Abstract
OBJECTIVE.
Central line–associated bloodstream infection (BSI) rates are a key quality metric for comparing hospital quality and safety. Traditional BSI surveillance may be limited by interrater variability. We assessed whether a computer-automated method of central line–associated BSI detection can improve the validity of surveillance.
DESIGN.
Retrospective cohort study.
SETTING.
Eight medical and surgical intensive care units (ICUs) in 4 academic medical centers.
METHODS.
Traditional surveillance (by hospital staff) and computer algorithm surveillance were each compared against a retrospective audit review using a random sample of blood culture episodes during the period 2004–2007 from which an organism was recovered. Episode-level agreement with audit review was measured with κ statistics, and differences were assessed using the test of equal κ coefficients. Linear regression was used to assess the relationship between surveillance performance (κ) and surveillance-reported BSI rates (BSIs per 1,000 central line–days).
RESULTS.
We evaluated 664 blood culture episodes. Agreement with audit review was significantly lower for traditional surveillance (κ [95% confidence interval (CI)] = 0.44 [0.37–0.51]) than computer algorithm surveillance (κ [95% CI] = 0.58 [0.52–0.64]; P = .001). Agreement between traditional surveillance and audit review was heterogeneous across ICUs (P = .01); furthermore, traditional surveillance performed worse among ICUs reporting lower (better) BSI rates (P = .001). In contrast, computer algorithm performance was consistent across ICUs and across the range of computer-reported central line–associated BSI rates.
CONCLUSIONS.
Compared with traditional surveillance of bloodstream infections, computer automated surveillance improves accuracy and reliability, making interfacility performance comparisons more valid.
Hospital-acquired central line–associated bloodstream infections (BSIs) commonly lead to adverse patient outcomes and are largely preventable.1,2 Central line–associated BSI rates, which are self-reported by hospitals, are perceived as a key performance measure used to compare patient safety between institutions.3 In 2012, the Center for Medicare and Medicaid Services (CMS) began reporting central line–associated BSI rates on its Hospital Compare website (http://www.medicare.gov/hospitalcompare) to facilitate public transparency with BSI rates and with the intention of levying future payment reductions for those hospitals reporting poorer rates (ie, pay for performance).4,5
Traditional surveillance of central line–associated BSIs uses standard case definitions and operational methodology, such as those supported by the Centers for Disease Control and Prevention (CDC) National Healthcare Safety Network (NHSN).6 Trained hospital staff (infection preventionists) interpret clinical events to judge whether certain rules for BSI are satisfied, such as whether a blood culture pathogen originated from the bloodstream versus an extravascular source (ie, a secondary bacteremia).
Earlier studies have detected significant variation in the performance of traditional BSI surveillance at the individual infection preventionist level7,8 as well as at a facility level;9 such variation complicates the comparison of hospitals on the basis of their publicly reported rates. The CDC has recommended audit review of BSI surveillance as a primary method to assess and improve the reliability of central line–associated BSI reporting at facilities.10
As a proposed alternative to traditional surveillance, automated computer algorithmic BSI detection11 is objective, is efficient, and can be implemented identically across multiple institutions, potentially making interinstitutional rate comparisons more reliable.12 Previously, computer algorithm surveillance was shown to have substantial agreement with expert review at a single institution.13 A multicenter assessment of computer algorithm surveillance reporting characteristics through audited review is needed to better compare its performance with that of traditional central line–associated BSI surveillance.
We performed a multi-institutional comparison of the performance of traditional surveillance versus computer algorithm central line–associated BSI surveillance using a retrospective audit review as the comparator. We evaluated performance at the individual BSI episode level as well as at the intensive care unit (ICU) level. We hypothesized that computer algorithm BSI surveillance would have better accuracy and reliability than traditional surveillance. If confirmed, such findings could have important implications for improving the current practice of public reporting and interinstitutional comparison of central line–associated BSI rates.
METHODS
This study was conducted at 4 academic medical centers (2 in Chicago, IL; 1 in Columbus, OH; 1 in St Louis, MO). One medical ICU and 1 surgical ICU from each medical center, for a total of 8 ICUs, contributed patient data. The study involved clinical data from January 1, 2004, through June 30, 2007. Retrospective audit review was performed from 2009 to 2010. This study was approved by the institutional review boards at each participating center.
At all medical centers, blood and other body site cultures were obtained as a part of usual clinical practice. Blood cultures were processed at each medical center’s microbiology laboratory using standard automated blood culture detection systems.
Traditional Surveillance
Data from routine central line–associated BSI surveillance were produced by infection preventionists at each medical center using NHSN definitions. All participants in traditional surveillance were blinded to participation in the study, with no knowledge of computer algorithm detection. Fifteen infection preventionists participated; all were registered nurses or microbiologists and had a median of 7 years of infection control experience (range, 0–30 years). All had received formal central line–associated BSI surveillance training, either provided by the CDC or by the Association for Professionals in Infection Control and Epidemiology.
Traditional surveillance was performed using standardized NHSN surveillance criteria (Table 1). During the study period (and until 2008), NHSN central line–associated BSI criteria also included a criterion for infection preventionists to categorize a single positive blood culture with a common skin commensal (Corynebacterium species, Bacillus species, Propionibacterium species, coagulase-negative Staphylococcus species, and Micrococcus species) as central line–associated BSI if clinical symptoms were present, the blood pathogen was not related to an infection at another site, and appropriate antimicrobials were prescribed by a physician. After 2008, this criterion was removed as part of the NHSN definition of a central line–associated BSI. To make the analysis applicable to post-2008 NHSN surveillance activity, study personnel retrospectively reclassified any blood culture episodes containing only a single culture result of a single common skin commensal as “not central line–associated BSI.” No blood culture episodes required recoding based on this reclassification scheme.
TABLE 1.
National Healthcare Safety Network Definition for Laboratory-Confirmed Bloodstream Infection Used by Audit Reviewer, 2009–2010
| Criterion | Definition |
|---|---|
| 1 | Patient has a recognized pathogen cultured from at least 1 blood culture, and pathogen cultured from blood is not related to an infection at another site OR |
| 2 | Patient has at least 1 of the following signs or symptoms: fever (temperature, >38°C), chills, or hypotension; positive cultures are not related to infection at another site; and common commensala is cultured from at least 2 blood cultures obtained on separate occasions |
NOTE. Central line–associated bloodstream infection is defined as a laboratory-confirmed bloodstream infection not present or incubating at admission, and the patient has a central line at the time or within 48 hours of the time that blood culture was obtained.25 Note that the current 2014 central line–associated bloodstream infection definition uses the same criteria as above, except that a 2-calendar-day rule is used instead of 48 hours.
Includes Corynebacterium species (diphtheroids), Bacillus species, Propionibacterium species, coagulase-negative Staphylococcus species, and Micrococcus species.
Computer Algorithm Surveillance
A previously validated computer algorithm was used to approximate NHSN central line–associated BSI surveillance criteria.13 The algorithm retrospectively identified blood culture episodes that fulfilled algorithmic criteria for central line–associated BSIs within ICUs at each medical center using data available from the electronic health record (see Figure 1 for algorithm steps and data inputs; computer programming code is available at http://bsi.cchil.org). All 4 medical centers automated the computer algorithm surveillance, although 3 centers required a preparatory step of manual data abstraction of central line presence before running the computer program. The computer algorithm defined a blood culture episode as an eligible blood culture plus all subsequent cultures within a 5-day period; each episode had 1 of the following possible 4 determinations: “central line–associated BSI,” “primary bloodstream infection, not central line associated,” “secondary bloodstream infection,” or “contaminant.”
FIGURE 1.
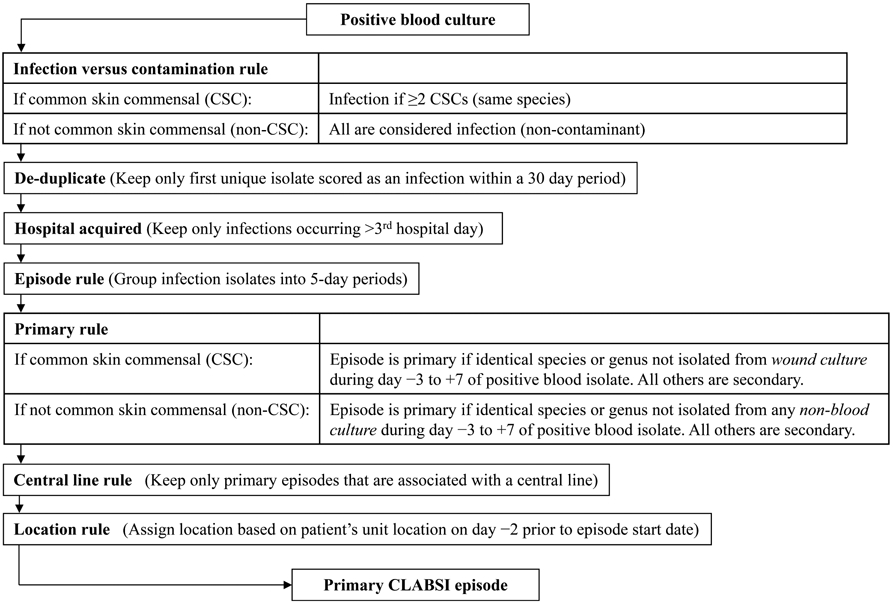
Schematic of computer algorithm adapting National Healthcare Safety Network criteria for central line–associated bloodstream infection (CLABSI) surveillance. Common skin commensals (CSCs) are defined as diphtheroids, Bacillus species, Propionibacterium species, coagulase-negative Staphylococcus species, or Micrococcus species. The computer algorithm required recovery of the same common skin commensal from 2 separate blood cultures within 2 consecutive hospital days in an episode. Calendar date of admission to the hospital is considered hospital day 1. Active surveillance screening cultures and catheter tip cultures were not considered to avoid misclassifying episodes as secondary. Central line presence included any duration of use and was assessed on first day of a primary bloodstream infection episode through 2 hospital days before the episode.
Sampling for Audit Review and Primary Analysis
For all institutions, to preserve the relative proportion of available blood culture episodes in each respective ICU type, we performed stratified random sampling of blood culture episodes within medical and surgical ICUs. We limited our sampling on the basis of a quota, such that the number of blood culture episodes reviewed by the audit reviewer at each institution was 140. To direct the limited resources of the audit reviewers toward blood culture episodes with potentially ambiguous categorization, sampled episodes that were already categorized as “not central line–associated BSI” by the infection preventionist and “contaminant” (specifically, single common skin commensals) by the computer algorithm were presumed to have an audit reviewer categorization of “negative;” these episodes were retained in the final random sample for analysis but did not count toward the quota at each institution. Thus, inclusion of these episodes allowed each center to variably expand its study sample beyond the 140-episode review quota.
Audit Reviewer
A single infection preventionist from each medical center who had not previously performed central line–associated BSI surveillance at the study ICUs during the study period performed a retrospective audit of the sampled blood culture episodes from the study cohort. The audit reviewers were all registered nurses with formal central line–associated BSI surveillance training and 2–9 years of infection control experience. The audit reviewers additionally received a standardized study orientation and investigator-led training by telephone. To encourage adherence to NHSN surveillance criteria, they used a surveillance worksheet that prompted application of NHSN central line–associated BSI surveillance criteria (Table 1) in a step-wise, structured format. The audit reviewers, blinded to traditional and computer algorithm surveillance determinations, categorized each episode as either “central line–associated BSI” or “not central line–associated BSI.”
Analysis
We calculated the level of agreement (κ statistic) between traditional surveillance and audit review and between the computer algorithm and audit review. We used the test of equal κ coefficients to assess for differences in overall κ estimates as well as to assess for heterogeneity of ICU-specific κ values across study ICUs.
To test for significant differences in κ heterogeneity among study ICUs for traditional surveillance versus computer algorithm surveillance, we calculated the distance from the mean level of agreement for each ICU compared with the pooled mean. After confirming that these differences were normally distributed, we compared the distribution of the differences using the paired Student t test.
To assess for a relationship between an ICU’s surveillance performance and its BSI rates, we created a robust linear regression of each ICU’s κ for traditional surveillance/audit agreement versus its overall central line–associated BSI rate as determined using traditional surveillance. We similarly created a linear regression using the computer algorithm/audit agreement versus the computer algorithm–determined ICU-specific central line–associated BSI rate. The central line–associated BSI rate was defined as the number of central line–associated BSIs divided by 1,000 central line–days. Central line–days were obtained from daily counts provided by each ICU’s nursing unit using CDC methods;6 these counts were used as denominators to calculate rates for both infection preventionist and computer algorithm measures of central line–associated BSI rates. We performed analysis to detect outliers and violations of model assumptions. For all analyses, we used Stata (version 10) or SAS (version 9.1.3).
RESULTS
Blood Culture Episodes
Among the 8 ICUs, we identified 1,251 blood culture episodes during the study period; a random sample of 664 episodes (53% of the total) was selected. Microbiological characteristics of the random sample of blood culture episodes are presented in Table 2. Most were monomicrobial; common skin commensals (Bacillus species, coagulase-negative Staphylococcus species, Corynebacterium species, Micrococcus species, and Propionibacterium species) were the most common organism type recovered, followed by Enterococcus species, Enterobacteriaceae, Candida species, and Staphylococcus aureus. The proportion of the study sample categorized as central line–associated BSI varied by method, as follows: traditional surveillance, 145 (22%); computer algorithm, 266 (40%); and audit review, 213 (32%).
TABLE 2.
Characteristics of the Random Sample of Blood Culture Episodes
| Variable | No. (%) of episodes (n = 664) |
|---|---|
| Facility, ICU | |
| A | |
| MICU | 86 (13) |
| SICU | 107 (16) |
| B | |
| MICU | 77 (12) |
| SICU | 79 (12) |
| C | |
| MICU | 100 (15) |
| SICU | 59 (9) |
| D | |
| MICU | 77 (12) |
| SICU | 79 (12) |
| Unique species | |
| 1 | 529 (80) |
| 2 | 106 (16) |
| ≥3 | 29 (4) |
| Organism characteristics | |
| Gram positive only | 421 (63) |
| Gram negative only | 116 (17) |
| Fungal only | 48 (7) |
| Other (mixed infection) | 79 (12) |
| Episodes with ≥1 of following species recovereda | |
| Common skin commensal species | 347 (52) |
| Enterococcus species | 98 (15) |
| Enterobacteriaciae | 90 (14) |
| Candida species | 77 (12) |
| Staphylococcus aureus | 62 (9) |
| Pseudomonas aeruginosa | 46 (7) |
NOTE. ICU, intensive care unit; MICU, medical intensive care unit; SICU, surgical intensive care unit.
Episodes can have more than 1 species if polymicrobial; therefore, percentages may total greater than 100%.
Comparison of Agreement with Audit Review
The computer algorithm had a significantly higher rate of agreement with the audit review, compared with that of traditional surveillance (computer algorithm, κ [95% confidence interval (CI)] = 0.58 [0.52–0.64]; traditional surveillance, κ [95% CI] = 0.44 [0.37–0.51]); P = .001; Figure 2).
FIGURE 2.
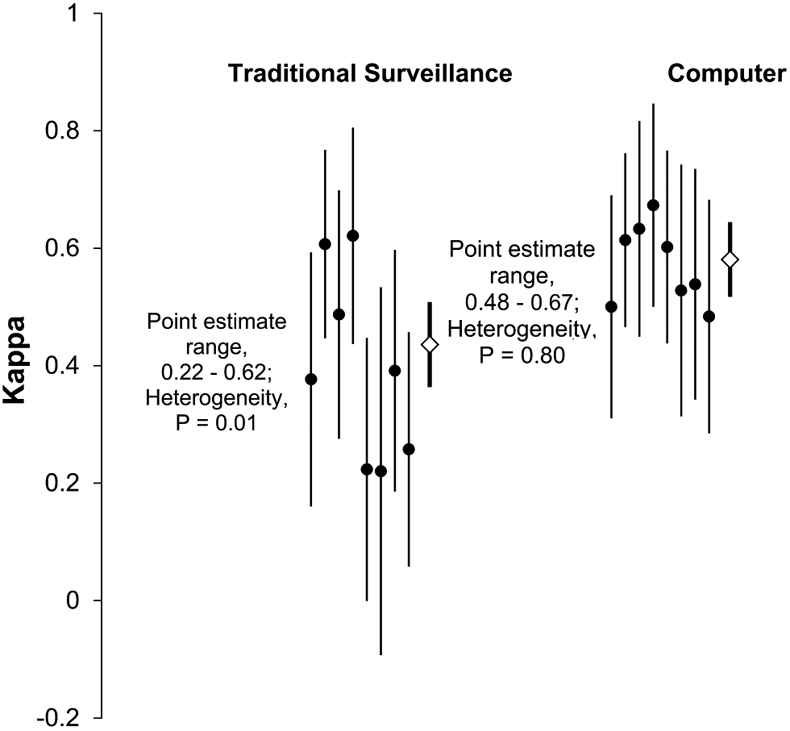
Distribution of κ estimates for traditional surveillance and computer algorithm surveillance, both compared with audit review. Solid circles represent κ point estimates of individual intensive care units. Diamonds represent summary point estimates. Vertical lines represent 95% confidence intervals of the κ estimates.
Comparison of Variation in Performance
The consistency of central line–associated BSI agreement with the audit review was better for the computer algorithm compared to the traditional surveillance determinations (P = .04 for difference in κ heterogeneity). There was significant heterogeneity among the κ values for the traditional surveillance/audit review (range, 0.22–0.62; P = .01), whereas κ values for the computer algorithm/audit review were homogeneous (range, 0.48–0.67; P = .80; Figure 2).
Relationship between Performance (Audit Agreement) and Central Line–Associated BSI Rates
For traditional surveillance, we found a dependent relationship between performance (agreement with audit review) and central line–associated BSI rates (linear regression, slope = 0.16; r2 = 0.77; P = .001; Figure 3A). ICUs with lower traditional central line–associated BSI rates were associated with poorer agreement with the audit review; conversely, ICUs with higher traditional central line–associated BSI rates had better agreement with audit review.
FIGURE 3.
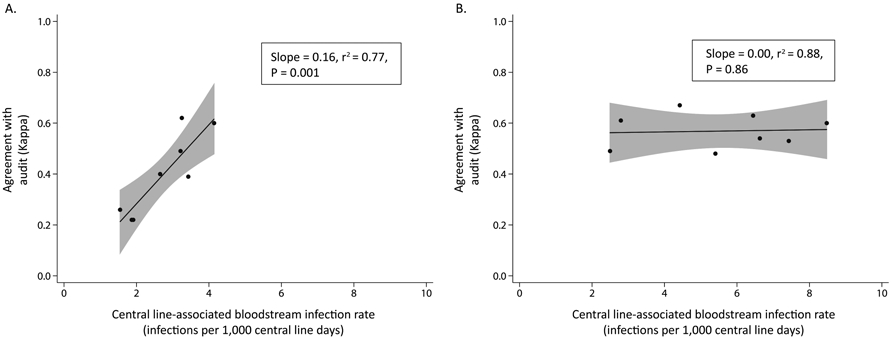
Relationship between performance of central line–associated bloodstream infection (CLABSI) surveillance (κ) and CLABSI rates at intensive care units, by surveillance method. Shaded regions represent the 95% confidence intervals around the fitted lines. A, Traditional surveillance. B, Computer algorithm surveillance.
In contrast, the computer algorithm performance was stable and independent of the algorithm-defined central line–associated BSI rate for ICUs (slope = 0.00; r2 = 0.88; P = .86; Figure 3B). The slopes of the respective linear models for traditional surveillance and computer algorithm surveillance (Figure 3) were significantly different (P = .001).
DISCUSSION
In this multicenter study, we found that computer algorithm detection of central line–associated BSI performed better than traditional surveillance. With retrospective audit as a comparator, the computer algorithm had better overall agreement across ICUs. Importantly for interfacility benchmarking and public reporting, computer algorithmic detection resulted in a consistent level of agreement with the audit review across ICUs, independent of the algorithmic central line–associated BSI rate reported. For traditional surveillance, we found inconsistent performance, as evidenced by variation in traditional surveillance agreement with the audit review across ICUs. Of particular concern, ICUs with lower (more favorable) traditional central line–associated BSI rates had worse agreement with the audit review, indicating that lower central line–associated BSI rates may be explained at least in part by variability in surveillance rather than simply differences in infection rates.
Evidence of significant facility-level variability in central line–associated BSI surveillance has been previously described in an ecologic study by Lin et al9 comparing ICU-level BSI surveillance rates against an objective reference standard. A subsequent study by Mayer et al7 of traditional central line–associated BSI surveillance found that, when multiple infection preventionists reviewed identical patient records, only 55% uniformly agreed with each other (overall κ = 0.42). Furthermore, with respect to reviewing identical sets of patient records, infection preventionists reported a wide range of central line–associated BSI rates (14%–39% of records reviewed), whereas a laboratory-based definition mimicking the current computer algorithm detected consistent central line–associated BSI rates (range, 36%–42%). Our current study, which confirms in a multifacility setting that traditional central line–associated BSI surveillance performs inconsistently when compared with audit review, fills a critical knowledge gap by demonstrating that computer algorithm surveillance performs more accurately and reliably across facilities.
Potential sources of variation in traditional surveillance have been recognized. First, the central line–associated BSI surveillance definition used during the study period (as well as currently) inherently requires some subjective judgment, particularly with respect to whether the primary source of a positive blood culture is from a vascular or extravascular infection site.14 Second, hospitals vary in their access to infection preventionist staffing, expertise, or informatics resources, which can lead to differences in surveillance intensity.15,16 Third, as hospitals enforce greater scrutiny over central line–associated BSI rates, the threshold for classifying a true central line–associated BSI may be raised for infection preventionists as well as for physicians or quality committees who may be internally involved in reviewing individual BSI cases.17-19 Efforts to decrease variation in surveillance practice and improve sensitivity of central line–associated BSI detection have focused on the use of retrospective audits, which are increasingly employed through state and territorial health departments.10,20 Such audits have found variable sensitivity in central line–associated BSI reporting.18,20,21 Continued validation efforts may increase the accuracy of traditional surveillance; however, such efforts are costly, resource intensive, and difficult to sustain broadly.
For purposes of public reporting or performance measurement, computer algorithms can provide an efficient and reliable alternative to traditional surveillance of central line–associated BSIs, especially as hospital data becomes increasingly accessible electronically. Despite lacking subjective human judgment, this study and others suggest that use of computer algorithms does not degrade accuracy.13 Importantly, a simulation study demonstrated that, even if accuracy is compromised through use of objective criteria, the enhanced reliability results in more accurate rankings of hospital-specific infection rates.12
There are practical barriers to implementing computer algorithms for infection surveillance. First, in this study and in others,9,22 computer algorithm–derived infection rates are generally higher than traditional surveillance infection rates; thus, meaningful interinstitution comparisons of rates would require that institutions be compared with others using the same surveillance strategy. Second, computer algorithm surveillance requires that hospitals store medical information, such as microbiology laboratory results, in a manner available for computer analysis; this is currently not universally available. Third, like any diagnostic test method, the positive predictive value of the algorithms will decrease as the true disease prevalence is reduced. To assess generalizability, the performance of computer algorithm surveillance would need to be evaluated over a broader range of ICUs with varying degrees of BSI burden.
Our study has several limitations. First, audit review at each institution was performed internally by a blinded individual from the same institution. Although the audit review was enhanced by a prereview training session and provision of a step-wise surveillance worksheet to adhere to NHSN surveillance standards, it is possible that the individual shared similar judgment patterns with colleagues performing in the traditional surveillance arm. Such a bias would have favored agreement between traditional surveillance and audit review, yet we found agreement that favored computer algorithm surveillance over traditional surveillance. Second, audit review, like computer algorithm surveillance, was performed retrospectively and thus limited to documented medical information, whereas traditional surveillance was performed prospectively and potentially enhanced through incorporation of unwritten clinical information obtained through discussion with clinical staff. However, NHSN surveillance criteria do not explicitly incorporate clinician opinion into BSI surveillance decision-making, and such information may be a source of variability or bias.16,23 Lastly, we were not able to qualitatively assess reasons for disagreement between traditional surveillance and audit review, because for the traditional surveillance review, we had no retained record of the decision making process.
Computer algorithm surveillance can improve the reliability and accuracy of central line–associated BSI surveillance, enhancing the credibility of one of the most important measures of hospital-acquired infection. Furthermore, increased automation of hospital infection surveillance can favorably shift hospital infection control resources from counting infections to preventing infections. Wide-scale adoption of the computer algorithm in a consistent and verifiable manner across reporting sites will be feasible in the future as clinical data become standardized.11,24 Healthcare and public health leaders should continue to support capacity building and further evaluation of automated methods of healthcare-associated infection detection.
ACKNOWLEDGMENTS
We thank Onofre Donceras, Edward Goodwin, Lisa Hines, and Amy Richmond for their roles as audit reviewers.
Financial support.
This study was funded by Centers for Disease Control and Prevention (CDC) cooperative agreements U01-CI000327, U01-CI000328, and U01-CI000333 from the CDC Prevention Epicenter Grants.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention or the Agency for Toxic Substances and Disease Registry.
Footnotes
Potential conflicts of interest. All authors report no conflicts of interest relevant to this article. All authors submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest, and the conflicts that the editors consider relevant to this article are disclosed here.
Presented in part: Fifth Decennial International Conference on Healthcare-Associated Infections; Atlanta, Georgia; March 18–22, 2010 (Abstract 80).
REFERENCES
- 1.Pronovost P, Needham D, Berenholtz S, et al. An intervention to decrease catheter-related bloodstream infections in the ICU. N Engl J Med 2006;355(26):2725–2732. [DOI] [PubMed] [Google Scholar]
- 2.Klevens RM, Edwards JR, Richards CL Jr, et al. Estimating health care-associated infections and deaths in U.S. hospitals, 2002. Public Health Rep 2007;122(2):160–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kahn KL, Weinberg DA, Leuschner KJ, Gall EM, Siegel S, Mendel P. The national response for preventing healthcare-associated infections: data and monitoring. Med Care 2014;52(2 suppl 1):S25–S32. [DOI] [PubMed] [Google Scholar]
- 4.Centers for Medicare and Medicaid Services (CMS) Media Relations (press release). CMS gives consumers access to more details about infection rates at America’s hospitals. Baltimore, MD: CMS, 2012. http://www.cms.gov/Newsroom/MediaReleaseDatabase/Press-releases/2012-Press-releases-items/2012-02-07.html. Accessed February 25, 2014. [Google Scholar]
- 5.Medicare program; hospital inpatient prospective payment systems for acute care hospitals and the long-term care hospital prospective payment system and fiscal year 2014 rates; quality reporting requirements for specific providers; hospital conditions of participation; payment policies related to patient status. Fed Regist 2013;78:50495. [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. National Healthcare Safety Network (NHSN) Manual, Patient Safety Component Protocol. Atlanta, GA: Division of Healthcare Quality Promotion, Centers for Disease Control and Prevention, 2008:13. [Google Scholar]
- 7.Mayer J, Greene T, Howell J, et al. Agreement in classifying bloodstream infections among multiple reviewers conducting surveillance. Clin Infect Dis 2012;55(3):364–370. [DOI] [PubMed] [Google Scholar]
- 8.McBryde ES, Brett J, Russo PL, Worth LJ, Bull AL, Richards MJ. Validation of statewide surveillance system data on central line–associated bloodstream infection in intensive care units in Australia. Infect Control Hosp Epidemiol 2009;30(11):1045–1049. [DOI] [PubMed] [Google Scholar]
- 9.Lin MY, Hota B, Khan YM, et al. Quality of traditional surveillance for public reporting of nosocomial bloodstream infection rates. JAMA 2010;304(18):2035–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Centers for Disease Control and Prevention. National Healthcare Safety Network (NHSN) validation guidance and toolkit 2012: validation for central line–associated bloodstream infection (CLABSI) in ICUs. http://www.cdc.gov/nhsn/toolkit/validation-clabsi/index.html. Accessed February 25, 2014.
- 11.Hota B, Lin M, Doherty JA, et al. Formulation of a model for automating infection surveillance: algorithmic detection of central-line associated bloodstream infection. J Am Med Inform Assoc 2010;17(1):42–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rubin MA, Mayer J, Greene T, et al. An agent-based model for evaluating surveillance methods for catheter-related bloodstream infection. AMIA Annu Symp Proc 2008:631–635. [PMC free article] [PubMed] [Google Scholar]
- 13.Trick WE, Zagorski BM, Tokars JI, et al. Computer algorithms to detect bloodstream infections. Emerg Infect Dis 2004;10(9):1612–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Worth LJ, Brett J, Bull AL, McBryde ES, Russo PL, Richards MJ. Impact of revising the National Nosocomial Infection Surveillance System definition for catheter-related bloodstream infection in ICU: reproducibility of the National Healthcare Safety Network case definition in an Australian cohort of infection control professionals. Am J Infect Control 2009;37(8):643–648. [DOI] [PubMed] [Google Scholar]
- 15.Stone PW, Dick A, Pogorzelska M, Horan TC, Furuya EY, Larson E. Staffing and structure of infection prevention and control programs. Am J Infect Control 2009;37(5):351–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dixon-Woods M, Leslie M, Bion J, Tarrant C. What counts? an ethnographic study of infection data reported to a patient safety program. Milbank Q 2012;90(3):548–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fraser TG, Gordon SM. CLABSI rates in immunocompromised patients: a valuable patient centered outcome? Clin Infect Dis 2011;52(12):1446–1450. [DOI] [PubMed] [Google Scholar]
- 18.Backman LA, Melchreit R, Rodriguez R. Validation of the surveillance and reporting of central line–associated bloodstream infection data to a state health department. Am J Infect Control 2010;38(10):832–838. [DOI] [PubMed] [Google Scholar]
- 19.Sexton DJ, Chen LF, Anderson DJ. Current definitions of central line–associated bloodstream infection: is the emperor wearing clothes? Infect Control Hosp Epidemiol 2010;31(12):1286–1289. [DOI] [PubMed] [Google Scholar]
- 20.Fridkin SK, Olmsted RN. Meaningful measure of performance: a foundation built on valid, reproducible findings from surveillance of health care-associated infections. Am J Infect Control 2011;39(2):87–90. [DOI] [PubMed] [Google Scholar]
- 21.Kainer M, Mitchell J, Frost B, Soe M. Validation of central line associated bloodstream Infection data submitted to the National Healthcare Safety Network—a pilot study by the Tennessee Department of Health. In: Program and abstracts of Fifth Decennial International Conference on Healthcare-Associated Infections. Atlanta, GA: Centers for Disease Control and Prevention, 2010. [Google Scholar]
- 22.Woeltje K, McMullen K, Butler AM, Goris A, Doherty J. Electronic surveillance for healthcare-associated central line–associated bloodstream infections outside the intensive care unit. Infect Control Hospital Epidemiol 2011;32(11):1086–1090. [DOI] [PubMed] [Google Scholar]
- 23.Grove WM, Zald DH, Lebow BS, Snitz BE, Nelson C. Clinical versus mechanical prediction: a meta-analysis. Psychol Assess 2000;12:19–30. [PubMed] [Google Scholar]
- 24.Wright MO, Fisher A, John M, Reynolds K, Peterson LR, Robicsek A. The electronic medical record as a tool for infection surveillance: successful automation of device-days. Am J Infect Control 2009;37(5):364–370. [DOI] [PubMed] [Google Scholar]
- 25.National Healthcare Safety Network. Central line–associated bloodstream infection (CLABSI) event. 2013. http://www.cdc.gov/nhsn/PDFs/pscManual/4PSC_CLABScurrent.pdf. Accessed May 6, 2013. [Google Scholar]


