Abstract
Ancient DNA (aDNA) has played a major role in our understanding of the past. Important advances in the sequencing and analysis of aDNA from a range of organisms have enabled a detailed understanding of processes such as past demography, introgression, domestication, adaptation and speciation. However, to date and with the notable exception of microbiomes and sediments, most aDNA studies have focused on single taxa or taxonomic groups, making the study of changes at the community level challenging. This is rather surprising because current sequencing and analytical approaches allow us to obtain and analyse aDNA from multiple source materials. When combined, these data can enable the simultaneous study of multiple taxa through space and time, and could thus provide a more comprehensive understanding of ecosystem-wide changes. It is therefore timely to develop an integrative approach to aDNA studies by combining data from multiple taxa and substrates. In this review, we discuss the various applications, associated challenges and future prospects of such an approach.
Keywords: palaeogenomes, sedaDNA, integrative, ecosystem shifts, extinction
1. Introduction
The development of ancient DNA (aDNA) as a scientific tool can be divided into three phases. First came the realization that DNA could be recovered from ancient remains and thus offer a temporal dimension to genetic analyses that modern data alone cannot provide [1]. This was followed by a period when most studies were focused on recovering DNA from different taxa and placing them into a phylogenetic context. Technical advances during this period, most notably the development of the PCR method [2] and use of silica for DNA extractions [3], paved the way for studies on the genetic relationships between extinct species and their extant relatives (e.g. flightless ratites [4]). However, several of these early studies, such as those on Cretaceous remains [5], are today considered the result of contamination, and therefore erroneous (e.g. [6]).
The second phase was catalysed by a series of seminal studies that made use of population-level datasets of short mitochondrial DNA sequences to investigate within-species demographic histories (e.g. [7]) as well as the origin of domestic species (e.g. [8]). These studies revealed a general pattern of dynamic history during the Late Quaternary, often characterized by population replacements and losses of genetic diversity. During this phase, it was also demonstrated that short barcode sequences recovered from ancient sediments or faeces could be used to examine the composition of prehistoric plant and animal communities or the diet of ancient taxa [9,10].
The third phase was initiated by the emergence of new DNA sequencing technologies and their application to aDNA [11]. This enabled aDNA to mature into a tool useful for a broad spectrum of scientific disciplines. The development of high-throughput sequencing methods also enabled the emergence of robust studies of ancient pathogens [12] and their importance for human prehistory [13], microbiomes [14] as well as the high-resolution reconstruction of past ecological communities from sedimentary aDNA (e.g. [15–17]). The first publications of complete prehistoric human and Neanderthal genomes [18,19] opened the floodgates for studies using ancient genomes (palaeogenomics), especially to trace human gene flow across continents [20]. The past decade has also seen an increase in the use of palaeogenomics to study population change, gene flow and extinction dynamics in wild and domestic animals [21,22]. Overall, the recent analyses of large-scale palaeogenomic datasets have been highly successful in investigating species-specific population histories.
2. Using palaeogenomics to investigate single-species histories
Palaeogenomics has been used to investigate species' histories, including changes in population size and gene flow. Bayesian coalescent methods have been used to reconstruct past changes in female effective population size (Ne) from mitochondrial genomic data [23], whereas sequentially Markovian coalescent (SMC) methods have made demographic analyses from single ancient nuclear genomes routine (e.g. Neanderthals [24]; woolly mammoths [21]).
The increasing availability of dated genomes from modern and ancient human populations [20] and domesticated species (e.g. horses [22]; canids [25]) has allowed for the inference of ancestral relationships between populations using ordination methods, such as principal component analysis (PCA) or, more recently, factor analyses (FA) [26], the latter of which properly accounts for sample age and temporal drift.
The generation of ancient genomic data has also spurred the development of methods to detect admixture between closely related species, including when hybridizing species are extinct [27]. For example, the sequencing of the first Neanderthal genome indicated that non-African modern human genomes comprise approximately 2% Neanderthal DNA [19]. These methods are now routinely used in palaeogenomics studies and have also contributed to a recent surge in studies on hybridization in a wide variety of modern taxa including insects, plants, mammals, birds and fish, and indicate that ancient admixture between related populations and species was commonplace (reviewed in [28]).
Selection and domestication studies have also benefited from the inclusion of palaeogenomic data. The temporal dimension provided by aDNA can allow for the study of changes in allele frequencies ‘in real time’ [29]. For instance, palaeogenomic data from Early Neolithic and Bronze Age Eurasian humans enabled a deeper understanding of the genetic basis of lactase persistence [30]. Palaeogenomics is beginning to provide valuable contributions to the study of natural selection in extinct taxa and has been used to investigate genetic changes associated with adaptations to cold climates [23,31], predatory lifestyle, behaviour and morphology [32] or the roles of natural selection and genomic diversity in extinction [33]. Finally, the inclusion of palaeogenomic data has been also necessary for studies on domestic species in which wild or past domestic lineages are currently extinct, such as horses [34].
3. Sedimentary ancient DNA adds another dimension
aDNA recovered directly from lake, cave, permafrost, archaeological or other environmental sediments (sedaDNA) is a rapidly evolving tool that holds much promise. As sediments, and the aDNA incorporated within them, are often deposited gradually and continuously over time, they can be used to reconstruct past ecological communities at fine taxonomic and temporal resolution and provide local first and last appearance dates (FADs, LADs) for taxa independent of the completeness of the body fossil record (e.g. [35,36]). Similarly, the recovery of aDNA from associated unidentifiable bulk fossil fragments can supplement sedaDNA data extracted directly from sediment (e.g. [37]). Integration of these data can, therefore, provide a detailed record of community changes that occurred across times of arrival and extinction of keystone taxa, such as mammalian herbivores.
The first reported recovery of sedaDNA was the bacterial profiling of lake sediment [38], with the first evidence for plant and animal sedaDNA reported from caves and permafrost [9,39]. Subsequently, the majority of studies have used PCR-based DNA metabarcoding methods to amplify sedaDNA molecules of interest from individual broad taxonomic groups (e.g. plants or mammals [40–42]). Advanced methods that sequence entire sedaDNA molecules, and thereby allow for aDNA damage authentication (see also §5a), have only recently been applied. These methods include shotgun metagenomics, whereby any molecules in the sedaDNA mixture are randomly sequenced (e.g. [15,36,43–46]), and target enrichment, in which sedaDNA molecules of interest are selectively enriched prior to sequencing (e.g. barcode or mitochondrial loci [16,47–49]). Detailed descriptions of these methods applied to sedaDNA have been recently reviewed elsewhere [50,51].
The recovery, analysis and interpretation of sedaDNA poses significant challenges, in part due to the complex mixture of ancient ecosystem DNA present in a sediment sample. Nonetheless, progress is rapidly being made to address these issues, which we detail in §5a, and many valuable contributions from sedaDNA have already been made. For example, detailed plant community reconstructions now exist for sites from multiple regions (e.g. [17,52]) and interglacial periods [41], hominin and human sedaDNA has been recovered (e.g. [47,48,53]), LADs have been refined for extinct megafauna (e.g. [36]), and FADs have been established for taxa arriving in a variety of contexts, from newly deglaciated landscapes [15] to island invasions [35]. Several studies have integrated sedaDNA findings with other palaeoecological proxy data to provide additional validation and/or contextualization (e.g. [36,52]). However, multi-site comparative sedaDNA studies (e.g. [42,54]) are still rare.
With the application of shotgun metagenomics and target enrichment approaches, it is now possible to recover haplotypic and genomic information directly from sedaDNA [43–45,47–49], which enables the exploration of population-level changes and has the potential to detect the arrival or disappearance of alleles and lineages in a region, as recently showcased for Neanderthals from a cave in Spain [48]. This expansion of sedaDNA into environmental palaeogenomics, together with the integration of sedaDNA and traditional palaeogenomic data derived from body fossils [55], will open up new approaches to understanding past biodiversity changes that are inaccessible with other palaeoecological proxies.
4. Integrating data from humans, animals and sediments
Genetic studies on modern-day samples have successfully integrated genomic and/or epigenomic data from multiple unrelated taxa (the multi-taxon approach) to address a range of questions in evolutionary biology (e.g. [56,57]), such as inferring the distribution of pathogens linked to early human migrations [58] using comparative phylogeographic approaches.
Generating multi-taxon datasets in palaeogenetics has been limited by sparse fossil records, the degraded nature of aDNA, contamination with modern DNA, sequencing costs and computational resources [59,60]. Although palaeogenetic data from multiple taxa have been used to contrast demographic histories (e.g. [61,62]), recent genomic studies have inferred the genetic ancestries and histories of multiple mammalian taxa from a single Pleistocene cave sedaDNA sample [44,45]. Another study inferred a clear parallel between dog and human lineage diversification by overlaying their population histories [25]. To our knowledge, this is the first study that used a multi-taxon approach by quantitatively coanalysing palaeogenomes of two coeval and cospatial species and thus paves the way towards a multi-taxon approach in aDNA studies.
In spite of the numerous technological and computational challenges in palaeogenomics, the increasing number of ancient genomes from wider geographical and deeper time scales (e.g. [27]) will enable the genomic history of numerous species to be unravelled, whereas sedaDNA will allow for direct evidence of the timing and extent of associated past ecological changes. However, appropriate statistical frameworks to quantitatively coanalyse intra-taxon/inter-taxa genomic patterns across time and space are required to overcome the inherent heterogeneity in such datasets (i.e. species from different spatio-temporal contexts) that may bias data interpretation. For instance, integrating distributional, demographic and coalescent modelling (iDDC) with approximate Bayesian computation (ABC) has been proposed as a methodological transition to coanalyse species datasets under biologically informed hypotheses [63].
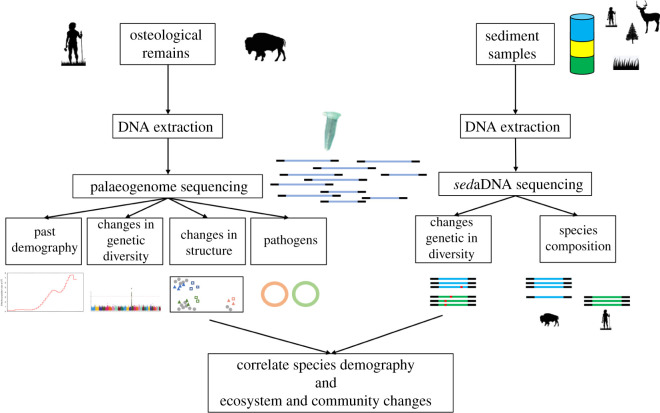
We here propose that recent advances in the generation and analysis of high-throughput sequencing data provide new opportunities to formally integrate multi-taxon knowledge from palaeogenomic and sedaDNA data into a cohesive picture of human–animal–environment interactions in the past (figure 1). In the following subsections, we discuss how aDNA data integration could provide new insights into the interaction between humans, wildlife and domesticated animals, and changes in their immediate environment. We also give an overview of the technical challenges and future prospects for the promising development of integrative approaches to build comprehensive and coherent datasets within a holistic aDNA evolutionary perspective.
Figure 1.
Workflow for the integration of multi-taxon palaeogenomes and sedaDNA. Silhouettes are from PhyloPic.org. (Online version in colour.)
(a) . Consequences of human arrival on wildlife
Thanks to their ability to adapt to a wide range of climatic and geographical conditions, humans have impacted ecosystems globally through hunting, domestication, sedentarization, and land and resource exploitation. Anatomically modern humans (AMHs) originated in Africa at least 200 000 years before present (ka BP), and expanded outside the continent within the past 100 ka BP [64], reaching North America by at least 16 ka BP and Polynesia around 1.0–0.7 ka BP [65]. Furthermore, changes in human technology that allowed for more efficient hunting or to target a specific species, such as the development of hunting tools used by Clovis hunters, are thought to have accelerated demographic declines in wild populations [66]. Similarly, the dispersal of Neolithic farmers from the Fertile Crescent across Europe and the introduction of their agricultural practises and domestic livestock from approximately 11 ka BP [67] followed by their sedentarization may have induced important changes in the environment [68].
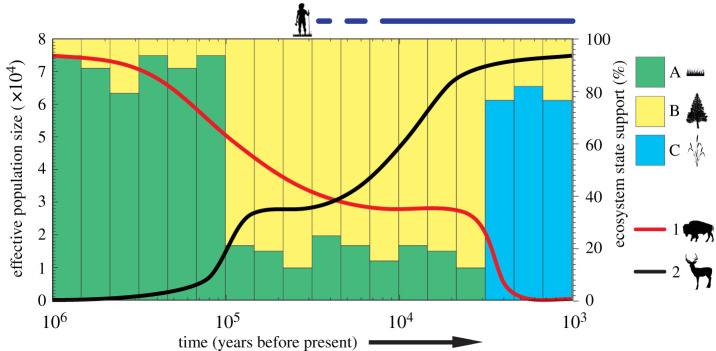
It has been suggested that human arrivals had significant impacts on previously unoccupied areas and were characterized by a number of extinctions as a result of overhunting and/or the introduction of non-native predators, particularly in island ecosystems [69,70]. In order to further elucidate the effects of human arrival on taxa, it is essential to refine the timing of first human presence in different regions. sedaDNA is a potentially valuable tool for detecting human FADs when macrofossil remains are sparse. Fine-scale information regarding human arrival and migrations could then be used to correlate the timing of human arrival with demographic declines in fauna inferred from palaeogenomic data. Multi-taxon demographic reconstructions using palaeogenomes can, for instance, be used to establish whether native taxa were impacted synchronously by human arrival (figure 2), and whether their extinction pattern is better explained by differences in life-history traits or body mass (e.g. megafauna [71]). Such information may also help elucidate whether wildlife populations that may have already been declining due to external factors (e.g. climate change) were more vulnerable to the arrival of human populations.
Figure 2.
Conceptual illustration of joint analysis of aDNA from multiple substrates. Red and black lines depict hypothetical changes in effective population size (Ne) inferred from the palaeogenomes of two distinct taxa (e.g. from a PSMC analysis). Filled bar colours represent three different ecosystem states (A–C) derived from sedaDNA. In this example, there are two distinct ecosystem state shifts. The Ne of taxon 1 is in decline prior to the ecosystem state shift from A to B. Its Ne remains stable after humans appear (dark blue line) but crashes during the shift from ecosystem state B to C. By contrast, the Ne of taxon 2 rapidly increases during the first ecosystem state shift (A–B), and again increases after the appearance of humans. The Ne of taxon 2 is unaffected by the second ecosystem state shift (B–C). Silhouettes are from PhyloPic.org. (Online version in colour.)
Similarly, sedaDNA could be used to test for ecosystem changes and examine the impact of human arrival on the abundance of another species in real time, in an approach similar to Gelabert et al. [45]. For example, early human populations may have competed with cave-dwelling species for shelter (e.g. [72]). Here, sedaDNA could be used to test whether humans and other cave fauna co-occur or are mutually exclusive.
(b) . Correlating human and animal demographies
An important question that remains to be addressed is whether there are tipping points of human population densities that could trigger significant declines in the demography of prey species. For instance, while human arrival in northeastern Siberia probably did not impact woolly rhinoceros demography, subsequent changes in human population density, which are currently unknown, may have had such an impact [23]. Thus, future work needs to focus not only on the effect of arrival, but also on correlating human and wildlife demographic trajectories thereafter.
Comparative analyses of palaeogenomic data could allow for this question to be tested by examining the impacts of human interference on species demography (e.g. using SMC; figure 2). Furthermore, larger multi-taxon datasets would enable testing of correlations among species, using different estimates of genetic diversity (e.g. FST, inbreeding), and provide evidence for anthropogenic impacts on wildlife. For example, palaeogenomic data indicate that human demographic events are correlated with dog population history and that the expansion of steppe pastoralists in Eurasia caused a complete replacement of European domesticated dog genetic diversity [25]. Such an approach could also be used to test to what extent local hunting pressures impacted the population dynamics of the extinct Baltic Harp seal [73].
Ideally, a combination of sedaDNA and palaeogenomes from fossil remains would allow examination of inter-taxon interactions across entire ecosystems. For instance, human dispersal into Australia and North America may have led to megafaunal extinctions, declines and range shifts which could be examined in time and space with these types of data in combination with modelling of interactions among taxa and changes in Ne through time (e.g. predator–prey models) [74]. Because megafaunal extinctions are often complex and multifactorial, a multi-taxon palaeogenomics approach will be especially valuable for assessment of the respective roles of human and non-human environmental changes in species extinction. In this regard, we stress that ethical considerations, engagement with indigenous communities, as well as careful interpretation of the narrative stemming from these discoveries, will be essential to avoid any potential stigmatisation of indigenous peoples [75].
(c) . Cascading effects of species extinctions
Species extinctions can have cascading effects on the physical and trophic structure of ecosystems, as well as the diversity and evolution of species. These effects could be examined by comparing demographic trajectories of several species simultaneously from multiple aDNA sources (e.g. sedaDNA, subfossil remains).
The extinction of ecologically important species can cause ecosystem state shifts. For example, extinct megaherbivores such as woolly mammoth and woolly rhinoceros are thought to have maintained a mosaic of open and shrub habitats characterized by high plant diversity [76], through grazing, soil fertilization and seed dispersal [77]. Consequently, the extinction of these large herbivores may have led to a shift towards dense and closed vegetation, a reduction in diversity and the extinction of species that had coevolved with these dispersers [78]. Moreover, large-bodied herbivores play an important role in maintaining connectivity between habitat patches through seed and nutrient dispersal [79]. sedaDNA could help identify changes in plant and invertebrate diversity, whereas genome-wide palaeogenetic data recovered from remains could allow for the testing of changes in connectivity (i.e. gene flow) between patches, thereby indicating whether such changes coincided with megaherbivore extinctions.
Single-species extinctions can also affect trophic interactions by triggering a number of secondary extinctions. For instance, the extinction of prey species can induce the disappearance of its predator [80]. Conversely, the extinction of an apex predator can lead to mesopredator release via reduced mortality and competition [81]. Furthermore, because apex predators regulate herbivore populations [82], the extinction of these predators could lead to changes in herbivore abundance, thereby altering trophic cascades and habitat structure and vegetation. A combination of sedaDNA and comparisons of population trajectories from subfossil remains would enable the testing of secondary extinctions and mesopredator release hypotheses (figure 2).
Another important consequence of species extinction is that it can trigger adaptive evolution in other species. For example, the extinction of carnivores could trigger a change in body size of herbivore prey species, similar to what has been proposed for the evolution of island herbivores in the absence of predators [83]. Conversely, the extinction of large prey species may have caused body size reduction in predators and scavengers [84]. Examining temporal changes in adaptive variation from scavenger and predator remains based on demographic reconstruction of its extinct prey could thus be used to test whether a reduction in body size has a genetic basis and whether it coincides with the extinction of their prey.
Finally, because many megafaunal species represented important food, building material, tool and artefactual resources for humans (e.g. [85]), thereby contributing to shaping human cultures [86], megafaunal extinction may have triggered migrations and perhaps even local extinction of human populations, as well as dietary shifts [87]. Decline or extinction of important prey species may even have contributed to cultural shifts towards new hunting strategies and subsequently domestication [88]. Using multiple aDNA sources, it should be possible to test whether the extinction of specific megafauna triggered changes in human demography, culture (e.g. changes in diet) and/or population turnovers. Moreover, examining extinction-driven cascading effects constitutes a ‘natural experiment’ that can be used to test whether particular ecosystems are under bottom-up or top-down control, a question that is still heavily debated in ecology.
(d) . Impact of domestic species on wild animals
As human populations expanded into new regions and impacted wild populations through hunting and habitat change, domesticated species that they brought with them also had a significant impact on these new ecosystems. For instance, since dingos were used to assist human hunting of small and large prey, their introduction in Australia probably contributed to the extinction of the thylacine and the Tasmanian devil on the mainland [89,90]. Similarly, the use of hunting dogs on other continents may have led to a higher hunting success and increased pressure on ungulate populations (e.g. ibex, gazelle [91]). Conversely, the replacement of hunter–gatherer populations by Neolithic farmers bringing domesticated taxa with them may have led to a relaxation of hunting pressure on wildlife. A combination of sedaDNA with demographic reconstructions for native wild species would thus enable testing of whether these declines or extinctions continued or stopped with the introduction of domesticated animals and farming cultures.
Another direct consequence of the introduction of domestic species to new ecosystems is introgression between domestic taxa and their wild counterparts both in modern (e.g. wild boar [92]) and ancient times (e.g. horses [22]; wolves [25]). Comparing genomes from a wild population and the domesticated species prior to and after the arrival of the latter could help resolve whether and when introgression occurred.
The introduction of domesticated species can also have indirect impacts on native fauna, with, for instance, the spread of both parasites and pathogenic microbes from domestic dogs to several wild canid species [93]. It is thus likely that similar transfers of diseases and pathogens occurred upon first contact between domesticated and wild animals. Consequently, comparing pathogens found in domesticated and related or unrelated wild taxa using a metagenomics approach could be used to test the hypothesis that the earliest domesticated arrivals were vectors of diseases into wild populations.
(e) . Human-driven landscape change
The dispersal and subsequent sedentarization of human populations had a severe impact on landscapes and ecosystems. These effects were most significant during the Neolithic transition, following the shift from hunter–gatherer to farming cultures (e.g. [94]). This shift entailed a steady decline and fragmentation of forested areas through land clearing as well as a profound alteration of aquatic ecosystems through irrigation and wetland draining [40], which likely had important effects on animal species [95], plant communities [94] and associated trophic networks.
Integrating aDNA data from sediments, bones, coprolites and other archaeological remains with data from more traditional methods (e.g. radiocarbon dating, pollen, macrofossils) could help infer the timing of human arrival and provide a comprehensive understanding of the effect of humans on the landscape. Moreover, because the Neolithic transition occurred at a time when the climate in Europe changed and sea levels were rising [96], this integrative approach could enable to disentangle the roles of human activities and climate change in the transformation of Holocene landscapes.
For instance, deforestation, grazing by domestic animals, and other human impacts in Iceland and Iberia during historical periods led to severe erosion, soil depletion and desertification [97,98]. An integrative approach targeting aDNA from plants, vertebrates and soil microorganisms could help unravel the cascading effects of deforestation and erosion on ecosystems. Furthermore, this approach could indicate whether changes in the genetic diversity of forest species coincided with an increase in human-induced landscape change or hunting. Similarly, combining aDNA from aquatic animal and microorganism remains could elucidate how human alterations of waterways due to irrigation and drainage affected aquatic plant and animal populations. Other prospects for aDNA are to investigate other types of human activities, such as the creation and development of man-made soils (i.e. anthrosols) that occur around the world, as well as to test whether the ‘elm decline’ approximately 6.3 ka BP [99] was caused by a fungal disease or human overexploitation.
5. Technical challenges and future prospects
(a) . Challenges inherent to palaeogenomics and sedaDNA research
aDNA research has rapidly advanced over the past 3 decades and challenges associated with DNA damage and modern contamination [59,60] have since been mitigated to a great extent. Yet, the presence of damaged exogenous DNA such as fragmented and deaminated DNA from bacteria and other non-target organisms may show false similarity to the reference genome used and become erroneously incorporated into the target sequence [59,60], thereby leading to incorrect inferences.
Significant challenges specific to sedaDNA research remain, whereby the sedaDNA composition of a sample is subject to intrinsic biases that need to be considered during analysis and integration with other data. For instance, because DNA preservation is reduced in warm and wet environments compared to dry and cold locations, the comparability of time scales and extents of detection across ecosystems may be limited. Generalizable and scalable approaches will, therefore, need to be developed to ensure robust harmonization of datasets using, for example, data quality metrics (e.g. [42]). Sampling from multiple comparable locations and using biological replicates and negative controls is essential to ensure proper characterization of a target area and to reduce taxonomic bias and the influence of contamination with modern DNA (e.g. [49,100]). Issues of taxonomic bias and sample heterogeneity are further confounded by a paucity of knowledge on sedaDNA taphonomy (the processes by which DNA is transported from an organism into an environmental archive [101]) and preservation, although experimental studies are beginning to address these unknowns (e.g. [102]). Post-depositional vertical movement of DNA via leaching [103], which could potentially lead to erroneous temporal interpretations, can be assessed by comparing sedaDNA data to other proxy and/or contextual information, although recent results from lake (e.g. [36,104]) and cave systems (e.g. [49]) do not find evidence of leaching. Furthermore, molecular biology protocols can be impacted by variations in substrate composition and the co-extraction of inhibitors, although new specialist methods, such as the cold-spin DNA extraction method [16], are beginning to mitigate these issues.
As the proportion of targeted and/or identified sedaDNA molecules may be very low, it is necessary for contamination to be monitored and sedaDNA assignments to be verified and authenticated, where possible (e.g. [46,105]). Although contamination with modern DNA may be excluded by examining aDNA damage patterns, sources of false positive taxonomic assignment could occur from other aDNA molecules that may be short, with low information content, and/or from genomically conserved regions that are shared across taxa. Although the impact of short aDNA molecules in single taxon palaeogenomics datasets, for example, can be characterized and mitigated (e.g. [27,106]), this is not yet true for multi-taxon sedaDNA mixtures and so new quality control methods will be required to determine and reduce false positive taxonomic assignments. An additional insidious source of taxonomic misassignment is incomplete reference databases, which are often sparsely populated and biased towards human-related taxa (exceptions include some databases used for metabarcoding; see [50] for examples). Although the phylogenetic intersection analysis (PIA) has recently been developed to mitigate taxonomic misassignments caused by incomplete reference databases [107], this approach could be developed further by, for example, incorporating geographical and ecological information to probabilistically determine and refine assignments. However, we caution that these data are also often unknown or incomplete for many taxa. Finally, bioinformatic pipelines, taxonomic classifications [42] and estimates of DNA damage [100] need to be standardized to avoid misrepresentation of species and their incorrect interpretation and association with related palaeogenomes at a shared spatio-temporal scale.
(b) . Issues with integrating data from varied sources
While comparative genomics frameworks have been proposed to obtain a better understanding of evolutionary processes, such as connectivity based on modern data [108], there are still several pitfalls when integrating aDNA from multiple sources. First, palaeogenomics or sedaDNA studies are often limited in sample size and the completeness of datasets, which may constrain models and reduce statistical power. Yet, integrative models can provide a generalized framework for meta-dimensional analysis [109], either using correlation or Bayesian-based models, and can be adapted to more generalistic assumptions for data integration [109]. Furthermore, unsupervised factor transformation methods or deep learning approaches have recently been widely applied for integrating heterogeneous data [110,111] and might be a good fit for particular complex scenarios.
Second, correlations among species demographies may not necessarily indicate a real impact of one species on another. This represents a significant challenge when testing for anthropogenic impacts on wildlife, for instance, because Ne is often smaller than census size (NC) [112]. Thus, a small Ne does not mean that NC did not reach the threshold necessary to induce a faunal decline. This challenge is analogous to the disconnect between Y chromosome and mtDNA Ne when comparing male and female demography. Using several time points to estimate relative changes in Ne could, however, circumvent this issue.
Finally, there may be a temporal disconnect between demographic trajectories among species. For instance, because Ne will be reduced more slowly than NC, there may be an observable time-lag in demographic reconstructions. One may thus erroneously exclude or infer a causal link between a change in the Ne of one species and a change in the abundance of another species. Such a disconnect can also be expected between changes in abundance in one species and a genome-wide or phenotypic response in another. For instance, while predator release may induce an increase in body size of the prey, this phenotypic change may take time, which is a function of Ne and generation time, and thus also result in a time-lag.
(c) . Future prospects
Future analytical developments will facilitate the integration of different forms of palaeogenetic data. Multi-taxon evolutionary dynamics may uncover molecular signatures of population differentiation as well as infer shared population histories. Recently, cross-species analyses have focused on how different ecologically connected species in the same ecosystems perceive and adapt to changes in the environment through time [108]. Whole-genome sequencing of selected individuals from different related populations across specific ecosystems have been used to infer micro- and macro-evolutionary connectivity patterns [108,113]. Extending such a framework to palaeogenomics and sedaDNA studies could add useful information on evolutionary dynamics across species and through time in a given region. Similarly, the analysis of inter-species dynamics using aDNA based on a multispecies coalescent model [111] is becoming more popular. For instance, a multi-taxon application of such coalescent models may allow for the joint inference of demographic and evolutionary parameters such as mutation rate, selection, and population expansions or contractions [114]. The integrated use of palaeogenomic and sedaDNA data would thus become more relevant to disciplines such as ecology, in order to infer whether particular ecosystems are under bottom-up or top-down control and could be incorporated in integrated population models or species distribution models [115,116]. The inclusion of temporal and spatially scaled integrated aDNA will help to improve these models, especially when assessing biodiversity changes or species population trends over time.
Supplementary Material
Acknowledgements
The authors are grateful to the staff at Tovetorp Research Station for their support during the writing of this manuscript. We thank two anonymous reviewers for their helpful comments on the text.
Contributor Information
Anders Götherström, Email: anders.gotherstrom@arklab.su.se.
Love Dalén, Email: love.dalen@nrm.se.
Peter D. Heintzman, Email: peter.d.heintzman@uit.no.
Data accessibility
This article does not contain any additional data.
Authors' contributions
L.D., A.G. and P.D.H. conceived the review; all authors drafted the manuscript; N.D., E.E. and P.D.H. drew the figures; N.D. and P.D.H. revised the manuscript. All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Competing interests
We declare we have no competing interests.
Funding
We acknowledge support from the Carl Tryggers Foundation (Grant CTS 19: 257 to N.D.), the Swedish Research Council (VR; 2017-04647 to L.D.; 2019-00849 to A.G.), FORMAS (2015-676, 2017-00704 and 2018-01640 to L.D.), the Bolin Centre for Climate Research (to E.L.), the Strategic Research Area (SFO) programme of the Swedish Research Council through Stockholm University (to E.M.-S.), the Stockholm University's Paired PhD Student Programme (to N.B.) and the European Union's Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement (no. 885088 to I.N.M.).
References
- 1.Higuchi R, Bowman B, Freiberger M, Ryder OA, Wilson AC. 1984DNA sequences from the quagga, an extinct member of the horse family. Nature 312, 282-284. ( 10.1038/312282a0) [DOI] [PubMed] [Google Scholar]
- 2.Mullis K, Faloona F, Scharf S, Saiki R, Horn G, Erlich H. 1986Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Cold Spring Harb. Symp. Quant. Biol. 51, 263-273. ( 10.1101/sqb.1986.051.01.032) [DOI] [PubMed] [Google Scholar]
- 3.Höss M, Pääbo S. 1993DNA extraction from Pleistocene bones by a silica-based purification method. Nucleic Acids Res. 21, 3913-3914. ( 10.1093/nar/21.16.3913) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cooper A, Mourer-Chauviré C, Chambers GK, Von Haeseler A, Wilson AC, Pääbo S. 1992Independent origins of New Zealand moas and kiwis. Proc. Natl Acad. Sci. USA 89, 8741-8744. ( 10.1073/pnas.89.18.8741) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woodward W, Weyand N, Bunnell M. 1994DNA sequence from Cretaceous period bone fragments. Science 266, 1229-1232. ( 10.1126/science.7973705) [DOI] [PubMed] [Google Scholar]
- 6.Zischler H, Höss M, Handt O, Von Haeseler A, Van Der Kuyl AC, Goudsmit J. 1995Detecting dinosaur DNA. Science 268, 1192-1193. author reply 1194. ( 10.1126/science.7605504) [DOI] [PubMed] [Google Scholar]
- 7.Shapiro B, et al. 2004Rise and fall of the Beringian steppe bison. Science 306, 1561-1565. ( 10.1126/science.1101074) [DOI] [PubMed] [Google Scholar]
- 8.Leonard JA. 2002Ancient DNA evidence for old world origin of new world dogs. Science 298, 1613-1616. ( 10.1126/science.1076980) [DOI] [PubMed] [Google Scholar]
- 9.Willerslev E, et al. 2003Diverse plant and animal genetic records from Holocene and Pleistocene sediments. Science 300, 791-795. ( 10.1126/science.1084114) [DOI] [PubMed] [Google Scholar]
- 10.Poinar HN, Hofreiter M, Spaulding WG, Martin PS, Stankiewicz BA, Bland H, Evershed RP, Possnert G, Pääbo S. 1998Molecular coproscopy: dung and diet of the extinct ground sloth Nothrotheriops shastensis. Science 281, 402-406. ( 10.1126/science.281.5375.402) [DOI] [PubMed] [Google Scholar]
- 11.Poinar HN, et al. 2006Metagenomics to paleogenomics: large-scale sequencing of mammoth DNA. Science 311, 392-394. ( 10.1126/science.1123360) [DOI] [PubMed] [Google Scholar]
- 12.Bos KI, et al. 2011A draft genome of Yersinia pestis from victims of the Black Death. Nature 478, 506-510. ( 10.1038/nature10549) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rasmussen S, et al. 2015Early divergent strains of Yersinia pestis in Eurasia 5,000 years ago. Cell 163, 571-582. ( 10.1016/j.cell.2015.10.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Warinner C, Speller C, Collins MJ, Lewis CM. 2015Ancient human microbiomes. J. Hum. Evol. 79, 125-136. ( 10.1016/j.jhevol.2014.10.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pedersen MW, et al. 2016Postglacial viability and colonization in North America's ice-free corridor. Nature 537, 45-49. ( 10.1038/nature19085) [DOI] [PubMed] [Google Scholar]
- 16.Murchie TJ, et al. 2020Optimizing extraction and targeted capture of ancient environmental DNA for reconstructing past environments using the PalaeoChip Arctic-1.0 bait-set. Quat. Res. 99, 305-328. ( 10.1017/qua.2020.59) [DOI] [Google Scholar]
- 17.Clarke CL, et al. 2019Persistence of arctic-alpine flora during 24,000 years of environmental change in the Polar Urals. Sci. Rep. 9, 19613. ( 10.1038/s41598-019-55989-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rasmussen M, et al. 2010Ancient human genome sequence of an extinct Palaeo-Eskimo. Nature 463, 757-762. ( 10.1038/nature08835) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Green RE, Krause J, Briggs AW, Maricic T. 2010A draft sequence of the Neandertal genome. Science 328, 710-722. ( 10.1126/science.1188021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skoglund P, Malmström H, Raghavan M, Storå J, Hall P, Willerslev E, Gilbert MTP, Götherström A, Jakobsson M. 2012Origins and genetic legacy of Neolithic farmers and hunter-gatherers in Europe. Science 336, 466-469. ( 10.1126/science.1216304) [DOI] [PubMed] [Google Scholar]
- 21.Palkopoulou E, et al. 2015Complete genomes reveal signatures of demographic and genetic declines in the woolly mammoth. Curr. Biol. 25, 1395-1400. ( 10.1016/j.cub.2015.04.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fages A, et al. 2019Tracking five millennia of horse management with extensive ancient genome time series. Cell 177, 1419-1435. ( 10.1016/j.cell.2019.03.049) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lord E, et al. 2020Pre-extinction demographic stability and genomic signatures of adaptation in the woolly rhinoceros. Curr. Biol. 30, 3871-3879. ( 10.1016/j.cub.2020.07.046) [DOI] [PubMed] [Google Scholar]
- 24.Prüfer K, et al. 2014The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505, 43-49. ( 10.1038/nature12886) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bergström A, et al. 2020Origins and genetic legacy of prehistoric dogs. Science 370, 557-564. ( 10.1126/science.aba9572) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.François O, Jay F. 2020Factor analysis of ancient population genomic samples. Nat. Commun. 11, 4661. ( 10.1038/s41467-020-18335-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Der Valk T, et al. 2021Million-year-old DNA sheds light on the genomic history of mammoths. Nature 591, 265-269. ( 10.1038/s41586-021-03224-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taylor SA, Larson EL. 2019Insights from genomes into the evolutionary importance and prevalence of hybridization in nature. Nat. Ecol. Evol. 3, 170-177. ( 10.1038/s41559-018-0777-y) [DOI] [PubMed] [Google Scholar]
- 29.Dehasque M, et al. 2020Inference of natural selection from ancient DNA. Evol. Lett. 4, 94-108. ( 10.1002/evl3.165) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burger J, et al. 2020Low prevalence of lactase persistence in Bronze Age Europe indicates ongoing strong selection over the last 3,000 years. Curr. Biol. 30, 4307-4315.e13. ( 10.1016/j.cub.2020.08.033) [DOI] [PubMed] [Google Scholar]
- 31.Lynch VJ, Bedoya-Reina OC, Ratan A, Sulak M, Drautz-Moses DI, Perry GH, Miller W, Schuster SC. 2015Elephantid genomes reveal the molecular bases of woolly mammoth adaptations to the Arctic. Cell Rep. 12, 217-228. ( 10.1016/j.celrep.2015.06.027) [DOI] [PubMed] [Google Scholar]
- 32.Barnett R, et al. 2020Genomic adaptations and evolutionary history of the extinct scimitar-toothed cat, Homotherium latidens. Curr. Biol. 30, 5018-5025. ( 10.1016/j.cub.2020.09.051) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murray GGR, et al. 2017Natural selection shaped the rise and fall of passenger pigeon genomic diversity. Science 358, 951-954. ( 10.1126/science.aao0960) [DOI] [PubMed] [Google Scholar]
- 34.Orlando L. 2020Ancient genomes reveal unexpected horse domestication and management dynamics. Bioessays 42, e1900164. ( 10.1002/bies.201900164) [DOI] [PubMed] [Google Scholar]
- 35.Ficetola GF, et al. 2018DNA from lake sediments reveals long-term ecosystem changes after a biological invasion. Sci. Adv. 4, eaar4292. ( 10.1126/sciadv.aar4292) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Graham RW, et al. 2016Timing and causes of mid-Holocene mammoth extinction on St. Paul Island, Alaska. Proc. Natl Acad. Sci. USA 113, 9310-9314. ( 10.1073/pnas.1604903113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seersholm FV, et al. 2020Rapid range shifts and megafaunal extinctions associated with late Pleistocene climate change. Nat. Commun. 11, 2770. ( 10.1038/s41467-020-16502-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coolen MJL, Overmann J. 1998Analysis of subfossil molecular remains of purple sulfur bacteria in a lake sediment. Appl. Environ. Microbiol. 64, 4513-4521. ( 10.1128/aem.64.11.4513-4521.1998) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hofreiter M, Mead JI, Martin P, Poinar HN. 2003Molecular caving. Curr. Biol. 13, R693-R695. ( 10.1016/j.cub.2003.08.039) [DOI] [PubMed] [Google Scholar]
- 40.Giguet-Covex C, et al. 2014Long livestock farming history and human landscape shaping revealed by lake sediment DNA. Nat. Commun. 5, 3211. ( 10.1038/ncomms4211) [DOI] [PubMed] [Google Scholar]
- 41.Crump SE, et al. 2021Ancient plant DNA reveals High Arctic greening during the Last Interglacial. Proc. Natl Acad. Sci. USA 118, e2019069118. ( 10.1073/pnas.2019069118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rijal DP, et al. 2021Sedimentary ancient DNA shows terrestrial plant richness continuously increased over the Holocene in northern Fennoscandia. Sci. Adv. 7, eabf9557. ( 10.1126/sciadv.abf9557) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lammers Y, Heintzman PD, Alsos IG. 2021Environmental palaeogenomic reconstruction of an Ice Age algal population. Commun. Biol. 4, 220. ( 10.1038/s42003-021-01710-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pedersen MW, et al. 2021Environmental genomics of Late Pleistocene black bears and giant short-faced bears. Curr. Biol. 31, 2728-2736. ( 10.1016/j.cub.2021.04.027) [DOI] [PubMed] [Google Scholar]
- 45.Gelabert P, et al. In press. Genome-scale sequencing and analysis of human, wolf and bison DNA from 25,000 year-old sediment. Curr. Biol. ( 10.1016/j.cub.2021.06.023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith O, Momber G, Bates R, Garwood P, Fitch S, Pallen M, Gaffney V, Allaby RG. 2015Archaeology. Sedimentary DNA from a submerged site reveals wheat in the British Isles 8000 years ago. Science 347, 998-1001. ( 10.1126/science.1261278) [DOI] [PubMed] [Google Scholar]
- 47.Slon V, et al. 2017Neandertal and Denisovan DNA from Pleistocene sediments. Science 356, 605-608. ( 10.1126/science.aam9695) [DOI] [PubMed] [Google Scholar]
- 48.Vernot B, et al. 2021Unearthing Neanderthal population history using nuclear and mitochondrial DNA from cave sediments. Science 372, eabf1667. ( 10.1126/science.abf1667) [DOI] [PubMed] [Google Scholar]
- 49.Zavala EI, et al. 2021Pleistocene sediment DNA reveals hominin and faunal turnovers at Denisova Cave. Nature 595, 399-403. ( 10.1038/s41586-021-03675-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Capo E, et al. 2021Lake sedimentary DNA research on past terrestrial and aquatic biodiversity: overview and recommendations. Quaternary 4, 6. ( 10.3390/quat4010006) [DOI] [Google Scholar]
- 51.Edwards ME. 2020The maturing relationship between Quaternary paleoecology and ancient sedimentary DNA. Quat. Res. 96, 39-47. ( 10.1017/qua.2020.52) [DOI] [Google Scholar]
- 52.Crump SE, Miller GH, Power M, Sepúlveda J, Dildar N, Coghlan M, Bunce M. 2019Arctic shrub colonization lagged peak postglacial warmth: molecular evidence in lake sediment from Arctic Canada. Glob. Change Biol. 25, 4244-4256. ( 10.1111/gcb.14836) [DOI] [PubMed] [Google Scholar]
- 53.Braadbaart F, et al. 2020Heating histories and taphonomy of ancient fireplaces: a multi-proxy case study from the Upper Palaeolithic sequence of Abri Pataud (Les Eyzies-de-Tayac, France). J. Archaeol. Sci. Rep. 33, 102468. ( 10.1016/j.jasrep.2020.102468) [DOI] [Google Scholar]
- 54.Willerslev E, et al. 2014Fifty thousand years of Arctic vegetation and megafaunal diet. Nature 506, 47-51. ( 10.1038/nature12921) [DOI] [PubMed] [Google Scholar]
- 55.Mitchell KJ, Rawlence NJ. 2021Examining natural history through the lens of palaeogenomics. Trends Ecol. Evol. 36, 258-267. ( 10.1016/j.tree.2020.10.005) [DOI] [PubMed] [Google Scholar]
- 56.Thomas JW, et al. 2003Comparative analyses of multi-species sequences from targeted genomic regions. Nature 424, 788-793. ( 10.1038/nature01858) [DOI] [PubMed] [Google Scholar]
- 57.Rincon-Sandoval M, Betancur-R R, Maldonado-Ocampo JA. 2019Comparative phylogeography of trans-Andean freshwater fishes based on genome-wide nuclear and mitochondrial markers. Mol. Ecol. 28, 1096-1115. ( 10.1111/mec.15036) [DOI] [PubMed] [Google Scholar]
- 58.Monot M, et al. 2009Comparative genomic and phylogeographic analysis of Mycobacterium leprae. Nat. Genet. 41, 1282-1289. ( 10.1038/ng.477) [DOI] [PubMed] [Google Scholar]
- 59.Prüfer K, Stenzel U, Hofreiter M, Pääbo S, Kelso J, Green RE. 2010Computational challenges in the analysis of ancient DNA. Genome Biol. 11, R47. ( 10.1186/gb-2010-11-5-r47) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Peyrégne S, Prüfer K. 2020Present-day DNA contamination in ancient DNA datasets. Bioessays. 42, 2000081. ( 10.1002/bies.202000081) [DOI] [PubMed] [Google Scholar]
- 61.Lorenzen ED, et al. 2011Species-specific responses of Late Quaternary megafauna to climate and humans. Nature 479, 359-364. ( 10.1038/nature10574) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stiller M, et al. 2010Withering away–25,000 years of genetic decline preceded cave bear extinction. Mol. Biol. Evol. 27, 975-978. ( 10.1093/molbev/msq083) [DOI] [PubMed] [Google Scholar]
- 63.Papadopoulou A, Lacey Knowles L. 2016Toward a paradigm shift in comparative phylogeography driven by trait-based hypotheses. Proc. Natl Acad. Sci. USA 113, 8018-8024. ( 10.1073/pnas.1601069113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nielsen R, Akey JM, Jakobsson M, Pritchard JK, Tishkoff S, Willerslev E. 2017Tracing the peopling of the world through genomics. Nature 541, 302-310. ( 10.1038/nature21347) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wilmshurst JM, Anderson AJ, Higham TFG, Worthy TH. 2008Dating the late prehistoric dispersal of Polynesians to New Zealand using the commensal Pacific rat. Proc. Natl Acad. Sci. USA 105, 7676-7680. ( 10.1073/pnas.0801507105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wroe S, Field J, Fullagar R, Jermin LS. 2004Megafaunal extinction in the late Quaternary and the global overkill hypothesis. Alcheringa Austral. J. Palaeontol. 28, 291-331. ( 10.1080/03115510408619286) [DOI] [Google Scholar]
- 67.Ammerman AJ, Cavalli-Sforza LL. 1984The Neolithic transition and genetics of populations in Europe. Princeton, NJ: Princeton University Press. ( 10.1515/9781400853113) [DOI] [Google Scholar]
- 68.Asouti E, Fuller DQ. 2013A contextual approach to the emergence of agriculture in Southwest Asia: reconstructing early Neolithic plant-food production. Curr. Anthropol. 54, 299-345. ( 10.1086/670679) [DOI] [Google Scholar]
- 69.Saltré F, et al. 2016Climate change not to blame for late Quaternary megafauna extinctions in Australia. Nat. Commun. 7, 10511. ( 10.1038/ncomms10511) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Collins CJ, et al. 2014Extinction and recolonization of coastal megafauna following human arrival in New Zealand. Proc. R. Soc. B 281, 20140097. ( 10.1098/rspb.2014.0097) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Johnson CN, Isaac JL. 2009Body mass and extinction risk in Australian marsupials: the ‘Critical Weight Range’ revisited. Austral. Ecol. 34, 35-40. ( 10.1111/j.1442-9993.2008.01878.x) [DOI] [Google Scholar]
- 72.Gretzinger J, et al. 2019Large-scale mitogenomic analysis of the phylogeography of the Late Pleistocene cave bear. Sci. Rep. 9, 10700. ( 10.1038/s41598-019-47073-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Glykou A, Lõugas L, Piličiauskienė G, Schmölcke U, Eriksson G, Lidén K. 2021Reconstructing the ecological history of the extinct harp seal population of the Baltic Sea. Quat. Sci. Rev. 251, 106701. ( 10.1016/j.quascirev.2020.106701) [DOI] [Google Scholar]
- 74.Aoki K. 2020A three-population wave-of-advance model for the European early Neolithic. PLoS ONE 15, e0233184. ( 10.1371/journal.pone.0233184) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Handsley-Davis M, Kowal E, Russell L, Weyrich LS. 2021Researchers using environmental DNA must engage ethically with Indigenous communities. Nat. Ecol. Evol. 5, 146-148. ( 10.1038/s41559-020-01351-6) [DOI] [PubMed] [Google Scholar]
- 76.Johnson CN. 2009Ecological consequences of Late Quaternary extinctions of megafauna. Proc. R. Soc. B 276, 2509-2519. ( 10.1098/rspb.2008.1921) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Malhi Y, Doughty CE, Galetti M, Smith FA, Svenning J-C, Terborgh JW. 2016Megafauna and ecosystem function from the Pleistocene to the Anthropocene. Proc. Natl Acad. Sci. USA 113, 838-846. ( 10.1073/pnas.1502540113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Anderson SH, Kelly D, Ladley JJ, Molloy S, Terry J. 2011Cascading effects of bird functional extinction reduce pollination and plant density. Science 331, 1068-1071. ( 10.1126/science.1199092) [DOI] [PubMed] [Google Scholar]
- 79.Berti E, Svenning J. 2020Megafauna extinctions have reduced biotic connectivity worldwide. Glob. Ecol. Biogeogr. 29, 2131-2142. ( 10.1111/geb.13182) [DOI] [Google Scholar]
- 80.Leonard JA, Vilà C, Fox-Dobbs K, Koch PL, Wayne RK, Van Valkenburgh B. 2007Megafaunal extinctions and the disappearance of a specialized wolf ecomorph. Curr. Biol. 17, 1146-1150. ( 10.1016/j.cub.2007.05.072) [DOI] [PubMed] [Google Scholar]
- 81.Ritchie EG, Johnson CN. 2009Predator interactions, mesopredator release and biodiversity conservation. Ecol. Lett. 12, 982-998. ( 10.1111/j.1461-0248.2009.01347.x) [DOI] [PubMed] [Google Scholar]
- 82.Estes JA, et al. 2011Trophic downgrading of planet Earth. Science 333, 301-306. ( 10.1126/science.1205106) [DOI] [PubMed] [Google Scholar]
- 83.Damuth J. 1993Cope's rule, the island rule and the scaling of mammalian population density. Nature 365, 748-750. ( 10.1038/365748a0) [DOI] [PubMed] [Google Scholar]
- 84.Galetti M, et al. 2018Ecological and evolutionary legacy of megafauna extinctions. Biol. Rev. Camb. Philos. Soc. 93, 845-862. ( 10.1111/brv.12374) [DOI] [PubMed] [Google Scholar]
- 85.Pitulko VV, Pavlova EY, Nikolskiy PA. 2015Mammoth ivory technologies in the Upper Palaeolithic: a case study based on the materials from Yana RHS, Northern Yana-Indighirka lowland, Arctic Siberia. World Archaeol. 47, 333-389. ( 10.1080/00438243.2015.1030508) [DOI] [Google Scholar]
- 86.Pryor AJE, Beresford-Jones DG, Dudin AE, Ikonnikova EM, Hoffecker JF, Gamble C. 2020The chronology and function of a new circular mammoth-bone structure at Kostenki 11. Antiquity 94, 323-341. ( 10.15184/aqy.2020.7) [DOI] [Google Scholar]
- 87.Nikolskiy P, Pitulko V. 2013Evidence from the Yana Palaeolithic site, Arctic Siberia, yields clues to the riddle of mammoth hunting. J. Archaeol. Sci. 40, 4189-4197. ( 10.1016/j.jas.2013.05.020) [DOI] [Google Scholar]
- 88.Larson G, Fuller DQ. 2014The evolution of animal domestication Ann. Rev. Ecol. Evol. Syst. 45, 115-136. ( 10.1146/annurev-ecolsys-110512-135813) [DOI] [Google Scholar]
- 89.Johnson CN, Wroe S. 2003Causes of extinction of vertebrates during the Holocene of mainland Australia: arrival of the dingo, or human impact? The Holocene 13, 941-948. ( 10.1191/0959683603hl682fa) [DOI] [Google Scholar]
- 90.Koungoulos L, Fillios M. 2020Hunting dogs down under? On the Aboriginal use of tame dingoes in dietary game acquisition and its relevance to Australian prehistory. J. Anthropol. Archaeol. 58, 101146. ( 10.1016/j.jaa.2020.101146) [DOI] [Google Scholar]
- 91.Guagnin M, Perri AR, Petraglia MD. 2018Pre-Neolithic evidence for dog-assisted hunting strategies in Arabia. J. Anthropol. Archaeol. 49, 225-236. ( 10.1016/j.jaa.2017.10.003) [DOI] [Google Scholar]
- 92.Goedbloed DJ, et al. 2013Genome-wide single nucleotide polymorphism analysis reveals recent genetic introgression from domestic pigs into Northwest European wild boar populations. Mol. Ecol. 22, 856-866. ( 10.1111/j.1365-294X.2012.05670.x) [DOI] [PubMed] [Google Scholar]
- 93.De Almeida Curi NH, Araújo AS, Campos FS, Lobato ZIP, Gennari SM, Marvulo MFV, Silva JCR, Talamoni SA. 2010Wild canids, domestic dogs and their pathogens in Southeast Brazil: disease threats for canid conservation. Biodivers. Conserv. 19, 3513-3524. ( 10.1007/s10531-010-9911-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Woodbridge J, Fyfe RM, Roberts N, Downey S, Edinborough K, Shennan S. 2014The impact of the Neolithic agricultural transition in Britain: a comparison of pollen-based land-cover and archaeological 14C date-inferred population change. J. Archaeol. Sci. 51, 216-224. ( 10.1016/j.jas.2012.10.025) [DOI] [Google Scholar]
- 95.Ersmark E, et al. 2019Genetic turnovers and northern survival during the last glacial maximum in European brown bears. Ecol. Evol. 9, 5891-5905. ( 10.1002/ece3.5172) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Turney CSM, Brown H. 2007Catastrophic early Holocene sea level rise, human migration and the Neolithic transition in Europe. Quat. Sci. Rev. 26, 2036-2041. ( 10.1016/j.quascirev.2007.07.003) [DOI] [Google Scholar]
- 97.Church MJ, Dugmore AJ, Mairs KA, Millard AR, Cook GT, Sveinbjarnardóttir G, Ascough PA, Roucoux KH. 2007Charcoal production during the Norse and early medieval periods in Eyjafjallahreppur, Southern Iceland. Radiocarbon 49, 659-672. ( 10.1017/s0033822200042557) [DOI] [Google Scholar]
- 98.López-Merino L, Cortizas AM, Reher GS, López-Sáez JA, Mighall TM, Bindler R. 2014Reconstructing the impact of human activities in a NW Iberian Roman mining landscape for the last 2500 years. J. Archaeol. Sci. 50, 208-218. ( 10.1016/j.jas.2014.07.016) [DOI] [Google Scholar]
- 99.Parker AG, Goudie AS, Anderson DE, Robinson MA, Bonsall C. 2002A review of the mid-Holocene elm decline in the British Isles. Prog. Phys. Geogr. Earth Environ. 26, 1-45. ( 10.1191/0309133302pp323ra) [DOI] [Google Scholar]
- 100.Ruppert KM, Kline RJ, Rahman MS. 2019Past, present, and future perspectives of environmental DNA (eDNA) metabarcoding: a systematic review in methods, monitoring, and applications of global eDNA. Glob. Ecol. Conserv. 17, e00547. ( 10.1016/j.gecco.2019.e00547) [DOI] [Google Scholar]
- 101.Giguet-Covex C, et al. 2019New insights on lake sediment DNA from the catchment: importance of taphonomic and analytical issues on the record quality. Sci. Rep. 9, 14676. ( 10.1038/s41598-019-50339-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kanbar HJ, Olajos F, Englund G, Holmboe M. 2020Geochemical identification of potential DNA-hotspots and DNA-infrared fingerprints in lake sediments. Appl. Geochem. 122, 104728. ( 10.1016/j.apgeochem.2020.104728) [DOI] [Google Scholar]
- 103.Haile J, et al. 2007Ancient DNA chronology within sediment deposits: are paleobiological reconstructions possible and is DNA leaching a factor? Mol. Biol. Evol. 24, 982-989. ( 10.1093/molbev/msm016) [DOI] [PubMed] [Google Scholar]
- 104.Sjögren P, Edwards ME, Gielly L, Langdon CT, Croudace IW, Merkel MKF, Fonville T, Alsos IG. 2017Lake sedimentary DNA accurately records 20 Century introductions of exotic conifers in Scotland. New Phytol. 213, 929-941. ( 10.1111/nph.14199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Weiß CL, Dannemann M, Prüfer K, Burbano HA. 2015Contesting the presence of wheat in the British Isles 8,000 years ago by assessing ancient DNA authenticity from low-coverage data. eLife 4, e10005. ( 10.7554/eLife.10005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.De Filippo C, Meyer M, Prüfer K. 2018Quantifying and reducing spurious alignments for the analysis of ultra-short ancient DNA sequences. BMC Biol. 16, 121. ( 10.1186/s12915-018-0581-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cribdon B, Ware R, Smith O, Gaffney V, Allaby RG. 2020PIA: more accurate taxonomic assignment of metagenomic data demonstrated on sedaDNA from the North Sea. Front. Ecol. Evol. 8, 84. ( 10.3389/fevo.2020.00084) [DOI] [Google Scholar]
- 108.Gagnaire P. 2020Comparative genomics approach to evolutionary process connectivity. Evol. Appl. 13, 1320-1334. ( 10.1111/eva.12978) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Huang S, Chaudhary K, Garmire LX. 2017More is better: recent progress in multi-omics data integration methods. Front. Genet. 8, 84. ( 10.3389/fgene.2017.00084) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Smith O, Dunshea G, Sinding M-HS, Fedorov S, Germonpre M, Bocherens H, Gilbert MTP. 2019Ancient RNA from Late Pleistocene permafrost and historical canids shows tissue-specific transcriptome survival. PLoS Biol. 17, e3000166. ( 10.1371/journal.pbio.3000166) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rannala B, Yang Z. 2017Efficient Bayesian species tree inference under the multispecies coalescent. Syst. Biol. 66, 823-842. ( 10.1093/sysbio/syw119) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tenesa A, Navarro P, Hayes BJ, Duffy DL, Clarke GM, Goddard ME, Visscher PM. 2007Recent human effective population size estimated from linkage disequilibrium. Genome Res. 17, 520-526. ( 10.1101/gr.6023607) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Duranton M, Allal F, Fraïsse C, Bierne N, Bonhomme F, Gagnaire P-A. 2018The origin and remolding of genomic islands of differentiation in the European sea bass. Nat. Commun. 9, 2518. ( 10.1038/s41467-018-04963-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Degnan JH, Rosenberg NA. 2009Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends Ecol. Evol. 24, 332-340. ( 10.1016/j.tree.2009.01.009) [DOI] [PubMed] [Google Scholar]
- 115.Isaac NJB, et al. 2020Data integration for large-scale models of species distributions. Trends Ecol. Evol. 35, 56-67. ( 10.1016/j.tree.2019.08.006) [DOI] [PubMed] [Google Scholar]
- 116.Plard F, Fay R, Kéry M, Cohas A, Schaub M. 2019Integrated population models: powerful methods to embed individual processes in population dynamics models. Ecology 100, e02715. ( 10.1002/ecy.2715) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This article does not contain any additional data.