Abstract
A formidable challenge for global change biologists is to predict how natural populations will respond to the emergence of conditions not observed at present, termed novel climates. Popular approaches to predict population vulnerability are based on the expected degree of novelty relative to the amplitude of historical climate fluctuations experienced by a population. Here, we argue that predictions focused on amplitude may be inaccurate because they ignore the predictability of environmental fluctuations in driving patterns of evolution and responses to climate change. To address this disconnect, we review major findings of evolutionary theory demonstrating the conditions under which phenotypic plasticity is likely to evolve in natural populations, and how plasticity decreases population vulnerability to novel environments. We outline key criteria that experimental studies should aim for to effectively test theoretical predictions, while controlling for the degree of climate novelty. We show that such targeted tests of evolutionary theory are rare, with marine systems being overall underrepresented in this venture despite exhibiting unique opportunities to test theory. We conclude that with more robust experimental designs that manipulate both the amplitude and predictability of fluctuations, while controlling for the degree of novelty, we may better predict population vulnerability to climate change.
Keywords: fluctuating environments, phenotypic plasticity, climate change, marine biology, evolutionary theory
1. Introduction
Marine and terrestrial ecosystems are facing rapid rates of environmental change caused by anthropogenic carbon dioxide (CO2) emissions. The rate of increase in atmospheric CO2 over the past century is two-to-three orders of magnitude faster than that seen in the past 300 million years, suggesting that this challenge may be unprecedented for extant species [1–3]. The rapid rate of environmental change may cause conditions not found at present to arise by the end of the twenty-first century, referred to as ‘novel climates’, and some twentieth-century climates may disappear [4–7]. The fundamental challenge of global change biology is to predict how the emergence of such novel climates will affect natural populations.
The effects of climate change have largely been quantified by testing biological sensitivity to specific environmental stressors. Yet, it may be logistically infeasible to take this approach for every stressor and putatively sensitive species. Rather, recent effort has attempted to identify general environmental features that could increase the susceptibility of a population to climate change. For example, the ecological literature has used the degree of expected climate novelty during the next century as an index of a population's vulnerability to environmental change. The degree of climate novelty is based on how extreme the climate is relative to the historical amplitude of fluctuations and can be calculated in different ways—as an index of ‘local novelty’ relative to historical conditions at that site, or as an index of ‘global novelty’ relative to historical conditions in a regional or global pool of data [4]. Ultimately, inferences drawn from this approach suggest that populations inhabiting regions currently and/or historically exposed to reduced amplitude of climate variability (e.g. those in tropical and polar regions) are more vulnerable to climate change because the same magnitude of change in mean conditions is more likely to induce climate novelty [4,7].
However, climate novelty-based estimates of population vulnerability may be inaccurate because they do not take into account other aspects of fluctuating environments, particularly the predictability of fluctuations, which evolutionary theory predicts will largely shape the phenotypic variation present within natural populations. This phenotypic variation will ultimately dictate if, and how, populations persist as environmental conditions extend beyond those observed historically [8–10]. As predictability is not a component of environmental variability considered by climate novelty-based approaches, there is a disconnect between these estimates and theoretical expectations. To resolve this disconnect, and thereby better infer how populations will respond to the emergence of novel climates, theoretical predictions must be empirically validated.
The goal of this synthesis is to bridge the disconnect among the climate novelty literature, the theory of evolution in fluctuating environments and predictions of population vulnerability to climate change. We first ask: what are the characteristics of fluctuating environments that influence a population's likelihood of persistence under climate novelty? To address this, we review theoretical predictions regarding how different aspects of fluctuating environments influence the evolution of phenotypic plasticity in natural populations, and how plasticity may in turn influence persistence under novel conditions. Next, we suggest key criteria that experimental studies should strive to meet in order to effectively test theoretical predictions. We highlight the strengths and weaknesses of different case studies in the terrestrial and marine literature. We show that although marine ecosystems are underrepresented in robust empirical tests of theoretical predictions, there are unique aspects of these environments that can be leveraged for robust tests of the theory. For example, marine environments tend to be less variable than terrestrial systems and dominated by conditions that change over longer timescales, resulting in generally higher environmental autocorrelation, and therefore predictability [16,17]. Furthermore, coastal topography and shoreline orientation can lead to fine-scale spatial and temporal variation in climate change stressors, presenting environmental gradients over which to test theoretical predictions [18–20]. We conclude by highlighting key empirical gaps that should be addressed in order to improve predictions for determining population persistence under climate change.
2. The evolution of phenotypic plasticity in fluctuating environments
Heterogeneity in the environment maintains phenotypic and genetic variation in natural populations [21,22]. Depending upon how the environment varies over space and time, populations may evolve to produce a series of genetically differentiated individuals, each exhibiting phenotypes that remain fixed across the range of observed environmental conditions [11]. Alternatively, populations may consist of phenotypically plastic genotypes, which produce different phenotypic trait values in response to different environmental conditions [11,12]. Each of these strategies can be modelled with a reaction norm, which describes how the phenotype of a single genotype changes as a function of the environment [23,24]. Reaction norms can take any form, but for the sake of simplicity, we consider the case in which the reaction norm is linear across different states of the environment. Linear reaction norms contain a parameter dictating the intercept of the reaction norm, which corresponds to a genotype's trait value in the mean environmental state (oftentimes referred to as the ‘breeding value’). An additional parameter controls the reaction norm slope, which corresponds to how a genotype alters its trait value as a function of the environment, and thus represents the degree of phenotypic plasticity [24]. At the population level, the total phenotypic variation is characterized by the variation in reaction norm intercepts and slopes, as well as variation in the environment [25]. Given the heritable genetic variation in both reaction norm slope and intercept, selection acts on each character as the environment changes. Therefore, although selection within any environmental state acts directly on phenotypes expressed there, it is equally valid to view selection as acting simultaneously on variation in the slope and elevation of reaction norms [25]. Note that the reaction norm approach described here is mathematically equivalent to the character state approach, another commonly used model of phenotypic plasticity (see [24–26]). Quantitative models of reaction norms and fluctuating environments are provided in the electronic supplementary material.
A primary goal of reaction norm models is to determine the conditions under which phenotypic plasticity is advantageous. In such models, the fitness of a given genotype in each environment depends on the width of stabilizing selection in that environment and how far away the genotype's expressed phenotype is from the optimal phenotype. The optimal phenotype is usually assumed to map directly to the state of the fluctuating environment [7,14,15,25,27]. The temporal structure of the environment, and hence the optimal phenotype, is characterized by the variance around the mean state (fluctuation amplitude) and the autocorrelation between time points [17]. Predictability then is the correlation between an environmental cue sensed by an organism that influences the development of its phenotype and the optimal phenotype at the time of selection [11,13]. If the environmental variable providing the cue that elicits a phenotypic response is the same as that which causes selection on the phenotype, the correlations in the environment between time steps dictate the degree of environmental predictability [28]. However, in nature, the environment causing selection may be different from the environment causing the expression of the phenotype [13,29]. This consideration, and a more nuanced definition of predictability, is described in box 1. Ultimately, evolution in fluctuating environments proceeds such that the population's mean phenotype follows changes in the optimal phenotype over time, albeit with some lag, via the evolution of reaction norm intercept and/or slope [7,33].
Box 1. Defining versus measuring environmental predictability and the relevant timescale.
The numerous definitions of, and methods to quantify, environmental predictability have made it challenging to properly understand and translate the idea from theoretical to empirical studies. When considering phenotypic plasticity, a quantitative and biological definition of predictability is the correlation between an environmental cue sensed by an organism that influences the development of its phenotype and the optimal phenotype at the time of selection [11,13]. Thus, the relevant timescale for measuring predictability is the time between development and selection on the phenotype. Because it is challenging to directly quantify both the cue and agent of selection in situ, empiricists often estimate the autocorrelation in an environmental variable and assume that it provides the cue and the agent of selection (i.e. the variables determining the optimal phenotype) [30,31]. Correlations in the environment between the time of development of the phenotype and selection can then be used as a proxy for environmental predictability for a given study system. In the case in which the same environmental variable is expected to provide the cue and cause selection, the autocorrelation of the variable's time series across the relevant timescale can provide an empirical metric of predictability, which is consistent with how predictability is parameterized in theoretical models of the evolution of plasticity [12,28,32]. Note that it is the timescale between receiving the cue and selection acting on the new phenotype that matters, which may or may not have any correspondence to a species' generation time [29]. This means that plasticity in developmental traits can still evolve even when the environment undergoes white noise (i.e. fluctuates unpredictably) across generations, so long as the environmental cue at the time of development is correlated with the environment at the time of selection within a generation [13,29]. Thus, a lack of autocorrelation in a time series between generations does not preclude biological predictability in the study system.
It is critical to note that the evolution of plasticity in predictably fluctuating environments is contingent upon the ability of an organism to both sense the cue and adjust its phenotype fast enough in response to that cue before selection occurs [13]. These contingencies suggest how patterns of plasticity may still differ among species that are exposed to the same level of empirically inferred predictability. To illustrate, consider a scenario in which the presence of a chemical cue produced by a crab (predator) during larval shell development in a snail (prey) is strongly correlated with the likelihood of predation during the snail's juvenile stage. Given a predation event, juvenile snails with thicker shells exhibit increased survival to adulthood compared to juvenile snails with thinner shells. Snails that develop thinner shells, however, exhibit higher relative fitness in the absence of crab predation, presumably because resources were not diverted towards making a thicker shell. For a species of snail that can both sense the cue and increase its shell thickness before predation occurs, shell thickness plasticity is expected to evolve adaptively. However, for a species of snail that cannot accrete shell matrix fast enough to match the optimal shell thickness to survive a predator attack, adaptive plasticity will not evolve, even though the cue is predictable.
(a) . The influence of fluctuation amplitude and predictability on phenotypic plasticity
A consensus from theoretical models is that plasticity is favoured in environments that are both variable and predictable. Plasticity does not evolve or is lost in environments that are constant or vary unpredictably [11]. Predictably fluctuating environments provide reliable cues for plastic genotypes to effectively track changes in the optimal phenotype over time or space, thereby elevating fitness relative to non-plastic genotypes [7,11,12,32]. The evolved reaction norm slope (degree of plasticity) is proportional to the correlation between the environmental variable acting as the cue and the optimal phenotype during selection (a measure of predictability, see box 1; [12,28,32]). Reductions in plasticity, or fixed phenotypic strategies such as bet-hedging, are associated with unpredictably fluctuating environments, as these strategies will reduce the fitness costs associated with mismatches between the developed and optimal phenotype over multiple generations, and increase the long-term likelihood of population persistence [11,15].
For models in which predictability is not considered, where it is assumed that cues are perfect predictors of selection, the degree of phenotypic plasticity evolves to match the degree of variation in the environment [23,34]. Under this scenario of perfectly reliable cues, the optimal phenotype can be produced in each state of the environment, and larger amplitude fluctuations lead to increased plasticity [23,27,35]. However, cues are rarely, if ever, perfect predictors of future selection. Plasticity is therefore rarely perfect and is limited in natural populations by cue reliability [32]. The amplitude of environmental fluctuations then determines the strength of selection on plasticity, dictating the rate at which the population evolves to its expected equilibrium level of plasticity (which is set by the environmental predictability, [14]). We illustrate theoretical expectations regarding the interplay of amplitude and predictability, and experimental considerations when testing this theory, in box 2. Furthermore, in box 2, we discuss how to design experiments that disentangle the historical effects of climate amplitude and predictability on population responses to novel climates.
Box 2. Disentangling the effects of fluctuation amplitude, predictability and novelty in reaction norm evolution.
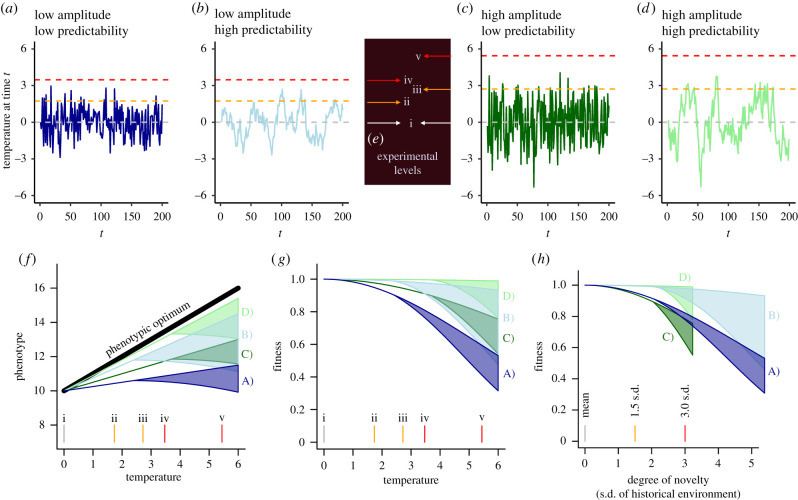
Empirically testing how organisms evolve in response to fluctuating environments, and how this evolution in turn affects their persistence under potential future climates, is a critical but challenging step in predicting population persistence under climate change. Ultimately, the goal of any experiment should be to establish how past selection has shaped the reaction norm(s) of a population, and subsequently query how populations that have evolved under different regimes of fluctuating selection respond to novel environments. This venture necessitates disentangling the relative contribution of fluctuation amplitude and predictability on reaction norm evolution. To that end, in table 1 we suggest criteria that empirical studies should meet in order to effectively accomplish this challenging goal. These criteria may be adopted within an experimental evolution setting or in a field-based approach where populations that are locally adapted to distinct regimes of fluctuating selection pressures are collected and compared in a laboratory-based experiment. To visualize these criteria in an experimental context and illustrate theoretical predictions, we consider a hypothetical example of four independent populations within a species of phytoplankton that have evolved under different histories of temperature fluctuations. In this hypothetical example, evolution is manipulated in the laboratory via experimental evolution, but the same design could be applied to populations collected from different fluctuating regimes in the wild (see literature review for examples). The temperature is perfectly correlated with the cue, which affects the development of phenotypes and is proportional to the optimal phenotype. Therefore, the autocorrelation of the temperature time series provides a measure of predictability (box 1). It is once again important to note that the specific timescale over which temperature fluctuates is only relevant within the context of the biology of the system, specifically the period between when the phenotype under selection develops and when selection on that phenotype occurs. In all populations, the mean temperature is the same, but populations experience either low or high amplitude and either low or high predictability, creating four scenarios (box 2, figure 1a–d, Criteria 1): (a) low amplitude–low predictability, (b) low amplitude–high predictability, (c) high amplitude–low predictability and (d) high amplitude–high predictability. If researchers instead compare different populations within a species that have different patterns of historical environmental fluctuations (such as those shown in (a–d)), the response of each population to climate change can be inferred from the experimental assays in (f–h). Note that climate novelty metrics would predict that populations (c) and (d) would be the least vulnerable to climate change (because they have the highest amplitude), while evolutionary theory predicts that populations (b) and (d) would evolve the most plasticity and therefore be the least vulnerable to climate change (because they have the highest predictability).
Table 1.
Experimental criteria for effectively testing theories of reaction norm evolution.
| Criterion 1 (Orthogonal manipulation and historical measurements): study populations are evolved in, or collected from, environments that vary orthogonally in the degree of both amplitude and predictability of environmental fluctuations, with similar means. |
| Criterion 2 (Assessment levels): experimental levels at which populations are assessed encompass a range of both observed and novel conditions with respect to the environments in which focal populations have evolved. The degree of novelty (based on the amplitude of historical fluctuations) is controlled for in the experimental assessment levels. |
| Criterion 3: the reaction norms for each population are quantified. |
| Criterion 4: fitness across the experimental levels is quantified. |
We propose that experiments seeking to disentangle the effects of amplitude and predictability, while controlling for the degree of novelty, should include five experimental levels as indicated in figure 1e (Criteria 2, and shown in figure 1f, g and h) for assessing reaction norms (Criteria 3) and fitness (Criteria 4). Figure 1f shows hypothetical reaction norms that evolved in each treatment, based on general theoretical predictions that the degree of plasticity scales with environmental predictability. Population reaction norms in novel environments are unknown, and possible values are indicated by the broadly shaded regions in figure 1f (see 4. Persistence in novel environments). The population average fitness or population growth rate (figure 1g) is calculated in this case by assuming no cost to plasticity and determined by (i) the distance between the average population phenotype and the optimal phenotype (solid black line in figure 1f) and (ii) the width of the stabilizing selection function (not shown, but assumed to be the same in all cases and narrow relative to the amplitude of fluctuations, see electronic supplementary material, R code). In this hypothetical example, at extreme temperatures fitness is high for populations B and D that evolved in a more predictable environment, as the evolved plasticity allows them to more closely track the optimal phenotype, but highest for population D because it experienced the strongest selection due to the larger fluctuations (figure 1g). When rescaled to the degree of novelty (standard deviations of historical environment in figure 1h), populations B and D have similarly high fitness per unit change.
Box 2 figure 1.
(a–d) Examples of time series with different amplitudes and predictabilities. The horizontal orange and red lines represent 1.5 and 3.0 standard deviations (σ) of the historical environment, respectively (e.g. degree of novelty). The timescale could be small (e.g. hours or days) or large (e.g. years), but should be relevant to the timescale between development of the phenotype and selection (see box 1). (e) Experimental levels for the assessment of reaction norms and fitness under future environments, which would allow for both absolute and relative degree of novelty comparisons. (f–d) Theoretical expectations for evolved reaction norms and patterns of fitness across temperature conditions that are informed by the time series data and experimental levels depicted in (a–d). (f) Hypothetical examples of reaction norms that evolve in each of the four time series in A–D, following the theoretical prediction that predictability is the primary determinant of reaction norm slope. The transparent shaded areas represent uncertainty in the value that the phenotype would have in novel conditions (greater than 2σ of the historical temperature). The black line shows the phenotypic optimum for different values of the environment. (g) Fitness based on stabilizing selection acting on each phenotype, assuming no cost to plasticity. (h) The same plot as (g), but the x-axis was rescaled to represent the degree of novelty (σ) of the historical environment, showing how populations that evolved in environments with similar predictability may have similar fitness as a function of novelty. Source code for the generation of this figure is provided in electronic supplementary material, file S1.
3. Empirical progress
Robust empirical validation is necessary to determine the validity, and generality, of the theory of evolution in fluctuating environments. This venture is by no means trivial, as either maintaining experimental systems with fluctuating conditions, or quantifying them in natural habitats, can be logistically challenging. Still, given the wide recognition of the potential importance of fluctuating selection on patterns of phenotypic variation, a range of studies have begun to test various aspects of the theory outlined above. These targeted studies have largely focused on disentangling the contribution of amplitude and predictability on reaction norm evolution and how changes in the predictability of environmental fluctuations affect phenotypic variation in natural populations. For example, Manenti et al. [36] showed that the predictability, rather than the amplitude, of temperature fluctuations shaped patterns of thermal tolerance and plasticity in the fruit fly, Drosophila simulans [36]. A suite of additional studies has directly tested how the autocorrelation of fluctuating environments influence patterns of phenotypic variation in species as diverse as springtails [37], bactiverous protozoans [38], nematodes [39], RNA viruses [40] and bacteria [41]. Targeted tests of the evolutionary theory appear to be largely confined to terrestrial systems. It is only within the last year that studies have conducted effective tests of theoretical predictions using marine populations [20,42–44]. The generality of theoretical predictions hinges upon more effectively including the range of systems exposed to patterns of fluctuating selection in empirical research. To better understand why there is a paucity of rigorous tests of theoretical predictions in marine environments, we conducted a literature review of experimental designs used by researchers in marine systems. The goal of the review was to identify aspects of experimental design that have been neglected, therefore limiting effective tests of evolutionary theory. Furthermore, we also assessed the extent to which studies have tested how evolution in fluctuating environments influences responses to novel conditions (those beyond the range of historic fluctuations), as this has been rarely tested experimentally with designs like those suggested in box 2.
(a) . Quantifying effective tests of evolutionary theory in marine systems
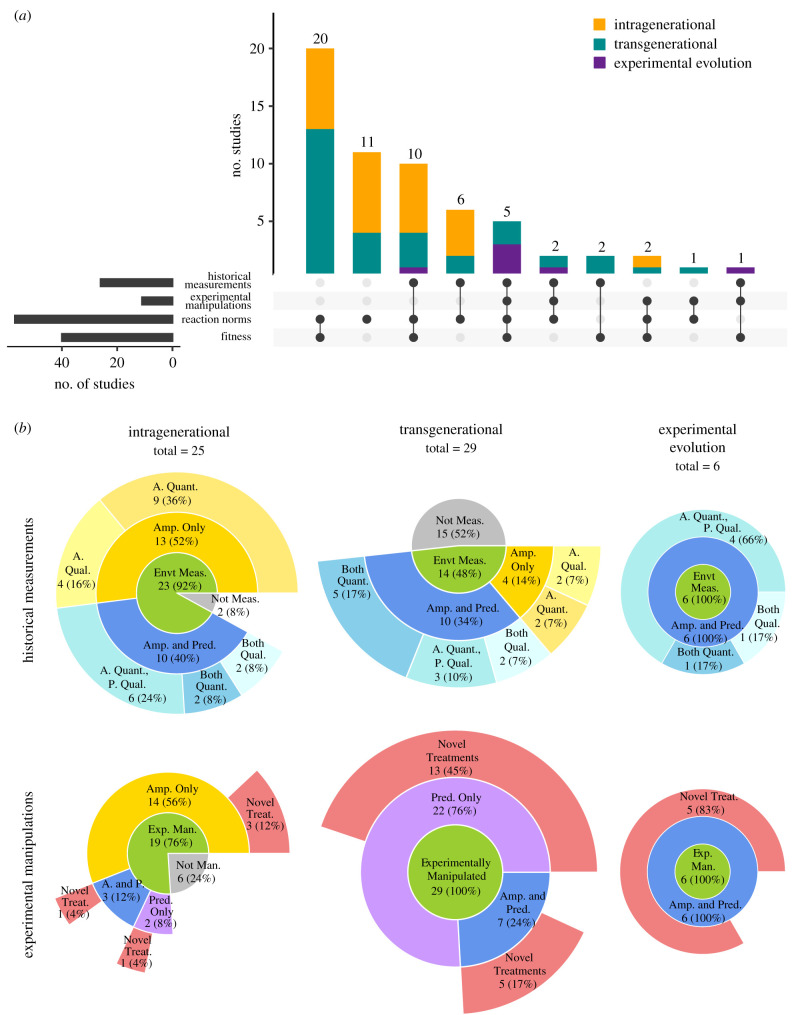
As published studies were not designed with box 2 in mind, and we did not find any studies that met all four criteria therein, we assessed the extent to which studies (i) quantified historical amplitude and/or predictability (historical measurements), (ii) manipulated predictability, amplitude and novelty (experimental manipulations), (iii) measured reaction norms and (iv) measured fitness (figure 1). ‘Historical measurements' refers to the conditions under which focal populations evolved and thus may refer to time-series oceanographic data collected in situ, or to the direct manipulation of the environment in which focal populations evolve (as in an experimental evolution study).
Figure 1.
(a) The number of empirical studies (intragenerational, transgenerational and experimental evolution approaches) included in our literature review, as well as which pertinent aspects of experimental design they incorporated. (b) Sunburst charts display the intragenerational (left column), transgenerational (centre column) and experimental evolution (right column) studies that were assessed to determine the proportion that measured, and in the case of experimental evolution studies, controlled, amplitude and/or predictability in the historical environment (top row) and that manipulated experimental levels of amplitude and/or predictability for comparative evaluations (bottom row). Studies that experimentally manipulated amplitude and/or predictability may have incorporated novel treatment(s). The number of studies within each chart category is displayed with its percentage of the total number of studies in parentheses. Abbreviations: Amp. = A = amplitude, Envt = environment, Man. = manipulated, Meas. = measured, Pred. = P = predictability, Qual. = qualified, Quant. = quantified, Treat. = treatment. (Online version in colour.)
Our literature search found 60 studies in marine systems that met our basis for inclusion: focal populations of marine organisms exposed to environmentally variable (in amplitude and/or predictability) conditions paired with measurements of trait plasticity and/or components of fitness (electronic supplementary material, file S2) (note that as our literature search was conducted before the publication of the studies described above [20,42–44], these were not included in our analysis). Of the studies included in our analysis, 25 were categorized as intragenerational (i.e. within a single generation), 29 as transgenerational (see electronic supplementary material, Methods for context on how these were categorized) and six as experimental evolution studies in single-celled organisms with short generation times. No studies compared populations that evolved under, or were collected from, environments that represented all four scenarios in an orthogonal design (low amplitude–low predictability, low amplitude–high predictability, high amplitude–low predictability and high amplitude–high predictability; box 2, figure 1a–d, Criteria 1) (note that some designs could have manipulated both amplitude and predictability, but still not have an orthogonal manipulation of amplitude and predictability because they were confounded in the design). While most studies reported reaction norms or measurements of fitness components, most of these studies did not measure and/or manipulate historical levels of both amplitude and predictability (figure 2a), and few chose experimental levels for evaluation that included novel climates. A total of 43 studies (72%) reported historic environmental measurements that could be used to infer the selective environment in which study populations evolved, though explicit quantification of the amplitude and predictability of the time-series data reported was rare (figure 2b). When an aspect of the environment was measured or manipulated for intragenerational studies, it was more commonly amplitude and not predictability (figure 2b left column, see electronic supplementary material, for an interpretation of predictability in transgenerational studies). In conclusion, while our analysis identified numerous studies assessing the influence of fluctuating environments on phenotypic variation in natural populations, explicit tests of how predictability and amplitude interact to shape patterns of plasticity and responses to novel climates are, as of yet, exceedingly rare in the marine literature.
Figure 2.
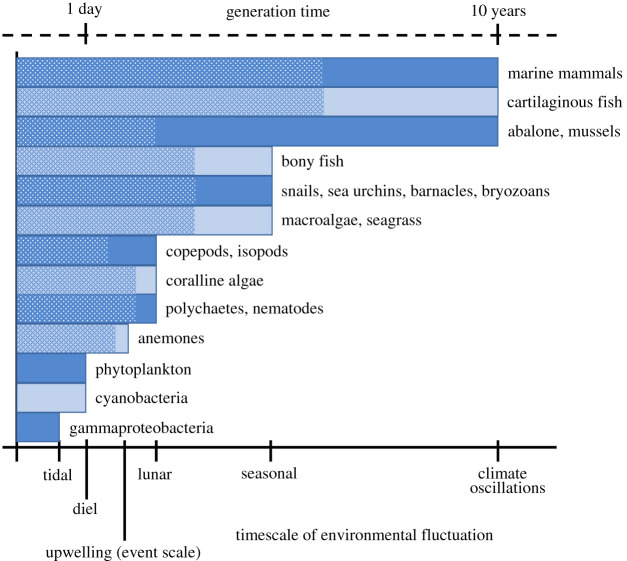
The frequency of environmental fluctuations that is relevant to selection depends on the time between when fitness-associated traits develop and when selection occurs. For many marine species, this period occurs during the early developmental and the larval stages (length of stippled portion), when major bottlenecks and selective mortality among genotypes is usually observed [45–47]. Plasticity in larval traits is likely to be advantageous when the environment of development (stippled portion) is correlated with the environment of selection, even if the environment fluctuates randomly between generations. (Online version in colour.)
Why there is a lack of marine studies that effectively meet the criteria in box 2 is an open question. One reason may be that research on novel climates in marine systems has focused on the amplitude of fluctuations [4], and empirical studies have followed suit. For example, Schaum & Collins [48] tested the relationship between variable environments and the evolution of plasticity by experimentally evolving clonal lineages of a single cell alga in low- and high-amplitude pCO2 conditions [48]. The work indeed demonstrated the evolution of plasticity in the high-amplitude treatment. Yet, because the design did not manipulate or measure predictability, the extent to which predictability affected their results cannot be inferred. Another reason is that there may be a perception that designing experiments that manipulate both predictability and amplitude in marine systems is prohibitively challenging. Indeed, most studies assessing marine population responses to climate change conditions have been conducted under static treatment conditions, despite recognition that such a design may limit ecological inference [19]. Deciding how to manipulate fluctuating environments is difficult because the appropriate frequency of fluctuations is dependent upon the biology of the study system (e.g. developmental time varies dramatically between micro- and macroscopic species), potentially confining research groups to focus on one or a few species and a single-selective pressure. However, once an experimental system is developed, the experimental design proposed in box 2 may consist of no more treatments than studies that assess responses to multiple environmental stressors in a fully factorial manner [49]. Recent experimental work has begun to demonstrate the feasibility of designing experiments that manipulate both the predictability and amplitude in marine systems [42,43]. Particularly noteworthy is the recent study by Leung et al. [42], who elegantly showed that reduced plasticity can evolve by empirically decreasing predictability. They subjected populations of a unicellular alga to randomly, but ecologically realistic, fluctuating salinities for approximately 500 generations while keeping the mean and variance in salinity constant. Lower levels of morphological plasticity (in traits associated with salinity tolerance) were observed in less predictable environments. As such experimental designs become more readily developed, we anticipate more targeted tests of evolutionary theory to emerge.
(b) . Considerations for future studies in marine systems
Despite the paucity of effective experimental tests of theory in marine systems, such habitats provide ample and unique opportunities to explore how fluctuating selection can affect population responses to novel environments. Specifically, numerous fluctuating selective pressures occurring at different temporal scales influence marine life histories (figure 2; [31,50]). On interannual timescales, the North Atlantic Oscillation, the El Niño Southern Oscillation and the North Pacific Gyre Oscillation can drive coupled changes in temperature, nutrients and salinity (among other parameters) [51–53]. In estuaries, seasonal changes in rainfall can drive large fluctuations in temperature, salinity, pH and dissolved oxygen [54,55]. On more rapid scales, tides drive shifts in ocean water levels and current velocities that ultimately cause changes in temperature and other selective agents at diurnal, semidiurnal and other frequencies [19]. Climate change will impact these, and other, marine environmental fluctuations in dynamic ways, while also causing shifts in mean conditions [19,56]. When events related to climate change (e.g. heat waves) are superimposed on natural environmental variability the amplitude of fluctuations may increase driving populations into novel conditions [57,58]. Finally, marine environments face unique climate change stressors to those that impact terrestrial systems. For example, two primary consequences of climate change—acidification and deoxygenation—are unique to aquatic environments and present additional selective agents to consider in empirical studies [56].
Studies testing evolutionary theory must be grounded in ecologically relevant environmental data that aim to sufficiently document and/or manipulate ‘what the organism sees’, which may not always be captured by what we can measure. It is therefore necessary to link pertinent life-history information (i.e. the time between cue detection and selection) with the predominant frequencies, amplitudes and predictability of fluctuating selection pressures experienced by focal populations (box 2; [31]). Thus, the relevant timescale(s) for predictability for a species could include timescales that range from very short (e.g. between development and selection within a generation) to very long (across generations) (figure 2). Approaches comparing replicated populations locally adapted to distinct regimes of fluctuating selection in a controlled laboratory setting may be particularly fruitful. This is indeed feasible in coastal marine systems, where topography and shoreline orientation lead to mesoscale (e.g. 10 km's) differences in waves, winds and tides that drive divergent patterns of environmental variability between habitats [18]. Identifying replicate populations that persist throughout such gradients would provide an opportunity to link patterns of environmental fluctuations with standing variation in reaction norms in a laboratory-based setting. For example, Bitter et al. [20] quantified differences in reaction norms between populations of a subtidal mussel and associated reductions in gene expression plasticity with less predictable fluctuations in seawater pH [20]. Such an approach relies on measures of the ‘historical environment’ in which focal populations likely evolved, and thus the existence of continuous environmental time-series data (which can be sparse in marine systems). Alternatively, studies may experimentally manipulate variable environments to generate treatments that vary orthogonally in fluctuation amplitude and predictability. Genetically diverse populations may then be evolved in these environments and subsequently assessed to link reaction norms and fitness to theoretical expectations, both within and outside the range of conditions in which populations were evolved.
4. Persistence in novel environments
As quantified in our literature search, how evolution in historically fluctuating environments will influence a population's response to the novel conditions expected under climate change remain largely unexplored. While we present an experimental design in box 2 that can give important insights into this difficult question, the results are unlikely to be as clean-cut as we show in figure 2f–h. Ultimately, persistence within novel environments will depend upon the ability of organisms to track environmental changes via existing adaptive plasticity in the short term and to adapt to new environments via natural selection on reaction norms in the longer term [59]. As highlighted above, theory predicts that the evolution of plasticity is influenced more strongly by the extent to which an environment fluctuates predictably, rather than how widely the environment fluctuates around the mean environmental state (amplitude). Thus, for determining the potential accuracy of climate novelty-based predictions of population vulnerability, the key issue to resolve is how reaction norms that have evolved under different histories of fluctuating environments influence current fitness and future evolution under novel environmental states. Ultimately, this is an empirical question that depends on the pattern of selection, the standing variation in reaction norms, and their underlying genetic basis and heritability. Nevertheless, theoretical models have begun to explore if, and how, plasticity may be expressed and affect persistence in novel environments.
The theory generally predicts that plasticity can increase population persistence in response to abrupt shifts in mean conditions [10,14,15,60]. Plasticity increases population persistence in novel climates when reaction norms are considered to be physiologically continuous, meaning that the same loci contribute to plasticity in both the novel and historical environment [60], and under the condition that the phenotypic optimum changes smoothly with the environment [61]. However, the advantage of plasticity when the mean environment shifts can still depend on whether the dynamics of environmental fluctuation themselves are changed: an increase in the amplitude or reduction in the predictability of environmental fluctuations can reduce the likelihood of population persistence because of plasticity-associated costs [9,14].
Another factor that affects whether plasticity will increase population persistence during abrupt shifts in mean conditions is the presence, and influence, of cryptic variation [14,15]. Cryptic genetic variation refers to standing genetic variation that does not contribute to the phenotypes expressed under contemporary conditions but takes on a functional role that modifies the phenotype following exposure to novel conditions [62]. A release of cryptic variation may drive a disconnect between genotype fitness pre- and post-environmental change if phenotypes in new environments are caused by different, and uncorrelated, loci to those that cause phenotypes in historical environments [15,24]. Furthermore, selection on cryptic variation in reaction norm slope can lead to the evolution of mean plasticity within a population, either increasing or decreasing fitness in the novel environment [14,15]. Ultimately, there is considerable uncertainty in how reaction norms may be expressed in response to novel environments, and this is an area in need of empirical data following the design proposed in box 2.
5. Conclusion
A fundamental goal of climate change biology is to infer how the sensitivity of populations to environmental change may vary as a function of the historic conditions under which the populations evolved. Estimates of population vulnerability to climate change based solely on the expected time to the emergence of climate novelty are likely inaccurate because they disregard the extent to which past evolution has shaped phenotypic plasticity. Experimental designs that disentangle the confounding effects of climate novelty and the historical amplitude and predictability of climate fluctuations, as we have recommended here, are necessary to make more robust predictions about population vulnerability. The experiments we propose can test the theoretical expectation that the predictability of fluctuations determines the degree of plasticity in natural populations and evaluate if that plasticity favours persistence in novel climates. Marine systems are underrepresented, yet potentially fruitful, in testing these theoretical predictions. The key components of theory that need empirical validation are determining the role of environmental predictability in shaping phenotypic variation in natural populations (as previously suggested in [30,45]), and how reaction norms respond to novel conditions. Addressing this knowledge gap will hinge upon developing a consensus on how predictability should, and can, be measured from an organismic perspective in natural settings. While several studies have begun to test theoretical predictions, these are largely confined to microbial systems and experimental evolution-based approaches. Extending the range of taxa studied will be critical for determining the extent to which general environmental features can be used to effectively predict population vulnerability to climate change.
Supplementary Material
Data accessibility
All data curated for the literature review are available as electronic supplementary files [63].
Authors' Contributions
This manuscript was initially conceptualized by M.C.B., S.C.B. and K.E.L., with inputs from J.M.W., H.G.D., S.C.D., C.D.K., L.M.K., K.J.N., E.B.R. and S.S. Funding for project support was obtained by K.E.L. Methods were devised and data were curated and analysed by J.M.W., H.G.D., C.D.K., L.M.K., K.J.N., E.B.R., S.S. and S.C.B. Data were visualized by K.E.L., J.M.W. and E.B.R. M.C.B. wrote the original draft of the manuscript, with input from all authors. All authors reviewed and edited the manuscript.
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Competing interests
The authors declare no competing interests.
Funding
This work was initiated as part of the NSF Research Coordinated Network for Evolution in Changing Seas and funded by NSF-OCE 1764316.
References
- 1.Caldeira K, Wickett ME. 2003Anthropogenic carbon and ocean pH. Nature 425, 365. ( 10.1038/425365a) [DOI] [PubMed] [Google Scholar]
- 2.Hoegh-Guldberg O, et al. 2007Coral reefs under rapid climate change and ocean acidification. Science 318, 1737-1742. ( 10.1126/science.1152509) [DOI] [PubMed] [Google Scholar]
- 3.Hönisch B, et al. 2012The geological record of ocean acidification. Science 335, 1058-1063. ( 10.1126/science.1208277) [DOI] [PubMed] [Google Scholar]
- 4.Williams JW, Jackson ST, Kutzbach JE. 2007Projected distributions of novel and disappearing climates by 2100 AD. Proc. Natl Acad. Sci. USA 104, 5738-5742. ( 10.1073/pnas.0606292104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams JW, Jackson ST. 2007Novel climates, no-analog communities, and ecological surprises. Front. Ecol. Environ. 5, 475-482. ( 10.1890/070037) [DOI] [Google Scholar]
- 6.Radeloff VC, et al. 2015The rise of novelty in ecosystems. Ecol. Appl. 25, 2051-2068. ( 10.1890/14-1781.1) [DOI] [PubMed] [Google Scholar]
- 7.Lotterhos KE, Láruson AJ, Jiang L-Q. In press.Novel and disappearing climates in the global surface ocean from 1800 to 2100. Sci. Rep. ( 10.1038/s41598-021-94872-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Botero CA, Weissing FJ, Wright J, Rubenstein DR. 2015Evolutionary tipping points in the capacity to adapt to environmental change. Proc. Natl Acad. Sci. USA 112, 184-189. ( 10.1073/pnas.1408589111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nadeau CP, Urban MC, Bridle JR. 2017Climates past, present, and yet-to-come shape climate change vulnerabilities. TREE 32, 786-800. ( 10.1016/j.tree.2017.07.012) [DOI] [PubMed] [Google Scholar]
- 10.Scheiner SM, Barfield M, Holt RD. 2020The genetics of phenotypic plasticity. XVII. Response to climate change. Evol. Appl. 13, 388-399. ( 10.1111/eva.12876) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moran NA. 1992The evolutionary maintenance of alternative phenotypes. Am. Nat. 139, 971-989. ( 10.1086/285369) [DOI] [Google Scholar]
- 12.Gavrilets S, Scheiner SM. 1993The genetics of phenotypic plasticity. V. Evolution of reaction norm shape. J. Evol. Biol. 6, 31-48. ( 10.1046/j.1420-9101.1993.6010031.x) [DOI] [Google Scholar]
- 13.Reed TE, Waples RS, Schindler DE, Hard JJ, Kinnison MT. 2010Phenotypic plasticity and population viability: the importance of environmental predictability. Proc. R. Soc. B 277, 3391-3400. ( 10.1098/rspb.2010.0771) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lande R. 2009Adaptation to an extraordinary environment by evolution of phenotypic plasticity and genetic assimilation. J. Evol. Biol. 22, 1435-1446. ( 10.1111/j.1420-9101.2009.01754.x) [DOI] [PubMed] [Google Scholar]
- 15.Ashander J, Chevin LM, Baskett ML. 2016Predicting evolutionary rescue via evolving plasticity in stochastic environments. Proc. R. Soc. B 283, 20161690. ( 10.1098/rspb.2016.1690) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steele JH, Brink KH, Scott BE. 2019Comparison of marine and terrestrial ecosystems: suggestions of an evolutionary perspective influenced by environmental variation. ICES J. Mar. Sci. 76, 50-59. ( 10.1093/icesjms/fsy149) [DOI] [Google Scholar]
- 17.Ruokolainen L, Lindén A, Kaitala V, Fowler MS. 2009Ecological and evolutionary dynamics under coloured environmental variation. TREE 24, 555-563. ( 10.1016/j.tree.2009.04.009) [DOI] [PubMed] [Google Scholar]
- 18.Booth JAT, McPhee-Shaw EE, Chua P, Kingsley E, Denny M, Phillips R, Bograd SJ, Zeidberg LD, Gilly WF. 2012Natural intrusions of hypoxic, low pH water into nearshore marine environments on the California coast. Cont. Shelf Res. 45, 108-115. ( 10.1016/j.csr.2012.06.009) [DOI] [Google Scholar]
- 19.Kroeker KJ, Bell LE, Donham EM, Hoshijima U, Lummis S, Toy JA, Willis-Norton E. 2020Ecological change in dynamic environments: accounting for temporal environmental variability in studies of ocean change biology. Glob. Change Biol. 26, 54-67. ( 10.1111/gcb.14868) [DOI] [PubMed] [Google Scholar]
- 20.Bitter MC, Kapsenberg L, Silliman K, Gattuso J-P, Pfister CA. 2021Magnitude and predictability of pH fluctuations shape plastic responses to ocean acidification. Am. Nat. 97, 486-501. ( 10.1086/712930) [DOI] [PubMed] [Google Scholar]
- 21.Lewontin RC, Cohen D. 1969On population growth in a randomly varying environment. Proc. Natl Acad. Sci. USA 62, 1056-1060. ( 10.1073/pnas.62.4.1056) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Felsenstein J. 1976The theoretical population genetics of variable selection and migration. Annu. Rev. Genet. 10, 253-280. ( 10.1146/annurev.ge.10.120176.001345) [DOI] [PubMed] [Google Scholar]
- 23.Via S, Lande R. 1985Genotype–environment interaction and the evolution of phenotypic plasticity. Evolution 9, 505-522. ( 10.1111/j.1558-5646.1985.tb00391.x.) [DOI] [PubMed] [Google Scholar]
- 24.Via S, Gomulkiewicz R, De Jong G, Scheiner SM, Schlichting CD, Van Tienderen PH. 1995Adaptive phenotypic plasticity: consensus and controversy. TREE 10, 212-217. ( 10.1016/S0169-5347(00)89061-8) [DOI] [PubMed] [Google Scholar]
- 25.de Jong G. 1995Phenotypic plasticity as a product of selection in a variable environment. Am. Nat. 145, 493-512. ( 10.1086/285752) [DOI] [PubMed] [Google Scholar]
- 26.van Tienderen PH, Koelewijn H. 1994Selection on reaction norms, genetic correlations and constraints. Genet. Res. 64, 115-125. ( 10.1017/S0016672300032729) [DOI] [PubMed] [Google Scholar]
- 27.Scheiner SM. 1993Genetics and evolution of phenotypic plasticity. Annu. Rev. Ecol. Syst. 24, 35-68. ( 10.1146/annurev.es.24.110193.000343) [DOI] [Google Scholar]
- 28.Chevin LM, Collins S, Lefèvre F. 2013Phenotypic plasticity and evolutionary demographic responses to climate change: taking theory out to the field. Funct. Ecol. 27, 967-979. ( 10.1111/j.1365-2435.2012.02043.x) [DOI] [Google Scholar]
- 29.Scheiner SM. 2013The genetics of phenotypic plasticity. XII. Temporal and spatial heterogeneity. Ecol. Evol. 3, 4596-4609. ( 10.1002/ece3.792) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burgess SC, Marshall DJ. 2011Temperature-induced maternal effects and environmental predictability. J. Exp. Biol. 214, 2329-2336. ( 10.1242/jeb.054718) [DOI] [PubMed] [Google Scholar]
- 31.Burgess SC, Marshall DJ. 2014Adaptive parental effects: the importance of estimating environmental predictability and offspring fitness appropriately. Oikos 123, 769-776. ( 10.1111/oik.01235) [DOI] [Google Scholar]
- 32.Tufto J. 2000The evolution of plasticity and nonplastic spatial and temporal adaptations in the presence of imperfect environmental cues. Am. Nat. 156, 121-130. ( 10.1086/303381) [DOI] [PubMed] [Google Scholar]
- 33.Tufto J. 2015Genetic evolution, plasticity, and bet-hedging as adaptive responses to temporally autocorrelated fluctuating selection: a quantitative genetic model. Evolution 69, 2034-2049. ( 10.1111/evo.12716) [DOI] [PubMed] [Google Scholar]
- 34.Lynch M, Gabriel W. 1987Environmental tolerance. Am. Nat. 129, 283-303. ( 10.1086/284635) [DOI] [Google Scholar]
- 35.Kassen R. 2002The experimental evolution of specialists, generalists, and the maintenance of diversity. J. Evol. Biol. 15, 173-190. ( 10.1046/j.1420-9101.2002.00377.x) [DOI] [Google Scholar]
- 36.Manenti T, Sørensen JG, Moghadam NN, Loeschcke V. 2014Predictability rather than amplitude of temperature fluctuations determines stress resistance in a natural population of Drosophila simulans. J. Evol. Biol. 27, 2113-2122. ( 10.1111/jeb.12463) [DOI] [PubMed] [Google Scholar]
- 37.Pike N, Tully T, Haccou P, Ferrière R. 2004The effect of autocorrelation in environmental variability on the persistence of populations: an experimental test. Proc. R. Soc. Lond. B 271, 2143-2148. ( 10.1098/rspb.2004.2834) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gonzalez A, Holt RD. 2002The inflationary effects of environmental fluctuations in source–sink systems. Proc. Natl Acad. Sci. USA 99, 14 872-7. ( 10.1073/pnas.232589299) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dey S, Proulx SR, Teotónio H. 2016Adaptation to temporally fluctuating environments by the evolution of maternal effects. PLoS Biol. 14, e1002388. ( 10.1371/journal.pbio.1002388) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wieczynski DJ, Turner PE, Vasseur DA. 2018Temporally autocorrelated environmental fluctuations inhibit the evolution of stress tolerance. Am. Nat. 191, 195-207. ( 10.1086/697200) [DOI] [PubMed] [Google Scholar]
- 41.Karve SM, Daniel S, Chavhan YD, Anand A, Kharola SS, Dey S. 2015Escherichia coli populations in unpredictably fluctuating environments evolve to face novel stresses through enhanced efflux activity. J. Evol. Biol. 28, 1131-1143. ( 10.1111/jeb.12640) [DOI] [PubMed] [Google Scholar]
- 42.Leung C, Rescan M, Grulois D, Chevin LM. 2020Reduced phenotypic plasticity evolves in less predictable environments. Ecol. Lett. 23, 1664-1672. ( 10.1111/ele.13598) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rescan M, Grulois D, Ortega-Aboud E, Chevin LM. 2020Phenotypic memory drives population growth and extinction risk in a noisy environment. Nat. Ecol. Evol. 4, 193-201. ( 10.1038/s41559-019-1089-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sasaki MC, Dam HG. 2020Genetic differentiation underlies seasonal variation in thermal tolerance, body size, and plasticity in a short-lived copepod. Ecol. Evol. 10, 12 200-12 210. ( 10.1002/ece3.6851) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Launey S, Hedgecock D. 2001High genetic load in the Pacific oyster Crassostrea gigas. Genetics 159, 255-265. ( 10.1093/genetics/159.1.255) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnson DW, Christie MR, Moye J. 2010Quantifying evolutionary potential of marine fish larvae: heritability, selection, and evolutionary constraints. Evolution 64, 2614-2628. ( 10.1111/j.1558-5646.2010.01027.x) [DOI] [PubMed] [Google Scholar]
- 47.Bitter MC, Kapsenberg L, Gattuso J-P, Pfister CA. 2019Standing genetic variation fuels rapid adaptation to ocean acidification. Nat. Commun. 10, 1-10. ( 10.1038/s41467-019-13767-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schaum CE, Collins S. 2014Plasticity predicts evolution in a marine alga. Proc. Biol. Sci. 281, 20141486. ( 10.1098/rspb.2014.1486) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boyd PW, et al. 2018Experimental strategies to assess the biological ramifications of multiple drivers of global ocean change—a review. Glob. Change Biol. 24, 2239-2261. ( 10.1111/gcb.14102) [DOI] [PubMed] [Google Scholar]
- 50.Barneche DR, Burgess SC, Marshall DJ. 2018Global environmental drivers of marine fish egg size. Glob. Ecol. Biogeogr. 27, 890-898. ( 10.1111/geb.12748) [DOI] [Google Scholar]
- 51.Lorenzo ED, et al. 2008North Pacific Gyre oscillation links ocean climate and ecosystem change. Geophys. Res. Lett. 35, 1-6. ( 10.1029/2007GL032838) [DOI] [Google Scholar]
- 52.Parnell PE, Miller EF, Cody CEL, Dayton PK, Carter ML, Stebbinsd TD. 2010The response of giant kelp (Macrocystis pyrifera) in southern California to low-frequency climate forcing. Limnol. Oceanogr. 55, 2686-2702. ( 10.4319/lo.2010.55.6.2686) [DOI] [Google Scholar]
- 53.Bell TW, Allen JG, Cavanaugh KC, Siegel DA. 2020Three decades of variability in California's giant kelp forests from the Landsat satellites. Remote Sens. Environ. 238, 110811. ( 10.1016/j.rse.2018.06.039) [DOI] [Google Scholar]
- 54.Melzner F, Thomsen J, Koeve W, Oschlies A, Gutowska MA, Bange HW, Hansen HP, Körtzinger A. 2013Future ocean acidification will be amplified by hypoxia in coastal habitats. Mar. Biol. 60, 1875-1888. ( 10.1007/s00227-012-1954-1) [DOI] [Google Scholar]
- 55.Waldbusser GG, Salisbury JE. 2014Ocean acidification in the coastal zone from an organism's perspective: multiple system parameters, frequency domains, and habitats. Annu. Rev. Mar. Sci. 6, 221-247. ( 10.1146/annurev-marine-121211-172238) [DOI] [PubMed] [Google Scholar]
- 56.Bindoff NL, et al. 2019Changing ocean, marine ecosystems, and dependent communities. In IPCC special report on the ocean and cryosphere in a changing climate (eds Pörtner H-O et al.). Cambridge, UK: Cambridge University Press. [Google Scholar]
- 57.Takeshita Y, et al. 2015Including high-frequency variability in coastal ocean acidification projections. Biogeosciences 12, 5853-5870. ( 10.5194/bg-12-5853-2015) [DOI] [Google Scholar]
- 58.Oliver ECJ, Perkins-Kirkpatrick SE, Holbrook NJ, Bindoff NL. 2018Anthropogenic and natural influences on record 2016 marine heat waves. Bull. Am. Meteorol. Soc. 99, S44-S48. ( 10.1175/BAMS-D-17-0093.1) [DOI] [Google Scholar]
- 59.Hoffmann AA, Sgrò CM. 2011Climate change and evolutionary adaptation. Nature 70, 479-485. ( 10.1038/nature09670) [DOI] [PubMed] [Google Scholar]
- 60.de Jong G. 2005Evolution of phenotypic plasticity: patterns of plasticity and the emergence of ecotypes. New Phytol. 166, 101-118. ( 10.1111/j.1469-8137.2005.01322.x) [DOI] [PubMed] [Google Scholar]
- 61.Chevin L-M, Hoffmann AA. 2017Evolution of phenotypic plasticity in extreme environments. Phil. Trans. R. Soc. B 372, 20160138. ( 10.1098/rstb.2016.0138) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Paaby AB, Rockman MV. 2014Cryptic genetic variation: evolution's hidden substrate. Nat. Rev. Genet. 15, 247-258. ( 10.1038/nrg3688) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bitter MC, et al. 2021Fluctuating selection and global change: a synthesis and review on disentangling the roles of climate amplitude, predictability and novelty. FigShare. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Bitter MC, et al. 2021Fluctuating selection and global change: a synthesis and review on disentangling the roles of climate amplitude, predictability and novelty. FigShare. [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
All data curated for the literature review are available as electronic supplementary files [63].