Summary
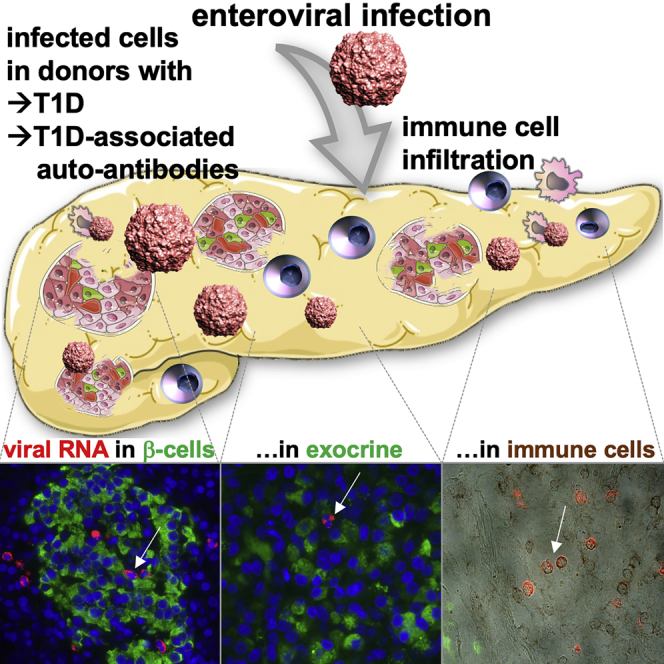
Enteroviral infections have been associated with autoimmunity and type 1 diabetes (T1D), but reliable methods to ascertain localization of single infected cells in the pancreas were missing. Using a single-molecule-based fluorescent in situ hybridization (smFISH) method, we detected increased virus infection in pancreases from organ donors with T1D and with disease-associated autoantibodies (AAb+).
Although virus-positive β cells are found at higher frequency in T1D pancreases, compared to control donors, but are scarce, most virus-positive cells are scattered in the exocrine pancreas. Augmented CD45+ lymphocytes in T1D pancreases show virus positivity or localization in close proximity to virus-positive cells. Many more infected cells were also found in spleens from T1D donors.
The overall increased proportion of virus-positive cells in the pancreas of AAb+ and T1D organ donors suggests that enteroviruses are associated with immune cell infiltration, autoimmunity, and β cell destruction in both preclinical and diagnosed T1D.
Keywords: type 1 diabetes, T1D, autoimmunity, enterovirus, smFISH, pancreas, coxsackievirus, β-cell, autoantibodies, AAb
Graphical abstract
Highlights
Enterovirus-infected cells are significantly increased in AAb+ and T1D pancreases
Most of the virus-positive cells are scattered within the exocrine pancreas
Virus-positive β cells are rare but more in T1D compared to control donors
Also elevated in T1D donors, there is more infection in spleens than in pancreases
By highly sensitive, single-molecule-based fluorescent in situ hybridization for viral RNA, Geravandi et al. found significantly increased enterovirus-infected cells in pancreases from organ donors with type 1 diabetes (T1D) and with disease-associated autoantibodies (AAb+), which were identified as exocrine, endocrine, and immune cells and scattered within the pancreas.
Introduction
Elevated apoptotic signaling cascades, hyper-expression of histocompatibility leukocyte antigen (HLA)-I clusters, as well as islet infiltration by T cells and other immune cells represent key pathological hallmarks of type 1 diabetes (T1D), leading to interferon response and T-cell-mediated β cell destruction, impaired insulin secretion, and severe hyperglycemia.1 A complex genetic susceptibility with polymorphisms in immune response, viral response, and β cell function genes are strong risk factors for disease development. Overall, approximately 50 loci have been linked to T1D risk.2,3 Despite the genetic predisposition, the rapid rise in the incidence of childhood T1D worldwide, at an estimated average annual increase of 3.9% over the past decades, is too high to result only from genetic causes;4, 5, 6 we also know from monozygotic twins that genetics do not fully explain T1D, given that disease concordance does not reach 100% (30%–70% range).7 Earlier studies have linked T1D risk to diverse environmental factors, including epidemiological, pathological, and in vitro studies that have implicated enteroviruses as initiators of autoimmunity and β cell failure in genetically susceptible individuals.1,8, 9, 10 Enteroviral infection could indeed affect the pancreas, and numerous studies show that β cells are highly susceptible to enteroviral infection in vitro, with subsequent activation of the interferon response pathway, upregulation of interferon (IFN)-inducible genes, massive cytokine and chemokine production, β cell death, T cell activation, and thus the triggering of autoimmunity and T1D.11, 12 Previous studies of pancreases from deceased patients with T1D have reported increased prevalence of enterovirus infections by examining the presence of the viral capsid protein (VP1)8 by immunohistochemistry. Yet detection of enterovirus RNA in pancreas has remained challenging, in part due to limited access to suitable pancreas specimens and sensitive methodology to detect RNA. Importantly, in the Diabetes Virus Detection (DiViD) study, enterovirus was found in pancreas biopsies of newly diagnosed patients with T1D.13 As part of the collaborative efforts of the Network for Pancreatic Organ Donors with Diabetes (nPOD)-Virus Group, an international collaborative effort that investigates the role of viruses in T1D, we aimed at examining the presence of enterovirus in pancreas samples from organ donors with and without T1D from the well-characterized human pancreatic donor tissue established by nPOD.14 To this end, we developed a robust and highly sensitive method to detect very low amounts of virus RNA and potentially even replication-deficient virus in pancreas specimens.15 We have previously established an adapted single-molecule in situ hybridization (smFISH) method that can detect single RNA molecules in situ using short (∼20 nt) fluorescently labeled oligonucleotides, which anneal to common regions of the RNA genome of the coxsackievirus family. We successfully detected viral RNA in both cultured cells and formalin-fixed, paraffin-embedded (FFPE) samples tissue sections, in combination with classical immunostaining. Short probes were able to detect viral infection at lower viral loads than classical immunostaining and even PCR.16,17 For initial validation, we performed several blinded concordance studies using pancreases from control and T1D patients with residual β cells, obtained from the nPOD and the Exeter (UK) cohorts.17
Here, we report results from an in situ analysis of pancreatic virus infiltration of nPOD organ donors without diabetes (control), with T1D, and with T1D-associated autoantibodies (AAb+) without diabetes, possibly representing individuals at increased risk of future T1D. The aims of this study were (1) to identify whether there is a correlation of pancreatic enteroviral RNA expression, autoantibody positivity, and T1D and (2) to localize and characterize enteroviral infection in the pancreas.
Results and discussion
smFISH detected increased enterovirus RNA in the pancreas of T1D and AAb+ donors
With smFISH, we successfully detected enteroviral RNA in FFPE fixed pancreases. As many of us undergo mostly asymptomatic enteroviral infections throughout life, we had initially expected to find viral RNA in all pancreases. The proportion of donors expressing enterovirus RNA in pancreatic cells were 64% (9/14) among controls, 90% (9/10) in AAb+ donors without T1D, and 100% (15/15) among T1D donors (Tables S1–S3 for donors’ demographic data).
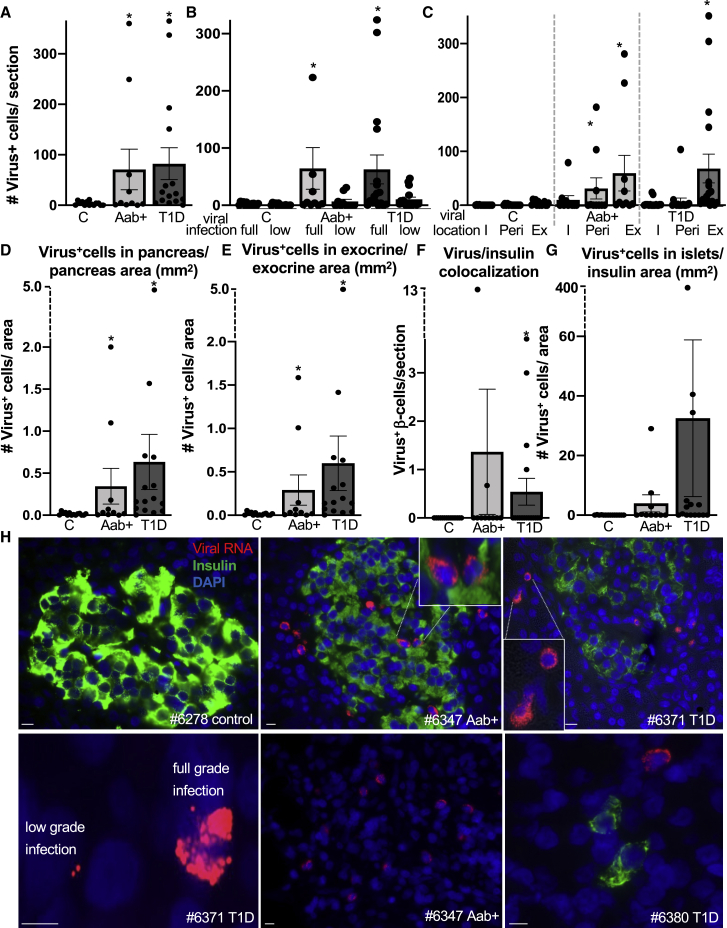
Although control donors had a mean of 3.7 ± 1 (SE) viral RNA-containing cells in their pancreases, there were 19- and 22-fold more in pancreases from AAb+ (mean 71 ± 40 [SE]; p = 0.03) and T1D donors (mean 82 ± 29 [SE]; p = 0.01), respectively, compared to the control group (Figure 1A).
Figure 1.
Pancreatic enteroviral RNA in AAb-positive and T1D patients within both the endocrine and exocrine pancreas
(A–H) Detection and quantification of viral RNA (red), insulin (green), and DAPI (blue) in FFPE pancreases from control donors without diabetes (n = 14), donors without diabetes but expressing T1D-associated autoantibodies (AAb+) (n = 10), and donors with T1D (n = 15).
(A–C) Data are presented as (A) mean number of all viral RNA+ cells throughout the whole pancreas section, (B) the mean number of full-grade (full; ≥10 single puncta/cell) or low-grade (low; 1–9 single puncta/cell) infected cells, and (C) their localization in the pancreas within insulin-containing islets (I), within the periphery of 3 cells of insulin-containing islets (Peri), and within the exocrine pancreas (Ex).
(D and E) All (D) viral mRNA+ cells were normalized to the pancreas area of the respective section and (E) viral mRNA+ cells in the exocrine area were normalized to the exocrine area of the respective section.
(F and G) Viral mRNA+ β cells (F) co-staining with insulin (absolute value) and viral mRNA+ cells (G) within islets normalized to islet area (insulin+ stained area in mm2).
(H) Representative microscopical pictures from immunostainings of control, AAb+, and T1D pancreatic sections. Stainings were performed in two technical replicas. Scale bars depict 10 μm. ∗p<0.05 to control.
The aggregated signal from all enteroviral probes bound to a single target molecule to produce a puncta signal requires >17 probes to bind before the observation of a fluorescence signal.18 This allows clear single-molecule viral RNA staining and the counting of the corresponding puncta. Based on the numbers of such puncta, we classified infections as low-grade (1–9 puncta per cell) and full-grade infection (≥10 puncta per cell; Figures 1B and 1H, lower left panel; Table S1). Although there was a significant increase in low-grade infected cells in T1D donors (9-fold), the numbers of full-grade infected cells were 26-fold and 29-fold higher in AAb+ and T1D pancreases, respectively, compared to controls (Figure 1B; Table S1). Moreover, 4/10 AAb+ and 9/15 T1D pancreases harbored more than 10 fully infected cells containing enteroviral RNA compared to 0/14 control pancreases (Table S1). A closer look at the virus-containing cells depicts their morphological distinction from their neighbors, with loss of structured membranes and cellular disintegration (Figures S1A, S1B, S2, and S3, lower right panels), as observed in response to viral infection.19
Initially, probes were designed to detect the positive-stranded RNA of the entire range of group B enteroviruses.17 In order to verify whether viruses have at least initially replicated in the cell, an additional probe set was designed to detect the negative-stranded RNA of conserved regions of the coxsackievirus B3 (CVB3) consensus-based sequence (M33854.1).17 Such proof-of-concept approach delivered very similar results in the pancreas, namely enteroviral RNA labeling within and in proximity of the endocrine pancreas as well as scattered in the exocrine part (Figures S1C–S1F).
Pancreatic enteroviral mRNA in AAb-positive and T1D organ donors in both the endocrine and exocrine pancreas
We found a significant increase of exocrine and endocrine cells of the pancreas carrying low- and full-grade virus infections in AAb+ and T1D, compared to controls, when we normalized to the whole pancreas area (Figure 1D). Moreover, virus-positive cells located in the exocrine pancreas normalized to the area of the exocrine pancreas were increased in AAb+ as well as in T1D pancreases compared to controls (Figures 1E and S2).
Double staining of viral RNA with the β cell marker insulin allowed us to localize and quantify the viral RNA within β cells. Despite variation and the low number of virus-positive β cells, these were significantly more numerous among residual β cells of T1D donors (Figures 1F and 1H), compared to control donors. There was no significant difference for AAb+ donors. Normalization of these virus-positive cells to the area of all remaining insulin-containing islets showed again a high variation and a non-significant tendency toward higher intra-islet viral RNA in T1D (Figures 1G and 1H). Based on availability of T1D donors, they were selected for the study to include donors with long (>3 years; n = 5) and short disease duration (<3 years; n = 10), with most of the donors presenting remaining insulin-containing islets (ICIs). With longer disease duration, there was a significant reduction in the number of ICIs, although the trend toward lesser virus-containing cells was not significant (p = 0.07; Figure S3A).
Quantification of β cells in the pancreas also confirmed the expected β cell loss in T1D: the loss in number of islets per section; the reduction in absolute β cell volume per section; as well as the percentage of insulin containing to all pancreatic cells (Figure S3B–S3D). We also found a reduction in the proportion of β cells relative to whole pancreas area in non-T1D AAb+ organ donors from this cohort (Figures S3C and S3D), which has not been seen before in similarly small cohorts,20, 21, 22, 23 especially not in adult donors, who show little evidence of β cell loss.24 Donor ages across studies may explain the difference; our study analyzed younger AAb+ donors (mean age of 20 years; Tables S2 and S3) than previous ones (mean age >30).20, 21, 22, 23
These observations clearly show that enteroviruses do not exclusively target β cells but are also found in the exocrine pancreas. The observation of virus-positive cells in the exocrine pancreas may provide a pathophysiological link to previous studies reporting reduced pancreas weight in AAb+ and T1D organ donors; moreover, living relatives of T1D patients with autoantibodies were reported to have a 25% reduction in pancreas weight.25,26 Moreover, acinar loss has been reported in the T1D pancreas.27,28 Altogether, we speculate that viral infections may contribute to pancreatic both endocrine and exocrine abnormalities that are being reported throughout the natural history of T1D.
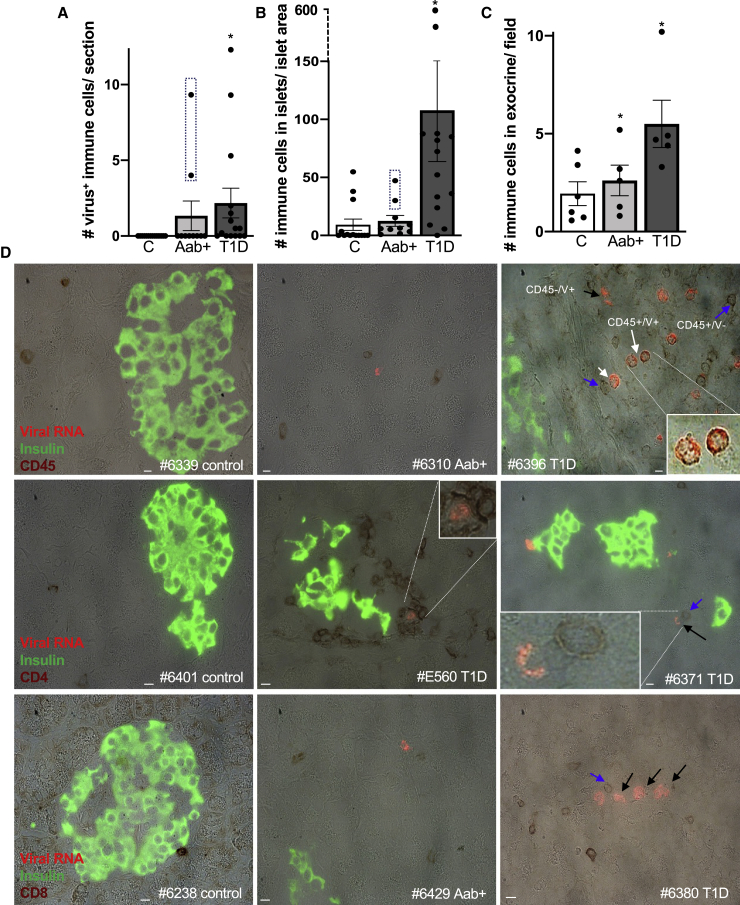
Viral RNA is increased in lymphocytic cells in T1D
Many virus-positive cells did not stain for insulin or glucagon (Figures S3E–S3H) nor were located within or near islets and did not stain for amylase, which marks exocrine cells (Figure S2). Thus, we evaluated their co-localization with markers of ductal and immune cells and their proximity to such cells. We did find a minority of CK-19-positive and viral-positive cells within pancreatic ducts (1–3 cells/pancreas in 4 AAb+ and 1–15 cells/pancreases in 6 T1D pancreases; data not shown), although no virus-positive ductal cells were observed in controls. We also found a significant increase of virus/CD45 co-positive cells in T1D pancreases, compared to nondiabetic controls, in whom no virus was found in CD45+ cells (Figure 2A). Virus/CD45 co-positive cells were identified in 9 out of 15 T1D cases. Of note, such virus/CD45 co-localization was only seen in pancreases of two of three donors with dual AAb+, but not among single AAb+ donors (Figure 2A). T1D pancreases and these two from dual AAb+ donors also presented a higher number of CD45+ cells within islets as well as in the exocrine pancreas, compared to controls. There was no significant increase in CD45-lymphocyte infiltration in pancreases from single AAb+ donors (Figures 2B and 2C) compared to controls. As individuals with multiple islet AAb have a higher risk of progression to T1D compared to those with a single AAb (∼80%–90% versus ∼10% in 5 years),24 the higher pancreatic CD45-lymphocyte counts may be an indicator of more aggressive pathology. As shown by representative examples (Figures 2D, S2, S3G, and S3H), virus-positive immune cells are often found in close proximity within three cell layers to insulin+ islet cells and in the exocrine pancreas. Moreover, many virus+/CD45− cells were located in proximity (within three cell layers) to CD45+ immune cells; further staining demonstrated some of these cells included CD4+ and CD8+ T cells (Figure 2D). Staining for negative-stranded enteroviral RNA confirmed viral replication in immune cells in T1D (Figure S1F). The observed increased virus load in pancreatic immune cells in dual AAb+ and in T1D donors may be a starting point of the disease; further studies on T1D at-risk donors are needed to confirm such hypothesis.
Figure 2.
Enteroviral RNA is increased in lymphocytic cells in T1D
(A–D) Detection and quantification of viral RNA (red), insulin (green), and CD45, CD4, or CD8 in bright field (dark red) in FFPE pancreases from autopsy from control donors without diabetes (A, B, and D: n = 14; C: n = 6), with T1D-associated autoantibodies (A, B, and D: AAb+, n = 10; C: n = 5), and with T1D (A, B, and D: n = 15; C: n = 5) and presented as (A) mean number of viral RNA+/CD45 co-positive cells throughout the whole pancreas section and (B) quantification of CD45 co-positive cells within insulin-containing islets and (C) within the exocrine pancreas. 15 randomly chosen parts of equal size of each pancreas section were analyzed. The dashed boxes in (A) and (B) mark two donors of the study cohort with dual AAb+, showing increased virus/CD45 co-localization (A) and CD45+ cells within islets (B), compared to controls and single AAb+ donors.
(D) Representative microscopical pictures from immunostainings of control, AAb+, and T1D pancreatic sections. White arrows show viral RNA+/CD45+ co-positive cells, black arrows viral RNA+/CD45− cells, and blue arrows viral RNA−/CD45+ cells next to a viral RNA+ cell. Stainings were performed in two technical replicas. Scale bars depict 10 μm. ∗p<0.05 to control.
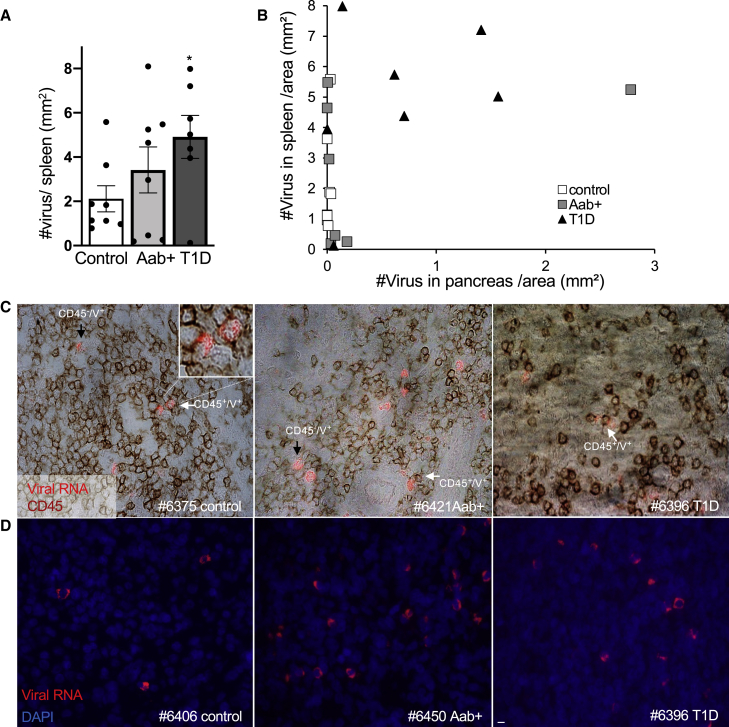
Viral RNA is increased in the spleen in T1D
Well known from animal models of diabetes, the spleen harbors autoreactive T cells, as splenocytes can cause disease in adoptive transfer experiments.29,30 Moreover, there is evidence that islet-infiltrating T cells from deceased patients are also present in the spleen.31,32 Therefore, we investigated whether enteroviral RNA is also carried by lymphocytes in the spleen. Indeed, viral RNA was distributed throughout the spleen sections from nondiabetic controls, AAb+, as well as T1D donors, with many more infected cells than seen in the pancreas (188-fold in controls and 8-fold higher both in AAb+ and T1D spleens, compared to the respective pancreases; Figure 3). Quantification of virus+ cells/spleen section area revealed increased virus+ cells in T1D donors compared to controls (Figure 3A); virus+ cells either co-stained with CD45 or were next to CD45+ cells (Figure 3C), similar to what has been observed in the pancreas. The number of virus load in spleen was independent from the pancreas, as we could not find a correlation between the number of overall virus+ cells in pancreas and spleen from the same patients (Figure 3B).
Figure 3.
Enteroviral RNA is increased in the spleen in T1D
(A and B) Detection and quantification of viral RNA (red), CD45 in bright field (dark red), and DAPI in blue in FFPE spleen sections from autopsy from a subset of the analyzed pancreas donors without diabetes (n = 8), with T1D-associated autoantibodies (AAb+) (n = 8), and with T1D (n = 7) and presented as (A) mean number of viral RNA+ cells/spleen section area and (B) a correlation of the mean number of viral RNA+ cells/spleen section area with the mean number of viral RNA+ cells/pancreas section area.
(C and D) Representative microscopical pictures from immunostainings of control, AAb+, and T1D pancreatic sections of viral RNA/CD45 double staining (C) and viral RNA/DAPI double staining (D). White arrows show viral RNA+/CD45+ co-positive cells and black arrows viral RNA+/CD45− cells next to CD45+ cells. Stainings were performed in two technical replicas. Scale bars depict 10 μm. ∗p<0.05 to control.
Increased enteroviral RNA found in the spleen in T1D is also in line with identified enteroviral RNA in peripheral blood mononuclear cells (PBMCs) from patients with recent onset as well as long-term T1D.33, 34, 35, 36 Enterovirus presence in T1D-PBMCs is linked with the T1D risk allele HLA-DR4, carried by all virus-positive patients in this previous study.35 Such correlations are difficult to predict from a small number of patients, as also in 4 out of 10 patients, no virus was identified in PBMCs in this previous study. Also in our cohort, the limited sample size prevented reaching conclusions about possible associations between splenic or pancreatic virus loads and T1D HLA-risk alleles. Although a double AAb+ donor with the virus load >200 infected cells in the pancreas had the high-risk alleles HLA-DRB1∗03 (DR3) and DQB1∗03:02 (DQ8), neither another double AAb+ donor nor another single AAb+ donor with >300 infected cells had the same high-risk alleles. Similarly, some T1D donors with large numbers of infected cells had the high-risk alleles, which contrasts to another donor with the same risk alleles but with a relatively lower number of infected cells (Table S3).
Overall, we found increased presence of enteroviral RNA-positive cells in the pancreas of organ donors with T1D-associated autoantibodies (without T1D) and with T1D, many of which had a short disease duration and presented remaining insulin-containing islets, suggesting an ongoing disease process. This was enabled through a highly sensitive and specific RNA detection down to a single RNA molecule in well-preserved FFPE pancreases with the advantage of spatial cellular localization of viral RNA at a single-cell level. Previous studies were limited to VP1 staining with much lower sensitivity,17 as well as to PCR, although with similar sensitivity, its limitation lies in the low accessibility and stability of nucleic acids through RNA degradation in the pancreas.
Enteroviral RNA-infected cells could be located within the endocrine and the exocrine pancreas; their appearance as well as the close proximity to immune cells suggest virus-induced immune cell attraction and infiltration. Viruses in multiple cell types in the pancreas highlight their various impacts on diabetes pathology, affecting the β cell, exocrine, and immune cells. What signal is transferred from an infected immune cell? One can speculate that it has either already transferred the virus to islets or will do so soon.
The presence of enteroviral RNA in nondiabetic AAb+ donors is in line with previous studies that have detected viral infection in children at risk for T1D.1,37, 38, 39 Multiple enteroviral infection rounds through early childhood may trigger the disease,39 and as seen here, viral sequences remain in the tissue for years after infection, having either been among the initiating or participating factors toward the damage of the pancreas and the vulnerable β cell.
Limitations of study
Although we could clearly observe increased enteroviral infection in spleens and pancreases in organ donors with T1D, the small sample size did not allow a correlation of virus load with other strong predictors of autoimmunity or T1D severity, e.g., with HLA phenotype, number of autoantibodies, disease duration, or age of T1D diagnosis. Furthermore, our broad approach in the enteroviral probe design did not enable identification of specific viral sequences. Using smFISH, larger studies are needed for the identification of the specific viral infection association with autoimmunity and T1D.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| guinea pig anti-insulin | Dako; now Agilent, Santa C#lara, CA, USA | #A0546; RRID:AB_2617169 |
| rabbit anti-glucagon | #A0565; RRID:AB_10013726 | |
| mouse anti-CD45 | #M0701; RRID:AB_2314143 | |
| rabbit anti-amylase | Abcam, UK | #21156; RRID:AB_446061 |
| rabbit anti-CD4 | #133616; RRID:AB_2750883 | |
| rabbit anti-CD8 | #4055; RRID:AB_304247 | |
| biotin- conjugated donkey anti-rabbit | Jackson Immuno Research, PA, USA |
#711-066-152; RRID:AB_2340594 |
| biotin- conjugated donkey anti-mouse | #715-065-150; RRID:AB_2307438 | |
| biotin- conjugated donkey anti- guinea pig | #706-066-148; RRID:AB_2340452 | |
| FITC-conjugated donkey anti-rabbit | #711-096-152; RRID:AB_2340597 | |
| FITC-conjugated donkey anti-guinea pig |
#706-096-148; RRID:AB_2340454 |
|
| Biological samples | ||
| Human FFPE pancreatic sections from organ donors | this paper; Kaddis et al.40 and Foulis et al.41 | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Sudan Black | Sigma-Aldrich | #199664 |
| Pepsin | Sigma-Aldrich | #MKCN5185 |
| Critical commercial assays | ||
| VECTASTAIN ABC Kit | Vector Labs, USA |
#PK-4000 |
| DAB Peroxidase Substrate Kit | #SK-4100 | |
| VECTASHIELD® with 4’,6-Diamidin-2-phenylindol (DAPI) |
#H-1200-10 |
|
| Oligonucleotides | ||
| Sequences of probes and respective viral target regions of probes sets CVB_1, CVB_2, CVB_3 | Designed with Stellaris® RNA FISH Probe Designer (Biosearch Technologies, Inc., Petaluma, CA) | This paper; See Table S3, Busse et al.17 |
| Software and algorithms | ||
| Vision Works LS Image Acquisition and Analysis software Version 6.8 | UVP BioImaging Systems, CA, USA | N/A |
| NIS-Elements software, v3.22.11 | Nikon GmbH, Germany | N/A |
| GraphPad Prism v8.4.3 | GraphPad | N/A |
| ImageJ (v.1.52(100) | NIH, USA | N/A |
| Other | ||
| Nikon MEA53200 | Nikon GmbH, Germany | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Kathrin Maedler (kmaedler@uni-bremen.de)
Materials availability
This study did not generate new unique reagents.
Experimental models and subject details
Organ donor cohort and virus detection in FFPE tissue samples
Our study of organ donors included 15 with T1D (average disease duration 4 years, range 0-23 years), 10 autoantibody-positive (AAb+) donors without diabetes, and 14 control donors (without diabetes, autoantibody-negative). Key demographic features are described in Table S2. Formalin-fixed paraffin-embedded pancreatic and spleen tissue sections were obtained from well-characterized organ donors from the network for Pancreatic Organ Donors with diabetes (nPOD)40 and from a UK cohort41. Detection of viral RNA was performed using custom Stellaris® FISH Probes, as described in detail in the supplemental methods and previously17.
Study approval
Ethical approval for the use of human pancreatic tissue had been granted by the Ethics Committee of the University of Bremen. The study complied with all relevant ethical regulations for work with human tissue for research purposes. Organ donors are not identifiable and anonymous, such approved analyses using tissue for research is covered by the NIH Exemption 4 (Regulation PHS 398).
Method details
Virus detection in FFPE tissue samples
Custom Stellaris® FISH Probes, each recognizing various enteroviral strains, labeled with Quasar 570 were purchased from Biosearch Technologies, Inc. (Petaluma, CA) as described previously17. Three probes sets were used for positive strand enteroviral RNA detection, CVB_1 designed on the CVB3 consensus-based sequence (M33854.1) and for CVB_2 and CVB_3, 106 genome sequences of viruses belonging to the enterovirus group B family were aligned and divided into three subgroups based on sequence similarities. For each subgroup probes were designed based on newly generated consensus sequences as described before42 and were distributed throughout the whole target genome. For negative strand enteroviral RNA detection, the CVB_1 set in the 3’-5’orientation was used.
Enterovirus mRNA detection was carried out by smFISH according to a previously established highly sensitive protocol17.
Deparaffinization of FFPE Tissue sections. Residual paraffin particles and wax crystals from the embedding procedure were removed by a series of Xylene washes (20 min at 70°C; 10 min at 70°C; 10 min at room temperature), followed by rehydration by ethanol (EtOH; 100%, 100%, 95%) for 10 min each and for 1 h in 70% EtOH at room temperature. Finally, sections were rehydrated by washing with RNase free water 2 times for 2 min. All steps were performed with constant steering.
Prehybridization. Sections were incubated with 0.2M HCl for 20 min at room temperature, transferred to a 50 mL tube containing prewarmed 2xSSC and incubated in a shaking water bath at 70°C for 15 min. The sections were then washed with PBS two times for 2 min at room temperature, then incubated with prewarmed (37°C) pepsin (Sigma) for 10 min at 37°C. The sections were then washed 2 times with PBS for 1 minute at room temperature then with 0.5% Sudan Black (Sigma-Aldrich) diluted in 70% EtOH for 20 min at room temperature in order to quench any remaining autofluorescence from islets. Sections were then washed with PBS (3 times for 5 min at room temperature), then 2 times with washing buffer (1xSSC,10% formamide) for 5 min at room temperature.
Hybridization. Stellaris® FISH Probes (viral RNA 0.25 μM) were diluted 1:100 in hybridization buffer (10% w/v Dextran sulfate, 10% formamide, 2xSSC). 50 μL of all three diluted probe sets were applied to the sections, covered with a glass and kept at 37°C overnight in a humidified chamber.
Post hybridization wash. Coverslip was removed using 2xSSC, 10% formamide prewarmed to 37°C. Sections were then washed with 37°C prewarmed solution in a shaking water bath at 37°C, centrifuged shortly between each wash in an empty tube to remove washing liquid. Washing steps were as follows: 2 times 2xSSC+10% formamide for 20 min, 2 times with 2xSSC for 15 min, followed by 2 times wash with 1xSSC for 15 min, then with 0.1xSSC for 15 min and lastly with 0.1xSSC for 5 min.
One day after smFISH, pancreas sections were co-stained by classical immunostaining43 for insulin (Dako#A0546), the exocrine acinar marker amylase (Abcam#21156), the general lymphocyte marker CD45 (Dako#M0701), and the T cell activation markers CD4 (Abcam#133616) and CD8 (Abcam#4055). All spleen sections were co-stained for CD45.
VECTASHIELD® antifade mounting medium (Vector laboratories) including 4’,6-Diamidin-2-phenylindol (DAPI) was immediately added and images were acquired with a Nikon Ti MEA53200 (NIKON GmbH, Düsseldorf, Germany) microscope. A 60x oil-immersion objective was used to acquire images of the Quasar 570 labeled probes. Control images were always taken with the FITC filter at the same exposure time to ensure no false-positive signals caused by a bleed-through from one channel to another.
Quantification and statistical analysis
Quantification of cells and tissues
Morphometrical measurement of enteroviral mRNA, insulin, CD4, CD8, CD45, CD68 and amylase and the respective pancreatic and splenic tissue area was carried out using a NikonTiMEA53200 (NIKON GmbH, Düsseldorf, Germany) microscope. NIS-Elements BR software (NIKON GmbH, Düsseldorf, Germany) and ImageJ (NIH, USA) were used for image analysis. Number of virus infected cells (single infected with 1-10 puncta or fully infected with ≥ 10 puncta), number of islets and immune cells were counted manually throughout the whole sections. Immune cells within the exocrine part of the pancreas were counted in 15 randomly selected areas of equal size for each pancreas slide. Mean percentage of the β-cell area per pancreas was calculated as the ratio of insulin-positive to whole pancreatic tissue area. The exocrine area was calculated as whole insulin-positive area subtracted from whole pancreas area. “Islet periphery” was defined as signal localization within 3 cell layers of insulin containing islets, and “close proximity” as signal localization within 3 cell layers to the respective islet cells or immune cells.
Statistical analyses
All biological replica referred to “n” for each individual human pancreas, which are means of two technical replica from independent staining analyses and presented as means ± SEM. Mean differences were determined by Student’s t tests. A p value < 0.05 was considered statistically significant. Investigators were blinded to the cases during staining and microscopical analysis.
Acknowledgments
This research was only possible with the support of the Network for Pancreatic Organ Donors with Diabetes (nPOD), a collaborative type 1 diabetes research project sponsored by JDRF. Organ Procurement Organizations (OPOs) partnering with nPOD to provide research resources are listed at https://www.jdrfnpod.org/for-partners/npod-partners/. We express our deep gratitude to the donors and their families. We also thank our colleagues from the nPOD viral working group (nPOD-V) for encouragement and discussion throughout this study and especially Irina Kusmartseva (University of Florida, Miami) for help with donor procurement for this study and Richard E. Lloyd (Baylor College of Medicine) for kindly providing the enterovirus image for the graphical abstract. We acknowledge Amin Ardestani for being a great mentor for all undergrad and grad students in the lab and his great suggestions for the normalization of the results in this study. We would like to thank Katrischa Hennekens for excellent technical assistance and Alaa Issawi (all Uni Bremen) for establishing the virus-amylase co-staining. This study was supported by JDRF to the nPOD-Virus Group (JDRF SRA-5-2012-516 and JDRF 3-SRA-2017-492-A-N; head-PI: A.P.; University of Miami) and by the German Research Foundation (DFG; to K.M.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions
S.G. performed experiments, analyzed data, and wrote the paper. S.R. provided intellectual support and pathological specimen. A.P. provided intellectual support, pathological specimen, and demographic data and wrote the paper. K.M. designed and performed experiments, analyzed data, supervised the project, and wrote the paper.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We worked to ensure gender and ethnicity balance regarding the scientists who initiated and performed this study and regarding the assignment of organ donors for this study.
Published: August 17, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2021.100371.
Supplemental information
Data and code availability
Original/source data in the paper are available in Tables S1 and S3. This study did not generate any unique datasets or code.
References
- 1.Geravandi S., Liu H., Maedler K. Enteroviruses and T1D: is it the virus, the genes or both which cause T1D. Microorganisms. 2020;8:E1017. doi: 10.3390/microorganisms8071017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrett J.C., Clayton D.G., Concannon P., Akolkar B., Cooper J.D., Erlich H.A., Julier C., Morahan G., Nerup J., Nierras C., Type 1 Diabetes Genetics Consortium Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat. Genet. 2009;41:703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robertson C.C., Inshaw J.R.J., Onengut-Gumuscu S., Chen W.M., Santa Cruz D.F., Yang H., Cutler A.J., Crouch D.J.M., Farber E., Bridges S.L., Jr., Type 1 Diabetes Genetics Consortium Fine-mapping, trans-ancestral and genomic analyses identify causal variants, cells, genes and drug targets for type 1 diabetes. Nat. Genet. 2021;53:962–971. doi: 10.1038/s41588-021-00880-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hyöty H., Taylor K.W. The role of viruses in human diabetes. Diabetologia. 2002;45:1353–1361. doi: 10.1007/s00125-002-0852-3. [DOI] [PubMed] [Google Scholar]
- 5.Yeung W.C., Rawlinson W.D., Craig M.E. Enterovirus infection and type 1 diabetes mellitus: systematic review and meta-analysis of observational molecular studies. BMJ. 2011;342:d35. doi: 10.1136/bmj.d35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oikarinen M., Tauriainen S., Oikarinen S., Honkanen T., Collin P., Rantala I., Mäki M., Kaukinen K., Hyöty H. Type 1 diabetes is associated with enterovirus infection in gut mucosa. Diabetes. 2012;61:687–691. doi: 10.2337/db11-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jerram S.T., Leslie R.D. The genetic architecture of type 1 diabetes. Genes (Basel) 2017;8:E209. doi: 10.3390/genes8080209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morgan N.G., Richardson S.J. Enteroviruses as causative agents in type 1 diabetes: loose ends or lost cause? Trends Endocrinol. Metab. 2014;25:611–619. doi: 10.1016/j.tem.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 9.Hober D., Alidjinou E.K. Enteroviral pathogenesis of type 1 diabetes: queries and answers. Curr. Opin. Infect. Dis. 2013;26:263–269. doi: 10.1097/QCO.0b013e3283608300. [DOI] [PubMed] [Google Scholar]
- 10.Hober D., Sane F. Enteroviral pathogenesis of type 1 diabetes. Discov. Med. 2010;10:151–160. [PubMed] [Google Scholar]
- 11.Marroqui L., Lopes M., dos Santos R.S., Grieco F.A., Roivainen M., Richardson S.J., Morgan N.G., Op de Beeck A., Eizirik D.L. Differential cell autonomous responses determine the outcome of coxsackievirus infections in murine pancreatic α and β cells. eLife. 2015;4:e06990. doi: 10.7554/eLife.06990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schulte B.M., Lanke K.H., Piganelli J.D., Kers-Rebel E.D., Bottino R., Trucco M., Huijbens R.J., Radstake T.R., Engelse M.A., de Koning E.J. Cytokine and chemokine production by human pancreatic islets upon enterovirus infection. Diabetes. 2012;61:2030–2036. doi: 10.2337/db11-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krogvold L., Edwin B., Buanes T., Frisk G., Skog O., Anagandula M., Korsgren O., Undlien D., Eike M.C., Richardson S.J. Detection of a low-grade enteroviral infection in the islets of langerhans of living patients newly diagnosed with type 1 diabetes. Diabetes. 2015;64:1682–1687. doi: 10.2337/db14-1370. [DOI] [PubMed] [Google Scholar]
- 14.Laiho J.E., Oikarinen S., Oikarinen M., Larsson P.G., Stone V.M., Hober D., Oberste S., Flodström-Tullberg M., Isola J., Hyöty H. Application of bioinformatics in probe design enables detection of enteroviruses on different taxonomic levels by advanced in situ hybridization technology. J. Clin. Virol. 2015;69:165–171. doi: 10.1016/j.jcv.2015.06.085. [DOI] [PubMed] [Google Scholar]
- 15.Tracy S., Smithee S., Alhazmi A., Chapman N. Coxsackievirus can persist in murine pancreas by deletion of 5′ terminal genomic sequences. J. Med. Virol. 2015;87:240–247. doi: 10.1002/jmv.24039. [DOI] [PubMed] [Google Scholar]
- 16.Laiho J.E., Oikarinen M., Richardson S.J., Frisk G., Nyalwidhe J., Burch T.C., Morris M.A., Oikarinen S., Pugliese A., Dotta F., JDRF nPOD-Virus Group Relative sensitivity of immunohistochemistry, multiple reaction monitoring mass spectrometry, in situ hybridization and PCR to detect Coxsackievirus B1 in A549 cells. J. Clin. Virol. 2016;77:21–28. doi: 10.1016/j.jcv.2016.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Busse N., Paroni F., Richardson S.J., Laiho J.E., Oikarinen M., Frisk G., Hyöty H., de Koning E., Morgan N.G., Maedler K. Detection and localization of viral infection in the pancreas of patients with type 1 diabetes using short fluorescently-labelled oligonucleotide probes. Oncotarget. 2017;8:12620–12636. doi: 10.18632/oncotarget.14896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raj A., van den Bogaard P., Rifkin S.A., van Oudenaarden A., Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods. 2008;5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hodik M., Skog O., Lukinius A., Isaza-Correa J.M., Kuipers J., Giepmans B.N., Frisk G. Enterovirus infection of human islets of Langerhans affects β-cell function resulting in disintegrated islets, decreased glucose stimulated insulin secretion and loss of Golgi structure. BMJ Open Diabetes Res. Care. 2016;4:e000179. doi: 10.1136/bmjdrc-2015-000179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodriguez-Calvo T., Richardson S.J., Pugliese A. Pancreas pathology during the natural history of type 1 diabetes. Curr. Diab. Rep. 2018;18:124. doi: 10.1007/s11892-018-1084-3. [DOI] [PubMed] [Google Scholar]
- 21.Campbell-Thompson M., Fu A., Kaddis J.S., Wasserfall C., Schatz D.A., Pugliese A., Atkinson M.A. Insulitis and β-cell mass in the natural history of type 1 diabetes. Diabetes. 2016;65:719–731. doi: 10.2337/db15-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.In’t Veld P., Lievens D., De Grijse J., Ling Z., Van der Auwera B., Pipeleers-Marichal M., Gorus F., Pipeleers D. Screening for insulitis in adult autoantibody-positive organ donors. Diabetes. 2007;56:2400–2404. doi: 10.2337/db07-0416. [DOI] [PubMed] [Google Scholar]
- 23.Rodriguez-Calvo T., Ekwall O., Amirian N., Zapardiel-Gonzalo J., von Herrath M.G. Increased immune cell infiltration of the exocrine pancreas: a possible contribution to the pathogenesis of type 1 diabetes. Diabetes. 2014;63:3880–3890. doi: 10.2337/db14-0549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leslie R.D., Atkinson M.A., Notkins A.L. Autoantigens IA-2 and GAD in type I (insulin-dependent) diabetes. Diabetologia. 1999;42:3–14. doi: 10.1007/s001250051105. [DOI] [PubMed] [Google Scholar]
- 25.Campbell-Thompson M., Wasserfall C., Montgomery E.L., Atkinson M.A., Kaddis J.S. Pancreas organ weight in individuals with disease-associated autoantibodies at risk for type 1 diabetes. JAMA. 2012;308:2337–2339. doi: 10.1001/jama.2012.15008. [DOI] [PubMed] [Google Scholar]
- 26.Campbell-Thompson M.L., Filipp S.L., Grajo J.R., Nambam B., Beegle R., Middlebrooks E.H., Gurka M.J., Atkinson M.A., Schatz D.A., Haller M.J. Relative pancreas volume is reduced in first-degree relatives of patients with type 1 diabetes. Diabetes Care. 2019;42:281–287. doi: 10.2337/dc18-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wright J.J., Saunders D.C., Dai C., Poffenberger G., Cairns B., Serreze D.V., Harlan D.M., Bottino R., Brissova M., Powers A.C. Decreased pancreatic acinar cell number in type 1 diabetes. Diabetologia. 2020;63:1418–1423. doi: 10.1007/s00125-020-05155-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kusmartseva I., Beery M., Hiller H., Padilla M., Selman S., Posgai A., Nick H.S., Campbell-Thompson M., Schatz D.A., Haller M.J. Temporal analysis of amylase expression in control, autoantibody-positive, and type 1 diabetes pancreatic tissues. Diabetes. 2020;69:60–66. doi: 10.2337/db19-0554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wicker L.S., Miller B.J., Mullen Y. Transfer of autoimmune diabetes mellitus with splenocytes from nonobese diabetic (NOD) mice. Diabetes. 1986;35:855–860. doi: 10.2337/diab.35.8.855. [DOI] [PubMed] [Google Scholar]
- 30.Höglund P., Mintern J., Waltzinger C., Heath W., Benoist C., Mathis D. Initiation of autoimmune diabetes by developmentally regulated presentation of islet cell antigens in the pancreatic lymph nodes. J. Exp. Med. 1999;189:331–339. doi: 10.1084/jem.189.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Codina-Busqueta E., Scholz E., Muñoz-Torres P.M., Roura-Mir C., Costa M., Xufré C., Planas R., Vives-Pi M., Jaraquemada D., Martí M. TCR bias of in vivo expanded T cells in pancreatic islets and spleen at the onset in human type 1 diabetes. J. Immunol. 2011;186:3787–3797. doi: 10.4049/jimmunol.1002423. [DOI] [PubMed] [Google Scholar]
- 32.Seay H.R., Yusko E., Rothweiler S.J., Zhang L., Posgai A.L., Campbell-Thompson M., Vignali M., Emerson R.O., Kaddis J.S., Ko D. Tissue distribution and clonal diversity of the T and B cell repertoire in type 1 diabetes. JCI Insight. 2016;1:e88242. doi: 10.1172/jci.insight.88242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alidjinou E.K., Chehadeh W., Weill J., Vantyghem M.C., Stuckens C., Decoster A., Hober C., Hober D. Monocytes of patients with type 1 diabetes harbour enterovirus RNA. Eur. J. Clin. Invest. 2015;45:918–924. doi: 10.1111/eci.12485. [DOI] [PubMed] [Google Scholar]
- 34.Dechaumes A., Bertin A., Sane F., Levet S., Varghese J., Charvet B., Gmyr V., Kerr-Conte J., Pierquin J., Arunkumar G. Coxsackievirus-B4 infection can induce the expression of human endogenous retrovirus W in primary cells. Microorganisms. 2020;8:E1335. doi: 10.3390/microorganisms8091335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schulte B.M., Bakkers J., Lanke K.H., Melchers W.J., Westerlaken C., Allebes W., Aanstoot H.J., Bruining G.J., Adema G.J., Van Kuppeveld F.J., Galama J.M. Detection of enterovirus RNA in peripheral blood mononuclear cells of type 1 diabetic patients beyond the stage of acute infection. Viral Immunol. 2010;23:99–104. doi: 10.1089/vim.2009.0072. [DOI] [PubMed] [Google Scholar]
- 36.Yin H., Berg A.K., Tuvemo T., Frisk G. Enterovirus RNA is found in peripheral blood mononuclear cells in a majority of type 1 diabetic children at onset. Diabetes. 2002;51:1964–1971. doi: 10.2337/diabetes.51.6.1964. [DOI] [PubMed] [Google Scholar]
- 37.Rodriguez-Calvo T. Enteroviral infections as a trigger for type 1 diabetes. Curr. Diab. Rep. 2018;18:106. doi: 10.1007/s11892-018-1077-2. [DOI] [PubMed] [Google Scholar]
- 38.Stene L.C., Oikarinen S., Hyöty H., Barriga K.J., Norris J.M., Klingensmith G., Hutton J.C., Erlich H.A., Eisenbarth G.S., Rewers M. Enterovirus infection and progression from islet autoimmunity to type 1 diabetes: the Diabetes and Autoimmunity Study in the Young (DAISY) Diabetes. 2010;59:3174–3180. doi: 10.2337/db10-0866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vehik K., Lynch K.F., Wong M.C., Tian X., Ross M.C., Gibbs R.A., Ajami N.J., Petrosino J.F., Rewers M., Toppari J., TEDDY Study Group Prospective virome analyses in young children at increased genetic risk for type 1 diabetes. Nat. Med. 2019;25:1865–1872. doi: 10.1038/s41591-019-0667-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaddis J.S., Pugliese A., Atkinson M.A. A run on the biobank: what have we learned about type 1 diabetes from the nPOD tissue repository? Curr. Opin. Endocrinol. Diabetes Obes. 2015;22:290–295. doi: 10.1097/MED.0000000000000171. [DOI] [PubMed] [Google Scholar]
- 41.Foulis A.K., Liddle C.N., Farquharson M.A., Richmond J.A., Weir R.S. The histopathology of the pancreas in type 1 (insulin-dependent) diabetes mellitus: a 25-year review of deaths in patients under 20 years of age in the United Kingdom. Diabetologia. 1986;29:267–274. doi: 10.1007/BF00452061. [DOI] [PubMed] [Google Scholar]
- 42.Skog O., Ingvast S., Korsgren O. Evaluation of RT-PCR and immunohistochemistry as tools for detection of enterovirus in the human pancreas and islets of Langerhans. J. Clin. Virol. 2014;61:242–247. doi: 10.1016/j.jcv.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ardestani A., Li S., Annamalai K., Lupse B., Geravandi S., Dobrowolski A., Yu S., Zhu S., Baguley T.D., Surakattula M. Neratinib protects pancreatic beta cells in diabetes. Nat. Commun. 2019;10:5015. doi: 10.1038/s41467-019-12880-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Original/source data in the paper are available in Tables S1 and S3. This study did not generate any unique datasets or code.