Abstract
Summary
PRRT is a receptor-targeted radiation-based therapy for NETs. The key challenges in its deployment are prediction of efficacy and toxicity, patient selection and response optimization. This manuscript reviews the molecular profile of NETs and the strategies and tools used to predict, monitor and assess PRRT toxicity. The few tumor gene mutations that can be evaluated (such as ATM/DAXX) are limited to pancreatic NETs. Transcriptomic and gene-based assays are effective in the prediction of radiotherapy response in other cancers. A blood-based 8 gene assay – the PPQ – has an overall accuracy of 95% for predicting responses to PRRT in NETs. There are currently no molecular markers that predict PRRT toxicity. Candidate molecular targets include 7 radiation-susceptibility SNPs. Blood transcriptomic evaluations and a combination of gene expression and specific SNPs, assessed by machine learning with tumor-specific algorithms, may yield molecular tools to enhance PRRT efficacy and safety.
Research strategy and selection criteria
A review of the literature based on the recommendations of systematic reviews and meta-analyses outlined by PRISMA and by the Cochrane Diagnostic Test Accuracy Working Group was undertaken (December 2019) using MEDLINE (PubMed.gov).
We identified 203 studies of which 7 had appropriate data for evaluation. All included appropriate data to assess whether molecular profiling was useful in PRRT, with a focus on prediction or response and/or toxicity. The final reference list was generated on the basis of relevance to the broad scope of this review.
Introduction
Background of PRRT:
Peptide receptor radiotherapy (PRRT) is a therapeutic strategy for metastatic or non-resectable neuroendocrine tumors (NETs) that involves systemic administration of a radiolabeled octreotide derivative which preferentially targets the neoplastic expression of somatostatin receptors (SSTR). The clinical benefit of PRRT in the NETTER 1 trial1 led to approval of 177Lu-DOTATATE for the treatment of well-differentiated gastroenteropancreatic (GEP) NETs. NETTER 1 was undertaken in well-differentiated G1 and G2 midgut NETs, that were metastatic or locally advanced and exhibited disease progression at entry. PRRT exhibited a marked superiority versus high dose somatostatin analogues (SSA) consistent with the importance of using an appropriate targeted and biologically active therapy.
Efficacy and toxicity:
Patient selection is based on criteria such as tumor histology (histological/cytological confirmation of NET), imaging (PET/CT with 68Ga-labeled somatostatin analogs [68Ga-SSA-PET] demonstrating tumor receptor binding) and safety measures (hematological, hepatic and renal function)2,3. FDG-PET can serve as a prognostic parameter but is not included as a stratification factor2,3.
PRRT is generally administered over 6–8 months and usually comprises 4 cycles of 177Lu-DOTATATE (25–30GBq). Some centers use more individualized approaches: either a mix of radio-ligands (177Lu- and 90Y-peptides), addition of chemotherapy (chemo-radiotherapy) or different treatment cycles and administered activity. 177Lu-labelled peptides are often preferred due to better tolerance than 90Y-peptides. Therapeutic efficacy defined as “disease control” includes disease stabilization, partial or complete responses2,3. Evaluation post-therapy is based on comparisons with pre-therapy imaging (CT/MRI and 68Ga-SSA-PET) 2,3.
Although PRRT prolongs progression-free survival (PFS), approximately 15–30% of patients will exhibit disease progression during therapy, and another 10–15% will progress early (6 months to 1 year) after treatment1–4. Although generally well-tolerated, PRRT may be associated with adverse events (e.g., for 177Lu-DOTATATE: subacute hematological toxicity (~10% G3-G4), myelodysplastic syndrome (2–4%) and renal failure (<0.4% grade 4)5.
Two areas are critical for future optimization of PRRT. First, better patient selection and pretreatment stratification. This requires robust predictive markers of response. Risk-based strategies, including PRRT combinations, targeted drugs, liver embolization, or chemotherapy, would be the result. Moreover, better methods are required for the prediction and early identification of toxicity, particularly myelotoxicity.
Outcome variables:
Treatment response represents the balance between the intrinsic aggressiveness of the tumor, the efficacy of the agent, medical status, immune response and adverse events. Several factors therefore influence outcome. Some, like grade or extent, provide “prognostic” information and cannot predict therapeutic responsiveness. Predictive features identify the likelihood of responding favorably to a medical intervention, irrespective of prognostic factors. Predictive features are usually derived from clinical trials that compare treatment to a control arm in subjects with and without the biomarker. Unfortunately, these factors are sometimes confused and prognostic factors are often erroneously considered predictive.
Precision medicine and tumor molecular profiling:
In the emerging era of precision medicine, criteria to assure drug efficacy and patient safety are critical. A balanced evaluation of cost-benefit ratios of high value therapies is also of utmost importance.
Increasingly, molecular biomarkers for predictive disease modeling and patient stratification are emerging, driven by the realization that genomic information facilitates understanding the architecture of disease. For instance, scoring systems or nomograms are used to assess risk in prostate cancer (https://www.mdcalc.com/ucsf-capra-score-prostate-cancer-risk), (https://www.mskcc.org/nomograms/prostate) and are FDA-approved molecular tools. Neoplasia including uveal melanoma (gene expression score risk stratification/imaging-treatment)6, breast (MammaPrint/treatment-decisions7), colon (prognostic subtyping8) and lung cancer (subtyping, treatment stratification)9 are successful applications of molecular profiling tools. These are also being developed for predicting radiation response (e.g., radio-sensitivity index [RSI]10) or radio-toxicity (e.g., radio-pathogenic SNPs in prostate cancer11).
The molecular characterization of NETs has lagged behind. Recent molecular observations in small bowel, pancreatic and bronchopulmonary NETs have pushed the development of effective targeted therapy12–15. In particular, strategies that can define the molecular genomic status of a tumor and predict its susceptibility to PRRT have been developed.
This manuscript reviews current NET molecular literature that could be used to develop predictive tools for response and toxicity. We focus on providing context for individualized molecular profiling in PRRT (Appendix Figure 1, Table 1) and examine molecular signatures and other radiation therapies (e.g., external beam radiation) to identify whether these provide relevant information for NETs. In addition, clinical parameters e.g., grade, are evaluated to contrast with the utility of molecular profile data. Overall, we explore how current practice can be augmented by the use of novel genomic tools to optimize radionuclide therapy in NETs.
Table 1.
Relevant PRRT-related studies – molecular-based profiling, prognosis and prediction
| Category | Radiotherapy | Parameters | Type of Response | Type of study | Patient number | Primary Site | Refs |
|---|---|---|---|---|---|---|---|
| Prognostic | 177 Lu-PRRT | Various clinical parameters including SSR uptake | PFS | P, two-arm | 221 | Midgut | 1 |
| Prognostic | None | FDG-glucose uptake (imaging) | PFS/OS | NHx | 100 | All NET sites | 51 |
| Prognostic | 177 Lu-PRRT | Various including CgA, Ki-67 etc | PFS/OS | R | 74 | GEP-NET | 55 |
| Predictive | 177 Lu-PRRT/ 90 Y-PRRT, mixed, and chemotherapy | PPQ, transcriptomics, grade, CgA, SSR etc | PFS | P, single arm* | 54 | All NET sites | 61 |
| Predictive | 177 Lu-PRRT/ 90 Y-PRRT, mixed, and chemotherapy | PPQ, transcriptomics, grade, CgA, SSR etc | PFS | P, single arm* | 158 | All NET sites | 62 |
| Interventional | 177 Lu-PRRT/ 90 Y-PRRT, mixed, and chemotherapy | NETest, CgA | PFS | P, single arm | 122 | All NET sites | 60 |
| Toxicity | 177 Lu-PRRT/ 90 Y-PRRT, mixed, and chemotherapy | Various clinical and pathobiological parameters | PFS/OS | R | 807 | All NET sites | 5 |
NHx = natural history (follow-up); (P), prospective study; (R), Retrospective study.
comparisons were made with separate study cohorts of SSA treatment and long-term follow-up
Results and Context
Radiobiology of radiation and PRRT:
From a radiobiological perspective, PRRT “kills” cells based on the absorbed dose, the type of radiation (e.g. beta-particles from 177Lu or 90Y, versus alpha-particles from 225Ac) and the intrinsic tumor radio-sensitivity16. Tumor doses that induce significant tumor shrinkage range from 10–340Gy using 177Lu-DOTATATE17.
Radio-sensitivity varies with the cell cycle; it is highest during mitosis and lowest during S-phase. This was recognized in 1906 when Bergonie and Tribondeau enunciated their “law” (“X-rays are more effective on cells which have a greater reproductive activity”)18. Radio-resistance conversely relates to lack of a substantial population of dividing tumor cells.
PRRT, like other radiotherapy, relies on radiation-induced DNA damage and suboptimal repair. Radioactive particles emitted by intracellularly trapped radio-peptides target DNA both directly and indirectly, through genotoxic oxidative stress, and induce single (SSB) or double-strand (DSB) breaks leading to apoptosis. Ionizing radiation from beta-emitters (Lu-177) principally induces SSB.
Several studies identify that radiation damage, irrespective of source (e.g., alpha-, beta-emitters) results in similar transcriptomic and pathway alterations19,20. Animal models (mice bearing human small intestinal NETs [GOT1] receiving 177Lu-DOTATATE) recapitulate these events (DNA damage/repair, proliferation, cell-cycle arrest, oxidative stress, apoptosis)21. Cellular radio-sensitivity is associated with defined processes and dysregulation of hundreds of genes (Table 2, Figure 1). A number of radiation-sensitizing and resistance gene assays have thus been developed in certain malignancies e.g., breast, esophageal adenocarcinoma22,23.
Table 2.
Genomic factors related to radiation responsiveness
| Proliferation | Ploidy/S-phase studies |
|---|---|
| Genomic | Chromosomal abnormalities e.g., chromosomal loss or amplification, copy number variation (CNV), or translocations |
| Mutations | Tumor burden, specific targeted mutations e.g., TP53 or ATM, or gene-fusions |
| GWAS | Single nucleotide polymorphisms (SNPs) |
| Transcription | Transcriptomics e.g., whole exome, mRNA, miRNA, lncRNA |
| Epigenome | Chromosomal modifications (transcriptome-targeting, or structural) e.g., methylation, acetylation |
GWAS = genome wide association studies, lncRNA = long non-coding RNA, mRNA = messenger RNA, miRNA = microRNA
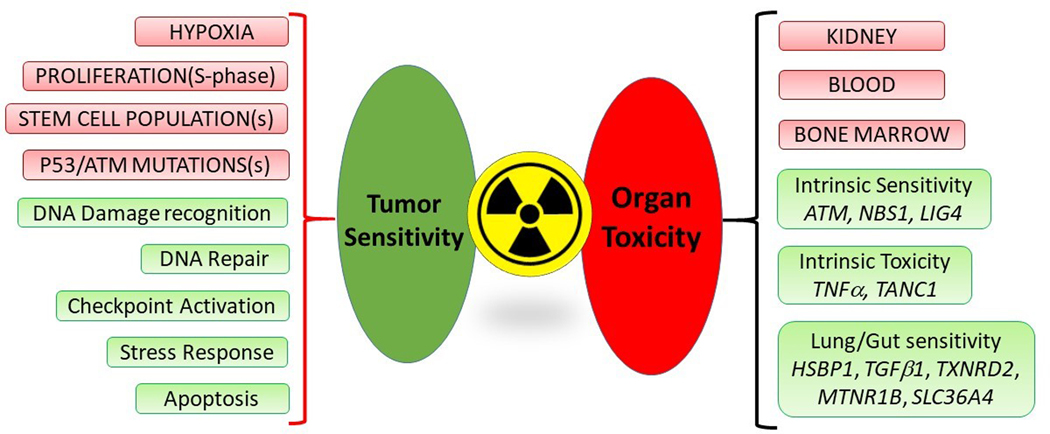
Figure 1. Radiation sensitivity and toxicity: pathobiological features, pathways and candidate genes.
Tumor features relevant to radiation sensitivity (pink – top left): include proliferation, hypoxia, “stemness” and mutations particularly in TP53 and the ATM genes. Molecular profiling studies have identified pathways common to radiation responsiveness (green – bottom left). These include DNA damage recognition and repair; senescence induction and apoptosis.
Radiation toxicity affects the kidneys, individual blood cell populations e.g., platelets, white cells and the bone marrow (pink – top right). Candidate factors related to toxicity (green – bottom right) include intrinsic sensitivity to radiation which may be related to SNPs or mutations in DNA damage/repair pathways including ATM, NBS1 and LIG4. A variety of genes validated as radiation-sensitive genes in other cancers have been identified as relevant to PRRT lung and gut sensitivity, that as they are. Furthermore, intrinsic radiation toxicity has been found for TNFα and TANC1 genes. None of these candidate genes have been evaluated for PRRT.
After radiation exposure, cells attempt to maintain their genetic integrity. This includes inhibition of progression from G1 to S-phase, activation of cell-cycle checkpoints (cell cycle arrest in G1, S and G2) and DNA repair and activation of transcription factors (P53 and NF-kB [nuclear factor-kappa B]). P53 is critical for radio-sensitivity: it is mutated in more than 50% of human cancers and loss of activity (mutation or phosphorylation) reduces radio-sensitivity24. DNA damage response is regulated by the PI3K kinase family: ATM (ataxia telangiectasia mutated), DNA-PK (DNA-dependent protein kinase), and ATR (Rad3-related kinase). Tumor oxygenation status is critical for efficient cell kill and hypoxia leads to radio-resistance. Hypoxia can also induce selection of clones resistant to apoptosis. This results in growth of tumor cells with expanded angiogenic potential, stem-like features and radio-resistance. Effective tumor cell kill results in decreasing tumor size, which is clinically recorded as “partial or complete response”. Re-growth depends on surviving cancer stem cells and the biological and genomic features related to radio-resistance.
The majority (80–90%) of well-differentiated NETs have a low proliferation rate and TP53 mutations are rare (<5%). In regard to hypoxia, GEP-NET hepatic metastases frequently exhibit areas of hypodensity on CT-imaging, which likely represent central tumor necrosis secondary to hypoxia, very little in general is known about “stemness”; the relevance of cancer stem cells in NETs is unclear. The four major areas (proliferation, TP53/ATM mutations, hypoxia, stemness) related to radiation-response in other tumors either suggest a radio-resistant phenotype or are poorly understood in NETs.
The NET molecular landscape:
DNA-ploidy evaluation studies of the NET genomic landscape were undertaken in the 1980s (Table 2). This was followed by more sophisticated, high-level analyses of DNA and its epigenome. Although no clear driver mutations were identified (especially for non-pancreatic NETs), a number of genomic aberrations and modifications related to malignancy and possibly be associated with response to therapy have been identified.
Ploidy/S-phase studies:
The low proliferative activity of NETs was noted in 1985, when 100% of intestinal carcinoid cases and 90% of pancreatic NETs were reported to exhibit diploid DNA and <2% S-phase nuclei25. The majority (87%) of rectal NETs are also diploid26. In bronchopulmonary NETs, diploidy was identified in 68% of typical carcinoids while aneuploidy was a feature of atypical histology27. Aneuploidy is a feature of malignancy and it is a prognostic marker; however, its relevance as a predictive marker in NETs remains unknown.
Chromosomal abnormalities:
In small intestinal NETs, segmental losses of chromosome 18 occur in ~ 78%28. Recent high-coverage target sequencing of 52 sporadic tumors identified allelic loss of BCL2, CDH19, DCC and SMAD4 (all chromosome 18) in 44%28. In pancreatic NETs, frequent losses of 1q, 3p (including VHL gene locus) and 11q (MEN1 and ATM gene loci) occurs and losses of 6q, 10q (PTEN locus) and 11p and gains on 7q and 9q have been noted29. In bronchopulmonary NETs, loss of 11q (including MEN1) is common (52%) as is loss of 10q and 13q (~30–40%)30.
Mutation studies:
NETs are mutationally “quiet” tumors with ~0.1 variants/106 nucleotides (mostly DNA base transitions – unlikely to affect protein activity)31. In small intestinal NETs, mutations in CDKN1B (P27, linked to MEN4; typically loss-of-function, truncating mutations) are identified in 8–10%32. Other mutations include APC (7.7%), CDKN2C (7.7%), BRAF, KRAS, PIK3CA and TP53 (3.8% each)28. Germline mutations in MUTYH33 and IPMK34 have also been identified in a few families.
In contrast, several recurrent mutations have been recognized in pancreatic NETs. Approximately 35–50% harbor MEN1 mutations35, typically truncating mutations involved in chromatin remodeling. ATRX (10%), SETD2 and MLL3 (both 5%) may also be inactivated13. Mutations in DAXX (an apoptotic regulator) occur in ~20%. DNA damage repair deficiencies occur in 11% manifested as mutations in MUTYH, CHEK2 and BRCA2 (both involved in homologous recombination). Additionally, inactivating mutations of negative regulators in the AKT/mTOR pathway have been noted in 10–12%.
In bronchopulmonary NETs, mutually exclusive mutations of histone covalent modifiers have been identified in ~40%15. These include mutations in MEN1 (9–11%) and TP53 (10%), eukaryotic translation initiation factor 1A, X-linked (EIF1AX) (9%), lysine methyltransferase 2C gene (KMT2C or MLL3) (8%), and AT-rich interaction domain 1 (ARID1A) (6–7%), mutations in the SETD family, in histone demethylases as well as in ATRP-dependent chromatin remodeling (SWI/SNF complex)15. Overall, chromatin remodeling is the pathway most affected (~50%) in these tumors36.
Transcriptomics:
Expression profiling has identified 2 subtypes of small intestinal NETs: serotonin- or mixed-amine production12 with distinctive gene expression profiles versus pancreatic neoplasia37. Global microRNA profiling of small intestinal and pancreatic NETs revealed no overlapping expression. Selected mRNA may be prognostic38.
Pancreatic NET gene expression has defined 2 subtypes that correlate with benign versus malignant behavior37. Somatostatin receptor 2 (SST2) mRNA is absent or low in insulinomas39. MicroRNA dysregulation including upregulation of mir-103 and mir-10740, and overexpression of mir-21 (proliferation/liver metastases) is noted40. Non-functional tumors fall into two major subtypes based on epigenomic and transcriptomic signatures (ARX-positive versus PDX1-positive)14; those that are ARX+PDX1− are almost exclusively associated with distant relapse, suggesting this was a prognostic molecular marker for this tumor subgroup.
Bronchopulmonary NETs comprise three molecular subgroups41. Aggressive disease and poor outcome is related to mutations in MEN1, loss of OTP and NKX2 and upregulation of ANGPTL3, ERBB4 (HER4) and UGT gene expression41. Thyroid transcription factor 1 (TTF1) is associated with outcome to PRRT but this is likely prognostic not predictive42.
A further application of gene expression data is circulating tumor marker signatures. The latter have been shown as clinically useful in diagnosis and have added value as liquid biopsies that provide direct measurements of NET proliferation, metabolism, epigenetic regulation, growth factor regulation and metastatic pathway signaling43.
Epigenetic regulation:
Significant epigenetic changes have been identified in small intestinal NETs. Differential promoter methylation of RASSF1A and CTNNB1 occur in metastatic tumors and increased methylation of TP73, CHFR and RUNX3 are reported44. Global hypomethylation is increased in liver metastases which exhibit increased expression of PI3K, ERBB1, PDGFRβ and mTOR signaling pathway components45. Genome methylation studies identified hyper-methylation of RASSF1A, CDKN2A and VHL genes and/or their promoter regions, and hypomethylation of ALU and LINE1 in pancreatic NETs46. DNA hypomethylation is associated with DAXX/ATRX loss and chromosomal instability47. In lung NETs, RASSF1 promoter hypermethylation and abnormal methylation of the p15INK4b gene have also been identified48.
Prediction of Tumor Response to PRRT
A number of parameters are used to predict response.
Dose of PRRT:
The relationship between the tumor-absorbed dose and RECIST 1.1 response has been prospectively demonstrated for metastatic pancreatic NETs (177Lu-DOTATATE). Tumor doses ranged from 10–340Gy with a Pearson correlation co-efficient with tumor reduction of 0.64 for tumors measuring 2.2–4cm and 0.91 for tumors >4cm in diameter17. A similar relationship was not demonstrable for small-intestinal metastases49. Dose-effect relationships for renal and bone marrow toxicity have been demonstrated in numerous studies. Nevertheless, a dose threshold is difficult to define suggesting individual patient sensitivity, possibly genomic5.
Utility of clinical evaluation and current tools:
Several clinical parameters have been studied. These include grade, extent, functional status, primary location and baseline metabolism (FDG PET/CT). None accurately predict response in a particular patient5. These factors are prognostic, not predictive (Figure 2, Table 3). The NETTER 1 study evaluated several factors as predictive markers1, none of which were associated with significant outcomes and the calculated hazard ratios were similar across all subgroups.Histological grade and Ki67 are “prognostic” markers but accuracy is limited by random biopsy, tumor heterogeneity, and inter-observer variability. Tissue biopsies are obtained from one site and do not represent disease status since metastatic biology differs from the primary and evolves over time. Ki67has e limited value in predicting PRRT. The NETTER 1 study demonstrated no value of G1 vs. G2 tumors for predicting PRRT1. Sorbye et al., recently evaluated PRRT in G3 tumors50. Although efficacy was evident, the mPFS (11–16 months) is considerably lower than G1/G2 tumors e.g., NETTER 1 (mPFS not reached). This suggests that proliferation index alone is not a useful marker for predicting PRRT and that other biological factors, as yet unidentified, determine the radio-sensitivity of NETs.
Figure 2. Clinical Factors commonly used to “predict” PRRT response.
A panoply of clinical factors has been evaluated as “predictors” of PRRT response. Factors that predict clinical outcomes and survival have been misperceived as predictors of PRRT response. Thus, performance status, primary location, and disease extent are all prognostic and unrelated to the prediction of PRRT response. Similarly, glucose-based metabolism (FDG-positive) and tumor grade are also prognostic features. Circulating monoanalyte biomarkers, if elevated, reflect tumor burden, and are prognostic. Other parameters such as somatostatin receptor imaging intensity or IHC SSTR expression provide evidence of target existence and properly are “inclusion” factors. They cannot predict PRRT response but represent indices of target acquisition likelihood.
CgA = chromogranin A; IHC = immunohistochemistry; KPS = Karnofsky performance score; NSE = neuron-specific enolase; ORR = objective response rate; OS = overall survival; PFS = progression free survival; SSTR = somatostatin receptor; IHC= immunohistochemical
(Figure adapted from Bodei et al. EJNMMI 2018; 45(7): 1155–69).
Table 3.
NET factors evaluated in PRRT studies
| FACTORS ASSOCIATED WITH A RESPONSE TO PRRT | ||||||
|---|---|---|---|---|---|---|
| Parameters | Criteria | Type of Response | Type of study | Patient number | Primary Site | Refs |
| Clinical | Reduced performance status | ORR | R | 310 | GEP-NETs | 2,75 |
| Extensive liver disease | TTP | R | 310 | GEP-NETs | 2 | |
| Bone metastases | TTP | R | 310 | GEP-NETs | 2 | |
| Functionality | TTP PFS |
R R |
310 61 |
GEP-NETs SI-NETs |
2
76 |
|
| Pathological | Ki-67 | PFS | R | 68 | P-NETs | 55 |
| Primary tumor site | ORR | R | 310 | GEP-NETs | 2 | |
| Radiation Absorbed Dose | Dose correlation with response | |||||
| P-NETs: demonstrated | ORR | P | 24 | P-NETs | 17 | |
| SI-NETs: no correlation (activity correlated) | ORR | P | 25 | SI-NETs | 49 | |
| Early per-cycle reduction in tumor dose | ORR | R | 27 | GEP-NETs | 77 | |
| PET/SPECT | Image-based heterogeneity | |||||
| SSR heterogeneity (asphericity) at pre-PRRT SPECT c/w poor response | ORR | R | 20 | GEP-NETs | 78 | |
| Textural features (entropy) at pre-PRRT SSR PET | PFS | R | 141 | NETs | 54 | |
| Intensity of uptake at SRI | ||||||
| Baseline OctreoScan uptake | ORR | R | 310 | GEP-NETs | 2 | |
| Baseline SUV max and SUV max/av at PET | ORR | R | 55 | GEP-NETs | 79 | |
| Early per-cycle reduction of SUV T/S , SUV max at PET | TTP | R | 31 | NETs | 80 | |
| Intensity of uptake at FDG PET | ||||||
| FDG positive lesions | PFS | R | 60 | P-NETs | 81 | |
| Discordant FDG and SRI lesions | ORR | R | 50 | GEP-NETs | 82 | |
| CT | TGR at baseline (e.g. >0.33%/month) | ORR | R | 39 | GEP-NETs | 83 |
| MR | DCE-MRI-derived pretreatment signal enhancement ratio at 40% to 60% radial distance | ORR | A | - | Human SI-NET cell line GOT1 | 21 |
| Mono/bi-analyte Biomarkers | CgA>600 ng/ml | PFS PFS |
R R |
22 61 |
BP-NETs SI-NETs |
75
76 |
| Inflammation-based index (serum C-reactive protein and albumin) | PFS PFS |
R P (failed correlation) |
55 43 |
NETs, PPGL NETs |
84
85 |
|
| Genomic multianalyte biomarkers | PPQ | PFS | P | 158 | GEP, BP-NETs | 62 |
(R), Retrospective study; (P), prospective study; (A), animal study; c/w, correlated with; SRI, somatostatin receptor imaging, either OctreoScan or 68Ga-SSA-PET/CT; TTP, time-to-progression.; TGR, tumor growth ratio; DCE, dynamic contrast-enhanced; PPQ, PRRT Predictive Quotient.
Extensive tumor load and reduced performance status (KPS ≤70) are associated with shorter PFS2. These are both prognostic factors. FDG avidity is also associated with shorter PFS (177Lu-DOTATATE), is a determinant of a poorer PFS irrespective of treatment modality51.
Tumor origin may have some relevance. Small intestine tumors, especially associated with the carcinoid syndrome (CS), have a lower response rate compared to certain pancreatic NETs e.g., gastrinoma4. However, there is evidence for symptomatic response in patients with CS and improved quality of life52. The poorer PFS likely reflects hepatic tumor burden. Alternatively, the differences in responses could reflect the generally lower replication/mitosis rates noted in small intestinal NETs.
Imaging biomarkers:
Somatostatin receptor expression intensity (imaging) provides a measure of tumor-absorbed dose. The probability of response, however, is only 60%, even with very intense OctreoScan uptake3. Moreover, the NETTER 1 study identified no significant difference between those with SSR Grade 4 uptake versus SSR<Grade 41. While somatostatin receptor expression is important as an inclusion factor (therapeutic target), it is the biology or molecular genomic characteristics of the tumor that determine sensitivity to PRRT. Recently, in patients with liver metastases, a 68Ga-DOTATOC SUVmax cut-off of 16.5, showed a 95% sensitivity and 60% specificity for response prediction53. The low specificity is problematic indicating that even individuals with a high SUVmax, may not respond to therapy. Tumor to liver SUV ratio >2.2 is a possible alternative but requires further validation.
Imaging evaluation is limited by the subjective nature of assessment. Recently, algorithmic qualitative and quantitative characterization (neural networking/AI) of CT or PET images, has been introduced to correlate tumor phenotype with genotype. These are effective in differentiating benign from malignant lesions, defining grading, and supporting tumor behavior prediction including therapy response. SSR-PET image heterogeneity, based on the textural feature “Entropy”, correlates with both PFS and overall survival (OS) (AUC of 0.60 and 0.70) after PRRT54. These novel strategies may advance the field if they can indeed provide predictive rather than just prognostic information.
Preliminary data in mice with GOT-1 NETs treated with 177Lu-DOTATATE suggest a correlation between pre-treatment DCE (dynamic contrast-enhanced)-MRI signal enhancement ratio at 40–60% radial distance, tumor response and CCD89 expression (a protein implicated in DNA damage repair, proliferation, and cell cycle arrest)21. Changes in diffusion after treatment correlated with response and with CATA, a protein involved in oxidative stress and apoptotic death21.
Current biomarkers:
Circulating tumor markers such as CgA and NSE have been evaluated extensively. Baseline CgA>600ng/ml (i.e., 6xULN) in one 177Lu-DOTATATE series constituted a risk factor for early progression. However, CgA is a surrogate marker for tumor burden, which is prognostic. Similar results were described for NSE; levels>15ng/ml correlated with a shorter OS. This likely indicates prognosis than evidence of PRRT-responsiveness55. The NETTER 1 study confirmed that secretory markers had no utility; there were no differences in outcome between individuals with CgA>2ULN vs <2ULN. Similarly, 5-HIAA (using 2ULN as a cut-off) had no relation to outcome1.
Molecular markers:
A number of molecular markers for PRRT-response prediction have been proposed and include: detection of specific mutations and methylated DNA, identification of chromosomal abnormalities or transcriptional alterations. These could be detected both at a tissue level and in the circulation, if assays were available. Tissue identification is problematic (access difficulty, tumor heterogeneity).
TP53 is not currently used as a predictive marker for radiation therapy. It is better regarded as an “inclusion” factor (i.e. evidence for wild-type TP53), similar to image-based SSR uptake. While TP53 is mutated in up to 50% of other cancers, this gene is mutated in <5% of grade 1 (G1) and grade 2 (G2) NETs, irrespective of anatomical origin. It is unlikely that P53 will have utility as a NET-predictive tool. However, in a high proportion of neuroendocrine carcinomas (60–100%) TP53 is mutated and these may require exclusion or combination therapy if PRRT is considered.
DNA damage repair genes and apoptosis regulation (e.g., pro-apoptotic DAXX) alterations occur in ~30% of pancreatic NETs and have been proposed as molecular targets. However, in animal models, DAXX heterozygosity did not induce sensitization to sub-lethal doses of ionizing radiation56.
ATM pathway alterations may be informative as increased radiation-responsiveness is a well-recognized relationship16. However, gene dysregulation is also associated with radiation toxicity so caution is required in resolving the role of ATM-markers in PRRT.
Chromatin remodeling genes exhibit high level of alterations including MEN1 (~40% lung NETs, 50–60% PNETs). There is a paucity of studies of PRRT in MEN1 syndrome but good palliative responses to 177Lu-DOTATATE have been reported57. Adequately powered, prospective studies are required to determine whether this gene is predictive or prognostic.
Differential methylation patterns are a consistent NET feature. However, their utility as predictive biomarkers has not been examined. Currently, epigenetic markers of radio-sensitivity or resistance have not been determined. Such markers are likely more relevant as prognostic factors.
Chromosomal imbalances and abnormalities are not currently standard tests except in hematological and fetal assays. While large-scale chromosomal instability is a common NET feature, and specific patterns of gain/loss have some prognostic value58, there is no evidence for predictive utility. Chromosomal assessment seems currently unlikely to provide predictive utility.
Transcriptional data is a useful resource to generate biomarkers with either prognostic or predictive value. Such information could be derived from the tumor or as a circulating biomarker. Molecular profiling in other cancers including tissue-based assays (MammaPrint, Oncotype Dx) suggests a viable precedent for assay development in NETs. In terms of radiobiology, a variety of tissue-based marker gene assays have been developed. These are explored in “Other transcriptome-assays”. In NETs, a circulating 51 NET-specific marker gene assay has been reported as an in vitro diagnostic tool. Genes are detectable in the different NET subtypes and the assay functions as a pan-tumor-biomarker. Circulating gene expression is robust, easy to detect and quantify, and is specifically derived from NET tissue and not from blood cell populations e.g. lymphocytes59. The tool is an effective NET therapy monitor, and test score alterations are useful as an interventional biomarker for PRRT60. Recently a predictive liquid biopsy assay for PRRT (PRRT Prediction Quotient or PPQ) has been developed using transcriptomics and grading61,62.
PRRT Prediction Quotient (PPQ):
Molecular Profiling to predict PRRT Efficacy:
Analysis of pre-treatment blood samples from patients undergoing PRRT identified different gene expression patterns in responders versus non-responders61. These reflected genes involved in growth factor signaling (GFS) and metabolism (M). Levels of gene expression (aggregating normalized values of GFS and M mRNA>5.9), were associated with an AUC of 0.74 for predicting lack of progression on PRRT (6 months after therapy). A separate analysis of histological grade indicated that G1/G2 and typical/atypical carcinoids were more likely to achieve stable disease or response (77%) than high grade tumors (50%)61. Two of the four high grade (G3) tumors in this series responded. Integration of gene expression and grading using logistic regression modeling allowed derivation of the PPQ that exhibited an AUC of 0.90 with a binary output of “responder (or PPQ-positive) and non-responder (PPQ-negative). This was associated with a predictive accuracy of 94% for response to PRRT (those achieving disease stabilization or demonstrating a partial response versus individual exhibiting progressive disease at the time of follow-up) in the test cohort.
A significant treatment effect (mPFS not reached) was identified for PPQ biomarker “positive” patients62. Conversely, PPQ-negative patients exhibited an mPFS of 8 months (from start of treatment; HR 36.4, p<0.0001). The sensitivity of the PPQ was 100% for responders with an NPV of 100% (non-responders).
A subsequent prospective evaluation at two independent sites validated the PPQ as an effective predictive biomarker62. Responders (stable disease or partial/complete response) were correctly predicted in 97% and non-responders in 100%. In responders, mPFS was not reached; in non-responders mPFS was 9.7 and 14 months, respectively. The HR was 18–92 (p<0.0001), with sensitivity and NPV of 94–97% and 83–93%62.
It should be noted that biomarkers may have both predictive and prognostic features. The association between a biomarker and outcome, regardless of treatment, should be evaluated. While this should ideally be undertaken in a randomized study, comparing results from single arm studies collected both prospectively and retrospectively are well-described and established approaches in biomarker validation63–65. Disappointingly the NETTER 2 study has determined that it will not include a predictive molecular biomarker in the evaluation of the efficacy of PRRT.
To determine the specificity of the PPQ as a predictive as opposed to prognostic, two additional cohorts (n=128 patients) of non-PRRT treated patients were examined: cohort #1, SSA treatment (n=28) was evaluated and PPQ measured before therapy. No differences were identified between those who had progression (47%) or stability (53%) on follow up at time. The PPQ was therefore not predictive for assessing SSA efficacy alone in the absence of a radioligand. Cohort #2 included 100 patients in a Registry study, none of whom received PRRT. The PPQ was not predictive of PFS and ineffective as a prognostic marker over the 18 months evaluated62.
These studies demonstrate that the PPQ functions specifically as a predictive biomarker for PRRT and correctly detected a radiation sensitivity fingerprint in the blood.
Other transcriptome-based radiation sensitivity assays:
Several gene tissue-based expression assays have been developed to predict radiation sensitivity in some other cancers. Clinically validated assays have been described by Eschrich et al.66 and Kim et al.23.
The signature proposed by Eschrich used a panel of 48 human cancer cell lines to develop a radio-sensitivity index (RSI). This is a 10 gene output which identifies radio-resistance based upon gene expression of the “Signal transduction and stress response signaling” pathway including RELA, IRF1 and JUN. In one study, 95% of breast cancer patients deemed radiosensitive were relapse-free 5-years after therapy compared to 77% predicted to be radiation-resistant22. A glioblastoma study reported 82% of the radiosensitive cohort alive at 12 months compared to 51% of the radiation-resistant group67. The overall predictive accuracy of the RSI ranged from 42%−57%. Those with a radiation-sensitive signature, however, exhibited longer relapse-free survival (95% vs. 75%) or OS (1 year: 84.1% vs. 53.7%, HR: 1.64).
Kim proposed a radio-sensitivity gene signature including 31 genes that captured information in the “Signal transduction and stress response” pathway as well as the “Cell cycle check-point activation” system. In a different study, neither signature accurately predicted radiation response in esophageal cancers suggesting there may be a neoplasia-specific signature68. These authors identified a 4 gene panel (CBR1, PAK2, RAB13 and TWF1) associated with cell adhesion and cellular cross-talk relevant to radiation sensitivity in this cancer. In 31 patients studied, this gene panel differentiated a responsive cohort with an OS not reached from a non-responsive cohort with an OS of ~565 days. The prediction accuracy was 81%.
These data identify that transcriptomic strategies have utility for developing tools to predict benefit from radiation therapy. The PPQ captures both growth factor and metabolomic genes that are specifically related to oxidative stress, metabolism and hypoxic signaling. A direct comparison of the PPQ genes with the Eschrich and Kim signatures identified significant overlap between genes measured in blood and those identified at a tissue level. This demonstrates that the blood PPQ signature captures radio-sensitive information derived from tumor tissue. It may be of interest to assess whether the NET PPQ is effective as a predictive tool in other endocrine and non-endocrine cancers treated with targeted radiation e.g., 131I-Na treated thyroid cancers or prostate cancers targeted by 177Lu-PSMA.
Prediction of PRRT Toxicity
Acute and delayed renal and hematologic toxicity result from exceeding the radiation threshold of individual organ tolerance. PRRT tolerance varies with the absorbed dose to specific organs (Figure 1). This is related both to the dwell-time in an excretory organ e.g., kidney, but also to binding of the radio-ligand to SSTR2 on non-tumor cells, such as lymphatic tissues, spleen, and in the hematological progenitors in the marrow.
Utility of clinical evaluation in the prediction of toxicity:
The role of clinical factors for predicting toxicity in NETs remains unclear. For the kidney, these may include long-standing and poorly controlled hypertension or diabetes. For the bone marrow, toxicity may be related to pre-exposure to alkylating chemotherapeutic agents. A review of 807 patients treated with PRRT using 177Lu, 90Y or a combination thereof with follow-up up to 180 months (1997–2013) from one center was evaluated to identify clinical factors that could be used to predict toxicity5. A series of regression analyses indicated that only 20–27% of severe nephrotoxicity (<2%) was modelled by clinical risk factors, with hypertension (co-efficient 0.14, p<0.0001) and low hemoglobin (co-efficient 0.21, p<0.0001) being the most relevant. Myelodysplastic syndrome occurred in ~2.5% although it was modelled by clinical data in 30%. Platelet toxicity grade (co-efficient 0.14, p=0.01) and increased time after PRRT (>1000 days) were relevant risk factors for MDS. Overall, known clinical risk factors provided a limited (<30%) risk estimate and were unable to predict toxicity in a particular individual. It seems likely that toxicity may represent an individual susceptibility, based upon an intrinsic genomic susceptibility in addition to residual reserve from prior treatments.
Genomic prediction of toxicity:
Tissue or intrinsic radio-sensitivity is substantially influenced by intrinsic factors encoded in our DNA. However, little is known regarding the genetic architecture of radio-sensitivity or the specific genomic variants underlying individual tissue responses to radiation. Currently, several studies e.g., the international, REQUITE study, are ongoing to prospectively determine and validate predictive models and biomarkers for radiotherapy for breast, lung and prostate cancer. Identifying intrinsic susceptibility to radiation exposure is a key unmet need in the field of radiation oncobiology, to reduce severe “unpredictable” long-term effects.
The Terra Incognita of Genomic Toxicity Prediction:
Several genes are involved in response to radiation injury, because homozygous mutations result in unusually severe reactions to radiation therapy. An example is the ATM gene: individuals with ataxia-telangiectasia exhibit an extreme propensity to radiation toxicity69. Other genes (NBS1, LIG4) may be involved in DNA damage response or development of fibrosis and are likewise considered candidate factors for radiation sensitivity/toxicity. However, the incidence of common adverse effects cannot be explained by high penetrant mutations (or genetic heterogeneity/homozygosity) as these occur too infrequently. An individual’s risk therefore is most likely determined by a series of common genetic variants (SNPs). Thousands of such variants may eventually be identified that determine radio-sensitivity.
The international Radio Genomics Consortium (RGC) was established in 2009 to develop assays to predict toxicity. This is a National Cancer Institute/NIH-supported Cancer multi-investigator, multinational Epidemiology Consortium (http://epi.grants.cancer.gov/Consortia/single/rgc.html). Molecular profiling using GWAS identified a number of SNPs associated with radiation injury including the ATM rs1801516 SNP70. In this study of 5456 breast and prostate cancer patients, a significantly increased risk of toxicity was identified for carriers of the minor (Asn) allele with odds ratios ranging from 1.2–1.5. In a separate meta-analysis, SNPs in the XRCC1 gene, specifically the rs25487 Arg399Gln polymorphism, increased the risk of acute radiation-induced side effects (odds ratios 1.29–1.49) but not late radiation-induced effects71. This suggests separate pathways may be pertinent to acute and irreversible toxicity.
Overall, seven SNPs have been confirmed as associated with late effects of radiotherapy. Genes range from TGFβ1 (growth factor signaling) to SLC36A4 (high affinity amino acid transporter) and are associated with toxicity-related endpoints including fibrosis and overall toxicity72. While these have been defined for external radiotherapy, they represent potential candidate factors for PRRT-related toxicity.
An alternative strategy is transcriptomic mining using interactome analysis and “omic cluster” interrogation. Gene expression profiles have been developed and validated for predicting the development of radiation-induced fibrosis in breast cancer73. The tissue expression of 9 genes CDC6, CXCL12, FAP, LMNB2, LUM, MXRA5, SOD2, SOD3, and WISP2, were evaluated in cultured normal fibroblasts and a risk score developed. This index had clinical utility (33% accuracy, 92% specificity) with an odds ratio of 7.373 and provides support for the feasibility of developing a transcriptome-based assay that can predict radiation-induced toxicity.
Despite these advances, the magnitude of the genetic contribution to radiation response likely will exceed our current understanding of individual risk variants and genes involved. Future deployment of mathematical analysis using machine learning methodology and deep neural network analysis may provide additional or complementary insights into myriad genes involved in defining an individual “toxicity-liable” profile for a specific radionuclide treatment.
Summary
Molecular profiling of NETs identifies few predictive markers for PRRT efficacy or toxicity (Figure 3). However, a blood RNA assay has been developed and validated as an accurate predictor of tumor response or stabilization in NETs treated with PRRT. Currently, no tool, exists for PRRT toxicity. We predict that transcriptomic evaluations or a combination of gene expression and specific SNPs, coupled to machine learning and using unique tumor-specific algorithmic constructs74, will yield a viable strategy to predict the safety of isotope therapy.
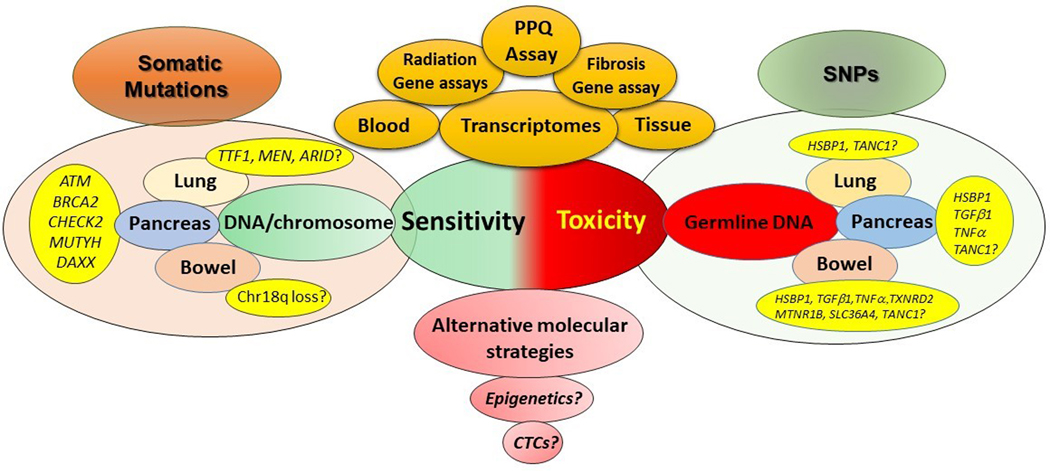
Figure 3. Current and future targets for predicting sensitivity and toxicity.
Sensitivity. PRRT-sensitivity prediction can currently be measured using the blood-based transcriptome assay (PPQ). Different tissue-based multigene radiation-sensitivity assays (10–31 genes) for other cancers have not yet been tested for PRRT. Molecular profiling indicates that somatic mutations of chromatin remodeling genes, DNA damage/repair and apoptosis may be viable targets for evaluation. For lung, this may include TTF1, MEN and ARID genes. In pancreatic NETs a series of mutations in ATM, BRCA2, CHECK2, MUTYH and DAXX might be relevant. Given the absence of mutations in small intestinal NETs, the most likely candidate is chromosome 18q loss.
Toxicity: Germline SNPs in a series of genes that are proven radiation sensitivity intrinsic factors require assessment. Fibrosis can be evaluated using a 9, circulating gene-based fibrosis assay.
Separately, epigenetic or CTC evaluation may provide alternative molecular strategies for either sensitivity or toxicity prediction.
Supplementary Material
Footnotes
Conflicts of interest
LB – grants and non-financial support from AAA-Novartis, non-financial support from Ipsen, non-financial support from Clovis Oncology, non-financial support from Curium.
RPB – Consultancy fees from ITG Isotope Technologies Garching, Ipsen Pharma, Novartis. He is shareholder of Telix Pharma, Clovis Oncology, BAMF Health and consultant/advisor of OctreoPharm Sciences GmbH, Advanced Accelerator Applications, and 1717 LSV.
KH – Consultancy fees from Endocyte, Bayer, Ipsen, AAA, Novartis, BTG, Sirtex, Curium, Amgen, Siemens Healthineers, GE Healthcare, Ymabs. Shareholder of Sofie Biosciences. Non-financial support from ABX. Grant support from BTG.
JS – Consultancy and speaker bureau from Lexicon, Ipsen, and Novartis, outside the submitted work
MC – Consultancy fees and Speaker honoraria from AAA, Ipsen, Novartis, Pfizer (all outside the submitted work).
IMM – Medical and scientific consultant for Wren Laboratories.
The other authors declared no conflicts of interest.
References
- 1.Strosberg J, El-Haddad G, Wolin E, et al. Phase 3 Trial of 177Lu-Dotatate for Midgut Neuroendocrine Tumors. N Engl J Med 2017; 376(2): 125–35. doi: 10.1056/NEJMoa1607427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kwekkeboom DJ, de Herder WW, Kam BL, et al. Treatment with the radiolabeled somatostatin analog [177 Lu-DOTA 0,Tyr3]octreotate: toxicity, efficacy, and survival. J Clin Oncol 2008; 26(13): 2124–30. [DOI] [PubMed] [Google Scholar]
- 3.Kwekkeboom DJ, Kam BL, van Essen M, et al. Somatostatin-receptor-based imaging and therapy of gastroenteropancreatic neuroendocrine tumors. Endocr Relat Cancer 2010; 17(1): R53–73. Print 2010 Mar. [DOI] [PubMed] [Google Scholar]
- 4.Baum RP, Kulkarni HR, Singh A, et al. Results and adverse events of personalized peptide receptor radionuclide therapy with (90)Yttrium and (177)Lutetium in 1048 patients with neuroendocrine neoplasms. Oncotarget 2018; 9(24): 16932–50. doi: 10.8632/oncotarget.24524. eCollection 2018 Mar 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bodei L, Kidd M, Paganelli G, et al. Long-term tolerability of PRRT in 807 patients with neuroendocrine tumours: the value and limitations of clinical factors. Eur J Nucl Med Mol Imaging 2015; 42(1): 5–19. doi: 10.1007/s00259-014-2893-5. Epub 2014 Oct 2. [DOI] [PubMed] [Google Scholar]
- 6.Aaberg TM Jr., Cook RW, Oelschlager K, Maetzold D, Rao PK, Mason JO 3rd. Current clinical practice: differential management of uveal melanoma in the era of molecular tumor analyses. Clin Ophthalmol 2014; 8:2449–60.(doi): 10.2147/OPTH.S70839. eCollection 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cardoso F, van’t Veer LJ, Bogaerts J, et al. 70-Gene Signature as an Aid to Treatment Decisions in Early-Stage Breast Cancer. N Engl J Med 2016; 375(8): 717–29. doi: 10.1056/NEJMoa1602253. [DOI] [PubMed] [Google Scholar]
- 8.Punt CJ, Koopman M, Vermeulen L. From tumour heterogeneity to advances in precision treatment of colorectal cancer. Nat Rev Clin Oncol 2017; 14(4): 235–46. doi: 10.1038/nrclinonc.2016.171. Epub Dec 6. [DOI] [PubMed] [Google Scholar]
- 9.Chen F, Zhang Y, Parra E, et al. Multiplatform-based molecular subtypes of non-small-cell lung cancer. Oncogene 2017; 36(10): 1384–93. doi: 10.038/onc.2016.303. Epub Oct 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eschrich SA, Pramana J, Zhang H, et al. A gene expression model of intrinsic tumor radiosensitivity: prediction of response and prognosis after chemoradiation. Int J Radiat Oncol Biol Phys 2009; 75(2): 489–96. doi: 10.1016/j.ijrobp.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kerns SL, Fachal L, Dorling L, et al. Radiogenomics Consortium Genome-Wide Association Study Meta-analysis of Late Toxicity after Prostate Cancer Radiotherapy. J Natl Cancer Inst 2019; 16(5490201). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kidd M, Modlin IM, Drozdov I. Gene network-based analysis identifies two potential subtypes of small intestinal neuroendocrine tumors. BMC Genomics 2014; 15:595.(doi): 10.1186/471-2164-15-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scarpa A, Chang DK, Nones K, et al. Whole-genome landscape of pancreatic neuroendocrine tumours. Nature 2017; 543(7643): 65–71. doi: 10.1038/nature21063. Epub 2017 Feb 15. [DOI] [PubMed] [Google Scholar]
- 14.Cejas P, Drier Y, Dreijerink KMA, et al. Enhancer signatures stratify and predict outcomes of non-functional pancreatic neuroendocrine tumors. Nat Med 2019; 25(8): 1260–5. doi: 10.038/s41591-019-0493-4. Epub 2019 Jul 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simbolo M, Mafficini A, Sikora KO, et al. Lung neuroendocrine tumours: deep sequencing of the four World Health Organization histotypes reveals chromatin-remodelling genes as major players and a prognostic role for TERT, RB1, MEN1 and KMT2D. J Pathol 2016; 22(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parliament MB, Murray D. Single nucleotide polymorphisms of DNA repair genes as predictors of radioresponse. Semin Radiat Oncol 2010; 20(4): 232–40. doi: 10.1016/j.semradonc.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 17.Ilan E, Sandstrom M, Wassberg C, et al. Dose response of pancreatic neuroendocrine tumors treated with peptide receptor radionuclide therapy using 177Lu-DOTATATE. J Nucl Med 2015; 56(2): 177–82. doi: 10.2967/jnumed.114.148437. Epub 2015 Jan 15. [DOI] [PubMed] [Google Scholar]
- 18.Bergonié J, Tribondeau L. De quelques résultats de la radiotherapie et essai de fixation d’une technique rationelle. Comptes Rendus des Séances de l’Academie des Sciences 1906; 143: 983–5. [Google Scholar]
- 19.Chauhan V, Howland M, Wilkins R. Identification of gene-based responses in human blood cells exposed to alpha particle radiation. BMC Med Genomics 2014; 7:43.(doi): 10.1186/755-8794-7-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edmondson DA, Karski EE, Kohlgruber A, et al. Transcript Analysis for Internal Biodosimetry Using Peripheral Blood from Neuroblastoma Patients Treated with (131)I-mIBG, a Targeted Radionuclide. Radiat Res 2016; 186(3): 235–44. doi: 10.1667/RR14263.1. Epub 2016 Aug 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Montelius M, Spetz J, Jalnefjord O, et al. Identification of Potential MR-Derived Biomarkers for Tumor Tissue Response to (177)Lu-Octreotate Therapy in an Animal Model of Small Intestine Neuroendocrine Tumor. Transl Oncol 2018; 11(2): 193–204. doi: 10.1016/j.tranon.2017.12.003. Epub 8 Jan 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eschrich SA, Fulp WJ, Pawitan Y, et al. Validation of a radiosensitivity molecular signature in breast cancer. Clin Cancer Res 2012; 18(18): 5134–43. Epub 2012 Jul 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim HS, Kim SC, Kim SJ, et al. Identification of a radiosensitivity signature using integrative metaanalysis of published microarray data for NCI-60 cancer cells. BMC Genomics 2012; 13:348.(doi): 10.1186/471-2164-13-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tchelebi L, Ashamalla H, Graves PR. Mutant p53 and the response to chemotherapy and radiation. Subcell Biochem 2014; 85:133–59.(doi): 10.1007/978-94-017-9211-0_8. [DOI] [PubMed] [Google Scholar]
- 25.Eriksson B, Oberg K, Wilander E, et al. Nuclear DNA distribution in neuroendocrine gastroenteropancreatic tumors before and during treatment. Acta Oncol 1989; 28(2): 193–7. doi: 10.3109/02841868909111246. [DOI] [PubMed] [Google Scholar]
- 26.Tsioulias G, Muto T, Kubota Y, et al. DNA ploidy pattern in rectal carcinoid tumors. Dis Colon Rectum 1991; 34(1): 31–6. doi: 10.1007/bf02050203. [DOI] [PubMed] [Google Scholar]
- 27.Jones DJ, Hasleton PS, Moore M. DNA ploidy in bronchopulmonary carcinoid tumours. Thorax 1988; 43(3): 195–9. doi: 10.1136/thx.43.3.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simbolo M, Vicentini C, Mafficini A, et al. Mutational and copy number asset of primary sporadic neuroendocrine tumors of the small intestine. Virchows Arch 2018; 473(6): 709–17. doi: 10.1007/s00428-018-2450-x. Epub 2018 Sep 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Capurso G, Lattimore S, Crnogorac-Jurcevic T, et al. Gene expression profiles of progressive pancreatic endocrine tumours and their liver metastases reveal potential novel markers and therapeutic targets. Endocr Relat Cancer 2006; 13(2): 541–58. [DOI] [PubMed] [Google Scholar]
- 30.Walch AK, Zitzelsberger HF, Aubele MM, et al. Typical and atypical carcinoid tumors of the lung are characterized by 11q deletions as detected by comparative genomic hybridization. Am J Pathol 1998; 153(4): 1089–98. doi: 10.16/S0002-9440(10)65653-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Banck MS, Kanwar R, Kulkarni AA, et al. The genomic landscape of small intestine neuroendocrine tumors. J Clin Invest 2013; 15(67963). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Francis JM, Kiezun A, Ramos AH, et al. Somatic mutation of CDKN1B in small intestine neuroendocrine tumors. Nat Genet 2013; 45(12): 1483–6. doi: 10.038/ng.2821. Epub 013 Nov 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dumanski JP, Rasi C, Björklund P, et al. A MUTYH germline mutation is associated with small intestinal neuroendocrine tumors. Endocr Relat Cancer 2017; 24: 427–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sei Y, Zhao X, Forbes J, et al. A Hereditary Form of Small Intestinal Carcinoid Associated With a Germline Mutation in Inositol Polyphosphate Multikinase. Gastroenterology 2015; 149(1): 67–78. doi: 10.1053/j.gastro.2015.04.008. Epub Apr 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiao Y, Shi C, Edil BH, et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science 2011; 331(6021): 1199–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Derks JL, Leblay N, Lantuejoul S, Dingemans AC, Speel EM, Fernandez-Cuesta L. New Insights into the Molecular Characteristics of Pulmonary Carcinoids and Large Cell Neuroendocrine Carcinomas, and the Impact on Their Clinical Management. J Thorac Oncol 2018; 13(6): 752–66. doi: 10.1016/j.jtho.2018.02.002. Epub Feb 14. [DOI] [PubMed] [Google Scholar]
- 37.Duerr EM, Mizukami Y, Ng A, et al. Defining molecular classifications and targets in gastroenteropancreatic neuroendocrine tumors through DNA microarray analysis. Endocr Relat Cancer 2008; 15(1): 243–56. [DOI] [PubMed] [Google Scholar]
- 38.Li SC, Essaghir A, Martijn C, et al. Global microRNA profiling of well-differentiated small intestinal neuroendocrine tumors. Mod Pathol 2013; 26(5): 685–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Missiaglia E, Dalai I, Barbi S, et al. Pancreatic endocrine tumors: expression profiling evidences a role for AKT-mTOR pathway. J Clin Oncol 2010; 28(2): 245–55. doi: 10.1200/JCO.2008.21.5988. Epub 2009 Nov 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roldo C, Missiaglia E, Hagan JP, et al. MicroRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. J Clin Oncol 2006; 24(29): 4677–84. [DOI] [PubMed] [Google Scholar]
- 41.Alcala N, Leblay N, Gabriel AAG, et al. Integrative and comparative genomic analyses identify clinically relevant pulmonary carcinoid groups and unveil the supra-carcinoids. Nat Commun 2019; 10(1): 3407. doi: 10.1038/s41467-019-11276-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ianniello A, Sansovini M, Severi S, et al. Peptide receptor radionuclide therapy with (177)Lu-DOTATATE in advanced bronchial carcinoids: prognostic role of thyroid transcription factor 1 and (18)F-FDG PET. Eur J Nucl Med Mol Imaging 2016; 43(6): 1040–6. doi: 10.07/s00259-015-3262-8. Epub 2015 Nov 27. [DOI] [PubMed] [Google Scholar]
- 43.Kidd M, Drozdov I, Modlin I. Blood and tissue net gene cluster analysis correlate, define hallmarks and predict disease status. Endocr Relat Cancer 2015; 2: 15–0092. [DOI] [PubMed] [Google Scholar]
- 44.Karpathakis A, Dibra H, Pipinikas C, et al. Prognostic Impact of Novel Molecular Subtypes of Small Intestinal Neuroendocrine Tumor. Clin Cancer Res 2016; 22(1): 250–8. doi: 10.1158/078-0432.CCR-15-373. Epub 2015 Jul 13. [DOI] [PubMed] [Google Scholar]
- 45.Karpathakis A, Dibra H, Pipinikas C, et al. Progressive epigenetic dysregulation in neuroendocrine tumour liver metastases. Endocr Relat Cancer 2017; 24(2): L21–L5. doi: 10.1530/ERC-16-0419. Epub 2017 Jan 3. [DOI] [PubMed] [Google Scholar]
- 46.Mafficini A, Scarpa A. Genomic landscape of pancreatic neuroendocrine tumours: the International Cancer Genome Consortium. J Endocrinol 2018; 236(3): R161–R7. doi: 10.1530/JOE-17-0560. Epub 2018 Jan 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roldo C, Missiaglia E, Hagan JP, et al. MicroRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. J Clin Oncol 2006; 24(29): 4677–84. Epub 2006 Sep 11. [DOI] [PubMed] [Google Scholar]
- 48.Pelosi G, Bianchi F, Hofman P, et al. Recent advances in the molecular landscape of lung neuroendocrine tumors. Expert Rev Mol Diagn 2019; 19(4): 281–97. doi: 10.1080/14737159.2019.1595593. Epub 2019 Mar 27. [DOI] [PubMed] [Google Scholar]
- 49.Jahn U, Ilan E, Sandstrom M, Garske-Roman U, Lubberink M, Sundin A. 177Lu-DOTATATE peptide receptor radionuclide therapy: dose response in small intestinal neuroendocrine tumors. Neuroendocrinology 2019; 10(000504001): 000504001. [DOI] [PubMed] [Google Scholar]
- 50.Sorbye H, Kong G, Grozinsky-Glasberg S. PRRT in high-grade gastroenteropancreatic neuroendocrine neoplasms (WHO G3). Endocr Relat Cancer 2020; 27(3): R67–R77. doi: 10.1530/ERC-19-0400. [DOI] [PubMed] [Google Scholar]
- 51.Binderup T, Knigge U, Loft A, Federspiel B, Kjaer A. 18F-fluorodeoxyglucose positron emission tomography predicts survival of patients with neuroendocrine tumors. Clin Cancer Res 2010; 16(3): 978–85. Epub 2010 Jan 26. [DOI] [PubMed] [Google Scholar]
- 52.Strosberg J. (177)Lutetium-Dotatate delays decline in quality of life in patients with midgut neuroendocrine tumors. Oncotarget 2018; 9(69): 33059. doi: 10.18632/oncotarget.25942. eCollection 2018 Sep 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kratochwil C, Stefanova M, Mavriopoulou E, et al. SUV of [68Ga]DOTATOC-PET/CT Predicts Response Probability of PRRT in Neuroendocrine Tumors. Mol Imaging Biol 2015; 17(3): 313–8. doi: 10.1007/s11307-014-0795-3. [DOI] [PubMed] [Google Scholar]
- 54.Werner RA, Lapa C, Ilhan H, et al. Survival prediction in patients undergoing radionuclide therapy based on intratumoral somatostatin-receptor heterogeneity. Oncotarget 2017; 8(4): 7039–49. doi: 10.18632/oncotarget.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ezziddin S, Attassi M, Yong-Hing CJ, et al. Predictors of long-term outcome in patients with well-differentiated gastroenteropancreatic neuroendocrine tumors after peptide receptor radionuclide therapy with 177Lu-octreotate. J Nucl Med 2014; 55(2): 183–90. doi: 10.2967/jnumed.113.125336. Epub 2014 Jan 16. [DOI] [PubMed] [Google Scholar]
- 56.Wasylishen AR, Estrella JS, Pant V, Chau GP, Lozano G. Daxx Functions Are p53-Independent In Vivo. Mol Cancer Res 2018; 16(10): 1523–9. doi: 10.158/41-7786.MCR-18-0281. Epub 2018 Jun 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Naik C, Basu S. Peptide Receptor Radionuclide Therapy with (177)Lu-DOTATATE for Metastatic Neuroendocrine Tumor Occurring in Association with Multiple Endocrine Neoplasia Type 1 and Cushing’s Syndrome. World J Nucl Med 2017; 16(2): 126–32. doi: 10.4103/1450-147.203068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yao JC, Garg A, Chen D, et al. Genomic profiling of NETs: a comprehensive analysis of the RADIANT trials. Endocr Relat Cancer 2019; 26(4): 391–403. doi: 10.1530/ERC-18-0332. Epub 2019 Jan 1. [DOI] [PubMed] [Google Scholar]
- 59.Chen F, Zhang Y, Gibbons DL, et al. Pan-Cancer Molecular Classes Transcending Tumor Lineage Across 32 Cancer Types, Multiple Data Platforms, and over 10,000 Cases. Clin Cancer Res 2018; 24(9): 2182–93. doi: 10.1158/078-0432.CCR-17-3378. Epub 2018 Feb 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bodei L, Kidd MS, Singh A, et al. PRRT neuroendocrine tumor response monitored using circulating transcript analysis: the NETest. Eur J Nucl Med Mol Imaging 2019; 14(10): 019–04601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bodei L, Kidd M, Modlin IM, et al. Measurement of circulating transcripts and gene cluster analysis predicts and defines therapeutic efficacy of peptide receptor radionuclide therapy (PRRT) in neuroendocrine tumors. Eur J Nucl Med Mol Imaging 2016; 43(5): 839–51. doi: 10.1007/s00259-015-3250-z. Epub 2015 Nov 23. [DOI] [PubMed] [Google Scholar]
- 62.Bodei L, Kidd MS, Singh A, et al. PRRT Genomic Signature in Blood for Prediction of 177Lu-octreotate Efficacy. EJNMMI 2018; 45(7): 1155–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li XJ, Hayward C, Fong PY, et al. A blood-based proteomic classifier for the molecular characterization of pulmonary nodules. Sci Transl Med 2013; 5(207): 207ra142. doi: 10.1126/scitranslmed.3007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vachani A, Pass HI, Rom WN, et al. Validation of a multiprotein plasma classifier to identify benign lung nodules. J Thorac Oncol 2015; 10(4): 629–37. doi: 10.1097/JTO.0000000000000447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pritzker KP. Predictive and prognostic cancer biomarkers revisited. Expert Rev Mol Diagn 2015; 15(8): 971–4. doi: 10.1586/14737159.2015.1063421. Epub 2015 Jul 1. [DOI] [PubMed] [Google Scholar]
- 66.Eschrich S, Zhang H, Zhao H, et al. Systems biology modeling of the radiation sensitivity network: a biomarker discovery platform. Int J Radiat Oncol Biol Phys 2009; 75(2): 497–505. doi: 10.1016/j.ijrobp.2009.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ahmed KA, Chinnaiyan P, Fulp WJ, Eschrich S, Torres-Roca JF, Caudell JJ. The radiosensitivity index predicts for overall survival in glioblastoma. Oncotarget 2015; 6(33): 34414–22. doi: 10.18632/oncotarget.5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang Q, Bing Z, Tian J, et al. Integrating radiosensitive genes improves prediction of radiosensitivity or radioresistance in patients with oesophageal cancer. Oncol Lett 2019; 17(6): 5377–88. doi: 10.3892/ol.2019.10240. Epub 2019 Apr 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gatti RA, Berkel I, Boder E, et al. Localization of an ataxia-telangiectasia gene to chromosome 11q22–23. Nature 1988; 336(6199): 577–80. doi: 10.1038/336577a0. [DOI] [PubMed] [Google Scholar]
- 70.Andreassen CN, Rosenstein BS, Kerns SL, et al. Individual patient data meta-analysis shows a significant association between the ATM rs1801516 SNP and toxicity after radiotherapy in 5456 breast and prostate cancer patients. Radiother Oncol 2016; 121(3): 431–9. doi: 10.1016/j.radonc.2016.06.017. Epub Jul 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhao J, Zhi Z, Zhang M, et al. Predictive value of single nucleotide polymorphisms in XRCC1 for radiation-induced normal tissue toxicity. Onco Targets Ther 2018; 11:3901–3918.(doi): 10.2147/OTT.S156175. eCollection 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kerns SL, Ostrer H, Rosenstein BS. Radiogenomics: using genetics to identify cancer patients at risk for development of adverse effects following radiotherapy. Cancer Discov 2014; 4(2): 155–65. doi: 10.1158/2159-8290.CD-13-0197. Epub 2014 Jan 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lyngholm CD, Overgaard J, Christiansen PM, Alsner J. Validation of a gene expression profile predictive of the risk of radiation-induced fibrosis in women treated with breast conserving therapy. Acta Oncol 2015; 54(9): 1665–8. doi: 10.3109/0284186X.2015.1069395. Epub 2015 Aug 12. [DOI] [PubMed] [Google Scholar]
- 74.Modlin IM, Kidd M, Drozdov IA, Bodei L. The use of Deep Learning and Neural Networks in Imaging - Welcome to the new Mathematical Milieu of Medicine. Neuroendocrinology 2019; 7(000504605): 000504605. [DOI] [PubMed] [Google Scholar]
- 75.Sabet A, Haug AR, Eiden C, et al. Efficacy of peptide receptor radionuclide therapy with (177)Lu-octreotate in metastatic pulmonary neuroendocrine tumors: a dual-centre analysis. Am J Nucl Med Mol Imaging 2017; 7(2): 74–83. eCollection 2017. [PMC free article] [PubMed] [Google Scholar]
- 76.Sabet A, Dautzenberg K, Haslerud T, et al. Specific efficacy of peptide receptor radionuclide therapy with (177)Lu-octreotate in advanced neuroendocrine tumours of the small intestine. Eur J Nucl Med Mol Imaging 2015; 42(8): 1238–46. doi: 10.007/s00259-015-3041-6. Epub 2015 Mar 26. [DOI] [PubMed] [Google Scholar]
- 77.Ezziddin S, Reichmann K, Yong-Hing C, et al. Early prediction of tumour response to PRRT. The sequential change of tumour-absorbed doses during treatment with 177Lu-octreotate. Nuklearmedizin 2013; 52(5): 170–7. doi: 10.3413/Nukmed-0581-13-05. Epub 2013 Aug 21. [DOI] [PubMed] [Google Scholar]
- 78.Wetz C, Apostolova I, Steffen IG, et al. Predictive Value of Asphericity in Pretherapeutic [(111)In]DTPA-Octreotide SPECT/CT for Response to Peptide Receptor Radionuclide Therapy with [(177)Lu]DOTATATE. Mol Imaging Biol 2017; 19(3): 437–45. doi: 10.1007/s11307-016-1018-x. [DOI] [PubMed] [Google Scholar]
- 79.Sharma R, Wang WM, Yusuf S, et al. (68)Ga-DOTATATE PET/CT parameters predict response to peptide receptor radionuclide therapy in neuroendocrine tumours. Radiother Oncol 2019; 141:108–115.(doi): 10.1016/j.radonc.2019.09.003.Epub Sep 18. [DOI] [PubMed] [Google Scholar]
- 80.Haug AR, Auernhammer CJ, Wangler B, et al. 68Ga-DOTATATE PET/CT for the early prediction of response to somatostatin receptor-mediated radionuclide therapy in patients with well-differentiated neuroendocrine tumors. J Nucl Med 2010; 51(9): 1349–56. Epub 2010 Aug 18. [DOI] [PubMed] [Google Scholar]
- 81.Sansovini M, Severi S, Ianniello A, et al. Long-term follow-up and role of FDG PET in advanced pancreatic neuroendocrine patients treated with 177Lu-D OTATATE. Eur J Nucl Med Mol Imaging 2016; 4: 4. [DOI] [PubMed] [Google Scholar]
- 82.Thapa P, Ranade R, Ostwal V, Shrikhande SV, Goel M, Basu S. Performance of 177Lu-DOTATATE-based peptide receptor radionuclide therapy in metastatic gastroenteropancreatic neuroendocrine tumor: a multiparametric response evaluation correlating with primary tumor site, tumor proliferation index, and dual tracer imaging characteristics. Nucl Med Commun 2016; 37(10): 1030–7. doi: 10.97/MNM.0000000000000547. [DOI] [PubMed] [Google Scholar]
- 83.Prasad V, Srirajaskanthan R, Toumpanakis C, et al. Lanreotide depot/autogel before, during, and after peptide receptor radionuclide therapy (PRRT) in advanced neuroendocrine tumors (NETs): Data from the PRELUDE study. Journal of Clinical Oncology 2018; 36(15_suppl): e16167-e. [Google Scholar]
- 84.Black JRM, Atkinson SR, Singh A, Evans J, Sharma R. The Inflammation-Based Index Can Predict Response and Improve Patient Selection in NETs Treated With PRRT: A Pilot Study. J Clin Endocrinol Metab 2019; 104(2): 285–92. doi: 10.1210/jc.2018-01214. [DOI] [PubMed] [Google Scholar]
- 85.Pauwels E, Van Binnebeek S, Vandecaveye V, et al. Inflammation-based index and (68)Ga-DOTATOC PET-derived uptake and volumetric parameters predict outcome in neuroendocrine tumor patients treated with (90)Y-DOTATOC. J Nucl Med 2019; 5(119): 236935. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.