Abstract
Acetylcholinesterase (EC 3.1.1.7), a key acetylcholine-hydrolyzing enzyme in cholinergic neurotransmission, is present in a variety of states in situ, including monomers, C-terminally disulfide-linked homodimers, homotetramers, and up to three tetramers covalently attached to structural subunits. Could oligomerization that ensures high local concentrations of catalytic sites necessary for efficient neurotransmission be affected by environmental factors? Using small-angle X-ray scattering (SAXS) and cryo-EM, we demonstrate that homodimerization of recombinant monomeric human acetylcholinesterase (hAChE) in solution occurs through a C-terminal four-helix bundle at micromolar concentrations. We show that diethylphosphorylation of the active serine in the catalytic gorge or isopropylmethylphosphonylation by the RP enantiomer of sarin promotes a 10-fold increase in homodimer dissociation. We also demonstrate the dissociation of organophosphate (OP)-conjugated dimers is reversed by structurally diverse oximes 2PAM, HI6, or RS194B, as demonstrated by SAXS of diethylphosphoryl-hAChE. However, binding of oximes to the native ligand-free hAChE, binding of high-affinity reversible ligands, or formation of an SP-sarin-hAChE conjugate had no effect on homodimerization. Dissociation monitored by time-resolved SAXS occurs in milliseconds, consistent with rates of hAChE covalent inhibition. OP-induced dissociation was not observed in the SAXS profiles of the double-mutant Y337A/F338A, where the active center gorge volume is larger than in wildtype hAChE. These observations suggest a key role of the tightly packed acyl pocket in allosterically triggered OP-induced dimer dissociation, with the potential for local reduction of acetylcholine-hydrolytic power in situ. Computational models predict allosteric correlated motions extending from the acyl pocket toward the four-helix bundle dimerization interface 25 Å away.
Keywords: acetylcholinesterase, small-angle X-ray scattering, SAXS, allosteric behavior, 4-helix bundle, organophosphate intoxication, paraoxon, oxime reactivation, molecular dynamics, enzyme structure, structure–function
Abbreviations: 4HB, 4-helix bundle; AChE, acetylcholinesterase; CBS, choline-binding site; F-2-F, face-to-face; hBChE, human butyrylcholinesterase; hNL, human neuroligin; MD, molecular dynamics; OP, organophosphate; PAS, peripheral anionic site; PDB, Protein Data Bank; POX, paraoxon; SAXS, small-angle X-ray scattering; SEC, size-exclusion chromatography; TR-SAXS, time-resolved SAXS
Acetylcholinesterase (EC 3.1.1.7; AChE), a key enzyme of cholinergic neurotransmission, is encoded by a single gene in vertebrates, yet in tissues it exists in diverse molecular forms (1, 2). AChE is found in different quaternary associations including monomers, homodimers covalently (-S-S-) linked via C-terminal cysteines, homotetramers (two reversibly associated dimers), and clusters of up to three tetramers covalently attached to distinct structural subunits (3). A likely evolutionary advantage for the clustering of large numbers of catalytic centers of this highly efficient biological catalyst, in neurons and muscle, is to achieve precise spatiotemporal control in the degradation of the neurotransmitter acetylcholine (4, 5). Disruption of the C-terminal region structure (triggered by covalent bonding, for example) may distort the dimerization interface and lead to dissociation of AChE dimers and disintegration of the clustered oligomers. That could disturb spatiotemporal hydrolysis of acetylcholine in tissue and compromise neurotransmission. In native AChE, catalytic properties of different molecular forms are generally assumed to be identical, in spite of slight variations in amino acid sequences at the C termini in some forms (6, 7). Recombinant AChE forms, truncated at the C terminus to remove intersubunit -S-S- bonding in order to form monomeric AChE, are commonly used for structural studies and show catalytic properties identical to those of native AChE (7, 8, 9, 10). No enzymatic cooperativity has been reported between subunits of oligomeric AChE (6). The active serine in the Ser203-Glu334-His447 catalytic triad in each catalytic monomer of the human AChE (hAChE) is located at the base of a ∼20 Å deep active center gorge. The gorge is narrow and curved and located in the center of the globular monomer immediately adjacent to the small acyl pocket (11).
The basic geometry of monomer association into homodimers, an essential element of AChE quaternary structure, is revealed in X-ray structures of insect, electric ray, and mammalian AChEs (8, 12). These structures show two C-terminally located α-helices from each monomer forming a stable “four-helix bundle” (4HB) dimerization interface, stabilized by hydrophobic intersubunit interactions. Crystal structures of disulfide-linked covalent homodimers (from tissue-derived AChE preparations; (12)) and of reversibly associated monomers (from recombinantly expressed AChE truncated at the C terminus; (8)) reveal nearly identical tertiary and quaternary structures.
Similar 4HB homodimers were also observed in X-ray structures of the structurally closely related α/β hydrolase-fold proteins, human butyrylcholinesterase (EC 3.1.1.8; hBChE; (13)), and human neuroligins 1 (hNL1; (14)) and 4 (hNL4; (15)), and in the cryo-EM structure of human thyroglobulin (16). Moreover, monomers of hAChE and of neuroligins were demonstrated to form functionally relevant 4HB heterodimers with thyroglobulin monomers (17, 18).
The lack of stability of noncovalent 4HB dimers in solution, leading to their dissociation, could significantly disrupt the physiological function of both neuroligins and thyroglobulin, and affect tissue localization of cholinesterases. Thus, there is the potential for considerable negative biological consequences associated with the dissociation of these 4HB dimers. A structural illustration on how destabilization in the dimeric structure at the 4HB interface could lead to nearly complete dimer dissociation in solution was reported for the Leu380Arg/Phe535Lys mutant of hAChE (9).
In the present study, we demonstrate, using small-angle X-ray scattering (SAXS), cryo-EM, and FPLC size-exclusion chromatography (SEC), that a construct of wildtype hAChE lacking the C-terminal cysteines forms reversible 4HB homodimers at micromolar concentrations. They are structurally identical to hAChE homodimers that are linked covalently by C-terminal S-S- bonds (8, 12). Exposure of this C-terminally truncated form of hAChE to paraoxon or the RP enantiomer of sarin, but not other organophosphates or a variety of structurally diverse reversible ligands, promotes dissociation of the homodimers in solution as indicated by SAXS. Dissociation of reversible hAChE homodimers upon covalent binding of paraoxon or the RP-sarin is likely the result of structural distortions in the impacted acyl pocket of the AChE catalytic gorge, which are allosterically transmitted to the dimer interface 25 Å away. The structural effect could be reversed by nucleophilic removal of the conjugated organophosphate. The biological significance of these organophosphate (OP)-induced structural changes lies in their potential for long-term loss of neurotransmitter turnover capacity in nervous tissue. Emerging structural details of the dissociation mechanism provide the first clear experimental evidence of allosteric communication between the buried active center of hAChE and the dimerization interface located on the surface of the globular hAChE monomer. Identification of this allosteric mechanism adds to the understanding of AChE catalysis and its structural dynamics. The corresponding knowledge can directly inform and enhance structure-based design of therapeutics to optimize treatment of OP intoxication.
Results
The catalytic subunit of truncated hAChE forms reversible homodimers in solution
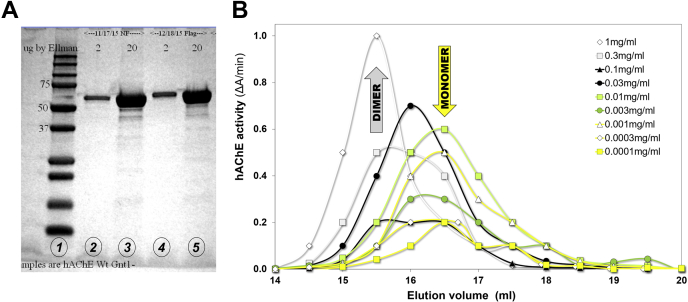
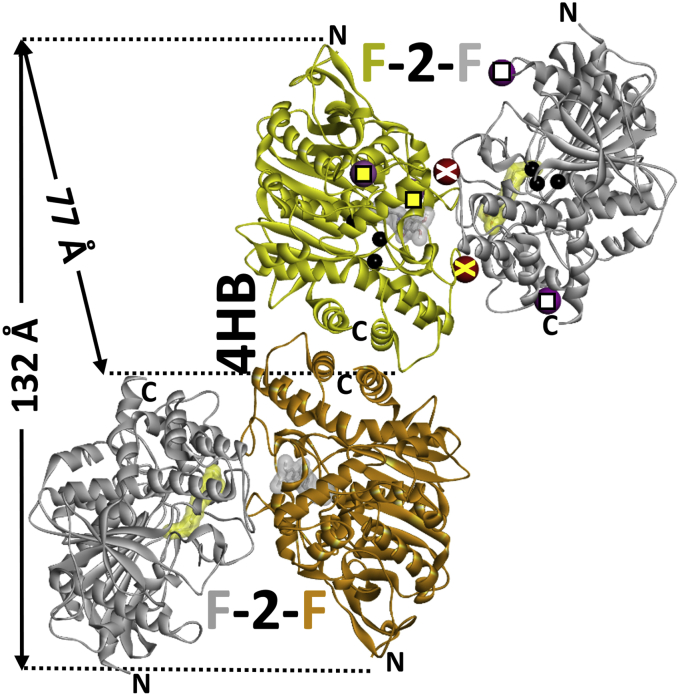
Monomeric catalytic subunit of hAChE, glycosylated at Asn265 and Asn464 and truncated at the C terminus, has a molecular weight of ∼65 kDa, similar to bovine serum albumin (Fig. 1A; (8)). When applied to a size-exclusion column at micromolar concentrations, this hAChE construct elutes earlier than expected (Fig. 1B) indicating hAChE may be forming higher-molecular-weight complexes. Upon dilution of hAChE to nanomolar concentrations, the elution peak emerges at larger volume values (Fig. 1B) close to those for bovine serum albumin, and to those expected for globular proteins of this size. This observation is consistent with the formation of reversible homodimers that dissociate to monomers when diluted to nanomolar concentrations. We performed FPLC SEC with a series of concentrations of recombinant monomeric hAChE in the range of 1.5 nM to 15 μM to assess concentration-dependent elution shifts. Peaks of hAChE were detected by measuring enzyme activity in elution fractions. Concentrations where peak shifts were observed indicated an estimated dimer dissociation constant of 1.4 to 2.4 μM (Table S1, Fig. S1). Three hAChE constructs were analyzed, which differed in the extent of the N-glycosylation (FLAG-tagged hAChE constructs expressed in either wildtype HEK293 or expressed in glycosylation-deficient Gnt1− HEK cells) and/or in the amino acid sequence of the N terminus (Gnt1− HEK-expressed Tyr337Ala/Phe338Ala (YAFA) hAChE double-mutant with FLAG-tag removed). The absence of any notable effect (Table S1) due to differences in the hAChE N-terminal sequence or the presence of equatorially located N-linked glycans appears to be consistent with the C-terminal 4HB being a primary dimerization interface in homodimer formation (Fig. 2). Thus, two crystallographic homodimers are observed in asymmetric subunits of the Gnt1− HEK-expressed wildtype hAChE (7). In solution, formation of a homodimer similar to the 4HB dimer and not to the face-to-face (F-2-F) dimer appears more likely. This notion is consistent with PISA server analysis of dimerization interfaces in this form of hAChE showing that the 4HB dimer was the more stable of the two homodimers ((11); Table S2).
Figure 1.
Monomeric hAChE, recombinantly expressed with a truncated C terminus, forms reversibly associating homodimers in the picomolar to micromolar concentration range.A, SDS-PAGE gel of hAChE expressed in Gnt1− HEK293 cells: lane 1, molecular size standards; lanes 2 and 3, 2 and 20 μg of hAChE devoid of N-terminal FLAG-tag sequence; lanes 4 and 5, 2 and 20 μg of hAChE containing N-terminal FLAG-tag sequence. B, FPLC–size-exclusion chromatography profiles of FLAG-tagged hAChE at nine discrete concentrations in the range from 1.5 nM to 15 μM (0.0001 mg/ml to 1 mg/ml). Positions of the homodimer and monomeric peaks are indicated by arrows. Representative profiles from five experiments with overlapping hAChE concentrations. Values of hAChE activity were corrected for dilution. hAChE, human acetylcholinesterase.
Figure 2.
Two crystallographic dimers (four-helix bundle, 4HB, and face-to-face, F-2-F) in the single asymmetric subunit of monomeric wildtype hAChE expressed in Gnt1−HEK cells, devoid of FLAG-tag. Shown is the complex of hAChE with donepezil (PDB ID: 6O4W). hAChE monomers are rendered as yellow, orange, or gray ribbon, and active center–bound donepezil is shown as a gray or yellow transparent surface. Sites of glycosylation (in the two topmost monomers) are indicated by yellow or white squares and crosses. Wildtype HEK293-expressed hAChE has three glycosylation sites per monomer, and in Gnt1− HEK-expressed hAChE sites indicated by crosses are not glycosylated. Location of the catalytic triad is shown by black spheres. Maximal crystallographic dimensions of monomer (77 Å) and four-helix bundle dimer (132 Å) are indicated. hAChE, human acetylcholinesterase.
The idea that wildtype hAChE associates predominantly as 4HB dimers in solution from monomers expressed in Gnt1− HEK is further supported by cryo-EM analysis (Fig. 3; Table S3). Consistent with the 1.4 to 2.4 μM range for homodimer Kd values (Table S1), analysis of cryo-EM images of ∼3.0 μM hAChE using 2D classification showed a mixed population of monomers and dimers (Fig. 3). Indeed, only the projected 4HB crystallographic dimer, but not the F-2-F dimer, matched the 2D class dimers (Fig. S2) and preliminary 3D density (Fig. S3) very well. Estimated maximal dimensions of monomer particles (79 Å) and dimer particles (132 Å) were in excellent agreement with the estimated crystallographic dimensions of 77 Å for monomer and 132 Å for 4HB dimer (Fig. 2). Moreover, we obtained a low-resolution map of dimer hAChE at 9.23 Å (0.143 FSC) that was used for fitting the 4HB dimer. The 3D map has been deposited in the EMDB/PDB under accession code EMD-24404.
Figure 3.
Cryo-EM analysis of hAChE.A, initial 2D classification after autopicking particles. B, representative 2D classes for the monomer hAChE. C, representative 2D classes for the dimer hAChE. D, X-ray structure (PDB ID 604W) four-helix bundle dimer projections compared with the selected 2D classes dimers. Scale bars, 50 Å. hAChE, human acetylcholinesterase.
The SAXS experiments performed at 7.5, 15, 30, and 60 μM hAChE concentrations (Table S4) clearly show the absence of aggregation at all concentrations. There is no upturn in the I(s) versus s plots at low s (Fig. S4A), and Guinier plots are linear throughout the entire Guinier region (smin to Rg∗s = 1.3), with no indication of upturn at low s. A plot of I(0) as a function of concentration shows a nonlinear increase in I(0) at higher concentrations, which is consistent with the ratio of homodimers to monomers increasing over this range of hAChE concentration. Multistate analysis at each concentration of hAChE using the FoXS server indicates 100% 4HB dimer at 30 μM and 60 μM hAChE, but a mixture of homodimer and monomer at lower concentrations of hAChE, with 13% and 31% monomer at 15 and 7.5 μM hAChE, respectively (Fig. S5). This monomer/dimer distribution as a function of concentration is consistent with a dimer dissociation constant in the 1.4 to 2.3 μM range (Table S1).
FoXS was also used to analyze the SAXS data at 30 μM hAChE (Fig. 4) to compare fits of the experimental I(s) versus s and P(r) curves to the predicted scattering curves for 4HB homodimer, F-2-F homodimer, and monomer. The best fit to the experimental I(s) versus s was clearly the predicted scattering of the 4HB homodimer (Fig. 4A). The predicted 4HB homodimer P(r) curve also closely resembles that of the experimental curve. In contrast, there are clear differences in the shape of the P(r) curve obtained from the SAXS experiments compared with the predicted P(r) curve for hAChE monomers, or F-2-F dimer (Fig. 4B). The monomeric form of hAChE should give a P(r) curve with a single symmetrical peak at ∼24 Å and a Dmax of around 72 Å, whereas the P(r) curve observed for SAXS data collected with hAChE shows a primary peak at ∼40 Å, a second peak at ∼85 Å, and a Dmax of ∼130 Å. The presence of a second peak in the P(r) profile, which corresponds to the intraparticle distances between subunits in the dimer, and a Dmax that is nearly twice that expected for the monomer, are indicative that hAChE predominantly forms a homodimer in solution at these concentrations. Further independent confirmation that the 4HB dimer is the homodimer form of apo hAChE in solution was obtained using FoXSDock. In this rigid-body docking analysis, tens of thousands of hAChE dimer models were fit to the experimental SAXS data of apo hAChE. The two best dimer models selected by FoXSDock were both 4HB dimers that were nearly identical to the 4HB crystal structures (Table S5, Fig. S6). None of the top ten dimer models generated by FoXSDock resembled the F-2-F dimer (Table S5, Fig. S6).
Figure 4.
Small-angle X-ray scattering intensity and calculated P(r) functions for hAChE. Profiles of the 30 μM (2 mg/ml) Gnt1− HEK-expressed wt apo hAChE lacking FLAG-tag (black circles) compared with FoXS-calculated predictions for hAChE monomer (blue line), 4HB hAChE dimer (green line), and F-2-F hAChE dimer (red line). A, scattering data intensity (black circles with error bars; upper panel) and Residual Intensity (“experimental - model”; lower panel). B, calculated P(r) functions compared with experimental data (error bars are smaller than the symbol size). Corresponding particle radii of gyration (Rg) were 40 Å (experimental), 24 Å (monomer), 33 Å (F-2-F dimer), and 40 Å (4HB dimer). Maximal particle dimensions (Dmax) were 130 Å (experimental), 72 Å (monomer), 100 Å (F-2-F dimer), and 130 Å (4HB dimer). P(r) curves/data were normalized to the highest value in each curve/data set. 4HB, four-helix bundle; F-2-F, face-to-face; hAChE, human acetylcholinesterase.
The value of the experimental SAXS Dmax (130 Å; Fig. 4) agrees well with the maximal dimer dimension determined by cryo-EM (132 Å, Fig. 3C) and with the 132 Å maximal dimension of the 4HB dimer formed in the 4HB crystal dimer (Fig. 2). Thus, hAChE homodimers formed in solution and in crystals show significant overall structural similarity. Furthermore, solution structures of hAChE homodimers were not noticeably influenced by (1) the presence or absence of a 14-amino-acid-long N-terminal FLAG-tag; (2) the difference in the extent and composition of glycans in the hAChE expressed in wildtype and in Gnt1− HEK293 cells; or (3) the Tyr337Ala/Phe338Ala amino acid substitutions within the active center gorge of hAChE. Their P(r) curves were nearly identical (data not shown).
Reversible ligand binding does not affect hAChE dimer–monomer equilibrium
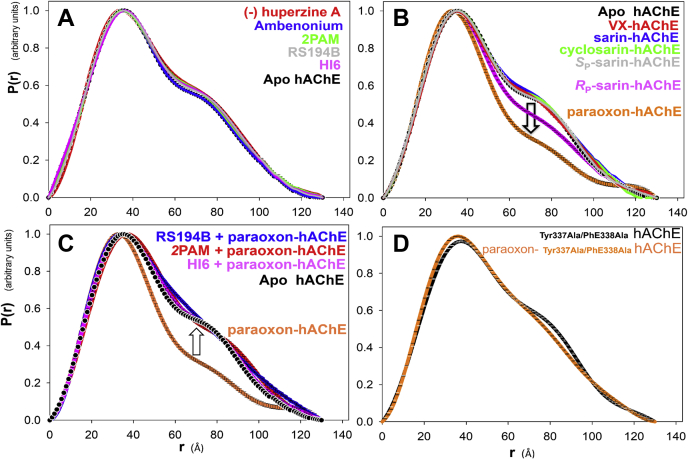
To investigate whether binding of reversible noncovalent ligands to hAChE can alter the enzyme solution structure we measured the SAXS profiles of a variety of reversible, noncovalent ligands (structures shown in Fig. S7) in complex with wildtype hAChE. High-affinity ligands, (-)huperzine A (Ki = 22 nM; (19)) and ambenonium (Ki = 250 pM; (19)) at 1.0 mM concentrations, and lower affinity aldoximes, 2PAM (Kd = 150 μM; (20)), HI6 (Kd = 38 μM; (20)), and RS194B (Ki = 157 μM; (21)) at 5 mM concentrations, were used to saturate all hAChE-binding sites in each hAChE–ligand complex. Figure 5A shows a P(r) profile overlay of wildtype apo hAChE with P(r) profiles recorded for each of the reversible ligand complexes. Reversible ligand complex formation did not reveal any significant effects on hAChE solution structure, in terms of Rg, positions or heights of the primary or the second peaks in the P(r) curve, or Dmax values. The five reversible ligands, being structurally diverse, occupy (when taken together) nearly all available binding sites within the 20 Å-deep active center gorge of hAChE, except for the acyl pocket (Fig. S8). These binding sites include Trp86 at the base of the gorge (where all ligands bind), Tyr337 and Phe338 of the choline-binding site (CBS; all ligands bind here as well), and Tyr72, Tyr124, Tyr341, Trp286, and Asp74 of the peripheral anionic site (PAS; where only HI6 and ambenonium bind). The X-ray crystal structure of the ambenonium∗AChE complex has not been solved yet, but biochemical data indicate it also binds to the PAS. Moreover, it is reasonable to assume that binding of this elongated bisquaternary compound (Fig. S7) spans Trp86 at the base of the gorge to the PAS at its opening, as seen in X-ray structures of complexes of a number of similar bisquaternary compounds such as decamethonium (1MAA), BW284c51 (6O50), HI6 (6CQU), TZ2PA6 (1Q84), as well as uncharged donepezil (6O4W). Thus, the dimerization equilibrium of hAChE does not appear to be affected by complexation with ligands occupying the main gorge volume, including the CBS and PAS. Interactions of ligands with residues of the hAChE Ω-loop and residues of the α-helical domains on the opposite side of the active center gorge, constituting the CBS and PAS, therefore do not seem to disturb the dimerization interface at the C terminus (Fig. S8). Of importance, none of these reversible ligands occupy the volume within the acyl pocket, as defined by the side chains of Phe295 and Phe297 (Fig. S8).
Figure 5.
Overlays of small-angle X-ray scattering P(r) profiles of diverse ligands incubated with wildtype and mutant hAChE.A, reversible complexes of (-)huperzine A (1.0 mM; red), ambenonium (1.0 mM; blue), 2PAM (5.0 mM; green), RS194B (5.0 mM; gray), and HI6 (5.0 mM; purple) with the 60 μM (4 mg/ml) wildtype hAChE. B, covalent OP inhibitors forming conjugates with 30 μM (2 mg/ml) hAChE, VX (red), cyclosarin (green), sarin (racemic mixture; blue), SP-sarin (gray), RP-sarin (purple), and paraoxon (orange). C, reversal in profiles of paraoxon-conjugated hAChE (30 μM; orange) upon 60 min incubation of the conjugate with 1 mM aldoxime reactivators 2PAM (red), HI6 (purple), or 5 mM RS194B (blue). D, paraoxon conjugate of 30 μM (2 mg/ml) Tyr337Ala/Phe338Ala double-mutant of hAChE (orange) and noninhibited, Tyr337Ala/Phe338Ala double-mutant (black). Profiles of noncomplexed, apo hAChE are shown as a dotted black line. The Gnt1− HEK-expressed wildtype hAChE devoid of FLAG-tag was used in experiments under A, B, and C. Error bars of the P(r) fit are shown with some curves and are the same size as the symbols. All curves were normalized to the highest P(r) value in each curve. hAChE, human acetylcholinesterase.
Organophosphate conjugation shifts the dimer–monomer equilibrium toward monomer
The SAXS profiles of hAChE prereacted with a variety of irreversible OP ligands to form covalent conjugates at the active site Ser203 show that several of the OP conjugates caused an increase in the height of the primary peak and a reduction in the second peak of the P(r) curve, although other OPs had no effect. These changes in the P(r) curve specifically caused by paraoxon (POX) and the RP enantiomer of sarin indicate a shift in the dimer–monomer equilibrium toward the monomer upon covalent binding (Fig. 5B). Multi-FoXS analysis of the SAXS data obtained with POX indicate that POX-hAChE at 30 μM concentration equilibrates between approximately 40% homodimer and 60% monomer compared with 100% homodimer for 30 μM apo hAChE (Fig. S5). This dimer/monomer ratio indicates a dimer dissociation constant for POX-hAChE about one order of magnitude higher than the one determined for apo hAChE (Table S1).
The ability of POX and RP-sarin to cause a change in the quaternary structure of hAChE can be explained by their chemical structures (Fig. S7) and by where they bind in the active site of hAChE compared with other OPs. All OPs listed in Figure 5B, except for paraoxon, are methylphosphonates. Upon covalent binding to Ser203, these OPs position their smallest substituents on the phosphorus atom, i.e., the methyl group, into the acyl pocket of the hAChE (Figure 6, Fig. S7). The structures of paraoxon and RP-sarin are different (Fig. S7). The phosphorus atom of paraoxon is symmetrically substituted and thus has to place one of its two ethoxy groups into the acyl pocket. In the case of RP-sarin, owing to its chirality, it has to approach and bind to Ser203 in such a way that its bulky isopropoxy substituent on the phosphorus atom orients toward the acyl pocket. Both the ethoxy and isopropoxy groups are larger than the methyl group of other OPs and larger than the methoxy group of the neurotransmitter acetylcholine. Thus, constraints of covalent bonding with Ser203, stabilization of phosphonyl oxygen in the oxyanion hole, and tetrahedral geometry around the conjugated phosphorus distinguish OPs that promote acyl pocket distortion upon binding. Another example of the impact of the constrained geometry in the active site is butyrylcholine turnover by AChE, which is about two orders of magnitude slower than that of the smaller acetylcholine (22) due to different sizes of their acyl group.
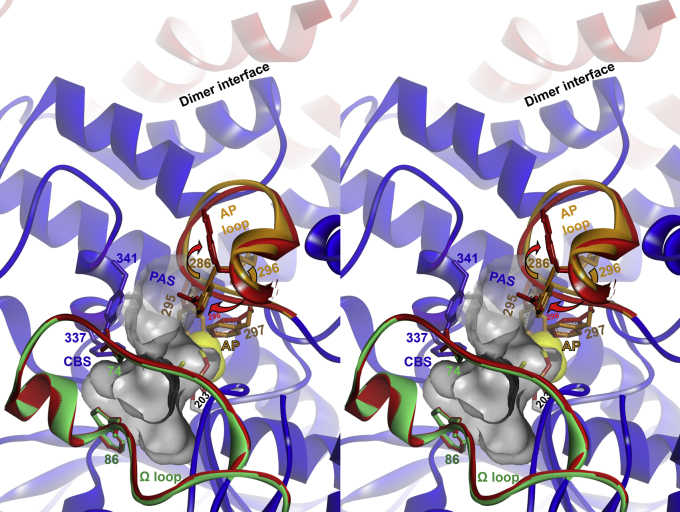
Figure 6.
Stereo overlay of X-ray structures of diethylphosphorylated hAChE obtained by conjugation with paraoxon (PDB ID:5HF5) and hAChE fromFigure S8(PDB ID:4EY5). Only selected loops and side chains from 5HF5 are shown in red (Ω loop and acyl pocket [AP] loop) for clarity. Conformational changes in conjugated hAChE are indicated by arrows, and the consequent enlargement in the acyl pocket volume is indicated by yellow surface. The diethylphosphoryl substituent of 5HF5 residing in the acyl pocket is rendered as sticks and colored by element (C = yellow, O = red). Selected residues of the AChE acyl pocket (AP; Phe295, Phe297), peripheral site (PAS; Asp74, Trp286, Tyr341), and choline binding site (CBS; Trp86, Tyr337) are also rendered as sticks. Locations of the active site Ser203, Ω loop (in green, covering the active center gorge), and dimer interface at the C terminus are also shown. hAChE, human acetylcholinesterase.
Incubation of the paraoxon diethylphosphoryl-hAChE conjugate with 1 mM (2PAM, HI6) or 5 mM (RS194B) aldoxime reactivators resulted in the reversal of P(r) profiles from the characteristic of a monomer–dimer ensemble to that of hAChE homodimer (Fig. 5C). Because structurally diverse aldoximes, including a monopyridinium monoxime (2PAM), a bispyridinium monoxime (HI6), and the zwitterionic oxime (RS194B), elicited a similar reversal in the P(r) profile of the paraoxon-hAChE conjugate, the changes clearly reflect the breakdown of the covalent bond of paraoxon conjugated with Ser203 rather than reversible aldoxime binding or nonspecific effects of millimolar aldoxime concentrations on the oligomerization state of OP-conjugated hAChE. Minor deviations compared with apo hAChE in the shape of P(r) profiles of the oxime-treated conjugate between 70 and 90 Å can be attributed to some residual OP-conjugate (Fig. 5C).
Substitution of hAChE Tyr337 and Phe338 with Ala enlarges the volume of the active center gorge and prevents OP-induced dimer dissociation
SAXS analysis indicates that paraoxon does not promote dimer dissociation of the Tyr337Ala/Phe338Ala double-mutant (Fig. 5D). The volume of the active center gorge is enlarged in this mutant by substitutions of bulky aromatic side chains with alanines, and covalently bound paraoxon fits well. The two mutations increase the volume of the active site gorge adjacent to the acyl-binding pocket by approximately 150 Å3. Similarity of the P(r) curves of the paraoxon-inhibited double mutant compared with that of apo-AChE (Fig. 5D) indicates that the larger volume in the mutant active site can accommodate paraoxon without causing structural distortion of the acyl pocket. Of interest, the large aromatic residues Tyr337 and Phe338 reside in the choline-binding site and their substitution does not directly increase the size of the nearby acyl pocket. It is likely that those substitutions allow the second ethoxy substituent of paraoxon, the one that points toward the two aromatic residues in wildtype hAChE, to move a small amount toward the two introduced alanines in the double mutant, consequently allowing relief of the acyl pocket steric strain. Thus, reducing general impaction in the active center gorge appears to relieve structural constraints toward the C-terminal dimerization interface and prevents dissociation of the dimer. In addition, it is possible that Tyr337 and Phe338 (attached to α-helices in the region 330–380) contribute in some manner to transmitting the conformational strain caused by paraoxon covalent binding toward the C-terminal α-helices of the dimerization interface (cf. Figs. S8–S11).
Time-resolved SAXS reveals approximate time frame of paraoxon-induced homodimer dissociation
Rapid time-resolved recording of successive SAXS measurements (TR-SAXS) during the early phase of covalent inhibition by paraoxon (Fig. 7) was enabled by the availability of a rapid stopped-flow mixing device and specialized X-ray detector at the BL-4 beamline of the Stanford Synchrotron Radiation Lightsource. This allowed us to follow the earliest kinetics of paraoxon-induced dimer dissociation. The TR-SAXS data indicate (Fig. 7) that conformational changes in hAChE triggered by active site Ser203 phosphorylation by paraoxon occur in the millisecond time frame. At the concentration of paraoxon used in these experiments (500 μM; Fig. 7), covalent inhibition of AChE by paraoxon occurs at the maximal rate and does not increase with concentration (23, 24, 25, 26). Our estimated first-order reaction rate constant (kobs=0.13 s−1; Fig. 7) is consistent with literature values of maximal rates of phosphorylation by paraoxon (0.15 s−1, (25); ∼0.05 s−1, (23)) and indicates that hAChE dimer dissociation kinetics parallel paraoxon conjugation with Ser203. Thus, OP-induced conformational changes in the active site of hAChE (Fig. 6) are not rate limiting for homodimer dissociation.
Figure 7.
Time-resolved small-angle X-ray scattering time course of changes in P(r) profiles of 50 μM hAChE during conjugation with 500 μM paraoxon, recorded every 20 ms during the first second of the reaction. Red curves indicate start (20 ms) and blue curves the last point (1000 ms) of the reaction. The rate constant of the reaction (kobs = 0.13 s−1) was obtained from the linear plot of the log of the ratio of P(r) values (at r1 = 71.5 Å and r2 = 34.7 Å) versus time (inset). Gray points represent the time course of 50 μM hAChE obtained in the absence of paraoxon. P(r) curves were not normalized. hAChE, human acetylcholinesterase.
Molecular dynamics simulations show correlated motions of the acyl pocket loop Arg296 and the dimer interface helices in paraoxon-conjugated hAChE
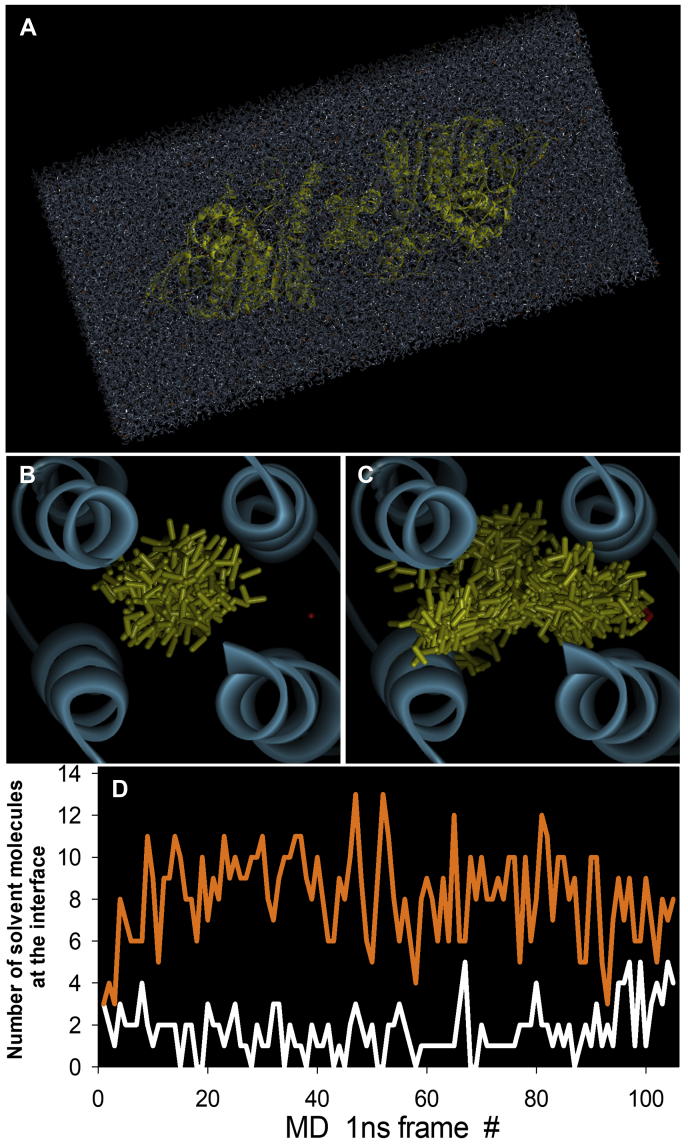
From the static X-ray structures (Fig. S12), there is no evidence for paraoxon-induced conformational change at the dimerization interface that could lead to dimer dissociation. This prompted us to study the atomic-level dynamic properties of native and paraoxon-conjugated hAChE dimers by molecular dynamics (MD) simulations using well-solvated X-ray structures as starting structures. The distance between the paraoxon-conjugated Ser203 in the active site and the dimer interface on the surface of the protein is approximately 25 Å. Comparative analysis of dimer trajectories of 110-ns MD simulations of apo-hAChE and paraoxon-hAChE did not reveal a single defined sequence of conformational changes that might be responsible for the effects observed in SAXS. Two general effects, however, were observed. The first one is that atomic fluctuations of Arg296 in the acyl pocket, the residue with the largest side-chain displacement upon paraoxon conjugation, correlated more strongly with atomic fluctuations of the dimer interface α-helices in the paraoxon-hAChE dimer compared with those in the apo hAChE dimer (Fig. 8). The negative values of correlations of MD fluctuations between the acyl pocket Arg296 and the 4HB interface in the paraoxon-conjugated hAChE suggest that the atoms in Arg296 and the 4HB interface oscillate toward each other and then back away, as opposed to a synchronized codisplacement. This dynamic coupling between the two sites indicates the existence of an allosteric pathway between the acyl pocket and dimer interface 25 Å away, eventually leading to changes in the geometry of the interface and dissociation of the paraoxon-conjugated dimers to monomers. Although neither of the hAChE homodimers effectively dissociated during the 110 ns of MD simulations, some initial signs of separation were noted for the paraoxon-conjugated homodimer. An inventory of the “solvent box” molecules able to penetrate the dimer interface indicates an increased propensity of the paraoxon-conjugated dimer to be solvated (Fig. 9, A–C). From the initial 42,259 solvent molecules surrounding each hAChE dimer (Fig. 9A) in the first 1 ns MD time frame, only three water molecules were found within each dimer interface (Fig. 9D). Later on, however, solvent was 4 to 5 times more often found in the paraoxon-hAChE dimer interface compared with apo hAChE. While the dimer interface of paraoxon-conjugated hAChE had solvent molecules bound at all times, solvent was completely absent in the apo-hAChE dimer interface 12% of the time. The average number of solvent molecules at the interface per 1-ns time frame was 1.7 ± 1.2 (ranging from 0 to 5) for apo-hAChE and 8.1 ± 2.2 (ranging from 3 to 10) for the paraoxon-conjugated dimer (Fig. 9D). The enhanced correlated conformational fluctuations of the paraoxon-conjugated dimer between the acyl pocket and the dimer interface α-helices (Fig. 8) thus seem likely to allow easier solvation of the largely hydrophobic dimerization interface and facilitate consequent dissociation of monomers.
Figure 8.
Molecular dynamics correlations of Arg276 of hAChE. Color-coded correlations of molecular dynamics fluctuations, between acyl pocket Arg296 (red) and remaining residues of the homodimer detected in 110-ns molecular dynamics simulations of (A) apo-hAChE (PDB ID: 4EY4) and (B) paraoxon-conjugated hAChE (PDB ID: 5HF5). The color coding scales in (A) and (B) were different in order to avoid “bleaching out” in coloring of dimer in (A). hAChE, human acetylcholinesterase.
Figure 9.
Solvent molecule inventory during the first 105 ns of the 110-ns MD simulations of hAChE dimer.A, box of 42, 259 solvent molecules (water, cations, and anions) surrounding the hAChE dimer during MD. B, the sum total of 183 solvent molecules (yellow sticks) found in 105, 1-ns time frames of MD at the interface of the apo hAChE. C, the sum total of 853 solvent molecules (yellow sticks) found in 105, 1-ns time frames of MD at the interface of paraoxon-conjugated hAChE. D, number of solvent molecules per 1-ns time frame found at interfaces of apo hAChE (white line) and paraoxon-conjugated hAChE (orange line). hAChE, human acetylcholinesterase; MD, molecular dynamics.
Discussion
The solution-based structural data of human AChE obtained in this study by four diverse experimental techniques (high-resolution SEC, cryo-EM, SAXS, and TR-SAXS) demonstrate that recombinantly expressed monomer, truncated at the C terminus and deficient in one (Asn350) of three N-glycosylation sites forms reversible homodimers in solution with dissociation constants in the 1.4 to 2.3 μM range. This range overlaps with local concentrations of AChE reported in tissue, from nanomolar in blood plasma to micromolar in synapses and nervous tissue (5). The data are consistent with dimerization occurring through a 4HB interface. Crystallographic structures of hAChE (28 of 39 deposited in Protein Data Bank [PDB]) are largely based on fully N-glycosylated hAChE truncated at the C terminus and expressed in a monomeric form. They reveal formation of a 4HB homodimer in the crystal packing. In our 11 recently deposited crystal structures based on the same hAChE construct as the one used in this study, we observe a F-2-F homodimer in addition to the 4HB dimer, for a total of two simultaneously formed homodimers in the crystal packing (11, 21, 27). So far, monomeric hAChE has not been observed in the crystalline state owing to high hAChE concentrations in the crystallization solutions, concentrations well above that of the dimer dissociation constant. The closest to the monomeric state was the X-ray structure of the Leu380Arg/Phe535Lys mutant of hAChE (9) that showed a severely distorted 4HB dimer (Table S2). Analysis of our SAXS and cryo-EM data indicates that the 4HB homodimer is the predominant structure in solution at micromolar concentrations; thus, distortion of the 4HB interface would be expected to impair formation of homodimer.
Of note, the SAXS data indicate that the 4HB dimer interface can be disrupted, and dissociation promoted, by covalent binding of paraoxon and RP-sarin enantiomer but not by several other racemic methyl-phosphono organophosphates or by a structurally diverse array of reversible noncovalent ligands. The ability of paraoxon and RP-sarin to disrupt the dimer interface is explained in part by their ability to distort the acyl-binding pocket in the catalytic gorge of hAChE. This pocket has evolved to accommodate the methyl group of acetylcholine and is too small for substituents larger than the methyl group without causing structural distortion of the enzyme. That was recognized early on (28) and later confirmed (22, 29) as the molecular basis for the exquisite substrate specificity of AChEs.
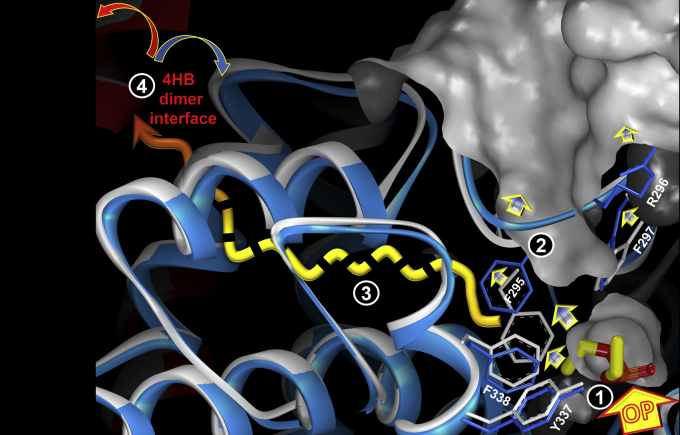
As is clearly seen and has been well described in crystal structures of AChEs from different species, all AChEs share a similar small acyl pocket that is distorted by large covalently bound ligands. This includes the “aged” conjugates of diisopropylfluorophosphate with Torpedo californica AChE (PDB ID: 2DFP) and mouse AChE (PDB ID: 2JGI), and in hAChE diethylphosphorylated by paraoxon (PDB ID: 5HF5; Figs. 6 and 10 and Figs. S9–S11). In these structures, the OP-conjugated acyl pocket loops were distorted, and the acyl pocket volume was enlarged in order to accommodate bulky isopropoxy or ethoxy groups. On the other hand, numerous X-ray structures of SP-methylphosphono-AChE enantiomer conjugates clearly show undistorted acyl pocket loop conformations by this smaller OP substituent. The exceptions are recent structures of Novichok A-234-conjugated hAChE (PDB IDs 6NTL and 6NTG) where the acyl pocket–bound ethoxy substituent does not cause any distortion owing to a 1.5-Å shift of the conjugated P atom (Figs. S9–S11). Thus, not only the size of the acyl pocket–bound substituent but also the propensity of the conjugated P atom to shift and remain at a distance from the acyl pocket determines the substrate specificity of AChE. This structural information is, however, sufficient to provide only a partial explanation for the P(r) profile shifts observed in SAXS (Fig. 5B). It seems clear that the large ethoxy group of paraoxon-conjugated hAChE, firmly held in the acyl pocket by covalent bonding to Ser203, was able to push away the aromatic rings of Phe295 and Phe297 and cause a ∼180° flip of the Arg296 side chain, thus enlarging the acyl pocket volume and causing distortion of the acyl pocket loop (Fig. 6; (30, 31)). What remains unclear, however, is how these changes in the acyl pocket loop could be transmitted to the dimer interface 25 Å away and what changes to the interface itself caused the two monomers to dissociate (Fig. 10). Some insights on the allosteric network can be gained by the PACCT analysis (Figs. S9 and S10) pointing at three α-helices in the residue region 330 to 380.
Figure 10.
Changes in tertiary and quaternary structures of hAChE triggered by covalent conjugation of paraoxon to the active Ser203 of hAChE yielding diethylphosphorylated hAChE (OP). Phases of structural change induced by conjugation: (1) covalent binding of OP; (2) local residue expansion around site of covalent binding (arrows indicate local expansions of gray solvent-accessible surface of native hAChE); (3) allosteric conformational transition toward 25-Å-distant dimer interface; (4) dissociation of “red” and “blue” monomers of the hAChE homodimer. More details are shown in Figures S9–S11. hAChE, human acetylcholinesterase.
Close visual inspection of the crystallographic dimerization interface does not reveal any difference between apo hAChE and paraoxon-conjugated-hAChE (Fig. S12). PISA server analysis (Table S2, (11)) furthermore showed that the two interfaces have very similar profiles of interactions and similar calculated monomer contact areas (988 Å2 for apo versus 1005 Å2 for the conjugate, i.e., only a 1.7% difference in contact area). The calculated free energies of solvation are also quite similar (−13.9 kcal/mol for apo versus −13.0 kcal/mol for the conjugate, i.e., a 6.5% difference). Moreover, 4HB homodimer packing is still preserved in crystals of both paraoxon- and RP-VX-conjugated hAChE (10, 31). Nevertheless, our SAXS data are consistent with an order of magnitude increase in dimer dissociation constant of POX-hAChE compared with apo hAChE 4HB dimer (Fig. S5). MD simulations of solvated hAChE homodimers indicate a propensity of the paraoxon-conjugated dimer to increase solvation of the dimerization interface, mediated by enhanced conformational dynamics at the interface that are correlated with altered acyl pocket loop dynamics. Dimer dissociation thus appears to be triggered by altered intramolecular dynamics of OP-conjugated hAChE and not statically by steric or electrostatic changes in the positions of amino acid side chains that form the dimer interface. Relaxation of dimeric structure in solution is also reflected in slight but consistent differences between crystallographic and solution-based FoXSDock-fitted 4HB dimers (Fig. S6, Table S5) evident in ∼20° relative rigid-body tilt of their monomeric vertical axes. In contrast, an example of a direct static disturbance of the dimer interface would be the “irreversible” monomerization reported for the Leu380Arg/Phe535Lys hAChE double-mutant (9), which is obvious both in solution and in the crystal packing. Dissociation to monomers of the wildtype hAChE that we observed here, however, is reversible, and homodimers can reassociate following nucleophilic removal of the conjugated phosphorus with oxime reactivators. We have thus revealed a dynamic communication between the hAChE active center and the 25-Å-distant dimerization interface promoted by spatial impaction in the acyl pocket. The OP-mediated communication of the acyl pocket with the dimer interface was compromised by either removing impaction from the acyl pocket, for example, in conjugates with SP-OP enantiomers, or by enlarging the nearby choline-binding site in the Tyr337Ala/Phe338Ala mutant, where “trimming” the size of residue side chains in the active site resulted in loss of the allosteric communication induced by paraoxon and RP-sarin conjugation.
The physiological consequences of OP-induced hAChE dimer dissociation are interesting to consider. Homodimers of hAChE in tissue are C-terminally disulfide bonded, and thus conjugation by diethylphosphoryl-based active forms of OP pesticides like paraoxon and chlorpyrifos oxon, or by RP- enantiomers of nerve agent OPs, would not be expected to lead to rapid dissociation of the homodimers. However, OP-induced dynamic relaxation of their quaternary structure, over an extended time period, could result in lower overall structural stability and compromised enzyme function. Once the conformation of the dimer is disturbed at the interface, access of reactivators could become difficult, and/or their alignment for efficient nucleophilic substitution compromised, assuming allosteric communication from the dimer interface to the catalytic site. Moreover, slow subunit dissociation, occurring as the disulfide bonds are broken, could lead to loss of catalytic subunits from their loci in the tissue. Conversely, prolonged structural disturbances at the dimerization interface, which might occur as the result of binding of ligands such as small molecules, peptides, or proteins could potentially affect proper functioning of the ∼25-Å-distant catalytic triad in an allosteric manner. Such ligand interactions with the dimer interface might occur physiologically with endogenous ligands or might represent a potential mechanism for pharmacological interventions that could facilitate reversal of OP intoxication.
Formation of C-terminal 4HB homodimers has been characterized for the closely related α/β-hydrolase fold proteins, including human butyrylcholinesterase (hBChE) both in crystals and in solution, catalytically inactive human neuroligins (hNLs), and human thyroglobulin, where dimerization is essential for function. Owing to differences in their respective amino acid sequences, the structures of those 4HB dimers, although similar, remain distinct. Dimers of hAChE and neuroligins appear more stable than dimers of hBChE, as inferred from their predicted energies of solvation (Table S2). Their quaternary structural geometries differ in the relative tilt of the vertical monomeric axes (Table S2; Fig. S13). Reduction in the number of glycosylation sites leads to an increase in the tilt and slightly improves the stability of crystallographic dimers. Binding of large surface peptidic ligands, such as snake toxin Fas2 to hAChE or neurexin to hNL, causes tilts of 28 to 38 degrees, destabilizing the dimers of hAChE but stabilizing those of neuroligins (Table S2), consistent with their function as cell adhesive structural proteins.
Small molecule ligand binding to hAChE can also affect the tilt in the crystal and reveal a predisposition for dimer destabilization in solution. Covalent inhibition by paraoxon or RP-VX causes a noticeable 5.2- to 5.5-degree monomer axes tilt in hAChE crystals (Table S2), unlike binding of donepezil and other reversible ligands, which have no effect. In equivalent OP conjugates with hBChE, where the volume of the acyl pocket is larger, the monomeric axes tilt is noticeably smaller (1.3–1.8 degrees; Table S2). This is consistent with the minimal dimer dissociation observed in SAXS of the Tyr337Ala/Phe338Ala hAChE mutant where the active center gorge volume is enlarged and similar in volume to hBChE.
Of interest, the geometries of crystallographic homodimers of hBChE are diverse and depend on glycosylation. Those fully glycosylated resemble the geometry observed in cryo-EM structures and appear more similar to dimers of hAChE. In the structure of thyroglobulin, where the geometry of 4HB dimers is constrained by other parts of this large protein, homodimers appear most similar to the geometry of dimers of partially glycosylated hBChE (only a 5.4° difference in axis tilt). Monomers in both hNL and in human thyroglobulin's AChE-like domain (Fig. S14) retain central solvent-accessible cavities that correspond to the active-center gorge in the hAChE. Diffusion of ligands into those cavities has the potential to create impaction within the cavities and thus alter their physiologically important dimerization states.
In summary, we describe here allosteric communication between the central gorge and the surface of an α/β-hydrolase-fold homodimer that has implications for understanding oligomer stability and catalytic activity in hAChE and provides a novel dynamic template for structure-based antidote and inhibitor design. These allosteric networks in a prototype member of α/β-hydrolase-fold proteins could better the understanding of the structural flexibility and dynamics of closely related and physiologically important noncatalytic members of this protein family.
Experimental procedures
Human AChE expression and purification
Gnt1− HEK293 mammalian cell culture was used to express hAChE (either wildtype or the Y337A/F338A double mutant) as an ∼60-kDa monomeric form protein truncated at C-terminal amino acid 547 and with a FLAG-tag placed at the N terminus. hAChE was purified as described (11). Briefly, the enzyme was eluted from the anti-FLAG affinity chromatography column (Sigma-Aldrich) either by proteolytic elution using cleavage site-specific Recombinant HRV 3C protease (Sinobiological.com) targeting the engineered Prescission protease recognition site or by elution with excess FLAG peptide. The N-terminal sequence of the proteolytically eluted hAChE was G-P-L-E-G-R- and that of FLAG peptide-eluted hAChE was D-Y-K-D-D-D-D-K-L-E-V-L-F-Q-G-P-L-E-G-R-. The amino acid sequence of the mature hAChE protein starts at E-G-R- and ends at the truncated C terminus with the sequence -S-A-T-D-T-L-D547.
Ligation of human AChE by reversible and covalent inhibitors
Reversible complexes of hAChE were prepared by adding reversible ligands to ∼30 μM hAChE in at least 10-fold stoichiometric excess, and at least 10-fold excess over the corresponding ligand Ki, immediately before each SAXS measurement.
Covalent OP-hAChE conjugates were formed before each SAXS experiment by adding millimolar concentration of OPs to 30 μM hAChE, allowing conjugates to form within ∼15 min incubation. Excess unreacted OP was removed by 1 min SEC in 1 ml Sephadex G-50 spin columns.
Nucleophilic reactivation of OP-hAChE conjugate formed by paraoxon was performed by incubating millimolar concentrations of aldoximes 2PAM, HI6, or RS194B with ∼30 μM OP-hAChE for 60 min, after which excess OP was removed by SEC.
Small-angle X-ray scattering analysis
SAXS experiments were used to assess the influence of reversible and nonreversible ligands on hAChE structure in solution. SAXS measurements using highly purified, monomeric human AChE (with or without N-terminal FLAG-tag) in both noninhibited and OP-covalently inhibited states were performed in 50 mM Tris/HCl, 100 mM NaCl, pH 7.4 at 22 °C. SAXS data were collected at the Stanford Synchrotron Radiation Lightsource (SSRL) BioSAXS beamline (BL 4-2) supervised by Dr Thomas Weiss and Dr Tsutomu Matsui. Data were reduced onsite at SSRL (Table S4). GNOM (32) was used to generate the P(r) curves. FoXS (http://modbase.compbio.ucsf.edu/foxs; (33, 34)) was used to generate theoretical scattering curves for the structure of apo hAChE monomer and 4B and F-2-F homodimers based on the crystal structure of apo hAChE (6O4X). MultiFoXS was used to fit dimer ensemble and monomer–dimer ensemble models to apo hAChE (3 mg/ml) and paraoxon-hAChE (3 mg/ml) SAXS data using monomer and dimer (4B and F-2-F) structures based on apo hAChE (6O4X). From the resulting monomer versus dimer ratios an approximate dissociation constant of hAChE homodimer was estimated, at two apo hAChE concentration points, 0.5 mg/ml and 1.0 mg/ml, or a single 2 mg/ml concentration (POX-hAChE) using the formula Kd = [monomer]2/[dimer]. FoXSDock (https://modbase.compbio.ucsf.edu/foxsdock) was used to generate thousands of homodimer models using the apo hAChE (6O4X) monomer structure and models that best fit the SAXS data for apo hAChE (3 mg/ml), as well as interface energy (33, 34, 35).
Time-resolved SAXS analysis
TR-SAXS experiments were used to monitor the rate of reaction of 500 μM paraoxon with 50 μM hAChE by following the change in amplitudes of P(r) curves (as a ratio of amplitude values at r = 71.5 Å and r = 34.7 Å) collected in subsequent 20-ms time bins during a 1000-ms total reaction period. Data were collected at the SSRL BL 4-2 configured with ∼1 ms dead time customized Bio-Logic four syringe stopped-flow mixer (SFM400) and Pilatus3 X 1M detector for rapid TR-SAXS data collection.
FPLC size-exclusion chromatography
High-resolution SEC was used to determine the dissociation constant of the hAChE homodimer. Purified hAChE preparations were run on a 24-ml SuperoseTM6 10/300 GL FPLC column at 0.5-ml/min flow rate in 50 mM Tris/HCl, 100 mM NaCl, pH 7.4 at 4 °C. Samples were applied in 100-μl volumes, and 0.5-ml fractions were collected. Three different hAChE samples were run in duplicate: N-terminally FLAG-tagged wildtype hAChE either fully glycosylated (HEK form) or expressed with reduced size of polysaccharide chains (Gnt1− hAChE) and the Tyr337Ala/Phe338Ala Gnt1− hAChE double mutant with no FLAG-tag. Relative concentrations of AChE in the fractions were determined by measuring catalytic activity using an end-point spectrophotometric Ellman assay (36). For the activity assay, samples were diluted to a final protein concentration of 0.01 μg/ml.
Cryo-electron microscopy
Cryo-EM grid preparation and data collection
Freshly prepared hAChE protein was purified using SEC. A 3-μl sample of 0.2 mg/ml (∼3.0 μM) in 10 mM Hepes, pH 7.4, 10 mM NaCl buffer was applied to a Quantifoil 1/2/1.3 grid (Ted Pella), glow discharged for 20 s at 15 mA (Pelco glow discharger), plunged with CP3 plunger (Gatan), and blotted for 3 s at 90% humidity (Scheme SI). A total of 963 images were collected on an Arctica 200-kV microscope with EPU on K2 Summit detector at 130,000-fold magnification at a pixel size of 1.04 Å with C2 aperture of 50 μm and objective aperture of 100 μm. A total exposure of 12 s was fractioned into 48 frames with each frame having an electron dose of 1.345 e/A2 (Table S3).
Cryo-EM data processing
Images were motion-corrected and dose-weighted using Motion Corr2 (37) (Scheme SI). CTF estimation was done using GCTF (38), and all procedures were performed in Relion 3.0 (39). Initial particle picking was done using the Laplacian–Gauss method in Relion. 2D classification showed a mixed population of monomers and dimers. Dimer and monomer particles were picked separately using 2D classes as templates, and multiple rounds of 2D classification were performed. A total dimer population with 2744 particles and monomer population with 99,596 particles were selected. The 4HB dimer from the X-ray structure 6O4W (Fig. 3) was projected to match 2D classes using EMAN2 (40). The dimer particles were exported to Crysoparc v 3.2 for ab initio model building and refinement using C2 symmetry. The 4HB dimer from the PDB 6O4W (Fig. 2) was fitted in the volume using volume fit in Chimera 1.13.1 (Fig. S3).
Molecular dynamics simulations
PDB-deposited datasets of native ligand-free hAChE (4EY4) and paraoxon-conjugated hAChE (paraoxon-hAChE; 5HF5) were used as initial structures in the MD simulations of the two dimeric enzymes. The missing residues of the surface loops 259 to 264 and 493 to 497 were rebuilt by MODELER 9v8 (41). MD simulations were carried out using NAMD 2.9 (42) and the CHARMM c36 force field for the protein (43). The force field parameters for paraoxon-conjugated Ser203 were obtained from the CGenFF software (44) and Quantum Mechanical calculations performed using Gaussian09 (45). The protonation states of all histidines were assigned based on visualization of their surrounding residues. All other protein residues were assigned to their canonical protonation state at neutral pH. Both dimer systems were immersed in a TIP3P water box with a minimal distance of 20 Å to any edge and periodic boundary conditions were applied. Chloride and sodium ions were added to neutralize the system and reach a concentration of 100 mM. Production MD simulations were performed in the constant particle number, pressure, and temperature ensemble. To allow for an integration time step of 2 fs in simulations, all bonds involving a hydrogen atom were constrained using the SHAKE algorithm (46). Short-range Coulomb interactions were cut off at 12 Å; long-range electrostatic interactions were calculated using the Particle-Mesh Ewald summation (47). Langevin dynamics (48) were used to control the temperature at 295 K with a damping coefficient of 2 ps−1. The Langevin piston method (49) was used to keep the pressure at 1 atm. Two independent simulations, each for ∼100 ns, were run for each dimeric system to interpret the OP-induced conformational changes detected by SAXS.
Cross-correlation analysis was applied to quantify the correlated atomic fluctuations between any pair of residues in both apo- and paraoxon-conjugated hAChE dimers. The cross-correlation coefficient Cij, between atoms i and j, is a measure of the correlated nature of their atomic fluctuations and is computed as follows:
where Δri and Δrj are the displacement vectors from the average position for atoms i and j, respectively. The angle brackets denote an average over the trajectory. The value of Cij ranges from −1 to 1 with the correlated (positive) residue pair moving in the same direction and the anticorrelated (negative) pair moving in the opposite direction.
Data availability
All data are contained within the manuscript or available to be shared upon request (please contact corresponding author). SAXS datasets of native ligand-free (“apo”) hAChE and paraoxon-conjugated hAChE have been deposited to SASBDB (Small Angle Scattering Biological Data Bank; https://www.sasbdb.org/) (50) under respective entry codes SASDL82 and SASDL92. The cryo-EM map has been deposited in the EMDB/PDB under accession code EMD-24404.
Supporting information
This article contains supporting information (11, 51).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
The highly professional and creative assistance of the Stanford Synchrotron Radiation Lightsource (SSRL) BioSAXS beamline (BL 4-2) scientists Dr Thomas Weiss, Dr Tsutomu Matsui, and Dr Ivan Rajković is highly appreciated. Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the NIH and the NIGMS, National Institutes of Health (P41GM103393). We are grateful to Dr Jonah Cheung, NYSBC, New York, for providing us with atomic coordinates of PDB ID 5HF5 well before their deposition to the PDB. We thank Dina Schneidman-Duhovny (dina@salilab.org) for her assistance in using MultiFoXS. This research was supported by the CounterACT Program, National Institutes of Health Office of the Director (NIH OD), the National Institute of Neurological Disorders and Stroke (NINDS) (grant numbers U01 NS083451 and R21 NS098998), and the UCSD Academic Senate grant BG084144. The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the National Institute of General Medical Sciences or National Institutes of Health.
Author contributions
D. K. B., P. T., A. K., Z. R. conceptualization; D. K. B., X. C., M. F., P. J., Z. R. methodology; D. K. B., X. C., M. F., Z. R. software; D. K. B., X. C., M. F., K.-Y. H., J. R., P. J., Z. R. validation; D. K. B., X. C., M. F., K.-Y. H., J. R., O. G., P. J., Z. R. formal analysis; D. K. B., X. C., M. F., K.-Y. H., J. R., O. G., P. J., A. K., Z. R. investigation; D. K. B., X. C., P. J., Z. R. resources; D. K. B., X. C., M. F., K.-Y. H., Z. R. data curation; D. K. B., X. C., M. F., P. J., A. K., Z. R. writing-original draft; D. K. B., X. C., M. F., P. T., P. J., A. K., Z. R. writing-review and editing; D. K. B., X. C., M. F., K.-Y. H., J. R., P. J., Z. R. visualization; D. K. B., X. C., Z. R. supervision; Z. R. project administration; Z. R. funding acquisition.
Edited by Wolfgang Peti
Supporting information
References
- 1.Schumacher M., Camp S., Maulet Y., Newton M., MacPhee-Quigley K., Taylor S.S., Friedmann T., Taylor P. Primary structure of Torpedo californica acetylcholinesterase deduced from its cDNA sequence. Nature. 1986;319:407–409. doi: 10.1038/319407a0. [DOI] [PubMed] [Google Scholar]
- 2.Maulet Y., Camp S., Gibney G., Rachinsky T.L., Ekström T.J., Taylor P. Single gene encodes glycophospholipid-anchored and asymmetric acetylcholinesterase forms: Alternative coding exons contain inverted repeat sequences. Neuron. 1990;4:289–301. doi: 10.1016/0896-6273(90)90103-m. [DOI] [PubMed] [Google Scholar]
- 3.Taylor P. The cholinesterases. J. Biol. Chem. 1991;266:4025–4028. [PubMed] [Google Scholar]
- 4.Taylor P., Camp S., Radić Z. In: Encyclopedia of Neuroscience. Squire L.R., editor. Academic Press; Oxford: 2009. Acetylcholinesterase; pp. 5–7. [Google Scholar]
- 5.Blotnick-Rubin E., Anglister L. Fine localization of acetylcholinesterase in the synaptic cleft of the vertebrate neuromuscular junction. Front. Mol. Neurosci. 2018;11:123. doi: 10.3389/fnmol.2018.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor P., Li Y., Camp S., Rachinsky T.L., Ekström T., Getman D., Fuentes M.E., Vellom D.C., Radić Z. Structure and regulation of expression of the acetylcholinesterase gene. Chem. Biol. Interact. 1993;87:199–207. doi: 10.1016/0009-2797(93)90043-x. [DOI] [PubMed] [Google Scholar]
- 7.Radić Z., Taylor P. In: Toxicology of Organophosphate and Carbamate Compounds. Gupta R., editor. Elsevier; Amsterdam: 2006. Structure and function of cholinesterases; pp. 161–186. [Google Scholar]
- 8.Bourne Y., Taylor P., Radić Z., Marchot P. Structural insights into ligand interactions at the acetylcholinesterase peripheral anionic site. EMBO J. 2003;22:1–12. doi: 10.1093/emboj/cdg005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bester S.M., Adipietro K.A., Funk V.L., Myslinski J.M., Keul N.D., Cheung J., Wilder P.T., Wood Z.A., Weber D.J., Height J.J., Pegan S.D. The structural and biochemical impacts of monomerizing human acetylcholinesterase. Protein Sci. 2019;28:1106–1114. doi: 10.1002/pro.3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bester S.M., Guelta M.A., Cheung J., Winemiller M.D., Bae S.Y., Myslinski J., Pegan S.D., Height J.J. Structural insights of stereospecific inhibition of human acetylcholinesterase by VX and subsequent reactivation by HI-6. Chem. Res. Toxicol. 2018;31:1405–1417. doi: 10.1021/acs.chemrestox.8b00294. [DOI] [PubMed] [Google Scholar]
- 11.Gerlits O., Ho K.Y., Cheng X., Blumenthal D., Taylor P., Kovalevsky A., Radić Z. A new crystal form of human acetylcholinesterase for exploratory room-temperature crystallography studies. Chem. Biol. Interact. 2019;309:108698. doi: 10.1016/j.cbi.2019.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sussman J.L., Harel M., Frolow F., Oefner C., Goldman A., Toker L., Silman I. Atomic structure of acetylcholinesterase from Torpedo californica: A prototypic acetylcholine-binding protein. Science. 1991;253:872–879. doi: 10.1126/science.1678899. [DOI] [PubMed] [Google Scholar]
- 13.Nicolet Y., Lockridge O., Masson P., Fontecilla-Camps J.C., Nachon F. Crystal structure of human butyrylcholinesterase and of its complexes with substrate and products. J. Biol. Chem. 2003;278:41141–41147. doi: 10.1074/jbc.M210241200. [DOI] [PubMed] [Google Scholar]
- 14.Araç D., Boucard A.A., Ozkan E., Strop P., Newell E., Südhof T.C., Brunger A.T. Structures of neuroligin-1 and the neuroligin-1/neurexin-1 beta complex reveal specific protein-protein and protein-Ca2+ interactions. Neuron. 2007;56:992–1003. doi: 10.1016/j.neuron.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 15.Fabrichny I.P., Leone P., Sulzenbacher G., Comoletti D., Miller M.T., Taylor P., Bourne Y., Marchot P. Structural analysis of the synaptic protein neuroligin and its beta-neurexin complex: Determinants for folding and cell adhesion. Neuron. 2007;56:979–991. doi: 10.1016/j.neuron.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coscia F., Taler-Verčič A., Chang V.T., Sinn L., O’Reilly F.J., Izore T., Renko M., Berger I., Rappsilber J., Turk D., Lowe J. The structure of human thyroglobulin. Nature. 2020;578:627–630. doi: 10.1038/s41586-020-1995-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X., Lee J., Di Jeso B., Treglia A.S., Comoletti D., Dubi N., Taylor P., Arvan P. Cis and trans actions of the cholinesterase-like domain within the thyroglobulin dimer. J. Biol. Chem. 2010;285:17564–17573. doi: 10.1074/jbc.M110.111641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park Y.N., Arvan P. The acetylcholinesterase homology region is essential for normal conformational maturation and secretion of thyroglobulin. J. Biol. Chem. 2004;279:17085–17089. doi: 10.1074/jbc.M314042200. [DOI] [PubMed] [Google Scholar]
- 19.Radić Z., Taylor P. Interaction kinetics of reversible inhibitors and substrates with acetylcholinesterase and its fasciculin 2 complex. J. Biol. Chem. 2001;276:4622–4633. doi: 10.1074/jbc.M006855200. [DOI] [PubMed] [Google Scholar]
- 20.Radić Z., Kalisiak J., Fokin V.V., Sharpless K.B., Taylor P. Interaction kinetics of oximes with native, phosphylated and aged human acetylcholinesterase. Chem. Biol. Interact. 2010;187:163–166. doi: 10.1016/j.cbi.2010.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorecki L., Gerlits O., Kong X., Cheng X., Blumenthal D.K., Taylor P., Ballatore C., Kovalevsky A., Radić Z. Rational design, synthesis, and evaluation of uncharged, “smart” bis-oxime antidotes of organophosphate-inhibited human acetylcholinesterase [published correction appears in J Biol Chem. 2020 May 8;295(19):6784] J. Biol. Chem. 2020;295:4079–4092. doi: 10.1074/jbc.RA119.012400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vellom D.C., Radić Z., Li Y., Pickering N.A., Camp S., Taylor P. Amino acid residues controlling acetylcholinesterase and butyrylcholinesterase specificity. Biochemistry. 1993;32:12–17. doi: 10.1021/bi00052a003. [DOI] [PubMed] [Google Scholar]
- 23.Kousba A.A., Sultatos L.G., Poet T.S., Timchalk C. Comparison of chlorpyrifos-oxon and paraoxon acetylcholinesterase inhibition dynamics: Potential role of a peripheral binding site. Toxicol. Sci. 2004;80:239–248. doi: 10.1093/toxsci/kfh163. [DOI] [PubMed] [Google Scholar]
- 24.Radić Z., Gibney G., Kawamoto S., MacPhee-Quigley K., Bongiorno C., Taylor P. Expression of recombinant acetylcholinesterase in a baculovirus system: Kinetic properties of glutamate 199 mutants. Biochemistry. 1992;31:9760–9767. doi: 10.1021/bi00155a032. [DOI] [PubMed] [Google Scholar]
- 25.Rosenfeld C.A., Sultatos L.G. Concentration-dependent kinetics of acetylcholinesterase inhibition by the organophosphate paraoxon. Toxicol. Sci. 2006;90:460–469. doi: 10.1093/toxsci/kfj094. [DOI] [PubMed] [Google Scholar]
- 26.Cochran R., Kalisiak J., Küçükkilinç T., Radic Z., Garcia E., Zhang L., Ho K.Y., Amitai G., Kovarik Z., Fokin V.V., Sharpless K.B., Taylor P. Oxime-assisted acetylcholinesterase catalytic scavengers of organophosphates that resist aging. J. Biol. Chem. 2011;286:29718–29724. doi: 10.1074/jbc.M111.264739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gerlits O., Kong X., Cheng X., Wymore T., Blumenthal D.K., Taylor P., Radić Z., Kovalevsky A. Productive reorientation of a bound oxime reactivator revealed in room temperature X-ray structures of native and VX-inhibited human acetylcholinesterase. J. Biol. Chem. 2019;294:10607–10618. doi: 10.1074/jbc.RA119.008725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Augustinsson K.-B., Nachmansohn D. Distinction between acetylcholine-esterase and other choline ester-splitting enzymes. Science. 1949;110:98–99. doi: 10.1126/science.110.2847.98. [DOI] [PubMed] [Google Scholar]
- 29.Radić Z., Pickering N.A., Vellom D.C., Camp S., Taylor P. Three distinct domains in the cholinesterase molecule confer selectivity for acetyl- and butyrylcholinesterase inhibitors. Biochemistry. 1993;32:12074–12084. doi: 10.1021/bi00096a018. [DOI] [PubMed] [Google Scholar]
- 30.Cheung J., Rudolph M.J., Burshteyn F., Cassidy M.S., Gary E.N., Love J., Franklin M.C., Height J.J. Structures of human acetylcholinesterase in complex with pharmacologically important ligands. J. Med. Chem. 2012;55:10282–10286. doi: 10.1021/jm300871x. [DOI] [PubMed] [Google Scholar]
- 31.Franklin M.C., Rudolph M.J., Ginter C., Cassidy M.S., Cheung J. Structures of paraoxon-inhibited human acetylcholinesterase reveal perturbations of the acyl loop and the dimer interface. Proteins. 2016;84:1246–1256. doi: 10.1002/prot.25073. [DOI] [PubMed] [Google Scholar]
- 32.Svergun D.I. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 1992;25:495–503. [Google Scholar]
- 33.Schneidman-Duhovny D., Hammel M., Tainer J.A., Sali A. Accurate SAXS profile computation and its assessment by contrast variation experiments. Biophys. J. 2013;105:962–974. doi: 10.1016/j.bpj.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schneidman-Duhovny D., Hammel M., Tainer J.A., Sali A. FoXS, FoXSDock and MultiFoXS: Single-state and multi-state structural modeling of proteins and their complexes based on SAXS profiles. Nucleic Acids Res. 2016;44:W424–W429. doi: 10.1093/nar/gkw389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schneidman-Duhovny D., Hammel M., Sali A. Macromolecular docking restrained by a small angle X-ray scattering profile. J. Struct. Biol. 2011;173:461–471. doi: 10.1016/j.jsb.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ellman G.L., Courtney K.D., Andres V., Jr., Feather-Stone R.M. A new and rapid colorimetric determination of acetylcholinesterase activity. Biochem. Pharmacol. 1961;7:88–95. doi: 10.1016/0006-2952(61)90145-9. [DOI] [PubMed] [Google Scholar]
- 37.Zheng S.Q., Palovcak E., Armache J.P., Verba K.A., Cheng Y., Agard D.A. MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;4:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scheres S.H.W. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang G., Peng L., Baldwin P.R., Mann D.S., Jiang W., Rees I., Ludtke S.J. EMAN2: An extensible image processing suite for electron microscopy. J. Struct. Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 41.Šali A., Blundell T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 42.Phillips J., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R., Kalé L., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Best R.B., Zhu X., Shim J., Lopes P.E., Mittal J., Feig M., MacKerell A.D., Jr. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ1 and χ2 dihedral angles. J. Chem. Theory Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vanommeslaeghe K., Hatcher E., Acharya C., Kundu S., Zhong S., Shim J., Darian E., Guvench O., Lopes P., Vorobyov I., MacKerell A.D., Jr. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frisch M.J., Trucks G.W., Schlegel H.B., Scuseria G.E., Robb M.A., Cheeseman J.R., Scalmani G., Barone V., Petersson S.A., Nakatsuji H., Li X., Caricato M., Marenich A., Bloino J., Janesko B.G. Gaussian, Inc.; Wallingford, CT: 2016. Gaussian 09, Revision A.02. [Google Scholar]
- 46.Ryckaert J.-P., Ciccotti G., Berendsen H. In numerical integration of the Cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 1977:327–341. [Google Scholar]
- 47.Darden T., York D., Pedersen L. Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 48.Pastor R.W., Brooks B.R., Szabo A. An analysis of the accuracy of Langevin and molecular dynamics algorithms. Mol. Phys. 1988;65:1409–1419. [Google Scholar]
- 49.Feller S.E., Zhang Y., Pastor R.W. Constant pressure molecular dynamics simulation: The Langevin piston method. J. Chem. Phys. 1995;103:4613–4621. [Google Scholar]
- 50.Kikhney A.G., Borges C.R., Molodenskiy D.S., Jeffries C.M., Svergun D.I. Sasbdb: Towards an automatically curated and validated repository for biological scattering data. Protein Sci. 2020;29:66–75. doi: 10.1002/pro.3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hajizadeh N.R., Franke D., Jeffries C.M., Svergun D.I. Consensus Bayesian assessment of protein molecular mass from solution X-ray scattering data. Sci. Rep. 2018;8:7204. doi: 10.1038/s41598-018-25355-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the manuscript or available to be shared upon request (please contact corresponding author). SAXS datasets of native ligand-free (“apo”) hAChE and paraoxon-conjugated hAChE have been deposited to SASBDB (Small Angle Scattering Biological Data Bank; https://www.sasbdb.org/) (50) under respective entry codes SASDL82 and SASDL92. The cryo-EM map has been deposited in the EMDB/PDB under accession code EMD-24404.