Summary
Adoptive transfer of T cells expressing a transgenic T cell receptor (TCR) has the potential to revolutionize immunotherapy of infectious diseases and cancer. However, the generation of defined TCR-transgenic T cell medicinal products with predictable in vivo function still poses a major challenge and limits broader and more successful application of this “living drug.” Here, by studying 51 different TCRs, we show that conventional genetic engineering by viral transduction leads to variable TCR expression and functionality as a result of variable transgene copy numbers and untargeted transgene integration. In contrast, CRISPR/Cas9-mediated TCR replacement enables defined, targeted TCR transgene insertion into the TCR gene locus. Thereby, T cell products display more homogeneous TCR expression similar to physiological T cells. Importantly, increased T cell product homogeneity after targeted TCR gene editing correlates with predictable in vivo T cell responses, which represents a crucial aspect for clinical application in adoptive T cell immunotherapy.
Keywords: T cell receptor engineering, TCR, TCR transgenic T cells, TCR editing, CRISPR/Cas9 mediated engineering, orthotopic TCR replacement, OTR, predictable functionality, homogenous TCR expreession
Graphical abstract

Highlights
T cell product function and safety is directly affected by editing method
Conventional TCR editing introduces variability through uncontrolled gene insertion
Targeted TCR editing results in homogenous and physiological TCR expression
T cell product homogeneity enables predictable in vivo function for clinical use
Müller et al. perform in-depth comparison of conventional TCR editing methods with CRISPR/Cas9-mediated orthotopic TCR replacement. Analysis of TCR transgene genomic insertion, mRNA transcription, and protein surface expression demonstrate that targeted TCR gene editing facilitates the production of homogenous TCR-transgenic T cell products with predictable in vivo functionality.
Introduction
Adoptive T cell therapy (ACT) can specifically restore T cell-mediated immunity to fight infectious diseases or cancer.1,2 The administration of autologous tumor-infiltrating lymphocytes3 or transfer of pathogen-specific T cells that were ex vivo isolated from an HLA-matched donor4 represent particularly successful approaches. However, while these approaches have proven their efficacy and safety, they are also largely restricted by the often limited availability and accessibility of antigen-specific, HLA-matched T cells from patient- or healthy donor-repertoires5, 6, 7. This problem could be solved by genetically engineering TCR-transgenic T cells for clinical application since the desired TCR and the HLA-matched T cell can be taken from different sources. Furthermore, through combining optimal T cell phenotype,8, 9, 10, 11 highest degree of HLA-matching,12 and TCR-intrinsic features such as antigen specificity or avidity,13,14 TCR-engineered T cells provide unique therapeutic opportunities. Thousands of patients have so far been safely treated with antigen-specific receptor transgenic T cell products. The clinical success, however, is until now rather limited.15, 16, 17
One aggravating factor is the complexity of TCR engineering. Interactions between the endogenous and the introduced transgenic receptor can result in T cell products with inferior functionality.18,19 The transgenic TCR competes with the endogenous TCR for a limited amount of CD3 molecules, which can decrease the surface expression of both.20 Furthermore, because of the TCR’s heterodimeric structure, chains of the transgenic and the endogenous receptor can form mispaired TCR variants, which again decreases surface expression of the desired transgenic TCR and also poses a safety hazard, because mispaired TCRs can exert off-target toxicity.21, 22, 23, 24 In the past, these problems in TCR engineering were tackled through many different approaches. Conventional lenti- or retroviral systems that enable high transgene integration efficiency are widely used for TCR editing in basic research18, 19, 20 and clinical studies.15, 16, 17 Furthermore, increased transgenic TCR surface expression and decreased levels of mispairing were achieved with TCR constructs consisting of murine constant regions25,26 with an additional disulfide bond27,28 or other stabilizing modifications of the TCR chains.29, 30, 31 However, complete elimination of mispairing and undisturbed expression of the transgenic TCR was only achieved via the full KO of the endogenous TCR.22,32, 33, 34
While interference through the endogenous TCR can be eliminated, conventional editing still largely lacks control over the quantity and site of transgenic TCR integration events. Such untargeted editing might affect T cell product safety and functionality. Upon conventional editing, advanced analysis of single-cell vector copy numbers (VCN) revealed up to 44 transgene integrations in a primary T cell.35 Both the US Food and Drug Administration (FDA) as well as the European Medicines Agency (EMA) highlight that the risk of gene-modified cell therapies via insertional oncogenesis36 should be reduced through the limitation of VCN.37 Accordingly, clinical studies that reported VCN for antigen-specific receptor transgenic T cells documented an average copy number between 1 and 2.38, 39, 40 However, even with low VCN, viral transduction results in close-to-random transgene integration41 and requires constitutively active extrinsic gene promoters for transgene expression. While a low copy number may be desired for safety reasons, it may also limit TCR transcription levels and protein surface expression, which could ultimately compromise the functionality of conventionally engineered T cell products.19,42,43 Today, novel advancements in the field of genetic engineering enable controlled TCR gene editing through targeted knockin (KI) of a transgenic receptor into the TCR locus.32,44,45 So-called orthotopic TCR replacement (OTR) simultaneously results in the removal of endogenous TCR chains and transgenic TCR transcription under the control of the endogenous TCR promoter.
In this report, we systematically compare targeted OTR technology to untargeted conventional TCR editing (both in absence of the endogenous TCR) in order to investigate differential consequences on the magnitude, variability, and interrelatedness of transgenic TCR surface expression and functionality. Because TCR-intrinsic characteristics such as epitope-ligand specificity and avidity can bias general conclusions, we built up and used a library of 51 unique antigen-specific TCRs for this study. First, we performed TLA and single-cell RNA sequencing (scRNAseq) to investigate TCR transgene integration and transcription. After conventional editing, TCR transgenes were dispersed over the whole genome, including exonic regions and cancer genes, thereby representing not only a safety hazard but also a source for T cell product variability. Accordingly, scRNAseq data revealed highly heterogenous TCR transcription that was also in correlation to VCN. In sharp contrast, OTR resulted in targeted TCR transgene insertion into the TCR locus as well as homogeneous TCR transcription. Next, we demonstrate that the heterogeneity in TCR transcription directly translates into variable TCR surface expression with consequences for functionality. First, we performed a functional comparison between OTR and conventional editing with a clinically relevant low TCR copy number and observed that OTR T cells have consistently higher TCR surface expression and slightly enhanced T cell functionality in vitro and in vivo. Second, the increased homogeneity of transgenic TCR transcription and surface expression after OTR led to more predictable in vivo functionality of T cell products compared to conventionally edited, heterogeneous TCR-transgenic T cells. These findings demonstrate that the method of TCR engineering can significantly shape T cell product functionality. Targeted TCR gene editing via OTR offers a high genetic safety profile and also produces defined T cell products with more predictable functionality compared to conventional editing. Thereby, OTR T cell products align well with the usual requirements of clinically approved medicinal products.
Results
Targeted TCR editing results in homogeneous TCR transcription
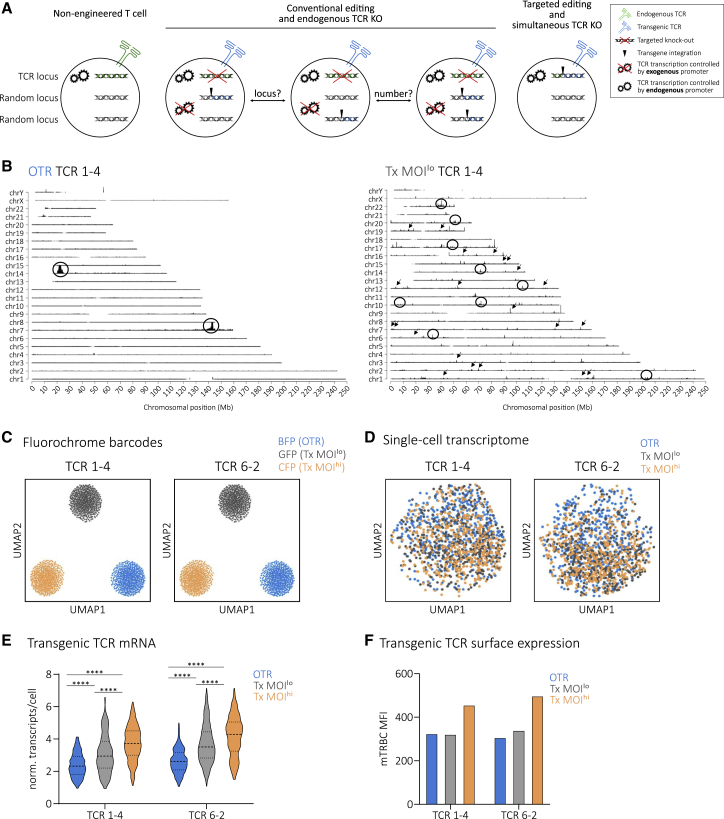
Conventional editing, such as through lenti-/retroviral transduction or transposon systems, is a rather uncontrolled process during which one or multiple transgene copies integrate at least semi-randomly into the genome.46,47 In contrast, OTR enables tightly controlled replacement of the endogenous TCR with transgenic TCR expression under the endogenous TCR promoter32,45 (Figure 1A). To investigate these aspects of TCR engineering and dissect their individual effects on T cell biology, we first established a platform for reliable TCR sequence identification (Figure S1) to generate a comprehensive library of CMV-specific TCRs. Performing experiments with more than one TCR decreases the risk of bias through individual TCR-intrinsic characteristics. OTR was performed via homology-directed repair (HDR) of a full TCR αβ construct into the first exon of the TCR α constant region (TRAC) and additional TCR β-chain KO (Figure S2). Conventional editing was performed via retroviral transduction with the commonly used MP71 vector that enables high TCR transgene expression.48 Full TCR αβ KO was also performed after conventional editing to exclude bias originating from endogenous TCR expression (see Figure S3 for how variability of TCR engineered T cells through competition and mispairing is removed through full TCR αβ KO, as shown for 19 different A1/pp50-specific TCRs). Furthermore, two different levels of virus multiplicity of infection (MOI) were used to achieve either integration of only one TCR copy according to Poisson-statistics42 (Tx MOIlo) or a high copy number (Tx MOIhi). In a first step, we investigated TCR transgene integration sites via TLA49 with two different A2/pp65-specific TCRs. For OTR samples, we detected two integration sites (Figure 1B; Figure S4). Primarily, the full TCR transgene was inserted at the intended target site in the first exon of TRAC through HDR. In addition, homology-independent partial transgene integration occurred at the intended double-strand break in the TCR β constant region (TRBC1/2) locus, as observed before32. In sharp contrast to that, conventional editing resulted in a large number of integration sites dispersed over the whole genome. We detected 37 and 465 integration sites for TCR 1-4 Tx MOIlo (Figure 1B) and TCR 6-2 Tx MOIlo (Figure S4), respectively. TLA indicated a high heterogeneity of the investigated T cell products so that many integration sites were below the detection limit. Hence, the detected number of integration sites most certainly represent underestimates. Bioinformatic analysis revealed that 27 (TCR 1-4 Tx MOIlo) and 196 gene regions (TCR 6-2 Tx MOIlo) were hit by transgene insertion (mostly intronic). Three (TCR 1-4 Tx MOIlo) and 35 (TCR 6-2 Tx MOIlo) integrations occurred in “cancer genes.” Interestingly, both Tx MOIlo samples showed integrations into introns of the cancer genes CHST11 and RUNX1. In addition, we also performed whole genome sequencing for untargeted determination of off-target events, representing an important quality control for regulatory authorities when producing GMP-conform OTR T cell products for clinical use. Variant analyses of these data did not reveal relevant off-target events in two separate donors and with both TRAC and TRBC guide RNAs (unpublished data). Next, we investigated individual impacts of low versus high copy number and untargeted versus targeted TCR integration on the transcriptome of edited T cells via scRNAseq. To minimize biases through batch effects, we additionally performed multiplexing through a co-transduction of three different fluorochromes (blue, cyan, and green fluorescent protein [BFP, CFP, and GFP]), which served as barcodes for the three different editing approaches. Fluorochrome barcodes were successfully retrieved from sequencing data for both TCR pools and could be used for demultiplexing (Figure 1C). In terms of the global transcriptome, we could not observe any substantial differences between editing methods (Figure 1D) or between TCRs (Figure S5A). However, editing methods resulted in significantly different transgenic TCR transcription (Figure 1E) and surface expression (Figure 1F and Figure S6). Remaining endogenous TCR transcription levels, potentially including frameshifted transcripts that might be subject of quick mRNA-degradation, were consistent between editing groups and slightly lower than levels observed after OTR (Figure S5B). In comparison to conventional editing, OTR resulted in a more homogeneous TCR transcription at a level that closely paralleled remaining endogenous TCR mRNA content. Within the two conventionally edited samples, higher virus MOI resulted in higher transgenic TCR mRNA (Figure 1E) and protein (Figure 1F; Figure S6) levels, but both samples showed increased heterogeneity compared to OTR T cells (Figure 1e; Figure S5C). House-keeping genes such as GAPDH, VIM, and MALAT1 were expressed uniformly irrespective of editing methods (Figure S5d). The frequency distribution (as visualized by changing widths of violin plots in Figure 1E) of transgenic TCR expression after Tx MOIlo indicates that a fraction of cells might have received more than one transgene copy, which would be in line with previously reported single-cell VCN distributions and Poisson-statistics.35 Another potential source of transgene expression variability is the observed heterogeneity of insertion sites (Figure 1B), since different loci have different chromatin accessibility which affects gene transcription.50
Figure 1.
Targeted TCR editing results in homogeneous TCR transcription
(A) Schematic illustration of conventional and targeted TCR editing (both with full endogenous TCR KO) and their differential transgene integration profiles in comparison to a non-engineered, physiological T cell. Conventionally edited T cells might differ in TCR copy number and TCR transgene integration site. For the sake of clarity, individual TCR α and β gene loci are not shown.
(B) TLA coverage across the human genome for integration of TCR 1–4. Circles indicate more abundant and arrows indicate examples of less abundant integration sites. TRAC locus is located in chromosome 14; TRBC locus in chromosome 7. Similar results were obtained with an independent primer set.
(C) Usage of fluorochrome barcodes for demultiplexing of samples. Visualization via uniform manifold approximation and projection (UMAP), n = 482 cells per barcode.
(D) UMAP on whole transcriptome data compared by editing method for two A2/pp65-specific TCRs, n = 450 cells per editing method.
(E) Quantification of transgenic TCR transcription at day 5 post editing. Violin plots indicate frequency distribution; median and quartiles are shown with dashed lines. Statistical testing by two-way ANOVA (∗∗∗∗p < 0.001 for editing method, ∗∗∗∗p < 0.001 for TCR, n = 256 cells per editing method) followed by Tukey’s multiple comparisons test, ∗∗∗∗p < 0.0001.
(F) TCR protein surface expression of cells in (B–E) via antibody staining of the murine constant TCR β-chain (mTRBC) within transgenic TCRs.
In summary, conventional editing generates a certain risk of mutagenesis and results in variable TCR transcription levels through random genomic integration of an undefined transgene copy number per cell. In contrast to that, targeted TCR editing via OTR offers a higher safety profile through defined TCR genomic integration that also results in homogeneous TCR expression.
Uncontrolled TCR editing impacts TCR surface expression and functionality
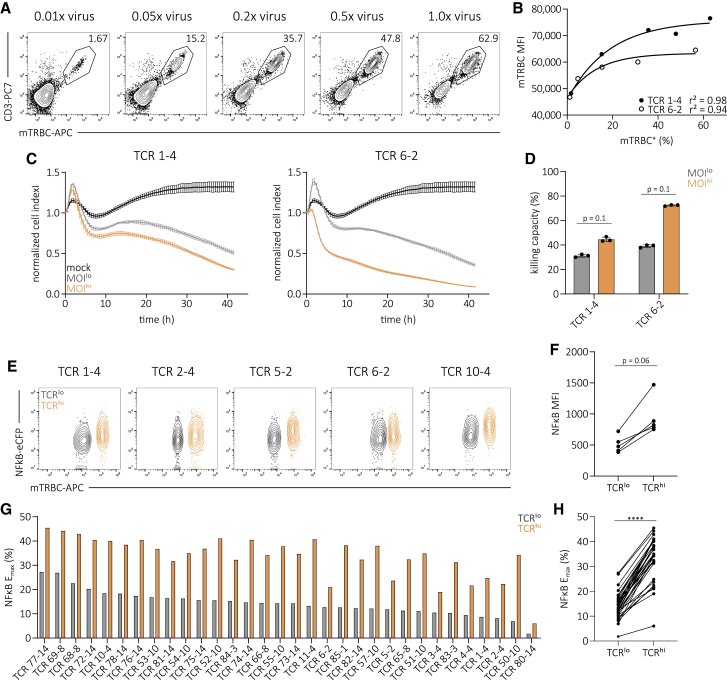
Having observed variable TCR integration and transcription after conventional editing, we speculated that this could also impact the functionality of TCR redirected T cell products.19,43 To test this hypothesis, we titrated virus MOIs for transduction of two A2/pp65-specific TCRs into endogenous TCR-KO primary T cells in order to simulate variable transgene integration numbers. Editing efficiency and TCR surface expression positively correlated with virus MOI (Figures 2A and 2b; Figures S7A and S7B). We then sorted equal numbers of TCR-transgenic Tx MOIlo (0.01× virus) and Tx MOIhi (1.0× virus) CD8+ T cells by flow cytometry and tested their functionality after a short time of in vitro culture. We could observe that increased transgenic TCR surface expression of Tx MOIhi T cells resulted in distinctly higher cytotoxic capacity (Figures 2C and 2D) and peptide sensitivity measured via intracellular cytokine staining (ICCS) (Figure S7C). Additional experiments with a clinically used lentiviral vector (pCDH-EF1) confirmed the described correlation of copy number, editing efficiency, TCR surface expression and functionality (Figures S8A–8C). Furthermore, we could not identify different phenotypic signatures between MOIlo versus MOIhi, as well as retro- versus lentiviral transduction (Figures S8D and S8E). To exclude potential TCR-intrinsic biases, we introduced 32 A2/pp65-specific TCRs each into an endogenous TCR-negative Jurkat triple-parameter reporter cell line (TPRKO) that allows for high throughput functional screening of TCR signaling.51,52 We observed that cells with high TCR surface expression (TCRhi) also showed increased levels of nuclear factor-kappa B (NFκB) reporter activity upon antigen-specific stimulation (five representative TCRs shown in Figures 2E and 2F). Maximum NFκB reporter activity in response to antigen (Emax) of 32 TCRs was significantly increased in TCRhi cells compared to TCRlo cells (Figures 2G and 2H), as was peptide sensitivity (Figure S9). In summary, uncontrolled conventional TCR editing—through variable VCN and undefined genomic integration—produces T cells with heterogeneous transgenic TCR surface expression that directly translates into variable functionality.
Figure 2.
Uncontrolled TCR editing impacts TCR surface expression and functionality
(A) Transgenic TCR surface expression of the A2/pp65-specific TCR 1-4. Editing was performed in human T cells via retroviral transduction with a titration of virus dose and additional elimination of the endogenous TCR. Numbers indicate percentages of CD8+.
(B) Correlation of editing efficiency to transgenic TCR surface expression (mTRBC MFI). Each dot represents one of two indicated TCRs. Fitting by non-linear regression.
(C) Killing of peptide-pulsed target cells over time by two A2/pp65-specific TCR-transgenic flow cytometry-sorted T cell products that were generated with a low (gray) or high (orange) virus MOI (0.01× virus and 1.0× virus, respectively; cell products depicted in A and B and Figure S7A). Lines illustrate the mean of three replicates ± SD.
(D) Quantification of killing capacity (as percentage of maximum killing, area under the curve normalized to mock control) measured in (C) for both TCRs and editing methods. Statistical testing by Mann-Whitney test. Depicted are replicates and mean ± SD.
(E) Flow cytometry plots of TCR surface expression and NFκB reporter activity of five representative A2/pp65-specific TCRs in the TPRKO cell line52 after antigen-specific stimulation. Editing was performed via retroviral transduction.
(F) NFκB mean fluorescence intensity (MFI) of data shown in (E).
(G) NFκB Emax of 32 A2/pp65-specific TCRs corresponding to data in (E). Bars represent mean of three replicates.
(H) Direct comparison between cells with low and high TCR surface of data shown in (G). Statistical testing by Wilcoxon matched-pairs signed rank test, ∗∗∗∗p < 0.0001. Data are representative of two independent experiments.
Targeted TCR editing enables homogeneous TCR surface expression and increased functionality
Antigen-specific receptor redirected T cell products in past and present clinical trials have so far been generated via conventional editing with low virus MOIs38, 39, 40 in order to minimize risks originating from multiple (random) transgene integrations. We therefore set out to perform VCN-standardized measurements of functional capacity by comparing T cell products that were either generated via OTR or conventional editing with statistically one TCR integration. For this, we sorted CD8+ A2/pp65-specific TCR-transgenic T cells by flow cytometry and cultivated them in an antigen-free manner. While TCR RNA levels were generally lower upon OTR a few days after editing (Figures 1E and 1F), we observed in several independent experiments and donor T cells that TCR surface expression and pMHC stainability was higher after OTR compared to transduction with low virus MOIs when cells were cultured for more than a week (Figures 3A and 3B; Figures S10A and S10B). This possibly reflects a delayed accumulation of TCR surface protein after OTR, which is from then on maintained at a consistent level. Corresponding to more homogeneous TCR RNA expression, TCR surface expression was significantly less variable in OTR T cell products, as indicated by a lower cell-to-cell surface expression variability (Figure S10C). We then used these cells to study antigen-specific cytokine release and cytotoxicity. OTR T cells showed a distinct expression pattern of interferon-γ (IFNγ) and interleukin-2 (IL-2) measured via ICCS and a consistently enhanced peptide sensitivity for all tested TCRs (Figures 3C and 3D). We also observed consistently higher cytotoxic capacities of OTR T cells (Figures 3E and 3F). Additional experiments with another donor confirmed these results (Figure S11). In summary, in a clinically relevant, VCN-standardized comparison of OTR to conventional editing, OTR T cell products consistently showed higher TCR surface expression and functional capacity.
Figure 3.
Targeted TCR editing enables homogeneous TCR surface expression and increased functionality
(A) TCR surface expression of four A2/pp65-specific TCRs after flow cytometry-sorting and two weeks of in vitro culture. Editing was performed in human T cells via OTR (blue) or retroviral transduction with a low virus MOI (gray) and in both cases additional elimination of the endogenous TCR.
(B) Quantification of TCR surface expression levels for each TCR individually and pooled for comparison of editing groups. Each dot represents measurement of one TCR at one of two time points (day 10 after sort indicated by open circles, day 14 after sort indicated by filled circles). Statistical testing by two-tailed paired Student’s t test, ∗∗∗p < 0.001.
(C) Functional cytokine response of T cell products shown in (A). Dose-dependent release of IFNγ (top left) and IL-2 (bottom left) after antigen-specific stimulation for one representative TCR. Corresponding half-maximum effective concentration (EC50) curves (right).
(D) Quantification of IFNγ (top left) and IL-2 (bottom left) EC50 values of four individual A2/pp65-specific TCRs. Depicted are replicates and mean ± SD. Statistical testing by Mann-Whitney tests. Direct comparison of IFNγ and IL-2 EC50 between editing groups (right). Here, each dot represents the mean EC50 of three replicates for one TCR. Statistical testing by Wilcoxon matched-pairs signed rank test.
(E) Killing of peptide-pulsed target cells over time of one representative A2/pp65-specific TCR-transgenic T cell product (shown in A, B, C, and D). Lines illustrate the mean of three replicates ± SD.
(F) Quantification of killing capacity (as percentage of maximum killing, area under the curve normalized to mock control) for all four TCRs and editing methods. Depicted are replicates and mean ± SD. Statistical testing by Mann-Whitney tests. Direct comparison between editing groups. Here, each dot represents the mean of three replicates for each TCR. Statistical testing by Wilcoxon matched-pairs signed rank test.
Targeted TCR gene editing results in more predictable in vivo T cell function
We observed in multiple experiments and a large set of different TCRs and donors (Figure 1; Figures S10C, S11B, and S12) that OTR generates more homogeneous TCR-transgenic T cell products in comparison to conventional editing. As we also collected evidence for a direct relationship between surface expression and in vitro functionality (see Figures 2 and 3), we wondered whether OTR would result in more predictable in vivo functionality and thereby ultimately enable the generation of more standardized T cell products for immunotherapy.
Upon adoptive T cell transfer, not all transferred T cells are recovered and recruited into the immune response, as reflected by the recovery rate in single-cell transfer experiments.53 The transferred cells that ultimately form the immune response are therefore a subpopulation of the initially transferred T cell product. Accordingly, we hypothesized that the variable TCR expression between single cells of a transferred T cell product should also lead to functional variability between recruited cell populations in different hosts (Figure 4A). To investigate this, we introduced four different fully human A2/pp65-specific TCRs into T cells either via OTR or conventional editing. In addition to TCR editing, we further introduced four different fluorochrome transgenes by retroviral transduction in order to color barcode each TCR.54 This allowed us to perform polyclonal adoptive transfer experiments, thereby reducing technical mouse-to-mouse variability for comparisons between TCRs.55 Five days post editing, we pooled defined numbers of all four TCR-transgenic fluorochrome-positive T cell products (1 × 105 cells per TCR) that were either generated via OTR or conventional editing and transferred the cells together with human IL-2 via intraperitoneal injection (i.p.) into irradiated human HLA-A∗02 transgenic NOD.Cg-PrkdcscidIl2rgtm1WjlTg(HLA-A/H2-D/B2M)1Dvs/SzJ (NSG/HHD) mice56 that were subsequently infected with a transgenic murine CMV (mCMV) strain that presents the human CMV (hCMV) HLA-A∗02-restricted pp65 epitope (NLV) (mCMV-ie2-ANLV) (Figure 4B). This model allows for in vivo investigation of human TCR-transgenic T cells in a system with natural CMV tropism.57 Again, fluorescence intensity (FI) of individual cells after pMHC-multimer staining was consistently less variable in OTR T cells (Figure 4C). Eight days after adoptive T cell transfer, we isolated livers of infected mice and analyzed T cell responses. The additional fluorochrome color barcode enabled us to differentiate between TCRs so that we could compare individual TCR population sizes between different mice (Figure 4D; for gating strategy see Figure S13A). Of note, TCR-transgenic T cells derived of the same T cell product contributed to the overall population in different mice in a variable manner when generated via conventional editing, but not when generated by OTR, as quantified by inter-host recruitment variability (Figure 4E; Figure S13B). To corroborate this finding and exclude a potential bias through specialties of fully human TCR constructs, we further performed a second experiment with four A2/pp65-specific murinized TCR constructs (i.e., murine constant regions) and again observed significantly reduced inter-host recruitment variability of OTR T cells compared to conventionally engineered T cells (Figure 4F; Figure S13C). Having observed these functional in vivo effects originating from heterogeneity within a T cell product, we further investigated the variability between different T cell infusion products. For this, we transgenically expressed five A2/pp65-specific TCRs either via OTR or via MOIlo retroviral transduction into endogenous TCR-KO T cells derived from three different donors. This revealed distinctly increased inter-donor variability after conventional editing (Figure S14). To determine the effect of different editing methods on in vivo recruitment side-by-side, we again performed co-transfer experiments (1 × 105 cells per TCR) in the NSG/HHD model, but now with fluorochromes serving as barcodes for the editing method (Figure S15A). Seven days after infection, OTR T cells showed consistently higher recruitment compared to conventionally MOIlo edited T cells (Figure S15B and S15C). Finally, we also assessed in vivo protectivity of differentially edited T cells transferred into individual mice (2 × 106 sorted CD8+ transgenic TCR+ T cells) (Figure S16A). In the NSG-HHD model system, OTR T cells showed a slightly increased protective capacity compared to conventionally edited T cells (Figures S16B–S16D). More noteworthy, however, variability between different recipients was again decreased in the OTR group (Figure S16E) as observed before (Figures 4E and 4F). We conclude that in contrast to uncontrolled conventional editing, targeted TCR insertion facilitates the production of highly defined, homogeneous T cell products with predictable in vivo functionality.
Figure 4.
Targeted TCR gene editing results in more predictable in vivo T cell function
(A) Heterogeneous TCR expression within a TCR-transgenic T cell product in combination with T cell recruitment after adoptive T cell transfer may cause differential in vivo T cell product functionality in different patients.
(B) Experimental setup.
(C) pMHC-multimer FI of four fully human A2/pp65-specific TCRs. Editing was performed either via OTR (blue) or retroviral transduction (gray) and additional elimination of the endogenous TCR (accounts for both methods). Each dot represents one single cell. For each TCR and editing method, 100 randomly drawn cells derived from a larger population are displayed (left). Quantification of the corresponding cell-to-cell pMHC-multimer FI variability (right). Statistical testing by Wilcoxon matched-pairs signed rank test.
(D) Analysis of TCR-transgenic T cell responses at day 8 in the liver of sacrificed animals. Depicted are representative responses in two mice per editing group. Numbers denote percentages of human CD8+pMHC+ cells.
(E) Quantification of TCR-transgenic T cell response variability between individual mice. For each TCR and editing method, the percentage deviation of total T cell recruitment in one mouse compared to the mean recruitment in all mice is depicted as one dot (left). Bars indicate minimal to maximal deviation from mean. Quantification of inter-host variability of T cell responses by editing group (right). Each dot represents one TCR. Statistical testing by Wilcoxon matched-pairs signed rank test; n = 4-5 mice per group.
(F) Repetition of the experiment shown in (E) with four murinized A2/pp65-specific TCRs in T cells of different donor origin than shown in (E). Statistical testing by Wilcoxon matched-pairs signed rank test; n = 5 mice per group.
Discussion
First reports on CRISPR/Cas9-mediated replacement of the endogenous TCR with either a chimeric antigen receptor (CAR)44 or a TCR45 were perceived as important proof-of-concept studies in the fields of T cell engineering and ACT. Targeted transgene integration not only promised to offer an increased safety profile but is also highly appealing as OTR T cell products should closely resemble physiological, unedited T cells. Indeed, placing an antigen-specific receptor under endogenous TCR transcriptional control facilitates near-physiological TCR regulation32. Despite these advancements, many questions on how OTR T cell products differ from conventionally edited T cells have remained unresolved. This comparison is of fundamental importance because conventional editing via viral transduction is currently the most widely used method in basic research and for the production of clinically applied T cell products.
In this study, we re-expressed 51 CMV-specific TCRs via OTR and conventional editing and investigated the consequences of differential genetic TCR integration profiles on the magnitude, variability, and interrelatedness of transgenic TCR surface expression and functionality. We measured TCR transgene integration site, transcription, surface expression, and T cell product functionality after conventional editing as well as after OTR. Thereby, we could directly relate defined transgene integration via OTR to a more homogeneous, physiological TCR transcription as well as to a less variable surface expression and functionality. In case of conventional editing, both variable VCN and random transgene integration independently increase the variability of TCR transcription, surface expression, and functionality. In fact, it is well known that copy number affects editing efficiency and transgene expression19,42 and that differential accessibility of a transgene at a specific genomic locus should have an impact on transgene transcription.50 Conventionally edited TCR redirected T cell products (without additional endogenous TCR-KO) have shown to be safe in many clinical trials.15, 16, 17 However, there are also reports of insertional mutations upon conventional editing in stem cells58 and CAR-T cells59 highlighting a certain need for caution. These might be unfortunate exceptions, and most clinical trials delivered very promising results. However, overall functionality of conventionally edited, clinically applied TCR-transgenic T cell products seems to be rather variable.15, 16, 17 Conventional editing in combination with an additional KO of the endogenous TCR is one option to decrease variability through diminishing the formation of mispaired TCR variants and ultimately enabling unbiased transgenic TCR surface expression,22,33,34 but does not enable controlled transgenic TCR integration and transcriptional regulation under the endogenous TCR promoter.
Here, we performed a standardized functional comparison between conventionally edited T cells with statistically one TCR integration and OTR (both in absence of the endogenous TCR). The aspect of variable VCN after conventional editing has not been taken into account by previous studies. Accordingly, these studies reported that OTR T cells exhibit similar functionality in vitro32,45,60 and similar or enhanced functionality in vivo.45,60 Here, we show with a set of different A2/pp65-specific TCRs that OTR consistently increased TCR surface expression as well as in vitro and in vivo functionality in comparison to conventional editing with a low virus MOI. In the future, more long-term in vivo protection experiments are needed to test whether OTR T cells show prolonged maintenance and enhanced effector function, as proposed by more physiological TCR regulation32 and OTR CAR-T cell data.44
We observed by scRNAseq as well as flow cytometry that OTR decreases cell-to-cell TCR expression variability. In contrast, conventional editing results in variable TCR surface expression that directly affects functionality and hence might be—at least to some extent—attributable to largely variable clinical results. Using a polyclonal transfer system to study different color barcoded TCRs side by side in vivo, we could show that this observed heterogeneity within the T cell product indeed introduces substantial T cell response variability. For clinical application, a highly defined and homogeneous T cell product that provides predictable in vivo function is of utmost importance. Targeted TCR gene editing via OTR facilitates the production of highly defined T cell products with an enhanced safety profile as well as increased and predictable functionality.
Limitations of the study
We employed state-of-the-art technologies to characterize overall 51 TCRs without bias from the endogenous TCR, investigate RNA expression of endogenous and transgenic TCRs after OTR or viral transduction on the single-cell level, analyze whole-genome integration sites of CRISPR/Cas9-mediated targeted integration and viral transduction, and monitor a polyclonal human T cell repertoire by color barcoding in an in vivo model. However, our study is not without limitations that should be mentioned. TLA analysis of TCR transgene integration after conventional editing and OTR did not provide information on the single-cell level. Furthermore, the frequency of integration events at a specific locus could only be determined qualitatively. For more detailed and long-term investigation of human TCR-transgenic T cell products, suitable in vivo models are currently missing. The here-applied immunodeficient mouse model does allow determination of human T cell recruitment and short- but not long-term T cell responses to CMV infection. Performing such experiments in syngeneic mouse models would be a promising alternative; however, protocols for OTR in primary murine T cells are currently not available.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-human TCR α/β FITC | BioLegend | #306706 |
| anti-human CD3 PC7 | Beckman Coulter | #737657 |
| anti-human CD8 PE | Invitrogen | #MHCD0804 |
| anti-murine TCR β-chain APC/Fire750 | BioLegend | #109246 |
| Streptavidin-BV421 | BioLegend | #405225 |
| Bacterial and virus strains | ||
| mCMV-ie2-ANLV | 61 | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Alt-R S.p. HiFi Cas9 Nuclease V3 | IDT DNA | https://eu.idtdna.com |
| Alt-R CRISPR-Cas9 crRNA | IDT DNA | https://eu.idtdna.com |
| Alt-R CRISPR-Cas9 tracrRNA | IDT DNA | https://eu.idtdna.com |
| Alt-R Cas9 Electroporation Enhancer | IDT DNA | https://eu.idtdna.com |
| P3 Primary Cell 24 cuvettes Nucleofector™ Kit | Lonza | V4XP-3024 |
| Critical commercial assays | ||
| Targeted locus amplification | Cergentis | https://www.cergentis.com |
| Chromium next GEM Single Cell VDJ V1.1. | 10X Genomics | Rev D |
| Deposited data | ||
| scRNA-seq | This paper | https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE179566 |
| Experimental models: cell lines | ||
| TPRKO | 52 | N/A |
| Experimental models: organisms/strains | ||
| NOD.Cg-PrkdcscidIl2rgtm1WjlTg(HLA-A/H2-D/B2M)1Dvs/SzJ (NSG/HHD) | 56 | JAX stock #014570 |
| Oligonucleotides | ||
| See Table S2 | This paper | N/A |
| Recombinant DNA | ||
| See Table S1 | This paper | N/A |
| pMP71 | Addgene | #108214 |
| pCDH-EF1 | Addgene | #72266 |
| Software and algorithms | ||
| FlowJo v10 | FlowJo, LLC | https://www.flowjo.com/ |
| GraphPad Prism | Graphpad | https://www.graphpad.com/ |
| SCANPY | 62 | https://scanpy.readthedocs.io/en/stable/ |
| Cell Ranger (V3.0.2) | 10X Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/installation |
| Other | ||
| Script for analysis of scRNA-seq data | This paper | https://github.com/SebastianJarosch/2021_Mueller_scRNA-OTR-vs-Tx |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dirk H. Busch (dirk.busch@tum.de).
Materials availability
TCR constructs generated in this study can be made available under appropriate materials transfer agreement. No other unique reagents were generated.
Experimental model and subject details
Human primary T cells from whole blood and cell culture
T cells for editing were isolated from whole blood via density gradient centrifugation using Biocoll (Biochrom, reference #L6115). Generally, T cells were cultured in RPMI 1640 (LifeTechnologies, reference #31870074) supplemented with 10% FCS (Sigma, reference #F7524), 50 μM 2-mercaptoethanol (Invitrogen, reference #31350-010), 1.34 mM L-glutamine (Sigma, reference #G8540), 5 mM HEPES (Roth, reference #HN77.4), 25 mg gentamycin (LifeTechnologies, reference #15750-037) and 1x penicillin/streptomycin (LifeTechnologies, reference #10378-016) and 180 IU ml-1 recombinant human IL-2 (Peprotech, reference #200-02) (‘RPMI’ hereafter) unless indicated otherwise. Sorted cells were cultured with 1x106 ml-1 irradiated (30 Gy) allogeneic feeder peripheral blood mononuclear cells (PBMC), 1 μg ml-1 PHA (ThermoFisher, reference #R30852801) and 180 IU ml-1 recombinant human IL-2.
Written informed consent was obtained from the donors, and usage of the blood samples was approved according to national law by the local Institutional Review Board (Ethikkommission der Medizinischen Fakultät der Technischen Universität München). Information about age and gender of donors is not available. The study conforms to the standards of the Declaration of Helsinki.
Human T cell reporter TPRKO cell line culture
TPRKO cell lines52 were cultured in RPMI 1640 (GIBCO, Thermo Fisher Scientific; Waltham, Massachusetts) supplemented with 10% FCS, 0.025% L-glutamine, 0.1% HEPES, 0.001% gentamycin, and 0.002% streptomycin (‘RPMI’ hereafter).
Mouse model
In vivo experiments were performed with human HLA-A∗0201 transgenic NOD.Cg-PrkdcscidIl2rgtm1WjlTg(HLA-A/H2-D/B2M)1Dvs/SzJ (NSG/HHD) mice56 and a chimeric murine CMV engineered to express the human CMV A2/pp65495-503 (NLV) peptide (mCMV-ie2-ANLV)57. Both, female and male individuals in the age of 15 to 22 weeks were used for experiments and evenly distributed over experimental groups. First, mice were irradiated (2 Gy) and subsequently TCR-transgenic T cell product (cell numbers as mentioned in main text) was injected intraperitoneally. About 24 h after infection, mice were infected with 5x103 PFU of mCMV-ie2-ANLV. Mice were sacrificed at day 9 after T cell transfer and their liver harvested. Intra-hepatic lymphocytes were subsequently stained with pMHC-multimer BV421, anti-human CD45 PC7 (eBiosciences, reference #25-9459-42), anti-human CD8 PE, anti-murine TCR β-chain APC/Fire750 and propidium iodide.
All animal experiments were approved by the district government of upper Bavaria (Department 5 – Environment, Health and Consumer Protection; ROB 55.2-2532.Vet_02-18-162).
Method details
TCR identification
CMV-seropositive donor PBMCs were stained with respective pMHC-multimer that was individually conjugated with two different fluorophores to achieve reliable double pMHC-multimer staining. Single cells positive for CD8, CD62L, CD45RO and pMHC-multimer were sorted in a 384 well plate and stimulated with 10 μg ml-1 plate bound anti-CD3 (Biolegend, reference #317302) and anti-CD28 (BD PharMingen, reference #348040) each. RPMI medium was supplemented with 200 IU ml-1 recombinant human IL-2 and 5 ng ml-1 recombinant human IL-15 (Peprotech, reference #200-15). Single cell derived clones were harvested between day 7 and 14 after sort for TCR amplification via TCR-SCAN RACE PCR63. Cells were resuspended in 2 μl reaction mix comprising 1.6 mM dNTPs (Agilent, reference #200415), 10 mM DTT (Sigma, reference #D9779-10G), 0.1 mg/mL BSA, 0.1 mg/mL tRNA (LifeTechnologies, reference #FD0596), 0.25% Igepal CA-630 (Sigma-Aldrich, reference #238570-100G), 0.8 U/μl RNAsin Plus (Promega, reference #N2611), 1 μM reverse transcription primers each (for sequences see Table S2) and 0.05 reactions/μl (rxn/μl) Affinityscript reverse transcriptase (Agilent, reference #600107) and 1x Affinityscript buffer. Reverse transcription was performed for 20 min at 51°C followed by 30 min at 70°C on a Biometra TAdvanced thermal cycler. For primer exonuclease I digest, 0.51 μl reaction mix comprising 1 U/μl Exonuclease I (LifeTechnologies, EN0581) and 1x Exonuclease I buffer was added. Exonuclease-I digest was performed for 30 min at 37°C and enzyme was inactivated for 20 min at 70°C on a Biometra TAdvanced thermal cycler. Oligo-dG tailing was performed by adding 7 μl reaction mix comprising 100 mM MgCl2, 10 mM DTT, 100 mM Tris (pH 7.5) and 20 mM dGTP (Sigma-Aldrich, reference #11934538001) and 0.75 U/μl terminal dNTP transferase (Promega, reference #M1875). Tailing reaction was performed for 45 min at 37°C and enzyme was inactivated for 20 min at 37°C on a Biometra TAdvanced thermal cycler. For anchor PCR, 40 μl of reaction mix comprising 1x HerculaseII reaction buffer, 0.20 mM dNTPs, 3% formamide, 0.02 rxn/μl HerculaseII DNA polymerase (Agilent, reference #600679) and 0.5 μM of each primer (for sequences see Table S2) was added. PCR was performed as follows: 94°C and 3 min for initial denaturation followed by 24 cycles of 94°C for 15 s, 60°C for 30 s, 72°C for 45 s and 72°C for 5 min as final extension step. 1 μl of product was transferred to nested PCR amplification in separate reactions for α- and β-chain. PCR conditions for nested PCR round I and II were identical to anchor PCR except for primers (for sequences see Table S2). PCR amplificates were subsequently sequenced on the Illumina MiSeq platform.
TCR DNA template design
DNA templates were designed in silico and synthesized by GeneArt (Life Technologies, Thermo Fisher Scientific) or Twist Bioscience. DNA constructs for retro-/lentiviral transduction had the following structure: Human Kozac sequence64 followed by TCR β (including as indicated either human TRBC or murine TRBC with additional cysteine bridge25,27,28), followed by P2A, followed by TCR α (including as indicated either human TRAC or murine TRAC with additional cysteine bridge25,27,28), cloned into the pMP71 vector (kindly provided by Wolfgang Uckert, Berlin, Addgene plasmid backbone #108214) or into the pCDH-EF1 vector (Addgene plasmid #72266). DNA constructs for CRISPR/Cas9-mediated homology-directed repair (HDR) had the following structure: 5¢ homology arm (300-400 bp), P2A, TCR β (as above), T2A, TCR α (as above), bGHpA tail, 3¢ homology arm (300-400 bp). The homology arm sequences of the TRBC locus were derived from TRBC1 and are highly homologous to TRBC2. All TCR constructs for retro-/lentiviral transduction and OTR are listed in Table S1.
Cas9 RNPs
crRNA sequences for gRNAs were 5′-GGAGAATGACGAGTGGACCC-3′ for TRBC65 (targeting both TRBC1 and TRBC2) and 5′-AGAGTCTCTCAGCTGGTACA-3′ for TRAC65.
80 μM tracrRNA (IDT DNA) and 80 μM crRNA (IDT DNA) were incubated at 95°C for 5 min, then cooled to RT on the bench top. 24 μM high fidelity Cas9 (IDT DNA) was added slowly to gRNA solution to yield RNPs with 12 μM Cas9 and 20 μM gRNA, as well as 20 μM electroporation enhancer (IDT DNA). RNPs were incubated for 15 min at RT and directly used for electroporation.
Double-stranded DNA production for HDR
Double-stranded DNA PCR products were used for electroporation and HDR. Vector DNA was amplified by PCR according to the following protocol: reaction mix contained 1xHerculase buffer, 0.5 mM dNTPs, 0.4 μM primer forward and reverse (for sequences see Table S2), 0.01 rxn/μl Herculase (Agilent, reference #600679) and 60 ng template DNA. Thermal cycling was performed as following: 95°C and 3 min for initial denaturation followed by 34 cycles of 95°C for 30 s, 62°C for 30 s, 72°C for 3 min and 72°C for 3 min as final extension step. PCR products were purified by Ampure XP beads (Beckman Coulter, reference #A63881) in a 1:1 ratio. All TCR DNA templates were titrated but generally electroporation of 1 μg DNA per 1x106 cells yielded best KI efficiencies.
CRISPR/Cas9-mediated KO and KI
Either column selected CD4+ and CD8+ T cells mixed in a 1:1 ratio or bulk PBMCs were activated for two days in RPMI with CD3/CD28 Expamer (provided by Juno Therapeutics, activation with plate-bound aCD3/aCD28 as described above is also possible and gives similar results), 300 IU ml-1 recombinant human IL-2, 5 ng ml-1 recombinant human IL-7 (Peprotech, reference #200-07) and 5 ng ml-1 IL-15. Expamer stimulus was removed by incubation with 1 mM D-biotin (Sigma, reference #D1411-1G). 1x106 cells were electroporated (pulse code EH100) with Cas9 ribonucleoprotein and DNA templates in 20 μl Nucleofector Solution P3 (Lonza, reference #V4SP-3096) with a 4D Nucleofector X unit (Lonza). After electroporation, cells were cultured in RPMI supplemented with 180 IU ml-1 IL-2 until a first FACS analysis on day five after editing.
Retroviral transduction
Virus packaging cell line RD114 was cultured in DMEM (LifeTechnologies, reference #10938025) supplemented with 10% FCS (Sigma, reference #F7524), 50 μM 2-mercaptoethanol (Invitrogen, reference #31350-010), 1.34 mM L-glutamine (Sigma, reference #G8540), 5 mM HEPES (Roth, reference #HN77.4), 25 mg gentamycin (LifeTechnologies, reference #15750-037) and 1x penicillin/streptomycin (LifeTechnologies, reference #10378-016). For the production of retroviral particles, RD114 were transfected with pMP71 expression vector (containing a TCR or fluorochrome) by calcium phosphate precipitation. For this, 15 μl of a 3.31 M MgCl2 was mixed with 18 μg vector DNA and filled up to a total of 150 μl with ddH2O. This mix was slowly added under vortexing to 150μl transfection buffer containing 274 mM NaCl, 9.9 mM KCl, 3.5 mM Na2HPO4 and 41.9 mM HEPES. Subsequently, transfection mix was added to RD114 cells and incubated for 6 h at 37°C, 5% CO2 followed by a complete medium exchange. Virus supernatant was harvested three days later and coated on retronectin (TaKaRa, reference # T100B) treated well plates via centrifugation at 2000 g, for 2 h at 32°C. Either column selected CD4+ and CD8+ T cells mixed in a 1:1 ratio or bulk PBMCs were activated for two days in RPMI with CD3/CD28 Expamer (as above described for CRISPR/Cas9-mediated KO and KI), 300 IU ml-1 IL-2, 5 ng ml-1 IL-7 and 5 ng ml-1 IL-15. Expamer stimulus was removed by incubation with 1 mM D-biotin. Activated T cells were transduced via spinoculation (1000 g, 10 min, 32°C) on virus-coated plates. TCR and/or fluorochrome transduction occurred 15 min after CRISPR/Cas9-mediated TCR KO editing.
Lentiviral Transduction
Virus packaging cell line HEK293T-lentiX was cultured in DMEM with 10% FCS and 1% penicillin/streptomycin/L-glutamine. For the production of lentiviral particles, HEK293T-lenitX cells were transfected with 550 ng pCDH-EF1 expression vector (containing a TCR) and the packaging plasmids pMD2.G (275 ng) and psPAX2 (550 ng) (Addgene #12259 and #12260). The DNA mix was slowly added to 4.25 μl FuGENE®HD (Promega, reference #E2311) in Opti-MEM I Reduced Serum Medium (ThermoFisher, reference #31985062). Subsequently, the transfection mix was applied to HEK293T-lentiX cells and incubated at 37°C, 5% CO2. Virus supernatant was harvested 48 h and 72 h later. The virus supernatant was combined and precipitated in 8.5% PEG-6000 and 0.3 M NaCl for 4 h at 4°C. Afterward, the virions were pelleted at 3500 rpm for 20 min at 4°C. The supernatant was decanted and the concentrated virus was used for T cell transduction. Either column selected CD4+ and CD8+ T cells mixed in a 1:1 ratio or bulk PBMCs were activated for two days in RPMI with CD3/CD28 Expamer (as above described for CRISPR/Cas9-mediated KO and KI), 300 IU ml-1 IL-2, 5 ng ml-1 IL-7 and 5 ng ml-1 IL-15. Expamer stimulus was removed by incubation with 1 mM D-biotin. Activated T cells were transduced using LentiBoostTM (Sirion Biotech). TCR and/or fluorochrome transduction occurred 15 min after CRISPR/Cas9-mediated TCR KO editing.
pMHC multimer and surface antibody staining
pMHC monomers were generated as previously described66. All pMHC-multimer reagents were generated by 45 min incubation of 4 μg biotinylated pMHC monomer with 1 μg streptavidin-BV421 (Biolegend, reference #405225) or Streptavidin-APC (eBioscience, reference #17-4317-82) in a total volume of 100 μl FACS buffer for staining of up to 1x107 cells. pMHC-multimer staining was performed separately from surface antibody staining for 45 min on ice. Surface antibody staining was performed for 25 min on ice subsequent to pMHC-multimer staining. Depending on the respective experiments, a subset of the following antibodies were used: anti-human TCR α/β FITC (BioLegend, reference #306706), CD3 PC7 (Beckman Coulter, reference #737657), CD8 PE (Invitrogen, reference #MHCD0804), anti-murine TCR β-chain APC (BioLegend, reference #109212), anti-murine TCR β-chain APC/Fire750 (BioLegend, reference #109246), CD62L FITC (Biolegend, reference #304804), CD45-RO PC7 (BioLegend, reference #304230), Lag3 FITC (Biolegend, reference #369308) and PD-1 APC (Invitrogen, reference #17-2799-42). Live/dead discrimination was performed with propidium iodide (LifeTechnologies, reference #P1304MP).
Antigen-specific activation and intracellular cytokine staining
One day before co-culture with T cells, K562 cells (retrovirally transduced to express the human MHC class-I molecule of interest) were irradiated (80 Gy) and loaded with peptide (10−12 M, 10−10 M, 10−9 M, 10−8 M, 10−7 M, 10−6 M, 10−5 M, 10−4 M) overnight at 37°C, 5% CO2. T cells were co-cultured in a 1:1 ratio with peptide-loaded K562 cells and 1x Golgi plug (BD PharMingen, reference #555029) for 4 h at 37°C. 25 ng ml-1 Phorbol-12-myristat-13-acetat (PMA) (Sigma, reference #P1585-1mg) and 1μg ml-1 ionomycin (Sigma, reference #I9657-1mg) were used for positive control. After co-culture, staining for live/dead discrimination was performed with ethidium-monoazide-bromide (LifeTechnologies, reference #E1374). Subsequently, surface marker antibody staining for CD8 (PE, Invitrogen) and anti-murine TCR β-chain APC/Fire750 (BioLegend, reference #109246) was performed and followed by permeabilization using Cytofix/Cytoperm (BD Biosciences, reference #554714), and intracellular staining of IFNγ FITC (BD PharMingen, reference #340449), TNFα PC7 (eBioscience, reference #25-7349-82) and IL-2 APC (BD PharMingen, reference #341116).
Generation and analysis of TCR-edited human T cell reporter lines
TCRs were introduced into the JE6.1 triple parameter reporter cell line (J-TPR)51,52 via retroviral transduction. Antigen-specific stimulation was performed using irradiated (80 Gy) and peptide pulsed (10−9 M, 10−8 M, 10−7 M, 10−6 M, 10−5 M, 10−4 M) K562 cells (retrovirally transduced to express the human MHC class-I molecule of interest). Effector and target cells were co-cultured in a 1:5 ratio for 18 h, at 37°C and 5% CO2. Subsequently, NFκB-eCFP reporter activity of J-TPR cells was analyzed on a flow cytometer.
Cytotoxic T lymphocyte assay
HLA-A∗0201 positive HepG2 cells were loaded with 10−10 M of A2/pp65495-503 (NLV) peptide (peptide&elephants, reference #EP04509_1). 4x104 peptide-pulsed HepG2 cells were plated per well in a collagen-coated 96 well E-Plate (ACEA Biosciences, reference #2801035). The plate was subsequently placed into a xCELLigence RTCA MP Real Time Cell Analyzer (ACEA Biosciences) and HepG2 cell growth was monitored every 15 min. After 18 h, 4x104 rested T cells were added per well containing HepG2 target cells. Mock edited (TCR transgene negative) T cells derived of the same donor were used as negative control. Effector and target cells were co-incubated for 48 h and cell growth/death was monitored every 15 min.
Flow cytometry
Acquisition of FACS samples was done on a Cytoflex (S) flow cytometer (Beckman Coulter). Flow sorting was conducted on a FACSAria III (BD Bioscience) or MoFlo Astrios EG (Beckman Coulter).
scRNA sequencing
TCRs were introduced either via CRISPR/Cas9-mediated KI or via retroviral transduction into endogenous TCR-KO primary T cells as described. T cells that underwent different TCR editing approaches (OTR, MOIlo, MOIhi) were barcoded via retroviral co-transduction with three different fluorochromes (BFP, CFP, GFP). Five days after editing, FACS sorted CD8+TCR+fluorochrome+ cells (8000 cells per editing method for each TCR) were centrifuged and the supernatant was carefully removed. Cells were resuspended in the mastermix plus 37.8 μl of water before 70 μl of the cell suspension was transferred to the chip. After each step, the integrity of the pellet was checked under the microscope to ensure that all cells are loaded onto the chip. From here on, the experiment was performed according to the manufacturer’s protocol (10x Genomics, Chromium next GEM Single Cell VDJ V1.1, Rev D). QC has been performed with a High sensitivity DNA Kit (Agilent, reference #5067-4626) on a Bioanalyzer 2100 as recommended in the protocol and libraries were quantified with the Qubit dsDNA HS assay kit (LifeTechnologies, reference #Q32851). All steps have been performed using RPT filter tips (Starlab, reference #S1183-1710, #SS1180-8710, #S1182-1730) and DNA LoBind tubes (Sigma, reference #EP0030108051, #EP0030108078, #EP0030124359). In order to amplify transgenic TCR sequences, the cDNA was amplified in two steps in parallel to the VDJ enrichment by using the 10x forward outer and inner primers as well as TCR- and/or editing method-specific reverse primers (see Table S2). In target enrichment 1 and 2 (protocol step 4.1 and 4.3) the elongation time was increased to 90 s in adaptation to the longer transduction products. In addition, the second amplification of the KI constructs was performed for 15 cycles instead of 10 in order to compensate for lower integration frequencies. The double-sided size selection (Step 4.4 of the 10x protocol) was adjusted to 0.4X and 0.8X for the transduction constructs and was kept at 0.5X and 0.8X for the KI constructs.
scRNA sequencing – Data processing
Combined fastq-files (transcriptome and transgenic TCR library) for each TCR were annotated against a custom reference containing all genes of the human genome (GRCh38), the fluorochromes (BFP, CFP, GFP) used for multiplexing and the respective TCR constructs using Cell Ranger (V3.0.2). Data analysis of the annotated count matrix was performed in SCANPY62. Cells were filtered to contain at least 200 genes and genes being present in less than 3 cells were excluded. 20% mitochondrial gene expression was allowed. Counts were normalized to 10.000 counts per cell and expression was log transformed. The number of counts, percent of mitochondrial genes, S and G2M phase scores were regressed out. In order to demultiplex subsamples according to fluorochrome expression, cells were filtered to express only one of the three fluorochromes and resulting leiden-clusters67 of the neighborhood graph were annotated according to fluorochrome expression. Highly variable genes (HVGs) were identified with mean values between 0.0125 and 3 and a minimal dispersion of 0.5. Expression values exceeding a standard deviation of 10 were clipped. The neighborhood graph was calculated for the 10 nearest neighbors and the first 7 components of the principal component analysis (PCA) for the HVGs. Fluorochromes and constructs have been excluded for the neighborhood embedding. Violin plots show the normalized raw gene expression. Scripts for analysis of scRNA sequencing data are provided here: https://github.com/SebastianJarosch/2021_Mueller_scRNA-OTR-vs-Tx.
Targeted locus amplification
TLA including sequencing was performed by Cergentis as previously described49. Two primer sets (provided in Table S2) were designed to target each transgene. Primer sets were used in individual TLA amplifications. PCR products were purified and library prepped using the Illumina Nextera flex protocol and sequenced on an Illumina sequencer. NGS reads were aligned to the transgene sequence and host genome (human hg19 reference sequence). Bioinformatic analysis of integration sites and detection of hit cancer genes via Enhort (unpublished, https://enhort.mni.thm.de/), Cosmic68 and Network of Cancer Genes69.
mCMV mutagenesis and virus stock production
mCMV-ie2-ANLV was generated by fusing AANLVPMVATV peptide at the C terminus of the ie2 protein. HCMV pp65495-503 epitope (NLVPMVATV) was preceded by two alanine residues that enhance peptide processing and presentation61. The recombinant virus is based on mCMV smith strain (pSM3fr-MCK-2fl clone 3.3)70. The peptide was inserted at the ie2 C terminus position 187,296 (NCBI accession: NC_004065) using en passant mutagenesis as described before71,72. Briefly, we generated the en passant mutagenesis cassette using pGP704 I-SceI-Kan plasmid and primers (ie2-pp65-fw and ie2-pp65-rv, see Table S2) that contain homology to the insertion site in the mCMV genome and pp65495-503 epitope sequence. The amplified cassette was transformed into electro-competent GS1783 bacteria that harbor mCMV BAC. After successful insertion of the cassette, the positive selection marker (kanamycin) was excised from the BAC by I-SceI restriction and homology mediated recombination. The resulting clones were screened with colony PCR and confirmed by Sanger sequencing of the target region. Virus was recovered by transfecting mouse embryo fibroblast (MEF) cells with virus BAC, and after reconstitution, mCMV-ie2-ANLV was propagated on M2-10B4 cells. mCMV virus stocks were prepared by pelleting supernatants of infected M2-10B4 cells (26,000 g for 3.5 h). Subsequently, the pellet was re-suspended in VSB buffer (0.05 M Tris-HCl, 0.012 M KCl, and 0.005 M EDTA, adjusted to pH 7.8) and purified by centrifugation through a 15% sucrose cushion in VSB buffer (53,000 g), and a subsequent slow centrifugation step (3000 g, 5 min) to remove cellular debris. The mCMV virus stocks were titrated on MEF cells and in vivo virus growth was compared to mCMVWT to ensure virus genome stability and rule out off target effects.
Quantification and statistical analysis
All flow cytometric data were analyzed with FlowJo v10. All statistical analyses were performed using GraphPad Prism and are represented as indicated in figure legends. Statistical tests were selected based on the dataset and are accordingly indicated in figure legends. A P value of < 0.05 was considered statistically significant.
Acknowledgments
We thank members of the Busch and Buchholz laboratories for experimental help and critical discussion, particularly F. Mohr, M. Plambeck, A. Hochholzer, F. Graml, S. Dötsch, E. d’Ippolito, and V. R. Buchholz. We also thank our flow cytometry unit, specifically L. Henkel, C. Angerpointner, and M. Schiemann. We appreciate fruitful discussions with C. Stemberger, M. Poltorak, and L. Germeroth (Juno Therapeutics). We are also grateful to O. Quitt, U. Protzer, K. Dennehy, and W. Uckert for providing cell lines and constructs. This work was supported by the German Center for Infection Research (DZIF) as well as by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) SFB- TRR 338/1 2021-45288190, SFB 1321/TP17, SFB 1054/B09, and SFB 1371/TP04.
Author contributions
T.R.M., K.S., and D.H.B. conceived the study. T.R.M., K.S., and D.H.B. designed and analyzed experiments. T.R.M., M.H., and K.S. developed the TCR library. M.N. provided a donor biobank for TCR isolation. M.E. generated pMHC-multimers. T.R.M performed TCR editing and in vitro experiments. B.B., K.Sch., A.M., and U.P. provided and generated lentivirus for transduction. P.S. developed and advised on experiments with J-TPR cell line. T.R.M, K.S., and J.L. performed in vivo experiments. M.Z.C and L.C-S. generated mCMV for in vivo experiments. S.G. and I.A. performed and advised on FACS sorting. T.K. and T.F. performed whole genome sequencing. T.R.M., S.J., and M.H. performed single-cell transcriptomics. S.J. performed bioinformatics analysis. T.R.M., K.S., and D.H.B. wrote the manuscript. All authors read and reviewed the manuscript.
Declaration of interests
D.H.B. is co-founder of STAGE Cell Therapeutics GmbH (now Juno Therapeutics, a Bristol-Myers Squibb Company) and T Cell Factory B.V. (now Kite, a Gilead Company). D.H.B. has a consulting contract with and receives sponsored research support from Juno Therapeutics.
Published: August 17, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2021.100374.
Contributor Information
Kilian Schober, Email: kilian.schober@uk-erlangen.de.
Dirk H. Busch, Email: dirk.busch@tum.de.
Supplemental information
Data and code availability
Single-cell RNA sequencing data are available: GEO Series accession number GSE179566 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE179566)
Scripts for analysis of scRNA sequencing data are provided here: https://github.com/SebastianJarosch/2021_Mueller_scRNA-OTR-vs-Tx
References
- 1.Billingham R.E., Brent L., Medawar P.B. Quantitative studies on tissue transplantation immunity. II. The origin, strength and duration of actively and adoptively acquired immunity. Proc. R. Soc. London. Ser. B, Biol. Sci. 1954;143:58–80. doi: 10.1098/rspb.1954.0054. [DOI] [PubMed] [Google Scholar]
- 2.June C.H., Riddell S.R., Schumacher T.N. Adoptive cellular therapy: a race to the finish line. Sci. Transl. Med. 2015;7:280ps7. doi: 10.1126/scitranslmed.aaa3643. [DOI] [PubMed] [Google Scholar]
- 3.Rosenberg S.A., Spiess P., Lafreniere R. A new approach to the adoptive immunotherapy of cancer with tumor-infiltrating lymphocytes. Science. 1986;233:1318–1321. doi: 10.1126/science.3489291. [DOI] [PubMed] [Google Scholar]
- 4.Riddell S.R., Watanabe K.S., Goodrich J.M., Li C.R., Agha M.E., Greenberg P.D. Restoration of viral immunity in immunodeficient humans by the adoptive transfer of T cell clones. Science. 1992;257:238–241. doi: 10.1126/science.1352912. [DOI] [PubMed] [Google Scholar]
- 5.Scheper W., Kelderman S., Fanchi L.F., Linnemann C., Bendle G., de Rooij M.A.J., Hirt C., Mezzadra R., Slagter M., Dijkstra K. Low and variable tumor reactivity of the intratumoral TCR repertoire in human cancers. Nat. Med. 2019;25:89–94. doi: 10.1038/s41591-018-0266-5. [DOI] [PubMed] [Google Scholar]
- 6.Schumacher T.N., Scheper W., Kvistborg P. Cancer Neoantigens. Annu. Rev. Immunol. 2019;37:173–200. doi: 10.1146/annurev-immunol-042617-053402. [DOI] [PubMed] [Google Scholar]
- 7.Ljungman P., Brand R., Einsele H., Frassoni F., Niederwieser D., Cordonnier C. Donor CMV serologic status and outcome of CMV-seropositive recipients after unrelated donor stem cell transplantation: an EBMT megafile analysis. Blood. 2003;102:4255–4260. doi: 10.1182/blood-2002-10-3263. [DOI] [PubMed] [Google Scholar]
- 8.Berger C., Jensen M.C., Lansdorp P.M., Gough M., Elliott C., Riddell S.R. Adoptive transfer of effector CD8+ T cells derived from central memory cells establishes persistent T cell memory in primates. J. Clin. Invest. 2008;118:294–305. doi: 10.1172/JCI32103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graef P., Buchholz V.R., Stemberger C., Flossdorf M., Henkel L., Schiemann M., Drexler I., Höfer T., Riddell S.R., Busch D.H. Serial transfer of single-cell-derived immunocompetence reveals stemness of CD8(+) central memory T cells. Immunity. 2014;41:116–126. doi: 10.1016/j.immuni.2014.05.018. [DOI] [PubMed] [Google Scholar]
- 10.Stemberger C., Graef P., Odendahl M., Albrecht J., Dössinger G., Anderl F., Buchholz V.R., Gasteiger G., Schiemann M., Grigoleit G.U. Lowest numbers of primary CD8(+) T cells can reconstitute protective immunity upon adoptive immunotherapy. Blood. 2014;124:628–637. doi: 10.1182/blood-2013-12-547349. [DOI] [PubMed] [Google Scholar]
- 11.Gattinoni L., Speiser D.E., Lichterfeld M., Bonini C. T memory stem cells in health and disease. Nat. Med. 2017;23:18–27. doi: 10.1038/nm.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neuenhahn M., Albrecht J., Odendahl M., Schlott F., Dössinger G., Schiemann M., Lakshmipathi S., Martin K., Bunjes D., Harsdorf S. Transfer of minimally manipulated CMV-specific T cells from stem cell or third-party donors to treat CMV infection after allo-HSCT. Leukemia. 2017;31:2161–2171. doi: 10.1038/leu.2017.16. [DOI] [PubMed] [Google Scholar]
- 13.Nauerth M., Weißbrich B., Knall R., Franz T., Dössinger G., Bet J., Paszkiewicz P.J., Pfeifer L., Bunse M., Uckert W. TCR-ligand koff rate correlates with the protective capacity of antigen-specific CD8+ T cells for adoptive transfer. Sci. Transl. Med. 2013;5:192ra87. doi: 10.1126/scitranslmed.3005958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang S.Q., Parker P., Ma K.Y., He C., Shi Q., Cui Z., Williams C.M., Wendel B.S., Meriwether A.I., Salazar M.A., Jiang N. Direct measurement of T cell receptor affinity and sequence from naïve antiviral T cells. Sci. Transl. Med. 2016;8:341ra77. doi: 10.1126/scitranslmed.aaf1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morgan R.A., Dudley M.E., Wunderlich J.R., Hughes M.S., Yang J.C., Sherry R.M., Royal R.E., Topalian S.L., Kammula U.S., Restifo N.P. Cancer regression in patients after transfer of genetically engineered lymphocytes. Science. 2006;314:126–129. doi: 10.1126/science.1129003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robbins P.F., Morgan R.A., Feldman S.A., Yang J.C., Sherry R.M., Dudley M.E., Wunderlich J.R., Nahvi A.V., Helman L.J., Mackall C.L. Tumor regression in patients with metastatic synovial cell sarcoma and melanoma using genetically engineered lymphocytes reactive with NY-ESO-1. J. Clin. Oncol. 2011;29:917–924. doi: 10.1200/JCO.2010.32.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chodon T., Comin-Anduix B., Chmielowski B., Koya R.C., Wu Z., Auerbach M., Ng C., Avramis E., Seja E., Villanueva A. Adoptive transfer of MART-1 T-cell receptor transgenic lymphocytes and dendritic cell vaccination in patients with metastatic melanoma. Clin. Cancer Res. 2014;20:2457–2465. doi: 10.1158/1078-0432.CCR-13-3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Loenen M.M., de Boer R., Hagedoorn R.S., van Egmond E.H.M., Falkenburg J.H.F., Heemskerk M.H.M. Optimization of the HA-1-specific T-cell receptor for gene therapy of hematologic malignancies. Haematologica. 2011;96:477–481. doi: 10.3324/haematol.2010.025916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okamoto S., Mineno J., Ikeda H., Fujiwara H., Yasukawa M., Shiku H., Kato I. Improved expression and reactivity of transduced tumor-specific TCRs in human lymphocytes by specific silencing of endogenous TCR. Cancer Res. 2009;69:9003–9011. doi: 10.1158/0008-5472.CAN-09-1450. [DOI] [PubMed] [Google Scholar]
- 20.Ahmadi M., King J.W., Xue S.-A., Voisine C., Holler A., Wright G.P., Waxman J., Morris E., Stauss H.J. CD3 limits the efficacy of TCR gene therapy in vivo. Blood. 2011;118:3528–3537. doi: 10.1182/blood-2011-04-346338. [DOI] [PubMed] [Google Scholar]
- 21.Bendle G.M., Linnemann C., Hooijkaas A.I., Bies L., de Witte M.A., Jorritsma A., Kaiser A.D.M., Pouw N., Debets R., Kieback E. Lethal graft-versus-host disease in mouse models of T cell receptor gene therapy. Nat. Med. 2010;16:565–570. doi: 10.1038/nm.2128. 1p, 570. [DOI] [PubMed] [Google Scholar]
- 22.Provasi E., Genovese P., Lombardo A., Magnani Z., Liu P.-Q., Reik A., Chu V., Paschon D.E., Zhang L., Kuball J. Editing T cell specificity towards leukemia by zinc finger nucleases and lentiviral gene transfer. Nat. Med. 2012;18:807–815. doi: 10.1038/nm.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bunse M., Bendle G.M., Linnemann C., Bies L., Schulz S., Schumacher T.N., Uckert W. RNAi-mediated TCR knockdown prevents autoimmunity in mice caused by mixed TCR dimers following TCR gene transfer. Mol. Ther. 2014;22:1983–1991. doi: 10.1038/mt.2014.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Loenen M.M., de Boer R., Amir A.L., Hagedoorn R.S., Volbeda G.L., Willemze R., van Rood J.J., Falkenburg J.H.F., Heemskerk M.H.M. Mixed T cell receptor dimers harbor potentially harmful neoreactivity. Proc. Natl. Acad. Sci. USA. 2010;107:10972–10977. doi: 10.1073/pnas.1005802107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cohen C.J., Zhao Y., Zheng Z., Rosenberg S.A., Morgan R.A. Enhanced antitumor activity of murine-human hybrid T-cell receptor (TCR) in human lymphocytes is associated with improved pairing and TCR/CD3 stability. Cancer Res. 2006;66:8878–8886. doi: 10.1158/0008-5472.CAN-06-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sommermeyer D., Uckert W. Minimal amino acid exchange in human TCR constant regions fosters improved function of TCR gene-modified T cells. J. Immunol. 2010;184:6223–6231. doi: 10.4049/jimmunol.0902055. [DOI] [PubMed] [Google Scholar]
- 27.Cohen C.J., Li Y.F., El-Gamil M., Robbins P.F., Rosenberg S.A., Morgan R.A. Enhanced antitumor activity of T cells engineered to express T-cell receptors with a second disulfide bond. Cancer Res. 2007;67:3898–3903. doi: 10.1158/0008-5472.CAN-06-3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuball J., Dossett M.L., Wolfl M., Ho W.Y., Voss R.-H., Fowler C., Greenberg P.D. Facilitating matched pairing and expression of TCR chains introduced into human T cells. Blood. 2007;109:2331–2338. doi: 10.1182/blood-2006-05-023069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Voss R.H., Thomas S., Pfirschke C., Hauptrock B., Klobuch S., Kuball J., Grabowski M., Engel R., Guillaume P., Romero P. Coexpression of the T-cell receptor constant α domain triggers tumor reactivity of single-chain TCR-transduced human T cells. Blood. 2010;115:5154–5163. doi: 10.1182/blood-2009-11-254078. [DOI] [PubMed] [Google Scholar]
- 30.Bethune M.T., Gee M.H., Bunse M., Lee M.S., Gschweng E.H., Pagadala M.S., Zhou J., Cheng D., Heath J.R., Kohn D.B. Domain-swapped T cell receptors improve the safety of TCR gene therapy. eLife. 2016;5:1–24. doi: 10.7554/eLife.19095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomas S., Mohammed F., Reijmers R.M., Woolston A., Stauss T., Kennedy A., Stirling D., Holler A., Green L., Jones D. Framework engineering to produce dominant T cell receptors with enhanced antigen-specific function. Nat. Commun. 2019;10:4451. doi: 10.1038/s41467-019-12441-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schober K., Müller T.R., Gökmen F., Grassmann S., Effenberger M., Poltorak M., Stemberger C., Schumann K., Roth T.L., Marson A., Busch D.H. Orthotopic replacement of T-cell receptor α- and β-chains with preservation of near-physiological T-cell function. Nat. Biomed. Eng. 2019;3:974–984. doi: 10.1038/s41551-019-0409-0. [DOI] [PubMed] [Google Scholar]
- 33.Mastaglio S., Genovese P., Magnani Z., Ruggiero E., Landoni E., Camisa B., Schiroli G., Provasi E., Lombardo A., Reik A. NY-ESO-1 TCR single edited stem and central memory T cells to treat multiple myeloma without graft-versus-host disease. Blood. 2017;130:606–618. doi: 10.1182/blood-2016-08-732636. [DOI] [PubMed] [Google Scholar]
- 34.Legut M., Dolton G., Mian A.A., Ottmann O.G., Sewell A.K. CRISPR-mediated TCR replacement generates superior anticancer transgenic T cells. Blood. 2018;131:311–322. doi: 10.1182/blood-2017-05-787598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Santeramo I., Bagnati M., Harvey E.J., Hassan E., Surmacz-Cordle B., Marshall D., Di Cerbo V. Vector Copy Distribution at a Single-Cell Level Enhances Analytical Characterization of Gene-Modified Cell Therapies. Mol. Ther. Methods Clin. Dev. 2020;17:944–956. doi: 10.1016/j.omtm.2020.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aiuti A., Cossu G., de Felipe P., Galli M.C., Narayanan G., Renner M., Stahlbom A., Schneider C.K., Voltz-Girolt C. The committee for advanced therapies’ of the European Medicines Agency reflection paper on management of clinical risks deriving from insertional mutagenesis. Hum. Gene Ther. Clin. Dev. 2013;24:47–54. doi: 10.1089/humc.2013.119. [DOI] [PubMed] [Google Scholar]
- 37.Sadelain M. Insertional oncogenesis in gene therapy: how much of a risk? Gene Ther. 2004;11:569–573. doi: 10.1038/sj.gt.3302243. [DOI] [PubMed] [Google Scholar]
- 38.Maude S.L., Laetsch T.W., Buechner J., Rives S., Boyer M., Bittencourt H., Bader P., Verneris M.R., Stefanski H.E., Myers G.D. Tisagenlecleucel in Children and Young Adults with B-Cell Lymphoblastic Leukemia. N. Engl. J. Med. 2018;378:439–448. doi: 10.1056/NEJMoa1709866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rapoport A.P., Stadtmauer E.A., Binder-Scholl G.K., Goloubeva O., Vogl D.T., Lacey S.F., Badros A.Z., Garfall A., Weiss B., Finklestein J. NY-ESO-1-specific TCR-engineered T cells mediate sustained antigen-specific antitumor effects in myeloma. Nat. Med. 2015;21:914–921. doi: 10.1038/nm.3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Linette G.P., Stadtmauer E.A., Maus M.V., Rapoport A.P., Levine B.L., Emery L., Litzky L., Bagg A., Carreno B.M., Cimino P.J. Cardiovascular toxicity and titin cross-reactivity of affinity-enhanced T cells in myeloma and melanoma. Blood. 2013;122:863–871. doi: 10.1182/blood-2013-03-490565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Monjezi R., Miskey C., Gogishvili T., Schleef M., Schmeer M., Einsele H., Ivics Z., Hudecek M. Enhanced CAR T-cell engineering using non-viral Sleeping Beauty transposition from minicircle vectors. Leukemia. 2017;31:186–194. doi: 10.1038/leu.2016.180. [DOI] [PubMed] [Google Scholar]
- 42.Kustikova O.S., Wahlers A., Kuhlcke K., Stahle B., Zander A.R., Baum C., Fehse B. Dose finding with retroviral vectors: correlation of retroviral vector copy numbers in single cells with gene transfer efficiency in a cell population. Blood. 2003;102:3934–3937. doi: 10.1182/blood-2003-05-1424. [DOI] [PubMed] [Google Scholar]
- 43.Viola A., Lanzavecchia A. T cell activation determined by T cell receptor number and tunable thresholds. Science. 1996;273:104–106. doi: 10.1126/science.273.5271.104. [DOI] [PubMed] [Google Scholar]
- 44.Eyquem J., Mansilla-Soto J., Giavridis T., van der Stegen S.J.C., Hamieh M., Cunanan K.M., Odak A., Gönen M., Sadelain M. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature. 2017;543:113–117. doi: 10.1038/nature21405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roth T.L., Puig-Saus C., Yu R., Shifrut E., Carnevale J., Li P.J., Hiatt J., Saco J., Krystofinski P., Li H. Reprogramming human T cell function and specificity with non-viral genome targeting. Nature. 2018;559:405–409. doi: 10.1038/s41586-018-0326-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Izsvák Z., Ivics Z. Sleeping beauty transposition: biology and applications for molecular therapy. Mol. Ther. 2004;9:147–156. doi: 10.1016/j.ymthe.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 47.Field A.-C., Vink C., Gabriel R., Al-Subki R., Schmidt M., Goulden N., Stauss H., Thrasher A., Morris E., Qasim W. Comparison of lentiviral and sleeping beauty mediated αβ T cell receptor gene transfer. PLoS ONE. 2013;8:e68201. doi: 10.1371/journal.pone.0068201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Engels B., Cam H., Schüler T., Indraccolo S., Gladow M., Baum C., Blankenstein T., Uckert W. Retroviral vectors for high-level transgene expression in T lymphocytes. Hum. Gene Ther. 2003;14:1155–1168. doi: 10.1089/104303403322167993. [DOI] [PubMed] [Google Scholar]
- 49.de Vree P.J.P., de Wit E., Yilmaz M., van de Heijning M., Klous P., Verstegen M.J.A.M., Wan Y., Teunissen H., Krijger P.H.L., Geeven G. Targeted sequencing by proximity ligation for comprehensive variant detection and local haplotyping. Nat. Biotechnol. 2014;32:1019–1025. doi: 10.1038/nbt.2959. [DOI] [PubMed] [Google Scholar]
- 50.Cao J., Cusanovich D.A., Ramani V., Aghamirzaie D., Pliner H.A., Hill A.J., Daza R.M., McFaline-Figueroa J.L., Packer J.S., Christiansen L. Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science. 2018;361:1380–1385. doi: 10.1126/science.aau0730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jutz S., Leitner J., Schmetterer K., Doel-Perez I., Majdic O., Grabmeier-Pfistershammer K., Paster W., Huppa J.B., Steinberger P. Assessment of costimulation and coinhibition in a triple parameter T cell reporter line: Simultaneous measurement of NF-κB, NFAT and AP-1. J. Immunol. Methods. 2016;430:10–20. doi: 10.1016/j.jim.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 52.Müller T.R., Schuler C., Hammel M., Köhler A., Jutz S., Leitner J., Schober K., Busch D.H., Steinberger P. A T-cell reporter platform for high-throughput and reliable investigation of TCR function and biology. Clin. Transl. Immunology. 2020;9:e1216. doi: 10.1002/cti2.1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Buchholz V.R., Flossdorf M., Hensel I., Kretschmer L., Weissbrich B., Gräf P., Verschoor A., Schiemann M., Höfer T., Busch D.H. Disparate individual fates compose robust CD8+ T cell immunity. Science. 2013;340:630–635. doi: 10.1126/science.1235454. [DOI] [PubMed] [Google Scholar]
- 54.Grassmann S., Pachmayr L.O., Leube J., Mihatsch L., Andrae I., Flommersfeld S., Oduro J., Cicin-Sain L., Schiemann M., Flossdorf M., Buchholz V.R. Distinct Surface Expression of Activating Receptor Ly49H Drives Differential Expansion of NK Cell Clones upon Murine Cytomegalovirus Infection. Immunity. 2019;50:1391–1400.e4. doi: 10.1016/j.immuni.2019.04.015. [DOI] [PubMed] [Google Scholar]
- 55.Schober K., Voit F., Grassmann S., Müller T.R., Eggert J., Jarosch S., Weißbrich B., Hoffmann P., Borkner L., Nio E. Reverse TCR repertoire evolution toward dominant low-affinity clones during chronic CMV infection. Nat. Immunol. 2020;21:434–441. doi: 10.1038/s41590-020-0628-2. [DOI] [PubMed] [Google Scholar]
- 56.Shultz L.D., Saito Y., Najima Y., Tanaka S., Ochi T., Tomizawa M., Doi T., Sone A., Suzuki N., Fujiwara H. Generation of functional human T-cell subsets with HLA-restricted immune responses in HLA class I expressing NOD/SCID/IL2r γ(null) humanized mice. Proc. Natl. Acad. Sci. USA. 2010;107:13022–13027. doi: 10.1073/pnas.1000475107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thomas S., Klobuch S., Podlech J., Plachter B., Hoffmann P., Renzaho A., Theobald M., Reddehase M.J., Herr W., Lemmermann N.A.W. Evaluating Human T-Cell Therapy of Cytomegalovirus Organ Disease in HLA-Transgenic Mice. PLoS Pathog. 2015;11:e1005049. doi: 10.1371/journal.ppat.1005049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hacein-Bey-Abina S., Von Kalle C., Schmidt M., McCormack M.P., Wulffraat N., Leboulch P., Lim A., Osborne C.S., Pawliuk R., Morillon E. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 59.Fraietta J.A., Nobles C.L., Sammons M.A., Lundh S., Carty S.A., Reich T.J., Cogdill A.P., Morrissette J.J.D., DeNizio J.E., Reddy S. Disruption of TET2 promotes the therapeutic efficacy of CD19-targeted T cells. Nature. 2018;558:307–312. doi: 10.1038/s41586-018-0178-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Albers J.J., Ammon T., Gosmann D., Audehm S., Thoene S., Winter C., Secci R., Wolf A., Stelzl A., Steiger K. Gene editing enables T-cell engineering to redirect antigen specificity for potent tumor rejection. Life Sci. Alliance. 2019;2:1–10. doi: 10.26508/lsa.201900367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dekhtiarenko I., Ratts R.B., Blatnik R., Lee L.N., Fischer S., Borkner L., Oduro J.D., Marandu T.F., Hoppe S., Ruzsics Z. Peptide Processing Is Critical for T-Cell Memory Inflation and May Be Optimized to Improve Immune Protection by CMV-Based Vaccine Vectors. PLoS Pathog. 2016;12:e1006072. doi: 10.1371/journal.ppat.1006072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wolf F.A., Angerer P., Theis F.J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19:15. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dössinger G., Bunse M., Bet J., Albrecht J., Paszkiewicz P.J., Weißbrich B., Schiedewitz I., Henkel L., Schiemann M., Neuenhahn M. MHC multimer-guided and cell culture-independent isolation of functional T cell receptors from single cells facilitates TCR identification for immunotherapy. PLoS ONE. 2013;8:e61384. doi: 10.1371/journal.pone.0061384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nakagawa S., Niimura Y., Gojobori T., Tanaka H., Miura K. Diversity of preferred nucleotide sequences around the translation initiation codon in eukaryote genomes. Nucleic Acids Res. 2008;36:861–871. doi: 10.1093/nar/gkm1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ren J., Liu X., Fang C., Jiang S., June C.H., Zhao Y. Multiplex genome editing to generate universal CAR T cells resistant to PD1 inhibition. Clin. Cancer Res. 2017;23:2255–2266. doi: 10.1158/1078-0432.CCR-16-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Effenberger M., Stengl A., Schober K., Gerget M., Kampick M., Müller T.R., Schumacher D., Helma J., Leonhardt H., Busch D.H. FLEXamers: A Double Tag for Universal Generation of Versatile Peptide-MHC Multimers. J. Immunol. 2019;202:2164–2171. doi: 10.4049/jimmunol.1801435. [DOI] [PubMed] [Google Scholar]
- 67.Traag V.A., Waltman L., van Eck N.J. From Louvain to Leiden: guaranteeing well-connected communities. Sci. Rep. 2019;9:5233. doi: 10.1038/s41598-019-41695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tate J.G., Bamford S., Jubb H.C., Sondka Z., Beare D.M., Bindal N., Boutselakis H., Cole C.G., Creatore C., Dawson E. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019;47(D1):D941–D947. doi: 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Repana D., Nulsen J., Dressler L., Bortolomeazzi M., Venkata S.K., Tourna A., Yakovleva A., Palmieri T., Ciccarelli F.D. The Network of Cancer Genes (NCG): a comprehensive catalogue of known and candidate cancer genes from cancer sequencing screens. Genome Biol. 2019;20:1. doi: 10.1186/s13059-018-1612-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jordan S., Krause J., Prager A., Mitrovic M., Jonjic S., Koszinowski U.H., Adler B. Virus progeny of murine cytomegalovirus bacterial artificial chromosome pSM3fr show reduced growth in salivary Glands due to a fixed mutation of MCK-2. J. Virol. 2011;85:10346–10353. doi: 10.1128/JVI.00545-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chaudhry M.Z., Casalegno-Garduno R., Sitnik K.M., Kasmapour B., Pulm A.K., Brizic I., Eiz-Vesper B., Moosmann A., Jonjic S., Mocarski E.S., Cicin-Sain L. Cytomegalovirus inhibition of extrinsic apoptosis determines fitness and resistance to cytotoxic CD8 T cells. Proc. Natl. Acad. Sci. USA. 2020;117:12961–12968. doi: 10.1073/pnas.1914667117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chaudhry M.Z., Messerle M., Čičin-Šain L. In: Yurochko A.D., editor. 2021. Construction of Human Cytomegalovirus Mutants with Markerless BAC Mutagenesis; pp. 133–158. (Human Cytomegaloviruses: Methods and Protocols). Springer, US. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single-cell RNA sequencing data are available: GEO Series accession number GSE179566 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE179566)
Scripts for analysis of scRNA sequencing data are provided here: https://github.com/SebastianJarosch/2021_Mueller_scRNA-OTR-vs-Tx