Abstract
Introduction
The coronavirus disease 2019 (COVID-19) pandemic is a global public health emergency causing a disparate burden of death and disability around the world. The viral genetic variants associated with outcome severity are still being discovered.
Methods
We downloaded 155 958 severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) genomes from GISAID. Of these genomes, 3637 samples included useable metadata on patient outcomes. Using this subset, we evaluated whether SARS-CoV-2 viral genomic variants improved prediction of reported severity beyond age and region. First, we established whether including genomic variants as model features meaningfully increased the predictive power of our model. Next, we evaluated specific variants in order to determine the magnitude of association with severity and the frequency of these variants among SARS-CoV-2 genomes.
Results
Logistic regression models that included viral genomic variants outperformed other models (area under the curve = 0.91 as compared with 0.68 for age and gender alone; P < 0.001). We found 84 variants with odds ratios greater than 2 for outcome severity (17 and 67 for higher and lower severity, respectively). The median frequency of associated variants was 0.15% (interquartile range 0.09–0.45%). Altogether 85% of genomes had at least one variant associated with patient outcome.
Conclusion
Numerous SARS-CoV-2 variants have 2-fold or greater association with odds of mild or severe outcome and collectively, these variants are common. In addition to comprehensive mitigation efforts, public health measures should be prioritized to control the more severe manifestations of COVID-19 and the transmission chains linked to these severe cases.
Lay summary: This study explores which, if any, SARS-CoV-2 viral genomic variants are associated with mild or severe COVID-19 patient outcomes. Our results suggest that there are common genomic variants in SARS-CoV-2 that are more often associated with negative patient outcomes, which may impact downstream public health measures.
Keywords: molecular epidemiology, epidemiologic surveillance, virulence, communicable disease control, COVID-19, SARS-CoV-2, human
INTRODUCTION
Since the coronavirus disease 2019 (COVID-19) pandemic emerged, humans have faced unprecedented disruption from the newfound obligate parasite. Within the USA alone, the unwelcome guest has already caused an estimated 2.5 million years of life lost [1]. Beyond the USA, the global burden is substantial and growing, but it is not uniform; continents, nations, communities, families and patients are all affected differently. Understanding the basis for this variability is an important global health priority.
One of the most common measures to describe the severity of COVID-19 is the infection fatality ratio (IFR) or the number of deaths for every infection. There are widespread differences in IFR between studies and this heterogeneity is not due to chance alone [2–4]. One meta-analysis of 26 studies estimated an IFR of 0.68% (0.53–0.82%) while cautioning this was likely an ‘underestimate’ and another meta-analysis with 61 studies estimated a median IFR of 0.26%, outside the range of the first meta-analysis [2, 3].
Other studies have suggested the variability in infection fatality ratio is more consistent when infections are stratified by age but is more likely to vary between studies when considering the population above age 65 [4–6]. One recent meta-analysis estimated that the IFR of COVID-19 increases progressively with age, estimating an IF of 0.4% at age 55, 1.4% at age 65, 4.6% at age 75, and 15% at age 85. The authors posit that the age distribution of the population, in combination with the age-specific prevalence of COVID-19, could explain the majority of the geographical variation in population IFR [4].
Although vulnerable age groups should be protected as recommended by the U.S. Centers for Disease Control and Prevention (CDC), there are likely more factors at play in the IFR than age alone. Indeed, specific underlying health conditions (e.g. obesity, heart conditions, chronic kidney disease, etc.) as well as other health statuses such as smoking can dramatically increase COVID-19 risk profiles. Interestingly, even within the same hospital systems, there are strong time trends in case fatality rate after adjusting for baseline characteristics/risk [7] even at the national scale [8], which may be attributed to more efficacious treatments. Unless baseline risk adjustments were inadequate, there are likely time trends in IFR, which would also help reconcile the disparate estimates noted in the two meta-analyses cited above.
Another possibility is severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virulence could differ across geographic locations because some local strains have differential virulence. Previous reports have suggested that the evolution of attenuated strains is expected for RNA viruses [9]. Moreover, it has been observed that respiratory pathogens, in contrast to vector-borne diseases, typically evolve toward lower virulence because healthier hosts tend to engage in more social contact [10].
Using publicly available data from Global Initiative on Sharing Avian Influenza Data (GISAID), we interrogate the relationship between SARS-CoV-2 variants and associated patient outcomes in the GISAID metadata. GISAID stores SARS-CoV-2 genomic data and sequencing metadata—including sequencing technology and assembly method—as well as some patient metadata, including high-level patient outcomes, region of origin, age, gender and date of collection. We used high-level patient metadata submitted by GISAID users to differentiate between severe and mild patient outcomes. We utilized a logistic regression model in order to better understand how viral genomic SARS-CoV-2 variants are linked with COVID-19 patient outcomes. Despite a lack of data on specific comorbidities or other risk factors, we found several statistically significant associations between genomic variants and COVID-19 severity. This study may help to lay the groundwork for other researchers to use statistical models with viral genomic variants in order to study functional impacts of mutations, acutely and longitudinally, and ultimately inform downstream public health responses.
METHODS
Variant alignment and variant calling
SARS-CoV-2 genome sequences were obtained from GISAID (Global Initiative on Sharing All Influenza Data) on 21 October 2020 [11, 12]. GISAID sequences were filtered to include those of human origin. FASTA sequences were aligned to the reference sequence, Wuhan-Hu-1 (NCBI: NC_045512.2; GISAID: EPI_ISL_402125) using Minimap2 (version 2.17) [13]. Resulting VCF (Variant Call Format) files were annotated using SnpEff (version 5.0) and filtered using SnpSift [14, 15]. The shell scripts used for variant alignment and variant calling, along with the Python scripts used to perform the steps described below, are available here: https://github.com/Digital-Biobank/covid_variant_severity/ (11 July 2021, date last accessed).
Metadata preprocessing and cohort building
Raw GISAID patient data was parsed from a JSON file using Python (version 3.8.2). Patient outcomes that were unclear or empty were not included in our analyses. Of 155 958 samples available in GISAID, a subset of 3637 samples was used for our analyses (Supplementary Fig. 1). Patient outcomes were then aggregated into positive (‘Mild’) outcomes or negative (‘Severe’) outcomes (detailed in Supplementary Fig. 1). Briefly, ‘Mild’ outcomes included: Outpatient, Asymptomatic, Mild, Home/Isolated/Quarantined and Not Hospitalized. ‘Severe’ outcomes included: Hospitalized (including severe, moderate and stable) and Deceased (Death). The age variable was normalized using MinMaxScaler (bound between 0 and 1) in scikit-learn. The gender variable was converted into binary. Indicator (dummy) variables were used for region, clade and variants. The number of indicator variables is equal to the number of unique categories minus one (k − 1).
Variant and metadata modeling
Annotated VCF files were parsed, pivoted to wide format, and joined with GISAID patient data using Pandas (version 1.0.3) [16]. Scikit-learn (version 0.23.2) [17] was used to fit logistic regression models with the L1 penalty (Lasso regularization) and the default regularization strength (C = 1) to the patient (rows) and variant (columns). Data were split into five cross-validation folds (80% train set and 20% test size per fold) by the Scikit-learn stratified K-fold cross-validation generator. Models were persisted as pickle files using joblib (version 0.14.1).
Plotting
Scatter and bar plots were created using Pandas (version 1.0.3) [16], Matplotlib (version 3.2.1) [18] and Seaborn (version 0.10.1) [19]. Genome position tracks were added to scatterplots using DNA Features Viewer (version 3.0.3) [20]. ROC curves were plotted using Scikit-learn (version 0.23.2) [17], and Matplotlib [18].
Statistical analysis
A total of five logistic regression models were trained using different input features. For each model, Scikit-learn(version 0.23.2) [17] was used to calculate the area under the curve (AUC), a measure of goodness of fit of a binary classification model. AUC confidence intervals and P-values values and diagnostic odds ratios (OR) were calculated using Numpy (version 1.19.5) [21]. Numpy was also used to calculate diagnostic ORs with confidence intervals and P-values for each of the five logistic regression models. The Scikit-learn implementation of logistic regression does not provide ORs or P-values for individual variables. ORs and Chi-square test P-values for the association of variants with ‘Severe’ outcomes (Supplementary Table 1) were calculated from variant count data using Statsmodels (version 0.12.1) and Scipy (version 1.5.0), respectively [22]. Variant frequency was calculated using Pandas [16].
RESULTS
Sample population characteristics
We collected a total of 155 958 viral genomes along with clinical metadata (Supplementary Fig. S1). The metadata included numerous entries whereby the severity of the condition could not readily be discerned. For example, ‘recovering’, ‘recovered and released’ and ‘mild symptoms inpatient for observation’ were found in the raw data and were not included. The full downloaded dataset included 148 121 entries with empty or unknown clinical observations and 4200 entries for which clinical severity could not be classified. From the remaining 3637 sequences with clear severity indications, we generated two classes (Supplementary Fig. S2) by recoding the observational metadata into consistent terminology and creating a ‘Severe’ class of ‘deceased’, ‘hospitalized’, ‘ICU’, and ‘pneumonia’ (n = 2,870); and a ‘Mild’ class of ‘outpatient’, ‘mild’, ‘epidemiology study’, ‘asymptomatic’, ‘screening’ and ‘stable in quarantine’ (n = 767). 85% of these genomes had at least one variant associated with patient outcome. Viral sequences were obtained from the six major geographical regions in GISAID between January and October 2020 (shown in Supplementary Fig. S3).
SARS-CoV-2 variants associated with ‘severe’/‘mild’ outcome categories
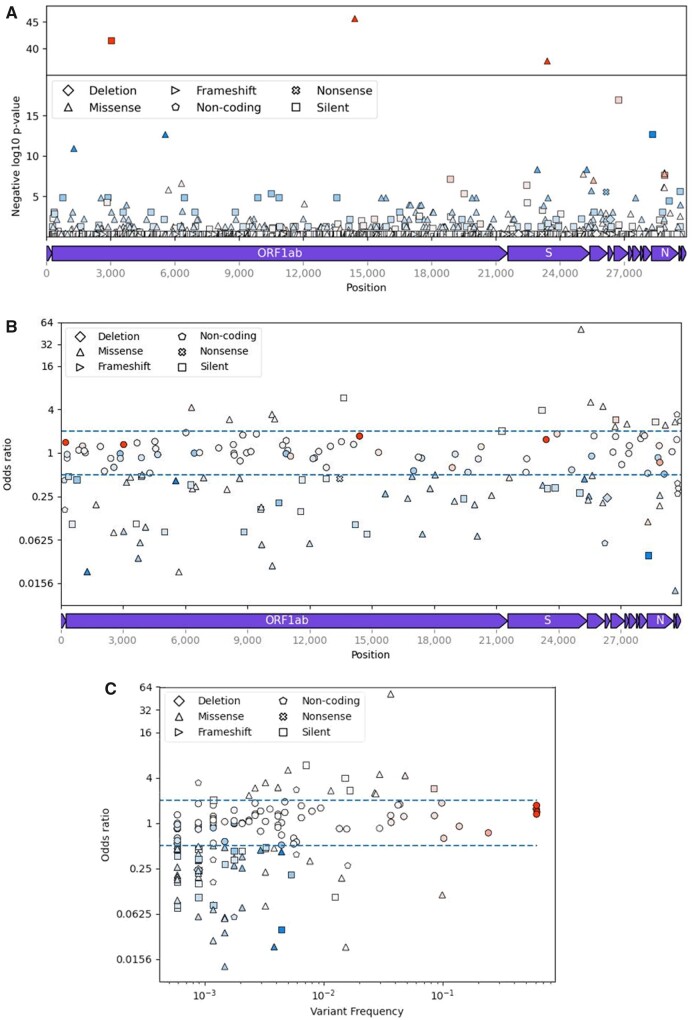
The overwhelming majority of variants in the SARS-CoV-2 genomes assessed were rare, with only 12 common variants with at least a 5% alternative allele frequency (Fig. 1C). Two of these common variants, C26735T (geneM: Y71Y) and C28311T (geneN: P13L) were associated with ‘Severe’ or ‘Mild’ outcomes, as measured by having an OR of greater than 2 or ≤0.5, respectively. We also observed 84 of 157 rare variant associations with ‘Severe’ or ‘Mild’ outcomes. Collectively, 17 variants were associated with ‘Severe’ classification with at least an OR of 2, while we found 67 associations with ‘Mild’ classification (OR ≤ 0.5). The ORs, confidence intervals and P-values of the 40 variants with the highest (n = 20) or lowest (n = 20) ORs are reported in Supplementary Table 1. The variants associated with outcomes were distributed across the genome, including the strongest ‘Severe’ association within the C-terminal end of the spike protein (Fig. 1B). The majority of variants characterized here were transitions (121, 71%), with 47% of those transitions (79) being C > T (Supplementary Fig. S4).
Figure 1.
Overview of SARS-CoV-2 variants selected from GISAID data (n = 155 958). (A) Negative log10 P-values of variant association (chi-square test) with ‘Severe’ outcome group (hospitalized, deceased, etc.) plotted against position of variants (n = 4484) in the SARS-CoV-2 genome. (B) ORs (log2 scale) of ‘Severe’ versus ‘Mild’ (outpatient, asymptomatic, etc.) outcome groups plotted against the positions of variants with ORs not equal to one (n = 168) in the SARS-CoV-2 genome. (C) ORs (log2 scale) of ‘Severe’ versus ‘Mild’ outcome groups plotted against log10 frequency of variants (n = 168) in the patient subpopulation (n = 3363) without missing variables. The shape of each data point corresponds to a mutation type (triangle pointing upward: missense, pentagon: noncoding, square: silent, x: nonsense, diamond: deletion, triangle point right: frameshift). Additionally, the color of each data point shows whether a given variant is found primarily in Mild (blue) or Severe (red) status patients. The intensity of the colors shows the frequency of the variant in either Mild (darker blue) or Severe (darker red) status patients. The genome track at the bottom of A and B do not include noncoding regions.
Predicting clinical outcomes for patients based upon clinical metadata and viral genomics
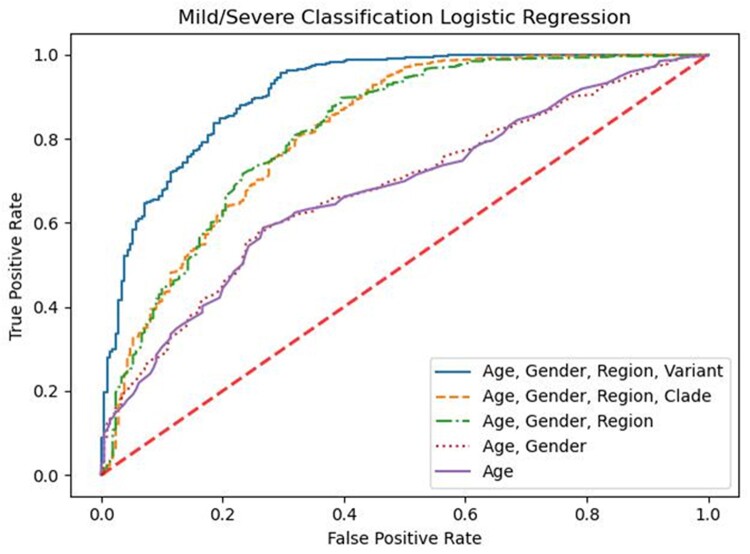
Age and gender have been previously reported to be predictive of clinical outcomes [23]. Our logistic regression models predicting ‘Severe’ outcomes confirm these prior associations (Fig. 2). We also evaluated the inclusion of features including region, viral clade and viral genomic variants. In order to get a general understanding of the performance of each of our logistic regression models, we evaluated the area under the curve (AUC), the probability that a randomly selected patient who had some outcome (e.g. severe COVID-19) actually had a higher risk score than a patient who did not have that outcome. We found that the AUC for predictions based on age or age and gender are moderately better than random chance at 0.677 (0.642–0.712) and 0.679 (0.644–0.714), respectively (Supplementary Table 3). We hypothesized that viral genomic variants could also contribute to severity classification. When accounting for the region of collection, we observed an increased AUC to 0.817 (0.817–0.818). Moreover, we found that adding clade to an age/gender/region model resulted in a slight improvement of accuracy (81% vs 86%, respectively) with a nearly identical AUC of 0.818 (0.817–0.818) (Fig. 2). While the difference between region and clade appears insignificant, adding clade-level information increased the predictive ability of our model beyond age and gender alone. We then considered whether variant-level information would further improve model performance. We found that substituting clade with 4499 genomic variants in the model increased the AUC to 0.911 (0.910–0.911), significantly improving predictability for clinical outcomes. Interestingly, only 168 of the 4,999 variants had nonzero coefficients in the logistic regression model with L1 penalty, compared to 1438 variants with nonzero coefficients in a logistic regression model without any penalty. In addition to the improvement in the AUC, we compared the accuracy of our predictions, which started at 81% for the age-only model and improved to 86% and 88% for each additional step in the model building before finally reaching a maximum at 91% accuracy for the age/gender/region/variant model (Supplementary Table 3). To assess the robustness of our model performance assessment, we obtained accuracy values for five cross-validation folds for each model. The standard deviation of the five accuracy values for the age/gender/region/variant model was less than 1% (0.0064), indicating that the modeling results are not dependent on any particular split of the data.
Figure 2.
Comparison of nested logistic regression models. Models are labeled based on the predictor variables (purple solid line: age; red dotted line: [age, gender], green dash-dotted line: [age, gender, region], orange dashed line: [age, gender, region, clade], blue solid line: [age, gender, region, variant]) used to predict whether SARS-CoV-2 patients (n = 3386) belong to ‘Severe’ (hospitalized, deceased) or ‘Mild’ (outpatient, asymptomatic, etc.) outcome groups. The diagonal dash red line represents a theoretical unskilled estimator, which can be described as a random guess.
Classifications based solely on age or on age and gender resulted in insignificant ORs in our models (Supplementary Table 3). Both models had the same diagnostic OR (4.4), confidence interval (2.8–6.0) and P-value (0.072). However, the other three models all had significant ORs with P-values less than 0.0001. Consistent with the AUC results, the OR was greatest for the full model (age/gender/region/variant) followed by age/gender/region/clade and finally age/gender/region (ORs: 12.3 (11.8–12.8), 8.4 (8.0–8.9) and 8.0 (7.6–8.4), respectively). Similarly, the negative likelihood ratio for the full model displayed a large reduction in the likelihood of a patient classified as ‘Mild’ developing ‘Severe’ symptoms (−LR = 0.039) as compared to a moderate reduction in the post-test outcomes for the age-only or age and gender models (−LR = 0.231 for both models).
DISCUSSION
We demonstrate that including viral genomic variants can substantially improve the classification of COVID-19 patient outcomes as compared with models using only age and region. Moreover, in our models, we observe that some individual variants are particularly important with substantial associations with severity and that collectively these variants are not rare.
Associations between viral genomic variants and patient outcomes are expected. Consistent with known patterns in the evolution of virulence in RNA viruses [9, 10, 24], we would expect many common strains have differing association with patient severity by this point in the pandemic by chance alone and even as sampling is more likely to occur with severe outcomes, variants correlated with mild outcomes are still being identified. Indeed, as more variants of interest (VOI) and variants of concern (VOC) are identified, patterns of convergent evolution are coming into view and analytic models that are able to accommodate viral genomic variants may be better suited to informing public health policies.
Though respiratory pathogens often evolve toward lower virulence, there have been historical exceptions [25]. Modeling future fitness landscapes suggests that even partial isolation of symptomatic cases can substantially reduce deaths with less transmission in the short term. Importantly, this isolation can also potentially alter the evolutionary path by favoring less virulent strains [25]. Alterations in virulence can happen with a small number of selections. For example, Enterococcusfaecalis evolved from a pathogen to a commensal strain in 15 passages in a worm model, but most of the worm phenotype changed after just 5 rounds of bacterial selection [26]. Mouse studies using serial passaging of a mouse-modified SARS-CoV-2 virus have demonstrated higher strain virulence and a corresponding linear decrease in body weight after 10 passages. Notably, the virus acquired five nonsynonymous mutations throughout those serial passages [27].
Based upon these findings and the potentially large number of passages in the human outbreak, it can be expected that significant evolution could occur in the SARS-CoV-2 genome. Ultimately, natural and vaccine-driven evolution could result in the circulation of SARS-CoV-2 strains that have differential effects on immune escape, virulence, transmission, severity, etc. Indeed, a recent evaluation of within-host variation in 41 multiply sampled individuals demonstrated little concordance of minor allele frequencies between measurements, suggesting that such evolution occurs regularly and could be a source of stochastic variation emergence [28].
Others have previously found and characterized individual variants with in vitro assays [29] or provided correlates of severity with any change in protein coding [30], or genomic correlates of mortality [31]. We have taken a comprehensive approach to describe all variants associated with the mild or severe outcome regardless of whether it is synonymous. While there are challenges in identifying signatures of selection in noncoding regions [32], empiric tests of selection and structural modeling can identify regions under selection without using the ratio of nonsynonymous to synonymous mutations [32]. Studies of selection in SARS-CoV-2 have also advised not to underestimate the role of synonymous substitutions [33]. Beyond RNA molecule interactions, there also appear to be selective pressures on which codon is used for an amino acid, which could be attributed to tRNA abundance [34] or could be related to broader patterns of host RNA editing involving deamination and similar mechanisms [35–37]. Alternatively, a variant correlated with a severity might represent an epiphenomenon that is linked to multiple variants that each have a smaller association with severity.
Regardless of the applicability of these explanations for why synonymous changes could be important indicators of virulence, we wanted to take an agnostic approach to characterize all variants correlated with severity so that they could be further resolved with additional study and additional surveillance (particularly among asymptomatic cases). These studies, in addition to comprehensive prognostic study, could better clarify how unexpected a patient’s severity is as compared or combined with additional risk factors. For example, one mutation we identified, C13620T (ORF1ab: D4452D) is associated with 5.8 times the odds of severe disease (Supplementary Table 2). Although it does not result in any change in an amino acid of the NSP12 (RNA-dependent RNA polymerase), it could result in altered expression. Because NSP12 is required for the transcription of all viral RNA in coronaviruses [38], increased replication could increase virulence.
Other mutations were nonsynonymous. We identified a previously reported spike mutation, G25088T (S: V1176F), as an important indicator for COVID-19 disease severity (OR = 49, Supplementary Table 2). G25088T (S: V1176F) is part of a trio of Spike mutations in the VOI P.2/20J strain first detected in Brazil in April 2020. Recent protein modeling studies have indicated that both mutations cause favorable energetic changes that result in a more flexible Spike protein and can change RBD-ACE2 binding. Importantly, both mutations have been associated with higher mortality rates, and are therefore expected to have significant impacts on public health [39, 40]. Other mutations were observed less commonly, but could still be relevant for understanding higher viral pathogenesis. The G26144T causes an amino acid change G251V in Orf3a and in our model was associated with 4.4 times the odds of severe disease (Supplementary Table 2). Protein trafficking is a complex multifactorial process [41] and Orf3a functions as a modulator of the trafficking properties of the spike protein of SARS-1 and is dependent on the protein-protein interaction of Orf3a and S [42]. A structural analysis of the G26144T (ORF3a: G251V) in Orf3a of SARS-CoV-2 results in significant changes in the overall protein structure and weaker affinity for both the S and M proteins with Orf3a. One possible outcome of the weaker Orf3a-S and Orf3a-M interaction could be an increased Orf3a-TRAF3 interaction resulting in increased activation of the NLRP3 inflammasome by promoting TRAF3-dependent ubiquitination of ASC [43, 44]. Thus, the altered protein–protein interaction of the G251V Orf3a may impact the trafficking of Orf3a resulting in a higher propensity for inflammatory cytokine activation. Interestingly, at the time of writing, G251V has not yet been associated with VOI or VOC.
We also identified that the C28311T (N: P13L) mutation was associated with a lower OR of 0.11 (Supplementary Table 2). This mutation lies within the probe of the N1 assay in the CDC’s PCR assay [45], creating a P13L change in the nucleocapsid protein. A previous study evaluated how this mutation may alter protein–protein interaction and proposed it impacted virus stability, potentially contributing to lower pathogenesis [46]. At this time, P13L has not yet been associated with VOI or VOC. Additional follow-up studies will help to illuminate the effects of these variants on viral fitness, infectivity, host response and evolutionary trajectory.
Identifying genetic variants associated with outcomes could provide mechanistic understanding of the viral life cycle [47]. Efforts such as the CDC’s annual influenza surveillance rely upon understanding those key genetic variants to predict the seasonal intensity and attempt to develop effective countermeasures (vaccines) (https://www.cdc.gov/flu/weekly/overview.htm).
Although deep molecular insights are important, they are not necessary for public health applications. The CDC offers symptom-based criteria for prioritizing testing and symptom-based criteria are also included in recommendations for prioritizing contact tracing (https://www.cdc.gov/coronavirus/2019-ncov/php/contact-tracing/contact-tracing-plan/contact-tracing.html# (11 July 2021, date last accessed)). By prioritizing cases and contacts with symptoms (as compared with asymptomatic cases and their asymptomatic contacts) in addition to the recommended global mitigation efforts, there could be relative selective pressure against strains that are more likely to cause symptoms. This could favor the emergence of attenuated strains over the long term.
The COVID-19 pandemic demonstrated the limitations of the global healthcare system in intensive care units, mechanical ventilators and emerging therapeutics and other medical countermeasures [48, 49]. Early in the outbreak, cities such as New York became inundated with infections and their ability to adequately sort and treat patients was quickly overwhelmed [50]. The existence of a rapid and accurate tool that could help identify COVID-19 patients or clusters that are more likely to experience severe symptoms or require intensive medical resources (e.g. inpatient hospitalization and ventilation) may be able to help healthcare systems allocate resources to the regions with the most critical needs. Therefore, by providing a molecular risk factor for more severe outcomes, these findings could help prioritize limited treatment supplies to those at greatest risk, particularly as therapeutic interventions for infectious disease often need to be given early in the disease course (e.g. empiric antivirals for influenza).
There are limitations with our analyses. First, the SARS-CoV-2 genomes uploaded to GISAID are not necessarily representative of all circulating genomes, which can introduce a selection or sampling bias into our analyses based on region, patient severity, or other unmeasured factors. Indeed, some of the mutations associated with mild outcomes in our dataset are rare and the associations we measured could potentially be spurious. In Supplementary Fig. S3, we show the region-specific sampling patterns over time by our patient severity categorization. We sought to mitigate these limitations by eliminating the categories that had ambiguous severity (e.g. ‘live’ or ‘recovered’), and adjusting the associations for known confounders. Second, we use a total of 4499 variants in our model from 3386 sequences, which can run the risk of producing a model that is overfit, due to a potential imbalance of coefficients and datapoints. To assess the risk of overfitting, we plotted a Learning Curve (Supplementary Fig. S5) to further investigate whether our model was overfit. If the model was overfit, we would expect to see a large differential in between the training and test set scores, indicating that it was not generalizable to new data. Because the training set score converges on the test set score when the number of training examples is above 2000, which is the case for our data set, these results indicated to us that there is minimal overfitting of the model as long as a sufficient number of rows are included in the training set. Thirdly, the lack of specific data in GISAID on underlying health conditions is also a confounding variable in our analyses, as we could not control for the prevalence of any comorbidities. The lack of this information may obscure the true contribution of viral genomic variants, as we cannot stringently stratify our data set by systemic health and social disparities that are known to significantly impact COVID-19 patient outcomes. Given these caveats, we do not seek to make causal claims about any specific viral genomic variant. Ultimately, in aggregate, these variants are predictive of outcome and the candidates we identify can be further studied using molecular and other methods.
In summary, we have demonstrated that some SARS-CoV-2 genomic variants are strong predictors of COVID-19 disease severity, and these variants appear to be commonly circulating. This study provides a rationale for prioritizing control efforts for cases and populations manifesting with unusually high severity, consistent with symptom-based criteria for testing used by the CDC. Longitudinal monitoring of genomic variants within a novel pathogen such as SARS-CoV-2 will be important for understanding drivers and effects of its evolution and ultimately, its spread or control.
Supplementary data
Supplementary data is available at EMPH online.
Conflict of interest: None declared.
Disclaimer
The views expressed in this article are those of the authors and do not necessarily reflect the official policy or position of the Air Force, the Department of Defense, or the U.S. Government. Clearance PAIRS CASE #2020-0613.
Supplementary Material
Acknowledgements
The authors gratefully acknowledge the contributors, originating and submitting laboratories of the sequences from the GISAID EpiCoV Database (Elbe and Buckland-Merrett, 2017; Shu and McCauley, 2017), the basis of this research. A detailed list of contributing labs to GISAID is available in the Supplementary Information.
Funding
This study was funded/supported by the United States Air Force, Air Force Research Laboratory, 711 HPW, Wright Patterson, AFB. Contract Support was provided by Booz Allen Hamilton.
REFERENCES
- 1. Elledge SJ. 2.5 Million person-years of life have been lost due to COVID-19 in the United States. medRxiv 2020. DOI: 10.1101/2020.10.18.20214783. [DOI]
- 2. Meyerowitz-Katz G, Merone L. A systematic review and meta-analysis of published research data on COVID-19 infection-fatality rates. medRxiv 2020. DOI: 10.1101/2020.05.03.20089854. [DOI] [PMC free article] [PubMed]
- 3. Ioannidis JP. The infection fatality rate of COVID-19 inferred from seroprevalence data. Bulletin of the World Health Organization 2020. DOI: 10.2471/BLT.20.265892. [DOI] [PMC free article] [PubMed]
- 4. Levin AT, Hanage WP, Owusu-Boaitey N. et al. Assessing the age specificity of infection fatality rates for COVID-19: systematic review, meta-analysis, and public policy implications. medRxiv 2020. DOI: 10.1101/2020.07.23.20160895. [DOI] [PMC free article] [PubMed]
- 5. O'Driscoll M, Dos Santos GR, Wang L. et al. Age-specific mortality and immunity patterns of SARS-CoV-2 infection in 45 countries. medRxiv 2020. DOI: 10.1101/2020.08.24.20180851.
- 6. Onder G, Rezza G, Brusaferro S.. Case-fatality rate and characteristics of patients dying in relation to COVID-19 in Italy. JAMA 2020; 323:1775–6. [DOI] [PubMed] [Google Scholar]
- 7. Horwitz LI, Jones SA, Cerfolio RJ. et al. Trends in Covid-19 risk-adjusted mortality rates in a single health system. J Hospital Med 2021; 16:90–2. [DOI] [PubMed] [Google Scholar]
- 8. Dennis J, McGovern A, Vollmer S. et al. Improving COVID-19 critical care mortality over time in England: a national cohort study. medRxiv 2020; 2020. DOI: 10.1101/2020.07.30.20165134.
- 9. Armengaud J, Delaunay‐Moisan A, Thuret JY. et al. The importance of naturally attenuated SARS‐CoV‐2 in the fight against COVID‐19. Environ Microbiol 2020; 22:1997–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ewald PW. Evolution of virulence. Infect Dis Clin N Am 2004; 18:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Elbe S, Buckland-Merrett G.. Data, disease and diplomacy: GISAID's innovative contribution to global health. Glob Challeng 2017; 1:33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shu Y, McCauley J.. GISAID: global initiative on sharing all influenza data–from vision to reality. Eurosurveillance 2017; 22:30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 2018; 34:3094–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cingolani P, Platts A, Wang LL. et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012; 6:80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cingolani P, Patel VM, Coon M. et al. Using Drosophila melanogaster as a model for genotoxic chemical mutational studies with a new program SnpSift. Front Genetics 2012; 3:35.; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McKinney W. Data structures for statistical computing in python. In Proceedings of the 9th Python in Science Conference. Austin, TX, 2010, pp. 51–6. http://conference.scipy.org/proceedings/scipy2010/pdfs/mckinney.pdf (11 July 2021, date last accessed).
- 17. Pedregosa F, Varoquaux G, Gramfort A. et al. Scikit-learn. Machine learning in Python. J Mach Learn Res 2011; 12:2825–30. [Google Scholar]
- 18. Hunter JD. Matplotlib: a 2D graphics environment. Comput Sci Eng 2007; 9:90–5. [Google Scholar]
- 19. Waskom M, Botvinnik O, Gelbart M. et al. mwaskom/seaborn: v0. 11.0 (Sepetmber 2020), 2020. DOI: 10.5281/zenodo.4019146.
- 20. Zulkower V, Rosser S. DNA Features Viewer, a sequence annotations formatting and plotting library for Python. bioRxiv 2020. DOI: 10.1101/2020.01.09.900589. [DOI] [PubMed]
- 21. Harris CR, Millman KJ, van der Walt SJ. et al. Array programming with NumPy. Nature 2020; 585:357–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Virtanen P, Gommers R, Oliphant TE, SciPy 1.0 Contributors et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat Methods 2020; 17:261–72.; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Matsushita K, Ding N, Kou M. et al. The relationship of COVID-19 severity with cardiovascular disease and its traditional risk factors: a systematic review and meta-analysis. Global Heart 2020; 15:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Holmes EC. The Evolution and Emergence of RNA Viruses. New York, NY, USA: Oxford University Press, 2009. [Google Scholar]
- 25. Rochman ND, Wolf YI, Koonin EV. Evolution of human respiratory virus epidemics. medRxiv 2020.11.23; 2020:20237503. DOI: 10.1101/2020.11.23.20237503.
- 26. King KC, Brockhurst MA, Vasieva O. et al. Rapid evolution of microbe-mediated protection against pathogens in a worm host. ISME J 2016; 10:1915–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Leist SR, Dinnon IIK, Schäfer A. et al. A mouse-adapted SARS-CoV-2 induces acute lung injury and mortality in standard laboratory mice. Cell 2020; 183:1070–85.e12.; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lythgoe KA, Hall M, Ferretti L, et al. ; on behalf of the Oxford Virus Sequencing Analysis Group (OVSG). SARS-CoV-2 within-host diversity and transmission. Science 2021; 372:eabg0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yao H-P, Lu X, Chen Q. et al. Patient-derived mutations impact pathogenicity of SARS-CoV-2. CELL-D-20-01124 2020. DOI: 10.2139/ssrn.3578153.
- 30. Nagy A, Pongor S, Gyorffy B. Different mutations in SARS-CoV-2 associate with severe and mild outcome. medRxiv 2020. DOI: 10.1101/2020.10.16.20213710. [DOI] [PMC free article] [PubMed]
- 31. Hahn G, Wu CM, Lee S. et al. Mutations in SARS-CoV-2 spike protein and RNA polymerase complex are associated with COVID-19 mortality risk. bioRxiv 2020. DOI: 10.1101/2020.11.17.386714.
- 32. Berrio A, Gartner V, Wray GA.. Positive selection within the genomes of SARS-CoV-2 and other coronaviruses independent of impact on protein function. PeerJ 2020; 8:e10234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Velazquez-Salinas L, Zarate S, Eberl S. et al. Positive selection of ORF1ab, ORF3a, and ORF8 genes drives the early evolutionary trends of SARS-CoV-2 during the 2020 COVID-19 pandemic. Front Microbiol 2020; 11:550674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dilucca M, Forcelloni S, Georgakilas AG. et al. Codon usage and phenotypic divergences of SARS-CoV-2 genes. Viruses 2020; 12:498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Simmonds P. Rampant C→ U hypermutation in the genomes of SARS-CoV-2 and other coronaviruses: causes and consequences for their short-and long-term evolutionary trajectories. MSphere 2020; 5:e00408–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Matyášek R, Kovařík A.. Mutation patterns of human SARS-CoV-2 and Bat RaTG13 coronavirus genomes are strongly biased towards C> U transitions, indicating rapid evolution in their hosts. Genes 2020; 11:761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Di Giorgio S, Martignano F, Torcia MG. et al. Evidence for host-dependent RNA editing in the transcriptome of SARS-CoV-2. Sci Adv 2020; 6:eabb5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Subissi L, Posthuma CC, Collet A. et al. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc Natl Acad Sci USA 2014; 111:E3900–E3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Farkas C, Mella A, Haigh JJ. Large-scale population analysis of SARS-CoV2 whole genome sequences reveals host-mediated viral evolution with emergence of mutations in the viral Spike protein associated with elevated mortality rates. medRxiv 2020. DOI: 10.1101/2020.10.23.20218511.
- 40. Turoňová B, Sikora M, Schürmann C. et al. In situ structural analysis of SARS-CoV-2 spike reveals flexibility mediated by three hinges. Science 2020; 370:203–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gibbons TF, Storey SM, Williams CV. et al. Rotavirus NSP4: cell type-dependent transport kinetics to the exofacial plasma membrane and release from intact infected cells. Virol J 2011; 8:278– 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tan Y-J. The Severe Acute Respiratory Syndrome (SARS)-coronavirus 3a protein may function as a modulator of the trafficking properties of the spike protein. Virol J 2005; 2:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Siu KL, Yuen KS, Castano‐Rodriguez C. et al. Severe acute respiratory syndrome Coronavirus ORF3a protein activates the NLRP3 inflammasome by promoting TRAF3‐dependent ubiquitination of ASC. FASEB J 2019; 33:8865–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Issa E, Merhi G, Panossian B. et al. SARS-CoV-2 and ORF3a: nonsynonymous mutations, functional domains, and viral pathogenesis. Msystems 2020; 5:e00266–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lu X, Wang L, Sakthivel SK. et al. US CDC real-time reverse transcription PCR panel for detection of severe acute respiratory syndrome coronavirus 2. Emerg Infectious Dis 2020; 26:1654–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Oulas A, Zanti M, Tomazou M. et al. Generalized linear models provide a measure of virulence for specific mutations in SARS-CoV-2 strains. bioRxiv 2020. DOI: 10.1101/2020.08.17.253484. [DOI] [PMC free article] [PubMed]
- 47. Geoghegan JL, Holmes EC.. The phylogenomics of evolving virus virulence. Nat Rev Genetics 2018; 19:756–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Grasselli G, Pesenti A, Cecconi M.. Critical care utilization for the COVID-19 outbreak in Lombardy, Italy: early experience and forecast during an emergency response. JAMA 2020; 323:1545–6. [DOI] [PubMed] [Google Scholar]
- 49. White DB, Lo B.. A framework for rationing ventilators and critical care beds during the COVID-19 pandemic. JAMA 2020; 323:1773–4. [DOI] [PubMed] [Google Scholar]
- 50. Chin V, Samia NI, Marchant R. et al. A case study in model failure? COVID-19 daily deaths and ICU bed utilisation predictions in New York State. Eur J Epidemiol 2020; 35:733–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.