Abstract
Aims: To explore the contagiousness and new SARS-CoV-2 mutations in pediatric COVID-19.
Methods: This cohort study enrolled all pediatric patients admitted to 8 hospitals in Zhejiang Province of China between 21 January and 29 February 2020, their family members and close-contact classmates. Epidemiological, demographic, clinical and laboratory data were collected. Bioinformatics was used to analyze the features of SARS-CoV-2. Individuals were divided into 3 groups by the first-generation case: Groups 1 (unclear), 2 (adult), and 3 (child). The secondary attack rate (SAR) and R0 were compared among the groups.
Results: The infection rate among 211 individuals was 64% (135/211). The SAR in Groups 2 and 3 was 71% (73/103) and 3% (1/30), respectively; the median R0 in Groups 2 and 3 was 2 (range: 1-8) and 0 (range: 0-1), respectively. Compared with adult cases, the SAR and R0 of pediatric cases were significantly lower (p<0.05). We obtained SARS-CoV-2 sequences from the same infant's throat and fecal samples at a two-month interval and found that the new spike protein A958D mutation detected in the stool improved thermostability theoretically.
Conclusions: Children have lower ability to spread SARS-CoV-2. The new A958D mutation is a potential reason for its long residence in the intestine.
Keywords: Contagiousness, Pediatrics, SARS-CoV-2, Evolutionary tree, New A958D mutation
Introduction
As the number of pediatric COVID-19 cases increases, exploring the contagiousness of these patients is important for prevention and control, especially in families, kindergartens, and schools. However, data about the contagiousness of pediatric COVID-19 is limited. In Zhejiang Province of China, imported cases from Hubei Province occurred in January 2020 and were followed by second-generation cases in families and communities. The epidemic was essentially controlled by the end of February 2020 by isolating and treating cases and by tracking and screening family members and close-contact persons. As of 7 May 2020, a total of 1268 cases (including adults and children) had been diagnosed in Zhejiang Province, China, and 48210 close contacts were traced; 1 adult died (http://med.china.com.cn/content/pid/176850/tid/1026). The clear epidemiologic chain of transmission at the early stage of the COVID-19 pandemic is amenable to the investigation of the infectivity of pediatric cases in Zhejiang Province, outside of Hubei Province, China.
Most pediatric COVID-19 patients present mild or no symptoms, and some cases may be accompanied by digestive tract symptoms (CDC COVID-19 Response Team, 2020; Lu et al., 2020). Some children have no digestive tract symptoms with persistent fecal nucleic acid positivity (Xu et al., 2020). The virus can be isolated from stool samples at the early stages of the disease, and some mutations were detected (Jin et al., 2020). The genomic and epidemiological surveillance around the world is highlighted in order to rapidly determine the ongoing virus evolution (Cella et al., 2021). Despite the abundant SARS-CoV-2 variability (Lokman et al., 2020), one key question remains with regard to whether these mutations have any functional impact on clinical features. For example, researchers have found that the Europe-prevalent D614G mutation of the S protein may increase the infectivity of SARS-CoV-2 (Korber et al., 2020). In general, the duration of intestinal excretion of viral nucleic acids varies greatly among patients, and exploring SARS-CoV-2 mutations in the stool of patients who excrete viral nucleic acids for a long period may facilitate the detection of viral mutations with potential clinical impact.
Methods
Study population
We conducted a retrospective cohort study at 8 hospitals (i.e., authors' units) in Zhejiang Province, China, between 21 January and 29 February 2020. The study population included all pediatric cases (0-18 years of age) with laboratory-confirmed COVID-19 admitted to these hospitals, and family members and close-contact classmates of the pediatric patients. The exclusion criteria were as follows: first-generation cases of the family including both adults and children; eligible participants exposed to other COVID-19 patients. The study population was divided into 3 groups according to the first-generation case. Group 1, the first-generation case was unclear, and all family members were from the epidemic area; Group 2, the first-generation case involved an adult (>18 y); Group 3, the first-generation case was a child (≤18 y).
The diagnosis, clinical type, and field epidemiological investigation of COVID-19 were based on guidelines issued by the National Health Commission of the People's Republic of China (http://www.nhc.gov.cn/yzygj/s7653p/202002/3b09b894ac9b4204a79db5b8912d4440.shtml; http://www.nhc.gov.cn/xcs/zhengcwj/202001/c67cfe29ecf1470e8c7fc47d3b751e88.shtml). SARS-CoV-2 was screened in all patients who had cough, fever, and radiographic presentation of ground-glass opacities at the initial assessment, an epidemiological history of travel, residence in epidemic areas, or close contact with confirmed COVID-19 cases within 14 days. Uninfected individuals were defined as those with a negative nucleic acid test before and after 14 days of isolation as well as negative antibody results during the follow-up period. A COVID-19 case was defined as an individual with laboratory-confirmed SARS-CoV-2 infection by positive RT-PCR results or by positive IgG and IgM results. The first-generation case is usually the first case of a clustered epidemic; if asymptomatic infections or infections during the incubation period occur, it is necessary to conduct a comprehensive analysis and judgment based on epidemiological investigation and laboratory test results. The judgment of second-generation cases meets the following three conditions: only contact with first-generation cases within 14 days before onset; never been to Wuhan and surrounding areas, communities with case reports, or countries where the epidemic situation is severe; no history of suspicious exposure such as hospital visits. The contact history included people with a history of contacting a confirmed or suspected case infected with SARS-CoV-2 within 14 days prior to disease onset.
Epidemiological data (mainly including activity track and exposure history) and demographic, clinical, laboratory and microbiological data were collected and analyzed through on-site epidemiological investigations and electronic medical records. Our study was approved by the ethics committee of The Children's Hospital of Zhejiang University School of Medicine, and written consent was obtained from the guardians of the patients.
Etiological detection
The procedures for sample collection, RT-PCR analysis, and interpretation of results have been previously described (Huang et al., 2020). Real-time polymerase chain reaction (RT-PCR) kits were used to perform nucleic acid testing for SARS-CoV-2 (Shanghai Zhijiang Biotechnology Co., Ltd., Shanghai, China). Rapid Antigen Detection Kits were employed to detect common respiratory viruses, including influenza A, influenza B, adenovirus and respiratory syncytial virus (Kaibili, Hangzhou Innovation Biotechnology Co., Ltd., Hangzhou, China). SARS-CoV-2 antibodies were detected with 2019-nCoV kits (WANTAI, Beijing WANTAI Biological Pharmacy Enterprise Co., Ltd. Beijing, China) .
SARS-CoV-2 culture and whole-genome sequencing
SARS-CoV-2 was isolated from throat or stool samples in a biosafety tertiary laboratory and cultured in Vero E6 cells. From the third day of culture, the cytopathic effect was observed daily by microscopy. On the fifth day, when the cytopathic effect was significant, RT-PCR was used to detect SARS-CoV-2 nucleic acid (Lu et al., 2020). The collected sample was sent to BGI for sequencing. A 200-μL virion suspension was frozen and thawed repeatedly 3 times, and 140 μL supernatant was used for RNA extraction with QIAamp Viral RNA Mini Kit (52904; Qiagen, Heiden, Germany), following the manufacturer's recommendations. Data analysis was conducted at BGI according to a published protocol (Lu et al., 2020).
Phylogenetic tree construction and genome analysis by bioinformatics
To perform phylogenetic tree construction and mutation analysis, 2019 SARS-CoV-2 genomic sequences were retrieved from the following databases (Zhao et al., 2020): NCBI GenBank of the United States, the National Microbiological Science Data Center (NMDC), the National Bioinformatics Center (CNCB), and the National Genomic Science Data Center (NGDC). The sequences were filtered using the following criteria: (1) sequence covered the full length of the viral genome; (2) no ambiguous sequences ('N' bases) or degenerate sequences; (3) no excessively large or high-density gaps after alignment. After filtering, 50 of the SARS-CoV-2 genomic sequences were included. These 50 genomic sequences, as well as one coronavirus sequence (MG772933) isolated from bats and one SARS coronavirus sequence (NC004718), were used as input information to construct a phylogenetic tree together with the isolate SARS-CoV-2-CHZJU in this study.
Multiple-sequence alignment of the SARS-CoV-2 and reference sequences was performed in muscle (Edgar RC., 2004). The phylogenetic tree was constructed using the maximum likelihood method with MEGA X software (Kumar et al., 2018). Variations in this isolate (SARS-CoV-2-CHZJU) were called using MN908947 as the reference sequence. The function of the variation series was annotated manually.
The cryo-EM structure of the spike protein of SARS-CoV-2 (PDB entry: 6 VXX) (Walls et al., 2020) was viewed with PyMOL (Janson et al., 2017); mutagenesis was also performed and visualized with PyMOL.
Statistical analysis
Continuous variables are expressed as medians (ranges). Kruskal-Wallis H tests were used for comparisons among 3 groups, and Wilcoxon rank-sum tests were used for comparisons between 2 groups. Count data are expressed as numbers (%), and Pearson's chi-squared test was applied for comparisons among groups. We judged a two-sided p value less than 0.05 as statistically significant. All data analyses were carried out using SPSS version 20.
Results
Pediatric COVID-19 cases have significantly lower contagiousness
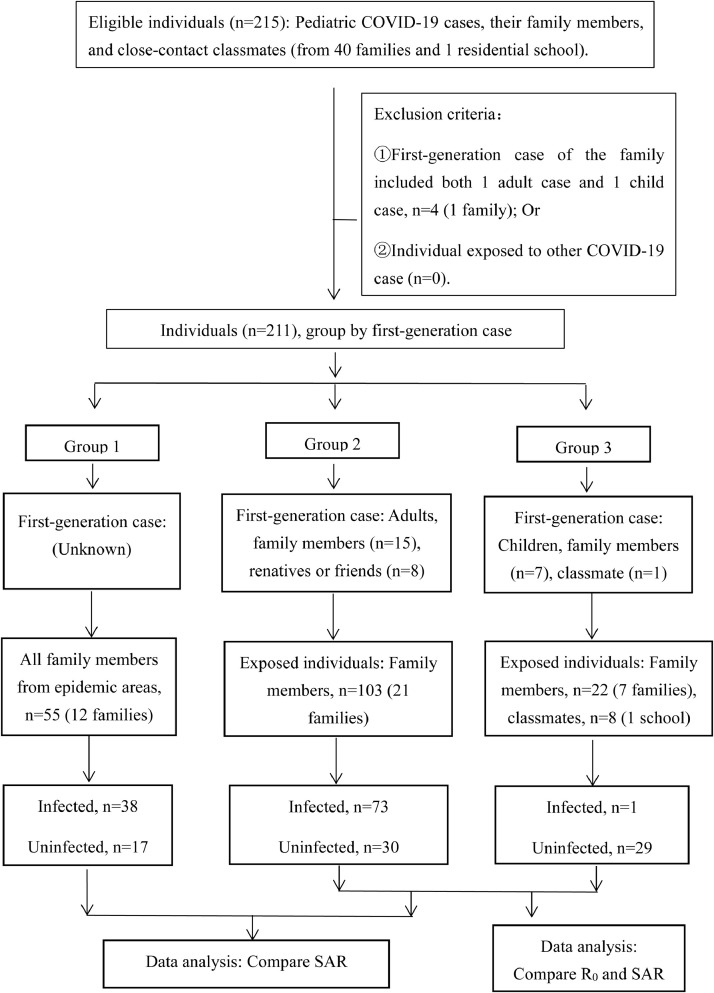
This study enrolled all 46 pediatric COVID-19 cases admitted to 8 hospitals between 21 January and 29 February 2020, including 45 pediatric cases in 40 families with 202 family members and 1 pediatric case with 8 close contacts (4 close-contact classmates and 4 roommates). No other students or staff at a residential school became infected due to contact with the pediatric patient (Figure 1 ). The study enrolled a total of 211 individuals from among 215 eligible individuals, and 4 were excluded according to the exclusion criteria (Figure 1). Among the 211 individuals, the total infection rate was 64% (135/211), and the total secondary attack rate (SAR) was 60% (112/188) (Table 1 ). SARs in Group 1, 2 and 3 were 69% (38/55), 71% (73/103) and 3% (1/30), respectively. The median R0 in Group 2 and 3 was 2 (range: 1-8) and 0 (range: 0-1), respectively (details on R0 in each family are listed in appendix pages 1-30). Statistical analysis revealed significant differences (p<0.05) in the SAR and R0 between Group 2 and 3, suggesting that the ability of children to spread disease is significantly lower than that of adults (Table 1). Moreover, the children in our study were often the last family members to develop symptoms. Between Group 1 and 2, there was no significant difference (p>0.05) in SAR between children and adults (Table 1), but we excluded families without pediatric cases. This result indicates that children are susceptible to SARS-CoV-2 infection but that their susceptibility is not equivalent to that of adults.
Figure 1.
Flow chart of the clinical research on the contagiousness of children and adults with COVID-19. This cohort study enrolled all 46 pediatric patients admitted to 8 hospitals in Zhejiang Province between 21 January and 29 February 2020. The study enrolled a total of 211 individuals, including 202 family members (from 40 families) and 1 pediatric case with 8 close contacts in a residential school. Four eligible individuals were excluded as the first-generation cases of this family, including both adult and child cases, and 211 individuals were divided into 3 groups according to the first-generation case. The secondary attack rate and R0 were compared among the groups.
Table 1.
Comparison of the contagiousness of COVID-19 between children and adults.
| Total | Group 1 | Group 2 | Group 3 | Pa value | |
| General information of first-generation case | |||||
| Age of first-generation case, years (Median, range) | / | Unknown | Adult (>18y) 55 (30-70) | Child (≦18y) 13.5 (10-18) | .. |
| First-generation case (n) | / | Unknown | 23 | 8 b | •• |
| Relationship with individuals (n) | / | Unknown | Family member (15), relative or friend (8) | family member (7), classmate (1) | •• |
| Exposure history of individuals | |||||
| Route of exposure | / | Epidemic area | Feast/Household | Household | •• |
| Time spent with family before symptoms onset of first-generation case, days (Median, range) | / | Unknown | 4 (1-14) | 7.5(4-20) | •• |
| The interval between second-generation case symptom onset and exposure to first-generation case, days (Median, range) | / | Unknown | 7(2-16) | 7 | •• |
| The interval between first case symptom onset and isolation, days (Median, range) | / | 1(1-4) | 2(1-6) | 1(1-3) | •• |
| General information of exposure individuals (Excluding family member of first-generation case) | |||||
| Family (n) | 41 | 12 | 21 | 8 b | •• |
| Exposure adult, n (%) | 122(65) | 35(64) | 68 (66) | 19(63) | 0.938 |
| Exposure child, n (%) | 66(35) | 20(36) | 35 (34) | 11(37) | •• |
| Male, n (%) | 88(47) | 28(51) | 49(48) | 11(37) | 0.442 |
| Female, n (%) | 100(53) | 27(49) | 54(52) | 19(63) | •• |
| Infection of exposure individuals (Excluding family member of first-generation case) | |||||
| Infected adult, n (SAR, %) | 74(61) | 24(69) | 49 (78) | 1c(5) | <0.001 (0.712) |
| Infected child, n (SAR, %) | 38(58) | 14(70) | 24 (69) | 0 | <0.001 (0.912) |
| Infected individual, n (SAR, %) | 112 (60) | 38(69) | 73 (71) | 1c(3) | <0.001 (0.815) |
| R0, (Median, range) | Unknown | Unknown | 2 (1- 8) d | 0 (0-1)d | 0.018 |
| Order of second-generation case onset | |||||
| First symptomatic case (n) | 32A+2C | 12A | 19A+2C | 1A | •• |
| Last symptomatic case (n) | 8A, 27C | 2A, 10C | 6A, 17C | 0 | •• |
Secondary attack rate = (number of confirmed cases in susceptible contacts during the incubation period)/(total number of susceptible contacts) × 100%
R0 = (number of confirmed COVID-19 cases among individuals)/(number of first-generation cases).
p values indicate the difference between 3 family groups, p values in parentheses indicate the difference between Group 1 and 2.
1 Student case in a residential school with 8 close contacts (including 4 classmates around her seat and 4 roommates).
This adult underwent splenectomy after trauma 10 years ago.
Details on R0 for each family are listed in appendix pages 1-29. Indicate: SAR = secondary attack rate; A = adult; C = Child.
Half of the fecal nucleic acid tests were still positive after being undetectable in throat swabs
Among the 46 cases of pediatric COVID-19, 46 (100%), 25 (54%), 17 (37%), 12 (26%), and 5 (11%) had epidemiological histories, fever, cough, runny nose, and diarrhea, respectively. In addition, 10 (22%) patients were asymptomatic, 21 (46%) had upper respiratory tract infection, and 15 (33%) had pneumonia (Table 2 ). After the throat swab nucleic acid test became negative, we tested for fecal nucleic acid and found 26 (57%) pediatric cases that were positive but without gastrointestinal symptoms. The median time for these fecal nucleic acid tests to become negative was 10 days. An infant with a mild infection was positive for fecal nucleic acid for 53 days after the throat swab became negative. There were no significant differences (P>0.05) among those with asymptomatic infection, upper respiratory infection and pneumonia with regard to whole blood cells, lymphocytes, C-reactive protein, alanine aminotransferase, time taken to become PCR negative and duration of hospitalization (Table 2).
Table 2.
Epidemiological and clinical features of pediatric COVID-19 stratified by three clinical types
| Total (n = 46) | Asympotamic infection (n=10) | Upper respiratory infection (n=21) | Pneumonia (n = 15) | Pa value | |
| Demographic and clinical characteristics of pediatric COVID-19 | |||||
| Male, n(%) | 27 (55) | 5(50) | 11(52) | 11(73) | 0.371 |
| Female, n(%) | 19(45) | 5(50) | 10(48) | 4(27) | •• |
| Age, years (Median, range) | 10(0.3-18) | 9.5(1-16) | 10(0.3-18) | 11(0.7-18) | 0.743 |
| Epidemiological history, n(%) | 46(100) | 10(100) | 21(100) | 15(100) | •• |
| Fever, n(%) | 25 (54) | 0 | 13 (62) | 12 (80) | <0.001 (0.245) |
| Cough, n(%) | 17(37) | 0 | 10 (48) | 7 (47) | 0.024 (0.955) |
| Runny nose/sniffle/sore throat, n(%) | 12(26) | 0 | 8 (38) | 4 (27) | 0.078 |
| Diarrhea, n(%) | 5 (11) | 0 | 3 (14) | 2 (13) | 0.457 |
| Tachypnea, n(%) | 1 (2) | 0 | 0 | 1 | •• |
| Underlying disease, n | 2 | 0 | 1b | 1c | •• |
| Laboratory and image characteristics of pediatric COVID-19 | |||||
| White blood cells [(4-10) × 10⁹cells/L], (Median, range) | 5.65 (3.30-12.50) | 5.00 (3.85-9.50) | 6.70 (3.30-11.90) | 5.20 (3.80-12.50) | 0.292 |
| Lymphocytes [(1.1-3.2) × 10⁹cells/L], (Median, range) | 2.05 (0.50-8.20) | 2.45 (1.20-6.60) | 2.10 (0.50-8.20) | 1.70 (0.80-4.10) | 0.127 |
| Ratio of neutrophils/lymphocytes | 1.50 (0.20-11.60) | 0.85 (0.30-2.30) | 1.50 (0.20-11.60) | 1.80 (0.50-6.00) | 0.180 |
| Alanine aminotransferase (<40 U/L), (Median, range) | 18.5(5-105) | 14(9-23) | 21(10-105) | 20(5-77) | 0.061 |
| C-reactive protein (<8 mg/L), (Median, range) | 2.4(0.2-43.1) | 1.1(0.2-5.9) | 2.2(0.2-5.8) | 2.8(0.4-43.1) | 0.154 |
| Procalcitonin (<0.46 ng/ml), (Median, range) | 0.06 (0.01-0.25) | 0.12 (0.02-0.25) | 0.05 (0.01-0.25) | 0.04 (0.01-0.14) | 0.215 |
| Pneumonia imaging, n(%) | 12(36) | 0 | 0 | 15(100) | •• |
| Ground-glass opacity, n(%) | 9 (24) | 0 | 0 | 9(60) | •• |
| Time to become PCR-negative and duration of hospitalization | |||||
| dDuration of positive nucleic acid by throat swab, days (Median, range) | 12(3-27) | 12(4-22) | 15(3-27) | 12(3-21) | 0.544 |
| PCR-positivity of anal swab after throat swab PCR becoming negative, n(%) | 26(57) | 8(80) | 13(42) | 5(33) | 0.056 |
| eDuration of positive fecal nucleic acid after throat swab PCR becoming negative, days (Median, range) | 10(3-53) | 9(4-23) | 10(3-53) | 12(10-31) | 0.263 |
| Duration of hospitalization, days (Median, range) | 20(3-33) | 20(15-27) | 22(3-33) | 15(5-29) | 0.102 |
p values indicate the difference among 3 groups, P values in parentheses indicate the difference between Group 2 and 3; the Chi-squared test was used for rate comparison, analysis of variance was used for mean comparison, and the rank sum test was used for median comparison
fatty liver disease, diabetes;
ankylosing spondylitis;
days from the first positive to the last positive nucleic acid test using a throat swab, followed by two consecutive negatives with an interval of at least 24 hours;
days from the first positive to the last positive of the nucleic acid test using an anal swab, followed by one negative result.
Whole-genome sequencing of SARS-CoV-2 using throat swab and fecal samples from the same infant with a two-month interval
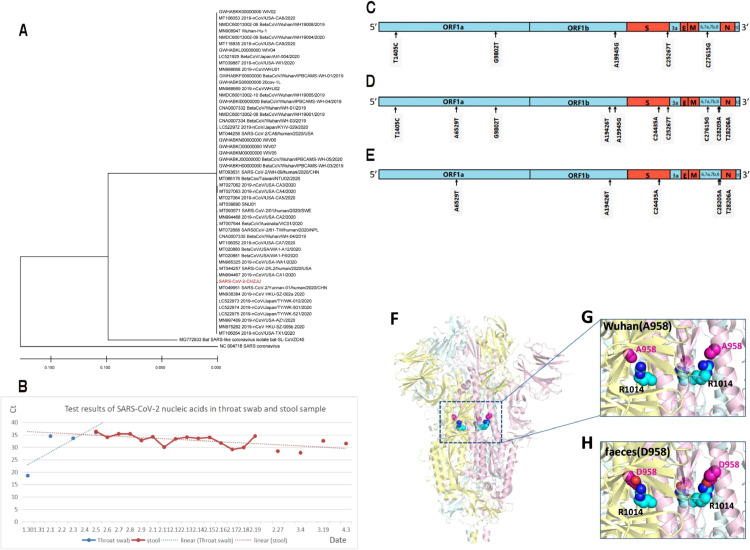
After the throat swab nucleic acid test became negative in pediatric cases, virus cultures based on stool samples were negative, even though 55% (18/33) of the stool samples continued to be positive for viral nucleic acid, indicating that the stool was not a source of infection in the later stage of the disease. In a mild case involving a 3-month-old infant, the duration of SARS-CoV-2 nucleic acid positivity based on a throat swab was 7 days, from 30 January to 5 February. Stool samples were positive for SARS-CoV-2 nucleic acid on 5 February and remained positive for 53 days, with a slightly decreasing Ct value (Figure 2 B). This downward trend in Ct value indicates an increase in viral load and that the virus was reproducing. The infant was isolated but did not receive any medicine as therapy, and the serum tested positive for both IgG and IgM against SARS-CoV-2 on 17 February. Influenza virus, adenovirus, respiratory syncytial virus and parainfluenza virus were not detected by rapid antigen testing using throat swabs. Throat swab specimens form the infant were collected on 1 February and fecal specimens on 9, 17, and 27 February, on 4 and 19 March, and on 3 April; SARS-CoV-2 cultures were carried out. Only the virus culture based on the early pharyngeal swab was positive; it was sent to BGI for whole-genome sequencing. The stool virus cultures were all negative. The Ct value for the stool sample obtained on 3 April was 31.5. However, as the stool culture did not show a cytopathic effect on the third day, the culture supernatant was sent for whole-gene sequencing, and fortunately, we obtained the whole-genome sequence.
Figure 2.
New mutations of SARS-CoV-2 were identified separately from throat swab and fecal samples from the same infant at a two-month interval. (A) Phylogenetic trees of genetic sequences. Red text indicates the coronavirus strains detected in the infant in the present study. (B) SARS-CoV-2 nucleic acid-specific Rdrp test results in different infant samples. The trendline was generated by using SARS-CoV-2 nucleic acid-specific Rdrp test results. (C.D. E) A summary of the nucleotide mutations in the strain (SARS-CoV-2-CHZJU) compared to the reference sequence (MN908947). C24435A and A19945G are missense variants; the others are synonymous variants. (C) Mutations in the strain isolated from throat samples. (D) Mutations in the sequence from stool samples. (E) Mutation of sequences between stool samples and throat samples at a two-month interval. (F.G. H) The structure and the A958D mutation of the spike protein. (F) An overview of the spike protein trimer and the locations of A958 and R1014. (G) A958 is close to R1014 in space. (H) Salt bridge formation between D958 and R1014 predicted by PyMOL. Three A/D958 residues are shown in magenta spheres, and three R1014 residues are shown in cyan spheres.
The evolutionary tree between SARS-CoV-2 sequences was highly similar
The phylogenetic evolutionary tree showed high similarity between the SARS-CoV-2 sequences isolated (Figure 2.A). The average difference between the sequences is 0.019%, and the sequence difference between this isolate (SARS-CoV-2-CHZJU) and the remaining 50 strains was found to be 0.08-0.10%. Compared to the outgroup, all SARS-CoV-2 strains clustered into one specific group. Among the two reference sequences used as outgroups, the bat-SL-CoVZC45 strain isolated from bats was closest to SARS-CoV-2, with an average sequence difference of 14%; the SARS coronavirus was slightly separated, with a sequence difference of 27.8%.
The new A958D mutation improves theoretically viral thermostability
Compared to the reference sequence (MN908947), a summary of the nucleotide mutations in the strain (SARS-CoV-2-CHZJU) is shown in Figure 2(C.D.E). A19945G and C24435A are missense variants; the others are synonymous variants. Intestine-derived SARS-CoV-2 mutations are presented in Figure 2.E. Among these mutations, the new A958D mutation of the spike protein (24435C>A mutation in the virus genome) is of special interest because it is the only one for which the 3D structure of the protein has been solved. The structure reveals that A958 is located in the middle of one α-helix of heptad repeat 1 (HR1) in the S2 domain of the spike protein (Figure 2.F). Very intriguingly, A958 is very close to R1014 from the neighbouring helix (Figure 2. G), and R1014 is stabilized by forming H-bonds with Q954 and Q1011 in the wild-type protein. The structure suggests that the A958D mutation will lead to strong salt bridge formation between D958 and R1014 (Figure 2. H), which we expect would dramatically improve the thermostability of the spike protein.
Discussion
SARS-CoV-2 is a highly contagious virus that has caused a global pandemic. To explore the contagiousness of pediatric cases, close-contact persons in families or schools were divided into 3 groups according to the first-generation case. The first-generation case had close contact with family members from 2 days before the onset of symptoms to 1 day after onset, and this contact period was within the contagious time of the first-generation case. There were no significant differences among the groups in age or sex. In the 21 families of Group 2, the median R0 was 2 (range: 1-8), which is consistent with previous reports [R0 = 2.2-3.5 (Chen et al., 2020; Viceconte and Petrosillo, 2020) and Steven Sanche reported R0 values up to 5.7 (Sanche et al., 2020)]. Among 30 close contacts of 8 pediatric cases in Group 3, only 1 adult was infected, and this adult was immunocompromised due to spleen resection 10 years prior. SARs and R0 in Group 2 were significantly higher (p<0.05) than those in Group 3, which suggests that compared to the adult case, the contagiousness of the pediatric case was much lower. Our results are consistent with the low transmission rate of children reported by Xue Li (Xue et al., 2020). Indeed, children have limited infectivity for those with normal immunity but can infect the immunocompromised.
Following infection, the viral load in pediatric respiratory secretions was high during the end of the incubation period and at the onset of symptoms (Vera et al., 2020; Maltezou et al., 2020). Children are young with less muscle mass, and the ability to produce droplets when coughing is lower in children than in adults. In addition, children tend to swallow respiratory secretions instead of expectorating, which reduces their ability to transmit virus. COVID-19 is similar to tuberculosis in that children are significantly less contagious than adults.
Our study shows that COVID-19 in children mainly comprises upper respiratory infections and pneumonia, followed by asymptomatic infection. This result is consistent with Wuhan's and national reports, but the proportion of asymptomatic infections in our study was higher (Lu et al., 2020; Dong et al., 2020). Fever, cough and runny nose are the main symptoms. Among pediatric COVID-19 patients, nearly one-third had pneumonia, and a quarter exhibited lung ground-glass opacities, though both rates were lower than those reported in Wuhan (Wang et al., 2020).
After the respiratory viral nucleic acid test became negative, fecal samples in half of the pediatric cases were still positive for nucleic acid. In a 3-month-old infant, the fecal nucleic acid test continued to be positive for 53 days after the pharyngeal swab became negative. The Ct value was approximately 30, suggesting that SARS-CoV-2 first multiplied in the respiratory tract, causing upper respiratory infection; after SARS-CoV-2 was swallowed, it multiplied in the intestine, resulting in asymptomatic infection of the digestive tract and excretion of viral nucleic acid for a long time, as primary human airway epithelial cells and gut enterocytes are permissive to SARS-CoV-2 infection (Parolin et al., 2021). Overall, the pediatric COVID-19 patients shed viral nucleic acid in the feces for a long time after the throat swab nucleic acid test became negative, which has also been reported previously (Xu et al., 2020).
SARS-CoV-2 enters host cells through the receptor angiotensin-converting enzyme-2 (ACE-2), and ACE-2 is highly expressed in lung AT2 cells, esophageal epithelial cells, stratified epithelial cells and absorptive enterocytes in the ileum and colon (Harmer et al., 2002; Lalitha Guruprasad, 2020). Other research teams have also isolated SARS-CoV-2 from stool samples (Jin et al., 2020). The intestines of children are susceptible to SARS-CoV-2 infection, which may be related to their immature function and imperfect intestinal flora.
The sequence isolated from the infant (SARS-CoV-2-CHZJU) is highly homologous to known strains. The sequence difference between this isolate (SARS-CoV-2-CHZJU) and the 50 known strains is 0.08-0.10%. This isolate (SARS-CoV-2-CHZJU) does not harbor many mutations, except for the orf1ab region (hypervariable region), which is highly variable in all SARS-CoV-2 strains worldwide. Compared to the reference sequence (MN908947), only five missense mutations (S, ORF3a, E, M, and ORF8) were found in the relatively conserved region, and two of these mutations (S and ORF8) occur frequently in multiple SARS-CoV-2 strains.
Compared to the sequence isolated from throat swabs, the 24435C>A mutation detected in stool leads to a missense A958D mutation in the S2 domain of the spike protein. Because A958 faces R1014 of the neighboring helix, we predict that the A958D mutation will inevitably lead to salt bridge formation between D958 and R1014, which will dramatically improve the thermostability of the spike protein. A958 is located on HR1 of the S2 subunit; therefore, it is possible that this mutation may also affect cell-cell fusion after viral attachment to the host cell receptor. We deduce that the new A958D mutation might have been a potential reason for the long residence of SARS-CoV-2 in the infant's intestines without apparent symptoms.
Conclusions
This study indicates that (1) children have a lower ability to spread COVID-19 and that the first-generation case of cluster outbreaks among children is usually an adult, as opposed to a child; (2) pediatric COVID-19 may have long fecal virus nucleic acid shedding times, but the stool is not a source of infection after throat swab PCR becomes negative; and (3) the new A958D mutation of the spike protein improves SARS-CoV-2 thermostability and affects cell-cell fusion potentially after viral attachment to the host cell receptor.
Funding
This study was supported by the National Science Foundation of China (82071812), Zhejiang University special scientific research fund for COVID-19 prevention and control, The role and mechanisms of macrophages in SARS-CoV-2-induced acute lung injury, and the National Science Foundation of Zhejiang Province, China (LY19H100003).
Conflicts of interest
The authors declare that they have no conflicts of interest.
Acknowledgments
We thank Professor Feng Ling and Ziping Miao from Zhejiang Provincial Center for disease control and prevention to provide support for field epidemiological investigations.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ijid.2021.08.036.
Appendix. Supplementary materials
References
- CDC COVID-19 Response Team Coronavirus Disease 2019 in Children - United States. MMWR Morb Mortal Wkly Rep. 2020;69(14):422–426. doi: 10.15585/mmwr.mm6914e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Zhang L, Du H. SARS-CoV-2 Infection in Children. N Engl J Med. 2020;382(17):1663–1665. doi: 10.1056/NEJMc2005073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Li X, Zhu B. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat Med. 2020;26(4):502–505. doi: 10.1038/s41591-020-0817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X, Lian JS, Hu JH. Epidemiological, clinical and virological characteristics of 74 cases of coronavirus-infected disease 2019 (COVID-19) with gastrointestinal symptoms. Gut. 2020;69(6):1002–1009. doi: 10.1136/gutjnl-2020-320926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cella E, Benedetti F, Fabris S. SARS-CoV-2 Lineages and Sub-Lineages Circulating Worldwide: A Dynamic Overview. Chemotherapy. 2021;66(1-2):3–7. doi: 10.1159/000515340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokman SM, Rasheduzzaman M, Salauddin A. Exploring the genomic and proteomic variations of SARS-CoV-2 spike glycoprotein: A computational biology approach. Infect Genet Evol. 2020;84 doi: 10.1016/j.meegid.2020.104389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell. 2020;182(4):812–827. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Wang Y, Li X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao WM, Song SH, Chen ML. The 2019 novel coronavirus resource. Yi Chuan. 2020;42(2):212–221. doi: 10.16288/j.yczz.20-030. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K.MEGA X. Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181(2):281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janson G., Zhang C., Prado M.G., Paiardini A. PyMod 2.0: improvements in protein sequence-structure analysis and homology modeling within PyMOL. Bioinformatics. 2017;33:444–446. doi: 10.1093/bioinformatics/btw638. [DOI] [PubMed] [Google Scholar]
- Chen TM, Rui J, Wang QP, Zhao ZY, Cui JA, Yin L. A mathematical model for simulating the phase-based transmissibility of a novel coronavirus. Infect Dis Poverty. 2020;9(1):24. doi: 10.1186/s40249-020-00640-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viceconte G, Petrosillo N. COVID-19 R0: Magic number or conundrum? Infect Dis Rep. Infect Dis Rep. 2020;12(1):8516. doi: 10.4081/idr.2020.8516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanche S, Lin YT, Xu C, Romero-Severson E, Hengartner N, Ke R. High Contagiousness and Rapid Spread of Severe Acute Respiratory Syndrome Coronavirus 2. Emerg Infect Dis. 2020;26(7):1470–1477. doi: 10.3201/eid2607.200282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltezou HC, Magaziotou I, Dedoukou X. Children and Adolescents With SARS-CoV-2 Infection: Epidemiology, Clinical Course and Viral Loads. Pediatr Infect Dis J. 2020;39(12):e388–e392. doi: 10.1097/INF.0000000000002899. [DOI] [PubMed] [Google Scholar]
- Dong Y, Mo X, Hu Y. Epidemiology of COVID-19 Among Children in China. Pediatrics. 2020;145(6) doi: 10.1542/peds.2020-0702. [DOI] [PubMed] [Google Scholar]
- Wang D, Hu B, Hu C. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parolin C, Virtuoso S, Giovanetti M, Angeletti S, Ciccozzi M, Borsetti A. Animal Hosts and Experimental Models of SARS-CoV-2 Infection. Chemotherapy. 2021;66(1-2):8–16. doi: 10.1159/000515341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer D, Gilbert M, Borman R, Clark KL. Quantitative mRNA expression profiling of ACE 2, a novel homologue of angiotensin converting enzyme. FEBS Lett. 2002;532(1-2):107–110. doi: 10.1016/s0014-5793(02)03640-2. [DOI] [PubMed] [Google Scholar]
- Guruprasad Lalitha. Evolutionary relationships and sequence-structure determinants in human SARS coronavirus-2 spike proteins for host receptor recognition. Proteins. 2020;88(11):1387–1393. doi: 10.1002/prot.25967. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.