Abstract
Access to metabolic information in vivo using magnetic resonance (MR) technologies has generally been the niche of MR spectroscopy (MRS) and spectroscopic imaging (MRSI). Metabolic fluxes can be studied using infusion of substrates labeled with magnetic isotopes, with the use of hyperpolarization especially powerful. Unfortunately, these promising methods are not yet accepted clinically, where fast, simple, and reliable measurement and diagnosis are key. Recent advances in functional MRI and chemical exchange saturation transfer (CEST) MRI allow the use of water imaging to study oxygen metabolism and tissue metabolite levels. These, together with the use novel data analysis approaches such as machine learning, for all of these metabolic MR approaches, are increasing the likelihood of their clinical translation.
Introduction
Magnetic Resonance has a multitude of approaches for assessing molecular information in vivo. Until recently, the study of metabolism has been the niche of MRS or MRSI, both homonuclear and heteronuclear [1]. However, this has expanded to functional MRI and especially CEST MRI methods that indirectly assess molecular information through imaging of the water signal. In the following we briefly summarize recent advances in metabolic MR using MRS and MRI.
1. Proton MRS and MRSI
While MRS is viewed by many as a mature technique, in fact over the last two years there have been substantial new developments, many of which have important implications for clinical application of this methodology. The main focus for clinical MRS is in the area of brain tumors, and in particular diagnosis of isocitrate dehydrogenase (IDH) mutation status using measurements of 2-hydroxyglutarate (2HG). Recently it has also been demonstrated that spectral editing could be used to measure cystathionine as well as 2HG, which gave information on 1p19q chromosome deletion as well as IDH status [2], opening up the possibility for accurate presurgical molecular tumor diagnosis. Brain tumors are known to often be very spatially inhomogeneous, from the viewpoint of both pathophysiology and metabolism, and it is therefore important to measure the distribution of metabolites within the tumor and surrounding brain. For mapping of multiple brain metabolites at relatively high spatial resolution, recent developments using MRSI at 7 T have also been demonstrated [3]. This technology could have multiple clinical applications, but a particularly promising one is the use of total choline (tCho) maps (or maps of the tCho/N-acetylaspartate ratio) to guide radiotherapy [4]. Preliminary results of this approach in high grade glioma with localized dose-escalation have given promising results (Dr H. Shim, Emory University, personal communication).
In the past, many MRS or MRSI studies have been hampered to some degree by variable technical quality or disparate technical approaches. Over the last 2 years there has been a substantial effort to reach consensus on optimal techniques that should be used for human MRS studies, both in terms of data acquisition and analysis. The result of this effort has been a series of technical consensus articles that have been published in the last year [5–9]. A second area of substantial and recent development is the application of deep learning (DL) to the analysis of MRS data. Given the particular strength of DL in extracting information from data with low signal-to-noise ratio (SNR), this is expected to remain a growth area. Applications of DL to MRS lag behind MRI somewhat, with the translation of DL-MRI approaches to the imaging domain of MRSI one immediately accessible area. For example, a densely connected U-Net that uses additional information from high-resolution T1-weighted MR images was shown to be able to generate ‘super-resolution’ metabolic images [10]. In the spectral dimension, DL approaches have been used for post-processing steps including artifact identification and removal [11], accelerated spectral fitting [12], shot-to-shot frequency and phase correction [13], and replacement of Fourier transformation for truncated FIDs [14]. MRS quantification amounts to extracting the concentration of a small number of metabolites from a large number of spectral data points, a task that should be amenable to DL; significant recent progress has been made in this area, including the use of truncated MRS data (Fig. 1, [15]). An additional potential of DL lies in combining disparate information from different imaging modalities, for example, combining voxel-based morphometry analysis of structural MRI with MRS data to enhance the independent diagnostic power of each modality [16].
Fig. 1:
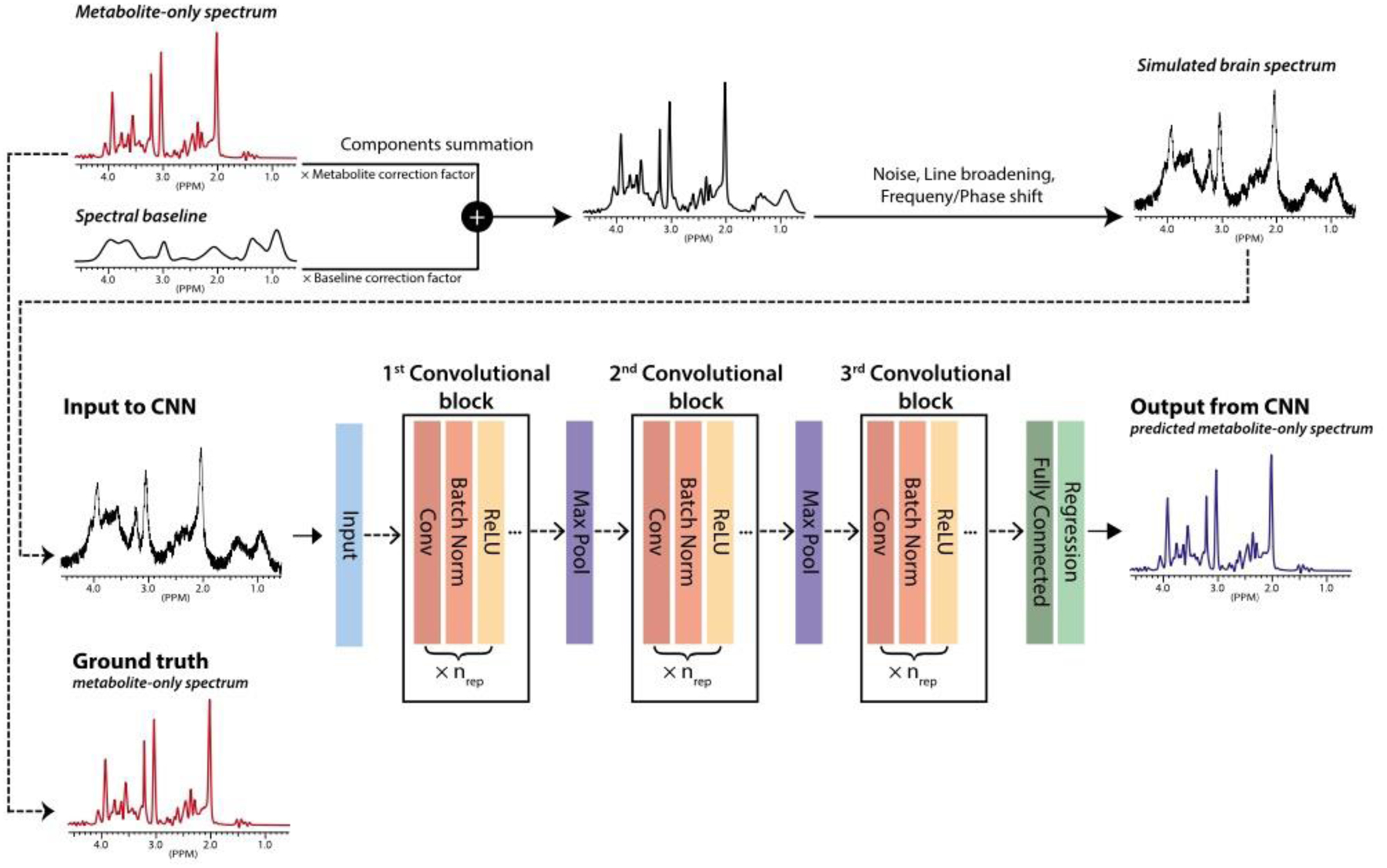
A schematic of the training of the convolutional neural network (CNN) to analyze brain proton MRS data. ‘Metabolite‐only’ spectra, simulated by linearly combining phantom spectra of individual compounds according to randomly selected relative concentration ratios, are used as the ground truth in the training of the CNN. To mimic in vivo brain spectra, the metabolite‐only spectra are combined with noise, line broadened, frequency and phase shifted, and a slowly varying baseline added. These simulated brain spectra are used as the input to the CNN in the training, whose goal is to determine the original metabolite-only spectra and the individual metabolite concentrations. Abbreviations: Conv, convolution; Batch Norm, batch normalization; Max Pool, max pooling; ReLU, rectified linear unit. Reproduced, with permission, from Lee H.H. and Kim H. [15].
2. Heteronuclear MRS and MRSI
2.1: Hyperpolarized MR
While 1H MRS is known to provide metabolite levels, the study of metabolic fluxes has relied on the infusion of substrates labeled with non-radioactive magnetic isotopes such as 13C, but these have even less sensitivity to detection due to their lower gyromagnetic ratio. The advance to human imaging and possible clinical relevance has come from performing metabolic imaging with injected hyperpolarized 13C-labelled substrates, where a more than 10,000 gain in sensitivity has enabled 13C MRSI of these substrates and their downstream metabolites. After a decade of preclinical research with a variety of isotopically labelled and hyperpolarized cell substrates [17–20], [1-13C]pyruvate has emerged as the most promising and widely used substrate. This is because it can be polarized to high levels (>60% at 7T), the 13C nucleus has a relatively long T1 (~30 s in vivo), and it undergoes rapid cell uptake and subsequent metabolism leading to substantial formation of downstream metabolites (lactate and CO2/HCO3−) within the short lifetime of the polarization (2–3 minutes). Pyruvate, which is at the end of the glycolytic pathway, stands at a metabolic crossroads, where it can undergo reduction by NADH to produce lactate, in the reaction catalyzed by lactate dehydrogenase (LDH), or it can enter the mitochondria and be oxidized in the TCA cycle with the production of CO2. The relative labeling of lactate and CO2/HCO3− provides, therefore, an index of glycolytic versus TCA cycle activity. For example, in tumors, where there are often high levels of lactate and LDH activity, relatively high levels of lactate labelling are observed (Fig. 2), whereas in more aerobic tissues, such as heart and brain, there is increased CO2/HCO3− labelling, which is decreased in ischemia.
Fig. 2:
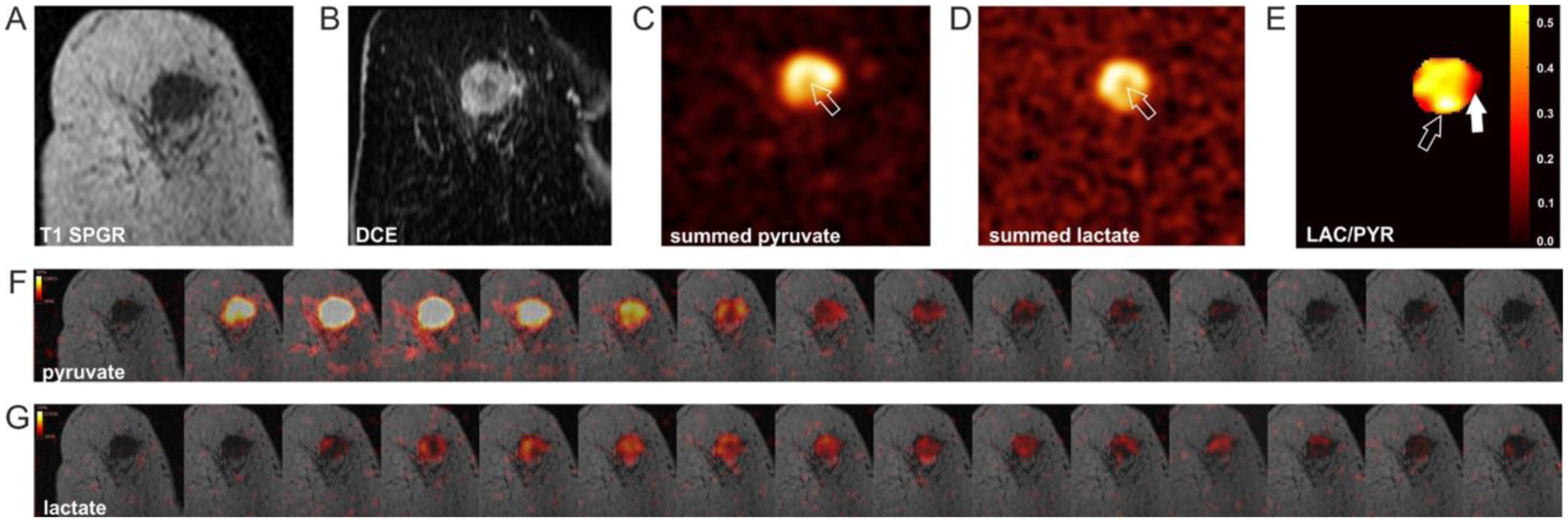
[1-13C]pyruvate and [1-13C]lactate images acquired following intravenous injection of hyperpolarized [1-13C]pyruvate in a breast cancer patient. (A) Coronal image, where the tumor can be visualized as a region of hypointensity. (B) Dynamic contrast enhanced (DCE) image, where the tumor can be visualized as a region of hyperintensity. 13C magnetic resonance spectroscopic images of (C) hyperpolarized [1-13C]pyruvate and (D) hyperpolarized [1-13C]lactate; arrows indicate a region with low signal intensity corresponding to a region of low enhancement on the contrast agent-enhanced image. (E) Map of the ratio of lactate and pyruvate signal intensities, showing intratumoral heterogeneity; open arrow indicates an area of low [1-13C]pyruvate signal but high lactate labeling; closed arrow indicates a tumor area with high [1-13C]pyruvate signal and intermediate lactate labelling. (F-G) Dynamic hyperpolarized [1-13C]pyruvate and [1-13C]lactate images acquired after intravenous injection of hyperpolarized [1-13C]pyruvate (temporal resolution = 4 s). Reproduced from Gallagher, F.A. et al. [35], with permission (https://creativecommons.org/licenses/by-nc-nd/4.0/legalcode).
The hyperpolarized [1-13C]pyruvate technology was translated to the clinic in 2013 with a study of metabolism in prostate cancer [21]. This substrate now stands on the threshold of wider clinical application with the publication of initial studies in brain [22,23] and heart [24,25] plus further studies of prostate cancer [26–30], brain [31–34], breast [35,36], pancreas [37] and kidney [38]. The important question now is how metabolic imaging with hyperpolarized [1-13C]pyruvate could change clinical practice? Early preclinical studies in cancer suggested that the technique could be used to provide early evidence of treatment response [39] and to determine tumor grade and disease progression [40] and some of the initial clinical studies support these ideas [26,27,29,31,36]. PET with 2-Deoxy-2-[18F]fluoroglucose (FDG) is already established in the clinic as a method for detecting tumors and monitoring treatment response and therefore it is important to ask what advantage imaging with hyperpolarized [1-13C]pyruvate might have. Several studies have directly compared the two techniques [33,41–45]. Importantly, FDG PET signal can detect a signal decrease after treatment with few false negative results (no response detected even though there was a treatment response), and therefore has good sensitivity for detecting response. However, FDG PET can give false positive results (decrease in signal not due to a treatment response), and thus has poor specificity. On the other hand, while hyperpolarized [1-13C]pyruvate imaging currently is less likely to detect low volume disease and does not have whole body capability, its capacity to evaluate effectively more of the glycolytic pathway may make it more specific for detecting treatment response under some circumstances [42,44].
Even more than PET, hyperpolarized 13C MRI is plagued by a lack of availability of specialized equipment (the need for a heteronuclear RF channel, not available on clinical scanners) and easy access to hyperpolarization (isotope lifetimes are shorter than PET). However, technical issues can always be addressed if the need is demonstrated and substrate availability could be addressed by centralized production of hyperpolarized contrast media. Removal of the stable radical, the source of the unpaired electron spins used in the dynamic nuclear polarization process, may allow storage and shipping of the hyperpolarized material at low temperature. Removal can be achieved by immobilizing the radical and physically separating it from the hyperpolarized 13C spins [46] or by generating the radical using u.v. irradiation and removing it by transiently raising the temperature [47].
2.2. Deuterated substrates
Recently, the field of magnetic-isotope-labeled substrates for MR studies of metabolism has received a boost by the demonstration that deuterated substrates can also be used to study metabolic pathways with a spatial resolution comparable to hyperpolarized MR [48,49]. Deuterium has the advantage of unlimited lifetime, but similarly requires specialized equipment (RF hardware) not available on clinical scanners. This field is very young, but the first results are promising, for instance for monitoring tumor metabolism (lactate production) after administration of [6,6-2H2]glucose, in a manner similar to hyperpolarized [1-13C]pyruvate [48,49]. Importantly, such metabolism can be detected indirectly through the proton spectrum [50,51], opening up the possibility to study metabolic pathways using deuterium-labelled substrates on standard human scanners [51], e.g. through isotopomer analysis. The promises and pitfalls of this approach were analyzed recently [52].
3. Chemical Exchange Saturation Transfer (CEST) MRI
CEST MRI is related to MRS but employs indirect detection of metabolites that contain groups with exchangeable protons such as hydroxyl, amide, amine, and guanidinium protons [53,54]. This is done by magnetically labeling these protons (using RF saturation or multi-pulse excitation [55]) and detecting a resulting saturation effect in the water signal. Using repetitive label-exchange events per acquisition, CEST MRI allows not only water detection, but also comes with a sensitivity enhancement of one or more orders of magnitude, allowing imaging of millimolar concentrations of metabolites. Importantly, mobile macromolecules such as proteins and glycogen can also be detected through labeling of aliphatic protons if these have a dipolar coupling with nearby exchangeable protons, through a process called exchange relayed nuclear Overhauser effect (rNOE). In principle any compound with exchangeable protons can be detected, but this unfortunately also comes with a detection specificity problem for individual metabolites in vivo due to overlap of these many broad resonances. There is also interference of background signals from direct water saturation (DS) and magnetization transfer contrast (MTC) from semi-solid tissue components, e.g. membranes, myelin sheets. In addition, the conventional CEST data analysis approach of asymmetry analysis with respect to the water resonance frequency further mixes signals and the use of different RF strengths (B1) changes the proportions of these different signal contributions [54,56–59]. Signal interpretation in terms of concentration is also not straightforward because CEST signals are often pH dependent. In the physiological range, exchange rates decrease with pH, which leads to a decrease in the detectability of amide and guanidinium protons, but an increase for OH and amine protons, because the signal intensities depend on the exchange regime at a particular field strength (B0). This may lead to erroneous interpretation in terms of metabolite changes if the pH is changing, for instance during ischemia or exercise, or in tumor environments. A typical example is for the amine protons of glutamate (Glu), because Glu concentration increases during early ischemia and the reduced pH increases the visibility of amine protons and it is unclear what is being measured. Images of the endogenous in vivo CEST signals are therefore often called “weighted”, to indicate contaminated signals. Specificity can be increased, for instance using difference experiments, e.g. from physical or physiological tasks or metabolite-specific drug treatment.
We focus here on recent literature for endogenous metabolites. The field has grown tremendously with many interesting applications in animal models, some having translated to humans. Multiple approaches are being developed to detect at least some species selectively, especially compounds with higher concentrations, such as glutamate (Glu) in the brain, creatine (Cr) and phosphocreatine (PCr) in muscle, and glycogen (Gly) in liver and muscle. For instance, there are many papers on glutamate CEST weighted (GluCESTw) MRI, confirming a major Glu signal contribution and also its reproducibility [60]. Recent data in humans [61], when not using asymmetry analysis and correcting for large background signals of DS and MTC, show a correct gray matter/white matter ratio for Glu, that corresponds with MRS measurements. GluCEST is showing potential for assessing neurodegenerative diseases such as Huntington’s [62], Alzheimer’s [63], Parkinson’s Disease/dopamine-deficiency [64], Multiple Sclerosis [65], and the presence of epileptic foci [66]. Unfortunately, due to the high exchange rate of amines, it appears that GluCEST may be limited to high field scanners (7T and above). The second-most studied metabolic pathway using CEST MRI is the creatine kinase reaction, especially in muscle [67–69]. During exercise, the PCr concentration decreases and Cr increases, but the sensitivity for measuring increased Cr concentrations is compromised by a concomitant pH decrease. At the same time, PCr-CEST signal decreases will be amplified by such pH decreases. It is increasingly recognized that the use of asymmetry analysis is not that useful for Cr/PCr CEST MRI and that the full spectrum should be studied. At some B0 and B1 field strengths, the CEST signals of Cr and PCr can be measured simultaneously [70,71] and the data interpreted correctly. Importantly, at high B0 the CrCEST signal dominates, while at low field (3T and below) the guanidinium protons of Cr are in the intermediate to fast exchange regime and disappear into the background. Interestingly, it was shown recently that for skeletal muscle in humans at 3T, B1 can be tuned so that PCr is the only distinguishable CEST signal in the high frequency range relative to water [72]. Using a deep learning algorithm to assess the effects of B0, exchange rate (pH dependent) and signal intensity, this allowed the effects of exercise to be monitored in vivo at high spatial and temporal resolution (Fig. 3). Another area of noteworthy progress has been the ability to study glycogen levels in vivo in liver using rNOE signals [73]. Overall, the relatively young CEST field is showing many interesting metabolic applications in animal models at higher field strengths and is increasingly being translated to humans at 7T and also 3T. We expect a continuous growth of the CEST field, especially judged from recent work using spectral analysis with machine learning approaches [72,74–77] and fast acquisition approaches with reduced sampling and increased sensitivity, such as compressed sensing and fingerprinting [78–82].
Fig. 3:
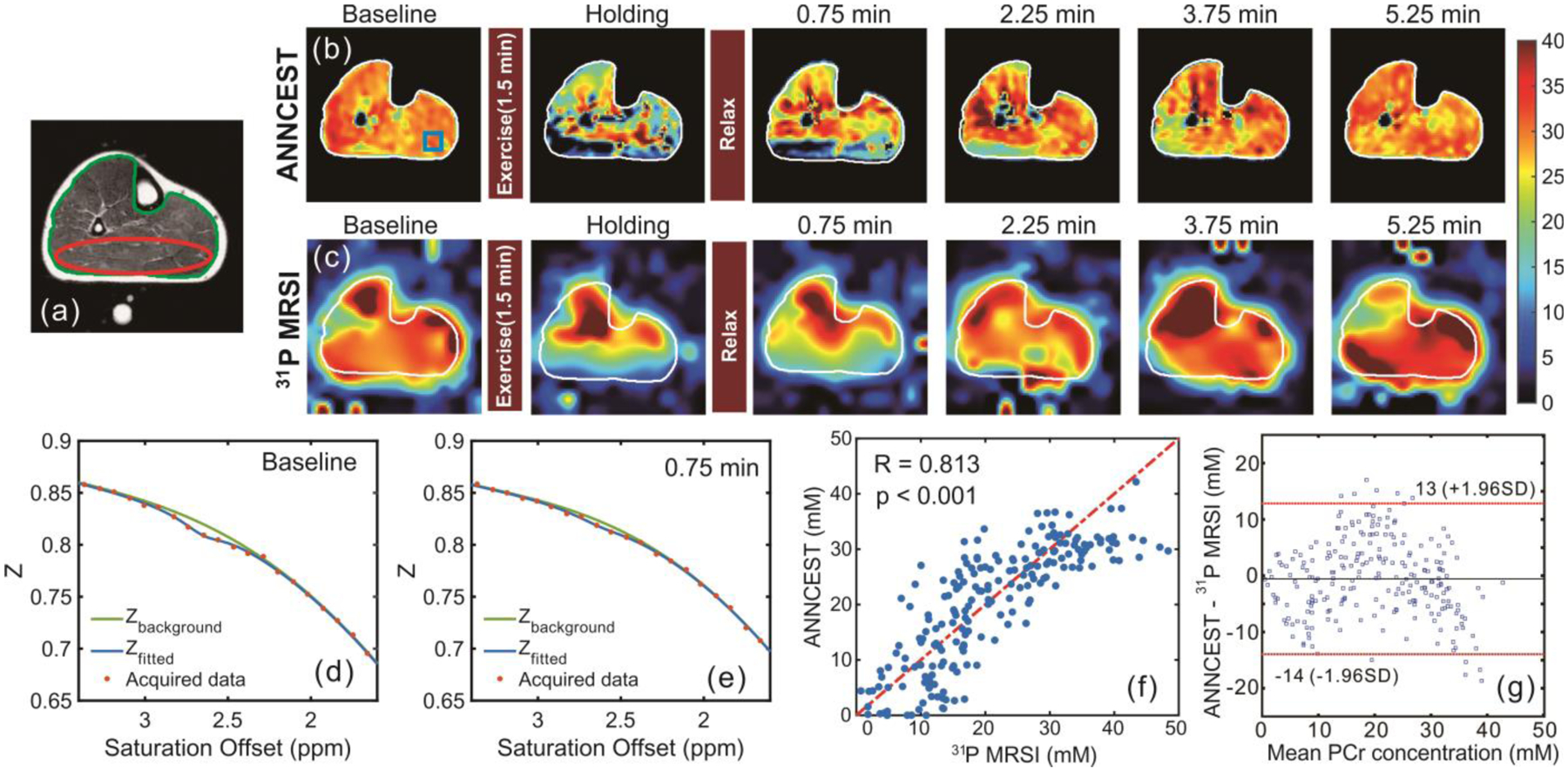
Maps of human skeletal muscle at 3 Tesla pre and post in-magnet plantar flexion exercise using PCr CEST analyzed with an artificial neural network (ANNCEST) compared with the same exercise observed using 31P spectroscopic imaging. (a) T2-weighted anatomical image. (b,c) PCr concentration maps (scale bar in mM) obtained by (b) ANNCEST and (c) 31P MRSI during the exercise process. The water saturation spectrum (Z-spectrum, in which Z is the signal during radiofrequency saturation normalized by the signal without such saturation, Z = Ssat/S0) at baseline (d) and post exercise (e) extracted from the blue rectangle region shown in (b). (f,g) Correlation and Bland-Altman analyses between concentrations obtained from ANNCEST and 31P MRSI for all four subjects (n = 202 voxels) from the red ellipse region shown in (a). Reproduced, with permission, from Chen L. et al. [72]
4. Functional MRI
Functional MRI (fMRI) based on the Blood-Oxygenation-Level-Dependent (BOLD) signal can be used to assess metabolic activity. The BOLD signal reflects a hemoglobin-oxygenation-based magnetic susceptibility effect that affects the transverse relaxation times (T2 or T2*) of intravascular and extravascular water protons [83–85]. The venous oxygenation fraction (Yv) of hemoglobin is determined by the tissue cerebral metabolic rate of oxygen (CMRO2), cerebral blood flow (CBF), and the total hemoglobin concentration in blood ([Hbtot], which is proportional to hematocrit (Hct). Importantly, focusing on the intravascular BOLD effect, CMRO2 can be quantified without stimulation or contrast agent using the Fick Principle:
where Ya is the arterial oxygenation saturation fraction and OEF the oxygen extraction fraction. Using MRI, whole-brain Yv can be measured either based on a calibrated relationship between blood T2 and oxygenation fraction ([86–88] and references therein) or magnetic susceptibility quantification of blood ([89–92] and references therein). As one of several rapid MRI pulse sequences, T2-Relaxation-Under-Spin-Tagging (TRUST) MRI [87] can measure global Yv within approximately 1 minute and has been validated with 15O-PET [93]. Recent advances in understanding the relationship between T2 and Y allow calibration of Y at most field strengths [94], while T1 can be used to noninvasively estimate Hct [95]. Knowing Ya (typically from pulse oximetry) and whole-brain CBF (e.g. from phase-contrast MRI, which can be used to determine CBF in less than 1 minute, [96,97]), CMRO2 can be calculated. Without CBF information, OEF can be calculated and used in clinical applications, as recently demonstrated for aging and neurodegenerative disease (Fig. 4, [98–100]).
Fig. 4.
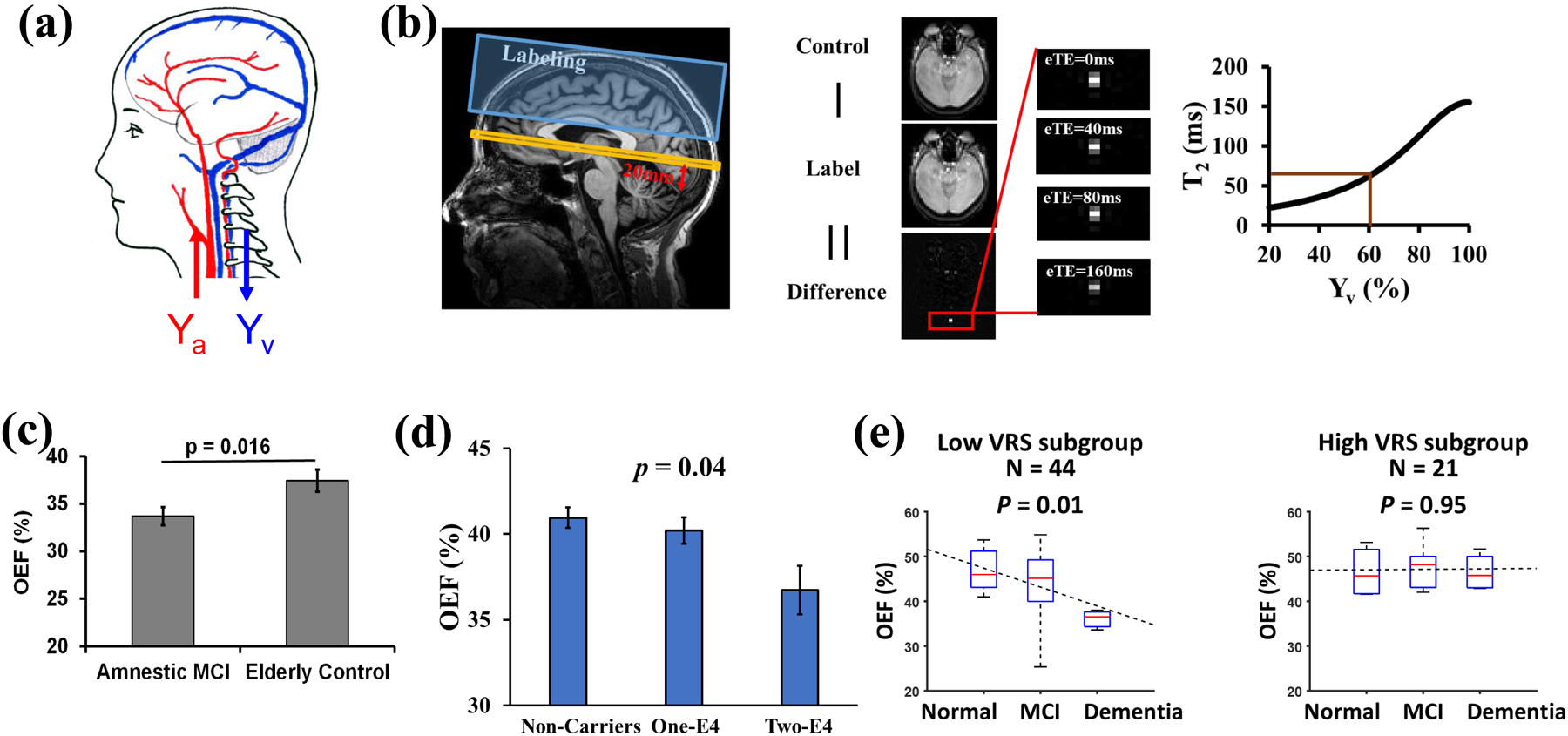
Example of OEF determination in neurodegenerative disease. (a) Illustration of arterial (Ya) and venous (Yv) oxygenation fraction in the brain. OEF = (Ya-Yv)/Ya. (b) Measurement of Yv with TRUST MRI. Left panel: Typical positions of imaging slice (yellow) and labeling slab (blue). The red arrow and label indicate that the imaging plane is placed to be parallel to the anterior commissure – posterior commissure (AC-PC) line and 20 mm above the sinus confluence point. Middle panel: Representative raw images of TRUST MRI. Right panel: Conversion from blood T2 to oxygenation fraction. (c) Diminished OEF in patients with mild cognitive impairment (MCI). Reproduced, with permission, from Thomas, B.P. et al. [100]. (d) OEF was decreased in cognitively normal older individuals who have a higher genetic risk (i.e. APOE4) to develop Alzheimer’s disease. Reproduced, with permission, from Lin, Z. et al. [99]. (e) Vascular risk factors (VRS) have an additional effect on OEF. Reproduced, with permission, from Jiang, D. et al. [98].
Regional measurements of OEF and CMRO2 are also receiving interest, for instance the ability to differentiate left versus right hemisphere, cortical versus subcortical regions, and major brain lobes. Given that CBF mapping techniques are available for MRI, the main efforts have been directed toward developing methods to measure regional Yv. For instance, vessel-specific T2-based methods are being developed that account for tissue partial volume effects [101,102]. With these newer techniques, Yv in major intracerebral draining veins, the vein of Galen, and straight sinus have been determined in adults and neonates [101,102]. Smaller draining veins have lower signal and greater anatomic variability across individuals and their Yv measurement is less established. An alternative approach to determine blood oxygenation with very high spatial resolution (1×1×1 mm3 or better) is quantitative susceptibility mapping (QSM) [89–92]. Here the actual oxygen-sensitive physical quantity underlying intravascular T2 changes, the tissue magnetic susceptibility χ, is calculated on a voxel-by-voxel basis from the local field in each voxel. The latter is generally determined from the MR signal phase measured by gradient echo imaging. This local field in an MRI voxel is a parameter that depends on the brain orientation as well as on the susceptibility of surrounding voxels, and therefore is difficult to measure reproducibly between subjects. However, the magnetic susceptibility that is reconstructed through an inverse calculation is brain-orientation independent and voxel specific. Using referencing to a standard tissue (e.g. cerebral spinal fluid, CSF, assuming to have zero susceptibility) and a separate calibration of magnetic susceptibility differences as a function of blood oxygenation, the blood oxygenation can be calculated non-invasively in vivo [89–92].
Voxel-by-voxel mapping of CMRO2 or OEF without the need for a physiological stimulation would be the holy grail of functional brain imaging, but when using the BOLD T2* effect these would require knowledge of the local venous cerebral blood volume (CBVv) and advanced BOLD models [83,85]. However, the reliability and practicality of these methods remain to be further investigated, and represent an active area of research which is beyond the scope of this review.
5. A look to the Future of Metabolic MR
The field of metabolic MR is undergoing rapid progress. While often criticized for its lack of clinical applicability, MRS may finally cross this hurdle through the recent availability of deep learning approaches for SNR improvement and, importantly, fast and reliable data interpretation on the scanner. Similarly, CEST MRI may, either in combination with MRS or independently, provide high resolution metabolic information using water imaging. The use of hyperpolarized magnetic isotopes for the study of active metabolism has seen a decade of strong progress and is on the threshold of clinical application but its applicability is still hampered by a lack of common availability. Newer deuterium-based approaches have potential, especially when using proton detection. Combination of these methods with PET on PET-MR scanners, which are becoming more widely available, and the use of fast and higher SNR data acquisition and analysis approaches that exploit machine learning makes clinical translation of combinations of these advanced methods for voxel-based metabolic analysis increasingly likely. The capability for regional CMRO2 determination may further enhance the clinical information content.
Acknowledgments
We acknowledge the reviewers for helpful suggestions with the text. We are grateful for funding from NIH/NIBIB (P41 EB015909, P41 EB031771, and R01 EB015032). Work in KMB’s lab is funded by grants from CRUK (C197/ A17242, C197/A16465, C9685/A25177) and the European Union (EC H2020: 831514, 858149). Dr. Edden is supported by NIH R01grants EB016089 and EB023963. Dr. Knutsson is supported by Swedish Research Council Grant/Award Number: 2015-04170; Swedish Cancer Society, Grants/Award Numbers: CAN 2015/251 and 2018/550; Swedish Brain Foundation, Grant/Award Number: FO2017-0236
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
REFERENCES
- 1.Bottomley PA, Griffiths JR: Handbook of magnetic resonance spectroscopy in vivo: MRS theory, practice and applications. John Wiley and Sons Ltd., Chichester, UK: (2016). [Google Scholar]
- 2.Branzoli F, Pontoizeau C, Tchara L, Di Stefano AL, Kamoun A, Deelchand DK, Valabregue R, Lehericy S, Sanson M, Ottolenghi C, Marjanska M: Cystathionine as a marker for 1p/19q codeleted gliomas by in vivo magnetic resonance spectroscopy. Neuro Oncol (2019) 21(6):765–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hangel G, Cadrien C, Lazen P, Furtner J, Lipka A, Heckova E, Hingerl L, Motyka S, Gruber S, Strasser B, Kiesel B et al. : High-resolution metabolic imaging of high-grade gliomas using 7T-CRT-FID-MRSI. Neuroimage Clin (2020) 28(102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gurbani S, Weinberg B, Cooper L, Mellon E, Schreibmann E, Sheriff S, Maudsley A, Goryawala M, Shu HK, Shim H: The brain imaging collaboration suite (brics): A cloud platform for integrating whole-brain spectroscopic MRI into the radiation therapy planning workflow. Tomography (2019) 5(1):184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bogner W, Otazo R, Henning A: Accelerated MR spectroscopic imaging-a review of current and emerging techniques. NMR in biomedicine (2020) e4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi IY, Andronesi OC, Barker P, Bogner W, Edden RAE, Kaiser LG, Lee P, Marjanska M, Terpstra M, de Graaf RA: Spectral editing in 1(H) magnetic resonance spectroscopy: Experts’ consensus recommendations. NMR in biomedicine (2020) e4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kreis R, Boer V, Choi IY, Cudalbu C, de Graaf RA, Gasparovic C, Heerschap A, Krssak M, Lanz B, Maudsley AA, Meyerspeer M et al. : Terminology and concepts for the characterization of in vivo MR spectroscopy methods and MR spectra: Background and experts’ consensus recommendations. NMR in biomedicine (2020) e4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Near J, Harris AD, Juchem C, Kreis R, Marjanska M, Oz G, Slotboom J, Wilson M, Gasparovic C: Preprocessing, analysis and quantification in single-voxel magnetic resonance spectroscopy: Experts’ consensus recommendations. NMR in biomedicine (2020) e4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oz G, Deelchand DK, Wijnen JP, Mlynarik V, Xin L, Mekle R, Noeske R, Scheenen TWJ, Tkac I, Experts’ Working Group on Advanced Single Voxel HM: Advanced single voxel (1)H magnetic resonance spectroscopy techniques in humans: Experts’ consensus recommendations. NMR in biomedicine (2020) e4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iqbal Z, Nguyen D, Hangel G, Motyka S, Bogner W, Jiang S: Super-resolution (1)H magnetic resonance spectroscopic imaging utilizing deep learning. Front Oncol (2019) 9(1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gurbani SS, Schreibmann E, Maudsley AA, Cordova JS, Soher BJ, Poptani H, Verma G, Barker PB, Shim H, Cooper LAD: A convolutional neural network to filter artifacts in spectroscopic MRI. Magn Reson Med (2018) 80(5):1765–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gurbani SS, Sheriff S, Maudsley AA, Shim H, Cooper LAD: Incorporation of a spectral model in a convolutional neural network for accelerated spectral fitting. Magn Reson Med (2019) 81(5):3346–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tapper S, Mikkelsen M, Dewey BE, Zollner HJ, Hui SCN, Oeltzschner G, Edden RAE: Frequency and phase correction of J-difference edited MR spectra using deep learning. Magn Reson Med (2021) 85(4):1755–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee H, Lee HH, Kim H: Reconstruction of spectra from truncated free induction decays by deep learning in proton magnetic resonance spectroscopy. Magn Reson Med (2020) 84(2):559–568. [DOI] [PubMed] [Google Scholar]
- 15.Lee HH, Kim H: Intact metabolite spectrum mining by deep learning in proton magnetic resonance spectroscopy of the brain. Magn Reson Med (2019) 82(1):33–48. [DOI] [PubMed] [Google Scholar]
- 16.Tsuda M, Asano S, Kato Y, Murai K, Miyazaki M: Differential diagnosis of multiple system atrophy with predominant parkinsonism and parkinson’s disease using neural networks. J Neurol Sci (2019) 401(19–26. [DOI] [PubMed] [Google Scholar]
- 17.Brindle KM: Imaging metabolism with hyperpolarized 13C-labeled cell substrates. J Am Chem Soc (2015) 137(20):6418–6427. [DOI] [PubMed] [Google Scholar]
- 18.Comment A, Merritt ME: Hyperpolarized magnetic resonance as a sensitive detector of metabolic function. Biochemistry (2014) 53(47):7333–7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hesketh RL, Brindle KM: Magnetic resonance imaging of cancer metabolism with hyperpolarized 13C-labeled cell metabolites. Current Opinion in Chemical Biology (2018) 45(187–194. [DOI] [PubMed] [Google Scholar]
- 20.Keshari KR, Wilson DM: Chemistry and biochemistry of 13C hyperpolarized magnetic resonance using dynamic nuclear polarization. Chemical Society reviews (2014) 43(5):1627–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nelson SJ, Kurhanewicz J, Vigneron DB, Larson PEZ, Harzstark AL, Ferrone M, van Criekinge M, Chang JW, Bok R, Park I, Reed G et al. : Metabolic imaging of patients with prostate cancer using hyperpolarized [1-13C]pyruvate. Science Translational Medicine (2013) 5(198):198ra108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grist JT, McLean MA, Riemer F, Schulte RF, Deen SS, Zaccagna F, Woitek R, Daniels CJ, Kaggie JD, Matys T, Patterson I et al. : Quantifying normal human brain metabolism using hyperpolarized 1-C-13 pyruvate and magnetic resonance imaging. Neuroimage (2019) 189(171–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee CY, Soliman H, Geraghty BJ, Chen AP, Connelly KA, Endre R, Perks WJ, Heyn C, Black SE, Cunningham CH: Lactate topography of the human brain using hyperpolarized 13C-mri. NeuroImage (2020) 204:116202. [DOI] [PubMed] [Google Scholar]
- 24.Cunningham CH, Lau JY, Chen AP, Geraghty BJ, Perks WJ, Roifman I, Wright GA, Connelly KA: Hyperpolarized 13C metabolic mri of the human heart: Initial experience. Circulation research (2016) 119(11):1177–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rider OJ, Apps A, Miller J, Lau JYC, Lewis AJM, Peterzan MA, Dodd MS, Lau AZ, Trumper C, Gallagher FA, Grist JT et al. : Noninvasive in vivo assessment of cardiac metabolism in the healthy and diabetic human heart using hyperpolarized 13C mri. Circulation research (2020) 126(6):725–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aggarwal R, Vigneron DB, Kurhanewicz J: Hyperpolarized [1-13C]-pyruvate magnetic resonance imaging detects an early metabolic response to androgen ablation therapy in prostate cancer. European Urology (2017) 72(6):1028–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen H-Y, Aggarwal R, Bok RA, Ohliger MA, Zhu Z, Lee P, Gordon JW, van Criekinge M, Carvajal L, Slater JB, Larson PEZ et al. : Hyperpolarized 13C-pyruvate mri detects real-time metabolic flux in prostate cancer metastases to bone and liver: A clinical feasibility study. Prostate Cancer and Prostatic Diseases (2020) 23(2):269–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen H-Y, Larson PEZ, Gordon JW, Bok RA, Ferrone M, van Criekinge M, Carvajal L, Cao P, Pauly JM, Kerr AB, Park I et al. : Technique development of 3D dynamic CS-EPSI for hyperpolarized 13C pyruvate mr molecular imaging of human prostate cancer. Magn Reson Med (2018) 80(5):2062–2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Granlund KL, Tee S-S, Vargas HA, Lyashchenko SK, Reznik E, Fine S, Laudone V, Eastham JA, Touijer KA, Reuter VE, Gonen M et al. : Hyperpolarized mri of human prostate cancer reveals increased lactate with tumor grade driven by monocarboxylate transporter 1. Cell Metabolism (2020) 31(1):105–114.e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larson PEZ, Chen HY, Gordon JW, Korn N, Maidens J, Arcak M, Tang S, Criekinge M, Carvajal L, Mammoli D, Bok R et al. : Investigation of analysis methods for hyperpolarized 13C-pyruvate metabolic MRI in prostate cancer patients. NMR in biomedicine (2018) 31(11):e3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Autry AW, Gordon JW, Chen H-Y, LaFontaine M, Bok R, Van Criekinge M, Slater JB, Carvajal L, Villanueva-Meyer JE, Chang SM, Clarke JL et al. : Characterization of serial hyperpolarized 13C metabolic imaging in patients with glioma. NeuroImage: Clinical (2020) 27(102323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mammoli D, Gordon J, Autry A, Larson PEZ, Li Y, Chen HY, Chung B, Shin P, Van Criekinge M, Carvajal L, Slater JB et al. : Kinetic modeling of hyperpolarized carbon-13 pyruvate metabolism in the human brain. IEEE transactions on medical imaging (2020) 39(2):320–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miloushev VZ, Granlund KL, Boltyanskiy R, Lyashchenko SK, DeAngelis LM, Mellinghoff IK, Brennan CW, Tabar V, Yang TJ, Holodny AI, Sosa RE et al. : Metabolic imaging of the human brain with hyperpolarized 13C pyruvate demonstrates 13C lactate production in brain tumor patients. Canc Res (2018) 78:3755–3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park I, Larson PEZ, Gordon JW, Carvajal L, Chen HY, Bok R, Van Criekinge M, Ferrone M, Slater JB, Xu D, Kurhanewicz J et al. : Development of methods and feasibility of using hyperpolarized carbon-13 imaging data for evaluating brain metabolism in patient studies. Magn Reson Med (2018) 80(3):864–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gallagher FA, Woitek R, McLean MA, Gill AB, Manzano Garcia R, Provenzano E, Riemer F, Kaggie J, Chhabra A, Ursprung S, Grist JT et al. : Imaging breast cancer using hyperpolarized carbon-13 mri. Proceedings of the National Academy of Sciences (2020) 117(4):2092–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woitek R, McLean MA, Gill AB, Grist JT, Provenzano E, Patterson AJ, Ursprung S, Torheim T, Zaccagna F, Locke M, Laurent M-C et al. : Hyperpolarized 13C mri of tumor metabolism demonstrates early metabolic response to neoadjuvant chemotherapy in breast cancer. Radiology: Imaging Cancer (2020) 2(4):e200017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stødkilde-Jørgensen H, Laustsen C, Hansen ESS, Schulte R, Ardenkjaer-Larsen JH, Comment A, Frøkiaer J, Ringgaard S, Bertelsen LB, Ladekarl M, Weber B: Pilot study experiences with hyperpolarized [1-(13)C]pyruvate mri in pancreatic cancer patients. Journal of magnetic resonance imaging: JMRI (2020) 51(3):961–963. [DOI] [PubMed] [Google Scholar]
- 38.Tran M, Latifoltojar A, Neves JB, Papoutsaki M-V, Gong F, Comment A, Costa ASH, Glaser M, Tran-Dang M-A, El Sheikh S, Piga W et al. : First-in-human in vivo non-invasive assessment of intra-tumoral metabolic heterogeneity in renal cell carcinoma. BJR Case Rep (2019) 5(3):20190003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Day SE, Kettunen MI, Gallagher FA, Hu DE, Lerche M, Wolber J, Golman K, Ardenkjaer-Larsen JH, Brindle KM: Detecting tumor response to treatment using hyperpolarized 13C magnetic resonance imaging and spectroscopy. Nature medicine (2007) 13(11):1382–1387. [DOI] [PubMed] [Google Scholar]
- 40.Albers MJ, Bok R, Chen AP, Cunningham CH, Zierhut ML, Zhang VY, Kohler SJ, Tropp J, Hurd RE, Yen YF, Nelson SJ et al. : Hyperpolarized 13C lactate, pyruvate, and alanine: Noninvasive biomarkers for prostate cancer detection and grading. Cancer research (2008) 68(20):8607–8615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gutte H, Hansen AE, Larsen MM, Rahbek S, Henriksen ST, Johannesen HH, Ardenkjaer-Larsen J, Kristensen AT, Hojgaard L, Kjaer A: Simultaneous hyperpolarized 13C-pyruvate mri and 18F-fdg pet (hyperpet) in 10 dogs with cancer. Journal of nuclear medicine: official publication, Society of Nuclear Medicine (2015) 56(11):1786–1792. [DOI] [PubMed] [Google Scholar]
- 42.Hesketh RL, Wang JZ, Wright AJ, Lewis DY, Denton AE, Grenfell R, Miller JL, Bielik R, Gehrung M, Fala M, Ros S et al. : Magnetic resonance imaging is more sensitive than PET for detecting treatment-induced cell death-dependent changes in glycolysis. Cancer research (2019) 79(14):3557–3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Menzel MI, Farrell EV, Janich MA, Khegai O, Wiesinger F, Nekolla S, Otto AM, Haase A, Schulte RF, Schwaiger M: Multimodal assessment of in vivo metabolism with hyperpolarized 1-C-13 mr spectroscopy and F-18-FDG pet imaging in hepatocellular carcinoma tumor-bearing rats. J Nucl Med (2013) 54(7):1113–1119. [DOI] [PubMed] [Google Scholar]
- 44.Ros S, Wright AJ, D’Santos P, Hu D-e, Hesketh RL, Lubling Y, Georgopoulou D, Lerda G, Couturier D-L, Razavi P, Pelossof R et al. : Metabolic imaging detects resistance to pi3kα inhibition mediated by persistent foxm1 expression in er+ breast cancer. Cancer Cell (2020) 38:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Witney T, Kettunen M, Day S, Hu D, Neves A, Gallagher F, Fulton S, Brindle K: A comparison between radiolabeled fluorodeoxyglucose uptake and hyperpolarized C-13-labeled pyruvate utilization as methods for detecting tumor response to treatment. Neoplasia (2009) 6:574–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ji X, Bornet A, Vuichoud B, Milani J, Gajan D, Rossini AJ, Emsley L, Bodenhausen G, Jannin S: Transportable hyperpolarized metabolites. Nature communications (2017) 8:13975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Capozzi A, Cheng T, Boero G, Roussel C, Comment A: Thermal annihilation of photo-induced radicals following dynamic nuclear polarization to produce transportable frozen hyperpolarized 13C-substrates. Nature communications (2017) 8:15757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.De Feyter HM, Behar KL, Corbin ZA, Fulbright RK, Brown PB, McIntyre S, Nixon TW, Rothman DL, de Graaf RA: Deuterium metabolic imaging (DMI) for MRI-based 3D mapping of metabolism in vivo. Sci Adv (2018) 4(8):eaat7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu M, Zhu XH, Zhang Y, Mateescu G, Chen W: Quantitative assessment of brain glucose metabolic rates using in vivo deuterium magnetic resonance spectroscopy. J Cereb Blood Flow Metab (2017) 37(11):3518–3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brindle KM, Brown FF, Campbell ID, Foxall DL, Simpson RJ: A 1h n.M.R. Study of isotope exchange catalysed by glycolytic enzymes in the human erythrocyte. Biochem J (1982) 202(3):589–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rich LJ, Bagga P, Wilson NE, Schnall MD, Detre JA, Haris M, Reddy R: (1)h magnetic resonance spectroscopy of (2)H-to-(1)H exchange quantifies the dynamics of cellular metabolism in vivo. Nat Biomed Eng (2020) 4(3):335–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Zijl PCM, Brindle KM: Spectroscopic measurements of metabolic fluxes. Nat Biomed Eng (2020) 4(3):254–256. [DOI] [PubMed] [Google Scholar]
- 53.Hoefemann M, Doring A, Fichtner ND, Kreis R: Combining chemical exchange saturation transfer and (1)H magnetic resonance spectroscopy for simultaneous determination of metabolite concentrations and effects of magnetization exchange. Magn Reson Med (2021) 85(4):1766–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Zijl PC, Sehgal AA: Proton chemical exchange saturation transfer (CEST), MRS, and MRI. eMagRes (2016) 5:1307–1332; 10.1002/9780470034590.emrstm9780470031482. [DOI] [Google Scholar]
- 55.Knutsson L, Xu J, Ahlgren A, van Zijl PCM: CEST, ASL, and magnetization transfer contrast: How similar pulse sequences detect different phenomena. Magn Reson Med (2018) 80(4):1320–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Consolino L, Anemone A, Capozza M, Carella A, Irrera P, Corrado A, Dhakan C, Bracesco M, Longo DL: Non-invasive investigation of tumor metabolism and acidosis by mri-cest imaging. Front Oncol (2020) 10:161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jones KM, Pollard AC, Pagel MD: Clinical applications of chemical exchange saturation transfer (CEST) MRI. Journal of magnetic resonance imaging: JMRI (2018) 47(1):11–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shaffer JJ Jr, Mani M, Schmitz SL, Xu J, Owusu N, Wu D, Magnotta VA, Wemmie JA: Proton exchange magnetic resonance imaging: Current and future applications in psychiatric research. Front Psychiatry (2020) 11:532606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van Zijl PCM, Lam WW, Xu J, Knutsson L, Stanisz GJ: Magnetization transfer contrast and chemical exchange saturation transfer mri. Features and analysis of the field-dependent saturation spectrum. Neuroimage (2018) 168:222–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nanga RPR, DeBrosse C, Kumar D, Roalf D, McGeehan B, D’Aquilla K, Borthakur A, Hariharan H, Reddy D, Elliott M, Detre JA et al. : Reproducibility of 2D gluCEST in healthy human volunteers at 7 T. Magn Reson Med (2018) 80(5):2033–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Debnath A, Hariharan H, Nanga RPR, Reddy R, Singh A: Glutamate-weighted CEST contrast after removal of magnetization transfer effect in human brain and rat brain with tumor. Mol Imaging Biol (2020) 22(4):1087–1101. [DOI] [PubMed] [Google Scholar]
- 62.Pepin J, de Longprez L, Trovero F, Brouillet E, Valette J, Flament J: Complementarity of gluCEST and (1)H-mrs for the study of mouse models of Huntington’s disease. NMR in biomedicine (2020) 33(7):e4301. [DOI] [PubMed] [Google Scholar]
- 63.Crescenzi R, DeBrosse C, Nanga RP, Byrne MD, Krishnamoorthy G, D’Aquilla K, Nath H, Morales KH, Iba M, Hariharan H, Lee VM et al. : Longitudinal imaging reveals subhippocampal dynamics in glutamate levels associated with histopathologic events in a mouse model of tauopathy and healthy mice. Hippocampus (2017) 27(3):285–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bagga P, Pickup S, Crescenzi R, Martinez D, Borthakur A, D’Aquilla K, Singh A, Verma G, Detre JA, Greenberg J, Hariharan H et al. : In vivo gluCEST MRI: Reproducibility, background contribution and source of glutamate changes in the mptp model of parkinson’s disease. Sci Rep (2018) 8(1):2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.O’Grady KP, Dula AN, Lyttle BD, Thompson LM, Conrad BN, Box BA, McKeithan LJ, Pawate S, Bagnato F, Landman BA, Newhouse P et al. : Glutamate-sensitive imaging and evaluation of cognitive impairment in multiple sclerosis. Mult Scler (2019) 25(12):1580–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Davis KA, Nanga RP, Das S, Chen SH, Hadar PN, Pollard JR, Lucas TH, Shinohara RT, Litt B, Hariharan H, Elliott MA et al. : Glutamate imaging (gluCEST) lateralizes epileptic foci in nonlesional temporal lobe epilepsy. Sci Transl Med (2015) 7(309):309ra161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Haris M, Singh A, Cai K, Kogan F, McGarvey J, Debrosse C, Zsido GA, Witschey WR, Koomalsingh K, Pilla JJ, Chirinos JA et al. : A technique for in vivo mapping of myocardial creatine kinase metabolism. Nat Med (2014) 20(2):209–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kogan F, Haris M, Debrosse C, Singh A, Nanga RP, Cai K, Hariharan H, Reddy R: In vivo chemical exchange saturation transfer imaging of creatine (CrCEST) in skeletal muscle at 3t. Journal of magnetic resonance imaging: JMRI (2014) 40(3):596–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kumar D, Nanga RPR, Thakuri D, Wilson N, Cember A, Martin ML, Zhu D, Shinohara RT, Qin Q, Hariharan H, Reddy R: Recovery kinetics of creatine in mild plantar flexion exercise using 3D creatine CEST imaging at 7 Tesla. Magn Reson Med (2021) 85(2):802–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen L, Barker PB, Weiss RG, van Zijl PCM, Xu J: Creatine and phosphocreatine mapping of mouse skeletal muscle by a polynomial and lorentzian line-shape fitting cest method. Magn Reson Med (2019) 81(1):69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chung JJ, Jin T, Lee JH, Kim SG: Chemical exchange saturation transfer imaging of phosphocreatine in the muscle. Magn Reson Med (2019) 81(6):3476–3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen L, Schar M, Chan KWY, Huang J, Wei Z, Lu H, Qin Q, Weiss RG, van Zijl PCM, Xu J: In vivo imaging of phosphocreatine with artificial neural networks. Nature communications (2020) 11(1):1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou Y, van Zijl PCM, Xu X, Xu J, Li Y, Chen L, Yadav NN: Magnetic resonance imaging of glycogen using its magnetic coupling with water. Proceedings of the National Academy of Sciences of the United States of America (2020) 117(6):3144–3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Glang F, Deshmane A, Prokudin S, Martin F, Herz K, Lindig T, Bender B, Scheffler K, Zaiss M: DeepCEST 3T: Robust MRI parameter determination and uncertainty quantification with neural networks-application to cest imaging of the human brain at 3T. Magn Reson Med (2020) 84(1):450–466. [DOI] [PubMed] [Google Scholar]
- 75.Kim B, Schar M, Park H, Heo HY: A deep learning approach for magnetization transfer contrast mr fingerprinting and chemical exchange saturation transfer imaging. Neuroimage (2020) 221:117165. [DOI] [PubMed] [Google Scholar]
- 76.Li Y, Xie D, Cember A, Nanga RPR, Yang H, Kumar D, Hariharan H, Bai L, Detre JA, Reddy R, Wang Z: Accelerating glucest imaging using deep learning for B0 correction. Magn Reson Med (2020) 84(4):1724–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zaiss M, Deshmane A, Schuppert M, Herz K, Glang F, Ehses P, Lindig T, Bender B, Ernemann U, Scheffler K: DeepCEST 9.4T chemical exchange saturation transfer mri contrast predicted from 3 T data - a proof of concept study. Magn Reson Med (2019) 81(6):3901–3914. [DOI] [PubMed] [Google Scholar]
- 78.Cohen O, Huang S, McMahon MT, Rosen MS, Farrar CT: Rapid and quantitative chemical exchange saturation transfer (CEST) imaging with magnetic resonance fingerprinting (MRF). Magn Reson Med (2018) 80(6):2449–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Heo HY, Han Z, Jiang S, Schar M, van Zijl PCM, Zhou J: Quantifying amide proton exchange rate and concentration in chemical exchange saturation transfer imaging of the human brain. Neuroimage (2019) 189:202–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kang B, Kim B, Schar M, Park H, Heo HY: Unsupervised learning for magnetization transfer contrast mr fingerprinting: Application to cest and nuclear overhauser enhancement imaging. Magn Reson Med (2021) 85(4):2040–2054. [DOI] [PubMed] [Google Scholar]
- 81.Perlman O, Herz K, Zaiss M, Cohen O, Rosen MS, Farrar CT: CEST-MR-fingerprinting: Practical considerations and insights for acquisition schedule design and improved reconstruction. Magn Reson Med (2020) 83(2):462–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhou Z, Han P, Zhou B, Christodoulou AG, Shaw JL, Deng Z, Li D: Chemical exchange saturation transfer fingerprinting for exchange rate quantification. Magn Reson Med (2018) 80(4):1352–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.He X, Yablonskiy DA: Quantitative BOLD: Mapping of human cerebral deoxygenated blood volume and oxygen extraction fraction: Default state. Magn Reson Med (2007) 57(1):115–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ogawa S, Menon RS, Tank DW, Kim SG, Merkle H, Ellermann JM, Ugurbil K: Functional brain mapping by blood oxygenation level-dependent contrast magnetic resonance imaging. A comparison of signal characteristics with a biophysical model. Biophys J (1993) 64(3):803–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.van Zijl PC, Eleff SM, Ulatowski JA, Oja JM, Ulug AM, Traystman RJ, Kauppinen RA: Quantitative assessment of blood flow, blood volume and blood oxygenation effects in functional magnetic resonance imaging. Nat Med (1998) 4(2):159–167. [DOI] [PubMed] [Google Scholar]
- 86.Li W, Xu X, Liu P, Strouse JJ, Casella JF, Lu H, van Zijl PCM, Qin Q: Quantification of whole-brain oxygenation extraction fraction and cerebral metabolic rate of oxygen consumption in adults with sickle cell anemia using individual T2 -based oxygenation calibrations. Magn Reson Med (2020) 83(3):1066–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lu H, Xu F, Grgac K, Liu P, Qin Q, van Zijl P: Calibration and validation of TRUST MRI for the estimation of cerebral blood oxygenation. Magn Reson Med (2012) 67(1):42–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wright GA, Hu BS, Macovski A: 1991 I.I. Rabi award. Estimating oxygen saturation of blood in vivo with mr imaging at 1.5 T. JMRI (1991) 1(3):275–283. [DOI] [PubMed] [Google Scholar]
- 89.Fan AP, Evans KC, Stout JN, Rosen BR, Adalsteinsson E: Regional quantification of cerebral venous oxygenation from mri susceptibility during hypercapnia. Neuroimage (2015) 104:146–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Haacke EM, Lai S, Reichenbach JR, Kuppusamy K, Hoogenraad FG, Takeichi H, Lin W: In vivo measurement of blood oxygen saturation using magnetic resonance imaging: A direct validation of the blood oxygen level-dependent concept in functional brain imaging. Hum Brain Mapp (1997) 5(5):341–346. [DOI] [PubMed] [Google Scholar]
- 91.Wehrli FW, Fan AP, Rodgers ZB, Englund EK, Langham MC: Susceptibility-based time-resolved whole-organ and regional tissue oximetry. NMR in biomedicine (2017) 30(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang J, Liu T, Gupta A, Spincemaille P, Nguyen TD, Wang Y: Quantitative mapping of cerebral metabolic rate of oxygen (CMRO2) using quantitative susceptibility mapping (qsm). Magn Reson Med (2015) 74(4):945–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jiang D, Deng S, Franklin CG, O’Boyle M, Zhang W, Heyl BL, Pan L, Jerabek PA, Fox PT, Lu H: Validation of T2 -based oxygen extraction fraction measurement with (15)O positron emission tomography. Magn Reson Med (2021) 85(1):290–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li W, van Zijl PCM: Quantitative theory for the transverse relaxation time of blood water. NMR in biomedicine (2020) 33(5):e4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li W, Grgac K, Huang A, Yadav N, Qin Q, van Zijl PC: Quantitative theory for the longitudinal relaxation time of blood water. Magn Reson Med (2016) 76(1):270–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Peng SL, Su P, Wang FN, Cao Y, Zhang R, Lu H, Liu P: Optimization of phase-contrast mri for the quantification of whole-brain cerebral blood flow. Journal of magnetic resonance imaging: JMRI (2015) 42(1126–1133). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xu F, Ge Y, Lu H: Noninvasive quantification of whole-brain cerebral metabolic rate of oxygen (cmro2) by mri. Magn Reson Med (2009) 62(1):141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jiang D, Lin Z, Liu P, Sur S, Xu C, Hazel K, Pottanat G, Darrow J, Pillai JJ, Yasar S, Rosenberg P et al. : Brain oxygen extraction is differentially altered by Alzheimer’s and vascular diseases. Journal of magnetic resonance imaging : JMRI (2020) 52(6):1829–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lin Z, Sur S, Soldan A, Pettigrew C, Miller M, Oishi K, Bilgel M, Moghekar A, Pillai JJ, Albert M, Lu H: Brain oxygen extraction by using mri in older individuals: Relationship to apolipoprotein e genotype and amyloid burden. Radiology (2019) 292(1):140–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Thomas BP, Sheng M, Tseng BY, Tarumi T, Martin-Cook K, Womack KB, Cullum MC, Levine BD, Zhang R, Lu H: Reduced global brain metabolism but maintained vascular function in amnestic mild cognitive impairment. J Cereb Blood Flow Metab (2017) 37(4):1508–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jiang D, Lu H, Parkinson C, Su P, Wei Z, Pan L, Tekes A, Huisman T, Golden WC, Liu P: Vessel-specific quantification of neonatal cerebral venous oxygenation. Magn Reson Med (2019) 82(3):1129–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Krishnamurthy LC, Mao D, King KS, Lu H: Correction and optimization of a T2-based approach to map blood oxygenation in small cerebral veins. Magn Reson Med (2016) 75(3):1100–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]


