Abstract
The epidermal growth factor receptor (EGFR) pathway is important in tumorigenesis of oropharyngeal carcinoma (OPC). However, the molecular mechanisms contributing to EGFR expression in OPC are not well-known. To detect relating factors and clinicopathological impact of EGFR protein expression in OPC, gene amplification/loss, point mutations including synonymous mutations, and promoter methylation of EGFR, and the viral genome load of human papillomavirus type 16 (HPV16)-E5, -E6, and -E7, after extracting HPV16-related OPCs with qPCR of HPV16-E6 and E7, were investigated in 74 OPC surgical cases, including 52 HPV-related (HPV-OPC) and 22 HPV-unrelated (nHPV-OPC). Immunohistochemical (IHC) data of EGFR expression (high, weak, and negative), validated by the qPCR of EGFR mRNA, were compared with molecular, viral, and clinicopathological data of patients. All nHPV-OPC cases were EGFR-IHC-high, whereas 21.2%, 65.4%, and 13.5% of HPV-OPC cases showed EGFR-IHC-high, -weak, -negative (p < 0.01), respectively. In HPV-OPC cases, EGFR-IHC-weak/negative status was related to promoter methylation of EGFR (p = 0.009), but not with gene amplification/loss or the point mutation of EGFR and was more often seen in HPV16-OPC cases (p = 0.049). Among HPV16-OPC cases, EGFR-IHC-weak/negative was related to high E6 expression. EGFR protein-loss was related to the tumor histology of non-keratinizing squamous cell carcinoma (SCC) (p = 0.035) but not with patient prognosis. In conclusion, decreased EGFR protein expression was more frequent in HPV-OPC than in nHPV-OPC and was related to EGFR methylation, infection of HPV16, and the viral genome load of HPV16-E6. Clinicopathologically, it was related to the tumor histology of non-keratinizing SCC.
Supplementary information
The online version of this article (10.1007/s12105-020-01261-w) contains supplementary material, which is available to authorized users.
Keywords: Epidermal growth factor receptor, Human papilloma virus, Oropharyngeal carcinoma, E5, E6, E7, Methylation, Non-keratinizing squamous cell carcinoma
Introduction
Head and neck cancer is the sixth most frequent cancer globally [1], and oropharyngeal carcinoma (OPC) accounts for approximately 10% of all head and neck cancers. OPC can be divided into human papillomavirus (HPV)-related OPC (HPV-OPC) and HPV-unrelated OPC (nHPV-OPC). These two groups differ in many aspects, including patient prognosis, tumor behavior, and tumor histology [2–4].
Following HPV status, the clinical impacts of epidermal growth factor receptor (EGFR) expression on head and neck squamous cell carcinoma (HNSCC) have been extensively studied. EGFR is a member of the ErbB tyrosine kinase family, which also includes HER2 (ErbB2), HER3 (ErbB3) and HER4 (ErbB4). Several ligands for EGFR including epidermal growth factor, transforming growth factor-alpha, and epiregulin, are known [5]. Before ligand-binding, EGFR exists as a monomer on the cell surface and, upon ligand binding, EGFR dimerizes, resulting in signal pathway activation by self-phosphorylation on tyrosine residues [5]. The activation of the EGFR signaling pathway depends on its ligand status, feedback from the downstream pathway, interaction from other ErbB family members, and EGFR status [5].
In HPV-related carcinoma, viral genome E6 and E7 are constitutively expressed since they are mandatory for carcinogenesis and the maintenance of malignancy [6–8]; therefore, quantitative PCR (qPCR) of these genes has been the gold standard method for detecting HPV-OPC [9–11], followed by p16INK4A immunohistochemistry (IHC). Although the role of viral genome E5 is more poorly characterized, it has been shown to increase the recycling of EGFR to the cell surface by inhibiting endosomal acidification in studies using cell lines [12, 13].
In OPC, several authors have shown increased EGFR expression in nHPV-OPC and decreased expression in HPV-OPC [14, 15]. Several authors have also discussed the prognostic value of EGFR protein expression, but the results are controversial [14, 16–18]. Thus far, few studies have investigated the molecular mechanisms that contribute to EGFR protein expression in OPC.
Hence, to clarify the factors contributing to EGFR protein expression and to further examine the clinicopathological impact of EGFR expression on OPC, gene amplification/loss, point mutations, including synonymous mutations, and promoter methylation of EGFR were investigated and compared with EGFR protein expression. Viral genome loads of E5, E6, and E7 of HPV type 16 (HPV16) were also analyzed to extract HPV-OPC associated with HPV16 infection (HPV16-OPC) and determine the relationship of each genome load with EGFR protein expression. By comparing EGFR expression and clinicopathological data, including tumor histology, the clinicopathological impacts of EGFR expression were investigated.
Materials and Methods
Materials
A total of 74 OPC cases (52 HPV- and 22 nHPV-OPCs), were enrolled in this study. All cases were operated at Department of Otorhinolaryngology, Juntendo University and pathological diagnoses were given as squamous cell carcinoma (SCC) or its variants between January 2004 and October 2019. The classification of HPV- or nHPV-OPC was conducted by immunohistochemistry (IHC) for p16INK4A, according to a previous study [19], where the OPC cases in which more than 70% of tumor cells showed positive for p16INK4A in both tumor nuclei and the cytoplasm were classified as HPV-OPCs. OPC cases without invasive carcinomas, with preoperative chemo- or radiotherapy, and of tumor recurrence were excluded from this study. For each case, a representative tumor slide from a formalin-fixed paraffin-embedded (FFPE) tissue block was selected for further study.
This study was approved by the Ethics Committee of Juntendo University, Tokyo, Japan (#2,018,131) on November, 2018, and was performed according to the Declaration of Helsinki. Informed consent was obtained from all subjects.
Clinicopathological Data Collection and Evaluation of Tumor Histology
Information regarding patients’ age/sex, smoking/drinking history, tumor site and size, TNM classification, tumor stage according to UICC criteria [20], overall survival (OS)/progression-free survival ratio (PFS), and histological type was collected. Smoking history was divided into two groups: never smoker/former smoker (for patients smoking 1 year or more before the surgery) or current smoker (for patients smoking within 1 year of the surgery). The drinking history was divided into positive (for patients drinking more than once a week) or negative.
The histology of the tumor was classified into keratinizing SCC (KSCC), non-keratinizing SCC, hybrid-SCC, basaloid SCC or lymphoepithelial carcinoma, according to previous studies [4, 21] and WHO classification [22], with the help of CK14 IHC. A brief explanation of the histological classification is shown in Supplementary table 1.
IHC for p16INK4A, CK14, and EGFR
For IHC, deparaffinized 4-µm sections from each tissue block were used. After exposure to 0.3% hydrogen peroxide for 10 min to block endogenous peroxide activity, IHC for p16INK4A, EGFR, and CK14 were performed. IHC for p16INK4A and CK14 was performed to determine whether the tumor was HPV-related and to confirm squamous differentiation of the tumor, respectively. Details of the primary antibodies used in this study are summarized in Supplementary Table 2. For EGFR IHC, only membranous staining was judged positive, according to a previous study [23] and it was categorized as (1) high, when circumferential and intense staining was seen for tumor cells, (2) weak, when weak staining was seen or only a part of the tumor cell membrane was positive, or (3) negative, when no staining was observed. When the tumor contains more than one histological type, IHC data were obtained from the predominant subtype.
Fluorescence Iin Situ Hybridization for EGFR Gene
To determine the relationship between EGFR IHC and the amplification/deletion of the EGFR gene, fluorescence in situ hybridization (FISH) was conducted for 11 and 6 HPV- and nHPV-OPC cases, respectively. These 17 cases were selected from recently operated cases and cases with few intratumoral lymphocytes for easier identification of EGFR/centromere of tumor cells. A total of 2, 5, 4, and 6 of HPV-OPC with EGFR-IHC-high, -weak, -negative, and nHPV-OPC were analyzed, respectively, and all 6 nHPV-OPC cases were EGFR-IHC-high. Unstained sections cut from tissue blocks of 4-µm thickness were prepared for each OPC case, and a 20 mm-sized representative area was analyzed. These unstained sections were sent to the Genetic Labolatory, CO., (Sapporo, Japan) for staining, using telomeric probes (Vysis® LSI EGFR Spectrum Orange) and centromeric probes (CEP® 7 Spectrum Green probe). For EGFR /centromere counting, 2–3 areas showing representative histopathological features were selected and 20 nuclei were evaluated using the Zeiss ApoTome.2 (Carl Zeiss, Jena, Germany).
DNA and RNA Extraction
Depending on the tumor size, one to four continuous FFPE tissue sections (5 µm, mounted on positively charged slides) were dissected for DNA and RNA extraction. The invasive tumor tissue showing the predominant histological type was collected into a 1.5 µl microcentrifuge tube using a sterile razor blade.
Isolation of DNA/RNA was conducted using DNeasy/RNeasy FFPE Kits (Qiagen, Melbourne, Australia), respectively, according to the manufacturer’s instructions. Thereafter, the extracted DNA/RNA was quantified using a Nanodrop 1000 spectrophotometer (Thermo Fisher Scientific, Lenexa, KS, USA).
qPCR of EGFR mRNA
To determine the relationship of EGFR IHC and the amount of EGFR mRNA, qPCR was conducted for 49 and 15 HPV-and nHPV-OPC cases, respectively, after eliminating OPC cases from before 2010. The extracted RNA was diluted to 125 ng/µl and reverse transcribed using SuperScript™IV VILO Master Mix (Thermo Fisher Scientific) according to the manufacturer’s instructions. qPCR was performed using TaqMan gene expression assay kits (Thermo Fisher Scientific) and a 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). All qRT-PCR reactions were performed in triplicate. The value of relative quantification (RQ-value) was obtained using the ΔΔCt method. The GAPDH gene served as an endogenous control for the normalization of expression levels, and the details of the probes used in this qPCR study are listed in Supplementary Table 3.
Mutational Analysis of the EGFR Gene
The extracted DNA (diluted to 50 ng/µl) was amplified by PCR using four pairs of primers encompassing exons 18–21 of EGFR (Supplementary Table 4). Isolated PCR products were then sequenced in both sense and antisense directions and analyzed by Sequencing Analysis V3.5.1 software (Applied Biosystems). Mutations were determined when the height of the mutated peak reached 20% of the height of the normal peak in both the sense and antisense directions. Mutations were analyzed by three independent researchers, YS, YF and MA.
Methylation-Specific PCR
To determine the presence of the promoter-region methylation of EGFR, methylation-specific PCR was conducted. Bisulfite modification was performed using an EZ DNA Methylation-Gold Kit (Zymo Research, Irvine, CA, USA). Then, the bisulfate-treated DNA was amplified using specifically designed primers for methylated and unmethylated alleles (Primer Express software v1.0, Applied Biosystems). The sequences of the primers, annealing temperature, and product size are listed in Supplementary Table 5. After amplification, the products were electrophoresed using 2% agarose gels, stained with ethidium bromide, and visualized under ultra-violet illumination. An Epitect PCR control DNA set (QIAGEN) was used as the control DNA.
Quantitative PCR of Viral Genome E5,E6,E7 of HPV16
The viral genome load of E5, E6, and E7 of HPV16 was investigated with the qPCR method for 49 HPV-OPC cases, utilizing GAPDH as a reference gene. These 49 cases were selected by excluding cases from before 2010. The primers and probes of these genes were designed according to a previous study [24, 25] to bind within the amplicon for each target gene. The fluorescent dye, FAM was conjugated to a probe. Cases that showed an exponential proliferation curve for either E6 or E7 and a ΔCT of no more than 8 were determined to be HPV16-OPC, and cases that did not meet these criteria were determined to be HPV of other type-related (HPVnon16-OPC), according to a previous study [25]. The RQ values of E5, E6, and E7 were compared to EGFR IHC results for these HPV16-OPC cases. The primer and probe details of our qPCR are in Supplementary Table 6.
Statistical Analysis
Statistical analyses were performed to compare the clinicopathological and molecular features between HPV-OPC and nHPV-OPC cases and between EGFR-IHC-high, -weak, and -negative HPV-OPC cases. The Kaplan-Meier method with Log-rank test and multivariate analysis were used to determine the relationship between EGFR-IHC data and disease-free survival ratio (OS + PFS). Among HPV16-OPC cases, RQ values of E5, E6, and E7 were compared between EGFR-IHC-high and weak/negative groups. GraphPad Prism8 software (GraphPad Software, San Diego, CA, USA) and Statmate V software (ATMS Co. Ltd, Chiba, Japan) were used in this study. Chi-square test and Fisher’s exact test were used to compare categorical data, and the Mann–Whitney U test and Kruskal–Wallis test was used for sequential data. An F test was used to compare the variance of E5, E6, and E7. A P value < 0.05 was considered statistically significant.
Results
Clinicopathological and EGFR-Related Molecular Features of OPC Cases
Clinicopathological features of our cohort are shown in Table 1; the data are shown by dividing HPV- and nHPV-OPC cases, and statistically significant differences between the HPV- and nHPV-groups are shown with a P value or otherwise with NS. Briefly, the smoking history, pathological stage, and tumor histological subtype were significantly different between HPV- and nHPV-OPC cases. Most of the nHPV-OPC cases (90.9%) showed KSCC histological subtype, while 63.5% of HPV-OPC cases showed subtypes other than KSCC. The representative tumor histology of HPV- and nHPV-OPC is shown in Fig. 1.
Table 1.
Clinicopathological features of HPV-OPC/nHPV-OPC cases
| HPV-OPC (n = 52) | nHPV-OPC (n = 22) | P value§ | |
|---|---|---|---|
| Age | NS* | ||
| Mean | 67.6 | 63.14 | |
| Range | (44–86) | (37–86) | |
| Sex—no. (%) | NS† | ||
| Male | 43 (82.7) | 14 (63.6) | |
| Female | 9 (17.3) | 8 (36.4) | |
| Smoking history—no. (%) | 0.013† | ||
| Never or former smoker | 38 (73.1) | 5 (22.7) | |
| Current smoker | 7 (13.5) | 6 (27.3) | |
| Uncertain | 7 (13.5) | 11 (50.0) | |
| Drinking history—no. (%) | NS† | ||
| Positive | 34 (65.4) | 9 (40.9) | |
| Negative | 10 (19.2) | 5 (22.7) | |
| Uncertain | 8 (15.4) | 8 (36.4) | |
| Tumor size | NS* | ||
| Mean (mm) | 28.3 | 25.68 | |
| Range | 6–60 | 12–53 | |
| Tumor site—no. (%) | NS‡ | ||
| Lateral wall | 39 (75.0) | 9 (40.9) | |
| Upper wall | 1 (1.9) | 3 (13.6) | |
| Anterior wall | 6 (11.5) | 2 (9.1) | |
| Posterior wall | 0 | 1 (4.5) | |
| Uncertain | 6 (11.5) | 7 (31.8) | |
| T factor—no. (%) | NS† | ||
| T1-2 | 40 (76.9) | 18 (81.8) | |
| T3-4 | 12 (23.1) | 4 (18.2) | |
| N factor—no. (%) | NS† | ||
| N0 | 15 (28.8) | 11 (50) | |
| N1-3 | 37 (71.1) | 11 (50) | |
| M factor—no. (%) | NS† | ||
| M0 | 52 (100) | 22 (100) | |
| M1 | 0 (0) | ||
| Stage—no. (%) | < 0.001† | ||
| I–II | 48 (92.3) | 10 (45.5) | |
| III–IV | 4 (7.7) | 12 (54.5) | |
| Prognosis—no. (%) | |||
| Overall survival | 51 (96.2) | 20 (90.9) | NS|| |
| Progression free survival | 38 (73.1) | 14 (63.3) | NS|| |
| Histological subtype—no. (%) | < 0.01‡ | ||
| Keratinizing SCC | 19 (36.5) | 20 (90.9) | |
| Hybrid SCC | 12 (23.1) | 1 (4.5) | |
| Non-keratinizing SCC | 19 (36.5) | 1 (4.5) | |
| Basaloid SCC | 1 (1.9) | 0 | |
| Lymphoepithelial carcinoma | 1(1.9) | 0 | |
| EGFR protein expression—no. (%) | < 0.001‡ | ||
| High | 11 (21.2) | 22 (100.0) | |
| Weak | 34 (65.4) | 0 | |
| Negative | 7 (13.5) | 0 | |
| EGFR mRNA (RQ score)¶ | |||
| EGFR protein overexpression | |||
| Median | 3.60 | 27.9 | < 0.01* |
| Mean | 13.27 | 132.55 | |
| EGFR protein weakly expression | |||
| Median | 3.12 | ||
| Mean | 3.79 | ||
| EGFR protein negative expression | |||
| Median | 1.16 | ||
| Mean | 1.18 | ||
| EGFR missense mutation—no. (%) | NS† | ||
| Present | 3 (5.8) | 0 | |
| Absent | 49 (94.2) | 22 (100.0) | |
| Q787Q synonymous mutation—no. (%) | NS† | ||
| Present | 9 (17.3) | 7 (31.8) | |
| Absent | 43 (82.7) | 15 (68.2) | |
| EGFR amplification—no.(%)¶ | NS† | ||
| Present | 0 | 0 | |
| Absent | 11 (100) | 6 (100) | |
| EGFR promoter methylation—no.(%)¶ | NS† | ||
| Present | 13 (25.5) | 2 (9.1) | |
| Absent | 38 (74.5) | 20 (90.9) | |
¶Data of EGFR mRNA (RQ score) are of 49 HPV-OPC and 15 nHPV-OPC cases. Data of EGFR amplification are of 11 HPV-OPC and 6 nHPV-OPC cases. Data of EGFR promoter methylation are of 51 HPV-OPC and 22 nHPV-OPC cases
§NS not significant and the superscript letter
*Mann-Whitney U test
†Fisher’s exact test
‡Chi-square test
||Log-rank test, respectively
Fig. 1.
Representative tumor histology of oropharyngeal carcinoma. a Keratinizing squamous cell carcinoma (KSCC). b Non-keratinizinig SCC. c Hybrid SCC with abrupt keratinization (Arrows). d Basaloid SCC (lower part of the view). e Lymphoepithelial carcinoma. (a–e Hematoxylin and eosin)
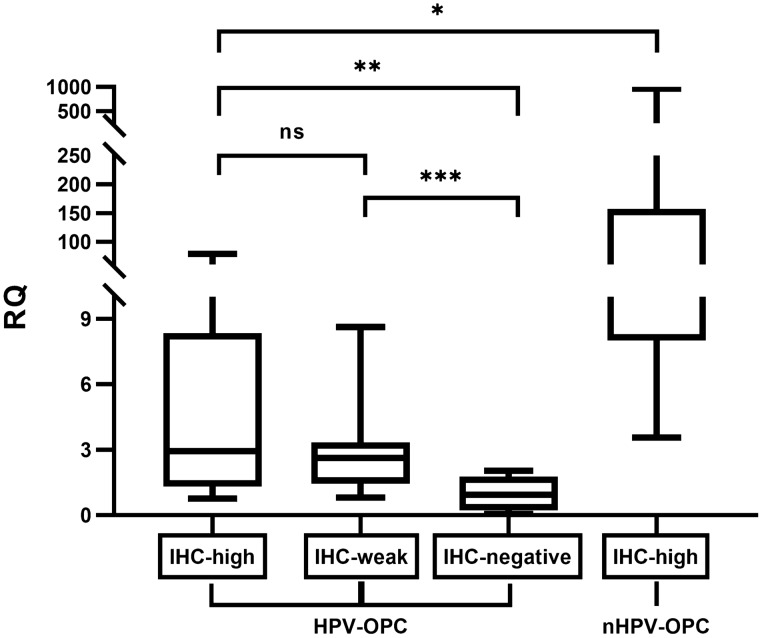
The results of our EGFR-IHC showed that all (100%) nHPV-OPC cases showed diffusely high expression for EGFR, while 21.2%, 65.4%, and 13.5% of HPV-OPC cases showed high, weak and negative expression, respectively, and EGFR expression was significantly different between HPV- and nHPV-OPC (p < 0.001) (Fig. 2; Table 1). In the qPCR of EGFR mRNA, a significantly higher RQ value was obtained for nHPV-OPC than for HPV-OPC (p < 0.01). Among HPV-OPC cases, the RQ value for HPV-OPC in descending order was with IHC-high, IHC-weak, and IHC-negative cases, respectively (p = 0.553 for IHC-high vs. IHC-weak, < 0.01 for IHC-weak vs. IHC-negative, and < 0.05 for IHC-high vs. IHC-negative, respectively), validating the IHC results (Fig. 3).
Fig. 2.
Representative immunohistochemistry (IHC) for EGFR. a nHPV-OPC case with EGFR-IHC-High. b HPV-OPC case with EGFR-IHC-high. c HPV-OPC case with EGFR-IHC-weak. d HPV-OPC case with EGFR-IHC-negative. (a–d Immunohistochemistry for EGFR)
Fig. 3.
Relationship between EGFR-immunohistochemistry (IHC) and quantitative PCR of EGFR mRNA. The box plot shows the relative quantification (RQ) of EGFR mRNA in EGFR-IHC-high, weak, and –negative groups of HPV-OPC and nHPV-OPC. Each graph shows the maximum value, the first quartile (the top of the rectangle), median (horizontal line near the middle of the rectangle, the third quartile (the bottom of the rectangle), and the minimum value. nHPV-OPC cases showed significantly higher RQ value than HPV-OPC (*P < 0.01). Among HPV-OPC cases, the RQ value for IHC-high group and IHC-weak group showed significantly higher RQ value than IHC-negative group (**P < 0.05 and ***P < 0.01, respectively)
The EGFR-related molecular features of OPC are summarized in Table 1. Briefly, the results of FISH analysis revealed that none of HPV- and nHPV-OPC cases showed gene amplification or deletion (Fig. 4). Missense mutations in the EGFR gene were detected in 5.8% of HPV-OPC and 0% of nHPV-OPC cases. Regarding the synonymous mutation of EGFR, the Q787Q mutation with base changes of CAG to CAA was the most frequent, and it was detected in 17.3% and 31.8% of HPV- and nHPV-OPC cases. The details of the mutations detected are shown in Supplementary Table 7. Methylation of the EGFR promoter region was detected in 25.5% and 9.1% of HPV- and nHPV-OPC cases, respectively.
Fig. 4.
Fluorescence in situ hybridization of EGFR gene. a nHPV-OPC case with EGFR-IHC-high. b HPV-OPC case with EGFR-IHC-high. c HPV-OPC case with EGFR-IHC-weak. d HPV-OPC case with EGFR-IHC-negative. (a–d green probe: centromere, orange probe: EGFR DNA)
Extraction of HPV16-OPC by qPCR of E6 and E7
Using qPCR of viral genome E6 and E7 of HPV16, 71.4% (35/49) of our HPV-OPC cases were determined to be HPV16-OPC.
Clinicopathological Factors Relating to EGFR Expression in HPV-OPC
EGFR-IHC showed consistently high expression in nHPV-OPC cases, and diverse expression in HPV-OPC cases (65.4%: weak and 13.5%: negative); clinicopathological data and EGFR-related molecular data were compared among EGFR-IHC-high, -weak, and -negative groups of HPV-OPC to identify factors related to EGFR expression. The EGFR-IHC-negative group was younger with a mean age of 60.9 years (p = 0.04) compared to the -high group (mean age = 71.8 years) and the -weak group (mean age = 67.6 years). None of EGFR-IHC-negative group showed KSCC as tumor histology, while 54.5% of the -high group and 52.9% of the -weak group showed KSCC as tumor histology (p = 0.035). No significant differences were observed in sex distribution, smoking/drinking history, tumor size, tumor site, TNM factor, stage, and patient prognosis (PFS and OS) between the groups (Fig. 5).
Fig. 5.
Relationship between EGFR-immunohistochemistry (IHC) and patient prognosis. a Overall survival rates according to immunohistochemical results for EGFR. b progression free survival rates according to immunohistochemical results for EGFR. There were no statistically significant relationships between EGFR expression (high, weak, and negative) and overall survival/progression free survival
Molecular Factors Relating to EGFR Expression in HPV-OPC
Methylation of the EGFR promoter region was more frequent in the EGFR-IHC-negative group (83.3%) than in the -high (18.2%) and -weak (17.6%) groups (p = 0.009). (Table 2).
Table 2.
Clinicopathological features of EGFR-high, weak, and negative cases in HPV-related oropharyngeal carcinoma
| HPV-related oropharyngeal carcinoma (n = 52) | P value§ | P value | |||
|---|---|---|---|---|---|
| EGFR-high (n = 11) | EGFR-weak (n = 34) | EGFR-negative (n = 7) | |||
| Age | 0.04† | ||||
| Mean | 71.8 | 67.6 | 60.86 | ||
| Range | (62–86) | (44–62) | (50–79) | ||
| Sex—no. (%) | NS‡ | ||||
| Male | 10 (90.9) | 28 (82.4) | 5 (71.4) | ||
| Female | 1 (9.1) | 6 (17.6) | 2 (28.6) | ||
| Smoking history—no. (%) | NS‡ | ||||
| Never or former smoker | 10 (90.9) | 23 (67.6) | 5 (71.4) | ||
| Current smoker | 1 (9.1) | 6 (17.6) | 0 | ||
| Uncertain | 0 | 5 (14.7)) | 2 (28.6) | ||
| Drinking history—no. (%) | NS‡ | ||||
| Positive | 7 (63.6) | 23 (67.6) | 3 (42.9) | ||
| Negative | 2 (18.2) | 6 (17.6) | 3 (42.9) | ||
| Uncertain | 2 (18.2) | 5 (14.7) | 1 (14.3) | ||
| Tumor size | – | NS† | |||
| Mean (mm) | 28.6 | 28.0 | 24.0 | ||
| Range | (22–45) | (6–50) | (10–40) | ||
| Tumor site—no. (%) | NS‡ | ||||
| Lateral wall | 7 (63.6) | 27 (79.4) | 5 (71.4) | ||
| Upper wall | 0 | 0 | 1 (14.3) | ||
| Anterior wall | 3 (27.3) | 3 (8.8)) | 0 | ||
| Posterior wall | 0 | 0 | 0 | ||
| Uncertain | 1 (9.1) | 5 (14.7) | 1 (14.3) | ||
| T factor—no. (%) | NS‡ | ||||
| T1-2 | 8 (72.7) | 26 (76.5) | 6 (85.7) | ||
| T3-4 | 3 (27.3) | 8 (23.5) | 1 (14.3) | ||
| N factor—no. (%) | NS‡ | ||||
| N0 | 6 (54.5) | 7 (20.6) | 2 (28.6) | ||
| N1-3 | 5 (45.5) | 27 (79.4)) | 5 (71.4) | ||
| M factor—no. (%) | NS‡ | ||||
| M0 | 11 (100.0) | 34 (100.0) | 7 (100) | ||
| M1 | 0 | 0 | 0 | ||
| Stage—no. (%) | NS‡ | ||||
| I–II | 10 (90.9) | 32 (94.1) | 6 (85.7) | ||
| III–IV | 1 (9.1) | 2 (5.9) | 1 (14.3) | ||
| Prognosis | |||||
| Overall survival (%) | 100 | 94.1 | 100 | NS|| | |
| Progression free survival (%) | 72.7 | 73.5 | 71.4 | NS|| | |
| Histology | NS‡ | 0.035a | |||
| Keratinizing SCC | 6 (54.5) | 18 (52.9) | 0 |

|
|
| Hybrid SCC | 1 (9.1) | 9 (26.5) | 1 (14.3) | ||
| Non-keratinizing SCC | 4 (36.4) | 5 (14.7) | 5 (71.4) | ||
| Basaloid SCC | 0 | 1 (2.9) | 1 (14.3) | ||
| Lymphoepithelial carcinoma | 0 | 1 (2.9) | 0 | ||
| CK14 protein expression | 0.037‡ | ||||
| Positive | 6 (54.5) | 23 (67.6) | 1 (14.3) | ||
| Negative | 5 (45.5) | 11 (32.4) | 6 (85.7) | ||
| EGFR missense mutation—no. (%) | NS‡ | ||||
| Present | 1 (9.1) | 1 (2.9) | 1 (14.3) | ||
| Absent | 10 (90.9) | 33 (97.1) | 6 (85.7) | ||
| Q787Q synonymous mutation—no.(%) | NS‡ | ||||
| Present | 0 | 7 (20.6) | 2 (28.6) | ||
| Absent | 11 (100.0) | 27 (79.4) | 5 (71.4) | ||
| EGFR amplification—no.(%)¶ | NS‡ | ||||
| Present | 0 | 0 | 0 | ||
| Absent | 2 (100.0) | 5 (100.0) | 4 (100) | ||
| EGFR promoter methylation—no.(%)¶ | 0.009‡ | ||||
| Present | 2 (18.2) | 6 (17.6) | 5 (83.3) | ||
| Absent | 9 (81.8) | 28 (82.4) | 1 (14.3) | ||
| Associated HPV type—no.(%)¶ | NS‡ | 0.049b | |||
| Type16 | 4 (40.0) | 27 (79.4) | 3 (60.0) | ||
| Other types | 6 (60.0) | 7 (20.6) | 2 (40.0) | ||
¶Data of EGFR amplification are of 2 EGFR-high, 5 EGFR-weak, and 4 EGFR-negative cases. Data of EGFR promoter methylation are of 11 EGFR-high, 34 EGFR-weak, and 6 EGFR-negative cases. Data of HPV type are of 10 EGFR-high, 34 EGFR-weak, and 5 EGFR-negative cases
§NS not significant and the superscript letter
†Kruskal–Wallis test
‡Chi-square test
||Log-rank test, respectively
aP value between EGFR expression and histology by dividing histology into keratinizing SCC and other SCCs
bP value between EGFR expression and associated HPV types by dividing EGFR expression into -high/weak and -negative
EGFR missense mutation were identified in 4.4%, 2.9% and 14.3% of EGFR-high, -weak and -negative groups, respectively (p = 0.580), and the synonymous mutation Q787Q was identified in 0, 20.6 and 28.6% of EGFR-high, -weak, and -negative groups (p = 0.109). Using FISH, EGFR amplification or loss was not identified in any HPV-OPC case, examined. The EGFR/centromere ratio of each case was shown in supplementary table 8. Hence, no evident relationship between EGFR protein expression and gene mutation, including synonymous mutation and amplification/loss, was detected.
HPV16-OPC, determined by its E6/E7 qPCR, was observed in 40.0% (4/10) of EGFR-IHC-high cases and 76.9% (30/39) of EGFR-IHC-weak/negative cases, and its frequency was significantly different considering the EGFR expression (p = 0.049) (Fig. 6).
Fig. 6.
Relationship between HPV type and EGFR-immunohistochemistry (IHC). Among EGFR-IHC-high group, 40% (4/10) and 60% (6/10) were HPV type 16-related and other HPV types-related, respectively. Among EGFR-IHC-weak/negative group, 77% (30/39) and 23% (9/30) were HPV type 16-related and other HPV types-related, respectively. HPV type 16-related OPC is more frequent in EGFR-IHC-weak/negative group than in EGFR-IHC-high group (P = 0.049)
.
Relationship of Viral Genome E5, E6, E7 with EGFR Expression in HPV16-OPC
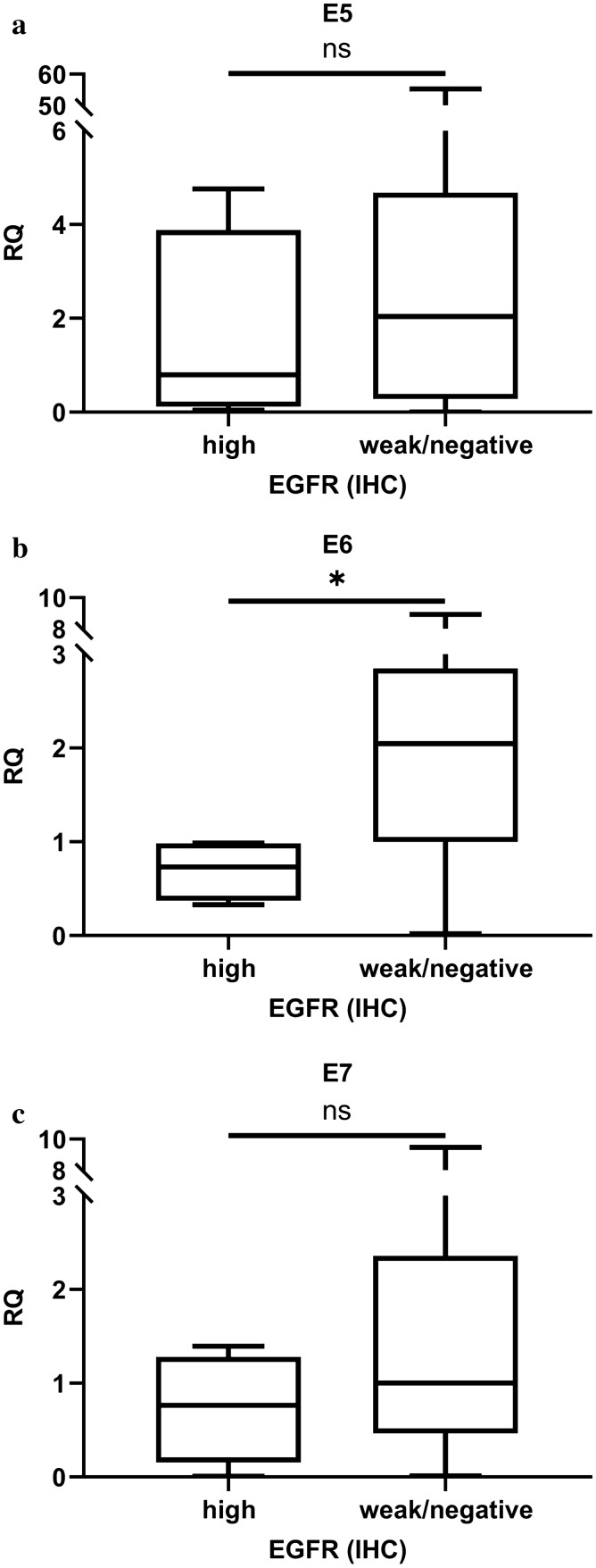
Among the 34 HPV16-OPC cases, the RQ values of E5, E6, and E7 were compared with EGFR-high and EGFR-weak/negative groups. All E5, E6, and E7 were higher in the EGFR-weak/negative group than in the EGFR-high group, while statistical significance was seen only for E6 (p = 0.519, 0.019, 0.262 for E5, E6 and E7, respectively) (Fig. 7).
Fig. 7.
Relationship of viral genome load of HPV-E5, E6, and E7and EGFR-immunohistochemistry (IHC). a No relationship was seen between the amount of HPV16-E5 mRNA and EGFR-IHC (p = 0.519). b The amount of HPV16-E6 mRNA was more in EGFR-IHC-weak/negative cases than in EGFR-IHC-high cases (p = 0.019). c No evident relationship was seen between the amount of HPV-E7 mRNA and EGFR-IHC. (p = 0.262)
In the F-test, the RQ value of E5 was more diverse compared to the RQs of E6 and E7 (P < 0.01 for E5 vs. E6, P < 0.01 for E5 vs. E7, and P > 0.05 for E6 vs. E7).
Discussion
There are five findings, in short, in the present study; (1) HPV-OPC cases, but not nHPV-OPC, often weakly or negatively express EGFR protein. (2) The loss and weakened expression of EGFR-protein relates to promoter methylation of EGFR, (3) to the infection to HPV type 16, and (4) to the high E6 expression among HPV16-OPC. (5) The loss of EGFR protein relates to non-KSCC histology.
Our IHC study showed that all nHPV-OPC cases exhibited high EGFR expression, whereas only 21.1% of HPV-OPC showed high EGFR expression. These IHC results were validated by qPCR; hence, the negative or weak expression of this protein, observed in some HPV-OPC cases, represents a lower amount of EGFR mRNA in these tumors.
HNSCC, in general, is reported to overexpress EGFR protein, and the EGFR pathway play a prominent role in tumorigenesis/tumor progression of HNSCC; thus, it is noteworthy that EGFR protein is often downregulated in HPV-OPC, although its pathway status in HPV-OPC was not assessed in this study. The EGFR pathway status depends not only on EGFR protein status but also on its ligand status and the feedback from downward factors. A few authors have also reported low EGFR expression in HPV-OPC. Lassen et al. showed 32.5% of HPV- OPC exhibited low expression of this protein [14], and Kumar et al. showed an inverse relationship between HPV titer and EGFR protein expression [15]. However, the exact mechanism for EGFR downregulation in HPV-OPC is not well-known.
In our analysis, the lack or weakened expression of EGFR protein was related to the methylation of the EGFR promoter region and viral type, that is more frequent for HPV16-OPC than HPVnon16-OPC. To our knowledge, no study has investigated EGFR methylation in OPC, and this is the first report to show the relationship between EGFR methylation and loss of protein expression. The relationship between methylation and carcinogenesis attributable to infections has often been reported, such as Helicobacter pylori-related gastric cancer, hepatitis B/C-related hepatocellular carcinoma, and HPV-related cervical cancer [26]. The relationship between the methylation of the EGFR promoter region and the loss of its protein expression and decrease in mRNA has been reported in colon cancer [27]. The relationship of EGFR promoter methylation investigated with the methylation specific PCR method and lower efficacy for EGFR-target therapy has also been reported in colorectal carcinoma [28].
Among HPV types known to be related to cancers, HPV16 accounts for most HPV-OPCs, followed by types 18 and 33 [29]. In the present study, 69% of HPV-OPC cases were HPV16-related and 31% were HPVnon16-related. Our study showed that HPV16-OPC tended to show weak or negative EGFR expression compared to that shown by HPVnon16-OPC. Thus far, many studies have been performed on the effect of HPV integration but have been mostly limited to HPV16; hence, biological differences between HPV16 and other HPV types are not well understood. To our knowledge, a few studies show the difference between the behavior of HPV16 and HPV18 as well as between HPV16 E6 and HPV18 E6, the former shows the difference in physical status (integrated form vs. episomal form) of HPV16 and HPV18, and the latter shows downregulating function of NHERF1 of HPV16-E6, leading to the cytoskeleton but not of HPV18-E6 [30, 31]. The difference in EGFR expression between HPV16- and HPVnon16-OPC seen in the present study may represent another difference between HPV16 and other types of biological activity, although it is beyond the scope of our study to see the underlying mechanism of the HPV genome in detail.
The present study investigated the relationship between EGFR protein expression and the viral load of HPV16-E5. Previous studies with cell lines have shown the relationship between HPV E5-encoded transcripts and EGFR expression [8, 32], suggesting that a high level of E5 results in the upregulation of EGFR by recycling EGFR to the cell surface by inhibiting endosomal acidification and other mechanisms. To our knowledge, there is one study that investigated its relation with human HPV-OPC tissues, showing only a slight relationship between E5 and EGFR level [25]. In the present study, the mean E5 level in the EGFR-IHC-high group was lower than that in the EGFR-weak/negative group, but the difference was not statistically significant. On the other hand, a high E6 level was related to the weak/negative expression of EGFR with statistical significance. High E7 levels also tended to relate to the weak/negative expression of EGFR, although the difference was not statistically significant. We consider that the relationship between E6 and EGFR expression seen this study may reflect the relationship between the viral amount of HPV16 and EGFR expression because both E6 and E7 showed similar relationships to EGFR expression and the load of HPV16-E6 and -E7 are reported to reflect the amount of integrated HPV genome in cervical cancers and HNSCC [33–35], However, it is also possible that weak or negative EGFR expression is derived from modified p53 or RB signal pathway since E6 is well-known to inhibit p53 pathway and E7 is known to inactivate RB protein [8, 36].
Other than HPV status and E5 expression, EGFR copy number and mutation have been investigated by a few authors for the relationship to EGFR expression. Recent comprehensive studies [37] have shown that the frequency of EGFR amplification is 15% for nHPV-OPC and 6% for HPV-OPC. However, the relationship between EGFR amplification and EGFR protein expression is controversial; several studies failed to demonstrate any correlation between them [38]. The present study investigated 11 and 6 HPV- and nHPV-OPC cases, all of which showed no amplification or deletion. Although our sample size for EGFR-FISH was small, we included IHC-high (n = 2), -weak (n = 5), -negative (n = 4) cases to analyze the contribution of gene amplification/deletion and EGFR IHC data, showing that there is no evidence of the contribution of gene amplification/deletion.
In the present study, missense mutations of EGFR were analyzed for exons 18–21, where the tyrosine kinase domain is located and the two most frequent mutations L858R and exon 19 in flame deletion are covered. Our cohort showed that EGFR missense mutations were present in 5.8% and 0% of HPV-OPC and nHPV-OPC, respectively. In HPV-OPC, 4.4% of EGFR-IHC-high/weak and 14.3% of EGFR-IHC-negative cases showed missense mutations for this gene, suggesting that an EGFR missense mutation at exons 18–21 is a rare event both for HPV- and nHPV-OPC, and no association with EGFR protein expression was seen. To our knowledge, there have been a few studies which investigated EGFR mutation in HNSCC that show the frequency of EGFR mutation to be 7.3% and 0% for HPV- and nHPV-OPC [39, 40], respectively, which is similar to our study.
Our mutational study of EGFR also detected that 17.3% and 31.8% of HPV- and nHPV-OPC cases, respectively, showed synonymous mutations, Q787Q. In HPV-OPC, 15.6% and 28.6% of EGFR-IHC-high/weak and -negative cases, respectively, showed this synonymous mutation, suggesting that the silence mutation is a relatively frequent event for OPC irrespective of HPV status, although this event did not show a relationship with EGFR protein expression. We examined the relationship between Q787Q mutation and EGFR expression since this type of silence mutation is reportedly related to the efficacy to tyrosine kinase inhibitors in several types of cancers, such as cervix, colon, lung, and HNSCC [41–43]. There is limited data, but in HNSCC, the enhanced efficacy of tyrosine kinase inhibitors caused by the Q787Q mutation is suggested to be unrelated to EGFR mRNA or protein expression [41, 44].
In the present study, we detected a strong relationship between tumor histology and EGFR expression in HPV-OPC. Regarding tumor histology of HPV-OPC, many authors have reported specific features such as frequent non-keratinizing SCC histology, peculiar nuclear features called basaloid, syncytial, vesicular, or undifferentiated [19], and abrupt keratinization, where keratinization or squamous maturation foci appears abruptly in non-keratinizing SCC [4, 45]. Although some authors speculated that the interaction of HPV oncoproteins with cell cycle mediators might result in the non-keratinizing/non-maturing morphology of HPV-OPC [4, 46], no published data were available on what kind of molecular mechanisms bring such specific histology to HPV-OPC. The strong relationship between tumor histology and EGFR expression observed in this study may seem reasonable when we consider that EGFR pathway activation is known to progress cell differentiation, as well as cell proliferation and migration.
Recently, many authors have investigated the prognostic impact of EGFR protein expression on HNSCC [1, 47], showing rather inconsistent results; For example, Sivarajah et al. showed that a high EGFR level of the tumor is related to poor prognosis, whereas Lassen et al. did not detect such a relationship [14, 18]. The present study did not show a relationship between EGFR expression and PFS or OS. Since there were no cases with previous neoadjuvant therapy, we could not see the relationship between EGFR expression and therapy effect.
In conclusion, the present study showed that weakened or loss of EGFR protein expression was frequent in HPV-OPC compared to nHPV-OPC, and it was related with EGFR methylation, infection of HPV16, viral genome load of HPV-16 E6, and non-keratinizing SCC histology. Gene amplification/loss, missense mutation, and nonsynonymous mutation were not related to EGFR protein expression. Further studies are necessary to understand how these molecular mechanisms modify EGFR expression.
Supplementary information
Below is the link to the electronic supplementary material.
Acknowledgements
We would like to thank Editage (www.editage.com) for English language editing.
Funding
This study was supported by a Grant-in-Aid from the Japan Society for the Promotion of Sciences (JSPS) KAKENHI (Grant # 17K16947 to M.F. and # 16K20279 to S.O.).
Compliance with Ethical Standards
Conflict of interest
The authors have no conflicts of interest to declare.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bossi P, Resteghini C, Paielli N, Licitra L, Pilotti S, Perrone F. Prognostic and predictive value of EGFR in head and neck squamous cell carcinoma. Oncotarget. 2016;7:74362–79. doi: 10.18632/oncotarget.11413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tan PF, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363:24–35. doi: 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen AM, Li J, Beckett LA, Zhara T, Farwell G, Lau DH, et al. Differential response rates to irradiation among patients with human papillomavirus positive and negative oropharyngeal cancer. Laryngoscope. 2013;123:152–7. doi: 10.1002/lary.23570. [DOI] [PubMed] [Google Scholar]
- 4.Chernock RD. Morphologic features of conventional squamous cell carcinoma of the oropharynx: ‘keratinizing’ and ‘nonkeratinizing’ histologic types as the basis for a consistent classification system. Head Neck Pathol. 2012;6(Suppl 1):41-7. doi: 10.1007/s12105-012-0373-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–37. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 6.Moody CA, Laimins LA. Human papillomavirus oncoproteins: pathways to transformation. Nat Rev Cancer. 2010;10:550–60. doi: 10.1038/nrc2886. [DOI] [PubMed] [Google Scholar]
- 7.Paget-Bailly P, Meznad K, Bruyere D, Perrard J, Herfs M, Jung AC, et al. Comparative RNA sequencing reveals that HPV16 E6 abrogates the effect of E6*I on ROS metabolism. Sci Rep. 2019;9:5938. doi: 10.1038/s41598-019-42393-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Estevao D, Costa NR, Gil da Costa RM, Medeiros R. Hallmarks of HPV carcinogenesis: the role of E6, E7 and E5 oncoproteins in cellular malignancy. Biochim Biophys Acta Gene Regul Mech. 2019;1862:153–62. doi: 10.1016/j.bbagrm.2019.01.001. [DOI] [PubMed] [Google Scholar]
- 9.Bishop JA, Ma XJ, Wang H, Luo Y, Illei PB, Begum S, et al. Detection of transcriptionally active high-risk HPV in patients with head and neck squamous cell carcinoma as visualized by a novel E6/E7 mRNA in situ hybridization method. Am J Surg Pathol. 2012;36:1874–82. doi: 10.1097/PAS.0b013e318265fb2b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smeets SJ, Hesselink AT, Speel EJ, Haesevoets A, Snijders PJ, Pawlita M, et al. A novel algorithm for reliable detection of human papillomavirus in paraffin embedded head and neck cancer specimen. Int J Cancer. 2007;121:2465–72. doi: 10.1002/ijc.22980. [DOI] [PubMed] [Google Scholar]
- 11.Shi W, Kato H, Perez-Ordonez B, Pintilie M, Huang S, Hui A, et al. Comparative prognostic value of HPV16 E6 mRNA compared with in situ hybridization for human oropharyngeal squamous carcinoma. J Clin Oncol. 2009;27:6213–21. doi: 10.1200/JCO.2009.23.1670. [DOI] [PubMed] [Google Scholar]
- 12.Mullick Chowdhury S, Manepalli P, Sitharaman B. Graphene nanoribbons elicit cell specific uptake and delivery via activation of epidermal growth factor receptor enhanced by human papillomavirus E5 protein. Acta Biomater. 2014;10:4494–504. doi: 10.1016/j.actbio.2014.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DiMaio D, Mattoon D. Mechanisms of cell transformation by papillomavirus E5 proteins. Oncogene. 2001;20:7866–73. doi: 10.1038/sj.onc.1204915. [DOI] [PubMed] [Google Scholar]
- 14.Lassen P, Overgaard J, Eriksen JG. Expression of EGFR and HPV-associated p16 in oropharyngeal carcinoma: correlation and influence on prognosis after radiotherapy in the randomized DAHANCA 5 and 7 trials. Radiother Oncol. 2013;108:489–94. doi: 10.1016/j.radonc.2013.08.036. [DOI] [PubMed] [Google Scholar]
- 15.Kumar B, Cordell KG, Lee JS, Worden FP, Prince ME, Tran HH, Titer HPV, et al. Bcl-xL and p53, sex, and smoking as indicators of response to therapy and survival in oropharyngeal cancer. J Clin Oncol. 2008;EGFR:3128–37. doi: 10.1200/JCO.2007.12.7662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindquist D, Ahrlund-Richter A, Tarjan M, Tot T, Dalianis T. Intense CD44 expression is a negative prognostic factor in tonsillar and base of tongue cancer. Anticancer Res. 2012;32:153–61. [PubMed] [Google Scholar]
- 17.Vainshtein JM, Spector ME, McHugh JB, Wong KK, Walline HM, Byrd SA, et al. Refining risk stratification for locoregional failure after chemoradiotherapy in human papillomavirus-associated oropharyngeal cancer. Oral Oncol. 2014;50:513–9. doi: 10.1016/j.oraloncology.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sivarajah S, Kostiuk M, Lindsay C, Puttagunta L, O’Connell DA, Harris J, et al. EGFR as a biomarker of smoking status and survival in oropharyngeal squamous cell carcinoma. J Otolaryngol Head Neck Surg. 2019;48:1. doi: 10.1186/s40463-018-0323-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singhi AD, Westra WH. Comparison of human papillomavirus in situ hybridization and p16 immunohistochemistry in the detection of human papillomavirus-associated head and neck cancer based on a prospective clinical experience. Cancer. 2010;116:2166–73. doi: 10.1002/cncr.25033. [DOI] [PubMed] [Google Scholar]
- 20.Brierley JD, Wittekind GMC, editors. Union for International Cancer Control (UICC): TNM classification of malignant tumours. 8. Hoboken: Wiley-Blackwell; 2017. pp. 23–24. [Google Scholar]
- 21.Lewis JS. Jr. Morphologic diversity in human papillomavirus-related oropharyngeal squamous cell carcinoma. Catch Me If You Can! Mod Pathol. 2017;30:44–53. doi: 10.1038/modpathol.2016.152. [DOI] [PubMed] [Google Scholar]
- 22.El-Naggar AKJC, Grandis JR, Takata T, Grandis J, Slootweg P, editors. WHO classification of head and neck tumours. 4. IARC: Lyon; 2017. [Google Scholar]
- 23.Reis-Filho JS, Milanezi F, Carvalho S, Simpson PT, Steele D, Savage K, et al. Metaplastic breast carcinomas exhibit EGFR, but not HER2, gene amplification and overexpression: immunohistochemical and chromogenic in situ hybridization analysis. Breast Cancer Res. 2005;7:R1028-35. doi: 10.1186/bcr1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang-Johanning F, Lu DW, Wang Y, Johnson MR, Johanning GL. Quantitation of human papillomavirus 16 E6 and E7 DNA and RNA in residual material from ThinPrep Papanicolaou tests using real-time polymerase chain reaction analysis. Cancer. 2002;94:2199–210. doi: 10.1002/cncr.10439. [DOI] [PubMed] [Google Scholar]
- 25.Um SH, Mundi N, Yoo J, Palma DA, Fung K, MacNeil D, et al. Variable expression of the forgotten oncogene E5 in HPV-positive oropharyngeal cancer. J Clin Virol. 2014;61:94–100. doi: 10.1016/j.jcv.2014.06.019. [DOI] [PubMed] [Google Scholar]
- 26.Hattori N, Ushijima T. Epigenetic impact of infection on carcinogenesis: mechanisms and applications. Genome Med. 2016;8:10. doi: 10.1186/s13073-016-0267-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geissler AL, Geissler M, Kottmann D, Lutz L, Fichter CD, Fritsch R, et al. ATM mutations and E-cadherin expression define sensitivity to EGFR-targeted therapy in colorectal cancer. Oncotarget. 2017;8:17164–90. doi: 10.18632/oncotarget.15211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scartozzi M, Bearzi I, Mandolesi A, Giampieri R, Faloppi L, Galizia E, et al. Epidermal growth factor receptor (EGFR) gene promoter methylation and cetuximab treatment in colorectal cancer patients. Br J Cancer. 2011;104:1786–90. doi: 10.1038/bjc.2011.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ndiaye C, Mena M, Alemany L, Arbyn M, Castellsague X, Laporte L, et al. HPV DNA, E6/E7 mRNA, and p16INK4a detection in head and neck cancers: a systematic review and meta-analysis. Lancet Oncol. 2014;15:1319–31. doi: 10.1016/S1470-2045(14)70471-1. [DOI] [PubMed] [Google Scholar]
- 30.Wang Q, Song R, Zhao C, Liu H, Yang Y, Gu S, et al. HPV16 E6 promotes cervical cancer cell migration and invasion by downregulation of NHERF1. Int J Cancer. 2019;144:1619–32. doi: 10.1002/ijc.31876. [DOI] [PubMed] [Google Scholar]
- 31.Badaracco G, Venuti A, Sedati A, Marcante ML. HPV16 and HPV18 in genital tumors: Significantly different levels of viral integration and correlation to tumor invasiveness. J Med Virol. 2002;67:574–82. doi: 10.1002/jmv.10141. [DOI] [PubMed] [Google Scholar]
- 32.Straight SW, Herman B, McCance DJ. The E5 oncoprotein of human papillomavirus type 16 inhibits the acidification of endosomes in human keratinocytes. J Virol. 1995;69:3185–92. doi: 10.1128/jvi.69.5.3185-3192.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim J, Kim BK, Jeon D, Lee CH, Roh JW, Kim JY, et al. Type-Specific Viral Load and Physical State of HPV Type 16, 18, and 58 as Diagnostic Biomarkers for High-Grade Squamous Intraepithelial Lesions or Cervical Cancer. Cancer Res Treat. 2020;52:396–405. doi: 10.4143/crt.2019.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hong D, Liu J, Hu Y, Lu XN, Li BH, Li Y, et al. Viral E6 is overexpressed via high viral load in invasive cervical cancer with episomal HPV16. Bmc Cancer. 2017;17:1–8. doi: 10.1186/s12885-017-3124-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chai RC, Lambie D, Verma M, Punyadeera C. Current trends in the etiology and diagnosis of HPV-related head and neck cancers. Cancer Med. 2015;4:596–607. doi: 10.1002/cam4.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yim EK, Park JS. The role of HPV E6 and E7 oncoproteins in HPV-associated cervical carcinogenesis. Cancer Res Treat. 2005;37:319–24. doi: 10.4143/crt.2005.37.6.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cancer Genome Atlas N Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–82. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Temam S, Kawaguchi H, El-Naggar AK, Jelinek J, Tang H, Liu DD, et al. Epidermal growth factor receptor copy number alterations correlate with poor clinical outcome in patients with head and neck squamous cancer. J Clin Oncol. 2007;25:2164–70. doi: 10.1200/JCO.2006.06.6605. [DOI] [PubMed] [Google Scholar]
- 39.Lee JW, Soung YH, Kim SY, Nam HK, Park WS, Nam SW, et al. Somatic mutations of EGFR gene in squamous cell carcinoma of the head and neck. Clin Cancer Res. 2005;11:2879–82. doi: 10.1158/1078-0432.CCR-04-2029. [DOI] [PubMed] [Google Scholar]
- 40.Chung CH, Ely K, McGavran L, Varella-Garcia M, Parker J, Parker N, et al. Increased epidermal growth factor receptor gene copy number is associated with poor prognosis in head and neck squamous cell carcinomas. J Clin Oncol. 2006;24:4170–6. doi: 10.1200/JCO.2006.07.2587. [DOI] [PubMed] [Google Scholar]
- 41.Tan DSW, Chong FT, Leong HS, Toh SY, Lau DP, Kwang XL, et al. Long noncoding RNA EGFR-AS1 mediates epidermal growth factor receptor addiction and modulates treatment response in squamous cell carcinoma. Nat Med. 2017;23:1167–75. doi: 10.1038/nm.4401. [DOI] [PubMed] [Google Scholar]
- 42.Qureshi R, Arora H, Biswas S, Perwez A, Naseem A, Wajid S, et al. Mutation analysis of EGFR and its correlation with the HPV in Indian cervical cancer patients. Tumour Biol. 2016;37:9089–98. doi: 10.1007/s13277-016-4789-4. [DOI] [PubMed] [Google Scholar]
- 43.Nagalakshmi, Jamil K, Rani P. Association of EGFR gene polymorphism in head and neck cancer patients with tobacco and alcohol consuming habits. Biol Med. 2013;5:69–77. [Google Scholar]
- 44.Taguchi T, Tsukuda M, Imagawa-Ishiguro Y, Kato Y, Sano D. Involvement of EGFR in the response of squamous cell carcinoma of the head and neck cell lines to gefitinib. Oncol Rep. 2008;19:65–71. [PubMed] [Google Scholar]
- 45.Fujimaki M, Fukumura Y, Mitani K, Kurisaki A, Yokoyama J, Ikeda K, et al. Histological subtypes and characteristic structures of HPV-associated oropharyngeal carcinoma: study with Japanese cases. Diagn Pathol. 2013;8:211. doi: 10.1186/1746-1596-8-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.El-Mofty SK. Histopathologic risk factors in oral and oropharyngeal squamous cell carcinoma variants: an update with special reference to HPV-related carcinomas. Med Oral Patol Oral Cir Bucal. 2014;19:e377-85. doi: 10.4317/medoral.20184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ang KK, Berkey BA, Tu X, Zhang HZ, Katz R, Hammond EH, et al. Impact of epidermal growth factor receptor expression on survival and pattern of relapse in patients with advanced head and neck carcinoma. Cancer Res. 2002;62:7350–6. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.