Abstract
Objective
The current study addresses the cellular complexity and plasticity of subcutaneous (inguinal) white adipose tissue (iWAT) in mice during the critical periods of perinatal growth and establishment.
Methods
We performed a large-scale single cell transcriptomic (scRNA-seq) and epigenomic (snATAC-seq) characterization of cellular subtypes (adipose stromal cells (ASC) and adipocyte nuclei) during inguinal WAT (subcutaneous; iWAT) development in mice, capturing the early postnatal period (postnatal days (PND) 06 and 18) through adulthood (PND56).
Results
Perinatal and adult iWAT contain 3 major ASC subtypes that can be independently identified by RNA expression profiles and DNA transposase accessibility. Furthermore, the transcriptomes and enhancer landscapes of both ASC and adipocytes dynamically change during postnatal development. Perinatal ASC (PND06) are highly enriched for several imprinted genes (IGs; e.g., Mest, H19, Igf2) and extracellular matrix proteins whose expression then declines prior to weaning (PND18). By comparison, adult ASC (PND56) are more enriched for transcripts associated with immunoregulation, oxidative stress, and integrin signaling. Two clusters of mature adipocytes, identified through single nuclei RNA sequencing (snRNA-seq), were distinctive for proinflammatory/immune or metabolic gene expression patterns that became more transcriptionally diverse in adult animals. Single nuclei assay for transposase-accessible chromatin (snATAC-seq) revealed that differences in gene expression were associated with developmental changes in chromatin accessibility and predicted transcription factor motifs (e.g., Plagl1, Ar) in both stromal cells and adipocytes.
Conclusions
Our data provide new insights into transcriptional and epigenomic signaling networks important during iWAT establishment at a single cell resolution, with important implications for the field of metabolic programming.
Keywords: Single cell RNA-Seq, Single cell ATAC-Seq, White adipose tissue, Development, Imprinted genes, Adipocytes
Abbreviations
- ASC
adipose tissue stromal cells
- ATAC-seq
assay for transposase-accessible chromatin sequencing
- CCA
canonical correlation analysis
- CPM
counts per million
- DARs
differentially accessible regions
- DEGs
differentially expressed genes
- DMEM
Dulbecco's Modified Eagle's Medium
- ECM
extracellular matrix
- EDTA
ethylenediaminetetraacetic acid
- FC
fold-change
- FF-BSA
fatty acid–free bovine serum albumin
- GO
gene ontology
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- HFD
high-fat diet
- IGs
imprinted genes
- IGN
imprinted gene network
- iWAT
inguinal white adipose tissue
- gWAT
gonadal white adipose tissue
- LinNeg SVC
lineage negative (immune-depleted) stromal vascular cells
- NaHCO3
sodium bicarbonate
- NH4Cl
ammonium chloride
- PBS
phosphate buffered saline
- PCA
principal component analysis
- PDGFRA
platelet-derived growth factor alpha
- PND
postnatal day
- scRNA-seq
single cell RNA sequencing
- snRNA-seq
single nuclei RNA sequencing
- snATAC-seq
single nuclei assay for transposase-accessible chromatin sequencing
- SVC
stromal vascular cells
- TF
transcription factor
- t-SNE
t-Distributed Stochastic Neighbor Embedding
- TBS
tris-buffered saline
- WAT
white adipose tissue
1. Introduction
White adipose tissue (WAT) is a complex organ that plays a central role in systemic energy balance through its interrelated metabolic, endocrine, and immune functions (reviewed in [[1], [2], [3], [4]]). Adipocytes, the parenchymal cells of adipose tissue, have diverse functions that include storage and mobilization of lipids. They also release endocrine signals that report energy status to the brain, regulating metabolic functions in peripheral organs [3,5,6]). Importantly, the metabolic character of white adipocytes is flexible, with cells capable of assuming distinct anabolic and catabolic/thermogenic phenotypes, often within the same adipose tissue depot [[7], [8], [9], [10], [11], [12], [13], [14], [15], [16]].
Interestingly, the vast majority of cells (>80%) within typical white adipose tissues are not adipocytes, but rather a diverse set of vascular and immune cells as well as “adipose tissue stromal cells” (ASC, or “stromal” cells) that include mesenchymal stem cells and committed adipocyte progenitors [[17], [18], [19]]. ASC were initially characterized based on expression of the surface markers PDGFRA, CD29, CD34, and Sca-1 [[20], [21], [22], [23]]. However, recent studies using unbiased single cell RNA-seq (scRNA-seq) in adult adipose tissue has revealed unanticipated complexity in both ASC and immune cell populations [19,[24], [25], [26], [27], [28], [29], [30], [31]]. Specifically, adult WAT contains 2–3 ASC populations that have distinct spatial distributions and are differentially poised for adipogenesis [19,[24], [25], [26]]. Using nomenclature we recently proposed [32], ASC1a are found within the adipose tissue parenchyma, are enriched for adipogenic transcription factors such as Pparg, and appear to be a direct adipocyte precursor [19,[24], [25], [26]]. ASC2 exhibit a fibroblastic/mesenchymal phenotype with latent adipogenic potential [24,25]. These cells form a connective tissue layer (“interstitial reticulum” or “fascia”) that surrounds the adipose depot in the iWAT [25,32]. An additional progenitor cell population (“Aregs”; ASC1b) is localized to perivascular regions in the iWAT [26], with both pro- and anti-adipogenic characteristics [25,26]. These stromal subtypes have also been referred to as adipose stem cells (ASC; ASC2), preadipocytes (PreA; ASC1a), and adipogenesis regulators (Aregs; ASC1b) [33].
Recent scRNA-seq analysis of adult WAT demonstrates that the composition and expression pattern of ASC populations can be altered by diverse nutritional and pharmacological challenges [19,27,31]. By comparison, very little is currently known regarding how the composition and expression profiles of ASC change during development. Adipose tissues are established during fetal development and/or the perinatal period, depending on the species and tissue location, and continue to expand throughout adolescence [[34], [35], [36], [37], [38]]. In mice, inguinal subcutaneous WAT (iWAT) is specified between E14.5–16.5 [[39], [40], [41]], with lipid accumulation beginning prior to birth and extending until about 8 weeks of age [36,40,41]. Interestingly, iWAT undergoes a period of catabolic remodeling, or “browning,” during the second postnatal week, in which brown adipocyte markers are transiently expressed [[11], [12], [13], [14], [15], [16]]. In contrast, gonadal (visceral) gWAT establishment and expansion mostly occurs postnatally [34,40,42].
In the current study, we performed a large-scale single cell transcriptomic and epigenomic characterization of cell types during iWAT establishment. We found that both the transcriptome and chromatin landscape of stromal cells and adipocytes dynamically change during development. Perinatal ASC (PND06-18) are characterized by high expression of several imprinted genes (IGs) and extracellular matrix/structural components, whereas adult stromal cells differentially express integrin signaling, oxidative stress/complement, and immune-related transcripts, some in a subtype-specific manner. Additionally, we identified two clusters of adipocyte nuclei in the iWAT that are distinguished by enrichment for inflammatory/immune and metabolic gene expression patterns that further evolve over time. These findings provide new insights into specific cell types and transcriptional events that are dynamically regulated during iWAT establishment.
2. Materials and methods
2.1. Animals and husbandry
Male and female C57BL/6J breeder mice (7–8 weeks of age) were purchased from Jackson Laboratories (Bar Harbor, ME). Animals were housed in temperature- and humidity-controlled rooms in an AALAC-approved animal facility at Wayne State University (Detroit, MI) and provided Teklad chow (Envigo; Indianapolis, Indiana) and deionized water ad libitum. At distinct developmental time points (postnatal day (PND) 06, PND18 (range 18–19), or PND56 (range 54–57)), male mice were euthanized by decapitation (PND06) or CO2 asphyxiation and cervical dislocation (PND18, 56) and inguinal fats pads were surgically removed, rinsed in phosphate buffered saline (PBS, pH 7.4), and processed as described below. All animal procedures were approved by the Institutional Animal Care and Use Committee at Wayne State University (Detroit, MI).
2.2. Isolation of adipocytes and stromal vascular cells (SVC) from iWAT
Stromal vascular cells (SVC) and adipocytes were isolated from inguinal white adipose tissue (iWAT) as described previously [19,20]. Briefly, following excision, fat pads (combined from 3 to 6 male offspring per time point) were minced and then dissociated in Hanks balanced salt solution (pH 7.4, Sigma–Aldrich, St. Louis, MO) containing 0.5% fatty acid–free bovine serum albumin (FF-BSA; Gemini Bio, West Sacramento, CA) and 2 mg/mL Type 2 collagenase (Worthington Biochemical Corp; Lakewood, NJ) at 37 °C for 30 min with gentle agitation. The digestate was passed through a 100 μM filter and rinsed with buffer (PBS containing 0.5 mM EDTA, 10 mM HEPES, 10% fetal bovine serum, and 0.5% FF-BSA, pH7.4). Adipocytes were then separated by centrifugation at 200×g for 5 min at room temperature. Floating cells (adipocytes) were collected by removing the infranatant by aspiration. Adipocytes were then washed twice with pre-heated (37 °C) Dulbecco's Modified Eagle's Medium (DMEM) containing 1% FF-BSA (Gemini Bio) prior to downstream applications. SVC were pelleted from the infranatant by centrifugation (500×g for 10 min at 4 °C), and red blood cells (RBC) were lysed in lysis buffer (155 mM NH4Cl, 14 mM NaHCO3, and 0.1 mM EDTA, pH 7.4) for 5 min at room temperature. SVC were then filtered (40 μM), resuspended in PBS containing 0.5% FF-BSA (MACS buffer, pH 7.4), and pelleted again by centrifugation (500×g for 10 min at 4 °C). Hematopoietic lineage–positive cells were depleted from the total SVC fraction by cell sorting using the lineage depletion kit (Miltenyi Biotec; Auburn, CA) and a magnetic separator according to manufacturer's instructions.
2.3. RNA isolation and bulk 3′-mRNA sequencing (bulk-seq)
Total RNA was isolated from adipocytes and LinNeg SVC using the Ambion PureLink RNA extraction kit (Ambion/Thermo Fisher Scientific; Waltham, MA). cDNA libraries were prepared from high quality RNA (500 ng) using the Quantseq 3′-mRNA-seq FWD kit (Lexogen; Greenland, NH) according to manufacturer's recommendations. Libraries (n = 3–5 per cell type/developmental time point) were sequenced on the Illumina HiSeq 4000 flow cell (Michigan State University, East Lansing, MI) using a single-end read format. Sequencing reads were then demultiplexed and aligned to the mouse chromosome (mm10) with STAR (v 2.5.2a) and read counts were quantified with HTSeq-count using the data analysis pipeline available on the Bluebee platform (https://www.bluebee.com). Each sample was sequenced at a depth of ~5–8 million reads.
2.4. Single cell RNA sequencing (scRNA-seq) and single nuclei RNA sequencing (snRNA-seq)
Single cell RNA-seq (scRNA-seq) was conducted on LinNeg SVC using the 10X Genomics 3′ Library & Gel Bead Kit (v2), according to manufacturer's instructions (10X Genomics; Santa Clara, CA). Briefly, following cellular fractionation and magnetic cell sorting, LinNeg SVC were resuspended in PBS containing 0.04% non-acetylated BSA (Thermo Fisher Scientific, Waltham, MA) and ~10,000 cells used as input to generate gel beads in emulsion using the 10X Chromium controller. Prior to loading, cells were counted with a hemocytometer and viability (>90%) was estimated using Trypan blue. For adipocytes, nuclei were isolated from the floating fraction using the Nuclei EZ isolation kit according to manufacturer's instructions (Sigma–Aldrich, St. Louis, MO). Briefly, adipocytes were incubated in 5 mL lysis buffer (5 min on ice) and nuclei were collected by centrifugation (500×g for 8 min at 4 °C). Sucrose (250 mM final concentration) was added to the lysates prior to centrifugation. Pellets were resuspended in Nuclei EZ lysis buffer containing 250 mM sucrose, incubated on ice for 5 min, and then pelleted again by centrifugation (500×g, 6 min). After the supernatant was decanted, nuclei were washed once with PBS containing 1% nuclease-free FF-BSA (PBS wash buffer; Gemini) and RiboLock RNase Inhibitors (Thermo Fisher; Waltham, MA), centrifuged, and resuspended in fresh PBS wash buffer for single nuclei processing. Approximately ~10,000 nuclei/sample were used as input for the 10X Chromium controller to generate nuclei–gel bead emulsions according to the 10X Genomics User Guide. After processing, libraries were quantified with the Kappa PCR kit and sequenced with the Illumina NextSeq500 flow cell (Wayne State University, Detroit, MI) with the following parameters: 26 cycles Read1, 58 cycles Read2, and 8 cycles i7 index. Raw sequencing data was demultiplexed using bcl2fastq (Illumina), sequencing reads aligned to the mm10 reference genome (or pre-mRNA mouse reference genome for snRNA-seq), and count matrices generated using the Cell Ranger 3.0.2 analysis pipeline (10X Genomics) with default settings. For the nuclear RNA data, due to higher ambient RNA contamination, the force cells command was additionally used (--force-cells=4000 (PND06 and PND18) or 5000 (PND56)) prior to further processing (see below). Each library was sequenced to a depth of ~25,000–30,000 mean reads per cell.
2.5. Single nuclei ATAC sequencing (snATAC-seq)
Developmental differences in chromatin accessibility were evaluated with the 10X Genomics snATAC-seq kit (v 1.0, 10X Genomics). The cell fractions used for snATAC-seq were prepared independently (i.e., from different animals) of the samples used for sc/snRNA-seq experiments, potentially leading to some differences in enrichment between preparations. Nuclei were isolated from stromal cells or adipocytes using the Nuclei EZ isolation kit (Sigma–Aldrich), modified according to the protocol described above. After the second incubation in Nuclei EZ buffer, nuclei were pelleted by centrifugation (500×g, 6 min), and pellets were resuspended in 10X Genomics resuspension buffer (10X Genomics). Nuclei (~12,000) were then tagmented with Tn5 transposase for 1 h at 37 °C, and transposed nuclei were processed with the 10X Genomics chromium controller according to the manufacturer's protocol (10X Genomics). After processing and sample indexing, libraries were sequenced with an Illumina NextSeq500 flow cell, (Wayne State University, Detroit, MI) using the following format: 34 cycles Read1, 34 cycles Read2, 8 cycles i7 index, and 16 cycles i5 index. Raw sequencing data were demultiplexed and sequences aligned to the mouse genome (mm10) using the Cell Ranger ATAC (v 1.2.0) pipeline (10X Genomics). Additional data processing and analysis steps are provided in the Bioinformatics and Data Analysis section.
2.6. Bioinformatics and Data Analysis
Raw read counts from bulk RNA sequencing (n = 3–5/tissue type/time point) were processed using the integrated Differential Expression and Pathway analysis (iDEP.90) ShinyAPP [43]. Briefly, read counts were filtered to exclude genes displaying fewer than 0.5 counts per million (CPM) in at least 40% of samples. Filtered samples were then used as input for k-means clustering, principal component analysis (PCA), and gene ontology. The number of k-means clusters was determined using the elbow method. Differentially expressed genes between individual groups were detected with DESeq2 using a false discovery rate (FDR) cutoff of P < 0.05 and minimal fold change of 3. Venn diagrams displaying differentially expressed genes were generated using Venny 2.1.0 [44] and additional heatmaps were made with ClustVis [45].
Individual gene count matrices for scRNA-seq and snRNA-seq from the Cell Ranger pipeline (see above) were first processed using the SoupX R package (v 1.0.1) to remove ambient RNA contamination following the detailed vignette (https://rawcdn.githack.com/constantAmateur/SoupX/204b602418df12e9fdb4b68775a8b486c6504fe4/inst/doc/pbmcTutorial.html) [46]. Briefly, for scRNA-seq (stromal cells), Fabp4 and Car3 were used to calculate the RNA “soup” (background) contamination using the “estimateNonExpressingCells” command. For snRNA-seq (adipocytes), because most of the nuclei were adipocytes and the contaminating RNA was primarily from adipocyte lysis, we set the contamination fraction at 20% using the “setContaminationFraction” command, as described in the vignette, without providing cell cluster information for either dataset. Corrected gene matrices were then imported to the Seurat R package (v 3.0.2) for filtering, normalization, and integration of cells using methods described previously [[47], [48], [49]]. Briefly, cells containing <500 or >5000 genes and >15% mitochondrial reads, nuclei containing <400 or >4000 genes and >10% mitochondrial reads, and red blood cells (when present) were filtered prior to downstream analysis. Cell populations were identified based on marker gene expression using the FindAllMarkers function in Seurat. Differential gene expression analysis was conducted using the Wilcox test with a log fold-change threshold of 0.5 for scRNA-seq in a minimum of 25% of cells and 0.4 for nuclei (minimum cells = 15%). Heterotrophic doublets were estimated using the R package DoubletFinder (https://github.com/chris-mcginnis-ucsf/DoubletFinder), with the pK set at 0.01 and a doublet estimation of 3–4% [50]. Identified doublets were removed from the datasets following integration. Additional pathway analyses were conducted using Enrichr (https://amp.pharm.mssm.edu/Enrichr/) [51] and graphs generated by GraphPad Prism (v 8.0.1; San Diego, CA). STRING analysis (v11) was conducted using the DEGs identified by scRNA-seq analysis (P < 0.05; PND06 vs. 56) and basic settings (full network, confidence = 0.4) [52]. Networks were exported to Cytoscape (v 3.8.2) for visualization and GO enrichment analysis with a redundancy cutoff of 0.8 [53].
Custom peak calling for snATAC-seq datasets was performed using the filtered fragment files as detailed in the SnapATAC vignette (https://github.com/r3fang/SnapATAC) [54]. Briefly, barcodes were filtered to retain high quality cells and datasets (SVC or adipocytes) were then integrated using Harmony to remove batch effects of developmental time [55]. Peaks were called following integration and cell clustering with MACS2 using the following settings: nomodel –shift 100 –ext 200 –qval 5e-2. Custom peaks were then used to create a cell-peak matrix using the Cellranger-ATAC reanalyze pipeline (10X Genomics, v 1.2) on aggregated libraries without depth correction. The custom cell-peak matrix was then imported into the Signac (v 1.1.0) and Seurat (v 3.2.0) R packages (https://github.com/timoast/signac; [[47], [48], [49],56]) for further filtering, integration, and analysis.
Aggregated cell-peak matrices from snATAC-seq were filtered in Signac (v 1.1.0) to include only high-quality nuclei using the following cutoffs: peak region fragments > 500 and <100,000; nucleosome signal < 5; transcription site enrichment > 2.5, percent reads in peaks > 60 (ASC) or >35 (adipocyte), and blacklist ratio < 0.025. The count matrix was normalized using term frequency–inverse document frequency (TF-IDF) and datasets were integrated with the R package Harmony [55] before being clustered and visualized using the shared nearest neighbor method in Seurat. Clusters were annotated based on enrichment of marker gene activity and further validated using integration and label transfer with our scRNA-seq and snRNA-seq datasets, as described [47]. Differentially accessible regions (DARs) were detected using the FindMarkers function in Seurat with the likelihood ratio test. The number of fragments overlapping peaks (peak_region_fragments) served as the latent variable, with significant peaks exhibiting a log fold-change threshold of 0.25 between conditions (i.e., clusters, time) in at least 15% of nuclei. DARs for each comparison were imported into the Genomic Regions Enrichment of Annotations Tool (GREAT; v 4.0.4; http://great.stanford.edu/public/html/) for annotation and gene ontology using the mm10 assembly, the whole genome as background, and default settings for associating genomic regions with genes [57]. Motif enrichment was conducted using the Seurat wrapper Signac (v 1.1.0). Briefly, a position frequency matrix (pfm) was constructed for each integrated library using the JASPAR2020 database, with mouse (10090) as the designated species and set to include all versions [58]. DNA motifs that were significantly enriched within differentially accessible regions (15% of nuclei; logFC > 0.25) were then detected using the FindMarker and FindMotif functions of Signac. Background peaks for each comparison were selected from a set of open (“accessible”) peaks that were matched for GC content. Significant peaks were then filtered to remove redundant motifs (retaining the most updated versions) and those exhibiting <1.65 fold-change between comparisons. Mouse motifs were named according to the nomenclature indexed in the JASPAR2020 database for Mus musculus (http://jaspar.genereg.net/). TF activity scores were also analyzed for visualization of enriched motifs using the Signac wrapper ChromVAR [59]. Plots for TF footprints and pseudo bulk genomic accessibility tracks were created using Signac, each according to the respective vignettes (https://satijalab.org/signac/articles/mouse_brain_vignette.html). For coverage tracks (bulk genomic accessibility), a scale factor of 106 was used. The R package ChIPseeker was used to annotate custom peaks with the “annotatePeak” function and default settings [60].
2.7. Data availability
Raw and processed datasets for bulk RNA-seq, scRNA-seq, and snATAC-seq were deposited into NCBI's Gene Expression Omnibus (GEO) under the Series accession number GSE180589.
3. Results
3.1. Bulk RNA-seq reveals time-dependent expression changes in stromal cells and adipocytes during iWAT development
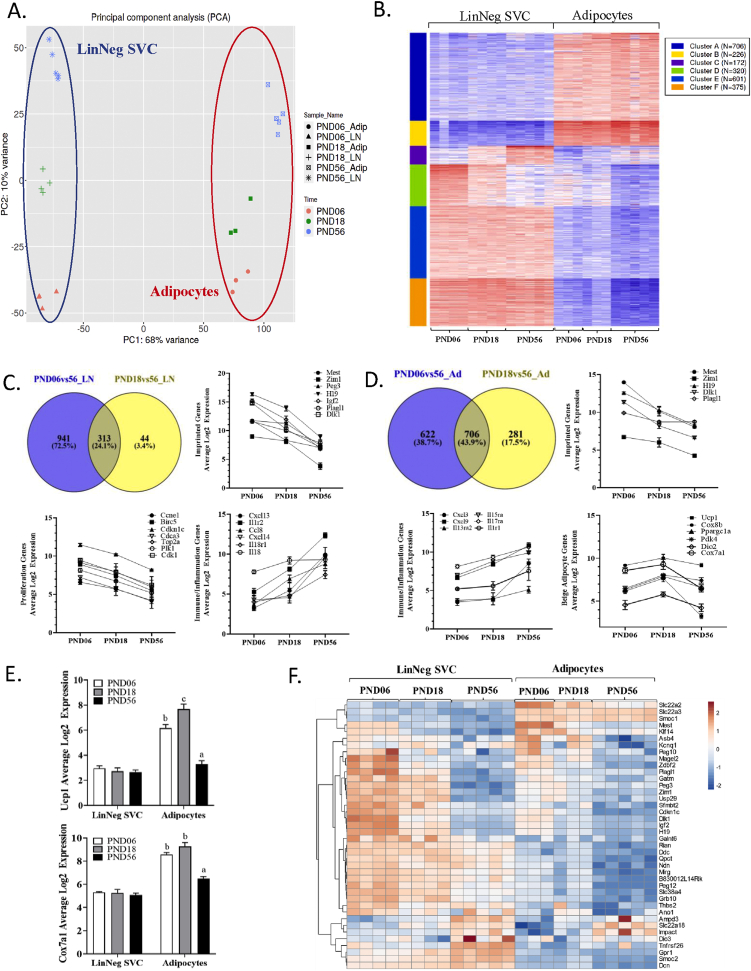
To identify transcriptional networks influencing development of white adipose tissue, we initially profiled time-dependent expression changes using bulk RNA-seq. Stromal vascular cells (SVC; LinNeg SVC) and adipocytes were isolated at different stages of development—early postnatal (PND06), prior to weaning (PND18), and adolescent/adult mice (PND56)—and global changes in gene expression within each fraction were compared. Principal component analysis (PCA) of the cellular fractions across development is depicted in Figure 1A. As shown, differences between the SVC and adipocytes were captured by PC1, which accounted for 68% of variability in gene expression. Interestingly, the effect of developmental time for both SVC and adipocytes was captured by PC2 (10%; Figure 1A). K-means clustering of the 2,400 most variable genes identified 6 different clusters (Figure 1B), 4 of which showed differential expression between cell fractions (adipocytes vs. LinNeg SVC) and 2 showed time-dependent changes (Figure 1B; Cluster C, D).
Figure 1.
Bulk RNA-seq of stromal vascular cells (LinNeg SVC) and adipocytes isolated from inguinal subcutaneous white adipose tissue (iWAT) of mice during postnatal development. Bulk RNA-seq (3′ end-seq) was performed on RNA isolated from stromal vascular cells (LinNeg SVC) and the floating adipocyte fraction of male C57BL/6J (B6) mice at postnatal days 06, 18, and 56 (PND06-56). (A) Principal component analysis (PCA) showing variance in gene expression due to differences among iWAT cell fractions (PC1, 68%) and developmental time (PC1, 10%; eclipses hand drawn). (B) Heatmap displaying k-means clustering of the 2,400 most variable genes across datasets. Clustering resolved 6 expression patterns. Data were centered and scaled by row (gene). Expression scale colors (low; blue) to (high; red). (C–D) Venn diagrams and expression plots depicting differential gene expression changes (DEGs; FDR P < 0.05, minimum fold-change > 3) in (C) stromal vascular cells (LinNeg SVC) and (D) adipocytes among developmental time points. PND56 samples were used as the reference group (denominator) for individual comparisons. (E) Log2 expression of Ucp1 and Cox7a1 mRNA in each cellular fraction. Different superscripts denote significant differences of developmental time on expression for each gene in the adipocyte fraction (P < 0.05). (F) Expression heatmap displaying imprinted genes. Clustering was performed using correlation distance and complete linkage. Values are scaled per gene (row).
Using a combination of KEGG, GO, and pathway analyses, genes most highly upregulated in the adipocyte fraction were strongly associated with mature adipocyte function and metabolism, as expected. For example, clusters A and B (Figure 1B) were highly enriched for mRNA transcripts involved in adipocyte function (i.e., Cebpa, Agpat2, Lep), fatty acid/triglyceride synthesis, and metabolic pathways (i.e., electron transport chain, glycolysis/gluconeogenesis, and branch-chain amino acid metabolism) (Suppl. Table 1; Suppl. Fig. 1). Genes that were more strongly expressed in the stromal cell compartment (Figure 1B; Clusters E, F) included those associated with cell motility, extracellular matrix (ECM) organization (metalloproteinases and structural/adhesion proteins), and secreted Wnt molecules, inhibitors, and their receptors (Suppl. Table 1). These gene patterns are consistent with known markers of mesenchymal progenitors (e.g., Pdgfra, Col1a1/a2) as well as other cell types likely captured with our fractionation method (i.e., endothelial cells, smooth muscle cells, and Schwann cells).
Two clusters (Figure 1B; Clusters C & D) showed patterns that were more dependent on time than cell type (Figure 1B). For example, genes in cluster C displayed a gradual increase in expression from the early perinatal period to adulthood, whereas expression of genes in cluster D exhibited the opposite pattern (Figure 1B). The temporal gene expression pattern was most striking for the LinNeg SVC, but less so for adipocytes. To further explore transcriptional differences across time, we compared the DEGs from each fraction using 56D as the reference group (denominator) (Figure 1C,D). As shown in Figure 1C, the greatest differential in gene expression was found when comparing PND06 to adult SVC (PND56, n = 1254, Figure 1C). By PND18, the number of DEGs decreased by ~70%, indicating a gradual shift towards adult gene expression patterns prior to weaning (n = 357, Figure 1C). In general, SVC during early development (PND06–18) showed higher expression of genes involved in cell cycle/cell proliferation (i.e., Ccnb1/2, Plk1), cell adhesion, and ECM organization (i.e., Thbs1, Cd44, Fbn2). By comparison, adult stromal cells were more enriched in immunoregulatory transcripts (i.e., cytokines/chemokines; Cxcl13, Cxcl14, Il18) and those regulating cell motility and migration (i.e., Ntf3, Egfr, Edn1; Figure 1C).
Progressive changes in gene expression were also observed in the floating adipocyte fraction (Figure 1D). Perinatal adipocytes (PND06) expressed higher levels of mRNA transcripts related to cell cycle/mitosis, ECM-receptor interaction (i.e., Col6a1/a2/a3, Col1a2), and focal adhesion, whereas adult adipocytes (PND56) were more highly enriched in transcripts involved in cytokine/chemokine signaling (i.e., Cxcl3, Cxcl9, Il15ra), leukocyte migration, complement and coagulation (i.e., C3, C2, C14), and xenobiotic/drug/glutathione metabolism (i.e., Gpx1, Gstm1, Cyp2e1, Hsd11b1). Although many of these markers were also expressed in the SVC fraction, we note that adipocyte expression tended to be similar to that of stromal cells at earlier time points and then deviate over time, potentially reflecting changes as adipocytes interact with the microenvironment and expand in size [61]. Previously identified markers of beige adipocytes (i.e., Ucp1, Cox8b, Ppargc1a) were also detected and tended to be higher at PND18, although only significant for Ucp1 and Cox7a1 (P < 0.05; Figure 1D,E).
Although not directly associated with a GO category, we also identified several imprinted genes (IGs) that were differentially regulated during early postnatal development (Figure 1C,D,F). Imprinted genes are expressed in a parent of origin manner, with multiple roles in perinatal growth and development [62]. Among 129 IGs in mice where the expressed allele is known (http://www.geneimprint.com), we identified 40 genes that were differentially expressed (FC > 3, P < 0.05) in either cell fraction (χ2 p-value < 0.00001; Figure 1F), with an equal distribution of maternally and paternally expressed genes. In general, IGs were more abundantly expressed in the LinNeg SVC fraction at PND06 (e.g., Dlk1, H19, Igf2), and levels declined sharply with aging (Figure 1C,D,F). One exception to this pattern was Mest, a gene associated with postnatal adipocyte lipid accumulation [36,63,64], which was more highly enriched in adipocytes at both PND06 and PND18 (Figure 1D,F). This overall expression pattern suggests that adipogenic differentiation may suppress IG expression in vivo.
3.2. Single cell RNA-seq reveals stromal cell heterogeneity during iWAT development
To determine whether time-dependent changes detected with bulk RNA-seq were due to developmental differences in the composition of cells and/or transcriptional differences within individual cell types, we profiled stromal cells at PND06, PND18, and PND56 using scRNA-seq (Figure 2A). Initial aggregation of stromal cells segregated datasets by time (Figure 2A, left), corresponding to differences in gene expression across development. To better identify ASC subtypes across time, we integrated the scRNA datasets from the three developmental timepoints (Figure 2A, right) [47,49]. Unsupervised clustering of the integrated datasets resolved ~13,000 cells that were projected onto 9 different clusters (Figure 2A, right). t-SNE plots of the unintegrated and integrated datasets separated by time are presented in Figure 2A (bottom). Similar to that previously observed [19,25], >85% of stromal cells expressed canonical ASC markers, including Pdgfra, Cd34, and Ly6a (Figure 2B; Suppl. Fig 2), and ASC subpopulations were further differentiated by marker gene expression (Figure 2B; Suppl. Fig 2). For example, Pi16 and Dpp4 were enriched in ASC2, Icam1, Adam12, and Col5a3 in ASC1a, while Clec11a and Vit were enriched in ASC1b (Figure 2B; Suppl. Fig 2). These markers are in line with the fibroblastic/mesenchymal (ASC2), preadipocyte (ASC1a), and “Areg” (ASC1b) populations recently described, although in general, marker genes for ASC1a at PND06 were less distinct than at later time points. A small population of proliferating/differentiating cells (i.e., Birc5, Top2a) was also present at PND06 and PND18, representing ~6–8% of stromal cells, whereas very few proliferating cells (<0.5%) were observed at 56 days of age. Proliferating cells form a distinct cluster owing to highly differential expression of proliferation markers. However, cells in this cluster express markers of ASC1 (i.e., Adam12, Col15a1) and ASC2 (i.e., Pi16, Dpp4), indicating that both ASC subtypes contribute to tissue expansion. Unfortunately, there were too few cells in this cluster to map a putative differentiation trajectory with high confidence. A feature heatmap and t-SNE expression plots of marker genes expressed in the integrated cell clusters are presented in Suppl. Fig 2.
Figure 2.
scRNA-seq of stromal vascular cells isolated from the iWAT of mice during early postnatal development. Single-cell transcriptomics was performed on stromal vascular cells isolated from the inguinal white adipose tissue of C57BL/6J mice at select developmental time points (PND 06, 18, 56). (A) t-SNE plots of unintegrated and integrated single cell libraries displaying cell clusters in aggregate (top) and integrated libraries separated by developmental time (bottom). (B) t-SNE plots displaying marker gene expression of cell clusters from the integrated scRNA-seq libraries. (C) Heatmap displaying changes in gene expression within the ACS clusters during early postnatal development. (D) Protein interaction network and gene ontology of upregulated DEGs (average logFC > 0.5; minimum of 25% of cells) detected in ASC1a and ASC2 when comparing perinatal (PND06) to adult (PND56) time points.
We queried some of the DEGs identified from bulk RNA-seq to confirm time-dependent changes across scRNA-seq datasets (Figure 2C) and found that proliferation markers (i.e., Top2a, Birc5) were highly induced in a small subset of cells at PND06 and PND18, imprinted genes were enriched in specific ASC populations at PND06, and immune-related genes were enriched at PND56 (Figure 2C). Because there was clear heterogeneity within ASC subpopulations, we compared both global and subtype-specific expression changes (PND06 vs. PND56; PND18 vs. PND56; Suppl. Fig. 3). As seen with bulk RNA-seq, many of the global DEGs detected at PND06 were more strongly enriched for transcripts involved in ECM structure/adhesion (i.e., Col1a1, Col1a2, Fbln2, Fbn2, Thbs1/3) as well as imprinted genes (i.e., H19, Dlk1, Igf2). Those with higher levels at PND56 included immune/defense genes and those involved in α6/β4 integrin signaling (i.e., Lamb2, Lamc1, Cdkn1a, Ar, Egfr), prostaglandin synthesis and regulation, and oxidative signaling (i.e., Gpx1, Junb, Mgst1, Mt1).
When comparing individual ASC subpopulations, we found that several DEGs detected at PND06 were often co-regulated in at least one other ASC (Suppl. Figure 3C,D). Similar results were observed when comparing PND18 to PND56, although fewer significant DEGs were detected overall (Suppl. Fig. 3D). ASC1b, which has been colocalized to the vasculature in iWAT [26], possessed the highest number of enriched transcripts (logFC > 0.5) early in development (Suppl. Fig. 3D). These included Meox2, Wisp1, and Ifitm1. Additionally, Dkk3, a secreted glycoprotein that stimulates SMC differentiation, was more abundant in this subtype at PND06 [65]. By PND18, the ASC2 population had the most DEGs (27%) compared to that of PND56 (Suppl. Fig. 3D). mRNA transcripts associated with phospholipid/eicosanoid metabolism (i.e., Pla1a, Smpd3, Ptgs2, Anxa3) and cytokine signaling (i.e., Il33, Il18, Il1r2) tended to increase more in this subset during development, whereas those involved in steroid metabolism (i.e., Akr1c18, Akr1c14), intercellular communication (Gap43), keratinocyte differentiation (Krtdap), and the anti-adipogenic factor (Cthrc1) were more enriched in ASC2 at PND06 [66]. Interestingly, the induction of phospholipid-metabolizing enzymes (i.e., Pla1a, Smpd3, Ptgs2, Anxa3) and several innate/immune-regulatory genes in adult animals were more distinctive for ASC2 and included cytokines mediating type I (Il18) and type 2 (Il18, Il33) immunity [67,68] as well as Il1r2, a decoy receptor that dampens IL-1 responses (Figure 2C) [69].
We next addressed changes in imprinted gene expression in LinNeg SVC during development (Figure 1C) and found that the expression pattern of IGs varied among different ASC populations (Figure 2C). For example, Mest, Dlk1, and the zinc finger transcription factor Zim1 were more ASC1a/b specific, whereas Cdkn1c was enriched in ASC2 at PND06 and PND18. Additionally, H19 and Igf2 were highly expressed in ASC1a and ASC2, while Peg3 and Plagl1 were expressed in all ASC at early time points (Figure 2C). By PND56, however, very little IG expression was detected. We further integrated our scRNA-seq dataset with that of Merrick et al. [25], who evaluated stromal cells at PND12. As shown in Suppl. Figure 4, global ASC as well as some of the development-specific changes were in concordance with our findings, especially for the inverse expression of IGs and immune-related genes.
Given the unique properties and localizations of ASC1a and ASC2, we also explored whether the IGs engage different networks (Figure 2D). We performed a STRING analysis [52] on DEGs identified within each ASC population (P < 0.05; PND06 vs. PND56; Figure 2D) during development and found that IGs were strongly associated with ECM proteins in both populations (Figure 2D). Although ~40% of DEGs were similarly expressed among the ASC populations, IGs engaged somewhat different protein networks (Figure 2D). For example, in ASC1a, IGs were more strongly connected to networks involved with GO processes including “biological adhesion” and “organ/tissue development”; whereas “extracellular structure organization” and “collagen fibril/supramolecular fiber organization” processes were more strongly enriched in ASC2 (Figure 2D). This likely corresponds to tissue location and the relative contribution of ASC populations towards adipogenesis (ASC1a) versus structural support/encapsulation of the fat pad (ASC2). Collectively, these data implicate both global and subtype-specific perinatal development differences in ASC that may be intrinsic to the subpopulation, influenced by spatial compartmentalization/function, and/or coordinately regulated by other presently undefined mechanisms.
3.3. Single nuclei RNA-seq (snRNA-seq) reveals transcriptionally heterogenous adipocyte nuclei during iWAT development
scRNA-seq of adipocytes is not currently feasible with the 10X Genomics platform; however, this limitation can be addressed by sequencing RNA in single adipocyte nuclei (i.e., snRNA-seq [27,30]). Therefore, we performed snRNA-seq of ~10,000 adipocyte nuclei during early postnatal development. In the absence of data integration, adipocytes resolved into 3 clusters corresponding to developmental gene expression patterns (Figure 3A, left), as in results with ASC populations (Figure 2A, left). To visualize common cell types across development, we integrated the datasets and identified mature adipocytes (~93% of total) that could be resolved into 2 clusters, referred to as Adip1 and Adip2 (Figure 3A, right). The remaining 7% were mostly contaminating endothelial cells that expressed markers for this cell type (i.e., Flt1, Cdh13, and Cldn5).
Figure 3.
snRNA-seq reveals transcriptionally heterogenous adipocyte nuclei in the iWAT of mice during early postnatal development. Single-nuclei RNA-seq was performed on nuclei isolated from inguinal adipocytes at select developmental time points (PND 06, 18, 56). (A) t-SNE plots of unintegrated and integrated single nuclei libraries showing cell clusters in aggregate (top left) and integrated libraries (top right) separated by developmental time (bottom). (B) t-SNE plots displaying general adipocyte and subpopulation-specific marker genes from integrated snRNA-seq libraries. (C) Pathway analysis of DEGs detected (logFC > 0.4; 15% of cells) when comparing the two adipocyte clusters (Adip1 vs. Adip2) at PND56. (D) Venn diagrams displaying DEGs (P < 0.05; average logFC > 0.4; min 15% of nuclei) detected in each adipocyte subpopulation when comparing early postnatal (PND06 or PND18) to adult (PND56) time points. (E) Violin plots displaying select time-dependent marker genes expressed within adipocyte nuclei subpopulations during postnatal development.
The adipocyte nuclei clusters expressed adipocyte-specific genes, including Adipoq, Acaca, Pnpla2, and Plin4, and could be further distinguished by the expression of immunomodulatory (Adip1) and metabolic-related genes (Adip2) (Figure 3B,C). For example, Adip1 was more highly enriched in transcripts associated with cytokine/chemokine/NFKB1 signaling (i.e., Nfkb1, Ccl2, Cxcl1, Ccl7). By comparison, Adip2 was more enriched in nuclear transcripts involved in adipogenesis/lipid metabolism (i.e., Dbi, Retn, G0s2, Pparg, Pck1, Apoe) as well as MAP kinase signaling (i.e., Jun, Fos), with some evidence of additional heterogeneity within this subset (Figure 3C). Collectively, Adip1 represented 70% of the total adipocyte population and Adip2 accounted for ~30%, with little distinction in proportions across development (Figure 3A).
We initially evaluated dynamic changes in adipocytes over time by comparing average expression differences with the reference point PND56 (Figure 3D, Suppl. Figure 5A,B). As observed with stromal cells, a majority of the DEGs occurred at PND06, when adipocytes were accumulating lipids and increasing in size (Suppl. Figure 5B). Gene ontology analysis suggests that global differences in gene expression between PND06 and PND56 reflect the distinct processes of tissue establishment and maintenance. Thus, PND06 adipocytes collectively express higher levels of transcripts for genes involved in ECM generation and remodeling (i.e., Lama4, Col5a3, Col6a1, Col6a2) as well as transcription regulation/RNA processing (i.e., Ncor1, Ewsr1, Dapk1, Fmr1, Clock, Plagl1). Additionally, Mageb18, a protein belonging to the melanoma antigen family that has anti-apoptotic properties [70], was highly expressed in Adip1 of neonates, but was silenced at PND56 (Figure 3E, Suppl. Fig. 6). By comparison, adult adipocytes more highly expressed genes associated with cytokine/growth factor signaling (i.e., Iigp1, Il6, Cxcl9, Il1r1, Jak1, Irf1, Cxcl1, Cxcl2, Egr1, Casp4, Jak2), oxidative stress (i.e., Birc3, Egfr, Hif1a, Sod2, Nfe2l2), and fatty acid/triglyceride synthesis (i.e., Acaca, Acacb, Acss2, Fasn, Dgat2) (Figure 3E, Suppl. Fig. 5 and 6).
Among adipocyte subpopulations, more mRNA transcripts discriminating Adip1 and Adip2 were detected at PND56 (Figure 3D), and a higher proportion of DEGs were found in the Adip1 fraction over time (Figure 3D, Suppl. Fig. 5C). DEGs included those involved in mRNA splicing/RNA processing and ubiquitin-mediated proteolysis at PND06 as well as inflammatory/defense/cytokine signaling at PND56. By comparison, the Adip2 cluster was differentiated by transcripts involved in fatty acid/glucose metabolism (i.e., G0s2, Pck1, Apoe), insulin signaling (i.e., Pde3b, Prkar2b), and Pparg signaling (i.e., Pparg, Ubc, Dbi) (Figure 3E), and transcripts associated with these categories were more abundantly enriched at PND18 and PND56 (Figure 3E). Violin plots of key marker genes illustrate how the gene expression patterns of Adip1 and Adip2 evolve over time. First, the differences between subpopulations grows, with Adip1 and Adip2 being less transcriptionally diverse at PND06 and more distinct at PND56 (Figure 3E, Suppl. Figure 5C). Expression of IGs was greatest at PND06 and nearly completely silenced by PND56. Over the same period, we observed upregulation of transcripts that distinguish the adipocyte subtypes, with Adip1 inducing those involved in oxidative stress, hypoxia, and inflammation (i.e., Ptx3, Hif1a, Nfkb1) and Adip2 upregulating gene transcripts involved in fatty acid/glucose metabolism (i.e., G0s2, Fasn, Apoe) as well as oxidative metabolism (i.e., Plin2, Cox8a, Pdk4). We note that several of the gene changes at PND56 were also observed in ASC (including an increase in the expression of Egfr, Cxcl1, and Il1r1) and other cytokines/chemokines and receptors. However, the upregulation of transcripts for these genes, along with receptors and downstream signaling targets, suggest further functionality and/or enhanced immune responsiveness in adult compared to perinatal adipose tissue.
Although mRNA markers (e.g., Ucp1, Cox7a1, Ppargc1a) for brown adipocytes were detected at PND06 and PND18 using bulk RNA-seq (Figure 1D,E), we did not find strong retention for these mRNAs in the nuclei. This may be due to rapid processing and/or transient expression of mRNA transcripts during development. However, we found the genes Pdk4 and Esrrg to be selectively expressed in a subset of Adip2 nuclei at PND18 (Suppl. Fig. 6B). Pdk4 is a downstream target of PPAR/adrenergic signaling and was also induced by CL treatment in “thermogenic” nuclei identified in the study by Rajbhandari et al. [[111], [112], [113]]. Additionally, Esrrg was recently shown to be a novel transcriptional regulator of beige adipocyte formation [71]. Further investigations to define and characterize this subset are underway. Nonetheless, our snRNA-seq data discriminate adipocyte nuclei in iWAT whose transcriptional heterogeneity evolves during development.
3.4. Chromatin landscape of stromal cells and adipocytes change during iWAT establishment
Because we found evidence of both global and subtype-specific gene expression changes, we next evaluated whether these changes were associated with alterations in the chromatin landscape and further addressed whether putative TF motifs mediating cell-dependent differences could be identified. Using snATAC-seq, we analyzed changes in chromatin accessibility in the stromal and adipocyte nuclei across postnatal development. Following filtering, integration, and clustering, ~15,000 stromal cells were resolved into 6 different clusters (Figure 4A). As shown, most total transposase-sensitive sites (~200,000) were located in distal intergenic and intronic regions, whereas ~20% of peaks fell into promoter regions (±3 kbp; Figure 4C). To help identify the snATAC-seq clusters, we constructed a gene activity matrix and applied methods for integration and label transfer using our scRNA-seq dataset [47,48] (Suppl. Fig. 7A; Figure 4A). In general, there was excellent concordance between clusters identified with peaks (Figure 4A (top)) and the cell types predicted based on gene activity (Suppl. Fig. 7A; Figure 4A (bottom); Figure. 4B).
Figure 4.
The chromatin landscape of adipose tissue stromal cells (ASC) changes during postnatal iWAT development. Stromal vascular cells (LinNeg SVC) were isolated from inguinal white adipose tissue of C57BL/6J mice at select developmental time points (PND 06, 18, 56). Nuclei were purified and snATAC-seq was performed using the 10X Genomics platform, as described in the Methods. (A) t-SNE plots of integrated snATAC-seq libraries displaying aggregated nuclei clusters identified from (top) the snATAC-seq peak matrix and (bottom) following integration and label transfer with our scRNA-seq dataset. (B) t-SNE plots displaying marker gene activity in the integrated snATAC-seq libraries. (C) Fragment length histogram of the first 10 Mbp on chromosome 1, transcription start site enrichment plot, and a pie chart displaying peak annotation distribution. (D) Line graph displaying the number of differentially accessible regions (DARs; P < 0.05; average logFC > 0.25; min 15% of nuclei) detected in ASC populations during postnatal development and gene ontology of the annotated DARs. (E) Pseudo-bulk accessibility tracks for select genes separated by ASC cluster and postnatal time point. Shaded areas (hand-drawn) indicate peaks that were differentially accessible.
We next compared differentially accessible regions (DARs) both globally (all ASC combined) and among individual ASC populations as a function of developmental time (vs. PND56) and found major differences in chromatin accessibility (Suppl. Fig. 8; Figure 4D). Globally, ~700 sites were significantly more “open,” or more accessible, at the early perinatal time point (PND06) while ~1000 sites were more accessible in adults (PND56; Suppl. Fig. 8A). By PND18, the vast majority of sites that were open at PND06 were no longer accessible, and there was a gradual acquisition (~50%) of adult pattern accessibility (Suppl. Fig. 8A). Similar trends were observed among individual ASC populations (Figure 4D; Suppl. Fig. 8B). Using the GREAT analysis tool [[111], [112], [113]], we annotated the differentially accessible peaks and performed gene ontology (GO). Although there was some overlap in GO categories, especially at PND56, many of the peaks were enriched for unique categories, including cell–cell adhesion (ASC1a), angiogenesis/vasculature development (ASC1b), and receptor signaling/MAPK pathways (ASC2; Figure 4D). Interestingly, at PND06, ASC2 and ASC1a peaks were also associated with mouse phenotypes of genetic imprinting and abnormal placental weight/size, whereas ASC1b peaks were associated more with vasculature/smooth muscle morphology (not shown). By PND56, peaks for ASC1a and ASC2 were enriched for GO categories associated with inflammation/immune signaling, whereas ASC1b was more strongly enriched for genes associated with cell motility/adhesion (Figure 4D). Genomic tracks for select regions are displayed in Figure 4E.
snATAC-seq libraries from floating adipocytes were processed using the same procedures as those for stromal cells. After filtering and integration, approximately 11,000 detected nuclei were clustered into 4 main groups, with ~75% of nuclei identified as adipocytes (Figure 5A, top). Although two clusters of adipocyte nuclei could initially be resolved from peak clustering and predicted with label transfer from snRNA-seq (Figure 5A, bottom), very few significant DARs discriminated the 2 clusters at any time point with the parameters used; therefore all adipocyte nuclei were combined for further analysis (Figure 5A, top). The reason for this discrepancy is currently unknown, although snATAC-seq data are much sparser than snRNA-seq or bulk-seq data and thus less sensitive at equivalent read depths. It is possible that a much greater sequencing depth would reveal more differences. Additionally, the different populations detected by snRNA-seq may be due to differences in occupancy of specific transcriptional regulators rather than the net overall effect of chromatin accessibility. However, this was not further explored in this study. Marker gene activities for the nuclei clusters are shown in Figure 5B, and a heatmap of top gene activities defining the integrated clusters is depicted in Suppl. Fig. 7B. Fragment quality metrics, transcription start site enrichment, and peak annotation distribution graphs for adipocyte nuclei are displayed in Figure 5C.
Figure 5.
Postnatal development influences chromatin accessibility in iWAT adipocytes. Adipocyte nuclei were isolated from the inguinal white adipose tissue of C57BL/6J male mice at postnatal days 06, 18, and 56 and processed for snATAC-seq using the 10X Genomics platform as described in the Methods. (A) t-SNE plots of integrated snATAC-seq libraries displaying cell clusters identified from (top) the peak matrix and (bottom) following integration and label transfer with our snRNA-seq dataset. Some differences in cluster labeling between datasets were due to differences in the presence of contaminating cells within each cell preparation. (B) t-SNE plots displaying marker gene activity from the integrated snATAC-seq libraries. (C) Fragment length histogram of the first 10 Mbp on chromosome 1, transcription start site enrichment plot, and a pie chart displaying peak annotation distribution. (D) Line graph displaying the number of differentially accessible regions (DARs; P < 0.05; average logFC > 0.25; min 15% of nuclei) detected in adipocyte nuclei across postnatal development and gene ontology of annotated DARs. (E) Pseudo-bulk accessibility tracks for select genes affected by postnatal development in adipocytes. Shaded areas (hand-drawn) indicate peaks that were differentially accessible.
In contrast to stromal cells, the vast majority (~90%) of DARs detected in adipocyte nuclei resulted from a gradual increase in chromatin accessibility between PND06 and PND56 (Figure 5D; Suppl. Fig. 9), with only a few peaks being more accessible at PND06. As shown in Figure 5D, adipocyte maturation was associated with increased accessibility in cis regulatory regions associated with inflammation/immune response, locomotion/cell motility, and estrogen and ROS metabolism. Bulk accessibility tracks for the imprinted genes Mest and Copg2, and the proinflammatory cytokine Il6 are displayed in Figure 5E.
We next looked for enrichment of transcription factor motifs in the DARs from both ASC and adipocytes as a function of developmental time. Despite a similar number of regions differentially accessible in ASC, ~2–5X more motifs were predicted within these DARs at PND06 compared to those found at PND56 (Suppl. Tables 2 and 3; Figure 6; P < 0.05, FC > 1.65). In several cases, variations in motif enrichment corresponded to transcription factor expression levels among ASC subtypes (e.g., Pparg in ASC1a and Creb5 in ASC2) and/or across development (e.g., Plagl1, Twist2, Hoxc9; Figure 6B). Interestingly, the zinc-finger protein PLAGL1 regulates several genes in the IGN [72,73], and the pattern of mRNA expression (Figure 6B) and enhancer accessibility (Figure 6A) suggests direct involvement in at least some of the postnatal decline in IG expression in ASC.
Figure 6.
Motif activity, gene expression, and transcription factor (TF) footprints within DARs from (A–C) stromal vascular and (D–F) adipocyte nuclei as detected by snATAC-seq. Nuclei were isolated from SVC and floating adipocytes in inguinal white adipose tissue of C57BL/6J mice at select developmental time points (PND 06, 18, 56). snATAC-seq was performed using the 10X Genomics platform, as described in the Methods. (A) Heatmap displaying per cell motif activity in ASC clusters, (B) scRNA-seq expression of transcription factors that bind enriched motifs within stromal cell clusters, and (C) TF footprints of select motifs in stromal cells. (D) Heatmap displaying per cell motif activity in adipocyte nuclei, (E) snRNA-seq expression of transcription factors that bind enriched motifs, and (F) TF footprints of select motifs in adipocyte nuclei affected by time.
At PND56, the most significantly enriched motif predicted within DARs was for the androgen receptor (Ar). The glucocorticoid receptor (NR3C1) and AR share similar DNA motif sequences, and both are also expressed in stromal cells by scRNA-seq (Figure 6B). However, only Ar mRNA increased in a development-dependent manner (Figure 6B), indicating that the potential for interacting functions with NR3C1 during development [74]. When comparing motif enrichment among ASC subtypes, Sp1- and Creb5-binding motifs were more specifically enriched in ASC2, Nfatc2 in ASC1b, and Pparg:Rxr in ASC1a at PND06 (Suppl. Table 2). At PND56, Atf and Creb5 were more ASC2-specific, and Stat5a/b, Bach1::Mafk, and Nfe2l2 motifs were significantly enriched in DARs within the ASC1a/b-populations (Suppl. Table 2).
Adipocyte nuclei exhibited a more limited number of predicted motifs within DARs (Suppl. Table 3). Among these, the androgen receptor (Ar), NR3C1, Smad2::Smad3, Bach1:Mafk, Stat proteins (i.e., Stat5a/b, Stat4, Stat6), NFKB1, and the oxidation-responsive Nfe2l2 (also known as NRF2) were more enriched in DARs from adults (Figure 6D). By comparison, motifs for Pparg:Rxr and Plagl1 were more highly enriched within DARs at PND06. Many of these transcription factors have been shown to play a role in adipogenesis (i.e., STAT5A, STAT5B, NR3C1), immune responses (i.e., NFKB1, BACH1, AR, NR3C1), and oxidative stress signaling (i.e., BACH1, NFKB1, NFE2L2) [[75], [76], [77], [78], [79], [80], [81], [82], [83]], consistent with gene expression changes found in scRNA-seq (Figure 6E). A heatmap of motif activities for both cellular fractions is displayed in Figure 6A,D. Additionally, TF footprints, an indicator of TF binding, are presented in Figure 6C,F. In general, footprints showed stronger binding to motifs at PND56 compared to PND06 in both cell fractions, except for Creb5 (Figure 6C). This may be due to transient expression of many transcription factors early in development as well as changes in chromatin architecture. A full listing of enriched motifs in the cellular fractions are presented in the Supplement (Suppl. Tables 2 and 3).
4. Discussion
In the current investigation, we used a large-scale single cell functional genomics approach to evaluate transcriptional networks involved in subcutaneous white adipose development. Although heterogeneity among stromal cell types has been previously reported [19,[24], [25], [26],30,31], our study advances knowledge of WAT development by showing the following: (1) the transcriptomes of adipose stromal cells change during iWAT establishment, (2) heterogenous populations of adipocyte nuclei arise early in the postnatal period and further evolve over time, and (3) the chromatin accessibility landscape of both stromal cells and adipocytes undergoes reorganization during the postnatal period. Collectively, these data provide a framework for understanding signaling events during WAT establishment at a single cell resolution.
The perinatal and early postnatal periods are considered critical times for WAT establishment [34]. Adipocyte cell number is thought to be “set” by late adolescence [84,85], and further expansion of subcutaneous depots occurs mostly by hypertrophic growth [34,40,86]. Indeed, we show that proliferation of ASC extends throughout a period up until weaning, after which few proliferating cells are observed. Additionally, we found that this critical period was positively associated with the expression of several imprinted genes. Imprinted genes are part of a large network (IGN) of both mono- and biallelically expressed genes that are often co-regulated and critical for normal growth and development [62,73,87,88]. A postnatal decline in imprinted gene expression has been reported in several tissues, including the liver, lung, and kidney, paralleling somatic growth deceleration [72,73,[89], [90], [91], [92]] but not appearing to involve changes in promoter methylation or loss of imprinting [90,93]. It has therefore been suggested that IGs are part of a common genetic program that regulates body/organ size [73,[89], [90], [91]].
We found that IGs were much more highly expressed in ASC compared to adipocytes, and that their expression was greatest during establishment of the inguinal fat pad. The activity of IGs has been mostly interpreted as a complex competition for the control of resources that affect the inheritance of maternal and paternal genomes [88,94,95]. Thus, maternally expressed alleles generally restrict growth, while paternally expressed alleles generally promote it [96]. In this context, it is interesting to note that the imprinted genes that are active in adipose tissue establishment and suppressed in adults are known to play important roles in preadipocyte proliferation and hypertrophy [63,97,98], parameters that could easily affect resource allocation and inclusive fitness. In one intriguing example, paternally expressed Mest and maternally expressed Klf14 are found within the same locus on chromosome 6 (7 in humans). Mest encodes a protein that belongs to the α/β-hydrolase superfamily of enzymes that promotes adipocyte hypertrophy [63,64], and expression is highly predictive of diet-induced fat accumulation in mice [98]. On the other hand, Klf14 is a transcriptional repressor that restrains preadipocyte differentiation in part by inhibiting the activity of SREBF1, a transcriptional activator of fatty acid synthesis [[99], [100], [101]]. Recently, a single nucleotide polymorphism in human KLF4 has been shown to regulate body fat distribution in a sex-specific fashion [[99], [100], [101]]. It is interesting that transcripts for both genes are enriched in mouse ASC1a/b and are essentially silenced by weaning. It is thus tempting to speculate that the expression of competing genes in the IGN during tissue establishment might set the stage for how the tissue adapts to energetic challenges in adults. Interestingly, IGs have also been linked to stem cell plasticity during WAT establishment [[99], [100], [101]]. Activating PDGFRA signaling within progenitor (perivascular) cells caused a switch from an adipogenic to a profibrotic phenotype that was associated with altered expression of several IGs [[99], [100], [101]]. Thus perinatal expression of the IGN is undoubtedly linked to numerous developmental processes.
In addition to IGs, we found general as well as ASC-intrinsic changes in expression profiles. For example, genes associated with cytokine signaling and remodeling of the ECM (i.e., Egfr, Cxcl1, Ar) increased more globally in stromal cells during postnatal development. Gene expression changes in ASC2, a subpopulation that comprises the fascia [25,32], were somewhat distinctive and included induction of components involved in phospholipase metabolism (i.e., Pla1a, Smpd3, Ptgs2, Anxa3) as well as cytokine signaling (i.e., Il33, Il18, Il1r2). Il33 in particular was recently shown to promote anti-inflammatory processes in the gWAT by inducing type 2 cytokines and thereby improving obesity-induced metabolic profiles [[99], [100], [101]]. These changes suggest regulatory functions of ASC2 that might shape immune cell recruitment and homeostasis during iWAT establishment.
Subcutaneous (inguinal) and retroperitoneal depots undergo a transient “browning” prior to weaning in mice [13,14,103]. Whether these adipocytes arise from distinctive progenitor cells or following adipocyte differentiation has not been fully resolved. However, developmentally-derived brown adipocytes can be reinstated in adults following acute adrenergic stimulation [7,8,[10], [11], [12]]. Despite identifying brown adipocyte markers in bulk RNA-seq at PND06 and PND18, we were unable to clearly differentiate a Ucp1+ adipocyte nuclei cluster by snRNA-seq. Furthermore, although bipotent ASC progenitors have been described during catabolic remodeling in the gWAT [[99], [100], [101]], a similar brown preadipocyte population (Ucp1+) was not resolved in the iWAT during development. Nonetheless, we identified at least 2 clusters of adipocyte nuclei following integration of datasets that become more transcriptionally diverse during adolescence (PND56). In particular, Adip1 was enriched for markers involved in immune, inflammatory, and growth factor signaling, whereas Adip2 expressed markers involved in glucose and fatty acid metabolism. Heterogeneous adipocyte populations detected by snRNA-seq were also recently described in adult iWAT and gWAT [27,30]. For example, Rajbhandari et al. [[99], [100], [101]] reported an enhanced thermogenic gene program in iWAT adipocytes following genetic deletion of Il10Ra. Our adipocyte clusters are similar to their “thermogenic adipocytes” (cluster 9) that express the general markers Ascl1 and Plin4 [[99], [100], [101]]. They also found a modest induction of Ucp1 and Cidea following cold-induced stress, indicating that this cluster captured some brown adipocytes. Sárvári and colleagues [[99], [100], [101]] isolated total nuclei from the gWAT of lean and HFD-fed mice and identified 3 subpopulations of adipocyte nuclei. Furthermore, they found a shift in the transcriptomes of adipocytes during obesity, including a downregulation of lipogenic genes and enhancement of genes related to stress and immune function.
Using snATAC-seq, we also demonstrate that postnatal changes in RNA were associated with changes in chromatin accessibility, although the overall effects were differentiation-dependent. In stromal cells, there was a notable difference in the enhancer landscape from early development (PND06) to adolescence/adulthood (PND56). This involved a sharp decline in accessibility of perinatal enhancers and a gradual acquisition of adult pattern accessibility. By comparison, most epigenetic changes in adipocytes were due to an increase in accessibility of enhancers with age. These findings suggest that most of the changes in chromatin remodeling that occur during adipogenesis [104] is already established in perinatal adipocytes, and further changes are likely due to the microenvironment.
Overall, the DARs in stromal cells at PND06 were also more diversified in predicted TF motifs, especially for ASC2, although not all associated genes were expressed at the time points evaluated. Plagl1 was highly expressed in ASC at early time points by bulk and scRNA-seq, and has been shown in part to regulate some genes within the IGN, such as the H19-Igf2 locus and ECM genes [72,73]. In adult mice, DARs were strongly enriched for androgen (Ar) and glucocorticoid receptor (GR, NR3C1) motifs in both ASC and adipocytes. Sex hormones are known to affect body fat composition and distribution, thereby influencing metabolic disease risk [[105], [106], [107], [108]]. In males, testosterone reduces fat mass in visceral and subcutaneous depots, although AR signaling may play a more dominant role in preventing expansion of subcutaneous fat [109,110]. Glucocorticoids promote adipogenesis and influence lipolysis [[111], [112], [113]]; however, overall effects of each receptor are likely dependent on abundance and ligand availability. In this regard, it is interesting that expression of Ar, but not Nr3c1, increased in both ASC and adipocytes with age (~3–6 fold). Additionally, the mRNA for HSD11β1, which converts cortisone to the active metabolite cortisol [114], increased in adipocytes as well as ASC1a at PND56, indicating that there may be age and spatial differences in nuclear receptor signaling. Finally, although our focus was on development-dependent changes, we also found evidence of cell-specific motif activity that may impart at least some of the ASC phenotypes previously described [25,26]. These include DNA binding motifs for Creb5 (ASC2), Nfatc2 (ASC1b), and Pparg::Rxra and FOS::JUN (ASC1a/b).
Collectively, our findings highlight transcriptional and epigenomic changes arising during establishment of subcutaneous WAT at a single cell resolution. Importantly, we find that IGs are highly expressed during early postnatal growth and are inversely related to expression of immunomodulatory genes. We also demonstrate the ability to deconvolve adipose tissue cell types based on chromatin accessibility. Furthermore, through integration with scRNA-seq, it is possible to extract probable regulatory motifs based on cell-specific gene expression. Future research to validate some of the findings and further explore development is warranted. The strength of single cell/nuclei functional genomics is the ability to retrospectively dissect cellular complexity without isolation of specific populations. However, in order to specify these populations/states prospectively, one would need to apply intersection genetics that combines multiple genetic and temporal controls. While there are emerging examples [115], these are currently limited to 2 genetic drivers and one temporal regulator. One can anticipate that improvements in gene editing technologies (CRISPR-Cas9) might overcome this barrier in the near future.
A potential limitation to this study is that the process of cell isolation could alter some gene expression patterns [116]. It is important to note, however, that the method of isolation was the same for all experiments and thus does not likely explain the developmental changes reported here. Additionally, the methods used to isolate and enrich for stromal cells and adipocytes inevitably resulted in some contamination by other cellular subtypes. While such contaminating cells are clearly resolved in single cell analyses, this minor contamination might have contributed to some of the gene expression changes observed in bulk RNA-seq analyses. Notwithstanding these limitations, our findings expand on previous work [13,36,41,102,[117], [118], [119]] and provide a resource that will lead to a better understanding of WAT development and metabolic disease origins.
Funding
This work was supported by the National Institute of Diabetes and Digestive and Kidney Disease (NIDDK) [JGG; R01DK062292] and the National Institute of Environmental Health Sciences [P30 ES020957].
Author contributions
E.A.R. performed experiments, analyzed data, and co-wrote the manuscript. V.D.R. and R.B.B. assisted in experiments and data collection. R.P.R. supervised and assisted in single cell data analysis (sc/nRNA-seq and snATAC-seq). J.G.G. conceived of the experiment, assisted in data analysis, and co-wrote the manuscript. All authors reviewed and edited the manuscript.
Acknowledgements.
The authors thank Dr. Douglas Ruden for his careful review of this manuscript and members of CIMER for useful suggestions.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2021.101307.
Conflict of interest
The authors declare no conflict of interest.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Multimedia component 1
Multimedia component 2
Multimedia component 3
Multimedia component 4
References
- 1.Choe S.S., Huh J.Y., Hwang I.J., Kim J.I., Kim J.B. Adipose tissue remodeling: its role in energy metabolism and metabolic disorders. Frontiers in Endocrinology. 2016;7:30. doi: 10.3389/fendo.2016.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luo L., Liu M. Adipose tissue in control of metabolism. Journal of Endocrinology. 2016;231(3):R77–R99. doi: 10.1530/JOE-16-0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang P., Mariman E., Renes J., Keijer J. The secretory function of adipocytes in the physiology of white adipose tissue. Journal of Cellular Physiology. 2008;216(1):3–13. doi: 10.1002/jcp.21386. [DOI] [PubMed] [Google Scholar]
- 4.Coelho M., Oliveira T., Fernandes R. Biochemistry of adipose tissue: an endocrine organ. Archives of Medical Science. 2013;9(2):191–200. doi: 10.5114/aoms.2013.33181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bodis K., Roden M. Energy metabolism of white adipose tissue and insulin resistance in humans. European Journal of Clinical Investigation. 2018;48(11) doi: 10.1111/eci.13017. [DOI] [PubMed] [Google Scholar]
- 6.Rosen E.D., Spiegelman B.M. Adipocytes as regulators of energy balance and glucose homeostasis. Nature. 2006;444(7121):847–853. doi: 10.1038/nature05483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenwald M., Perdikari A., Rulicke T., Wolfrum C. Bi-directional interconversion of brite and white adipocytes. Nature Cell Biology. 2013;15(6):659–667. doi: 10.1038/ncb2740. [DOI] [PubMed] [Google Scholar]
- 8.Lee Y.H., Kim S.N., Kwon H.J., Granneman J.G. Metabolic heterogeneity of activated beige/brite adipocytes in inguinal adipose tissue. Scientific Reports. 2017;7:39794. doi: 10.1038/srep39794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu J., Bostrom P., Sparks L.M., Ye L., Choi J.H., Giang A.H. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150(2):366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee K.Y., Luong Q., Sharma R., Dreyfuss J.M., Ussar S., Kahn C.R. Developmental and functional heterogeneity of white adipocytes within a single fat depot. The EMBO Journal. 2019;38(3) doi: 10.15252/embj.201899291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Contreras G.A., Lee Y.H., Mottillo E.P., Granneman J.G. Inducible brown adipocytes in subcutaneous inguinal white fat: the role of continuous sympathetic stimulation. American Journal of Physiology. Endocrinology and Metabolism. 2014;307(9):E793–E799. doi: 10.1152/ajpendo.00033.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y., Paulo E., Wu D., Wu Y., Huang W., Chawla A. Adipocyte liver kinase b1 suppresses beige adipocyte renaissance through class IIa histone deacetylase 4. Diabetes. 2017;66(12):2952–2963. doi: 10.2337/db17-0296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chabowska-Kita A., Kozak L.P. The critical period for brown adipocyte development: genetic and environmental influences. Obesity. 2016;24(2):283–290. doi: 10.1002/oby.21376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chabowska-Kita A., Trabczynska A., Korytko A., Kaczmarek M.M., Kozak L.P. Low ambient temperature during early postnatal development fails to cause a permanent induction of brown adipocytes. The FASEB Journal. 2015;29(8):3238–3252. doi: 10.1096/fj.15-271395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kozak L.P. The genetics of brown adipocyte induction in white fat depots. Frontiers in Endocrinology. 2011;2:64. doi: 10.3389/fendo.2011.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kozak L.P., Koza R.A., Anunciado-Koza R., Mendoza T., Newman S. Inherent plasticity of brown adipogenesis in white fat of mice allows for recovery from effects of post-natal malnutrition. PloS One. 2012;7(2) doi: 10.1371/journal.pone.0030392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eto H., Suga H., Matsumoto D., Inoue K., Aoi N., Kato H. Characterization of structure and cellular components of aspirated and excised adipose tissue. Plastic and Reconstructive Surgery. 2009;124(4):1087–1097. doi: 10.1097/PRS.0b013e3181b5a3f1. [DOI] [PubMed] [Google Scholar]
- 18.Prunet-Marcassus B., Cousin B., Caton D., Andre M., Penicaud L., Casteilla L. From heterogeneity to plasticity in adipose tissues: site-specific differences. Experimental Cell Research. 2006;312(6):727–736. doi: 10.1016/j.yexcr.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 19.Burl R.B., Ramseyer V.D., Rondini E.A., Pique-Regi R., Lee Y.H., Granneman J.G. Deconstructing adipogenesis induced by beta3-adrenergic receptor activation with single-cell expression profiling. Cell Metabolism. 2018;28(2):300–309. doi: 10.1016/j.cmet.2018.05.025. e304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee Y.H., Petkova A.P., Mottillo E.P., Granneman J.G. In vivo identification of bipotential adipocyte progenitors recruited by beta3-adrenoceptor activation and high-fat feeding. Cell Metabolism. 2012;15(4):480–491. doi: 10.1016/j.cmet.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodeheffer M.S., Birsoy K., Friedman J.M. Identification of white adipocyte progenitor cells in vivo. Cell. 2008;135(2):240–249. doi: 10.1016/j.cell.2008.09.036. [DOI] [PubMed] [Google Scholar]
- 22.Berry R., Rodeheffer M.S. Characterization of the adipocyte cellular lineage in vivo. Nature Cell Biology. 2013;15(3):302–308. doi: 10.1038/ncb2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee Y.H., Petkova A.P., Granneman J.G. Identification of an adipogenic niche for adipose tissue remodeling and restoration. Cell Metabolism. 2013;18(3):355–367. doi: 10.1016/j.cmet.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hepler C., Shan B., Zhang Q., Henry G.H., Shao M., Vishvanath L. Identification of functionally distinct fibro-inflammatory and adipogenic stromal subpopulations in visceral adipose tissue of adult mice. Elife. 2018;7 doi: 10.7554/eLife.39636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merrick D., Sakers A., Irgebay Z., Okada C., Calvert C., Morley M.P. Identification of a mesenchymal progenitor cell hierarchy in adipose tissue. Science. 2019;364(6438) doi: 10.1126/science.aav2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwalie P.C., Dong H., Zachara M., Russeil J., Alpern D., Akchiche N. A stromal cell population that inhibits adipogenesis in mammalian fat depots. Nature. 2018;559(7712):103–108. doi: 10.1038/s41586-018-0226-8. [DOI] [PubMed] [Google Scholar]
- 27.Sarvari A.K., Van Hauwaert E.L., Markussen L.K., Gammelmark E., Marcher A.B., Ebbesen M.F. Plasticity of epididymal adipose tissue in response to diet-induced obesity at single-nucleus resolution. Cell Metabolism. 2021;33(2):437–453. doi: 10.1016/j.cmet.2020.12.004. e435. [DOI] [PubMed] [Google Scholar]
- 28.Jaitin D.A., Adlung L., Thaiss C.A., Weiner A., Li B., Descamps H. Cell; 2019. Lipid-associated macrophages control metabolic homeostasis in a trem2-dependent manner. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hill D.A., Lim H.W., Kim Y.H., Ho W.Y., Foong Y.H., Nelson V.L. Distinct macrophage populations direct inflammatory versus physiological changes in adipose tissue. Proceedings of the National Academy of Sciences of the United States of America. 2018;115(22):E5096–E5105. doi: 10.1073/pnas.1802611115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rajbhandari P., Arneson D., Hart S.K., Ahn I.S., Diamante G., Santos L.C. Single cell analysis reveals immune cell-adipocyte crosstalk regulating the transcription of thermogenic adipocytes. Elife. 2019;8 doi: 10.7554/eLife.49501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cho D.S., Lee B., Doles J.D. Refining the adipose progenitor cell landscape in healthy and obese visceral adipose tissue using single-cell gene expression profiling. Life Sci Alliance. 2019;2(6) doi: 10.26508/lsa.201900561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rondini E.A., Granneman J.G. Single cell approaches to address adipose tissue stromal cell heterogeneity. Biochemical Journal. 2020;477(3):583–600. doi: 10.1042/BCJ20190467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferrero R., Rainer P., Deplancke B. Toward a consensus view of mammalian adipocyte stem and progenitor cell heterogeneity. Trends in Cell Biology. 2020;30(12):937–950. doi: 10.1016/j.tcb.2020.09.007. [DOI] [PubMed] [Google Scholar]
- 34.Holtrup B., Church C.D., Berry R., Colman L., Jeffery E., Bober J. Puberty is an important developmental period for the establishment of adipose tissue mass and metabolic homeostasis. Adipocyte. 2017;6(3):224–233. doi: 10.1080/21623945.2017.1349042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poissonnet C.M., Burdi A.R., Bookstein F.L. Growth and development of human adipose tissue during early gestation. Early Human Development. 1983;8(1):1–11. doi: 10.1016/0378-3782(83)90028-2. [DOI] [PubMed] [Google Scholar]
- 36.Kozak L.P., Newman S., Chao P.M., Mendoza T., Koza R.A. The early nutritional environment of mice determines the capacity for adipose tissue expansion by modulating genes of caveolae structure. PloS One. 2010;5(6) doi: 10.1371/journal.pone.0011015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poissonnet C.M., LaVelle M., Burdi A.R. Growth and development of adipose tissue. The Journal of Pediatrics. 1988;113(1 Pt 1):1–9. doi: 10.1016/s0022-3476(88)80520-1. [DOI] [PubMed] [Google Scholar]
- 38.Poissonnet C.M., Burdi A.R., Garn S.M. The chronology of adipose tissue appearance and distribution in the human fetus. Early Human Development. 1984;10(1–2):1–11. doi: 10.1016/0378-3782(84)90106-3. [DOI] [PubMed] [Google Scholar]
- 39.Sun C., Berry W.L., Olson L.E. PDGFRalpha controls the balance of stromal and adipogenic cells during adipose tissue organogenesis. Development. 2017;144(1):83–94. doi: 10.1242/dev.135962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Q.A., Tao C., Gupta R.K., Scherer P.E. Tracking adipogenesis during white adipose tissue development, expansion and regeneration. Nature Medicine. 2013;19(10):1338–1344. doi: 10.1038/nm.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Birsoy K., Berry R., Wang T., Ceyhan O., Tavazoie S., Friedman J.M. Analysis of gene networks in white adipose tissue development reveals a role for ETS2 in adipogenesis. Development. 2011;138(21):4709–4719. doi: 10.1242/dev.067710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Han J., Lee J.E., Jin J., Lim J.S., Oh N., Kim K. The spatiotemporal development of adipose tissue. Development. 2011;138(22):5027–5037. doi: 10.1242/dev.067686. [DOI] [PubMed] [Google Scholar]
- 43.Ge S.X., Son E.W., Yao R. iDEP: an integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinformatics. 2018;19(1):534. doi: 10.1186/s12859-018-2486-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oliveros J.C. 2007-2015. Venny. An interactive tool for comparing lists with Venn's diagrams. [Google Scholar]
- 45.Metsalu T., Vilo J. ClustVis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015;43(W1):W566–W570. doi: 10.1093/nar/gkv468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Young M.D., Behjati S. SoupX removes ambient RNA contamination from droplet-based single-cell RNA sequencing data. GigaScience. 2020;9(12) doi: 10.1093/gigascience/giaa151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd Comprehensive integration of single-cell data. Cell. 2019;177(7):1888–1902 e1821. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stuart T., Satija R. Integrative single-cell analysis. Nature Reviews Genetics. 2019;20(5):257–272. doi: 10.1038/s41576-019-0093-7. [DOI] [PubMed] [Google Scholar]
- 49.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nature Biotechnology. 2018;36(5):411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McGinnis C.S., Murrow L.M., Gartner Z.J. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 2019;8(4):329–337. doi: 10.1016/j.cels.2019.03.003. e324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(W1):W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Research. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fang R., Preissl S., Hou X., Lucero J., Wang X., Motamedi A. bioRxiv; 2019. Fast and accurate clustering of single cell epigenomes reveals cis-regulatory elements in rare cell types. [Google Scholar]
- 55.Korsunsky I., Millard N., Fan J., Slowikowski K., Zhang F., Wei K. Fast, sensitive and accurate integration of single-cell data with Harmony. Nature Methods. 2019;16(12):1289–1296. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stuart T., Srivastava A., Lareau C., Satija R. bioRxiv; 2020. Multimodal single-cell chromatin analysis with Signac. [Google Scholar]
- 57.McLean C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T., Lowe C.B. GREAT improves functional interpretation of cis-regulatory regions. Nature Biotechnology. 2010;28(5):495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fornes O., Castro-Mondragon J.A., Khan A., van der Lee R., Zhang X., Richmond P.A. JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Research. 2020;48(D1):D87–D92. doi: 10.1093/nar/gkz1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schep A.N., Wu B., Buenrostro J.D., Greenleaf W.J. chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nature Methods. 2017;14(10):975–978. doi: 10.1038/nmeth.4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu G., Wang L.G., He Q.Y. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015;31(14):2382–2383. doi: 10.1093/bioinformatics/btv145. [DOI] [PubMed] [Google Scholar]
- 61.Eguchi J., Yan Q.W., Schones D.E., Kamal M., Hsu C.H., Zhang M.Q. Interferon regulatory factors are transcriptional regulators of adipogenesis. Cell Metabolism. 2008;7(1):86–94. doi: 10.1016/j.cmet.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tycko B., Morison I.M. Physiological functions of imprinted genes. Journal of Cellular Physiology. 2002;192(3):245–258. doi: 10.1002/jcp.10129. [DOI] [PubMed] [Google Scholar]
- 63.Takahashi M., Kamei Y., Ezaki O. Mest/Peg1 imprinted gene enlarges adipocytes and is a marker of adipocyte size. American Journal of Physiology. Endocrinology and Metabolism. 2005;288(1):E117–E124. doi: 10.1152/ajpendo.00244.2004. [DOI] [PubMed] [Google Scholar]
- 64.Jura M., Jaroslawska J., Chu D.T., Kozak L.P. Mest and Sfrp5 are biomarkers for healthy adipose tissue. Biochimie. 2016;124:124–133. doi: 10.1016/j.biochi.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 65.Karamariti E., Zhai C., Yu B., Qiao L., Wang Z., Potter C.M.F. DKK3 (dickkopf 3) alters atherosclerotic plaque phenotype involving vascular progenitor and fibroblast differentiation into smooth muscle cells. Arteriosclerosis, Thrombosis, and Vascular Biology. 2018;38(2):425–437. doi: 10.1161/ATVBAHA.117.310079. [DOI] [PubMed] [Google Scholar]
- 66.Stohn J.P., Wang Q., Siviski M.E., Kennedy K., Jin Y.R., Kacer D. Cthrc1 controls adipose tissue formation, body composition, and physical activity. Obesity. 2015;23(8):1633–1642. doi: 10.1002/oby.21144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mahlakoiv T., Flamar A.L., Johnston L.K., Moriyama S., Putzel G.G., Bryce P.J. Stromal cells maintain immune cell homeostasis in adipose tissue via production of interleukin-33. Sci Immunol. 2019;4(35) doi: 10.1126/sciimmunol.aax0416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dahlgren M.W., Jones S.W., Cautivo K.M., Dubinin A., Ortiz-Carpena J.F., Farhat S. Adventitial stromal cells define group 2 innate lymphoid cell tissue niches. Immunity. 2019;50(3):707–722. doi: 10.1016/j.immuni.2019.02.002. e706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Peters V.A., Joesting J.J., Freund G.G. IL-1 receptor 2 (IL-1R2) and its role in immune regulation. Brain, Behavior, and Immunity. 2013;32:1–8. doi: 10.1016/j.bbi.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lin Y., Wen T., Meng X., Wu Z., Zhao L., Wang P. The mouse Mageb18 gene encodes a ubiquitously expressed type I MAGE protein and regulates cell proliferation and apoptosis in melanoma B16-F0 cells. Biochemical Journal. 2012;443(3):779–788. doi: 10.1042/BJ20112054. [DOI] [PubMed] [Google Scholar]
- 71.Muller S., Perdikari A., Dapito D.H., Sun W., Wollscheid B., Balaz M. ESRRG and PERM1 govern mitochondrial conversion in brite/beige adipocyte formation. Frontiers in Endocrinology. 2020;11:387. doi: 10.3389/fendo.2020.00387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Varrault A., Dantec C., Le Digarcher A., Chotard L., Bilanges B., Parrinello H. Identification of Plagl1/Zac1 binding sites and target genes establishes its role in the regulation of extracellular matrix genes and the imprinted gene network. Nucleic Acids Research. 2017;45(18):10466–10480. doi: 10.1093/nar/gkx672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Varrault A., Gueydan C., Delalbre A., Bellmann A., Houssami S., Aknin C. Zac1 regulates an imprinted gene network critically involved in the control of embryonic growth. Developmental Cell. 2006;11(5):711–722. doi: 10.1016/j.devcel.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 74.Spaanderman D.C.E., Nixon M., Buurstede J.C., Sips H.C., Schilperoort M., Kuipers E.N. Androgens modulate glucocorticoid receptor activity in adipose tissue and liver. Journal of Endocrinology. 2018;240(1):51–63. doi: 10.1530/JOE-18-0503. [DOI] [PubMed] [Google Scholar]
- 75.Park Y.K., Ge K. Glucocorticoid receptor accelerates, but is dispensable for, adipogenesis. Molecular and Cellular Biology. 2017;37(2) doi: 10.1128/MCB.00260-16. e00260-e00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nanbu-Wakao R., Morikawa Y., Matsumura I., Masuho Y., Muramatsu M.A., Senba E. Stimulation of 3T3-L1 adipogenesis by signal transducer and activator of transcription 5. Molecular Endocrinology. 2002;16(7):1565–1576. doi: 10.1210/mend.16.7.0862. [DOI] [PubMed] [Google Scholar]
- 77.Wakao H., Wakao R., Oda A., Fujita H. Constitutively active Stat5A and Stat5B promote adipogenesis. Environmental Health and Preventive Medicine. 2011;16(4):247–252. doi: 10.1007/s12199-010-0193-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lai J.J., Lai K.P., Zeng W., Chuang K.H., Altuwaijri S., Chang C. Androgen receptor influences on body defense system via modulation of innate and adaptive immune systems: lessons from conditional AR knockout mice. American Journal Of Pathology. 2012;181(5):1504–1512. doi: 10.1016/j.ajpath.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang X., Guo J., Wei X., Niu C., Jia M., Li Q. Bach1: function, regulation, and involvement in disease. Oxid Med Cell Longev. 2018;2018:1347969. doi: 10.1155/2018/1347969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhou Y., Wu H., Zhao M., Chang C., Lu Q. The Bach family of transcription factors: a comprehensive review. Clinical Reviews in Allergy and Immunology. 2016;50(3):345–356. doi: 10.1007/s12016-016-8538-7. [DOI] [PubMed] [Google Scholar]
- 81.John K., Marino J.S., Sanchez E.R., Hinds T.D., Jr. The glucocorticoid receptor: cause of or cure for obesity? American Journal of Physiology. Endocrinology and Metabolism. 2016;310(4):E249–E257. doi: 10.1152/ajpendo.00478.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schneider K.S., Chan J.Y. Emerging role of Nrf2 in adipocytes and adipose biology. Adv Nutr. 2013;4(1):62–66. doi: 10.3945/an.112.003103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tonelli C., Chio I.I.C., Tuveson D.A. Transcriptional regulation by Nrf2. Antioxidants and Redox Signaling. 2018;29(17):1727–1745. doi: 10.1089/ars.2017.7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Spalding K.L., Arner E., Westermark P.O., Bernard S., Buchholz B.A., Bergmann O. Dynamics of fat cell turnover in humans. Nature. 2008;453(7196):783–787. doi: 10.1038/nature06902. [DOI] [PubMed] [Google Scholar]
- 85.Knittle J.L., Timmers K., Ginsberg-Fellner F., Brown R.E., Katz D.P. The growth of adipose tissue in children and adolescents. Cross-sectional and longitudinal studies of adipose cell number and size. Journal of Clinical Investigation. 1979;63(2):239–246. doi: 10.1172/JCI109295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jeffery E., Church C.D., Holtrup B., Colman L., Rodeheffer M.S. Rapid depot-specific activation of adipocyte precursor cells at the onset of obesity. Nature Cell Biology. 2015;17(4):376–385. doi: 10.1038/ncb3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cassidy F.C., Charalambous M. Genomic imprinting, growth and maternal-fetal interactions. Journal of Experimental Biology. 2018;221(Pt Suppl 1) doi: 10.1242/jeb.164517. [DOI] [PubMed] [Google Scholar]
- 88.Millership S.J., Van de Pette M., Withers D.J. Genomic imprinting and its effects on postnatal growth and adult metabolism. Cellular and Molecular Life Sciences. 2019;76(20):4009–4021. doi: 10.1007/s00018-019-03197-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Finkielstain G.P., Forcinito P., Lui J.C., Barnes K.M., Marino R., Makaroun S. An extensive genetic program occurring during postnatal growth in multiple tissues. Endocrinology. 2009;150(4):1791–1800. doi: 10.1210/en.2008-0868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lui J.C., Finkielstain G.P., Barnes K.M., Baron J. An imprinted gene network that controls mammalian somatic growth is down-regulated during postnatal growth deceleration in multiple organs. American Journal of Physiology - Regulatory, Integrative and Comparative Physiology. 2008;295(1):R189–R196. doi: 10.1152/ajpregu.00182.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lui J.C., Forcinito P., Chang M., Chen W., Barnes K.M., Baron J. Coordinated postnatal down-regulation of multiple growth-promoting genes: evidence for a genetic program limiting organ growth. The FASEB Journal. 2010;24(8):3083–3092. doi: 10.1096/fj.09-152835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lui J.C., Baron J. Mechanisms limiting body growth in mammals. Endocrine Reviews. 2011;32(3):422–440. doi: 10.1210/er.2011-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Al Adhami H., Evano B., Le Digarcher A., Gueydan C., Dubois E., Parrinello H. A systems-level approach to parental genomic imprinting: the imprinted gene network includes extracellular matrix genes and regulates cell cycle exit and differentiation. Genome Research. 2015;25(3):353–367. doi: 10.1101/gr.175919.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wilkins J.F., Haig D. What good is genomic imprinting: the function of parent-specific gene expression. Nature Reviews Genetics. 2003;4(5):359–368. doi: 10.1038/nrg1062. [DOI] [PubMed] [Google Scholar]
- 95.Moore T., Haig D. Genomic imprinting in mammalian development: a parental tug-of-war. Trends in Genetics. 1991;7(2):45–49. doi: 10.1016/0168-9525(91)90230-N. [DOI] [PubMed] [Google Scholar]
- 96.Moore G.E., Ishida M., Demetriou C., Al-Olabi L., Leon L.J., Thomas A.C. The role and interaction of imprinted genes in human fetal growth. Philosophical Transactions of the Royal Society of London B Biological Sciences. 2015;370(1663):20140074. doi: 10.1098/rstb.2014.0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mortensen S.B., Jensen C.H., Schneider M., Thomassen M., Kruse T.A., Laborda J. Membrane-tethered delta-like 1 homolog (DLK1) restricts adipose tissue size by inhibiting preadipocyte proliferation. Diabetes. 2012;61(11):2814–2822. doi: 10.2337/db12-0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nikonova L., Koza R.A., Mendoza T., Chao P.M., Curley J.P., Kozak L.P. Mesoderm-specific transcript is associated with fat mass expansion in response to a positive energy balance. The FASEB Journal. 2008;22(11):3925–3937. doi: 10.1096/fj.08-108266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Magana M.M., Osborne T.F. Two tandem binding sites for sterol regulatory element binding proteins are required for sterol regulation of fatty-acid synthase promoter. Journal of Biological Chemistry. 1996;271(51):32689–32694. doi: 10.1074/jbc.271.51.32689. [DOI] [PubMed] [Google Scholar]
- 100.Latasa M.J., Moon Y.S., Kim K.H., Sul H.S. Nutritional regulation of the fatty acid synthase promoter in vivo: sterol regulatory element binding protein functions through an upstream region containing a sterol regulatory element. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(19):10619–10624. doi: 10.1073/pnas.180306597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Small K.S., Todorcevic M., Civelek M., El-Sayed Moustafa J.S., Wang X., Simon M.M. Regulatory variants at KLF14 influence type 2 diabetes risk via a female-specific effect on adipocyte size and body composition. Nature Genetics. 2018;50(4):572–580. doi: 10.1038/s41588-018-0088-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Iwayama T., Steele C., Yao L., Dozmorov M.G., Karamichos D., Wren J.D. PDGFRalpha signaling drives adipose tissue fibrosis by targeting progenitor cell plasticity. Genes & Development. 2015;29(11):1106–1119. doi: 10.1101/gad.260554.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Guerra C., Koza R.A., Yamashita H., Walsh K., Kozak L.P. Emergence of brown adipocytes in white fat in mice is under genetic control. Effects on body weight and adiposity. Journal of Clinical Investigation. 1998;102(2):412–420. doi: 10.1172/JCI3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Siersbaek R., Nielsen R., John S., Sung M.H., Baek S., Loft A. Extensive chromatin remodelling and establishment of transcription factor 'hotspots' during early adipogenesis. The EMBO Journal. 2011;30(8):1459–1472. doi: 10.1038/emboj.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Elbers J.M., Asscheman H., Seidell J.C., Gooren L.J. Effects of sex steroid hormones on regional fat depots as assessed by magnetic resonance imaging in transsexuals. American Journal of Physiology. 1999;276(2):E317–E325. doi: 10.1152/ajpendo.1999.276.2.E317. [DOI] [PubMed] [Google Scholar]
- 106.He Z., Rankinen T., Leon A.S., Skinner J.S., Tchernof A., Bouchard C. Plasma steroids, body composition, and fat distribution: effects of age, sex, and exercise training. International Journal of Obesity. 2018;42(7):1366–1377. doi: 10.1038/s41366-018-0033-1. [DOI] [PubMed] [Google Scholar]
- 107.Pasquali R., Casimirri F., Cantobelli S., Melchionda N., Morselli Labate A.M., Fabbri R. Effect of obesity and body fat distribution on sex hormones and insulin in men. Metabolism. 1991;40(1):101–104. doi: 10.1016/0026-0495(91)90199-7. [DOI] [PubMed] [Google Scholar]
- 108.Carr M.C. The emergence of the metabolic syndrome with menopause. Journal of Clinical Endocrinology & Metabolism. 2003;88(6):2404–2411. doi: 10.1210/jc.2003-030242. [DOI] [PubMed] [Google Scholar]
- 109.Sebo Z.L., Rodeheffer M.S. Testosterone metabolites differentially regulate obesogenesis and fat distribution. Mol Metab. 2021;44:101141. doi: 10.1016/j.molmet.2020.101141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sato T., Matsumoto T., Yamada T., Watanabe T., Kawano H., Kato S. Late onset of obesity in male androgen receptor-deficient (AR KO) mice. Biochemical and Biophysical Research Communications. 2003;300(1):167–171. doi: 10.1016/s0006-291x(02)02774-2. [DOI] [PubMed] [Google Scholar]
- 111.Xu C., He J., Jiang H., Zu L., Zhai W., Pu S. Direct effect of glucocorticoids on lipolysis in adipocytes. Molecular Endocrinology. 2009;23(8):1161–1170. doi: 10.1210/me.2008-0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Peckett A.J., Wright D.C., Riddell M.C. The effects of glucocorticoids on adipose tissue lipid metabolism. Metabolism. 2011;60(11):1500–1510. doi: 10.1016/j.metabol.2011.06.012. [DOI] [PubMed] [Google Scholar]
- 113.Campbell J.E., Peckett A.J., D'Souza A M., Hawke T.J., Riddell M.C. Adipogenic and lipolytic effects of chronic glucocorticoid exposure. American Journal of Physiology - Cell Physiology. 2011;300(1):C198–C209. doi: 10.1152/ajpcell.00045.2010. [DOI] [PubMed] [Google Scholar]
- 114.Dammann C., Stapelfeld C., Maser E. Expression and activity of the cortisol-activating enzyme 11beta-hydroxysteroid dehydrogenase type 1 is tissue and species-specific. Chemico-Biological Interactions. 2019;303:57–61. doi: 10.1016/j.cbi.2019.02.018. [DOI] [PubMed] [Google Scholar]
- 115.Han X., Zhang Z., He L., Zhu H., Li Y., Pu W. A suite of new Dre recombinase drivers markedly expands the ability to perform intersectional genetic targeting. Cell Stem Cell. 2021;28(6):1160–1176. doi: 10.1016/j.stem.2021.01.007. e1167. [DOI] [PubMed] [Google Scholar]
- 116.O'Flanagan C.H., Campbell K.R., Zhang A.W., Kabeer F., Lim J.L.P., Biele J. Dissociation of solid tumor tissues with cold active protease for single-cell RNA-seq minimizes conserved collagenase-associated stress responses. Genome Biology. 2019;20(1):210. doi: 10.1186/s13059-019-1830-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kodde A., Engels E., Oosting A., van Limpt K., van der Beek E.M., Keijer J. Maturation of white adipose tissue function in C57bl/6j mice from weaning to young adulthood. Frontiers in Physiology. 2019;10:836. doi: 10.3389/fphys.2019.00836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zietak M., Chabowska-Kita A., Kozak L.P. Brown fat thermogenesis: stability of developmental programming and transient effects of temperature and gut microbiota in adults. Biochimie. 2017;134:93–98. doi: 10.1016/j.biochi.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 119.Suwandhi L., Altun I., Karlina R., Miok V., Wiedemann T., Fischer D. Asc-1 regulates white versus beige adipocyte fate in a subcutaneous stromal cell population. Nature Communications. 2021;12(1):1588. doi: 10.1038/s41467-021-21826-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multimedia component 1
Multimedia component 2
Multimedia component 3
Multimedia component 4
Data Availability Statement
Raw and processed datasets for bulk RNA-seq, scRNA-seq, and snATAC-seq were deposited into NCBI's Gene Expression Omnibus (GEO) under the Series accession number GSE180589.