney ageing and CKD. Nat Rev Nephrol. 2015;11(5):264–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerarduzzi C, Kumar RK, Trivedi P, et al. Silencing SMOC2 ameliorates kidney fibrosis by inhibiting fibroblast to myofibroblast transformation. JCI Insight. 2017;2(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Srivastava A, Palsson R, Kaze A, et al. The Prognostic Value of Histopathologic Lesions in Native Kidney Biopsy Specimens: Results from the Boston Kidney Biopsy Cohort Study. J Am Soc Nephrol. 2018;29(8):2213–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Katz A, Caramori ML, Sisson-Ross S, et al. An increase in the cell component of the cortical interstitium antedates interstitial fibrosis in type 1 diabetic patients. Kidney Int. 2002;61(6):2058–2066. [DOI] [PubMed] [Google Scholar]
- 7.Nath KA. Tubulointerstitial changes as a major determinant in the progression of renal damage. Am J Kidney Dis. 1992;20(1):1–17. [DOI] [PubMed] [Google Scholar]
- 8.LeBleu VS, Taduri G, O’Connell J, et al. Origin and function of myofibroblasts in kidney fibrosis. Nat Med. 2013;19(8):1047–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meran S, Steadman R. Fibroblasts and myofibroblasts in renal fibrosis. Int J Exp Pathol. 2011;92(3):158–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tomasek JJ, Gabbiani G, Hinz B, et al. Myofibroblasts and mechano-regulation of connective tissue remodelling. Nat Rev Mol Cell Biol. 2002;3(5):349–363. [DOI] [PubMed] [Google Scholar]
- 11.Craciun FL, Bijol V, Ajay AK, et al. RNA Sequencing Identifies Novel Translational Biomarkers of Kidney Fibrosis. J Am Soc Nephrol. 2016;27(6):1702–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grgic I, Krautzberger AM, Hofmeister A, et al. Translational profiles of medullary myofibroblasts during kidney fibrosis. J Am Soc Nephrol. 2014;25(9):1979–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conway BR, O’Sullivan ED, Cairns C, et al. Kidney Single-Cell Atlas Reveals Myeloid Heterogeneity in Progression and Regression of Kidney Disease. J Am Soc Nephrol. published online, 2020/09/27, doi: 10.1681/asn.2020060806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson PC, Wu H, Kirita Y, et al. The single-cell transcriptomic landscape of early human diabetic nephropathy. Proc Natl Acad Sci U S A. 2019;116(39):19619–19625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hesketh EE, Vernon MA, Ding P, et al. A murine model of irreversible and reversible unilateral ureteric obstruction. J Vis Exp. 2014;(94): 52559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cochrane AL, Kett MM, Samuel CS, et al. Renal structural and functional repair in a mouse model of reversal of ureteral obstruction. J Am Soc Nephrol. 2005;16(12):3623–3630. [DOI] [PubMed] [Google Scholar]
- 17.Robinson-Cohen C, Littman AJ, Duncan GE, et al. Physical activity and change in estimated GFR among persons with CKD. J Am Soc Nephrol. 2014;25(2):399–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roshanravan B, Robinson-Cohen C, Patel KV, et al. Association between physical performance and all-cause mortality in CKD. J Am Soc Nephrol. 2013;24(5):822–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poussin C, Mathis C, Alexopoulos LG, et al. The species translation challenge-a systems biology perspective on human and rat bronchial epithelial cells. Sci Data. 2014;1:140009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levey AS, Stevens LA, Schmid CH, et al. A new equation to estimate glomerular filtration rate. Ann Intern Med. 2009;150(9):604–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inker LA, Schmid CH, Tighiouart H, et al. Estimating glomerular filtration rate from serum creatinine and cystatin C. N Engl J Med. 2012;367(1):20–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Francis ME, Eggers PW, Hostetter TH, et al. Association between serum homocysteine and markers of impaired kidney function in adults in the United States. Kidney Int. 2004;66(1):303–312. [DOI] [PubMed] [Google Scholar]
- 23.Greenland S. An application of logistic models to the analysis of ordinal responses. Biometrical Journal. 1985;27(2):189–197. [Google Scholar]
- 24.Brant R. Assessing proportionality in the proportional odds model for ordinal logistic regression. Biometrics. 1990;46(4):1171–1178. [PubMed] [Google Scholar]
- 25.Hu B, Shao J, Palta M. Pseudo-R 2 in logistic regression model. Statistica Sinica. 2006:847–860. [Google Scholar]
- 26.Bozdogan H. Model selection and Akaike’s information criterion (AIC): The general theory and its analytical extensions. Psychometrika. 1987;52(3):345–370. [Google Scholar]
- 27.Kramann R, Machado F, Wu H, et al. Parabiosis and single-cell RNA sequencing reveal a limited contribution of monocytes to myofibroblasts in kidney fibrosis. JCI Insight. 2018;3(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rocnik EF, Liu P, Sato K, et al. The novel SPARC family member SMOC-2 potentiates angiogenic growth factor activity. J Biol Chem. 2006;281(32):22855–22864. [DOI] [PubMed] [Google Scholar]
- 29.Liu P, Lu J, Cardoso WV, et al. The SPARC-related factor SMOC-2 promotes growth factor-induced cyclin D1 expression and DNA synthesis via integrin-linked kinase. Mol Biol Cell. 2008;19(1):248–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He X, Cheng R, Park K, et al. Pigment epithelium-derived factor, a noninhibitory serine protease inhibitor, is renoprotective by inhibiting the Wnt pathway. Kidney Int. 2017;91(3):642–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang JJ, Zhang SX, Lu K, et al. Decreased expression of pigment epithelium-derived factor is involved in the pathogenesis of diabetic nephropathy. Diabetes. 2005;54(1):243–250. [DOI] [PubMed] [Google Scholar]
- 32.Hui E, Yeung CY, Lee PC, et al. Elevated circulating pigment epithelium-derived factor predicts the progression of diabetic nephropathy in patients with type 2 diabetes. J Clin Endocrinol Metab. 2014;99(11):E2169–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hunt KJ, Jenkins AJ, Fu D, et al. Serum pigment epithelium-derived factor: Relationships with cardiovascular events, renal dysfunction, and mortality in the Veterans Affairs Diabetes Trial (VADT) cohort. J Diabetes Complications. 2019;33(10):107410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen H, Zheng Z, Li R, et al. Urinary pigment epithelium-derived factor as a marker of diabetic nephropathy. Am J Nephrol. 2010;32(1):47–56. [DOI] [PubMed] [Google Scholar]
- 35.Schneider DJ, Wu M, Le TT, et al. Cadherin-11 contributes to pulmonary fibrosis: potential role in TGF-beta production and epithelial to mesenchymal transition. Faseb j. 2012;26(2):503–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laherty CD, O’Rourke K, Wolf FW, et al. Characterization of mouse thrombospondin 2 sequence and expression during cell growth and development. J Biol Chem. 1992;267(5):3274–3281. [PubMed] [Google Scholar]
- 37.Daniel C, Amann K, Hohenstein B, et al. Thrombospondin 2 functions as an endogenous regulator of angiogenesis and inflammation in experimental glomerulonephritis in mice. J Am Soc Nephrol. 2007;18(3):788–798. [DOI] [PubMed] [Google Scholar]
- 38.Ruiz-Ortega M, Rayego-Mateos S, Lamas S, et al. Targeting the progression of chronic kidney disease. Nat Rev Nephrol. 2020;16(5):269–288. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.