Summary
This protocol describes a method to assess adipocyte numbers within a specific depot based on their inducible genomic label. By extracting DNA from a complete adipose tissue depot stemming from two transgenic mouse lines (Adipoq-CreERT2 x ROSA26-tdRFP and Ucp1-CreERT2 x ROSA26-tdRFP), the number of adipocytes can be determined based on the quantification of the recombined LoxPRed sites. This highly sensitive system allows for the quantification of white, brown, and brite/beige adipocytes in a spatially unbiased and size-independent manner.
For complete details on the use and execution of this protocol, please refer to Moser et al. (2021).
Subject areas: Cell culture, Cell isolation, Cell-based Assays, Cell separation/fractionation, Metabolism, Molecular Biology
Graphical abstract

Highlights
-
•
Employ transgenic mouse lines to label specific genomic sites in adipocytes
-
•
Isolate genomic DNA from a complete adipose tissue depot
-
•
Quantify adipocytes number, based on the genomic labels
-
•
The principle of the protocol can be adapted to other transgenic mouse lines and cells
This protocol describes a method to assess adipocyte numbers within a specific depot based on their inducible genomic label. By extracting DNA from a complete adipose tissue depot stemming from two transgenic mouse lines (Adipoq-CreERT2 x ROSA26-tdRFP and Ucp1-CreERT2 x ROSA26-tdRFP), the number of adipocytes can be determined based on the quantification of the recombined LoxPRed sites. This highly sensitive system allows for the quantification of white, brown, and brite/beige adipocytes in a spatially unbiased and size-independent manner.
Before you begin
This protocol was established to enable the quantification of adipocytes. With the Adipoq-tracer mouse line (Adipoq-CreERT2 x ROSA26-tdRFP), all adipocytes can be inducible labeled and quantified, while the Ucp1-tracer mouse line (Ucp1-CreERT2 x ROSA26-tdRFP) exclusively labels all brown and brite/beige adipocytes. By qPCR the Cre-driven LoxPRed recombination sites are specifically quantified and compared to the genomic DNA copy number (ApoB locus) [Figure 1] and thus the number of adipocytes and the total cell number are determined. In comparison to other methods, like histology or flow cytometry, this highly sensitive method allows for the quantification of adipocytes unbiased regarding their size or spatial location. Between 6 – 10 mice per condition are suggested to obtain reproducible results. Currently, this protocol is based on the quantification of the recombination sites obtained by the Ucp1-creERT2 or Adipoq-creERT2 driven ROSA26–tdRFP recombination (Luche et al., 2007). This method can be also be adapted to other Cre-driver mouse lines crossed to the ROSA26-tdRFP construct (Luche et al., 2007), hence enabling the quantification of other cell types than adipocytes. Further, by designing primers binding to another recombination site and generating a plasmid harboring the new recombination site of interest, the principle of the method can be also applied to other already existing transgenic mouse lines.
Figure 1.
Transgenic mouse lines
The transgenic Adipoq-tracer mouse line (Adipoq-CreERT2 × ROSA26-tdRFP) allows white, brown, and brite/beige adipocytes to be labeled by RFP, while the Ucp1-tracer mouse line (Ucp1-CreERT2 × ROSA26-tdRFP) exclusively labels brown and brite/beige adipocytes. The tamoxifen-inducible recombination site of the LoxPRed is used for the quantification of adipocytes.
The inducible CreERT2-LoxP system is a very useful tool for the temporal-controlled and tissue-specific recombination of DNA. However, at higher doses of tamoxifen, toxic effects were reported (Ye et al., 2015). With the very low dose of 2× 2 mg, we use 30%–80% less tamoxifen compared to others (Hesselbarth et al., 2015; Lee et al., 2015; Ye et al., 2015). However, we only established the minimal effective dose for the adipose tissue. If analyses of other tissues/cell types are planned, the ideal dose of tamoxifen has yet to be determined.
Induction of recombination
Timing: 0–1 h on 2 consecutive days
-
1.Oral gavage of 2 mg tamoxifen per animal per day.
-
a.100 μL of the tamoxifen solution (as described in the materials and equipment section) are orally gavaged with a gavage needle or a plastic feeding tube.
-
b.Repeat the step described in the section above the next day.
-
a.
-
2.
Full recombination can be observed latest two days after the last tamoxifen treatment.
Note: One dose of 2 mg tamoxifen was described to sufficiently induce full recombination in the main adipose tissue depots (Moser et al., 2021). Two applications on two consecutive days were chosen as a precaution in case technical issues with the oral gavage happen to occur during the first treatment with tamoxifen. The minimal dose required to sufficiently label all cells of interest has yet to be determined for other cell types/tissues.
Note: In this protocol tamoxifen ordered from Sigma Aldrich was used, however is can be also ordered from other companies (eg. VWR).
Tissue harvest
Timing: 0–30 min per mouse
-
3.
Euthanize the mouse using carbon dioxide.
-
4.Harvest the adipose tissue depots of interest. The fat depot nomenclature ascribed by de Jong et al (de Jong et al., 2015) was utilized. One lobe of each depot is to be used for the quantification of adipocytes.
-
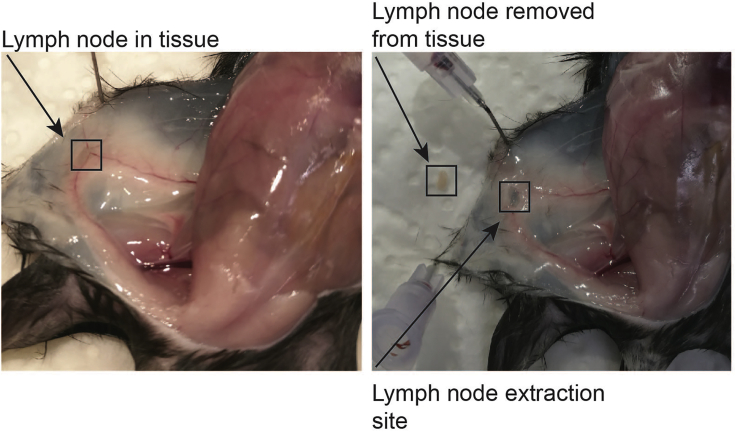
a.From the inguinal white adipose tissue depot, the popliteal lymph nodes should be carefully removed [Figure 2].
-
b.If two lobes per adipose tissue depot are available, the other lobe can be used for further analysis (e.g., histology or gene expression).
-
a.
-
5.
Place the adipose tissue samples in 2 mL Eppendorf tubes and snap-freeze them in liquid nitrogen.
-
6.
Store the samples at −20°C until further processing.
CRITICAL: It is critical to dissect the adipose tissue depots very precisely. Depots tightly connected to skeletal muscles must be removed especially carefully, as remnants of skeletal muscle can drastically influence total cell numbers [Figure 3].
Figure 2.
Lymph node in the inguinal white adipose tissue depot
As the lymph nodes contain a high number of immune cells that can heavily influence the number of total cells, said lymph nodes should be carefully removed from the samples during the dissection of the tissue.
Figure 3.
Skeletal muscle adhering to brown adipose tissue
Other tissues adhering to the tissue of interest can influence the number of total cells, thus they should be carefully removed from the samples during the dissection of the tissue.
Note: The tissue harvesting process can influence the results. When adhering to the classification of the depots by de Jong et al. (de Jong et al., 2015) there was no interpersonal variation observed (Moser et al., 2021). If this method is to be applied to other organs/cell types, similar schemes/guidelines for tissue harvesting are suggested.
Pause point: The samples can be stored at −20°C until further processing.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Tamoxifen | Sigma-Aldrich | Cat# T5648 |
| Fast SYBR™ Green Master Mix | Thermo Scientific | Cat# 4385610 |
| Experimental models: Organisms/strains | ||
| Ucp1-CreERT2 × ROSA26-tdRFP × Ucp1-DTR-GFP | Moser et al. | N/A |
| Adipoq-CreERT2 × ROSA26-tdRFP × Ucp1-DTR-GFP | Moser et al. | N/A |
| Oligonucleotides | ||
| qPCR primer LoxPRed and ApoB | Microsynth | Listed in materials and equipment |
| Plasmid pUC57recloxPRed-ApoB | Moser et al. | Listed in materials and equipment |
| Software and algorithms | ||
| QuantStudio Real-Time PCR software | Applied Biosystems | Version 1.3 |
| GraphPad Prism 9 | GraphPad Software | Version 9.0.1 |
| Other | ||
| Chow diet | Kliba-Nafag, Kaiseraugst | #2222 |
Materials and equipment
Tamoxifen Solution (20 mg/mL):
Please work in a biosafety cabinet while preparing the tamoxifen solution. Add 600 mg of Tamoxifen powder to a 50 mL falcon tube. Distribute the tamoxifen powder at the wall of the falcon tube. Carefully and slowly add 30 mL of sunflower oil to the tamoxifen powder. Wrap the falcon tube in aluminum foil, as tamoxifen is light-sensitive, and shake the solution for ca. 1 h until the tamoxifen powder is fully dissolved and no crystals are visible anymore. Dissolving can be eased by heating the solution to 37°C while shaking e.g., by putting it onto a shaker placed in an incubator Troubleshooting 1. When fully dissolved, prepare 1 mL aliquots and store them for up to 12 months at −20°C. Avoid repeated freeze/thaw cycles. When thawing the tamoxifen aliquots, make sure the tamoxifen is fully dissolved; no crystals should be visible.
50 mM NaOH
| Reagent | Final concentration | Amount |
|---|---|---|
| NaOH | 50 mM | 2 g |
| ddH2O | n/a | 1 L |
| Total | n/a | 1 L |
Store at 20°C–25°C with no time limit.
1 M Tris-HCl pH 8
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris | 1 M | 60.57 g |
| HCL | n/a | Until pH 8 is reached |
| ddH2O | n/a | 0.5 L |
| Total | n/a | 0.5 L |
Store at 20°C–25°C with no time limit.
Primer sequences
| Primer name | Primer sequence |
|---|---|
| Recombined LoxPRed forward | GCGCATGAACTCTTTGATGAC |
| Recombined LoxPRed reverse | TCGCGGTTGAGGACAAACTC |
| ApoB forward | GTCCAGGTTGAATCACGGGT |
| ApoB reverse | AGGATCCTGCAAGGTCAAGC |
Primer mix recombined LoxPRed:
| Primer name | Final concentration in mix | Amount |
|---|---|---|
| Recombined LoxPRed forward (100 μM Stock) | 2.5 μM | 10 μL |
| Recombined LoxPRed reverse (100 μM Stock) | 2.5 μM | 10 μL |
| ddH2O | n/a | 380 μL |
| Total | n/a | 400 μL |
Store at −20°C with no time limit. The final concentration of each primer in the qPCR reaction should be 250 nM.
Primer mix ApoB
| Primer name | Final concentration in mix | Amount |
|---|---|---|
| ApoB forward (100 μM Stock) | 2.5 μM | 10 μL |
| ApoB reverse (100 μM Stock) | 2.5 μM | 10 μL |
| ddH2O | n/a | 380 μL |
| Total | n/a | 400 μL |
Store at −20°C with no time limit. The final concentration of each primer in the qPCR reaction should be 250 nM.
Step-by-step method details
Preparation of genomic DNA
Timing: 2–3 h
Isolation of the genomic DNA from the adipose tissue depots
-
1.
Add two stainless steel metal beads per sample to the 2 mL Eppendorf tube.
-
2.
Add 1 mL of NaOH (50 mM) to the samples.
-
3.
Lyse the samples for 5–10 min with the TissueLyser LT (Qiagen). Carefully check that the tissue is fully lysed. If, not repeat the lysis for another 5 min.
-
4.
Shake the samples in a thermoshaker for 1 h at 1000 rpm and 92°C.
-
5.
Give the tubes a quick spin to remove any condensation from the lid. It is not necessary to let the tubes cool down to RT first.
-
6.
Add 250 μL of Tris-HCl (1 M, pH 8) to the samples to neutralize the pH.
-
7.Centrifuge the samples at 13000 rcf for 5 min at 4°C.
-
a.A fat cake will emerge on top of an aqueous phase and a pellet will be seen in the bottom of the tube [Figure 4].
-
b.The cooling of the centrifuge helps make the fat cake more stable and eases the separation of the fat from the aqueous phase.
-
a.
-
8.Carefully transfer 200 μL of the aqueous phase to a fresh tube.
-
a.If a lot of fat is sticking to the outside of the pipette tip, use a paper tissue to carefully remove the fat before transferring the sample to a fresh tube [Figure 5].
-
a.
-
9.
Centrifuge the samples again at 13000 rcf for 5 min at 4°C. Troubleshooting 2
-
10.
To obtain a 1:30 dilution of the genomic DNA, carefully transfer 2 μL of the aqueous phase from the samples to 58 μL of H2O.
CRITICAL: It is highly critical to remove all lipids from the sample before continuing with the next step.
Note: Other tissue lysing machines or methods e.g., the MagNA lyser (Roche), the BeadBug (Benchmark Scientific) or other hand held grinders can be used alternatively.
Pause point: The samples can be stored at −20°C until further processing.
Figure 4.
Three layers of the sample after centrifugation
On top of the sample, there will be a layer of fat followed by an aqueous phase and the beads and a pellet in the bottom of the tube. Carefully move through the layer of fat and transfer part of the aqueous phase to a fresh tube.
Figure 5.
Fat sticking to the outside of the pipet tip
If fat is sticking to the outside of the pipet tip, carefully remove the fat with a paper tissue before transferring the aqueous phase to a fresh tube.
Preparation of the standard curve
Timing: 0–1 h
To determine the exact number of recombined LoxPRed or ApoB molecules, a standard curve generated from a synthesized plasmid pUC57recloxPRed-ApoB needs to be prepared [Table 1].
-
11.
Prepare a standard curve from the pUC57recloxPRed-ApoB plasmid with the concentration depicted in [Table 2]. It is recommended to test the standard curve by qPCR at this point before assaying the samples. Troubleshooting 3
Note: The standard curve is essential to determine the absolute numbers of adipocytes and total cells. We recommend preparing 500 μL–1000 μL of each standard, so that the full study can be completed with the same standard. Aliquots of 100 μL can be prepared and stored at −20°C. The correct cell quantification depends heavily on stable plasmid concentrations of the individual solutions of the standard curve. The precise determination of the exact plasmid concentration before the preparation of the standard curve can be achieved using a NanoDrop microvolume spectrometer.
Note: The plasmid is available upon request from the lead contact. However, the principle of the method can be adapted either to any other vector backbone or to any other sequence of interest, if another recombination site is investigated.
Table 1.
recloxPRed-ApoB sequence of the pUC57 plasmid
| CTTGAAGCGCATGAACTCTTTGATGACGTCCTCGGAGGAGGCCAGCATGGATCCAGCGCTAGCTTGGCTGGAC GTAAACTCCTCTTCAGACCTAATAACTTCGTATAGCATACATTATACGAAGTTATGCGGCCGACCGGTAAGCTTA TCGATACCGTCGATCCCCACTGGAAAGACCGCGAAGAGTTTGTCCTCAACCGCGAGCTGTGGAGGTGGGGTC CAGGTTGAATCACGGGTTCTTCAGCACAATGCACAGTTCTCCAATGACCAAGAAGAAATACGGCTTGACCTTGC AGGATCCTTAGACGGAGCCGGAGAACCTGCGTGCAATCCATCTTGTTCAATGGCCGATCCCATGGCGGCACAG ATGAATTCTTAATAACTTCGTATAGCATACATTATACGAAGTTATGCGGCCGACCGGTAAGCTTATCGATACCGT CGATCCCCACTGGAAAGACCGCGAAGAGTTTGTCCTCAACCGC |
Table 2.
pUC57recloxPRed-ApoB plasmid standard curve
| Concentration [μg/μL] |
|---|
| 2.2 × 107 |
| 1.1 × 107 |
| 5.5 × 108 |
| 2.2 × 108 |
| 1.1 × 108 |
| 5.5 × 109 |
| 2.2 × 109 |
| 1.1 × 109 |
| 5.5 × 1010 |
| 2.2 × 1010 |
| 1.1 × 1010 |
Determination of recombined sites by qPCR
Timing: 0–2 h
The number of recombined LoxPRed or ApoB molecules are quantified by qPCR using a standard curve generated from a synthesized plasmid pUC57recloxPRed-ApoB in addition to primers that specifically bind at the ApoB or recombined LoxPRed site [Figure 6].
-
12.Perform a real time PCR with the samples and the standard curve using Fast SYBR Green according to the following protocol:
-
a.Reaction per well:Sample/Standard = 2 μL, Primer mix (as described in materials and equipment) = 1 μL, H2O = 2 μL, Fast SYBR Green Mastermix = 5 μL
-
a.
-
13.
Run the Fast SYBR Green program on your qPCR machine [Table 3].
-
14.
When exporting the data from the qPCR machine, it is important that the thresholds of LoxPRed samples and ApoB samples are set at the same value. However, be careful to set the threshold only within the geometric phase of the amplification blot.
Note: We established the protocol with Fast SYBR Green (ThermoScientific). However, also SYBR Green from other companies (e.g., KAPA SYBR® FAST, Roche) or other dyes/enzymes (e.g TagMan, ThermoScientific) compatible with qPCR can be used.
Figure 6.
Schematic description of the quantitative recombination analysis
The primer pair depicted in the upper part binds to the ApoB locus and serves to quantify the total number of cells in a depot. The second primer pair depicted in the lower part binds to the recombined transgene at the ROSA26 locus. The primer pair only produces a PCR product when LoxP Stop LoxP casette is genetically deleted by the Cre recombinase.
Table 3.
Fast SYBR green qPCR protocol
| Step | Temperature | Time | Cycles |
|---|---|---|---|
| Initial denaturation | 95°C | 20 s | |
| Denaturation | 95°C | 10 s | 40 cycles |
| Annealing and Extension | 60°C | 20 s |
Expected outcomes
Expected total cell numbers as well as recombined Adipoq+ or recombined Ucp1+ cell numbers for various adipose tissue depots under various housing conditions can be found in Moser et al., (Moser et al., 2021). The cell numbers at 20°C–22°C and after three weeks of cold exposure (8°C) can be found in the table below [Table 4].
Table 4.
Example of Ucp1+ and Adipoq+ cell numbers at 20°C–22°C and after one week of cold exposure (8°C)
| 20°C–22°C |
One week of cold exposure (8°C) |
|||
|---|---|---|---|---|
| Number of Ucp1+ cells | Number of Adipoq+ cells | Number of Ucp1+ cells | Number of Adipoq+ cells | |
| iBAT | 28.9 ± 7.5 M | 25.0 ± 1.1 M | 53.2 ± 5.2 M | 58.1 ± 3.2 M |
| ingWAT | 0.4 ± 0.1 M | 18.1 ± 0.8 M | 2.3 ± 0.7 M | n/a |
| eWAT | n/a | 6.9 ± 0.9 M | n/a | n/a |
M = 106. Data presented are SEM ± mean
Quantification and statistical analysis
Analysis of the qPCR data to determine the number of recombined LoxPRed sites and ApoB sites. A full example and a calculation template can be found online [Table S1].
-
1.Standard curve calculation:
-
a.The pUC57recloxPRed-ApoB plasmid is 3200 bp and the molecular weight of one bp is 650 [g/mol]. Thus, the molecular weight of the plasmid is 2.010 × 106 [g/mol]
-
b.To determine the molarity [M] of your plasmid, divide the concentration of your plasmid by the molecular weight of your plasmid,e.g., 2.2 × 10 7 [μg/μL] / 2.010 × 106 [g/mol] = 1.06 × 10−13 [M]
-
c.To calculate the number of molecules in 2 μL sample, multiply the molarity by the volume of the sample and the Avogadro constante.g., 1.06 × 10−13 [M] ∗ 2 × 10−6 [L] ∗ 6.02214076×1023= 1.28 × 105 molecules
-
d.Blot the average of your CT values obtained for the standard curve against the Log10(Molecules) to draw the standard curve
-
e.Calculate the standard curve for both the recombined LoxPRed and ApoB and determine the slope and y-axis intercept for both the LoxPRed and ApoB [Figure 7]
-
a.
Note: Ideally the slope of the standard curve should be −3.32, which indicates 100% efficiency. However, this is often difficult to achieve. If the range of the CT values is not too spread out, the removal of the highest/lowest standard point can help to bring the slope towards 100% efficiency. We generally accepted an efficiency of 93%–100%; if this was not achieved, the qPCR run was repeated. Troubleshooting 4
-
2.Calculation of the number ApoB sites
-
a.Take the average of the CT value replicates
-
b.Determine the Log10(Molecules) via the standard curveLog10(Molecules ApoB) = (average CT - y-axis intercept) / axis slope
-
c.Determine the number of ApoB molecules in 1 μLApoB molecules in 1 μL = (10ˆ(Log10(Molecules ApoB))/2
-
d.To get the number of cells in 1 μL, divide the number of ApoB molecules in 1 μL by two, as each cell harbors two ApoB alleles
-
e.To get the total number of cells, multiply the number of cells in 1 μL by the dilution factor of 30 and the total volume the tissue was lysed in (e.g., 1250 μL)
-
a.
-
3.Calculation of the number recombined LoxPRed sites
-
a.Take the average of the CT value replicates
-
b.Determine the Log10(Molecules rec.) via the standard curveLog10(Molecules rec.) = (average CT - y-axis intercept) / axis slope
-
c.Determine the number of recombined LoxPRed molecules in 1 μL rec. molecules in 1 μL = (10ˆ(Log10(Molecules rec.))/2
-
d.To get the number of recombined cells in 1 μL, divide the number of recombined LoxPRed molecules in 1 μL by two if the animal is homozygous for the LoxPRed allele. If the animal is heterozygous, do not divide the number of recombined LoxPRed molecules in 1 μL by two.
-
e.To get the total number of recombined cells, multiply the number of cells in 1 μL by the dilution factor of 30 and the total volume you lysed the tissue in (e.g., 1250 μL). Troubleshooting 5
-
a.
-
4.Determination of the percentage of recombined LoxPRed site
-
a.To determine the percentage of recombined LoxPRed sites, divide the number of recombined LoxPRed sites in 1 μL by the number of all cells present in 1 μL and multiply it by 100.
-
a.
Figure 7.
Example of the standard curves
(A) The standard curve generated from the pUC57recloxPRed-ApoB analyzed for ApoB.
(B) The standard curve generated from the pUC57recloxPRed-ApoB analyzed for LoxPRed.
Limitations
The protocol is currently limited to the analysis of recombination sites obtained by LoxPRed recombination sites. Thus, it can be only applied to mouse strains harboring the ROSA26–tdRFP construct (Luche et al., 2007). However, the principle can be applied to other mouse strains if the plasmid and the primers are adapted accordingly. The absolute cell numbers are determined by the standard curve. Therefore, variations or inaccuracies in the standard curve will drastically affect the results.
Further, the number of cells is greatly influenced by the tissue harvesting; thus, the tissues should ideally always be harvested in the same manner. When adhering to a clear tissue harvesting protocol, we observed no interpersonal variation among adipocyte cell numbers. Additionally, other tissue types that adhere to the tissue of interest should be carefully removed (e.g., in the case of analyzing adipose tissue depots adhering to skeletal muscles). Lastly, given that the presence of lymph nodes in the tissue depot of interest can dramatically influence the cell numbers, they should be carefully removed.
Troubleshooting
Problem 1
The tamoxifen is not dissolving in the oil.
Potential solution
Heat the solution to 37°C while shaking for 1 h e.g., on a shaker in an incubator or by taping the falcon tube onto a thermoshaker.
Problem 2
There is still a lot of fat in the genomic DNA solution after the second centrifugation step.
Potential solution
Repeat the centrifugation a third time before preparing the 1:30 dilution of the genomic DNA.
Problem 3
The slope of the standard curve is not equal to −3.32.
Potential solution
We recommend to not work with a serial dilution approach, because if an error is introduced in an early dilution then this error will be present in the whole series. We rather recommend to work with a series of dilutions e.g., diluting 2.2 × 107 by a 1:10 dilution to 2.2 × 108
Problem 4
The slope of the standard curve is not equal to −3.32 or 93%–100% efficiency is not achieved in the run with the samples.
Potential solution
Carefully check your duplicates of the standard curve, or eventually remove the highest/lowest points of the standard curve if the CT values of your sample nicely cluster around the middle part of the standard curve.
Problem 5
The recombined LoxPRed cell numbers are extraordinarily high.
Potential solution
Double check the genotype of the animal. If the animal is homozygous for the LoxPRed site, then divide the recombined LoxPRed cell numbers by two, as this animal is harboring two LoxPRed alleles.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Christian Wolfrum, christian-wolfrum@ethz.ch
Materials availability
Plasmids and animal lines are available from the lead contact upon request.
Data and code availability
This study did not generate/analyze dataset or codes.
Acknowledgments
The authors would like to thank Manuel Klug for his great support with the animal work and Ian Mitchell for editing this manuscript. The work was supported by the Swiss National Science Foundation, Switzerland.
Author contributions
C.M., L.G.S., and Y.R. established the method. C.M. wrote the manuscript and generated the figures, with the help of feedback from all other authors. C.W. supervised the study.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2021.100761.
Contributor Information
Caroline Moser, Email: caroline-moser@ethz.ch.
Christian Wolfrum, Email: christian-wolfrum@ethz.ch.
Supplemental information
References
- Hesselbarth N., Pettinelli C., Gericke M., Berger C., Kunath A., Stumvoll M., Blüher M., Klöting N. Tamoxifen affects glucose and lipid metabolism parameters, causes browning of subcutaneous adipose tissue and transient body composition changes in C57BL/6NTac mice. Biochem. Biophys. Res. Commun. 2015;464:724–729. doi: 10.1016/j.bbrc.2015.07.015. [DOI] [PubMed] [Google Scholar]
- de Jong J.M., Larsson O., Cannon B., Nedergaard J. A stringent validation of mouse adipose tissue identity markers. Am. J. Physiol. Endocrinol. Metab. 2015;308:E1085–E1105. doi: 10.1152/ajpendo.00023.2015. [DOI] [PubMed] [Google Scholar]
- Lee Y.H., Petkova A.P., Konkar A.A., Granneman J.G. Cellular origins of cold-induced brown adipocytes in adult mice. FASEB J. 2015;29:286–299. doi: 10.1096/fj.14-263038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luche H., Weber O., Nageswara Rao T., Blum C., Fehling H.J. Faithful activation of an extra-bright red fluorescent protein in “knock-in” cre-reporter mice ideally suited for lineage tracing studies. Eur. J. Immunol. 2007;37:43–53. doi: 10.1002/eji.200636745. [DOI] [PubMed] [Google Scholar]
- Moser C., Straub L.G., Rachamin Y., Dapito D.H., Kulenkampff E., Ding L., Sun W., Modica S., Balaz M., Wolfrum C. Quantification of adipocyte numbers following adipose tissue remodeling. Cell Rep. 2021;35:109023. doi: 10.1016/j.celrep.2021.109023. [DOI] [PubMed] [Google Scholar]
- Ye R., Wang Q.A., Tao C., Vishvanath L., Shao M., McDonald J.G., Gupta R.K., Scherer P.E. Impact of tamoxifen on adipocyte lineage tracing: Inducer of adipogenesis and prolonged nuclear translocation of Cre recombinase. Mol. Metab. 2015;4:771–778. doi: 10.1016/j.molmet.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate/analyze dataset or codes.