Summary
Bundibugyo virus (BDBV) is one of four ebolaviruses known to cause disease in humans. Bundibugyo virus disease (BVD) outbreaks occurred in 2007–2008 in Bundibugyo District, Uganda, and in 2012 in Isiro, Province Orientale, Democratic Republic of the Congo. The 2012 BVD outbreak resulted in 38 laboratory-confirmed cases of human infection, 13 of whom died. However, only 4 BDBV specimens from the 2012 outbreak have been sequenced. Here, we provide BDBV sequences from seven additional patients. Analysis of the molecular epidemiology and evolutionary dynamics of the 2012 outbreak with these additional isolates challenges the current hypothesis that the outbreak was the result of a single spillover event. In addition, one patient record indicates that BDBV’s initial emergence in Isiro occurred 50 days earlier than previously accepted. Collectively, this work demonstrates how retrospective sequencing can be used to elucidate outbreak origins and provide epidemiological contexts to a medically relevant pathogen.
Keywords: BDBV, BVD, Bundibugyo ebolavirus, Bundibugyo virus, Bundibugyo virus disease, EBOD, Ebola, Ebola disease, molecular epidemiology
Graphical abstract

Highlights
In 2012, BDBV was circulating earlier than previously appreciated
BDBV genomes from seven additional patients are sequenced
Molecular analyses indicate that multiple spillover events fueled the BVD outbreak
Elucidation of the epidemiology underlying the 2012 Bundibugyo virus disease outbreak in the Democratic Republic of the Congo has been challenging. Hulseberg et al. acquire additional genomic and phylodynamic data indicating that multiple Bundibugyo virus spillover events, some of which occurred much earlier than previously known, contributed to the outbreak.
Introduction
Bundibugyo virus (BDBV; Filoviridae: Ebolavirus: Bundibugyo ebolavirus) causes Bundibugyo virus disease (BVD), a subtype of Ebola disease (EBOD), in humans.1,2 BVD is characterized by an influenza-like prodrome (arthralgia, cough, fever, headaches, myalgia, and nausea) sometimes followed by severe diarrhea and vomiting, maculopapular rash, chest pain, and hemorrhagic manifestations, such as melena and bleeding from mucous membranes and puncture sites.3,4 Low serum concentrations of proinflammatory cytokines (e.g., interleukin-1 alpha [IL-1A], IL-1B, IL-6, and tumor necrosis factor) and high concentrations of anti-inflammatory IL-10 have been associated with lethal outcomes of BDBV infection.5 BVD survivors may suffer from sequelae such as arthralgia, blurred vision, retroorbital pain, and hearing loss.6 As is the case with other ebolaviruses, the reservoir host of BDBV remains unknown, and, accordingly, the ecology of the virus and its transmission mechanism into the human population are poorly understood.7
BDBV was first recognized as a cause of EBOD during a severe disease outbreak in Bundibugyo District, Western Uganda Administrative Region, Uganda, in August 2007.8 Generic PCR assays for known filoviruses failed to confirm the suspected etiology of a cluster of Marburg virus disease or Sudan virus disease cases in the region.3,8, 9, 10 In response, the US Centers for Disease Control and Prevention (CDC) obtained the complete viral genome of a novel etiological agent, BDBV (GenBank: FJ217161), from an isolate (200706291) recovered in Vero E6 cell cultures and developed a BDBV-specific diagnostic test.10 By the time the outbreak was declared over in February 2008, BDBV had caused 100 probable or confirmed cases of BVD and ∼40 deaths.4
In early August 2012, BDBV reemerged ∼400 km northwest of Bundibugyo District, in the Isiro Health Region, Haut-Uélé District, Province Orientale, Democratic Republic of the Congo (COD).11 The CDC, in collaboration with the COD Ministry of Health (MoH) and the Public Health Agency of Canada (PHAC), established a field laboratory in Isiro to assist with BVD diagnosis. On August 17, the day after the first laboratory confirmation of BDBV infection, the MoH declared an outbreak of BVD in Isiro. The outbreak continued in the region surrounding Isiro until the MoH declared it over on November 26, 2012, ∼42 days after the last positive PCR test result in mid-October.11,12 The lethality for laboratory-confirmed cases and deaths was 34% (38 cases and 13 deaths)9,11,13; Kratz et al. included probable and suspected cases and thus reported a slightly higher lethality of 46.8% (62 cases and 34 deaths).11
The four complete BDBV genomes obtained from the outbreak (GenBank: KC545393, KC545394, KC545395, and KC54396) confirmed the phylogenetic relatedness to the 2007 Ugandan BDBV isolate.13 Thus, to date, only these five complete BDBV genome sequences have been available to study an Ebola virus relative that has caused the deaths of many dozens of people. Here, we present an additional 11 complete or coding-complete BDBV genomes: six viral genomes directly sequenced from patient serum samples (not passaged; P0) and five genomes obtained after a single passage (P1) through cell culture. This latest set included two isolates that were obtained from specimens for which primary sequencing failed. We also resequenced one of the four P0 BDBV genomes originally obtained by the CDC (KC545394/case 120). Using a curated genomic dataset and additional epidemiological case information, molecular evidence posits two different lineages of BDBV co-circulating among infected individuals during the outbreak. The resulting data offer the most complete description to date of the evolutionary dynamics of the 2012 BVD outbreak.
Results
Outbreak index case and origins
As with most EBOD outbreaks, the definitive index case of the 2012 BVD outbreak remains unidentified. However, the earliest PCR-confirmed BDBV-containing sample originated from a clinic nurse in Isiro whose symptomatic disease began on June 28, 2012 (case 4; Table 1). This patient reported multiple potential exposures, including direct human contact with sick people, funeral attendance, and exposure to bats.12 There were a number of other laboratory-confirmed BVD cases after this first case; however, no other BDBV genome sequences were obtained prior to case 37 in August (Table 1).
Table 1.
Patient and sample characteristics from the 2012 BVD outbreak
| Case ID | GenBank number (isolate source) | Patient demographics | Occupation | Geographic location of sampling | Disease onset estimate | Clinical outcome | Date of death | Likely source of infection |
|---|---|---|---|---|---|---|---|---|
| 4 | MT680256 (P0) MT680248 (P1) | 41/F | clinic nurse | Isiro | June 28 | survived | NA | direct human contact (funeral) |
| 112 | KC545393 (P0) | 44/F | homemaker | Isiro | September 7 | deceased | NA | direct human contact (funeral) |
| 22 | MT680245 (P0) MT680262 (P1) | 27/F | homemaker | Isiro | September 7 | deceased | Sep 10 | direct human contact (funeral) |
| 37 | KC545396 (P0) | 18/F | student | Isiro | August 23 | deceased | Aug 31 | direct human contact (funeral) |
| 74 | MT680258 (P0) MT680250 (P1) | 77/M | tailor | Isiro | August 27 | survived | NA | direct human contact (funeral) |
| 120 | KC545394 (P0) MT742157 (P0) | 77/M | unknown | Vungba | September 5 | deceased | Sep 13 | unknown |
| 116 | MT680260 (P0) | 2/F | NA | Bédé | September 10 | survived | NA | direct human contact |
| 135∗ | MT680254 (P1) | 40/F | homemaker | Mambaya | September 12 | survived | NA | unknown |
| 130 | MT680261 (P0) | NA/F | unknown | Isiro | September 1 | deceased | unknown | unknown |
| 138∗ | MT680255 (P1) | 34/F | unknown | Isiro | September 20 | survived | NA | unknown |
| 122 | KC545395 (P0) | unknown | unknown | unknown | September 7 | survived | unknown | unknown |
F, female; M, male; NA, not applicable; P0, isolate from human serum; P1, isolate from single passage through cell culture.
Not included in curated dataset.
Analysis of genomic haplotypes
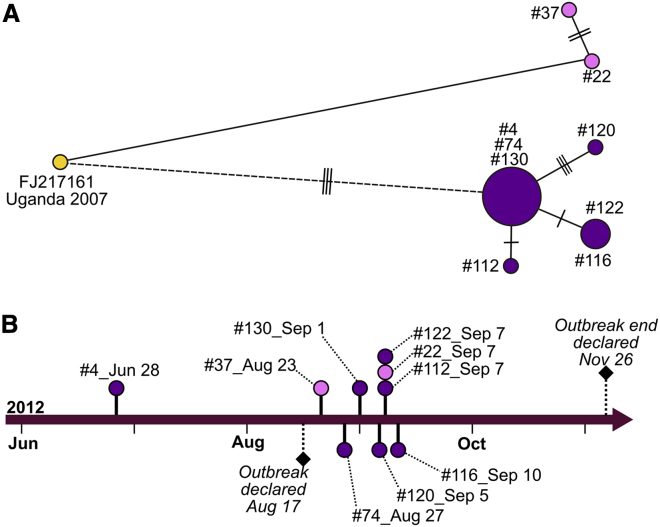
The genome sequences of the 2007 Ugandan BDBV isolate (GenBank: FJ217161) and 2012 Congolese BDBV isolate from case 4 differ by 230 nt over their complete genome (nucleotide identity, 98.7%; amino acid identity, 98.9%). Given case 4’s early date of symptom onset, we presume it to be ancestral to most of the remaining cases (Figure 1). Possible transmission links among the single representative 2007 outbreak genome sequence, case 4 in late June 2012, and case 37 in mid-August were estimated using a median-joining haplotype network (Figure 1). The haplotype network was inferred based on 12 single-nucleotide polymorphisms (SNPs) that were informative in the contextualization of inter- and lineage-specific, intra-outbreak evolution (Table 2). Two other P0 BDBV genomes, isolated from later cases (cases 74 and 130), share identical SNP profiles with case 4. Besides case 37 and case 22, all outstanding and sequenced BDBV haplotypes were linked to one or more individuals from the haplotype network cluster of cases 4, 74, and 130.
Figure 1.
Transmission and timeline of the 2012 BVD outbreak
(A and B) A medium-joining haplotype network of sequences from Bundibugyo virus isolates (A) and a scaled timeline of the 2012 outbreak (B). Labeled nodes in the haplotype network represent cases for available P0 sample sequences, and the size of each node correlates with the number of isolates sharing an SNP profile. Transmission line lengths reflect unscaled time, and transverse lines on transmission lines correspond to the number of segregating sites between the sequences of the linked nodes. Cases are grouped by color with the single representative isolate from 2007 shown in yellow, and two possible spillover events in 2012 colored in purple and pink. The dashed line edge describes the most likely temporal connection between the 2012 and 2007 outbreaks. In (B), the dates of symptomatic disease onset for sequenced, laboratory-confirmed BVD cases are plotted across time. Color schemes for the P0 samples reflect the separate spillover events depicted in (A). Official outbreak declaration and end dates are also shown.
Table 2.
SNPs identified in Bundibugyo virus genomes from the 2012 BVD outbreak in the Democratic Republic of the Congo
| Positiona | Reference base | Called base | NS mutation | Gene | Uganda 2007 | 22 P0 | 22 P1 | 37 P0 | 4 P0 | 4 P1 | 112 P0 | 74 P0 | 74 P1 | 120 P0 | 120b P0 | 116 P0 | 135 P1 | 130 P0 | 138 P1 | 122 P0 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 577 | C | T | NP | X | X | |||||||||||||||
| 5,928 | T | – | X | |||||||||||||||||
| 6,587 | T | C | GP | X | ||||||||||||||||
| 6,736 | T | C | L→P | GP | ||||||||||||||||
| 8,096 | T | C | X | X | ||||||||||||||||
| 8,287 | G | A | X | |||||||||||||||||
| 10,031 | G | A | X | X | ||||||||||||||||
| 10,798 | C | T | VP24 | X | X | X | X | |||||||||||||
| 11,138 | T | C | X | X | X | X | ||||||||||||||
| 11,271 | A | G | X | X | ||||||||||||||||
| 14,583 | T | C | L | X | X | X | X | |||||||||||||
| 15,019 | T | A | L | X | X |
An “X” denotes a consensus-level change at the referred position. NS, nonsynonymous; nc, noncoding; S, synonymous.
Reference genome used for SNP analysis is the consensus sequence from the 2012 outbreak; FJ217161 is the same sequence as KR063673 and KU182911.
The genome sequence from case 120 was previously sequenced by the CDC.
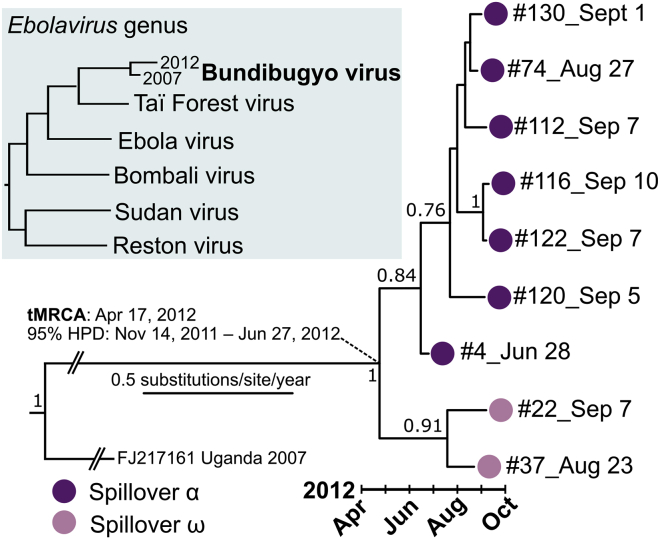
Despite the anchoring of the temporal outbreak in a central node around case 4, cases 22 and 37 fall outside of this main cluster (Figure 1A). Notably, the disease onset of case 37 occurred ∼2 weeks prior to the disease onset of case 22 (Figure 1B). Yet, the SNP profile from the sequenced isolate of case 22 is identical to the 2007 reference sequence among the informative SNPs. The sequenced isolate from case 37 has two additional SNPs at nucleotide positions 5,928 and 8,928, both mutations in noncoding regions. This results in a non-clocklike evolution among the 2007 reference sequence, case 37, and case 22 (Figure S1). Moreover, three more informative SNPs are shared between the 2007 reference sequence and all sequenced 2012 sequenced isolates, with the exception of cases 37 and 22 (Figure 1A; Table 2). A Bayesian-estimated, time-scaled, phylogenetic tree strongly supports a basal divergence between two groups of cases (posterior probability = 1; Figure 2). 2012 BDBV molecular lineages are indicted in both tables and figures as spillover α and spillover ω, which enables lineage-defining SNPs and non-clocklike evolution to be better represented in our data.
Figure 2.
Phylogenetic tree of the 2012 BVD outbreak
An inset shows the topology of the Ebolavirus genus with respect to the 2012 Bundibugyo virus spillover(s). A maximum-clade credibility tree was estimated using a single 2007 reference sequence and nine 2012 complete or coding-complete sequences. Tree branches are scaled by substitutions per site per year; the 2007 reference sequence branch was compressed for visual clarity and is not to scale. Posterior probabilities greater than 0.70 are shown at tree nodes. Multiple spillovers are shown using colored circles at tree tips and are labeled spillover α (violet) and spillover ω (pink). An estimated mean date and 95% highest posterior density (HPD) corresponding to the most recent common ancestor (tMRCA) of the include 2012 sequences are indicated by the diagonal dashed lines.
Available epidemiological information does not point to an obvious encounter with wildlife, such as exposure to bats or consumption of bushmeat, that would suggest that either case 22 or case 37 was a direct entry point into the human population for the virus. Given the number of cases that occurred around the time cases 22 and 37 became ill, it is highly likely that these were not the primary infections of this spillover. Case 22, the latter of the two cases, was a 27-year-old homemaker who became symptomatic on September 7. She reported possible direct human contact with sick patients and attending a funeral around August 18. Case 37 was an 18-year-old student from Isiro who became symptomatic on August 23. She, too, reported both and direct human contact with a sick patient (date unknown) and attending a funeral (around August 8–10). No additional epidemiological information connects these two cases, both of which were fatal.
Among the rest of the cases linked to case 4, we observed another identical SNP pattern when comparing case 116 and case 122 that might indicate direct transmission. Case 116, a 2-year-old who lived in Bédé (a village neighboring Isiro), became symptomatic on September 9. We do not have demographic data or the symptomatic onset date for case 122 (KC545395, originally sequenced13 to further investigate the possibility of a direct transmission line between these two cases).
In addition to sequencing the P0 isolates described above, we also obtained coding-complete or complete sequences for six P1 isolates, two of which (from cases 135 and 138) represent the only sequences available from these cases. Although we included the associated patient information and SNP data for P1 samples in Tables 1 and 2, we did not include them in the phylogenetic analyses. Our conservative approach was designed to avoid introducing artifacts of cell-culture adaptive mutations into the analyses given the very limited diversity observed in these genomes and the importance of a few SNPs for our conclusions.
Bayesian analysis
We identified 12 intra-outbreak segregating sites from the BDBV genome sequence variants in the sampled viral population during the 104-day outbreak period (June 28 to September 10). Using a single genomic sequence from the 2007 outbreak to anchor an inter-outbreak substitution rate calculation (based upon 230 segregating sites and a 1,774-day period from November 3, 2007 to September 10, 2012), we estimated a mean rate of 0.84 × 10−3 substitutions per site per year (95% highest posterior density [HPD], 0.11–2.1 × 10−3) for BDBV using a Bayesian approach. The estimated mean time to the most recent common ancestor (tMRCA) for the 2012 outbreak was April 17, 2012 (95% HPD, November 14, 2011 to June 27, 2012; Figure 2). An intra-outbreak substitution rate and tMRCA could not be precisely estimated at an informative interval using only 2012 outbreak sequences (95% HPD, 1.3 × 10−7–1.1 × 10−3), even when tree topology was constrained. A root-to-tip analysis without the inclusion of the 2007 sequence estimated a slower substitution rate and reduced temporal linearity for an intra-outbreak dataset (slope [rate] = 0.48 × 10−3; correlation coefficient = 0.52; Figure S1).
Discussion
In an effort to characterize the early transmission events of this outbreak, we sequenced BDBV genomes from 11 samples of the 2012 BVD outbreak in and around the town of Isiro, COD. This increase in publicly available genome data for BDBV, which now represent nearly one-third of all confirmed cases of the outbreak, offers deeper insight into the dynamics of the 2012 outbreak. From the resulting analyses, we inferred that a cluster of central infections in Isiro seeded multiple lines of transmission that spread BDBV into surrounding populations. These molecular-epidemiological findings provided further support to previous deductions made using classical epidemiology and limited molecular analysis based upon the four available BDBV sequences. These data also expanded the outbreak period to 50 days earlier than the official declaration.
In contrast to previous conclusions,13 the now-available genomic data challenge the hypothesis that a single BDBV introduction into the human population in September 2012 caused the outbreak. The contention lies in the existence of two separate branches between the ancestral 2007 outbreak in Uganda and cases 22 and 4 of the 2012 BVD outbreak in COD. The closest genomic link is from case 22, who became symptomatic on September 7. The isolate from this case had an SNP profile (considering the informative sites) identical to the reference BDBV for the 2007 outbreak. However, the earliest 2012 genome is from an isolate from case 4 (disease onset, June 28). To uphold the hypothesis of a single spillover, the genome of case 4’s BDBV isolate would have to mutate in a way that the exact three SNPs are changed to the ancestral sequence prior to infection of case 22.
Moreover, based on estimated mean substitution rates and the upper 95% HPD threshold of the tMRCA of June 27, the scenario of a single introduction would have placed the single spillover over 50 days before the official outbreak recognition. Other potential explanations are plausible, such as the two different clades (labeled Spillover ⍺ and Spillover ω in Figure 2) being the result of multiple introductions into the human population. The intra-outbreak root-to-tip analysis estimated a rate similar to the recently estimated Ebola virus (EBOV) inter-outbreak substitution rate14 and weak temporal linearity. However, given the paucity of the sampling of BDBV genomes circulating early in the outbreak, a definite answer is unlikely. Further sequencing of earlier samples could possibly detect mixed populations or a stepped progression between the isolates. In the 2-month period between the symptomatic onset dates for case 4 and case 22, there were six documented cases of EBOD in Isiro, five of which were laboratory-confirmed cases. Unfortunately, recovery of high-quality genomic sequences from any of those samples was not possible. That information would have been crucial to understanding the early transmission chains. In summary, while it is still possible that a single spillover event caused this outbreak as is indeed characteristic for many EBOD epidemics,15 the available data do not strictly support that hypothesis.
Also, there were key gaps in the epidemiological records. For example, many patients had reported close contact with bats and animals commonly hunted as bushmeat; however, dates and locations of exposure to these potential reservoirs for BDBV were often lacking. Records of familial relationships among cases was also notably missing, and, as transmission among family members is common, this knowledge may have reinforced suspected lines of direct transmission. Finally, in the months preceding the official outbreak declaration in Isiro, village chiefs reported “waves of funerals” and an “epidemic of death” near the villages where the parents of the first patient, case 4, lived.12 Any patients and their surrogates cited both funeral attendance and direct human contact as sources of potential virus exposure, ideal circumstances for the virus to spread through human populations. Again, however, there was limited information available to link attendees at specific funerals.
Still, one additional observation is the very low rate of change between case 4 and the rates of the cases that had identical profiles (cases 74 and 130). In highly sampled outbreaks of Ebolavirus disease, EBOV genomes isolated from persistently infected patients likewise exhibit a slow rate of change.16, 17, 18, 19, 20 This slow intra-outbreak viral evolutionary rate and the temporal gaps between sampled viruses is also consistent with a persistently and asymptomatically infected BVD survivor reintroducing the virus into the human population via sexual transmission. Since most (8/10) of the P0 genomes included in this study are from female patients (Table 1), establishment of lines of transmission between sexual partners is difficult to estimate due to limited epidemiological link and predicted onset date information availability.
In summary, from the available information, two scenarios could explain the findings. One scenario is that BDBV was introduced into the human population significantly earlier than the original analysis. The second scenario implies that BDBV was introduced via spillover from an unknown natural reservoir host into the human population more than once during the 2012 outbreak. This scenario would explain the lack of intermediary genomic information from infected humans defining a progressive gain and loss of the mutations. Multiple introductions greatly extend outbreak duration and expand the number of infected individuals contributing to an outbreak via the reservoir.15 Neither scenario is exclusive of the other; both are possible. Both scenarios point to the need to improve surveillance in the field.
The additional epidemiological links and genomic analyses provided here present two explanations as to how BDBV entered human populations near Isiro in 2012. Even with this fuller characterization of the outbreak, the complete natural history of the evolution of BDBV between the 2007 and 2012 outbreaks remains elusive. Since the 2012 outbreak, there has been no evidence of BDBV circulation in human populations or wildlife. Considering that nearly 16 years elapsed between EBOV outbreaks in COD (1978–1994),7 this may not be a rare occurrence for filovirus. On the other hand, the EBOV zoonotic spillovers since had significantly increased in frequency. However, we remain hopeful that the jump from natural reservoir to human populations is a rare and unlikely event for BDBV.
Limitations of the study
Clinical samples, contact tracing data, sample collection dates, and other information for BVD patients were sparse in months prior to the original August 2012 BVD outbreak declaration. Therefore, the possibility that there were earlier cases than case 4 cannot be experimentally validated. Ideally, suspected cases from June and July would be retrospectively tested for BDBV-specific antibodies or genomic RNA.
Another limitation was the lack of an in-country genomic sequencing center during the 2012 BVD outbreak, as advantages of such a capability were first demonstrated in Western Africa during the 2013–2016 Ebola virus disease outbreak.21 The lack of available complete BDBV genome sequences led to greater uncertainty of phylogenetic estimations. The availability of additional BDBV genomes sequenced directly from clinical isolates would have improved transmission network dynamics and may have further supported the multiple-spillover hypothesis.
STAR★Methods
Key resources table
Resource availability
Lead contact
Further information and correspondence request for resources relevant to this study should be directed to and will be fulfilled by the lead contact, Dr. Gustavo. F. Palacios, gustavo.f.palacios.civ@mail.mil.
Materials availability
No materials (reagents) were generated during the course of this study.
Data and code availability
All genomic sequences generated for this study were deposited in NCBI and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Raw sequence data will be shared by the lead contact upon request without restriction. No unique software/program code was generated for this study.
Experimental model and subject details
This study was conducted at the US Army Research Institute of Infectious Diseases as part of an ongoing EBOD response and EBOV surveillance effort under the project “Assessment of Human Clinical Samples from Viral Hemorrhagic Fevers of Known and Unknown Etiology” (HP-12-15). Informed consent was not obtained from BVD patients for this study but, as the work was deemed not human subject research, it was performed with the consent of the Human Use Committee under the supervision of the Office of Human Use Oversight at the US Army Research Institute of Infectious Diseases. All information related to or obtained from patients was anonymized for this report.
Human samples
Serum samples from 11 patients of the 2012 BVD outbreak around Isiro, Haut-Uélé District, Province Orientale, COD, were provided by Metabiota, Washington DC, USA (please see Table 1 for patient demographics). All sampled patients had presented with clinical signs of viral hemorrhagic fever and were laboratory-confirmed BDBV cases by quantitative PCR (cycle threshold < 24).
Cell lines
P1 BDBV isolates were passaged once through grivet (Chlorocebus aethiops) kidney epithelial Vero cell cultures to obtain sufficient genome coverage for standard drafts. Cells were exposed at a multiplicity of infection of 1. Infected and uninfected cells were maintained at 37°C, with 5% CO2 throughout the experiment.
Method details
Sample processing
RNA contained in BDBV samples was converted to cDNA and amplified using sequence-independent single-primer amplification as previously described.23 Amplified cDNA was quantified with a Qubit 3.0 fluorometer (Life Technologies, Carlsbad, CA, USA) and used as starting material for library preparation (Nextera XT DNA library preparation kit, Illumina, San Diego, CA, USA). Sequencing was performed on an Illumina MiSeq by using either V2 or V3 reagent kits (Illumina) with a minimum of 2 × 151 cycles per run.
Genome assembly
Amplification primers were removed from sequencing reads by using Cutadapt version 1.9.dev1.27 Low-quality reads/bases were filtered by using Prinseq-lite version 0.20.4 (-min_qual_mean 25 -trim_left 20 -min_len 50).32 BDBV genomes were assembled by aligning sequencing reads to the 2007 reference BDBV genome, Bundibugyo virus/H.sapiens-tc/UGA/2007/Butalya-811250 (RefSeq #NC_014373 and GenBank #FJ217161),22 using DNASTAR Lasergene NGen version v13.0.0.360 (DNAStar, Madison, WI, USA) as described previously.33 A new consensus sequence was generated using a combination of SAMtools v0.1.1931 and custom scripts. Only bases with Phred quality scores > 20 were used in consensus calling. A minimum of 3 × read-depth coverage, in support of the consensus, was required to make a base call; positions lacking this depth of coverage were treated as missing (i.e., called as “N”). All genomes in this study were deposited in GenBank (Tables 1 and S1).
Genetic analysis
Previously determined BDBV genomes sequences were downloaded from GenBank and RefSeq: one BDBV isolate sequence from the 2007–2008 BVD outbreak in Uganda (isolate 811250/Bun-038, 52-year-old male, lethal outcome: GenBank #FJ217161, #KU182911; RefSeq #NC_014373), and 4 BDBV isolate sequences from the 2012 BVD outbreak in COD.34 A median-joining haplotype network was constructed with the R package haploNet function from Pegas R package24 using all BDBV genome sequences and further modified in Inkscape (https://inkscape.org/) for publication quality.
Phylodynamics
For molecular evolution and rate analysis, all sequences, including the publicly available genomes from 2007 and 2012, were aligned using MAFFT, version 7.388.8.25 Genome sequences with significant deletions, duplicate sequences, and redundant in vitro isolates were removed from downstream analyses.
First, maximum-likelihood phylogenetic trees were estimated using PhyML 3.3,26 both with (i.e., inter-outbreak) and without (i.e., intra-outbreak) a 2007 BDBV genome sequence. The presence of positively correlated temporal signals were determined by correlating root-to-tip distances against time using TempEst28 with the best-fitting root option. Bayesian Evolutionary Analysis by Sampling Trees (BEAST), version 1.10.4, was used to generate Bayesian-inferred maximum-clade credibility trees and associated substitution rates. A Hasegawa–Kishino–Yano 85 (HKY85) nucleotide substitution model with gamma-distributed rate variation (+ G4)29 was used for path sampling/stepping stone coalescent model testing (default parameters) for each dataset (Table S1). Sequences were partitioned by coding and noncoding alignments, which shared a molecular clock, and a gamma rate prior distribution (shape = 0.001, scale = 1,000). Each analysis ran for 1.0 × 109 generations (10% discarded as burn-in) and were sampled every 10,000 generations. Tracer visualized each run to ensure convergence (effective sample size > 200), and TreeAnnotator 1.8.430 estimated a maximum-clade credibility tree using a posterior probability limit of 0.7.
Quantification and statistical analysis
BEAST version 1.10.4, was used to generate median Bayesian-inferred substitution rates as well as time to the most recent common ancestor estimations, and both their associated 95% highest posterior densities. Statistical results are summarized in the Bayesian Analysis section and Figure 2 (n = 10).
Acknowledgments
The authors thank Laura Bollinger and Anya Crane (Integrated Research Facility at Fort Detrick, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Fort Detrick, Frederick, MD) for critically editing the manuscript.
The content of this publication does not necessarily reflect the views or policies of the US Department of Defense, the US Department of Health and Human Services, the US Department of the Army, or the institutions and companies affiliated with the authors. This work was funded in part through the Global Emerging Infections Surveillance Section of the Armed Forces Health Surveillance Branch (ProMIS plans C0602_12_RD and P0108_13_RD; R.J.S.) This work was also supported in part through the current prime contract of Laulima Government Solutions with the US National Institutes of Health National Institute of Allergy and Infectious Diseases under contract no. HHSN272201800013C and Battelle Memorial Institute’s former prime contract with the National Institute of Allergy and Infectious Diseases under contract no. HHSN272200700016I. J.H.K. performed this work as an employee of Battelle Memorial Institute and subsequently as an employee of Tunnell Government Services, a subcontractor of Laulima Government Solutions, under contract no. HHSN272201800013C.
Author contributions
Conceptualization, G.F.P.; provided primary data, N.W., J.N.F., M.M., P.M., J.-J.M.-T., and R.J.S.; performed the experiments, E.R.N.; formal analysis, C.E.H., R.K., N.D.P., G.F.P., and J.H.K.; writing – review & editing, C.E.H., P.L., N.D.P., J.R., J.H., J.H.K., M.S.-L., G.F.P., and J.R.K.
Declaration of interests
The authors declare no competing interests.
Published: July 27, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2021.100351.
Supplemental information
References
- 1.Kuhn J.H., Adachi T., Adhikari N.K.J., Arribas J.R., Bah I.E., Bausch D.G., Bhadelia N., Borchert M., Brantsæter A.B., Brett-Major D.M. New filovirus disease classification and nomenclature. Nat. Rev. Microbiol. 2019;17:261–263. doi: 10.1038/s41579-019-0187-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization . 2018. International classification of diseases and related health problems (11th rev., ICD-11)https://icd.who.int/browse11/l-m/en#/http%3a%2f%2fid.who.int%2ficd%2fentity%2f694903163 [Google Scholar]
- 3.MacNeil A., Farnon E.C., Wamala J., Okware S., Cannon D.L., Reed Z., Towner J.S., Tappero J.W., Lutwama J., Downing R. Proportion of deaths and clinical features in Bundibugyo Ebola virus infection, Uganda. Emerg. Infect. Dis. 2010;16:1969–1972. doi: 10.3201/eid1612.100627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roddy P., Howard N., Van Kerkhove M.D., Lutwama J., Wamala J., Yoti Z., Colebunders R., Palma P.P., Sterk E., Jeffs B. Clinical manifestations and case management of Ebola haemorrhagic fever caused by a newly identified virus strain, Bundibugyo, Uganda, 2007-2008. PLoS ONE. 2012;7:e52986. doi: 10.1371/journal.pone.0052986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gupta M., MacNeil A., Reed Z.D., Rollin P.E., Spiropoulou C.F. Serology and cytokine profiles in patients infected with the newly discovered Bundibugyo ebolavirus. Virology. 2012;423:119–124. doi: 10.1016/j.virol.2011.11.027. [DOI] [PubMed] [Google Scholar]
- 6.Clark D.V., Kibuuka H., Millard M., Wakabi S., Lukwago L., Taylor A., Eller M.A., Eller L.A., Michael N.L., Honko A.N. Long-term sequelae after Ebola virus disease in Bundibugyo, Uganda: a retrospective cohort study. Lancet Infect. Dis. 2015;15:905–912. doi: 10.1016/S1473-3099(15)70152-0. [DOI] [PubMed] [Google Scholar]
- 7.Kuhn J.H., Amarasinghe G.K., Perry D.L. In: Fields Virology. Howley P.M., Knipe D.M., Whelan S.P.J., editors. Wolters Kluwer/Lippincott Williams & Wilkins; 2020. Filoviridae; pp. 449–503. [Google Scholar]
- 8.Wamala J.F., Lukwago L., Malimbo M., Nguku P., Yoti Z., Musenero M., Amone J., Mbabazi W., Nanyunja M., Zaramba S. Ebola hemorrhagic fever associated with novel virus strain, Uganda, 2007-2008. Emerg. Infect. Dis. 2010;16:1087–1092. doi: 10.3201/eid1607.091525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention . 2020. Ebola (Ebola virus disease). Years of Ebola virus disease outbreaks.https://www.cdc.gov/vhf/ebola/outbreaks/history/summaries.html [Google Scholar]
- 10.Towner J.S., Sealy T.K., Khristova M.L., Albariño C.G., Conlan S., Reeder S.A., Quan P.L., Lipkin W.I., Downing R., Tappero J.W. Newly discovered ebola virus associated with hemorrhagic fever outbreak in Uganda. PLoS Pathog. 2008;4:e1000212. doi: 10.1371/journal.ppat.1000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kratz T., Roddy P., Tshomba Oloma A., Jeffs B., Pou Ciruelo D., de la Rosa O., Borchert M. Ebola virus disease outbreak in Isiro, Democratic Republic of the Congo, 2012: signs and symptoms, management and outcomes. PLoS ONE. 2015;10:e0129333. doi: 10.1371/journal.pone.0129333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Epelboin A. CNRS-MNHN, Paris, France and World Health Organization; 2012. Rapport de mission anthropologique sur l’épidémie d’Ebola Isiro, R. D. Congo, 4 au 30 septembre 2012 [French] [Google Scholar]
- 13.Albariño C.G., Shoemaker T., Khristova M.L., Wamala J.F., Muyembe J.J., Balinandi S., Tumusiime A., Campbell S., Cannon D., Gibbons A. Genomic analysis of filoviruses associated with four viral hemorrhagic fever outbreaks in Uganda and the Democratic Republic of the Congo in 2012. Virology. 2013;442:97–100. doi: 10.1016/j.virol.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mbala-Kingebeni P., Aziza A., Di Paola N., Wiley M.R., Makiala-Mandanda S., Caviness K., Pratt C.B., Ladner J.T., Kugelman J.R., Prieto K. Medical countermeasures during the 2018 Ebola virus disease outbreak in the North Kivu and Ituri Provinces of the Democratic Republic of the Congo: a rapid genomic assessment. Lancet Infect. Dis. 2019;19:648–657. doi: 10.1016/S1473-3099(19)30118-5. [DOI] [PubMed] [Google Scholar]
- 15.Matson M.J., Chertow D.S., Munster V.J. Delayed recognition of Ebola virus disease is associated with longer and larger outbreaks. Emerg. Microbes Infect. 2020;9:291–301. doi: 10.1080/22221751.2020.1722036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blackley D.J., Wiley M.R., Ladner J.T., Fallah M., Lo T., Gilbert M.L., Gregory C., D’ambrozio J., Coulter S., Mate S. Reduced evolutionary rate in reemerged Ebola virus transmission chains. Sci. Adv. 2016;2:e1600378. doi: 10.1126/sciadv.1600378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christie A., Davies-Wayne G.J., Cordier-Lassalle T., Blackley D.J., Laney A.S., Williams D.E., Shinde S.A., Badio M., Lo T., Mate S.E., Centers for Disease Control and Prevention (CDC) Possible sexual transmission of Ebola virus - Liberia, 2015. MMWR Morb. Mortal. Wkly. Rep. 2015;64:479–481. [PMC free article] [PubMed] [Google Scholar]
- 18.Diallo B., Sissoko D., Loman N.J., Bah H.A., Bah H., Worrell M.C., Conde L.S., Sacko R., Mesfin S., Loua A. Resurgence of Ebola virus disease in Guinea linked to a survivor with virus persistence in seminal fluid for more than 500 days. Clin. Infect. Dis. 2016;63:1353–1356. doi: 10.1093/cid/ciw601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mate S.E., Kugelman J.R., Nyenswah T.G., Ladner J.T., Wiley M.R., Cordier-Lassalle T., Christie A., Schroth G.P., Gross S.M., Davies-Wayne G.J. Molecular evidence of sexual transmission of Ebola virus. N. Engl. J. Med. 2015;373:2448–2454. doi: 10.1056/NEJMoa1509773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schindell B.G., Webb A.L., Kindrachuk J. Persistence and sexual transmission of filoviruses. Viruses. 2018;10:683. doi: 10.3390/v10120683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kugelman J.R., Wiley M.R., Mate S., Ladner J.T., Beitzel B., Fakoli L., Taweh F., Prieto K., Diclaro J.W., Minogue T., US Army Medical Research Institute of Infectious Diseases. National Institutes of Health. Integrated Research Facility–Frederick Ebola Response Team 2014–2015 Monitoring of Ebola virus Makona evolution through establishment of advanced genomic capability in Liberia. Emerg. Infect. Dis. 2015;21:1135–1143. doi: 10.3201/eid2107.150522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuhn J.H., Andersen K.G., Bào Y., Bavari S., Becker S., Bennett R.S., Bergman N.H., Blinkova O., Bradfute S., Brister J.R. Filovirus RefSeq entries: evaluation and selection of filovirus type variants, type sequences, and names. Viruses. 2014;6:3663–3682. doi: 10.3390/v6093663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Djikeng A., Halpin R., Kuzmickas R., Depasse J., Feldblyum J., Sengamalay N., Afonso C., Zhang X., Anderson N.G., Ghedin E., Spiro D.J. Viral genome sequencing by random priming methods. BMC Genomics. 2008;9:5. doi: 10.1186/1471-2164-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paradis E. pegas: an R package for population genetics with an integrated-modular approach. Bioinformatics. 2010;26:419–420. doi: 10.1093/bioinformatics/btp696. [DOI] [PubMed] [Google Scholar]
- 25.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guindon S., Delsuc F., Dufayard J.-F., Gascuel O. Estimating maximum likelihood phylogenies with PhyML. Methods Mol. Biol. 2009;537:113–137. doi: 10.1007/978-1-59745-251-9_6. [DOI] [PubMed] [Google Scholar]
- 27.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011;17:10–12. [Google Scholar]
- 28.Rambaut A., Lam T.T., Max Carvalho L., Pybus O.G. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen) Virus Evol. 2016;2:vew007. doi: 10.1093/ve/vew007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baele G., Lemey P., Bedford T., Rambaut A., Suchard M.A., Alekseyenko A.V. Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Mol. Biol. Evol. 2012;29:2157–2167. doi: 10.1093/molbev/mss084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klinkenberg D., Backer J.A., Didelot X., Colijn C., Wallinga J. Simultaneous inference of phylogenetic and transmission trees in infectious disease outbreaks. PLoS Comput. Biol. 2017;13:e1005495. doi: 10.1371/journal.pcbi.1005495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmieder R., Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kugelman J.R., Wiley M.R., Nagle E.R., Reyes D., Pfeffer B.P., Kuhn J.H., Sanchez-Lockhart M., Palacios G.F. Error baseline rates of five sample preparation methods used to characterize RNA virus populations. PLoS ONE. 2017;12:e0171333. doi: 10.1371/journal.pone.0171333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burk R., Bollinger L., Johnson J.C., Wada J., Radoshitzky S.R., Palacios G., Bavari S., Jahrling P.B., Kuhn J.H. Neglected filoviruses. FEMS Microbiol. Rev. 2016;40:494–519. doi: 10.1093/femsre/fuw010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All genomic sequences generated for this study were deposited in NCBI and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Raw sequence data will be shared by the lead contact upon request without restriction. No unique software/program code was generated for this study.