Abstract
The three common alleles of the APOE gene, ɛ2/ɛ3/ɛ4, have been linked to human spatial orientation. We investigated the genetic role of APOE in developmental topographical disorientation (DTD), a lifelong condition that results in topographical disorientation. We genotyped the APOE ɛ2/ɛ3/ɛ4 alleles in a cohort of 20 unrelated DTD probands, and found allele frequencies not statistically different from the those seen in the population as a whole. Therefore, we found no evidence that DTD occurs preferentially on a genetic background containing any particular APOE allele, making it unlikely that these APOE alleles are contributing to the development of DTD.
Keywords: Cognition disorders, genotype, memory, navigation, spatial learning
INTRODUCTION
The gene APOE codes for the lipid-binding protein apolipoprotein E (ApoE), which binds to cholesterol and other lipids, and transports them between various cell and tissue types. ApoE is known to be involved in a wide range of processes, including lipoprotein metabolism, immunoregulation, and, importantly, cognition [1–3]. ApoE is associated with Alzheimer disease (AD), representing the main genetic factor determining individuals’ risk of occurrence of AD [2, 4–8]. There are three common isoforms of ApoE (ApoE2, ApoE3, and ApoE4), corresponding to three alleles of APOE (ɛ2, ɛ3, ɛ4), as defined by two single nucleotide polymorphisms at positions 2059 and 2179, affecting the amino acids encoded at positions 112 and 158 of the ApoE protein (Table 1; [3, 9, 10]). These polymorphisms affect the structure and function of the ApoE protein, with the different isoforms exhibiting altered lipid association and receptor binding characteristics [9]. E3 is the isoform most commonly seen in the population, while the E4 and E2 isoforms are associated with an increased and decreased risk for AD, respectively [5] (see [2] for a comprehensive review of APOE/ApoE).
Table 1.
Details of APOE alleles and corresponding ApoE isoforms
| APOEallele | DNA base at position 2059 of APOE | DNA base at position 2179 of APOE | ApoE Isoform | Amino acid at position 112 of ApoE | Amino acid at position 158 of ApoE | Frequency in Caucasian controls [5] |
| ɛ2 | T | T | E2 | Cys | Cys | 8.4% |
| ɛ3 | T | C | E3 | Cys | Arg | 77.9% |
| ɛ4 | C | C | E4 | Arg | Arg | 13.7% |
One important characteristic of AD is that patients get lost in familiar surroundings [11]. Such impairment is distinct from the general memory issue that is hallmark of the disease [12] and is present in nearly all AD patients from early on in the disease course [13]. This peculiar behavioral defect in AD patients raised the issue of specific effects that the E2/E3/E4 ApoE isoforms may have on the ability of the individuals to orient and navigate in the environment. Indeed, it has been shown that the E4 allele has an effect on brain activation during memory tasks in healthy adults [14], as well as on spatial memory behavior in healthy children, decades before AD might occur [15]. In searching for early indicators and mechanisms of AD’s neurocognitive pathology in young adults carrying the ɛ4 APOE allele (individuals who are therefore at risk of developing AD later in life), Kunz and colleagues [16] found evidence of a dysfunction of grid cells in the entorhinal cortex, a cell type well-known to be involved in the representation of a spatial layout and especially critical for effective orientation and navigation [17]. Specifically, in the ɛ4 carriers, the authors observed a reduction of grid-cell-like representations while undergoing a functional magnetic resonance imaging (fMRI) study, and a declined performance in spatial orientation while navigating a virtual arena. These authors also found that both of these changes were related to impaired spatial memory performance, although, as a group, the ɛ4 carriers did not exhibit reduced spatial memory when compared to participants homozygous for the ɛ3 allele.
In a different study, Konishi and colleagues also provided evidence of a significant link between the ApoE isoforms and spatial orientation, in which navigational behavior and functional and structural brain properties appeared to be influenced by APOE genotype in healthy young adults [18, 19]. Specifically, these authors found that ɛ2 carriers favored a navigation strategy dependent on the hippocampus (i.e., a spatial over a non-spatial/response strategy), and had increased gray matter volume of the hippocampus, as compared to ɛ4 carriers or participants homozygous for ɛ3, i.e., non-ɛ2 and non-ɛ4 carriers [18]. Later, in comparing ɛ4 carriers who favored a spatial navigation strategy (ɛ4-spatial) to ɛ4 carriers who favored a non-spatial navigation strategy (ɛ4-response), the authors found that ɛ4-response participants had a decreased gray matter volume compared to non-ɛ4 carriers, whereas ɛ4-spatial participants did not (i.e., their gray matter volumes were comparable to non-ɛ4 carriers). In addition, ɛ4-response participants had a decreased blood-oxygen-level-dependent signal (i.e., the indirect fMRI measure of neural activity) when compared to ɛ4-spatial participants [19]. Taken together, these studies suggest that the APOE gene might be a critical genetic determinant of spatial orientation abilities in the healthy population as well as among AD patients. Here, we sought to investigate the APOE ɛ2/ɛ3/ɛ4 genotypes in a group of individuals who suffer from a lifelong inability to orient and navigate in familiar surroundings, that is, individuals affected by developmental topographical disorientation (DTD) [20].
DTD refers to a highly-selective lifelong impairment in the ability of an individual to find their way around in extremely familiar surroundings such as the neighborhood where they live, the building where they have worked for many years, or in extreme cases, their own house [20–25]. This disability, which can be devastating, is present despite the fact that these individuals have well-preserved cognitive functions, no other neurological impairments and no brain injuries [26–28]. This behavioral defect appears most commonly to result from the inability of the individuals to form cognitive maps [21, 24, 26, 29]. A cognitive map is a mental representation of the environment including the landmarks within it and their spatial relationships and is formed as individuals familiarize themselves with their spatial surroundings, enabling them to reach any location from anywhere within the environment [30, 31]. Individuals affected by DTD are unable to form cognitive maps, and therefore never gain the ability to orient within a given environment, no matter how long they live in it [29]. Importantly, there is evidence found in a cohort of individuals with DTD of familial clustering of this trait, suggesting that genes might play a role in the development of DTD [20, 32].
The evidence that the three common APOE alleles may play a role in determining spatial orientation ability marks them as potential candidates for genetic variants contributing to the occurrence of DTD. Specifically, DTD may be more common among ɛ4 carriers, given the evidence of their grid cell dysfunction [16] and increased risk of developing AD. Also, DTD may be less common in ɛ2 carriers, given the evidence of a more likely use of a hippocampal effective spatial navigation strategy [18]. To test these hypotheses, we genotyped the common APOE alleles in a cohort of DTD individuals and their families.
MATERIALS AND METHODS
Participants
The cohort included 20 unrelated DTD probands, and 22 relatives of those probands (both with and without the DTD trait; see Table 2). Given that we reported participants’ APOE genotype status, which could pose a risk of harm in terms of worry and emotional distress, if such information were delivered without appropriate consent and counselling, it was important to mitigate the risk of participants identifying themselves. Therefore, age and sex are not reported. Since the outcome measure in our study is genotype—something participants were born with—age and sex does not affect the interpretation of results. The probands had either been part of one of two previous studies on DTD [24, 26], or were self-referred, having contacted us after hearing about DTD in the media or online. Each proband described getting lost in environments that most people would consider to be very familiar, such as the neighborhood where they grew up and/or had lived for many years, or their workplace, even after having worked there for several years. For all participants included in the study (probands and relatives), the DTD phenotype was confirmed using the online assessment adopted in a previous study [32], evaluating the four key inclusion criteria for DTD diagnosis [24]. The inclusion criteria consist of: (a) getting lost frequently in very familiar surroundings, (b) experiencing orientation difficulties consistently from childhood or adolescence (i.e., the stage at which one would expect an individual to begin independently navigating), (c) having no other cognitive complaints, and (d) having suffered no brain injuries or neurological conditions. In some cases, an interview was used in place of the online assessment to evaluate these same criteria. In addition, most participants completed the Cognitive Map Formation Test [32–36], which assesses an individual’s ability to form cognitive maps [35, 37] and which has been used in the past to confirm that individuals with DTD are indeed unable to form cognitive maps [20, 26, 36]. In our sample, 18 of the 20 probands failed to solve the Cognitive Map Formation Test in the 20 trials provided. One did not complete the test because it caused them to experience motion sickness, while another was eventually able to solve the test after 18 attempts (while the mean number of attempts required for non-DTD individuals is less than 10 [33, 34]). The study was approved by the local Ethics Research Board.
Table 2.
The cohort of unrelated DTD probands and their 22 relatives
| Family/Proband Number | Relationship to Proband* | DTD Phenotype | APOE genotype |
| 1 | Proband | DTD | ɛ2/ɛ3 |
| 2 | Proband | DTD | ɛ4/ɛ4 |
| Mother | DTD | ɛ3/ɛ4 | |
| Father | Not DTD | ɛ3/ɛ4 | |
| Sister | Not DTD | ɛ3/ɛ4 | |
| Daughter | Not DTD | ɛ3/ɛ4 | |
| Son | Not DTD | ɛ3/ɛ4 | |
| Son | Not DTD | ɛ3/ɛ4 | |
| 3 | Proband | DTD | ɛ2/ɛ3 |
| Daughter | DTD | ɛ2/ɛ4 | |
| Daughter | Not DTD | ɛ2/ɛ3 | |
| 4 | Proband | DTD | ɛ3/ɛ3 |
| Brother | Not DTD | ɛ2/ɛ3 | |
| Sister | Not DTD | ɛ2/ɛ3 | |
| Paternal First Cousin | DTD | ɛ3/ɛ3 | |
| Husband | Not DTD | ɛ3/ɛ3 | |
| Daughter | Not DTD | ɛ3/ɛ3 | |
| Daughter | DTD | ɛ3/ɛ3 | |
| 5 | Proband | DTD | ɛ3/ɛ3 |
| 6 | Proband | DTD | ɛ3/ɛ3 |
| 7 | Proband | DTD | ɛ3/ɛ4 |
| 8 | Proband | DTD | ɛ3/ɛ3 |
| 9 | Proband | DTD | ɛ3/ɛ3 |
| Sister | DTD | ɛ3/ɛ3 | |
| 10 | Proband | DTD | ɛ3/ɛ3 |
| Brother | DTD | ɛ3/ɛ3 | |
| Brother | Not DTD | ɛ3/ɛ3 | |
| Son | Not DTD | ɛ3/ɛ3 | |
| 11 | Proband | DTD | ɛ3/ɛ3 |
| 12 | Proband | DTD | ɛ3/ɛ3 |
| 13 | Proband | DTD | ɛ3/ɛ3 |
| 14 | Proband | DTD | ɛ3/ɛ3 |
| 15 | Proband | DTD | ɛ3/ɛ3 |
| 16 | Proband | DTD | ɛ3/ɛ3 |
| 17 | Proband | DTD | ɛ3/ɛ3 |
| Mother | DTD | ɛ3/ɛ3 | |
| Brother | Not DTD | ɛ3/ɛ3 | |
| Sister | DTD | ɛ3/ɛ3 | |
| Son | Not DTD | ɛ3/ɛ3 | |
| 18 | Proband | DTD | ɛ3/ɛ3 |
| 19 | Proband | DTD | ɛ3/ɛ3 |
| 20 | Proband | DTD | ɛ3/ɛ3 |
*Participant age and sex were not included so as to limit the potential for participants to be identified.
ApoE isoforms
To determine the particular ApoE isoforms present in individuals with DTD, we genotyped using a PCR method which detects the sequence variants at nucleotide positions 2059 and 2079 of APOE (Table 1), which are present in allele combinations to form haplotypes that correspond to the three common ApoE isoforms (ɛ2/ɛ3/ɛ4), as previously described [38].
The allele frequencies in the DTD cohort were compared to the allele frequencies observed in a large group of Caucasian controls (without dementia), as well as that expected in AD [2, 5], to determine whether or not any allele occurred more or less often in the DTD group than would be expected in a random sampling of individuals from the larger population. Statistical significance of allele frequency differences was evaluated using a chi-squared analysis as implemented in JASP Version 0.14.3. When possible, we also looked for co-segregation of the APOE alleles with the DTD trait within family members.
RESULTS
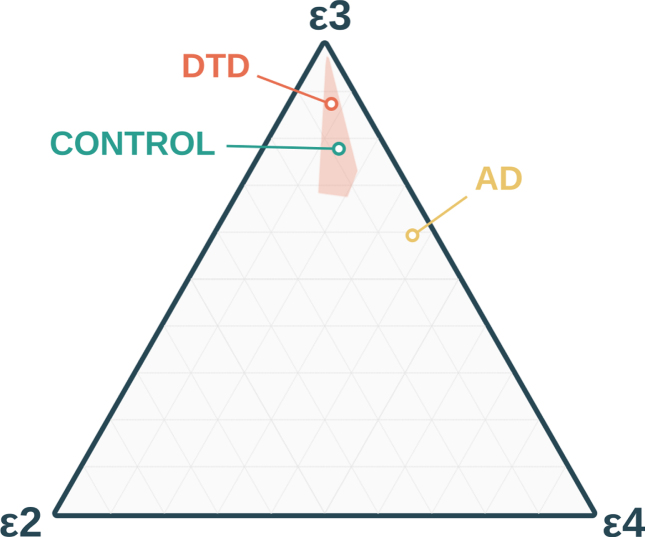
Among the 20 unrelated DTD probands, we observed 16 individuals with the ɛ3/ɛ3 genotype; two with ɛ2/ɛ3; one with ɛ3/ɛ4; and one with ɛ4/ɛ4 genotypes (Table 2). This pattern of allele frequencies did not appear to differ from that expected in the general population (χ2(2) = 2.340, p = 0.309), but were distinct from that expected in an AD population (χ2(2) = 15.642, p < 0.001), see Table 3 and Fig. 1. Additionally, when the less common ɛ2 and ɛ4 alleles were observed within a family, they did not co-segregate with either the DTD or non-DTD phenotype (Table 2).
Table 3.
APOE ɛ2/ɛ3/ɛ4 allele frequencies in DTD cohort and reference populations
Fig. 1.
Allele proportions of the present developmental topographical disorientation (DTD) cohort, as well as reference proportions from Caucasian control and Alzheimer’s disease (AD) populations [5]. The shaded area about the DTD point estimate represents a 95%confidence interval, estimated from independent binomial distributions.
DISCUSSION
To investigate whether APOE genetics was involved in the development of DTD, we determined the ɛ2/ɛ3/ɛ4 genotype in 20 unrelated individuals with DTD and found that the allele frequencies in this DTD group were similar to those seen in the general population (Table 3). These results indicate that the three major APOE isoforms do not determine DTD status, suggesting that if there is a genetic component to DTD, this would involve some other gene(s). Therefore, while evidence exists that APOE is involved in the development of spatial orientation abilities [16, 18, 19], other proteins/genes are likely involved as well. Our findings show that DTD can occur against a genetic background containing any of the three ɛ2/ɛ3/ɛ4 alleles. In other words, individuals with any of the three major APOE isoforms can have a total lack of developed spatial orientation abilities. Additionally, in an exome sequencing study of nine of these DTD probands, we did not identify rare protein-altering variants in APOE. Therefore, APOE cannot be the single major genetic determinant of spatial orientation abilities in humans. This is not at all surprising since most gene products work together with several (or many) other gene products in complex interaction networks in order to carry out their biological functions. This would especially be expected when looking at such a complex function as spatial orientation, which is already known to involve several brain regions and several cognitive processes. If a major genetic determinant of DTD is discovered in the future, it may be important to identify whether or not it interacts with APOE in any way.
Another way to investigate a possible relationship between APOE genetics and DTD would be to document the incidence of DTD and the ɛ2/ɛ3/ɛ4 genotype in a large population of individuals. In the future, such a large-scale study could eventually allow for a more definitive statement about whether ɛ2/ɛ3/ɛ4 genotype contributes to DTD risk. Indeed, such a large-scale study would allow for the investigation of all potential genetic contributors to the development of DTD. Here, in a small study of a DTD population, we have seen that the pattern of ɛ2/ɛ3/ɛ4 genotypes does not appear to differ from what would be expected in any population of this size, indicating, preliminarily, that ɛ2/ɛ3/ɛ4 genotype is at least not the major genetic determinant of DTD.
Interestingly, among individuals with the ɛ4 allele, brain changes appeared to be linked to navigation strategy [19]. ɛ4 carriers who used a non-spatial strategy were more likely to have the brain changes expected to be indicative of AD risk [19]. Individuals with DTD, being impaired in their ability to form cognitive maps, must rely on a non-spatial (e.g., a response) strategy to navigate. Therefore, future studies may focus on investigating the frequency of developing AD in DTD individuals, as well as the use of navigational strategies aimed at improving the spatial orientation skills in those with DTD, which may protect against AD-type brain changes in ɛ4 carriers. It would be critical to answer these questions in order to start defining the relationship between AD and the presence of a lifelong condition such as DTD.
Our study was limited by the sample size. Indeed, even if no DTD probands had the ɛ2 allele, our sample size would not be big enough to say that the ɛ2 allele frequency was significantly lower in our cohort than in the general population. However, our results show a similar pattern of APOE alleles in the DTD group as is seen in the general population, and, as such, any effect, if present, is going to be subtle. Therefore, future studies in a larger DTD cohort will be beneficial to continue exploring the genetics of the DTD trait, and will reveal whether or not such a subtle effect does indeed exist.
ACKNOWLEDGMENTS
We express our most sincere gratitude to all of the families that have participated in this study. This study was supported in part by the Natural Sciences and Engineering Research Council of Canada (NSERC) to GI, and the Canadian Institutes of Health Research (CIHR) to NTBH. SFB was partially supported by the Alberta Children’s Hospital Research Institute (ACHRI)/CIHR Training Program. FB was supported by NSERC.
CONFLICT OF INTEREST
The authors have no conflict of interest to report.
REFERENCES
- [1].Bu G (2009) Apolipoprotein E and its receptors in Alzheimer’s disease: Pathways, pathogenesis and therapy. Nat Rev Neurosci 10, 333–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Liu C-C, Kanekiyo T, Xu H, Bu G (2013) Apolipoprotein E and Alzheimer disease: Risk, mechanisms and therapy. Nat Rev Neurol 9, 106–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Mahley R (1988) Apolipoprotein E: Cholesterol transport protein with expanding role in cell biology. Science 240, 622–630. [DOI] [PubMed] [Google Scholar]
- [4].Corder E, Saunders A, Strittmatter W, Schmechel D, Gaskell P, Small G, Roses A, Haines J, Pericak-Vance M (1993) Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science 261, 921–923. [DOI] [PubMed] [Google Scholar]
- [5].Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, Myers RH, Pericak-Vance MA, Risch N, van Duijn CM (1997) Effects of age, sex, and ethnicity on the association between Apolipoprotein E genotype and Alzheimer disease: A meta-analysis. JAMA 278, 1349–1356. [PubMed] [Google Scholar]
- [6].Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, Jones N, Thomas C, Stretton A, Morgan AR, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Morgan K, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Love S, Kehoe PG, Hardy J, Mead S, Fox N, Rossor M, Collinge J, Maier W, Jessen F, Schürmann B, Heun R, Wiltfang J, Dichgans M, Frölich L, Hampel H, Hüll M, Rujescu D, Goate AM, Kauwe JSK, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, Deyn PPD, Broeckhoven CV, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Mühleisen TW, Nöthen MM, Moebus S, Jöckel K-H, Klopp N, Wichmann H-E, Carrasquillo MM, Pankratz VS, Younkin SG, Holmans PA, O’Donovan M, Owen MJ, Williams J (2009) Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet 41, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lambert J-C, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B, Letenneur L, Bettens K, Berr C, Pasquier F, Fiévet N, Barberger-Gateau P, Engelborghs S, De Deyn P, Mateo I, Franck A, Helisalmi S, Porcellini E, Hanon O, the European Alzheimer’s Disease Initiative Investigators, de Pancorbo MM, Lendon C, Dufouil C, Jaillard C, Leveillard T, Alvarez V, Bosco P, Mancuso M, Panza F, Nacmias B, Bossú P, Piccardi P, Annoni G, Seripa D, Galimberti D, Hannequin D, Licastro F, Soininen H, Ritchie K, Blanché H, Dartigues J-F, Tzourio C, Gut I, Van Broeckhoven C, Alpérovitch A, Lathrop M, Amouyel P (2009) Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat Genet 41, 1094–1099. [DOI] [PubMed] [Google Scholar]
- [8].Strittmatter WJ, Saunders, Ann M., Schmechel, Donald, Pericak-Vance M, Enghild J, Salvesen GS, Roses AD (1993) Apolipoprotein E: High-avidity binding to β-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci U S A 90, 1977–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hatters DM, Peters-Libeu CA, Weisgraber KH (2006) Apolipoprotein E structure: Insights into function. Trends Biochem Sci 31, 445–454. [DOI] [PubMed] [Google Scholar]
- [10].Weisgraber KH, Rall SC, Mahley RM (1981) Human E apoprotein heterogeneity. Cysteine-arginine interchanges in the amino acid sequence of the apo-E isoforms. J Biol Chem 256, 9077–9083. [PubMed] [Google Scholar]
- [11].Liu L, Gauthier L, Gauthier S (1991) Spatial disorientation in persons with early senile dementia of the Alzheimer type. Am J Occup Ther 45, 67–74. [DOI] [PubMed] [Google Scholar]
- [12].Vlček K, Laczó J (2014) Neural correlates of spatial navigation changes in mild cognitive impairment and Alzheimer’s disease. Front Behav Neurosci 8, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Monacelli AM, Cushman LA, Kavcic V, Duffy CJ (2003) Spatial disorientation in Alzheimer’s disease: The remembrance of things passed. Neurology 61, 1491–1497. [DOI] [PubMed] [Google Scholar]
- [14].Bookheimer SY, Strojwas MH, Cohen MS, Saunders AM, Pericak-Vance MA, Mazziotta JC, Small GW (2000) Patterns of brain activation in people at risk for Alzheimer’s disease. N Engl J Med 343, 450–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Acevedo SF, Piper BJ, Craytor MJ, Benice TS, Raber J (2010) Apolipoprotein E4 and sex affect neurobehavioral performance in primary school children. Pediatr Res 67, 293–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kunz L, Schroder TN, Lee H, Montag C, Lachmann B, Sariyska R, Reuter M, Stirnberg R, Stocker T, Messing-Floeter PC, Fell J, Doeller CF, Axmacher N (2015) Reduced grid-cell-like representations in adults at genetic risk for Alzheimer’s disease. Science 350, 430–433. [DOI] [PubMed] [Google Scholar]
- [17].Rowland DC, Roudi Y, Moser M-B, Moser EI (2016) Ten years of grid cells. Annu Rev Neurosci 39, 19–40. [DOI] [PubMed] [Google Scholar]
- [18].Konishi K, Bhat V, Banner H, Poirier J, Joober R, Bohbot VD (2016) APOE2 is associated with spatial navigational strategies and increased gray matter in the hippocampus. Front Hum Neurosci 10, 349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Konishi K, Joober R, Poirier J, MacDonald K, Chakravarty M, Patel R, Breitner J, Bohbot VD (2018) Healthy versus entorhinal cortical atrophy identification in asymptomatic APOE4 carriers at risk for Alzheimer’s disease. J Alzheimers Dis 61, 1493–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Burles F, Iaria G (2020) Behavioural and cognitive mechanisms of developmental topographical disorientation. Sci Rep 10, 20932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Bianchini F, Incoccia C, Palermo L, Piccardi L, Zompanti L, Sabatini U, Peran P, Guariglia C (2010) Developmental topographical disorientation in a healthy subject. Neuropsychologia 48, 1563–1573. [DOI] [PubMed] [Google Scholar]
- [22].Bianchini F, Palermo L, Piccardi L, Incoccia C, Nemmi F, Sabatini U, Guariglia C (2014) Where am I? A new case of developmental topographical disorientation. J Neuropsychol 8, 107–124. [DOI] [PubMed] [Google Scholar]
- [23].Conson M, Bianchini F, Quarantelli M, Boccia M, Salzano S, Di Vita A, Guariglia C (2018) Selective map-following navigation deficit: A new case of developmental topographical disorientation. J Clin Exp Neuropsychol 40, 940–950. [DOI] [PubMed] [Google Scholar]
- [24].Iaria G, Barton JJS (2010) Developmental topographical disorientation: A newly discovered cognitive disorder. Exp Brain Res 206, 189–196. [DOI] [PubMed] [Google Scholar]
- [25].Palermo L, Foti F, Ferlazzo F, Guariglia C, Petrosini L (2014) I find my way in a maze but not in my own territory! Navigational processing in developmental topographical disorientation. Neuropsychology 28, 135–146. [DOI] [PubMed] [Google Scholar]
- [26].Iaria G, Arnold AEGF, Burles F, Liu I, Slone E, Barclay SF, Bech-Hansen TN, Levy RM (2014) Developmental topographical disorientation and decreased hippocampal functional connectivity. Hippocampus 24, 1364–1374. [DOI] [PubMed] [Google Scholar]
- [27].Kim JG, Aminoff EM, Kastner S, Behrmann M (2015) A neural basis for developmental topographic disorientation. J Neurosci 35, 12954–12969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Nemmi F, Bianchini F, Piras F, Péran P, Palermo L, Piccardi L, Sabatini U, Guariglia C (2015) Finding my own way: An fMRI single case study of a subject with developmental topographical disorientation. Neurocase 21, 573–583. [DOI] [PubMed] [Google Scholar]
- [29].Iaria G, Burles F (2016) Developmental topographical disorientation. Trends Cogn Sci 20, 720–722. [DOI] [PubMed] [Google Scholar]
- [30].Epstein RA, Patai EZ, Julian JB, Spiers HJ (2017) The cognitive map in humans: Spatial navigation and beyond. Nat Neurosci 20, 1504–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Tolman EC (1948) Cognitive maps in rats and men. Psychol Rev 55, 189. [DOI] [PubMed] [Google Scholar]
- [32].Barclay SF, Burles F, Potocki K, Rancourt KM, Nicolson ML, Bech-Hansen NT, Iaria G (2016) Familial aggregation in developmental topographical disorientation (DTD). Cogn Neuropsychol 33, 388–397. [DOI] [PubMed] [Google Scholar]
- [33].Arnold AEGF, Burles F, Krivoruchko T, Liu I, Rey CD, Levy RM, Iaria G (2013) Cognitive mapping in humans and its relationship to other orientation skills. Exp Brain Res 224, 359–372. [DOI] [PubMed] [Google Scholar]
- [34].Burles F, Guadagni V, Hoey F, Arnold AEGF, Levy RM, O’Neill T, Iaria G (2014) Neuroticism and self-evaluation measures are related to the ability to form cognitive maps critical for spatial orientation. Behav Brain Res 271, 154–159. [DOI] [PubMed] [Google Scholar]
- [35].Iaria G, Chen J-K, Guariglia C, Ptito A, Petrides M (2007) Retrosplenial and hippocampal brain regions in human navigation: Complementary functional contributions to the formation and use of cognitive maps. Eur J Neurosci 25, 890–899. [DOI] [PubMed] [Google Scholar]
- [36].Iaria G, Bogod N, Fox CJ, Barton JJS (2009) Developmental topographical disorientation: Case one. Neuropsychologia 47, 30–40. [DOI] [PubMed] [Google Scholar]
- [37].Iaria G, Palermo L, Committeri G, Barton JJS (2009) Age differences in the formation and use of cognitive maps. Behav Brain Res 196, 187–191. [DOI] [PubMed] [Google Scholar]
- [38].Pantelidis P, Lambert-Hammill M, Wierzbicki AS (2003) Simple sequence-specific-primer-PCR method to identify the three main apolipoprotein E haplotypes. Clin Chem 49, 1945–1948. [DOI] [PubMed] [Google Scholar]
- [39].Barclay SF (2017) Using Next-Generation Sequencing to Identify Genes Mutated in Human Disorders (Unpub- lished doctoral thesis). University of Calgary, Calgary, AB. doi: 10.11575/PRISM/28740 [DOI]