Abstract
STUDY QUESTION
Are SARS-CoV-2 canonical cell entry machinery, consisting of ACE2, TMPRSS2, NRP1 and LY6E, or alternative potential cell entry machinery, consisting of BSG, ANPEP, CD209, CLEC4G, TMPRSS4, TMPRSS11A, FURIN, CTSB, CTSL and IFITM1, expressed in the human endometrium across the menstrual cycle?
SUMMARY ANSWER
Analysis of cell entry factors for SARS-CoV-2 by single-cell RNA-sequencing (scRNAseq) in the preconceptional human endometrium reveals low risk of infection.
WHAT IS KNOWN ALREADY
Gene expression datasets from bulk endometrial tissue show no significant expression of the SARS-CoV-2 receptor ACE2 and TMPRSS2. This is in contrast to reported expression of ACE2 at the single-cell level in the decidua and trophoblast cells at the maternal–fetal interface in early pregnancy, as well as vertical transmission of SARS-CoV-2 during pregnancy.
STUDY DESIGN, SIZE, DURATION
This analysis of SARS-CoV-2 cell entry machinery gene expression was conducted by scRNAseq in 73 181 human endometrial cells isolated from endometrial biopsies obtained from 27 donors across the menstrual cycle.
PARTICIPANTS/MATERIALS, SETTING, METHODS
ScRNAseq examined the expression of genes encoding cell entry machinery for SARS-CoV-2. The raw data were from a previously published dataset.
MAIN RESULTS AND THE ROLE OF CHANCE
ScRNAseq analysis showed no significant expression of ACE2 in stromal or unciliated epithelial cells in any phase of the menstrual cycle. TMPRSS2 was expressed in epithelial cells during the early proliferative and mid-secretory phases. Interestingly, the expression of NRP1 was observed in both stromal and epithelial cells across all phases of the menstrual cycle, and LY6E was highly expressed in stromal cells. In the mid-secretory phase, coexpression of ACE2 and TMPRSS2 was detected in 0.07% of luminal epithelial cells. No cells simultaneously expressed ACE2, NRP1 and TMPRSS2 at the time of embryo implantation. Focusing on non-canonical cell entry machinery, BSG was highly expressed in all cell types across the menstrual cycle and may interact with CTSB or CTSL proteases, but viral infection using this machinery has not yet been confirmed.
LARGE SCALE DATA
All raw data in this study can be found at NCBI’s Gene Expression Omnibus (series accession code GSE111976) and Sequence Read Archive (accession code SRP135922).
LIMITATIONS, REASONS FOR CAUTION
Our findings at the single-cell level imply low efficiency of SARS-CoV-2 endometrial infection using canonical receptors in a cohort of healthy reproductive-age women; however, infection of endometrial cells can only be assessed in the presence of the virus. All samples were processed for scRNAseq, so no samples are remaining to analyze protein expression or spatial transcriptomics.
WIDER IMPLICATIONS OF THE FINDINGS
Our results offer a useful resource to guide reproductive decisions when assessing risk of endometrial infection by SARS-CoV-2 during the preconceptional period in asymptomatic COVID-19 carriers.
STUDY FUNDING/COMPETING INTEREST(S)
This study was jointly supported by the March of Dimes, Chan Zuckerberg Biohub and MINECO/FEDER (SAF-2015-67164-R, to C.S.) (Spanish Government), and the European Union’s Horizon 2020 Framework Programme for Research and Innovation (Grant agreement 874867). W.W. was supported by the Stanford Bio-X Graduate Bowes Fellowship and Chan Zuckerberg Biohub. F.V. was supported by the Miguel Servet Program Type II of ISCIII (CPII18/00020) and the FIS project (PI18/00957). A patent disclosure has been filed for the study with the title ‘Methods for assessing endometrial transformation’ and the global patent number ‘EP 3807648 A2’ under the inventors S.R.Q., C.S., W.W. and F.V. C.S. is the Founder and Head of the Scientific Advisory Board of Igenomix SL. S.R.Q is the Director of Mirvie. I.M. is partially employed by Igenomix SL. B.R. has no interests to declare.
Keywords: COVID-19, SARS-CoV-2, ACE2, TMPRSS2, NRP1, scRNAseq
Introduction
The coronavirus disease 2019 (COVID-19) pandemic produced by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has impacted public health worldwide, including potential risks to couples aiming to conceive or already pregnant (Rasmussen et al., 2020). SARS-CoV-2 preferentially infects cells in the respiratory tract, heart, liver, brain and kidneys (Puelles et al., 2020), but knowledge of its direct tropism for reproductive organs involved in human pregnancy and subsequent neonatal health is limited. The endometrium acts as the maternal interface and possible infection gatekeeper, but there is conflicting evidence for (Zeng et al., 2020b) and against (Chen et al., 2020; Rasmussen et al., 2020) vertical transmission of SARS-CoV-2 during pregnancy.
SARS-CoV-2 uses angiotensin-converting enzyme 2 (ACE2) as the prime receptor for entry into host cells (Hoffmann et al., 2020), suggesting cells expressing ACE2 are most susceptible to viral infection (Zou et al., 2020). ACE2 is a member of the renin-angiotensin-aldosterone system (RAAS) that regulates blood pressure (Nakagawa et al., 2020). However, ACE2 expression alone is insufficient for cell entry of SARS-CoV-2. After viral-host tropism and adhesion of SARS-CoV-2 S protein to ACE2 on the cell surface, priming of the S protein between the S1 and S2 units is essential for fusion to the cell membrane and viral entry into the cell. This cleavage is efficiently performed by host serine protease TMPRSS2. The essential role of this protease in SARS-CoV-2 internalisation is confirmed by infection blockade after chemical treatment of lung cells with camostat mesylate or E-64d to inhibit TMPRSS2 (Hoffmann et al., 2020). In addition, recent evidence reveals that NRP1 acts as a cofactor to potentiate viral infectivity. The S protein of SARS-CoV-2 contains a cleavage site for the protease furin; NRP1 binds to furin-cleaved substrates and collaborates with ACE2 to promote viral entry and infectivity in cells that have low ACE2 expression (Cantuti-Castelvetri et al., 2020). Yet the cells have a mechanism to protect themselves from viral entrance, as LY6E restricts infection with the virus (Pfaender et al., 2020).
Recently, alternative receptors were proposed as possible players in infectivity of SARS-CoV-2, based on similarities between SARS-CoV-2 and other human coronaviruses. BSG is reported to play a specific role in viral entry in the absence of ACE2 in human cell lines (Wang et al., 2020a). Other cell surface proteins are described as main receptors that promote entry of other human coronavirus species, including ANPEP, CD209, CLEC4G (Yang et al., 2004; Gramberg et al., 2005) and DPP4, which is specific for MERS-CoV infection (Li et al., 2020b). Likewise, alternative cellular proteases may be used to prime the S protein. In the absence of TMPRSS2, Cathepsin B (CTSB) and cathepsin L (CTSL) also can cleave the S protein to facilitate viral entry. Proteases such as TMPRSS4, TMPRSS11A (Zang et al., 2020) and FURIN (Walls et al., 2020) also are proposed to cleave the S protein in other coronaviruses. In addition, host cells can use other specific proteins to restrict viral entrance. For example, IFITM can restrict entry of enveloped viruses like coronaviruses regardless of viral receptor expression (Huang et al., 2011).
The human endometrium expresses Angiotensins 1–7, which are cleaved by ACE2, and angiotensin receptor MAS. In addition, expression of angiotensins in the endometrium mirrors expression of ACE2 mRNA, with more abundant expression in epithelial cells compared with stromal cells (Vaz-Silva et al., 2009). During pregnancy, the human decidua has abundant expression of RAAS-related genes, prorenin (REN), prorenin receptor (ATP6AP2), AGT, ACE1, ACE2, AGTR1 and MAS, as demonstrated by quantitative PCR in specimens collected after cesarean section or spontaneous labor (Wang et al., 2012). Moreover, RAAS components including ACE2 are detected in decidua and fetal membranes in the human placenta, with potential roles in trophoblast invasion and angiogenesis (Marques et al., 2011; Pringle et al., 2011).
However, expression of SARS-CoV-2 cell entry machinery in the human endometrium is not well-characterised across the menstrual cycle, limiting understanding of the risk of potential transmission of SARS-CoV-2 during pregnancy. Here, we assessed, by single-cell RNA-sequencing (scRNAseq), the expression of SARS-CoV-2 cell entry machinery across the menstrual cycle, with a specific focus on the preconceptional period, using our published scRNAseq dataset (Wang et al., 2020b) to generate a ‘risk map’ for SARS-CoV-2 infection of the human endometrium.
Materials and methods
Population and sample collection
All human endometrium samples (N = 27) were collected in accordance with the Institutional Review Board (IRB) guidelines for Stanford University (IRB code IRB-35448) and IVI Valencia, Spain (1603-IGX-016-CS). Informed written consent was obtained from each donor in her natural menstrual cycle (no hormone stimulation) before an endometrial biopsy was performed. De-identified human endometrium was obtained from women aged 18–34 years, with regular menstrual cycles (3–4 days, every 28–30 days), with BMI ranging 19–29 kg/m2 (inclusive), negative serological tests for HIV, HBV, HCV and RPR, and a normal karyotype. Women with the following conditions were excluded from tissue collection: recent contraception (IUD in past 3 months; hormonal contraceptives in past 2 months), uterine pathology (endometriosis, leiomyoma or adenomyosis, or bacterial, fungal or viral infection) and polycystic ovary syndrome.
Endometrium tissue dissociation and population enrichment
A two-stage dissociation protocol was used to dissociate endometrium tissue and separate it into stromal fibroblast- and epithelium-enriched single-cell suspensions. Tissue was minced into pieces as small as possible and dissociated in Collagenase A1 (Sigma, St. Louis, MO, USA) overnight at 4°C in a 50-ml Falcon tube in a horizontal position. This primary enzymatic step dissociated stromal fibroblasts into single cells while leaving epithelial glands and lumen mostly undigested. The resulting tissue suspension was then briefly homogenised and left unagitated for 10 min in a 50-ml Falcon tube in a vertical position, during which epithelial glands and lumen sedimented as a pellet, and stromal fibroblasts stayed suspended in the supernatant. The supernatant was collected as the stromal fibroblast-enriched suspension. The pellet was washed twice in 50 ml DMEM to further remove residual stromal fibroblasts. The washed pellet was dissociated in 400 μl TrypLE Select (Gibco, ThermoFisher Scientific, Waltham, MA, USA) for 20 min at 37°C, during which homogenisation was performed via intermittent pipetting. DNaseI was added to the solution to digest extracellular genomic DNA. The digestion was quenched with 1.5 ml DMEM after a 5-min incubation. The resulting cell suspension was pipetted, filtered through a 50-μm cell strainer, and centrifuged at 1000 rpm for 5 min. The pellet was re-suspended as the epithelium-enriched suspension.
Fluidigm C1 single-cell capture, imaging and cDNA generation
Live cells were enriched using a MACS Dead Cell Removal kit (Miltenyi Biotec, Bergisch Gladbach, Germany). The resulting cell suspension was diluted in DMEM to a final concentration of 300–400 cells/μl before loading onto a medium Fluidigm C1 chip (South San Francisco, CA, USA) for mRNA sequencing. Live dead cell stain (Life Technologies, Carlsbad, CA, USA) was added directly into the cell suspension. Single-cell capture, mRNA reverse-transcription and cDNA amplification were performed on the Fluidigm C1 system using default scripts for mRNA sequencing. Detailed numbers of cells for each individual are included in Supplementary Table SI.
Fluidigm C1 scRNAseq library generation
Single-cell cDNA concentration and size distribution were analysed on a capillary electrophoresis-based automated fragment analyser (Advanced Analytical). Library preparation was performed using a Nextera XT DNA Sample Preparation kit (Illumina, San Diego, CA, USA) on a Mosquito HTS liquid handler (SPT Labtech, Hertfordshire, UK) following Fluidigm’s single-cell library preparation protocol, with a 4× scale-down of all reagents. Dual-indexed single-cell libraries were pooled and sequenced in pair-end reads on Nextseq (Illumina) to a depth of 1 × 106–2 × 106 reads per cell. bcl2fastq v2.17.1.14 was used to separate data for each single cell by using unique barcode combinations from the Nextera XT preparation and to generate *.fastq files.
Chromium 10x single-cell capture, cDNA generation and scRNAseq library generation
Live cells were enriched with a MACS Dead Cell Removal kit (Miltenyi Biotec), and live cells were washed twice with PBS to remove ambient RNA. The resulting epithelial and stromal cell portions were combined in a 1:1 ratio by concentration and loaded onto the Chromium Next GEM Chip G (10× Genomics, Pleasanton, CA, USA) for each donor. GEM generation and barcoding, reverse-transcription, cDNA generation, and library construction were done following the manufacturer’s protocol (Single Cell 3’ Reagent Kit v3.1, 10× Genomics). Dual-indexed single-cell libraries were pooled and sequenced in pair-end reads on Novaseq (Illumina). Detailed numbers of cells for each individual are included in Supplementary Table SII.
Quantification and statistical analysis
Fluidigm C1 dataset processing and quality metrics
Raw reads from FASTQ files were trimmed to 75 bp using fastqx 0.11.7, aligned with STAR 2.5 (Dobin et al., 2013) to Ensembl GRCh38.87 (dna.primary_assembly), and filtered for duplicates with MarkDuplicates (picard 2.9). Reads per gene were summed using HTSeq 0.7.0 (Anders et al., 2015) and Ensembl GTF for GRCh38.87 under the setting -m intersection-strict\-s no. For each cell, counts were normalised to log-transformed reads per million (log2(rpm + 1)) by the equation , where is cell and is gene . Quality control filtering was applied using the fraction of ERCCs. Cut-off was established at 5% of the null distribution of the ratio between ERCC reads and all detected reads. Null distribution was constructed using reads from empty capture sites.
Before dimensional reduction, over-dispersion of genes was calculated as , where is the squared variation of coefficient of gene i across cells of interest, and is the expected squared variation of coefficient given mean, fitted using non-ERCC counts. All pairwise distances between cells were calculated as: (1–Pearson’s correlation). Dimensional reduction was performed using the R implementation of tSNE (Rtsne 0.13).
Chromium 10× dataset
CellRanger 3.1.0 software (settings: expect-cells = 10 000) was used to process raw reads from FASTQ files, align to the reference genome (GRCh38-3.0.0), and generate a filtered UMI expression profile for each droplet. Raw UMI counts were downstream processed within Seurat package 3.1.2 (Stuart et al., 2019). Raw reads were normalised to log-transformed transcripts per million (log(TPM + 1)) by the equation , where is cell and is gene , using NormalizeData() function in Seurat.
10x quality filtering
Recovered cells from CellRanger were submitted to dimensional reduction, and each identified cell population was evaluated by quality control metrics that included: UMI counts, number of genes detected and percent of mitochondrial reads. Differentially expressed genes were obtained for each cluster compared to the rest of cells. Clusters with no uniquely expressed genes identified above threshold and poor quality metrics were removed. Also, clusters with combined expression of two distinct cell types were considered doublets. DoubletFinder 2.0.2 (McGinnis et al., 2019) was applied to remove homotypic doublet cell clusters. For unciliated epithelia and stromal fibroblasts, a Gaussian mixture model was fit on the distribution of number of genes detected (R package mixtools 1.1.0). For each cell type, the Gaussian distribution N(μ,σ2) with the lowest mean was identified, and a threshold (th) was calculated as th = μ + 2σ for N. Only cells (N = 71 032) with equal or higher number of genes detected than th were retained for downstream analysis.
Results
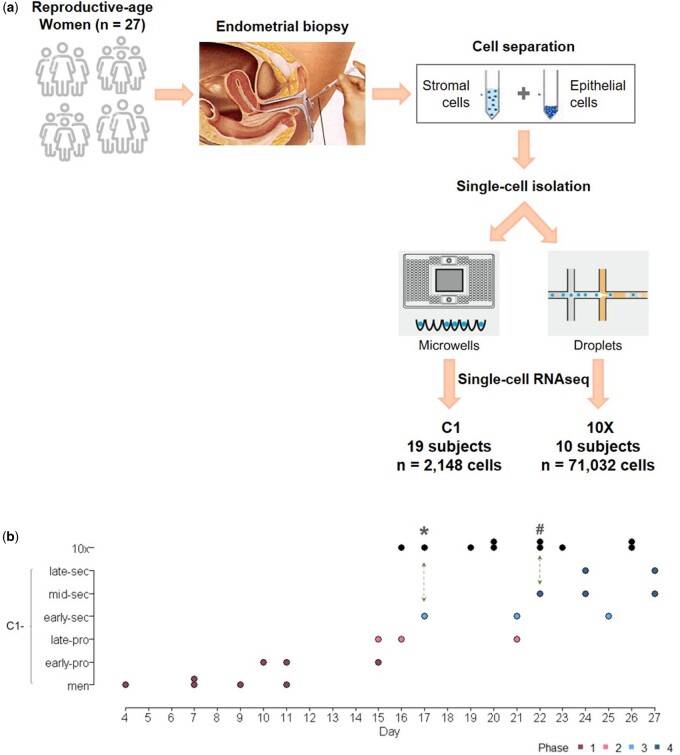
We investigated expression of SARS-CoV-2 canonical cell entry machinery genes ACE2, TMPRSS2, NRP1 and LY6E as well as alternative potential cell entry machinery genes BSG, ANPEP, CD209, CLEC4G, TMPRSS4, TMPRSS11A, FURIN, CTSB, CTSL and IFITM1 in endometrial samples obtained from 27 healthy reproductive-age women using scRNAseq. We collected scRNAseq data across the menstrual cycle in 19 participants using the Fluidigm C1 system, resulting in 2148 cells analysed. We also collected scRNAseq data from 10 participants in the preconceptional period using the 10× Chromium system, enabling analysis of an additional 71 032 cells (Fig. 1a). We collected both C1 and 10× data from two women, one in the mid-secretory phase and one in the early-secretory phase (Fig. 1b).
Figure 1.
Experimental design. (a) Representation of two methods for single-cell isolation and sequencing, showing number of cells analyzed for C1 and 10× datasets. (b) Distribution of samples across menstrual cycle days (number of days after onset of last menstrual bleeding) and endometrial phases (assigned based on scRNAseq data) for C1 (bottom) and 10× (top) datasets. Dots with * and # to the right are donors from whom both C1 and 10× data were collected. Phases 1–4: major endometrial phases identified in unciliated epithelia and stromal fibroblasts using whole transcriptomic scRNAseq data.
Expression of SARS-CoV-2 entry genes across the menstrual cycle
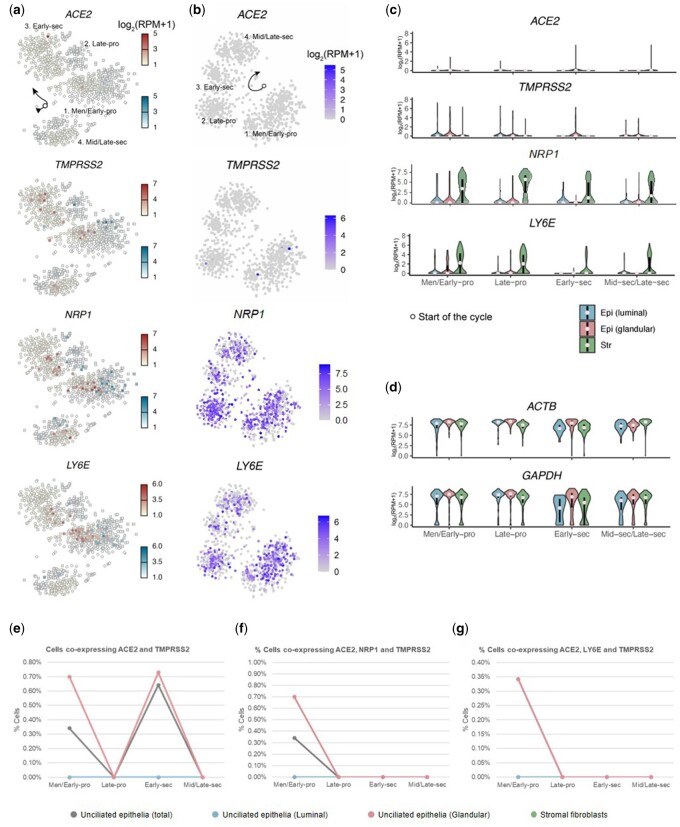
ScRNAseq analysis across the menstrual cycle (n = 2148 cells) showed that few stromal fibroblasts (∼0.5%) and unciliated epithelial cells (∼2%) expressed ACE2. In contrast, NRP1 expression was higher in both cell types (∼85% of stromal fibroblasts and ∼24% of unciliated epithelial cells) (Fig. 2a– c and Supplementary Fig. S1). TMPRSS2 was expressed in 16% of glandular epithelial cells during the early proliferative phase (Fig. 2a–c and Supplementary Fig. S1a). Expression of LY6E was mainly observed in stromal cells (Fig. 2a–c and Supplementary Fig. S1a). ACTB and GAPDH were used as housekeeping controls (Fig. 2d and Supplementary Fig. S1b).
Figure 2.
SARS-CoV-2 cell entry factor expression in human endometrial cells across the menstrual cycle. (a, b) Dynamics of abundance of cells expressing ACE2, TMPRSS2, NRP1 and LY6E across the menstrual cycle in luminal epithelial cells represented by blue squares and glandular epithelial cells represented by red circles (a) and stromal fibroblasts (b). (c) Quantification of data in panels a and b via violin plots. Dots are medians, and lower and higher ends of the black bars indicate 25th and 75th percentiles, respectively. (d) ACTB and GAPDH were used as housekeeping controls. (e) Percentage of cells that co-express ACE2 with TMPRSS2 across the menstrual cycle. (f) Percentage of cells that co-express ACE2 with TMPRSS2 and the enhancer NRP1 across the menstrual cycle. (g) Percentage of cells that co-express ACE2 with TMPRSS2 and LY6E inhibitor across the menstrual cycle. Data correspond to the C1 dataset with 2148 cells across the menstrual cycle.
To assess the full SARS-CoV-2 cell entry machinery, co-expression of the canonical receptor and protease as well as the enhancer and inhibitor was evaluated in endometrial cells. Co-expression of ACE2 and TMPRSS2 was residual, with maximum co-expression in 0.73% of cells analysed in the glandular epithelium during the early secretory phase (Fig. 2e). Triple co-expression of ACE2, NRP1 and TMPRSS2 was detected in up to 0.70% of cells, with maximum co-expression in the glandular epithelium during the early proliferative phase (Fig. 2f); co-expression of ACE2, LY6E, and TMPRSS2 was only observed in 0.35% of cells, with maximum co-expression also in the glandular epithelium during the early proliferative phase of the menstrual cycle (Fig. 2g).
Focusing on potential alternative cell entry machinery, there was no expression of ANPEP, CD209 and CLEC4G in any cell types and cycle phases (data not shown). DPP4 was expressed in the mid- and late-secretory phases (∼70% of unciliated epithelial cells), and BSG was expressed in both cell types across the menstrual cycle (between ∼20% and ∼80% of cells) (Supplementary Fig. S2a). Regarding new potential cellular proteases, no cells expressed TMPRSS11A (data not shown), while TMPRSS4 and FURIN were expressed across the menstrual cycle in unciliated epithelial cells (Supplementary Fig. S2b). Interestingly, CTSB and CTSL were expressed in both stromal and epithelial cells throughout the menstrual cycle, and CTSB expression was more abundant than CTSL. The percentage of CTSL-expressing stromal fibroblasts was higher than CTSL-expressing unciliated epithelial cells (Supplementary Fig. S2b). Like LY6E, IFITM1 was mainly expressed in stromal cells across the entire menstrual cycle (Supplementary Fig. S2c). The most extensive co-expression was with the BSG receptor and TMPRSS4 and CTSB proteases, specifically co-expression of BSG and TMPRSS4 in unciliated epithelial cells, but there was higher co-expression between BSG and CTSB in all cell types across the entire menstrual cycle (Supplementary Fig. S3a). Cell entry machinery described for MERS-CoV, DPP4 and FURIN, only showed positive co-expression in epithelial cells (∼10%) during the mid- and late-secretory phases (Supplementary Fig. S3b).
Expression of SARS-CoV-2 entry genes enriched in periconceptional phase
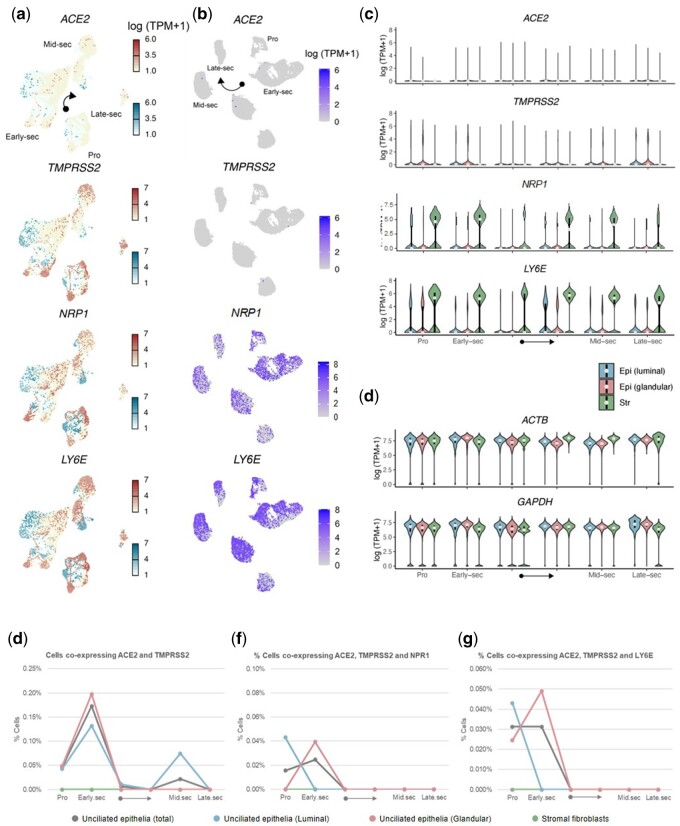
To focus on periconception when vertical transmission could begin, cells from the C1 dataset enriched with 71 032 cells during the preconceptional phase were analysed using 10× technology. ACE2 expression was low and detected in <1.5% of unciliated epithelial cells and <0.5% of stromal cells (Fig. 3a–c and Supplementary Fig. S4a). We observed higher expression of NRP1, which was detected in >20% of stromal cells across the cycle, reaching 51% of cells during the mid-secretory phase. TMPRSS2 was expressed in ∼10% of glandular epithelial cells, but expression of this gene was consistently low in stromal cells. LY6E was mainly expressed in stromal cells across the entire menstrual cycle (Fig. 3a and c and Supplementary Fig. S4a). ACTB and GAPDH were used as housekeeping controls (Fig. 3d and Supplementary Fig. S4b). Co-expression of ACE2 with TMPRSS2 during preconception was observed only in unciliated epithelia but was at low abundance (0.07% of luminal epithelial cells) during the window of implantation (Fig. 3e). No cells presented detectable ACE2, NRP1 and TMPRSS2 co-expression during the mid-secretory phase or adjacent timepoints (Fig. 3f), and cells at this stage also did not co-express ACE2, LY6E and TMPRSS2 (Fig. 3g).
Figure 3.
SARS-CoV-2 cell entry factor expression in human endometrial cells focused on the preconceptional period. (a, b) Dynamics of abundance of cells expressing ACE2, TMPRSS2, NRP1 and LY6E in luminal epithelial cells represented by blue squares and glandular epithelial cells represented by red circles (a) and stromal fibroblasts (b). (c) Quantification of data in panels a and b via violin plots. Dots are medians, and lower and higher ends of the black bars indicate 25th and 75th percentiles, respectively. (d) ACTB and GAPDH were used as housekeeping controls. (e) Percentage of cells that co-express ACE2 and TMPRSS2. (f) Percentage of cells that co-express ACE2, TMPRSS2 and the enhancer NRP1. (g) Percentage of cells that co-express ACE2 with TMPRSS2 and LY6E inhibitor. Data correspond to the C1 dataset with 2148 cells across the menstrual cycle, enriched with 71 032 cells from the 10× dataset during the preconceptional phase. Black arrows in panels c–g indicate menstrual cycle progression between early-secretory phase and mid-secretory phase.
As alternative cell entry machinery, there was no expression of CD209, CLEC4G and ANPEP (data not shown). BSG was expressed in both cell types (between ∼50% and ∼90% of cells) in the entire menstrual cycle, and DPP4 was expressed in mid- and late-secretory phases (∼80% of unciliated epithelial cells) (Supplementary Fig S5a). TMPRSS11A expression was not detected (data not shown), while TMPRSS4 was expressed across the menstrual cycle in unciliated epithelium, with higher expression in mid- and late-secretory phases (∼60% of cells). However, FURIN was mainly expressed during the early-secretory phase in unciliated epithelial cells (∼50%) (Supplementary Fig. S5b). In contrast, CTSB was highly expressed in both epithelial and stromal cells, with >70% of either cell type expressing this gene during the mid-secretory phase. CTSL was expressed in >20% of epithelial cells and >40% of stromal cells. IFITM1 showed expression in stromal cells mainly in early-secretory phase (Supplementary Fig. S5c). ACTB and GAPDH were used as housekeeping controls (Supplementary Fig. S5d). Focusing on BSG, co-expression with TMPRSS4 was high in unciliated epithelial cells in the mid-secretory phase (∼50%) and co-expression was also high with CTSB and CTSL in all cell types across the entire menstrual cycle, with highest co-expression between BSG and CTSB (between ∼50% and ∼80%) (Supplementary Fig. S6a). DPP4 and FURIN were co-expressed in epithelial cells (∼20%) during the mid- and late-secretory phases (Supplementary Fig. S6b).
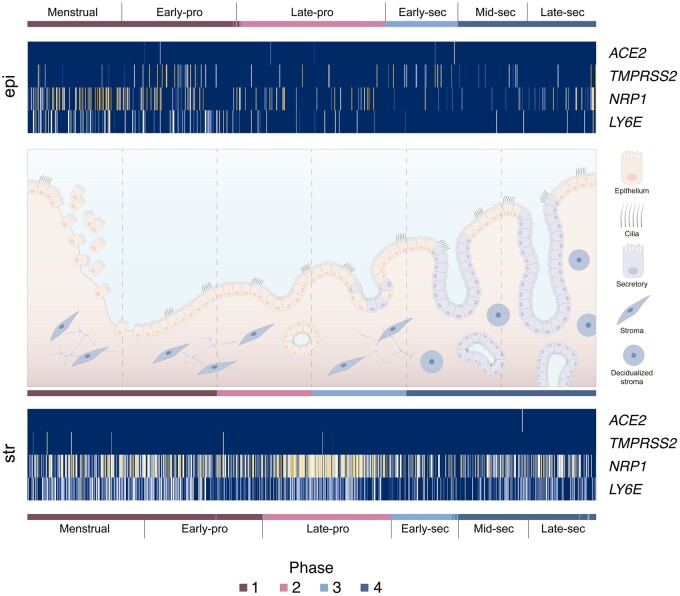
The transcriptomic dynamics of SARS-CoV-2 canonical cell entry machinery genes in epithelial and stromal cells were phase-independent, with ACE2 and TMPRSS2 present at low abundance. However, unlike in the epithelial compartment, NRP1 and LY6E were highly expressed in undecidualised and decidualised stromal cells (Fig. 4) and, therefore, may play a role in ACE2 non-expressing cells.
Figure 4.
Temporal transcriptome dynamics of SARS-CoV-2 entry genes across the human menstrual cycle. Expression of canonical SARS-CoV-2 cell entry machinery genes ACE2, TMPRSS2, NRP1 and LY6E for unciliated epithelia (epi) and stromal fibroblasts (str) across the human menstrual cycle. Columns are ordered by phase of the cycle. Data correspond to spatio-temporal analysis of the C1 dataset with 2148 cells across the menstrual cycle.
Discussion
The outbreak and third wave of the COVID-19 pandemic pose concerns to the general public, including couples wishing to conceive and pregnant women. There is evidence for (Zeng et al., 2020b) and against (Chen et al., 2020; Rasmussen et al., 2020) vertical transmission of SARS-CoV-2 originating from the uterus to the placenta and fetus during conception and pregnancy. Placental infection with SARS-CoV-2 has been suspected in a pregnant woman with symptomatic coronavirus disease who experienced a second-trimester miscarriage (Baud et al., 2020). Evidence also suggests potential vertical transmission in ∼9% of newborns from mothers infected with SARS-CoV-2 (Zeng et al., 2020a). Although these and additional reports have implicated intrauterine passage of the virus from the uterus/endometrium to the placenta and then the fetus (Dong et al., 2020; Egloff et al., 2020; Fenizia et al., 2020; Hu et al., 2020; Richtmann et al., 2020; Vivanti et al., 2020; Zeng et al., 2020a), other reports have not (Chen et al., 2020; Rasmussen et al., 2020).
Studies of gene expression in bulk tissue can lead to erroneous conclusions about the infective risk of specific organs, as demonstrated by the COVID-19 risk map at the single-cell level (Zou et al., 2020). For cells to be infected, the viral surface receptor and/or cofactors must be expressed in the same cell as the proteases. A previous observation analysing gene expression datasets from five publications that used bulk endometrial tissue showed no significant expression of ACE2 and TMPRSS2 (Henarejos-Castillo et al., 2020). This is in contrast to reported expression of the SARS-CoV-2 receptor gene ACE2 at the single-cell level in the decidua and trophoblast cells at the maternal–fetal interface in early pregnancy (Li et al., 2020a).
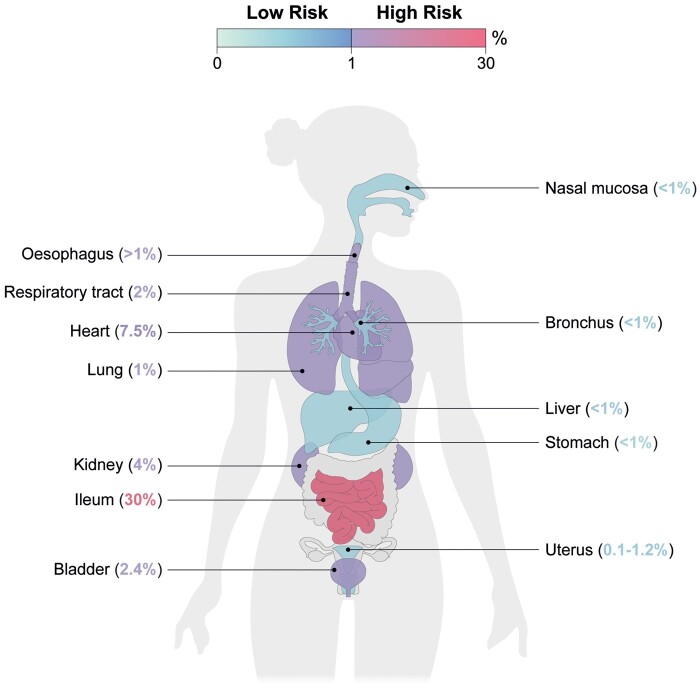
To determine the potential risk of SARS-CoV-2 infectivity of a given organ or tissue, single-cell transcriptomics have been used to report the abundance of ACE2-expressing cells as the virus’s prime receptor for infectivity, with a cut-off value of 1% of cells expressing this gene (Zou et al., 2020). Based on these data, a COVID-19 infection-related risk map was built, in which the nasal mucosa, bronchus, liver and stomach have <1% of cells expressing ACE2 and are considered low-risk for SARS-CoV-2 infection, whereas the percentage of cells expressing ACE2 in the lung (2%), esophagus (1%), ileum (30%), heart (7.5%), kidney (4%), and bladder (2.4%) places them at high risk of infection (Fig. 5). Interestingly, low expression of ACE2 in respiratory and olfactory cells supports the possibility that the virus requires cofactors to facilitate viral entry (Hikmet et al., 2020). That the main target organ for the virus is the lung and not the ileum suggests that cofactors such as NRP1 may play an important role in facilitating SARS-CoV-2 entry into cells (Cantuti-Castelvetri et al., 2020; Qi et al., 2020).
Figure 5.
Potential risk of SARS-CoV-2 endometrial infectivity measured using the abundance of ACE2-positive cells. Schematic representation of potential infectivity risk of SARS-CoV-2 using the percentage of cells positive for ACE2 in different organs of the human body.
Indeed, scRNAseq of lung tissue has revealed high expression of NRP1 in pulmonary epithelial cells, which may compensate for the low presence of ACE2 in these cells (Cantuti-Castelvetri et al., 2020). We found the maximum percentage of endometrial cells expressing ACE2 was generally <1% and only reached 1% positive cells in the epithelial compartment before the mid-secretory phase. Using this classification, the endometrium is predicted to have a low risk of infection (Fig. 5). However, we also analysed expression of the ACE2 enhancer NRP1. NRP1 expression is strikingly high in stromal cells independent of menstrual cycle phase. Our data suggest that presence of this new SARS-CoV-2 entry cofactor in this decidual compartment responsible for the maternal–fetal interface during pregnancy may translate to moderate infectivity potential, as it has been reported that the relative infection rate of SARS-CoV-2 in cells expressing NRP1 but not ACE2 is approximately five-times lower than in ACE2-expressing cells in the presence of high viral load (Cantuti-Castelvetri et al., 2020).
We also analysed a set of potential factors that could facilitate entrance of SARS-CoV-2 in the endometrium, as reported in major human organs (Singh et al., 2020). In this regard, we identified that the receptor gene BSG was highly expressed in all cell types across the menstrual cycle. The receptor gene DPP4, implicated in infection of MERS-CoV, is expressed in the mid- and late-secretory phases in epithelial cells (Li et al., 2020b). Cellular protease genes TMPRSS4 and FURIN are expressed in endometrial cells, opening the possibility that they may participate in viral infection. Interestingly, LY6E and IFITM1, described as cell protectors against entry of SARS-CoV family viruses, are mainly expressed in stromal cells, suggesting that these inhibitors may help prevent or reduce SARS-CoV-2 infection of the endometrium. However, these data should be considered with caution because there is not yet scientific evidence of an active role of alternative machinery in SARS-CoV-2 infection. Until now, the efficiency of BSG as a receptor for SARS-CoV-2 has only been shown in cultures of Vero and 293 T cells in vitro (Singh et al., 2020), while the roles of DPP4, TMPRSS4, FURIN and IFITM1 have only been proposed based on theoretical similarities of SARS-CoV-2 with SARS-CoV and MERS.
The main strength of our study is that single-cell analysis offers spatial and quantitative data, allowing us to determine whether individual endometrial cells simultaneously express the ACE2 receptor with the protease TMPRSS2 and new potential factors of viral entrance. TMPRSS2 can cleave adjacent cells expressing ACE2, but our results resolved this potential synergy. We found a low percentage of cells at risk of infection during the time at which an embryo could implant (0.07% expressing ACE2 with TMPRSS2) and during the late secretory phase (0% expressing ACE2 with TMPRSS2). Although widespread expression of BSG may imply that an alternative cell surface receptor of SARS-CoV-2 may be active in the endometrium at this time, this opens the possibility that BSG participates with CTSB and CTSL proteases, which are highly expressed in epithelial cells. However, the role of potential alternative machinery in SARS-CoV-2 cell infection has not yet been confirmed. NRP1 expression is strikingly high in stromal cells independent of menstrual cycle phase, suggesting infectivity potential during pregnancy. Finally, very few of cells of any type exhibited co-expression of ACE2/NRP1 or ACE2/NRP1/TMPRSS2 throughout the menstrual cycle. Altogether, our findings at the single-cell level suggest that the non-pregnant human endometrium is a low-risk organ for SARS-CoV-2 viral infection at least for canonical cell entry machinery. This study offers a useful resource to guide reproductive decisions when assessing the risk of endometrial infection by SARS-CoV-2 during the preconception period in asymptomatic COVID-19 carriers.
Data availability
All raw data in this study can be found at NCBI’s Gene Expression Omnibus (series accession code GSE111976) and Sequence Read Archive (Accession code SRP135922).
Authors’ roles
F.V. contributed to the conception and design of the study and the acquisition and interpretation of data and drafted the work. W.W. contributed to acquisition, analysis and interpretation of data and drafted the work. I.M. contributed to experimental design and data interpretation. B.R. contributed to data interpretation. S.R.Q. and C.S. contributed to conception and design of the study and the interpretation of data and drafted the work. All the authors have substantively revised the manuscript and approved the submitted version.
Funding
This study was jointly supported by the March of Dimes, Chan Zuckerberg Biohub and MINECO/FEDER (SAF- 2015-67164-R, to C.S.) (Spanish Government), and the European Union’s Horizon 2020 Framework Programme for Research and Innovation (Grant agreement number 874867). W.W. was supported by the Stanford Bio-X Graduate Bowes Fellowship and Chan Zuckerberg Biohub. F.V. was supported by the Miguel Servet Program Type II of ISCIII (CPII18/00020) and the FIS project (PI18/00957).
Conflict of interest
A patent disclosure has been filed for the study with the title ‘Methods for assessing endometrial transformation’ and the global patent number ‘EP 3807648 A2’ under the inventors S.R.Q., C.S., W.W. and F.V. C.S. is Founder and Head of the Scientific Advisory Board of Igenomix SL. S.R.Q is Director of Mirvie. I.M. is partially employed by Igenomix SL. B.R. has no interests to declare.
Supplementary Material
References
- Anders S, Pyl PT, Huber W.. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 2015;31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud D, Greub G, Favre G, Gengler C, Jaton K, Dubruc E, Pomar L.. Second-trimester miscarriage in a pregnant woman with SARS-CoV-2 infection. JAMA 2020;323:2198–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantuti-Castelvetri L, Ojha R, Pedro LD, Djannatian M, Franz J, Kuivanen S, van der Meer F, Kallio K, Kaya T, Anastasina M.. et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 2020;370:eabd2985–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Guo J, Wang C, Luo F, Yu X, Zhang W, Li J, Zhao D, Xu D, Gong Q.. et al. Clinical characteristics and intrauterine vertical transmission potential of COVID-19 infection in nine pregnant women: a retrospective review of medical records. Lancet 2020;395:809–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L, Tian J, He S, Zhu C, Wang J, Liu C, Yang J.. Possible vertical transmission of SARS-CoV-2 from an infected mother to her newborn. JAMA 2020;323:1846–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff C, Vauloup-Fellous C, Picone O, Mandelbrot L, Roques P.. Evidence and possible mechanisms of rare maternal–fetal transmission of SARS-CoV-2. J Clin Virol 2020;128:104447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenizia C, Biasin M, Cetin I, Vergani P, Mileto D, Spinillo A, Gismondo MR, Perotti F, Callegari C, Mancon A.. et al. Analysis of SARS-CoV-2 vertical transmission during pregnancy. Nat Commun 2020;11:5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramberg T, Hofmann H, Möller P, Lalor PF, Marzi A, Geier M, Krumbiegel M, Winkler T, Kirchhoff F, Adams DH.. et al. LSECtin interacts with filovirus glycoproteins and the spike protein of SARS coronavirus. Virology 2005;340:224–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henarejos-Castillo I, Sebastian-Leon P, Devesa-Peiro A, Pellicer A, Díaz-Gimeno P.. SARS-CoV-2 infection risk assessment in the endometrium: viral infection-related gene expression across the menstrual cycle. Fertil Steril 2020;114:223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikmet F, Méar L, Edvinsson Å, Micke P, Uhlén M, Lindskog C.. The protein expression profile of ACE2 in human tissues. Mol Syst Biol 2020;16:e9610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu N-H, Nitsche A.. et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020;181:271–280.e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Gao J, Luo X, Feng L, Liu W, Chen J, Benachi A, De Luca D, Chen L.. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) vertical transmission in neonates born to mothers with coronavirus disease 2019 (COVID-19) pneumonia. Obstet Gynecol 2020;136:65–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang I-C, Bailey CC, Weyer JL, Radoshitzky SR, Becker MM, Chiang JJ, Brass AL, Ahmed AA, Chi X, Dong L.. et al. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog 2011;7:e1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Chen L, Zhang J, Xiong C, Li X.. The SARS-CoV-2 receptor ACE2 expression of maternal–fetal interface and fetal organs by single-cell transcriptome study. PLoS One 2020a;15:e0230295–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhang Z, Yang L, Lian X, Xie Y, Li S, Xin S, Cao P, Lu J.. The MERS-CoV receptor DPP4 as a candidate binding target of the SARS-CoV-2 spike. iScience 2020b;23:101400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques FZ, Pringle KG, Conquest A, Hirst JJ, Markus MA, Sarris M, Zakar T, Morris BJ, Lumbers ER.. Molecular characterization of renin–angiotensin system components in human intrauterine tissues and fetal membranes from vaginal delivery and cesarean section. Placenta 2011;32:214–221. [DOI] [PubMed] [Google Scholar]
- McGinnis CS, Murrow LM, Gartner ZJ.. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst 2019;8:329–337.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa P, Gomez J, Grobe JL, Sigmund CD.. The renin–angiotensin system in the central nervous system and its role in blood pressure regulation. Curr Hypertens Rep 2020;22:7–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaender S, Mar KB, Michailidis E, Kratzel A, Boys IN, V'kovski P, Fan W, Kelly JN, Hirt D, Ebert N.. et al. LY6E impairs coronavirus fusion and confers immune control of viral disease. Nat Microbiol 2020;5:1330–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle KG, Tadros MA, Callister RJ, Lumbers ER.. The expression and localization of the human placental prorenin/renin–angiotensin system throughout pregnancy: roles in trophoblast invasion and angiogenesis? Placenta 2011;32:956–962. [DOI] [PubMed] [Google Scholar]
- Puelles VG, Lütgehetmann M, Lindenmeyer MT, Sperhake JP, Wong MN, Allweiss L, Chilla S, Heinemann A, Wanner N, Liu S.. et al. Multiorgan and renal tropism of SARS-CoV-2. N Engl J Med 2020;383:590–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi F, Qian S, Zhang S, Zhang Z.. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem Biophys Res Commun 2020;526:135–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen SA, Smulian JC, Lednicky JA, Wen TS, Jamieson DJ.. Coronavirus disease 2019 (COVID-19) and pregnancy: what obstetricians need to know. Am J Obstet Gynecol 2020;222:415–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richtmann R, Torloni MR, Oyamada Otani AR, Levi JE, Crema Tobara M, de Almeida Silva C, Dias L, Miglioli-Galvão L, Martins Silva P, Macoto Kondo M.. Fetal deaths in pregnancies with SARS-CoV-2 infection in Brazil: a case series. Case Rep Womens Health 2020;27:e00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M, Bansal V, Feschotte C.. A single-cell RNA expression map of human coronavirus entry factors. Cell Rep 2020;32:108175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, Hao Y, Stoeckius M, Smibert P, Satija R.. Comprehensive integration of single-cell data. Cell 2019;177:1888–1902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaz-Silva J, Carneiro MM, Ferreira MC, Pinheiro SVB, Silva DA, Silva AL, Witz CA, Reis AM, Santos RA, Reis FM.. The vasoactive peptide angiotensin-(1-7), its receptor Mas and the angiotensin-converting enzyme type 2 are expressed in the human endometrium. Reprod Sci 2009;16:247–256. [DOI] [PubMed] [Google Scholar]
- Vivanti AJ, Vauloup-Fellous C, Prevot S, Zupan V, Suffee C, Do Cao J, Benachi A, De Luca D.. Transplacental transmission of SARS-CoV-2 infection. Nat Commun 2020;11:3572–3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls AC, Park Y-J, Tortorici MA, Wall A, McGuire AT, Veesler D.. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020;181:281–292.e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Chen W, Zhang Z, Deng Y, Lian J-Q, Du P, Wei D, Zhang Y, Sun X-X, Gong L.. et al. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct Target Ther 2020a;5:283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Vilella F, Alamá P, Moreno I, Mignardi M, Isakova A, Pan W, Simón C, Quake SR.. Single-cell transcriptomic atlas of the human endometrium during the menstrual cycle. Nat Med 2020b;26:1644–1653. [DOI] [PubMed] [Google Scholar]
- Wang Y, Pringle KG, Sykes SD, Marques FZ, Morris BJ, Zakar T, Lumbers ER.. Fetal sex affects expression of renin–angiotensin system components in term human decidua. Endocrinology 2012;153:462–468. [DOI] [PubMed] [Google Scholar]
- Yang Z-Y, Huang Y, Ganesh L, Leung K, Kong W-P, Schwartz O, Subbarao K, Nabel GJ.. pH-dependent entry of severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein and enhanced by dendritic cell transfer through DC-SIGN. J Virol 2004;78:5642–5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang R, Gomez Castro MF, McCune BT, Zeng Q, Rothlauf PW, Sonnek NM, Liu Z, Brulois KF, Wang X, Greenberg HB.. et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci Immunol 2020;5:eabc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H, Xu C, Fan J, Tang Y, Deng Q, Zhang W, Long X.. Antibodies in infants born to mothers with COVID-19 pneumonia. JAMA 2020a;323:1848–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Xia S, Yuan W, Yan K, Xiao F, Shao J, Zhou W.. Neonatal early-onset infection with SARS-CoV-2 in 33 neonates born to mothers with COVID-19 in Wuhan, China. JAMA Pediatr 2020b;174:722–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou X, Chen K, Zou J, Han P, Hao J, Han Z.. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front Med 2020;14:185–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw data in this study can be found at NCBI’s Gene Expression Omnibus (series accession code GSE111976) and Sequence Read Archive (Accession code SRP135922).