Abstract
BACKGROUND
Artificial intelligence in colonoscopy is an emerging field, and its application may help colonoscopists improve inspection quality and reduce the rate of missed polyps and adenomas. Several deep learning-based computer-assisted detection (CADe) techniques were established from small single-center datasets, and unrepresentative learning materials might confine their application and generalization in wide practice. Although CADes have been reported to identify polyps in colonoscopic images and videos in real time, their diagnostic performance deserves to be further validated in clinical practice.
AIM
To train and test a CADe based on multicenter high-quality images of polyps and preliminarily validate it in clinical colonoscopies.
METHODS
With high-quality screening and labeling from 55 qualified colonoscopists, a dataset consisting of over 71000 images from 20 centers was used to train and test a deep learning-based CADe. In addition, the real-time diagnostic performance of CADe was tested frame by frame in 47 unaltered full-ranged videos that contained 86 histologically confirmed polyps. Finally, we conducted a self-controlled observational study to validate the diagnostic performance of CADe in real-world colonoscopy with the main outcome measure of polyps per colonoscopy in Changhai Hospital.
RESULTS
The CADe was able to identify polyps in the test dataset with 95.0% sensitivity and 99.1% specificity. For colonoscopy videos, all 86 polyps were detected with 92.2% sensitivity and 93.6% specificity in frame-by-frame analysis. In the prospective validation, the sensitivity of CAD in identifying polyps was 98.4% (185/188). Folds, reflections of light and fecal fluid were the main causes of false positives in both the test dataset and clinical colonoscopies. Colonoscopists can detect more polyps (0.90 vs 0.82, P < 0.001) and adenomas (0.32 vs 0.30, P = 0.045) with the aid of CADe, particularly polyps < 5 mm and flat polyps (0.65 vs 0.57, P < 0.001; 0.74 vs 0.67, P = 0.001, respectively). However, high efficacy is not realized in colonoscopies with inadequate bowel preparation and withdrawal time (P = 0.32; P = 0.16, respectively).
CONCLUSION
CADe is feasible in the clinical setting and might help endoscopists detect more polyps and adenomas, and further confirmation is warranted.
Keywords: Computer-assisted detection, Artificial intelligence, Deep learning, Colonoscopy, Clinical validation, Colorectal polyp
Core Tip: Our study indicated that the deep learning-based computer-assisted detection system trained from the dataset consisting of the largest number of polyps achieved high diagnostic performance on the test dataset of images, colonoscopy videos and clinical validation. This system might aid colonoscopists in finding more polyps and adenomas and deserves to be further validated in multicenter randomized trials.
INTRODUCTION
Colorectal cancers (CRC) are the third most prevalent cancer and the second highest cause of cancer deaths worldwide[1]. Colonoscopy is considered to be the gold standard for CRC screening[2] but cannot detect all colonic neoplasms[3,4]. Colonoscopy has been reported to miss 17%-48% of adenomas, which are considered to represent the causes of 50%-60% of interval cancers[5-7].
To detect more polyps, a wealth of auxiliary devices and technology have been invented to improve adenoma detection rates (ADRs) and reduce adenoma miss rates (AMRs)[8,9]. However, these devices cannot overcome the limitations of colonoscopists themselves, who could overlook the polyps flashing across the video screen[10]. The ability of colonoscopists to detect adenoma is also influenced by alertness, fatigue and monitoring and varies greatly among individual colonoscopists[11,12]. In addition, even colonoscopists with a high ADR may still miss adenomas because they may become less vigilant about inspecting the remaining colon after detecting one adenoma (one and done effect)[13].
Over the last two decades, computer-assisted polyp detection has been increasingly explored to improve inspection quality and reduce AMR[4,14]. Recently, artificial intelligence has made remarkable breakthroughs in medical fields with deep learning and convolutional neural networks (CNNs)[15,16]. Deep learning automatically makes use of CNNs, which logically imitate the structure and activity of brain neurons, to learn detailed features of medical images[17]. With sufficient learning materials, CNNs can reach even greater real-time detection accuracy than human experts, which suggests that computer-assisted detection systems (CADes) might serve as real-time “experts” to improve the quality of colonoscopies[15,18-20]. However, despite promising findings, most current CADes were established using a limited number of colonoscopic images from a single center, which might limit the robustness and generalizability of findings in wide practice[18,21]. Therefore, we trained and tested the CNN-based CADe using colonoscopic images of multicenter datasets as well as tested and validated its diagnostic performance in real-time colonoscopy videos and real-world colonoscopy.
MATERIALS AND METHODS
Overview of study design
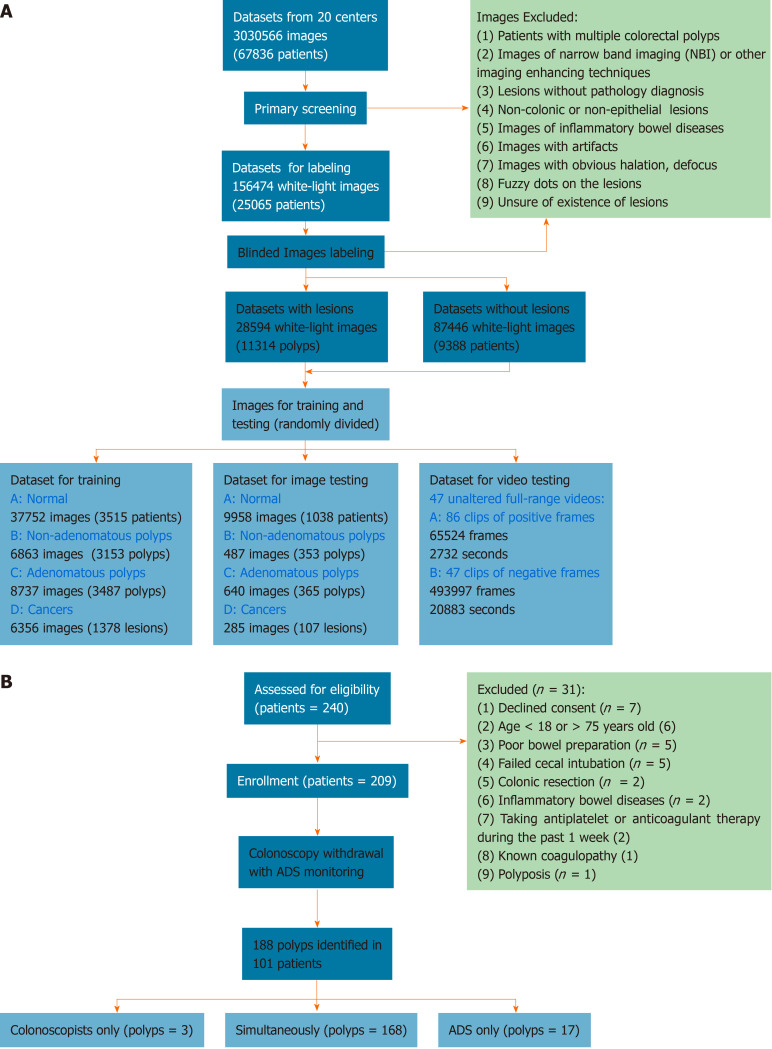
The colonoscopic records were retrospectively collected from 20 centers (Supplementary Table 1). Figure 1A illustrates the workflow of preprocessing images. Fifty-five qualified colonoscopists screened and labeled the images in a specific labeling system (Supplementary Video 1), and the labeled images were randomly divided into training and test datasets for the construction and testing of the CADe. The detailed preprocessing is illustrated in the Appendix A. To evaluate comprehensively ADe’s real-time diagnostic performance, 47 unaltered full-range colonoscopy videos of routine practice, including 86 histologically confirmed polyps, were collected (Supplementary Table 2, Appendix B), and a self-controlled observational study was conducted for clinical validation in the Changhai Endoscopy Center.
Figure 1.
Flowchart of image preprocessing and validation in clinical trials. A: Image preprocessing; B: Validation in clinical trials.
Construction of CADe
The CADe was built based on the You Only Look Once v2 deep learning framework. The training dataset was used to build the model, and the testing dataset was used to test the performances with the details illustrated in the Appendix C.
Validation of the CADe in clinical colonoscopy
Figure 1B shows the process of enrolling patients. Consecutive outpatients aged 18-75 years who were scheduled for screening, surveillance and diagnostic colonoscopies were invited to participate in the validation between November 1, 2018 and December 10, 2018. Exclusion criteria included declined consent, age < 18 or > 75 years, poor bowel preparation quality, failed cecal intubation, history of colonic resection, inflammatory bowel disease, antiplatelet or anticoagulant therapy during the past 1 wk, known coagulopathy and polyposis. Patients received 3 L polyethylene glycol as a split-dose bowel preparation and were either sedated with propofol or without. Twenty colonoscopists, including two trainees, with experience ranging from 100 to over 20000 procedures performed colonoscopies, and an additional colonoscopist served as the full-time observer. The ADRs of the included colonoscopists ranged from 14%-33% in a mixed-indications population. Water-based colonoscopy, antispasmodics, distal attachments, optical-enhanced imaging and image-guided devices were not used in the validation.
CADe was automatically initiated when the ileocecal valve was identified. When identifying any potential polyp, the CADe presented an alert rectangle surrounding polyps on a second monitor and sounded alert, which was audible to colonoscopists and the observer. However, colonoscopists only observed the regular monitor of colonoscopy and were unable to see directly the alert rectangle since the second monitor (showing CADe’s findings) was placed on their rear-right side. To view the findings identified by CADe, the colonoscopists had to turn back toward their right. When conducting withdrawal, the colonoscopists were required to offer immediately a verbal signal once they detected a potential polyp. The observer observed the two monitors, received alerts of CADe and signals from colonoscopists and finally determined the detection priority according to the following instructions.
Polyps identified in the study were finally confirmed by careful inspection of colonoscopists, biopsy and histologic examination, and the potential causes of false detection were determined by the discussion of colonoscopists and observers. Under the guidance of the observer, once hearing the alerts of the CADe, colonoscopists were required to recheck immediately all findings of the CADe, including observing the second monitor and carefully inspecting highlighted areas by the CADe. The duration of defining polyp-detection priority was confined to continuous withdrawal duration, and any process of observing the second monitor, seeking or unfolding polyps for rechecking, or performing biopsy and polypectomy was excluded. If the polyp was only identified by the CADe and colonoscopists could not localize the polyp before observing the second monitor, it was defined as type A (CADe only, missed by colonoscopists). If the polyp was reported by the colonoscopists and missed by the CADe before an unfolding or close view, it was defined as type B (colonoscopists only, missed by CADe). If the polyp was reported by both the colonoscopists and CADe during continuous withdrawal, it was defined as type C (colonoscopists and CADe). Therefore, types A + B + C included findings of the colonoscopists + CADe, whereas types B + C were classified as findings of the colonoscopists alone.
Outcome measures
For clinical validation, the main outcome measures were the mean polyps per colonoscopy and the sensitivity of CADe[22]. The mean adenomas per colonoscopy, false positives and negatives of CADe and potential causes were also analyzed. For the test dataset, the diagnostic performance was assessed based on the accuracy, sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) with definitions illustrated in the Appendix D.
Sample size and statistical analysis
The sample estimation was based on the number of polyps per colonoscopy using the comparison of paired quantitative data. In the pilot trial, we found that the CADe could help increase the identification of 0.1 polyps per colonoscopy with a standard derivation of 0.37 for the improvement and assumed a power of 0.9 with an α-error of 0.01 to determine the sample size. Therefore, 203 patients were at least required for clinical validation. Continuous data are presented as the means and standard deviations. The number of polyps identified by each detection method was compared using a Wilcoxon signed ranks test. A 2-sided McNemar test with a significance level of 0.05 was performed to compare the polyp detection rate (PDR), ADR and sensitivity between colonoscopists and colonoscopists + CADe. A 2-sided P value < 0.05 was considered statistically significant, whereas P values for multiple groups were compared with relevant levels after Bonferroni correction. All statistical analyses were performed using SPSS Statistics v21 (IBM, Armonk, NY, United States).
Ethics and study registration
This study received approval from the ethics committee of Shanghai Changhai Hospital and registered at ClinicalTrials.gov (identifier, NCT03761771) and was performed in accordance with the Declaration of Helsinki. The use of retrospective data (images and historical reports) and the study protocol was approved or exempted by the ethics committee of every hospital. Informed and written consent was obtained from all patients during clinical validation, and patients had the right to withdraw at any time. All authors had access to the study data and approved the final manuscript.
RESULTS
Polyp identification in the test dataset
Supplementary Table 3 shows the characteristics of the polyps in the test dataset. There were 285, 640 and 487 images of adenocarcinomas, adenomatous polyps and nonadenomatous polyps in the test datasets, respectively (Supplementary Table 3). Of the 285 adenocarcinomas, 17.2% (49/285) could not be classified by the Paris classification because advanced stages were suspected. A total of 99.2% (483/487) of the nonadenomatous polyps and 96.6% (618/640) of the adenomatous polyps were classified as sessile or flat type (Supplementary Table 3).
The mean time for polyp identification of CADe was 0.03 ± 0.01 s. The receiver operating characteristic curve (ROC) is presented in Figure 2. Based on the ROC curve, the optimal cutoff value of probability was determined to be 50%, which is indicated by the red dot in Figure 2. The CADe shows excellent diagnostic performance in identifying polyps with an area under the curve of 0.984. Overall, the CADe identified colorectal lesions with 95.0% sensitivity, 99.1% specificity, 93.7% PPV and 99.3% NPV (Table 1), and the highest sensitivity was noted for adenocarcinomas (97.2%, Table 1). Moreover, CADe exhibited the highest sensitivity for large polyps (96.7% for ≥ 10 mm) and the lowest sensitivity for diminutive polyps (94.1% for ≤ 5 mm, Table 1). Fecal fluid/bubble (24%), reflection of light (21%), difficult angle (19%) and shadow (11%) were considered to be the four most common causes of false negatives (Table 2, Supplementary Figure 1), whereas folds (19%), reflections of light (18%), fecal fluid/bubble (17%) and colonic/ileocecal valves (19%) were the most important causes of false positives (Table 2, Supplementary Figure 2).
Figure 2.
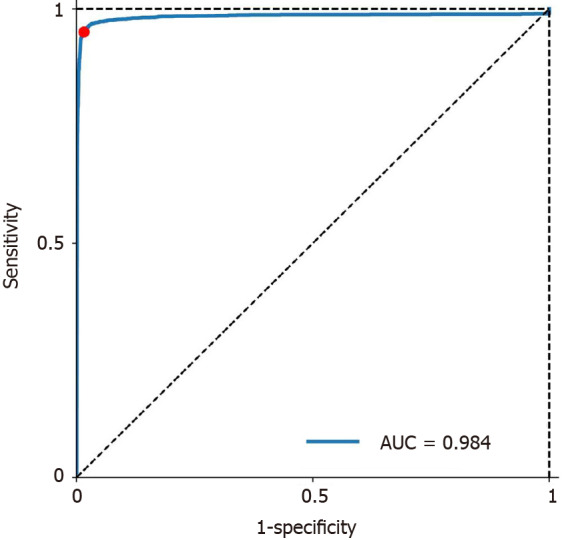
Receiver operating characteristic curve for localization of polyps. AUC: Area under the curve.
Table 1.
Performance of lesion detection and localization
|
|
Lesions
|
Sensitivity, %
|
Specificity, %
|
PPV, %
|
NPV, %
|
| Overall | 1412 | 95.0 (93.7-96.0) | 99.1 (98.9-99.3) | 93.7 (92.3-94.9) | 99.3 (99.1-99.4) |
| Pathology | |||||
| Non-adenomatous polyps | 487 | 93.4 (90.8-95.4) | 99.1 (98.9-99.3) | 83.5 (80.0-86.5) | 99.7 (99.5-99.8) |
| Adenomatous polyps | 640 | 95.2 (93.1-96.6) | 99.1 (98.9-99.3) | 87.1 (84.4-89.5) | 99.7 (99.5-99.8) |
| Adenocarcinomas | 285 | 97.2 (94.3-98.7) | 99.1 (98.9-99.3) | 75.5 (70.7-79.7) | 99.9 (99.8-100.0) |
| Size | |||||
| ≤ 5 mm | 713 | 94.1 (92.1-95.7) | 99.1 (98.9-99.3) | 88.2 (85.6-95.3) | 99.6 (99.4-99.7) |
| 6-9 mm | 396 | 95.2 (92.5-97.0) | 99.1 (98.9-99.3) | 80.7 (76.8-84.1) | 99.8 (99.7-99.9) |
| ≥ 10 mm | 303 | 96.7 (93.8-98.3) | 99.1 (98.9-99.3) | 76.5 (71.9-80.6) | 99.9 (99.8-99.9) |
| Paris classification | |||||
| Ip | 180 | 96.0 (91.8-98.3) | 99.1 (98.9-99.3) | 65.8 (59.7-71.4) | 99.9 (99.8-1.00) |
| Is | 581 | 96.1 (94.0-97.4) | 99.1 (98.9-99.3) | 86.1 (83.2-88.6) | 99.8 (99.6-99.8) |
| IIa | 525 | 92.6 (89.9-94.6) | 99.1 (98.9-99.3) | 84.4 (81.1-87.2) | 99.6 (99.5-99.7) |
| IIb | 30 | 100.0 (85.9-100.0) | 99.1 (98.9-99.3) | 25.0 (17.7-33.9) | 100.0 (99.9-100.0) |
| IIc | 2 | 100.0 (19.8-100.0) | 99.1 (98.9-99.3) | 2.2 (0.4-8.4) | 100.0 (99.9-100.0) |
| III | 32 | 96.9 (82.0-99.8) | 99.1 (98.9-99.3) | 25.6 (18.3-34.5) | 100.0 (99.9-100.0) |
| Unclassified | 62 | 96.8 (87.8-99.4) | 99.1 (98.9-99.3) | 40.0 (32.2-48.3) | 100.0 (99.9-100.0) |
PPV: Positive predictive value; NPV: Negative predictive value.
Table 2.
Potential causes of false negatives and false positives
|
Causes of false negatives
|
No. of images, n (%)
|
Causes of false positives1
|
No. of images, n (%)
|
| Test dataset | |||
| Fecal fluid/bubble | 17 (24) | Folds | 18 (19) |
| Reflections of light | 15 (21) | Reflections of light | 17 (18) |
| Difficult angle | 13 (19) | Fecal fluid/bubble | 16 (17) |
| Shadow | 8 (11) | Colonic valve | 11 (12) |
| Fuzzy image | 6 (9) | Ileocecal valve | 7 (7) |
| Color of background | 2 (3) | Fuzzy image | 5 (5) |
| Far distance | 2 (3) | Others2 | 11 (12) |
| Unclassified | 7 (10) | Unclassified | 10 (11) |
| Validation in clinical colonoscopy | |||
| Shadow | 1 (33.3) | Folds3 | 276 (59.0) |
| Difficult angle | 1 (33.3) | Reflections of light | 89 (19.0) |
| Depressed lesion | 1 (33.3) | Fecal fluid/bubble | 43 (9.2) |
| Normal structures4 | 46 (9.8) | ||
| Other lesions5 | 14 (3.0) | ||
Twelve lesions were considered to be missed by senior colonoscopists on colonoscopy.
Others included color change (3), anus (3), lesions under the mucous (2), melanosis coli (1), and bloodstain (1).
Sixteen folds were caused by suction.
Normal structures included vessels (20), ileocecal valve (12), anus (8), and lymphoid follicle (6).
Other lesions included ulcers (7), cysts under the mucous (4), and diverticulum (3).
Polyp identification in colonoscopy videos
The video dataset consisted of 56 adenomas and 30 nonneoplastic polyps, and 53, 27 and 6 polyps exhibited diameters of ≤ 5 mm, 6-9 mm and ≥ 10 mm, respectively (Supplementary Table 2). Most of the included polyps (71/86) exhibited flat or sessile morphology, and a small portion of polyps (13/86) were considered challenging to detect in routine practice (Supplementary Table 2).
For frame-based tests, a dataset of 86 video clips of positive frames (65524 frames and 2732 s in total) and 47 clips of negative frames (493997 frames and 20883 s) were constructed for test. Although CADe identified all 86 polyps, regarding frame-based analysis, there was an overall sensitivity of 92.2% and specificity of 93.6% for overall polyps and a sensitivity of 66.2% and specificity of 97.9% for “challenging” polyps (Supplementary Table 4, Supplementary Video 2).
Validation of the CADe in clinical colonoscopy
In total, 240 patients were recruited for validation, and 31 were excluded due to refusal of informed consent (7), age (6), poor bowel preparation (5), failed cecal intubation (5) and others (8) (Figure 1B). The baseline characteristics are shown in Supplementary Table 3, and 188 polyps were identified in 101 patients (Figure 1B), resulting in a 48.3% PDR. The withdrawal time of 107 procedures (52.2%) was shorter than 6 min (Supplementary Table 5).
The number of polyps identified by colonoscopists only, CADe only and both were three, 17 and 168, respectively, revealing a sensitivity of 98.4% (185/188) for CADe (Supplementary Table 6). When assisted by the CADe, the number of identified polyps and adenomas significantly increased (0.82 vs 0.90, P < 0.001; 0.30 vs 0.32, P < 0.05, respectively), although the PDR and ADR did not increase (P = 0.06 and P = 0.13, respectively, Tables 3 and 4). The diagnostic sensitivity of the CADe was significantly greater than that of colonoscopists [98.4% (185/188) vs 91.0% (171/188), P = 0.03, Supplementary Table 6]. A total of 468 false positives occurred in 209 colonoscopy withdrawals (2.2 false positives per colonoscopy withdrawal); folds (59.0%), reflections of light (19.0%), fecal fluid/bubble (9.2%) and normal structures (9.8%) were considered to be the common causes of false positives (Table 2).
Table 3.
Polyp detection between colonoscopists and colonoscopists + computer-assisted detection
|
|
Colonoscopists
|
Colonoscopists + CADe
|
P
value
|
||
| PDR, % | 45.9 | 48.3 | 0.06 | ||
| SSR, % | 3.83 | 4.78 | 0.50 | ||
| CRSR, % | 9.09 | 11.5 | 0.06 | ||
| Number of polyps, mean ± SD | 0.82 ± 1.20 | 0.90 ± 1.25 | < 0.001 | ||
| Pathology | |||||
| Non-adenomatous | 0.52 ± 0.94 | 0.58 ± 1.00 | 0.01 | ||
| Adenomatous | 0.30 ± 0.62 | 0.32 ± 0.64 | 0.025 | ||
| Age | |||||
| < 50 | 0.45 ± 0.70 | 0.51 ± 0.77 | 0.03 | ||
| ≥ 50 | 1.12 ± 1.42 | 1.22 ± 1.47 | 0.002 | ||
| Sex | |||||
| Male | 0.96 ± 1.28 | 1.05 ± 1.36 | 0.005 | ||
| Female | 0.66 ± 1.07 | 0.72 ± 1.11 | 0.01 | ||
| Location | |||||
| Proximal | 0.35 ± 0.69 | 0.39 ± 0.76 | 0.01 | ||
| Distal | 0.46 ± 0.89 | 0.51 ± 0.90 | 0.003 | ||
| Size | |||||
| ≥ 10 mm | 0.06 ± 0.28 | 0.06 ± 0.28 | 1 | ||
| 6-9 mm | 0.18 ± 0.52 | 0.19 ± 0.52 | 0.32 | ||
| ≤ 5 mm | 0.57 ± 0.95 | 0.65 ± 1.00 | < 0.001 | ||
| Morphology | |||||
| Flat | 0.67 ± 1.03 | 0.74 ± 1.08 | 0.001 | ||
| Subpedunculated | 0.11 ± 0.42 | 0.12 ± 0.42 | 0.16 | ||
| Pedunculated | 0.03 ± 0.21 | 0.03 ± 0.21 | 1 | ||
| Indications | |||||
| Screening | 0.76 ± 1.15 | 0.85 ± 1.21 | 0.03 | ||
| Surveillance | 1.19 ± 1.42 | 1.33 ± 1.53 | 0.03 | ||
| Diagnosis | 0.70 ± 1.10 | 0.75 ± 1.13 | 0.03 | ||
| Colonoscopes | |||||
| CF-Q290 | 0.78 ± 1.09 | 0.87 ± 1.17 | < 0.001 | ||
| CF-Q260 | 1.04 ± 1.58 | 1.15 ± 1.56 | 0.08 | ||
| CF-Q240 | 0.83 ± 1.64 | 0.83 ± 1.64 | 1 | ||
| Experience | |||||
| > 3000 | 1.24 ± 1.48 | 1.24 ± 1.48 | 1 | ||
| 1000-3000 | 0.79 ± 1.16 | 0.91 ± 1.26 | 0.003 | ||
| < 1000 | 0.54 ± 0.92 | 0.62 ± 0.99 | 0.03 | ||
| BBPS | |||||
| < 6 | 0.59 ± 1.19 | 0.62 ± 1.19 | 0.32 | ||
| ≥ 6 | 0.87 ± 1.19 | 0.96 ± 1.26 | < 0.001 | ||
| Withdrawal time | |||||
| < 6 min | 0.52 ± 0.97 | 0.54 ± 0.98 | 0.16 | ||
| ≥ 6 min | 1.13 ± 1.33 | 1.27 ± 1.39 | 0.001 | ||
CADe: Computer-assisted detection; BBPS: Boston bowel preparation score; PDR: Polyp detection rate; SSR: Sessile serrated adenoma/polyp detection rate; CRSR: Clinical serrated polyp detection rate.
Table 4.
Adenoma detection between colonoscopists and colonoscopists + computer-assisted detection
|
|
Colonoscopists
|
Colonoscopists + CADe
|
P
value
|
||
| ADR, % | 22.0 | 23.9 | 0.13 | ||
| Number of adenomas, mean ± SD | |||||
| 0.30 ± 0.62 | 0.32 ± 0.64 | 0.025 | |||
| Age | |||||
| < 50 | 0.14 ± 0.40 | 0.15 ± 0.41 | 0.32 | ||
| ≥ 50 | 0.43 ± 0.67 | 0.46 ± 0.69 | 0.045 | ||
| Sex | |||||
| Male | 0.38 ± 0.70 | 0.41 ± 0.73 | 0.045 | ||
| Female | 0.21 ± 0.48 | 0.22 ± 0.48 | 0.32 | ||
| Location | |||||
| Proximal | 0.12 ± 0.38 | 0.13 ± 0.39 | 0.16 | ||
| Distal | 0.17 ± 0.46 | 0.19 ± 0.47 | 0.08 | ||
| Size | |||||
| ≥ 10 mm | 0.06 ± 0.28 | 0.06 ± 0.28 | 1 | ||
| 6-9 mm | 0.11 ± 0.36 | 0.11 ± 0.36 | 1 | ||
| ≤ 5 mm | 0.12 ± 0.38 | 0.15 ± 0.42 | 0.025 | ||
| Morphology | |||||
| Flat | 0.20 ± 0.47 | 0.21 ± 0.50 | 0.045 | ||
| Subpedunculated | 0.07 ± 0.27 | 0.07 ± 0.28 | 0.32 | ||
| Pedunculated | 0.03 ± 0.21 | 0.03 ± 0.21 | 1 | ||
| Indications | |||||
| Screening | 0.29 ± 0.65 | 0.33 ± 0.66 | 0.08 | ||
| Surveillance | 0.43 ± 0.67 | 0.45 ± 0.67 | 0.32 | ||
| Diagnosis | 0.25 ± 0.56 | 0.26 ± 0.59 | 0.32 | ||
| Colonoscopes | |||||
| CF-Q290 | 0.31 ± 0.63 | 0.34 ± 0.65 | 0.025 | ||
| CF-Q260 | 0.23 ± 0.51 | 0.23 ± 0.51 | 1 | ||
| CF-Q240 | 0.25 ± 0.62 | 0.25 ± 0.62 | 1 | ||
| Experience | |||||
| > 3000 | 0.5 ± 0.72 | 0.5 ± 0.72 | 1 | ||
| 1000-3000 | 0.24 ± 0.59 | 0.27 ± 0.60 | 0.08 | ||
| < 1000 | 0.23 ± 0.53 | 0.26 ± 0.60 | 0.16 | ||
| BBPS | |||||
| < 6 | 0.19 ± 0.46 | 0.19 ± 0.46 | 1 | ||
| ≥ 6 | 0.32 ± 0.64 | 0.35 ± 0.66 | 0.025 | ||
| Withdrawal time | |||||
| < 6 min | 0.22 ± 0.57 | 0.22 ± 0.60 | 0.32 | ||
| ≥ 6 min | 0.38 ± 0.65 | 0.42 ± 0.65 | 0.045 | ||
CADe: Computer-assisted detection; BBPS: Boston bowel preparation score; ADR: Adenoma detection rate.
The diagnostic value of the CADe in polyps was observed in both age groups (P = 0.03 and P = 0.002, respectively), both sexes (P = 0.005 and P = 0.01, respectively), both hemi-colons (P = 0.01 and P = 0.003, respectively), all indications (all P = 0.03), inexperienced colonoscopists (< 1000 procedures) or general colonoscopists (1000-3000 procedures) (P = 0.003 and P = 0.03, respectively), and new generations of colonoscope (CF-290 and CF-260, P < 0.001 and P = 0.03, respectively), especially for diminutive (0.57 vs 0.65, P < 0.001) and flat polyps (0.67 vs 0.74, P = 0.001) (Table 3). Similarly, CADe also assisted colonoscopists in identifying more diminutive and flat adenomas (P = 0.025 and P = 0.045, respectively) in elderly (≥ 50 years, 0.43 vs 0.46, P = 0.045) and male patients (0.38 vs 0.41, P < 0.001), with the aid of a new-generation colonoscope (CF-290, P = 0.025) (Table 4). Notably, patients with adequate bowel preparation quality and withdrawal time showed a significantly increased number of polyps (P < 0.001 and P = 0.001) and adenomas (P = 0.025 and P = 0.045), whereas patients with < 6 Boston bowel preparation score and a < 6 min withdrawal time did not show any increase (Tables 3 and 4).
DISCUSSION
This CADe was developed and tested using over 70000 well-labeled colonoscopic images from 20 endoscopy centers, representing the dataset with the largest number of polyps. Notably, CADe did not merely perform well on the image tests of multicenter datasets and greater than 550 thousand frames of colonoscopy videos but also aided colonoscopists in identifying more polyps and adenomas in colonoscopy practice.
One of the first CADes systems validated in real-time colonoscopy was reported to identify polyps with 96% accuracy in the tests of labeled images[19], and consistently, the current CADe also reached a sensitivity of 95.0% for localizing polyps within a 30 ms constraint[23]. Notably, our findings further demonstrated that CADe assisted colonoscopists in detecting more adenomas in clinical practice but not merely in selected colonoscopy videos[19]. Compared with Yamada’s CADe[24], the current CADe also realized a high sensitivity in the dataset mainly consisting of nonpolypoid lesions in contrast to Yamada’s dataset, which was mainly based on polypoid lesions. However, the performance of the CADe seemed to decline in video tests (92.2% sensitivity and 93.6% specificity), which is consistent with previously reported CADes[20,24]. Further improvements in terms of process capability and colonoscopy video materials are warranted for CADes given that a great abundance and variety of random artifacts from the quick movement of videos with higher running speeds are indispensable to optimize further CADes in avoiding false positives and reducing time delays[19].
For clinical validation, several CADe systems in colonoscopy have been validated and are mainly focused on adenoma identification and quality control[25-31]. Wang et al[25,26,31] first conducted randomized controlled trials (RCTs) in an open-label or double-blind setting, most of which shared a large number of patients (both ~1000), rigorously illustrating that CADe effectively increased the ADR and adenomas per colonoscopy by identifying additional diminutive adenomas, including easy-to-miss adenomas for experienced colonoscopists and significantly reduced AMR[31]. Despite the demonstrated improvement of ADR and lower AMR in well-designed RCTs[25-31], our preliminary validation limited by the smaller sample size and observational design could not detect the difference in PDR or ADR and might calm our excessive excitement for AI. However, the observational and pragmatic design might reflect the realistic situation of colonoscopy practice and be beneficial for the real-world application of CADe. For example, in contrast to Wang’s studies, which filtered out alerts of CADe when the lumen was not inflated, the colonoscopists in our validation directly received all alerts from CADe and could better evaluate the real-world feasibility of CADe and the level of colonoscopists’ acceptance. We also analyzed all false positives rather than filtering out flashing-alert false positives given that the majority of the flashing alerts [folds (59.0%) and reflections of light (19.0%)] were clinically relevant to remind colonoscopists to inflate the lumen and carefully inspect the location of possibly missed polyps. Given that the exploration and application of CADe in colonoscopy is an emerging area and most studies were conducted in a single center, it is indispensable to explore prudently CADe’s feasibility and endoscopists’ acceptance in colonoscopy practice[32]. Presumably, the CADe should be further developed to identify different false positives to assess the bowel preparation quality and inspecting techniques as well as to remind the colonoscopist in real time to deal with the folds and residual fecal fluid, which may finally help to optimize high-quality colonoscopy.
Consistent with current work, Klare et al[29] also conducted an observational validation of their CADe with 55 clinical colonoscopies, indicating that their low-delay CADe might be feasible for real-time colonoscopy. However, their CADe detected no polyps before colonoscopists with a relatively greater false-positive frequency. Presumably, the construction of non-CNN algorithms and the design in which colonoscopists finally determine the criterion standard for polyp detection without rechecking the false positives of the CADe might lead to limitations[29]. Notably, our findings indicated that the diagnostic capability of CADe tended to be susceptible to inadequate withdrawal time or inadequate bowel preparation quality (P = 0.32 and P = 0.16) given that CADe could only detect polyps appearing in view during the colonoscopy but was unable to identify polyps covered by fecal fluid or hidden between the folds. Therefore, the bowel preparation and withdrawal time should also be considered even if the CADe is established to monitor colonoscopy withdrawal. The combination of an AI-assisted quality control system (monitoring withdrawal speed and bowel preparation quality) and CADe could provide more remarkable benefits in adenoma detection, even for large adenomas[28]. However, the additional detection of most CADes, including our current system, is confined to nonadenomatous polyps and diminutive adenomas, of which clinical significance was undetermined for interval cancers and cost-effectiveness analysis. Although promising preliminary findings seemed to appear, the CADes should be further validated in multicenter settings and community practices as well as optimized for satisfactory integration into clinical practice.
We defined a true positive with intersection over union (IoU) > 0.3, which was also adopted in previous reports[33]. In addition, Horie et al[34] considered the detection of CADe correct when the CADe could recognize merely a part of the esophageal cancer and false when identifying wide noncancerous areas occupying greater than 80% of the frame, whose standard was significantly lower than the level of 0.3 in IoU. In addition, Paul et al[33] proposed to use > 0.3 IoU in training and > 0.1 IoU to distinguish pulmonary nodules with the explanation of respecting the clinical need for coarse localization. However, there was no uniform standard for IoU, and more studies are warranted to discuss the optimal IoU or define other potential indicators to ensure the quality and comparability between CADes. Given that colonoscopists in our study were aware that their examinations were being monitored and reminded by sound of the CADe, the result might be vulnerable to the Hawthorne effect, which may explain our study’s significantly higher PDR (45.9%) compared to our previous report (PDR: 32.0-39.8%). Presumably, a fair proportion of polyps first detected by CADe or detected simultaneously by CADe and colonoscopists, which might have been ignored in real clinical practice, were additionally identified by colonoscopists in the current study. Although this weakness in our design may have prevented the benefit of CADe from being completely realized, more polyps and adenomas, especially those that were diminutive (< 5 mm) and flat, were still identified with the aid of CADe in the current study.
Compared with previous studies, our study has several strengths. First, our study retrospectively collected the datasets of 20 endoscopy centers, carefully screened patients with one polyp as well as the corresponding histological results, and deliberately labeled the images with blinding and repetition, finally constructing a large and high-quality dataset of colonoscopic images for training and testing. As a result, the dataset could represent a wider population and be ensured of the higher reliability of the original material, which is beneficial to reduce the risk of overfitting. Second, through pilot validation in clinical colonoscopy, our study not only indicated that CADe could help colonoscopists identify more polyps and adenomas but also preliminarily indicated the significance of quality control of colonoscopy (withdrawal time and bowel preparation quality) in CADe. Third, we analyzed the performance of CADe in various clinical scenarios and identified the most important causes of false negatives and positives, which might provide directions to optimize further the clinical application of the CADe.
Our study had several limitations. First, the CADe was completely established on selected retrospective white-light images without making use of the temporal coherence of videos, which contain many unclear and unfocused frames of images that could be not or mistakenly detected by CADe. As a result, the performance of CADe might be suboptimal with lower sensitivity or specificity in colonoscopy videos. Consistent with the concerns, the CADe’s performance was influenced by the unclear frames (too distant, unfocused and partially appeared) of seven challenging videos, which led to a testing sensitivity of less than 80%. Further efforts should be made to train the CADe with more consecutive videos to take advantage of the temporal coherence and improve diagnostic accuracy in clinical colonoscopy. Second, we did not identify a significant difference in PDR and ADR due to a limited sample size despite the fact that the polyps and adenomas identified per colonoscopy significantly increased. This information might provide a potential remedy to missed adenomas and the one-and-done effect of colonoscopists, which was demonstrated in recent tandem RCTs[31]. Finally, we artificially defined and classified the detection of polyps into three types. The validity of this classification was uncertain and might be influenced by the investigator’s bias (i.e. the delay between the recording time and actual time of detection on the part of the colonoscopists) despite the independent determination of observer using the objective measurement of time. As a result, the current study might not be completely adequate to demonstrate the clinical implementation of CADe due to the single-center setting with a limited sample size, and future large RCTs are warranted.
CONCLUSION
In summary, our study indicated that this deep learning-based CADe had been trained on the dataset consisting of the largest number of polyps from 20 centers and reached high diagnostic performance on the test dataset, colonoscopy videos and clinical validation with an acceptable frequency of false positives. This system might help colonoscopists identify more polyps and adenomas and deserves to be further validated in multicenter randomized trials.
ARTICLE HIGHLIGHTS
Research background
Artificial intelligence is an emerging area in the research and applications of digestive endoscopy. Current deep learning-based computer-assisted detection (CADe) for colorectal polyps is mainly constructed from single-center and limited-sample learning images.
Research motivation
Although several studies reported that CADes identified polyps in colonoscopic images, videos and exams in real time, the lack of large-sample and representative learning materials might limit the diagnostic performance in wide colonoscopy practice.
Research objectives
We aimed to train and test a CADe based on a multicenter high-quality dataset of polyps and preliminarily validated it in clinical colonoscopies.
Research methods
After retrospective collection and systematic screening and labeling, over 71000 images from 20 centers and 47 unaltered full-ranged videos were used to train and test a deep learning-based CADe. We also validated its diagnostic performance in real-world colonoscopy using a single-arm self-controlled observational study.
Research results
Our CADe identified polyps of images with 95.0% sensitivity and 99.1% specificity. For colonoscopy videos, all 86 polyps were detected with 92.2% sensitivity and 93.6% specificity for frame-by-frame analysis. The sensitivity of CAD in identifying polyps was 98.4% (185/188) in the prospective validation. Colonoscopists could detect more polyps (0.90 vs 0.82, P < 0.001) and adenomas (0.32 vs 0.30, P = 0.045) with the aid of CADe, particularly polyps < 5 mm and flat polyps/adenomas.
Research conclusions
A CADe based on multicenter high-quality colonoscopic images was constructed and might help endoscopists detect more polyps and adenomas.
Research perspectives
CADe is feasible in the clinical setting, and further confirmation is warranted.
ACKNOWLEDGEMENTS
We would like to thank Dr. Wei Qian for helping analyze our data and all the colonoscopists who participated in the validation of the proposed CADe system.
Footnotes
Institutional review board statement: The study was reviewed and approved by the ethics committees of Changhai Hospital.
Clinical trial registration statement: ClinicalTrials.gov, identifier NCT03761771.
Informed consent statement: Written informed consent was provided for all participants.
Conflict-of-interest statement: Wu JR, Chen LZ, Chang J and Song P were staff of Tencent Healthcare (Shenzhen) Co. LTD, which supported the work with provision of research grants, prototype software, prototype PC and GPU units. The other authors have no conflicts of interest to declare. The sponsor had no involvement in the design and conduct of the study; the collection, management, analysis, and interpretation of the data; the preparation, review, and approval of the manuscript; or the decision to submit the manuscript for publication.
Manuscript source: Unsolicited manuscript
Corresponding Author's Membership in Professional Societies: Chinese Society of Gastroenterology, No. 0920092010.
Peer-review started: February 19, 2021
First decision: March 28, 2021
Article in press: July 20, 2021
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Castro FJ S-Editor: Zhang H L-Editor: Filipodia P-Editor: Liu JH
Contributor Information
Sheng-Bing Zhao, Changhai Hospital, Second Military Medical University/Naval Medical University, Shanghai 200433, China.
Wei Yang, Tencent AI Lab, National Open Innovation Platform for Next Generation Artificial Intelligence on Medical Imaging, Shenzhen 518063, Guangdong Province, China.
Shu-Ling Wang, Department of Gastroenterology, Changhai Hospital, Second Military Medical University/Naval Medical University, Shanghai 200433, China.
Peng Pan, Department of Gastroenterology, Changhai Hospital, Second Military Medical University/Naval Medical University, Shanghai 200433, China.
Run-Dong Wang, Department of Gastroenterology, Changhai Hospital, Second Military Medical University/Naval Medical University, Shanghai 200433, China.
Xin Chang, Department of Gastroenterology, Changhai Hospital, Second Military Medical University/Naval Medical University, Shanghai 200433, China.
Zhong-Qian Sun, Tencent AI Lab, National Open Innovation Platform for Next Generation Artificial Intelligence on Medical Imaging, Shenzhen 518063, Guangdong Province, China.
Xing-Hui Fu, Tencent AI Lab, National Open Innovation Platform for Next Generation Artificial Intelligence on Medical Imaging, Shenzhen 518063, Guangdong Province, China.
Hong Shang, Tencent AI Lab, National Open Innovation Platform for Next Generation Artificial Intelligence on Medical Imaging, Shenzhen 518063, Guangdong Province, China.
Jian-Rong Wu, Tencent Healthcare (Shenzhen) Co. LTD., Shenzhen 518063, Guangdong Province, China.
Li-Zhu Chen, Tencent Healthcare (Shenzhen) Co. LTD., Shenzhen 518063, Guangdong Province, China.
Jia Chang, Tencent Healthcare (Shenzhen) Co. LTD., Shenzhen 518063, Guangdong Province, China.
Pu Song, Tencent Healthcare (Shenzhen) Co. LTD., Shenzhen 518063, Guangdong Province, China.
Ying-Lei Miao, Department of Gastroenterology, The First Affiliated Hospital of Kunming Medical University, Kunming 650000, Yunnan Province, China.
Shui-Xiang He, Department of Gastroenterology, The First Affiliated Hospital of Xi'an Jiaotong University, Xi'an 710061, Shaanxi Province, China.
Lin Miao, Institute of Digestive Endoscopy and Medical Center for Digestive Disease, The Second Affiliated Hospital of Nanjing Medical University, Nanjing 210011, Jiangsu Province, China.
Hui-Qing Jiang, Department of Gastroenterology, The Second Hospital of Hebei Medical University, Hebei Key Laboratory of Gastroenterology, Hebei Institute of Gastroenterology, Shijiazhuang 050000, Hebei Province, China.
Wen Wang, Department of Gastroenterology, 900th Hospital of Joint Logistics Support Force, Fuzhou 350025, Fujian Province, China.
Xia Yang, Department of Gastroenterology, No. 905 Hospital of The Chinese People's Liberation Army, Shanghai 200050, China.
Yuan-Hang Dong, Department of Gastroenterology, Changhai Hospital, Second Military Medical University/Naval Medical University, Shanghai 200433, China.
Han Lin, Department of Gastroenterology, Changhai Hospital, Second Military Medical University/Naval Medical University, Shanghai 200433, China.
Yan Chen, Department of Gastroenterology, Changhai Hospital, Second Military Medical University/Naval Medical University, Shanghai 200433, China.
Jie Gao, Department of Gastroenterology, Changhai Hospital, Second Military Medical University/Naval Medical University, Shanghai 200433, China.
Qian-Qian Meng, Department of Gastroenterology, Changhai Hospital, Second Military Medical University/Naval Medical University, Shanghai 200433, China.
Zhen-Dong Jin, Department of Gastroenterology, Changhai Hospital, Second Military Medical University/Naval Medical University, Shanghai 200433, China.
Zhao-Shen Li, Department of Gastroenterology, Changhai Hospital, Second Military Medical University/Naval Medical University, Shanghai 200433, China.
Yu Bai, Department of Gastroenterology, Changhai Hospital, Second Military Medical University/Naval Medical University, Shanghai 200433, China. baiyu1998@hotmail.com.
Data sharing statement
No additional data are available.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Løberg M, Kalager M, Holme Ø, Hoff G, Adami HO, Bretthauer M. Long-term colorectal-cancer mortality after adenoma removal. N Engl J Med. 2014;371:799–807. doi: 10.1056/NEJMoa1315870. [DOI] [PubMed] [Google Scholar]
- 3.Baxter NN, Goldwasser MA, Paszat LF, Saskin R, Urbach DR, Rabeneck L. Association of colonoscopy and death from colorectal cancer. Ann Intern Med. 2009;150:1–8. doi: 10.7326/0003-4819-150-1-200901060-00306. [DOI] [PubMed] [Google Scholar]
- 4.Robertson DJ, Lieberman DA, Winawer SJ, Ahnen DJ, Baron JA, Schatzkin A, Cross AJ, Zauber AG, Church TR, Lance P, Greenberg ER, Martínez ME. Colorectal cancers soon after colonoscopy: a pooled multicohort analysis. Gut. 2014;63:949–956. doi: 10.1136/gutjnl-2012-303796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heresbach D, Barrioz T, Lapalus MG, Coumaros D, Bauret P, Potier P, Sautereau D, Boustière C, Grimaud JC, Barthélémy C, Sée J, Serraj I, D'Halluin PN, Branger B, Ponchon T. Miss rate for colorectal neoplastic polyps: a prospective multicenter study of back-to-back video colonoscopies. Endoscopy. 2008;40:284–290. doi: 10.1055/s-2007-995618. [DOI] [PubMed] [Google Scholar]
- 6.Zhao S, Wang S, Pan P, Xia T, Chang X, Yang X, Guo L, Meng Q, Yang F, Qian W, Xu Z, Wang Y, Wang Z, Gu L, Wang R, Jia F, Yao J, Li Z, Bai Y. Magnitude, Risk Factors, and Factors Associated With Adenoma Miss Rate of Tandem Colonoscopy: A Systematic Review and Meta-analysis. Gastroenterology 2019; 156: 1661-1674. :e11. doi: 10.1053/j.gastro.2019.01.260. [DOI] [PubMed] [Google Scholar]
- 7.Adler J, Robertson DJ. Interval Colorectal Cancer After Colonoscopy: Exploring Explanations and Solutions. Am J Gastroenterol. 2015;110:1657–64; quiz 1665. doi: 10.1038/ajg.2015.365. [DOI] [PubMed] [Google Scholar]
- 8.Castaneda D, Popov VB, Verheyen E, Wander P, Gross SA. New technologies improve adenoma detection rate, adenoma miss rate, and polyp detection rate: a systematic review and meta-analysis. Gastrointest Endosc 2018; 88: 209-222. :e11. doi: 10.1016/j.gie.2018.03.022. [DOI] [PubMed] [Google Scholar]
- 9.Zhao S, Yang X, Wang S, Meng Q, Wang R, Bo L, Chang X, Pan P, Xia T, Yang F, Yao J, Zheng J, Sheng J, Zhao X, Tang S, Wang Y, Gong A, Chen W, Shen J, Zhu X, Yan C, Yang Y, Zhu Y, Ma RJ, Ma Y, Li Z, Bai Y. Impact of 9-Minute Withdrawal Time on the Adenoma Detection Rate: A Multicenter Randomized Controlled Trial. Clin Gastroenterol Hepatol. 2020 doi: 10.1016/j.cgh.2020.11.019. [DOI] [PubMed] [Google Scholar]
- 10.Aslanian HR, Shieh FK, Chan FW, Ciarleglio MM, Deng Y, Rogart JN, Jamidar PA, Siddiqui UD. Nurse observation during colonoscopy increases polyp detection: a randomized prospective study. Am J Gastroenterol. 2013;108:166–172. doi: 10.1038/ajg.2012.237. [DOI] [PubMed] [Google Scholar]
- 11.Atkins L, Hunkeler EM, Jensen CD, Michie S, Lee JK, Doubeni CA, Zauber AG, Levin TR, Quinn VP, Corley DA. Factors influencing variation in physician adenoma detection rates: a theory-based approach for performance improvement. Gastrointest Endosc 2016; 83: 617-26. :e2. doi: 10.1016/j.gie.2015.08.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rex DK, Cutler CS, Lemmel GT, Rahmani EY, Clark DW, Helper DJ, Lehman GA, Mark DG. Colonoscopic miss rates of adenomas determined by back-to-back colonoscopies. Gastroenterology. 1997;112:24–28. doi: 10.1016/s0016-5085(97)70214-2. [DOI] [PubMed] [Google Scholar]
- 13.Aniwan S, Orkoonsawat P, Viriyautsahakul V, Angsuwatcharakon P, Pittayanon R, Wisedopas N, Sumdin S, Ponuthai Y, Wiangngoen S, Kullavanijaya P, Rerknimitr R. The Secondary Quality Indicator to Improve Prediction of Adenoma Miss Rate Apart from Adenoma Detection Rate. Am J Gastroenterol. 2016;111:723–729. doi: 10.1038/ajg.2015.440. [DOI] [PubMed] [Google Scholar]
- 14.Bernal J, Tajkbaksh N, Sanchez FJ, Matuszewski BJ, Hao Chen, Lequan Yu, Angermann Q, Romain O, Rustad B, Balasingham I, Pogorelov K, Sungbin Choi, Debard Q, Maier-Hein L, Speidel S, Stoyanov D, Brandao P, Cordova H, Sanchez-Montes C, Gurudu SR, Fernandez-Esparrach G, Dray X, Jianming Liang, Histace A. Comparative Validation of Polyp Detection Methods in Video Colonoscopy: Results From the MICCAI 2015 Endoscopic Vision Challenge. IEEE Trans Med Imaging. 2017;36:1231–1249. doi: 10.1109/TMI.2017.2664042. [DOI] [PubMed] [Google Scholar]
- 15.Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, Thrun S. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gulshan V, Peng L, Coram M, Stumpe MC, Wu D, Narayanaswamy A, Venugopalan S, Widner K, Madams T, Cuadros J, Kim R, Raman R, Nelson PC, Mega JL, Webster DR. Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs. JAMA. 2016;316:2402–2410. doi: 10.1001/jama.2016.17216. [DOI] [PubMed] [Google Scholar]
- 17.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 18.Chen PJ, Lin MC, Lai MJ, Lin JC, Lu HH, Tseng VS. Accurate Classification of Diminutive Colorectal Polyps Using Computer-Aided Analysis. Gastroenterology. 2018;154:568–575. doi: 10.1053/j.gastro.2017.10.010. [DOI] [PubMed] [Google Scholar]
- 19.Urban G, Tripathi P, Alkayali T, Mittal M, Jalali F, Karnes W, Baldi P. Deep Learning Localizes and Identifies Polyps in Real Time With 96% Accuracy in Screening Colonoscopy. Gastroenterology 2018; 155: 1069-1078. :e8. doi: 10.1053/j.gastro.2018.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Misawa M, Kudo SE, Mori Y, Cho T, Kataoka S, Yamauchi A, Ogawa Y, Maeda Y, Takeda K, Ichimasa K, Nakamura H, Yagawa Y, Toyoshima N, Ogata N, Kudo T, Hisayuki T, Hayashi T, Wakamura K, Baba T, Ishida F, Itoh H, Roth H, Oda M, Mori K. Artificial Intelligence-Assisted Polyp Detection for Colonoscopy: Initial Experience. Gastroenterology 2018; 154: 2027-2029. :e3. doi: 10.1053/j.gastro.2018.04.003. [DOI] [PubMed] [Google Scholar]
- 21.Byrne MF, Chapados N, Soudan F, Oertel C, Linares Pérez M, Kelly R, Iqbal N, Chandelier F, Rex DK. Real-time differentiation of adenomatous and hyperplastic diminutive colorectal polyps during analysis of unaltered videos of standard colonoscopy using a deep learning model. Gut. 2019;68:94–100. doi: 10.1136/gutjnl-2017-314547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vinsard DG, Mori Y, Misawa M, Kudo SE, Rastogi A, Bagci U, Rex DK, Wallace MB. Quality assurance of computer-aided detection and diagnosis in colonoscopy. Gastrointest Endosc. 2019;90:55–63. doi: 10.1016/j.gie.2019.03.019. [DOI] [PubMed] [Google Scholar]
- 23.Wang P, Xiao X, Glissen Brown JR, Berzin TM, Tu M, Xiong F, Hu X, Liu P, Song Y, Zhang D, Yang X, Li L, He J, Yi X, Liu J, Liu X. Development and validation of a deep-learning algorithm for the detection of polyps during colonoscopy. Nat Biomed Eng . 2018;2:741–748. doi: 10.1038/s41551-018-0301-3. [DOI] [PubMed] [Google Scholar]
- 24.Yamada M, Saito Y, Imaoka H, Saiko M, Yamada S, Kondo H, Takamaru H, Sakamoto T, Sese J, Kuchiba A, Shibata T, Hamamoto R. Development of a real-time endoscopic image diagnosis support system using deep learning technology in colonoscopy. Sci Rep. 2019;9:14465. doi: 10.1038/s41598-019-50567-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang P, Berzin TM, Glissen Brown JR, Bharadwaj S, Becq A, Xiao X, Liu P, Li L, Song Y, Zhang D, Li Y, Xu G, Tu M, Liu X. Real-time automatic detection system increases colonoscopic polyp and adenoma detection rates: a prospective randomised controlled study. Gut. 2019;68:1813–1819. doi: 10.1136/gutjnl-2018-317500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang P, Liu X, Berzin TM, Glissen Brown JR, Liu P, Zhou C, Lei L, Li L, Guo Z, Lei S, Xiong F, Wang H, Song Y, Pan Y, Zhou G. Effect of a deep-learning computer-aided detection system on adenoma detection during colonoscopy (CADe-DB trial): a double-blind randomised study. Lancet Gastroenterol Hepatol. 2020;5:343–351. doi: 10.1016/S2468-1253(19)30411-X. [DOI] [PubMed] [Google Scholar]
- 27.Gong D, Wu L, Zhang J, Mu G, Shen L, Liu J, Wang Z, Zhou W, An P, Huang X, Jiang X, Li Y, Wan X, Hu S, Chen Y, Hu X, Xu Y, Zhu X, Li S, Yao L, He X, Chen D, Huang L, Wei X, Wang X, Yu H. Detection of colorectal adenomas with a real-time computer-aided system (ENDOANGEL): a randomised controlled study. Lancet Gastroenterol Hepatol. 2020;5:352–361. doi: 10.1016/S2468-1253(19)30413-3. [DOI] [PubMed] [Google Scholar]
- 28.Su JR, Li Z, Shao XJ, Ji CR, Ji R, Zhou RC, Li GC, Liu GQ, He YS, Zuo XL, Li YQ. Impact of a real-time automatic quality control system on colorectal polyp and adenoma detection: a prospective randomized controlled study (with videos). Gastrointest Endosc 2020; 91: 415-424. :e4. doi: 10.1016/j.gie.2019.08.026. [DOI] [PubMed] [Google Scholar]
- 29.Klare P, Sander C, Prinzen M, Haller B, Nowack S, Abdelhafez M, Poszler A, Brown H, Wilhelm D, Schmid RM, von Delius S, Wittenberg T. Automated polyp detection in the colorectum: a prospective study (with videos). Gastrointest Endosc 2019; 89: 576-582. :e1. doi: 10.1016/j.gie.2018.09.042. [DOI] [PubMed] [Google Scholar]
- 30.Repici A, Badalamenti M, Maselli R, Correale L, Radaelli F, Rondonotti E, Ferrara E, Spadaccini M, Alkandari A, Fugazza A, Anderloni A, Galtieri PA, Pellegatta G, Carrara S, Di Leo M, Craviotto V, Lamonaca L, Lorenzetti R, Andrealli A, Antonelli G, Wallace M, Sharma P, Rosch T, Hassan C. Efficacy of Real-Time Computer-Aided Detection of Colorectal Neoplasia in a Randomized Trial. Gastroenterology 2020; 159: 512-520. :e7. doi: 10.1053/j.gastro.2020.04.062. [DOI] [PubMed] [Google Scholar]
- 31.Wang P, Liu P, Glissen Brown JR, Berzin TM, Zhou G, Lei S, Liu X, Li L, Xiao X. Lower Adenoma Miss Rate of Computer-Aided Detection-Assisted Colonoscopy vs Routine White-Light Colonoscopy in a Prospective Tandem Study. Gastroenterology 2020; 159: 1252-1261. :e5. doi: 10.1053/j.gastro.2020.06.023. [DOI] [PubMed] [Google Scholar]
- 32.Ahmad OF, Soares AS, Mazomenos E, Brandao P, Vega R, Seward E, Stoyanov D, Chand M, Lovat LB. Artificial intelligence and computer-aided diagnosis in colonoscopy: current evidence and future directions. Lancet Gastroenterol Hepatol. 2019;4:71–80. doi: 10.1016/S2468-1253(18)30282-6. [DOI] [PubMed] [Google Scholar]
- 33.Jaeger PF, Kohl SAA, Bickelhaupt S, Isensee F, Kuder TA, Schlemmer H-P, Maier-Hein KH. Retina U-Net: Embarrassingly Simple Exploitation of Segmentation Supervision for Medical Object Detection. 2018 Preprint. Available from: arXiv: 1811.08661.
- 34.Horie Y, Yoshio T, Aoyama K, Yoshimizu S, Horiuchi Y, Ishiyama A, Hirasawa T, Tsuchida T, Ozawa T, Ishihara S, Kumagai Y, Fujishiro M, Maetani I, Fujisaki J, Tada T. Diagnostic outcomes of esophageal cancer by artificial intelligence using convolutional neural networks. Gastrointest Endosc. 2019;89:25–32. doi: 10.1016/j.gie.2018.07.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No additional data are available.