Abstract
Context
Central line–associated bloodstream infection (BSI) rates, determined by infection preventionists using the Centers for Disease Control and Prevention (CDC) surveillance definitions, are increasingly published to compare the quality of patient care delivered by hospitals. However, such comparisons are valid only if surveillance is performed consistently across institutions.
Objective
To assess institutional variation in performance of traditional central line–associated BSI surveillance.
Design, Setting, and Participants
We performed a retrospective cohort study of 20 intensive care units among 4 medical centers (2004-2007). Unit-specific central line–associated BSI rates were calculated for 12-month periods. Infection preventionists, blinded to study participation, performed routine prospective surveillance using CDC definitions. A computer algorithm reference standard was applied retrospectively using criteria that adapted the same CDC surveillance definitions.
Main Outcome Measures
Correlation of central line–associated BSI rates as determined by infection preventionist vs the computer algorithm reference standard. Variation in performance was assessed by testing for institution-dependent heterogeneity in a linear regression model.
Results
Forty-one unit-periods among 20 intensive care units were analyzed, representing 241 518 patient-days and 165 963 central line–days. The median infection preventionist and computer algorithm central line–associated BSI rates were 3.3 (interquartile range [IQR], 2.0-4.5) and 9.0 (IQR, 6.3-11.3) infections per 1000 central line–days, respectively. Overall correlation between computer algorithm and infection preventionist rates was weak (ρ=0.34), and when stratified by medical center, point estimates for institution-specific correlations ranged widely: medical center A: 0.83; 95% confidence interval (CI), 0.05 to 0.98; P = .04; medical center B: 0.76; 95% CI, 0.32 to 0.93; P = .003; medical center C: 0.50, 95% CI, −0.11 to 0.83; P = .10; and medical center D: 0.10; 95% CI −0.53 to 0.66; P = .77. Regression modeling demonstrated significant heterogeneity among medical centers in the relationship between computer algorithm and expected infection preventionist rates (P<.001). The medical center that had the lowest rate by traditional surveillance (2.4 infections per 1000 central line–days) had the highest rate by computer algorithm (12.6 infections per 1000 central line–days).
Conclusions
Institutional variability of infection preventionist rates relative to a computer algorithm reference standard suggests that there is significant variation in the application of standard central line–associated BSI surveillance definitions across medical centers. Variation in central line–associated BSI surveillance practice may complicate interinstitutional comparisons of publicly reported central line–associated BSI rates.
PUBLIC REPORTING OF HOSPITAL-specific infection rates is widely promoted as a means to improve patient safety.1,2 Central line–associated bloodstream infection (BSI) rates are considered a key patient safety measure because such infections are frequent,3 lead to poor patient outcomes,4 are costly to the medical system,5 and are preventable.6,7 Publishing infection rates on hospital report cards, which is increasingly required by regulatory agencies, is intended to facilitate interhospital comparisons that inform health care consumers and provide incentive for hospitals to prevent infections.8 Inter-hospital comparisons of infection rates, however, are valid only if the methods of surveillance are uniform and reliable across institutions.9,10
Most hospitals performing central line-associated BSI surveillance rely on infection preventionists (formerly known as infection control practitioners11) to manually perform central line–associated BSI surveillance. The infection preventionists apply surveillance case definitions published by the Centers for Disease Control and Prevention (CDC).12 These case definitions rely on a mixture of objective criteria (positive blood cultures) and subjective criteria (judging whether bacteremia is from a central line rather than secondary from an extravascular source, or determining whether recovery of a common skin commensal in the blood represents a true infection vs contamination).
In practice, surveillance is challenging because it is time consuming and lacks a gold standard for validation. Case finding by infection preventionists is effort dependent and can lack sensitivity, resulting in underreporting of infection rates.13 Furthermore, subjective components of surveillance criteria may be inconsistently applied by infection preventionists, reducing the validity of interhospital and potentially intrahospital comparisons.14 In spite of these threats to validity, traditional central line–associated BSI surveillance by infection preventionists has been rarely validated against external measures.13,15
The recent development of computer algorithms for central line–associated BSI surveillance provides an opportunity to establish an objective reference standard with which to benchmark infection preventionist determination of infection rates. A computer algorithm can be applied consistently among different institutions, using only objective criteria (microbiologic, pharmacy, and patient location records) in an identical and comprehensive manner.16,17 We used a computer algorithm that had been developed to approximate the prevailing CDC surveillance definition12 for central line–associated BSI surveillance from 1988 through the study period18; the algorithm was previously found to be as accurate as infection preventionist determinations when compared with an expert reference standard in a single institution.19
As a part of a research collaborative supported by the CDC Prevention Epicenter Program, we performed a study involving 4 medical centers, comparing ecologic (unit-period) central line–associated BSI rates determined by traditional infection preventionist surveillance with rates determined by a computer algorithm reference standard. Our study aims were to estimate the overall correlation between infection preventionist and computer algorithm ecologic rates, and to test for institution-dependent heterogeneity in this relationship. We also assessed whether institution-dependent variability, if found, would lead to relevant differences in the relative ranking of institutions based on reported central line–associated BSI rates.
METHODS
Four academic medical centers (2 in Chicago, Illinois; 1 in Columbus, Ohio; 1 in St Louis, Missouri) participated in this study. A convenience sample of 20 intensive care units (ICUs) across the 4 medical centers contributed electronic data and infection preventionist surveillance data that had been collected from 2004 to 2007. For analysis, infection rates for each ICU were aggregated into 12-month periods. These blocks could be nonconsecutive to exclude months when surveillance was not carried out by infection preventionists. Individual ICUs were asked to contribute 2 to 3 consecutive unit periods of data for the study, based on data availability.
All central line–associated BSI rates were expressed as (number of central line–associated BSI events)/(1000 central line–days). Central line–days were obtained from daily counts provided by each ICU’s nursing unit using CDC methods20; these counts were used as denominators to calculate rates for both infection preventionist and computer algorithm measures.
This study was powered to test the null hypothesis that there is no correlation between computer algorithm and infection preventionist central line–associated BSI rates. Using the parameters of an effect size (r) = 0.8 (based on prior estimation19), 2-sided α = .05, and β = .10, a sample size of 34 total unit-periods would be required.21
Each medical center obtained approval from its respective institutional review board for human subjects research; informed consent was waived, and 1 medical center excluded prisoners from review.
Infection Preventionist Review
As a part of routine infection control activity at each medical center, ICU-specific central line–associated BSI rates were prospectively measured by infection preventionists using CDC surveillance definitions applicable during the study period. All infection preventionists employed at each medical center were registered nurses, medical technologists, or microbiologists trained in infection control, and all were blinded to their participation in this study.
Each medical center’s infection control department worked independently, with unique organizational and informatics approaches for case finding and information gathering. Although each medical center used the hospital electronic medical record to generate lists of positive blood cultures for case finding, infection preventionists applied CDC central line–associated BSI definitions manually, without the assistance of computer-decision support.
Computer Algorithms
We used a computer algorithm that approximated the CDC surveillance definition used by infection preventionists during the study period. The algorithm (code available at http://bsi.cchil.org) retrospectively calculated central line–associated BSI rates within ICUs at each medical center, using clinical data in the electronic medical record. The algorithm evaluated laboratory-identified blood culture isolates in an objective fashion, using stepwise logic to make a central line–associated BSI determination (Figure 1); further details are published elsewhere.18
Figure 1. Schematic of Computer Algorithm for Central Line–Associated Bloodstream Infection Surveillance.
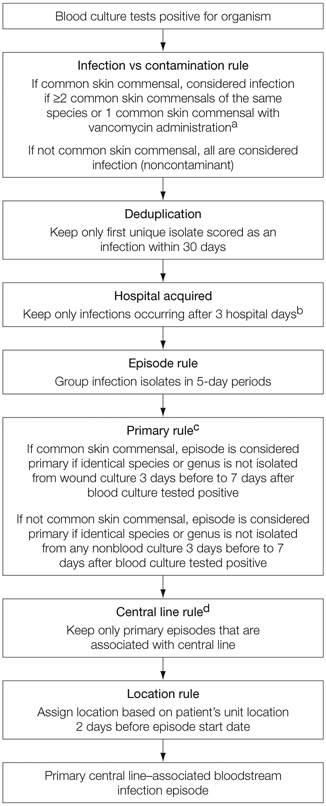
aCommon skin commensals are defined as diphtheroids, Bacillus, Propionibacterium, coagulase-negative Staphylococcus, or Micrococcus species.
bCalendar date of admission to the hospital is considered hospital day 1.
cActive surveillance screening cultures and catheter tip cultures were not considered to avoid misclassifying episodes as secondary.
dCentral line presence was assessed on the first day of a primary bloodstream infection episode and through 2 hospital days prior.
Common skin commensal organisms (defined as diphtheroids, Bacillus, Propionibacterium, coagulase-negative Staphylococcus, or Micrococcus species) recovered from blood cultures can represent either infection or blood culture contamination. The computer algorithm considered either the recovery of at least 2 positive common skin commensal cultures of the same species within 2 hospital days or a single positive common skin commensal culture with vancomycin administered within the 2 subsequent days as representing an infection.
Computer algorithms were distributed to each participating center in both logical representation and SQL (Structured Query Language) code.18 Each medical center adapted the code to its clinical database, and correct implementation was ascertained by verifying results from a standardized test data set.
Central line presence was determined by manual chart review at 3 medical centers; the fourth center was able to automate central line detection. Only blood culture episodes determined to be ICU-related and primary by the computer algorithm were manually reviewed. Central line data were abstracted from the nursing daily flow sheet; a central line was considered present if documented on the day of a positive blood culture result or up to 2 hospital days prior. Nursing flow sheets were available for all patients by chart review with complete ascertainment of central line status.
Statistics
We compared 12-month unit-period infection rates from infection preventionist and computer algorithm surveillance using several methods. Differences between the rates defined by the infection preventionist and computer algorithm were tested using the Wilcoxon signed-rank test, and strength of correlation was tested using the Spearman rank-order correlation. Next, we created linear regression models of infection preventionist rates vs the computer algorithm rates, stratified by medical center. In a final linear regression model with all medical centers combined, we tested for heterogeneity of slope and intercept using interaction terms with medical center as a nominal variable. All regression analyses were weighted to account for differences in the magnitude of the denominator (central line–days) among unit periods. The 95% confidence intervals (CIs) for central line–associated BSI rates were calculated assuming binomial distribution.
All analyses were performed using SAS version 9.1.3 (SAS Institute Inc, Cary, North Carolina) and Stata version 9.0 (StataCorp, College Station, Texas). P<.05 was considered statistically significant.
RESULTS
Twenty ICUs in 4 medical centers contributed 41 twelve-month unit periods, representing 241 518 patient-days and 165 963 central line–days and included 9 types of units: 5 medical, 4 surgical, 1 combined medical-surgical, 2 neurosurgical, 3 cardiac, 1 oncologic, 1 cardiothoracic surgery, 1 burn, 1 bone marrow transplant, and 1 trauma.
Across all unit periods, the median infection preventionist–measured central line–associated BSI rate was 3.3 infections per 1000 central line–days (interquartile range [IQR], 2.0-4.5; range, 0.4-8.5). The median rate determined by the computer algorithm was 9.0 per 1000 central line–days (IQR, 6.3-11.3; range, 2.0-21.5). The median rates for the infection preventionist and computer algorithm methods were significantly different (P<.001).
We calculated the Spearman rank correlation coefficients to determine how well the infection preventionist rates correlated with computer algorithm rates on a unit-period basis. When unit periods were analyzed in aggregate across medical centers, overall correlation (Spearman ρ) between computer algorithm and infection preventionist rates was weak, 0.34 (95% CI, 0.04-0.59; P = .03). However, when stratified by medical center, we found that the point estimates of the correlations varied widely (medical center A: ρ, 0.83; 95% CI, 0.05 to 0.98; P = .04; medical center B: ρ, 0.76; 95% CI, 0.32 to 0.93; P = .003; medical center C: ρ, 0.50; 95% CI, −0.11 to 0.83; P = .10; medical center D: ρ, 0.10; 95% CI, −0.53 to 0.66; P = .77).
In the linear regression models for each medical center (Figure 2), the computer algorithm rates were generally higher than the paired rate defined by the infection preventionist, and the slopes of the fitted linear regression lines were less than 1 (range, 0.28-0.63). The goodness-of-fit or R2, representing how closely the observations clustered around the regression line, varied widely between hospitals (Figure 2). The R2 values for each of the medical center regression lines were as follows: medical center A, 0.43; medical center B, 0.81; medical center C, 0.36; medical center D, 0.28. Higher R2 values represent more consistency between infection preventionist and computer algorithm determinations across unit-periods.
Figure 2. Linear Regression of Rates of Central Line–Associated Bloodstream Infection.
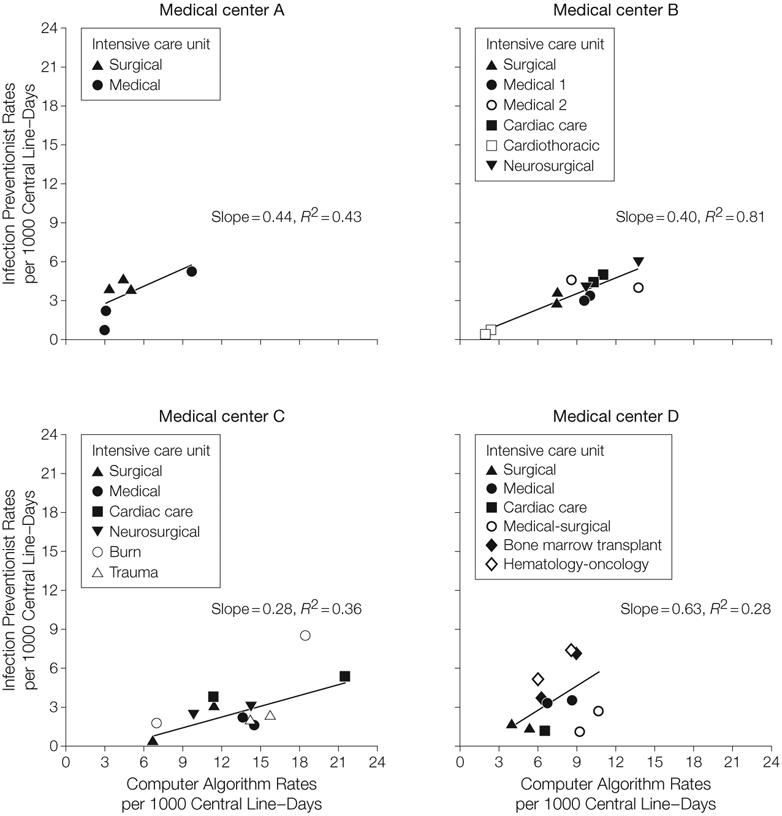
The line in each panel represents fitted values.
When we included all data into a single linear regression model, we found that although the slope did not differ significantly by medical center (P = .67), the intercept was significantly different across medical centers (P < .001). Because no differences in slope were detected, our final model (Figure 3) represented the study data with 4 parallel lines with equal slope (0.43) and different intercepts (range, −2.99 to 1.47) for each medical center. Thus, any given single computer algorithm rate would correspond to a different predicted infection preventionist surveillance rate at each institution (eg, for a hypothetical computer algorithm rate of 9 central line–associated BSIs per 1000 central line–days, the predicted infection preventionist rate would vary by hospital, from 1.1 infections for medical center C to 4.9 for medical center A).
Figure 3. Predicted Infection Preventionist Central Line–Associated Bloodstream Infection Rates.
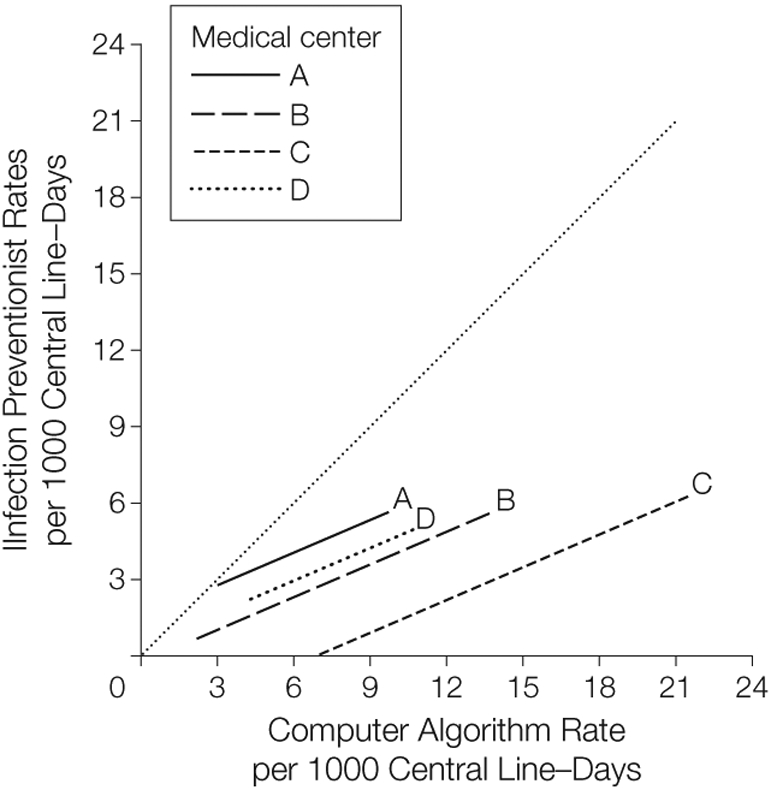
The dotted line that transects the graph from 0 represents a hypothetical line of perfect agreement (slope=1). Regression line equations that assign a common slope and varying intercepts: medical center A, y=1.47 + 0.43x; B, y=−0.28 + 0.43x; C, y = −2.99 + 0.43x; D, y=0.35 + 0.43x.
Relative rankings of the 4 medical centers differed depending on the surveillance method (Figure 4). Under infection preventionist surveillance, medical center C reported the lowest central line–associated BSI rate (pairwise comparison with the next lowest medical center D not statistically significant, P = .07). When reanalyzed with the computer algorithm, medical center C had the highest central line–associated BSI rate, and rates were statistically different across all 4 medical centers (P <.01).
Figure 4. Relative Ranking of 4 Medical Centers.
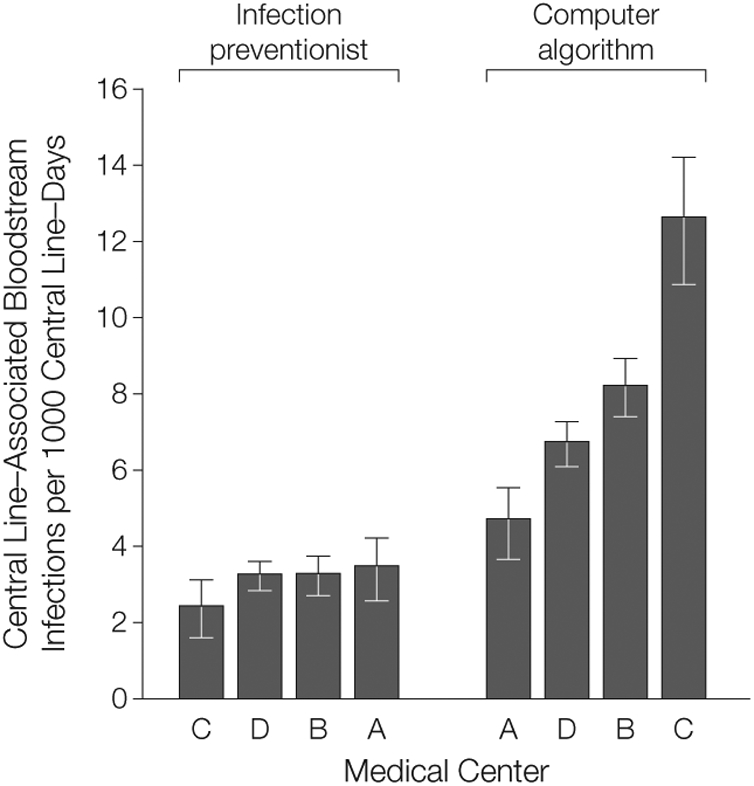
Within each surveillance group, medical centers are arranged in ascending order by central line–associated bloodstream infection rate. Error bars indicate 95% confidence intervals. Rates were calculated within each medical center by aggregating all intensive care unit rates. With infection preventionist surveillance, the following rates were determined: C, 2.4 (95% CI, 1.8-3.3); D, 3.2 (95% CI, 2.9-3.7); B, 3.3 (95% CI, 2.8-3.8); A, 3.5 (95% CI, 2.7-4.4). With the computer algorithm the following infection rates were determined: A, 4.7 (95% CI, 3.8-5.7); D, 6.7 (95% CI, 6.2-7.3); B, 8.2 (95% CI, 7.4-9.0); C, 12.6 (95% CI, 11.0-14.3).
COMMENT
We compared central line–associated BSI rates from infection preventionist surveillance with a computer algorithm reference standard in ICUs across 4 medical centers. We hypothesized that although we would find differences in the absolute infection rates estimated by infection preventionist surveillance as opposed to computer algorithm surveillance, we would find reasonable and uniform ecologic correlation between the 2 methods. Instead, we found weak overall correlation between the 2 methods and importantly, variable correlation when stratified by medical center.
Medical center–specific variation in infection preventionist surveillance was confirmed through linear regression modeling; we found that for a given computer algorithm central line–associated BSI rate, the expected rate defined by infection preventionist surveillance varied significantly by medical center. The center-specific variation markedly affected the rank order of institutions, such that the medical center with the lowest rate as reported by infection preventionists had the highest rate by the computer algorithm reference standard. Our findings suggest that there is local variation in central line–associated BSI surveillance performance at different medical centers, raising concern for the validity of interinstitutional rate comparisons.
In the past several years, public reporting of hospital infection rates has been increasingly promoted as a means of comparing patient safety among different institutions. In the United States, roughly one-quarter of all states have legislative mandates to report central line–associated BSI rates from ICUs.22 These data are used by patients, advocacy groups, and regulators to compare hospitals.22 Furthermore, hospital reimbursement is increasingly dependent on reported rates,23 creating potential financial incentives for hospitals to underreport rates. Our study highlights the potential fallibility of traditional surveillance methods using partially subjective criteria and underscores the need for cautious interpretation of these results until more reliable measures or validation against objective measures can routinely be performed.
Prior studies have analyzed the validity of infection preventionist central line–associated BSI surveillance through retrospective case review, although none to our knowledge has focused on variation in surveillance practice across medical centers. A pilot validation of the CDC surveillance definitions was performed in 1991 through1993 at 9 hospitals, with 1 ICU per hospital contributing a stratified random sample of 15 charts with central line–associated BSIs, as well as additional charts without them, for standardized review by trained infection preventionists and CDC epidemiologists.15 The study found that infection preventionist surveillance was 85% sensitive and 98% specific compared with the expert review. Although these favorable performance characteristics formed the evidence base for future surveillance, the study was performed before the onset of widespread public reporting and was not designed to look for differences in infection preventionist performance across medical centers.
More recently, poor sensitivity of surveillance conducted by infection preventionists was found during a retrospective review of a sample of blood cultures with potential central line–associated BSI in 6 Australian hospitals using a CDC-adapted surveillance system with the same criteria used in the present study. Compared with expert review, infection preventionist surveillance was found to have poor sensitivity (35%) and only marginal agreement (κ = 0.31).13
For interinstitutional comparison of infection rates, consistent surveillance (ie, reliability) among institutions may be even more important than accuracy. In a mathematical simulation comparing a purely objective approach (analogous to the computer algorithm used in the present study) with a partially subjective approach (performed by infection preventionists using clinical criteria) to rank institutions by central line–associated BSI rates, the objective method overestimated the true rate but was more accurate in ranking institutions by their rates.24 Thus, removing subjectivity (ie, clinical judgment) potentially leads to reduced accuracy but improved reliability, leading to more accurate rankings of institution-specific rates.
Discrepancies in surveillance practice may vary substantially across 2 domains, case finding and classification. Case finding of potential BSIs can be incomplete. For example, although a patient may have multiple blood cultures postive for central line–associated BSI throughout a prolonged ICU stay, not all blood cultures may be investigated, especially if the infection preventionist does not have a systematic method of tracking blood culture results. For classification, there is likely substantial variability in application of subjective aspects in the central line–associated BSI definition. For example, infection preventionists (and clinicians) will not always agree whether a positive blood culture originated from a central line or from an extravascular source such as an intra-abdominal abscess as a secondary BSI. In instances in which definitive information is not available, infection preventionists may differentially classify ambiguous cases.
To further explain the observed variability in rates, qualitative differences may exist at the institution level, such as local clinical culturing practice, quality of medical documentation, and strength of institutional oversight over infection prevention activities. Further qualitative assessments may be warranted to better understand the relative importance of institutional and episode-level sources of variability. All 4 medical centers are academic training institutions with strong interest in infection prevention and research; thus, we speculate that although institutional differences may exist, most of the variability observed in this study is explained by differences in infection preventionist performance of surveillance.
The use of computer algorithms for central line–associated BSI surveillance has been described in prior studies.19,25,26 The computer algorithm surveillance used in the current study was previously validated in a single-center study and was found to have substantial agreement with expert review (κ = 0.73).19 Distribution of the same computer algorithm to the 4 medical centers in the present study permitted the efficient estimation of an objective reference rate across a large number of ICUs and unit-periods, allowing for detection of clinically and statistically significant differences in infection preventionist surveillance performance across institutions. The computer algorithm has inherent limitations; in particular, it performs secondary infection determinations using information from nonblood cultures, which may not always be accurate. Furthermore, like the infection preventionist, it may be affected by the local propensity of hospitals to obtain clinical cultures. Future studies that analyze agreement at an episode level, with an additional reference standard, are needed to validate the computer algorithm for generalized use.
Our study has several limitations. First, although we use the computer algorithm as a reference standard, the true central line–associated BSI rate is unknown (and outside of simulation models, likely unknowable). We selected the computer algorithm as a reference standard because it could be consistently applied across institutions and because of its prior validation against expert review.19 To ensure uniform application, we tested each medical center’s installation of the computer algorithm by providing standardized input data sets to ensure that local installations of code produced identical results.18 Second, we studied agreement between ecologic rates at the unit-period level, not at the individual blood culture episode level. Even if there is excellent agreement at the ecologic level, there may be considerable disagreement on the significance of individual blood culture episode results. For the purpose of this study, because routine reporting and public ranking occurs at an ecologic level, we focused on the ICU as the unit of analysis rather than the individual patient. Third, although we used the CDC surveillance definitions contemporaneous to the study period, the CDC changed a component of the surveillance definitions in January 2008; the criterion that allowed a single positive blood culture caused by common skin commensals to represent an infection if appropriate antibiotic treatment was administered was removed.27 Whether the use of the new surveillance definition improves the correlation between computer algorithms and infection preventionist rates requires further study because a large subjective component of infection preventionist review (determination of primary vs secondary infection) remains.
Central line–associated BSI surveillance, whether performed by infection preventionists or computer algorithms, is not designed to guide or replace clinical diagnosis by physicians; rather, it is a specialized tool to provide consistency in monitoring rates over time and among institutions to inform infection prevention efforts. In this study, we found strong evidence of institutional variation in central line–associated BSI surveillance performance among medical centers. Inconsistent surveillance practice can have a significant effect on the relative ranking of hospitals, which threatens the validity of the metric used by both funding agencies and the public to compare hospitals. As central line–associated BSI rates gain visibility and importance—in the form of public report cards, infection reduction campaigns such as “Getting to Zero,”28 and financial incentives for reducing rates by private insurers and the Centers for Medicare & Medicaid Services23—we should seek and test surveillance measures that are as reliable and objective as possible.
Funding/Support:
This study was funded by CDC Co-operative Agreements U01-CI000327, U01-CI000328, and U01-CI000333 from the CDC Prevention Epicenter Grants.
Footnotes
Financial Disclosures: None reported.
Role of the Sponsor: The sponsors had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; and preparation, review, or approval of the manuscript.
Previous Presentation: Presented in part at the 18th Annual Scientific Meeting of the Society for Healthcare Epidemiology of America, Orlando, Florida, April 6, 2008 (Abstract 346).
Additional Contributions: We thank the infection preventionists who contributed their surveillance data to the analysis.
Contributor Information
Michael Y. Lin, Section of Infectious Diseases, Department of Medicine, Rush University Medical Center, Chicago, Illinois.
Bala Hota, Section of Infectious Diseases, Department of Medicine, Rush University Medical Center, and Division of Infectious Diseases, Department of Medicine, John H. Stroger, Jr, Hospital of Cook County, Chicago, Illinois.
Yosef M. Khan, Division of Infectious Diseases, College of Medicine, The Ohio State University, Columbus..
Keith F. Woeltje, Division of Infectious Diseases, Department of Medicine,Washington University School of Medicine, St Louis, Missouri.
Tara B. Borlawsky, Department of Biomedical Informatics, The Ohio State University, Columbus..
Joshua A. Doherty, Department of Medical Informatics, BJC Healthcare, St Louis, Missouri.
Kurt B. Stevenson, Division of Infectious Diseases, College of Medicine, The Ohio State University, Columbus..
Robert A. Weinstein, Section of Infectious Diseases, Department of Medicine, Rush University Medical Center, and Division of Infectious Disease, Department of Medicine, John H. Stroger, Jr, Hospital of Cook County, Chicago, Illinois.
William E. Trick, Department of Medicine, Rush University Medical Center, Department of Medicine, John H. Stroger, Jr, Hospital of Cook County, Chicago, Illinois.
REFERENCES
- 1.Fung CH, Lim YW, Mattke S, Damberg C, Shekelle PG. Systematic review: the evidence that publishing patient care performance data improves quality of care. Ann Intern Med. 2008;148(2):111–123. [DOI] [PubMed] [Google Scholar]
- 2.Weinstein RA, Siegel JD, Brennan PJ. Infection-control report cards—securing patient safety. N Engl J Med. 2005;353(3):225–227. [DOI] [PubMed] [Google Scholar]
- 3.Mermel LA. Prevention of intravascular catheter-related infections. Ann Intern Med. 2000;132(5): 391–402. [DOI] [PubMed] [Google Scholar]
- 4.Klevens RM, Edwards JR, Richards CL Jr, et al. Estimating health care-associated infections and deaths in US hospitals, 2002. Public Health Rep. 2007; 122(2):160–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perencevich EN, Stone PW, Wright SB, Carmeli Y, Fisman DN, Cosgrove SE; Society for Healthcare Epidemiology of America. Raising standards while watching the bottom line: making a business case for infection control. Infect Control Hosp Epidemiol. 2007; 28(10):1121–1133. [DOI] [PubMed] [Google Scholar]
- 6.Bleasdale SC, Trick WE, Gonzalez IM, Lyles RD, Hayden MK, Weinstein RA. Effectiveness of chlorhexidine bathing to reduce catheter-associated bloodstream infections in medical intensive care unit patients. Arch Intern Med. 2007;167(19):2073–2079. [DOI] [PubMed] [Google Scholar]
- 7.Pronovost P, Needham D, Berenholtz S, et al. An intervention to decrease catheter-related bloodstream infections in the ICU. N Engl J Med. 2006; 355(26):2725–2732. [DOI] [PubMed] [Google Scholar]
- 8.Werner RM, Asch DA. The unintended consequences of publicly reporting quality information. JAMA. 2005;293(10):1239–1244. [DOI] [PubMed] [Google Scholar]
- 9.Pronovost PJ, Miller M, Wachter RM. The GAAP in quality measurement and reporting. JAMA. 2007; 298(15):1800–1802. [DOI] [PubMed] [Google Scholar]
- 10.McKibben L, Horan TC, Tokars JI, et al. ; Healthcare Infection Control Practices Advisory Committee. Guidance on public reporting of healthcare-associated infections: recommendations of the Healthcare Infection Control Practices Advisory Committee. Infect Control Hosp Epidemiol. 2005;26(6):580–587. [DOI] [PubMed] [Google Scholar]
- 11.APIC announces new name for infection control profession. Infection Control Today. July11, 2008. http://www.infectioncontroltoday.com/news/2008/07/apic-announces-new-name-for-infection-control-pro.aspx. Accessed August 23,2010. [Google Scholar]
- 12.Garner JS, Jarvis WR, Emori TG, Horan TC, Hughes JM. CDC definitions for nosocomial infections, 1988. Am J Infect Control. 1988;16(3):128–140. [DOI] [PubMed] [Google Scholar]
- 13.McBryde ES, Brett J, Russo PL, Worth LJ, Bull AL, Richards MJ. Validation of statewide surveillance system data on central line-associated bloodstream infection in intensive care units in Australia. Infect Control Hosp Epidemiol. 2009;30(11):1045–1049. [DOI] [PubMed] [Google Scholar]
- 14.Worth LJ, Brett J, Bull AL, McBryde ES, Russo PL, Richards MJ. Impact of revising the National Nosocomial Infection Surveillance System definition for catheter-related bloodstream infection in ICU: reproducibility of the National Healthcare Safety Network case definition in an Australian cohort of infection control professionals. Am J Infect Control. 2009;37(8):643–648. [DOI] [PubMed] [Google Scholar]
- 15.Emori TG, Edwards JR, Culver DH, et al. Accuracy of reporting nosocomial infections in intensivecare-unit patients to the National Nosocomial Infections Surveillance System: a pilot study. Infect Control Hosp Epidemiol. 1998;19(5):308–316. [DOI] [PubMed] [Google Scholar]
- 16.Evans RS, Larsen RA, Burke JP, et al. Computer surveillance of hospital-acquired infections and antibiotic use. JAMA. 1986;256(8):1007–1011. [PubMed] [Google Scholar]
- 17.Tokars JI, Richards C, Andrus M, et al. The changing face of surveillance for health care-associated infections. Clin Infect Dis. 2004;39 (9):1347–1352. [DOI] [PubMed] [Google Scholar]
- 18.Hota B, Lin M, Doherty JA, et al. ; CDC Prevention Epicenter Program. Formulation of a model for automating infection surveillance: algorithmic detection of central-line associated bloodstream infection. J Am Med Inform Assoc. 2010;17(1): 42–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trick WE, Zagorski BM, Tokars JI, et al. Computer algorithms to detect bloodstream infections. Emerg Infect Dis. 2004;10(9):1612–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.National Healthcare Safety Network (NHSN) manual: patient safety component protocol. Atlanta, GA: Division of Healthcare Quality Promotion. http://www.cdc.gov/nhsn/library.html#psc. Accessed July 11, 2008. [Google Scholar]
- 21.Hulley SB, Cummings SR, Browner WS, Grady D, Hearst N, Newman TB. Designing Clinical Research. 2nd ed. Philadelphia, PA: Lippincott Williams&Wilkins; 2001. [Google Scholar]
- 22.Behind the data. How do patients fare?bloodstream infections. ConsumerReportsHealth.org [Web page]. March 2010. http://www.consumerreports.org/health/doctors-hospitals/hospital-infection/deadly-infections-hospitals-can-lower-the-danger/behind-the-data/index.htm. Accessed February 26, 2010. [Google Scholar]
- 23.Medicare fact sheet: new provisions to strengthen tie between Medicare payment and quality for inpatient stays in acute care hospitals in fiscal year 2011 and beyond. Washington, DC: Centers for Medicare & Medicaid Services; July30, 2010. [Google Scholar]
- 24.Rubin MA, Mayer J, Greene T, et al. An agent-based model for evaluating surveillance methods for catheter-related bloodstream infection. AMIA Annu Symp Proc. November2008:631–635. [PMC free article] [PubMed] [Google Scholar]
- 25.Yokoe DS, Anderson J, Chambers R, et al. Simplified surveillance for nosocomial bloodstream infections. Infect Control Hosp Epidemiol. 1998; 19(9):657–660. [DOI] [PubMed] [Google Scholar]
- 26.Woeltje KF, Butler AM, Goris AJ, et al. Automated surveillance for central line-associated bloodstream infection in intensive care units. Infect Control Hosp Epidemiol. 2008;29(9):842–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horan TC, Andrus M, Dudeck MA. CDC/NHSN surveillance definition of health care-associated infection and criteria for specific types of infections in the acute care setting. Am J Infect Control. 2008; 36(5):309–332. [DOI] [PubMed] [Google Scholar]
- 28.Edmond MB. Getting to zero: is it safe? Infect Control Hosp Epidemiol. 2009;30(1):74–76. [DOI] [PubMed] [Google Scholar]


