Abstract
While coding variants often have pleiotropic effects across multiple tissues, non-coding variants are thought to mediate their phenotypic effects by specific tissue and temporal regulation of gene expression. Here, we dissected the genetic and functional architecture of a genomic region within the FTO gene that is strongly associated with obesity risk. We show that multiple variants on a common haplotype modify the regulatory properties of several enhancers targeting IRX3 and IRX5 from megabase distances. We demonstrate that these enhancers impact gene expression in multiple tissues, including adipose and brain, and impart regulatory effects during a restricted temporal window. Our data indicate that the genetic architecture of disease-associated loci may involve extensive pleiotropy, allelic heterogeneity, shared allelic effects across tissues, and temporally-restricted effects.
One Sentence Summary:
Enhancer interactions regulate the activity of obesity-associated genes IRX3 and IRX5 in both adipose and brain tissue.
Although genome-wide association studies (GWAS) have contributed extensively to complex disease mapping, our understanding of the genetic architecture and molecular mechanisms underlying most disease associations remains incomplete (1, 2). Recent studies suggest pervasive pleiotropy of regulatory variants modulating gene expression across multiple tissues, impacting seemingly disparate disease phenotypes (3, 4). We set out to dissect the genetic architecture and phenotypic implications of a well-studied locus associated with human obesity. GWAS have identified common variants in the FTO gene as the strongest genetic association with obesity in humans (5). Much effort has been directed towards identifying the causal variant, gene, and tissues underlying this association. The associated region is within a large topologically associated domain (TAD) of approximately 2 Mb encompassing FTO, RPGRIP1L, and the IRXB cluster (including IRX3, IRX5, and IRX6) (6). As a consequence of this arrangement, the obesity-associated variants could impact the regulation of any or all of these genes. In fact, most of these genes have been independently implicated in body weight management phenotypes, leading to additional controversy within the field as to which of these genes mediate the genetic association with obesity (7–10). In addition, while compelling evidence implicates central nervous system phenotype such as food preference and feeding behavior underlying the association with body mass index (BMI) (5, 11), alternative models involving altered thermogenesis, autonomous to adipose tissue, have also been put forth as putative mechanisms (7, 8). To address these discrepancies, we applied an integrated approach to mechanistically dissect the genetic and functional architecture of the obesity GWAS signal emanating from the FTO locus.
To ascertain the pattern of long-range genomic interactions in the locus, we generated a comprehensive chromatin interaction map in cell types relevant to obesity. We performed in situ promoter capture Hi-C (PCHi-C) in human SGBS preadipocytes and in hypothalamic arcuate-like neurons derived from human induced pluripotent stem cells (hiPSCs). PCHi-C contact maps from both cell types, and additional 4C-seq data, revealed long-range interactions between the obesity-associated locus and promoters of IRX3 and IRX5, but not those of IRX6 or FTO/RPGRIP1L (Fig. 1A and fig. S1A). Similar results were obtained from an enhancer capture Hi-C dataset in primary human pre-adipocytes (fig. S1B) (12). Because this locus is highly conserved between humans and mice (fig. S2A), we engineered a mouse model (mmFtoΔ20) harboring a 20,204 bp deletion spanning the orthologous obesity-associated interval in Fto (fig. S2B). Using fluorescence in situ hybridization (FISH) as an orthogonal assay to PCHi-C, we interrogated the 3D organization of this region in vivo, in mouse brains from mmFtoΔ20 heterozygous animals. Designing fosmid-based probes for regions encompassing the Fto/Rpgrip1l, Irx3, Irx5, Irx6 promoters, and the Fto obesity-associated locus, as well as the region directly adjacent to the 20 kb deletion (Fig. 1B), we determined the pattern of interactions between the obesity-associated region and genes in the Fto-Irxb locus. Consistent with our PCHi-C and 4C-seq results, FISH data from WT alleles in cerebellum revealed significantly increased colocalization (≤200 nm) of the Irx3 and Irx5 promoters with the Fto obesity-associated interval in WT alleles, compared with deletion alleles and cortex cells that do not express Irx3 and Irx5. We obtained similar colocalization data in lung cells, supporting our previous observations that the obesity-associated interval harbors lung enhancers (7). Conversely, the 20 kb deletion had no impact on the distance between the Fto/Rpgri1L or Irx6 promoters and the Fto obesity-associated interval (Fig. 1C and D, fig. S2C). These observations support a model of chromatin compaction at this locus with the obesity-associated region physically interacting with both IRX3 and IRX5 in humans and mice.
Fig. 1. Regulatory architecture of obesity-associated noncoding elements within FTO.
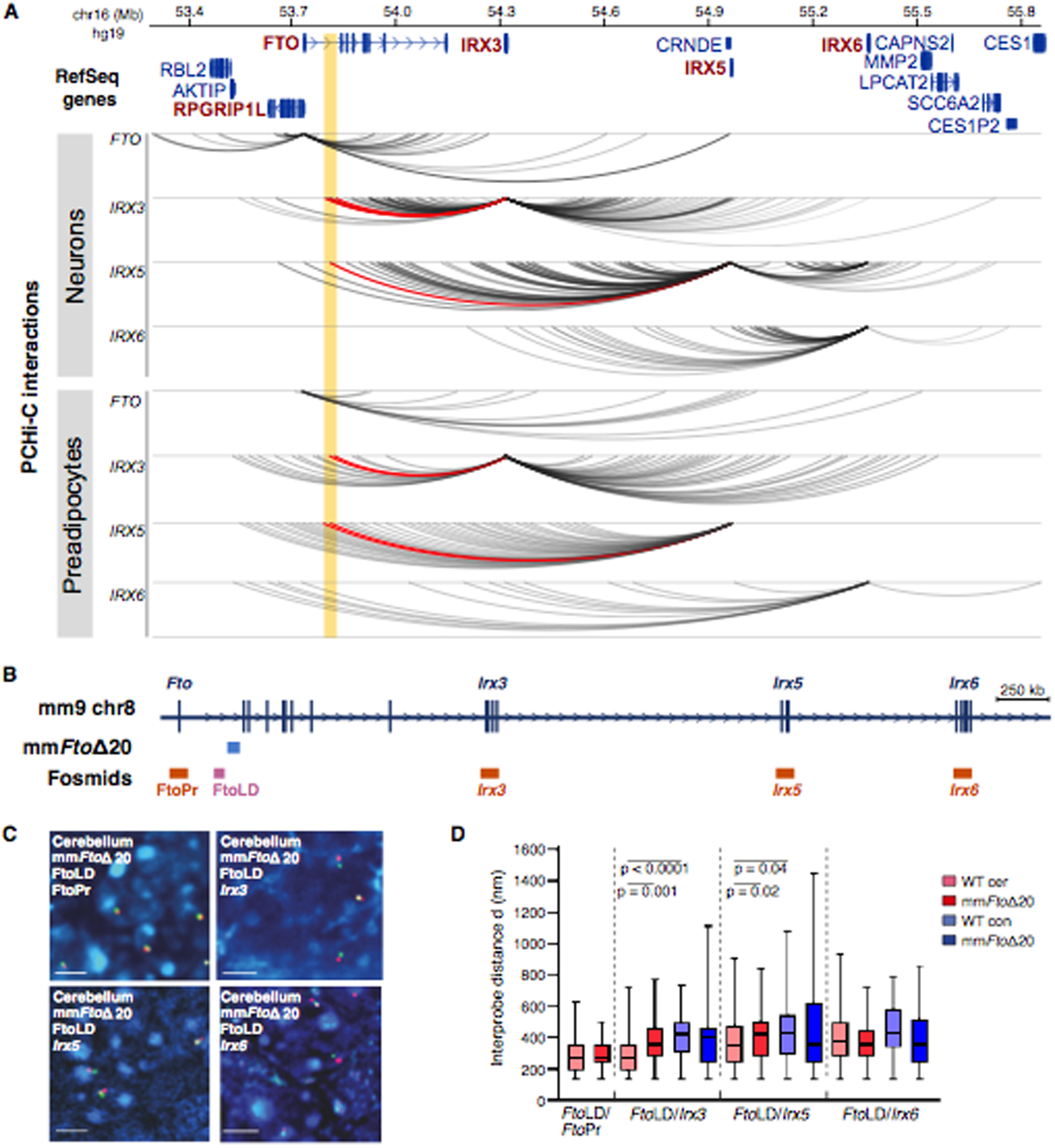
(A) PCHi-C interactions emanating from the FTO, IRX3, IRX5, and IRX6 promoters in human SBGS cells (preadipocytes) and iPSC-derived hypothalamic arcuate-like neurons. The yellow strip highlights the obesity-associated interval. PCHi-C interactions are presented as gray colored arcs. Red arcs highlight interactions of IRX3 and IRX5 promoters with obesity-associated region. (B) 1.1 Mb region analyzed by FISH in mouse cerebellum encompassing Fto, Irx3, Irx5, and Irx6 genes. Fragment deleted in the mmFtoΔ20 mouse is indicated in blue. Fosmids used for analysis in D are indicated in orange and light pink. (C) 3D-FISH with Fto, Irx3, Irx5, and Irx6 probes (red) and directly distal FTO obesity-associated interval (green), counterstained with DAPI (blue). Bars, 5 μm. (D) Box plots represent the distribution of interprobe distances (d in nm) between different probe combinations in Irx3-expressing (cerebellum: cer) and non-expressing (cortex: con) brain tissue of mmFtoΔ20 heterozygous mice. Lines represent median. Statistical significance of differences between data sets was examined using Mann Whitney U tests. n= 50 – 60 WT and mmFtoΔ20 alleles each per slide. Abbreviation: FtoLD (FTO obesity-associated interval); FtoPr (Fto promoter).
We next explored the biological relevance of these observations in vivo. We genetically engineered germline null (−/−) and heterozygous (+/−) alleles for Irx3, Irx5, and Irx6 in mice (fig. S3). At 20 weeks, Irx3−/− animals displayed a 15–20% blunting in weight gain compared to the WT control littermates, as well as a reduction of total fat mass (10–15%), activation of molecular markers of browning in white adipose tissue (WAT), and improved glucose tolerance, mimicking phenotypes we have previously shown (fig. S4) (7). While the most striking feature of Irx5−/− mice is early postnatal lethality, Irx5 heterozygous mice (Irx5+/−) were viable and thrived. Similar to Irx3−/−, Irx5+/− mice exhibited an anti-obesity phenotype with 15–20% weight reduction, loss of body fat mass (5%), activation of browning in WAT, and improved glucose tolerance (fig. S5). Irx6 knockout (Irx6−/−) mice showed none of these metabolic phenotypes (fig. S6). Altogether, our in vivo mouse models support our chromatin conformation data implicating Irx3 and Irx5, but not Irx6, as potentially mediating the genetic association with obesity.
The phenotypic impact of IRX3 and IRX5 on adipocyte biology has been described (7, 8). Specifically, a SNP (rs1421085) modulates IRX3 and IRX5 expression in preadipocytes and regulates an adipose thermogenesis program (8). These data, however, do not provide an immediate explanation for the well-described association of variants within FTO with eating behavior and, more specifically, eating preferences, such as increased caloric intake (5, 11, 13). Toward that end, we have previously shown that the hypothalamic expression of a dominant-negative IRX3 isoform in mice phenocopies the organismal level metabolic phenotypes seen in germline Irx3 null mice (7). To interrogate the impact of Irx3 in molecular and physiological brain phenotypes associated with obesity, we performed transcriptomic analysis (RNA-seq) on hypothalami from Irx3−/− mice and WT littermates. Gene ontology (GO) enrichment analysis showed that, of the 359 up-regulated genes, at least 103 are involved in neurodevelopment and cellular processes, such as cell communication and synaptic signaling, consistent with the well-known roles of Irx3 in brain development (fig. S7A) (14–16). Using the ToppGene Suite database, we investigated GO categories for disease links and found that the top ranked diseases associated with these differentially expressed genes are obesity, diabetes, and impaired glucose tolerance (Fig. 2A, fig. S7A and B and Table S1), supporting the notion that Irx3 expression in brain may coordinate a genetic program involved in metabolism. To examine whether Irx3 plays a role in food intake or macronutrient preference, we subjected a cohort of adult Irx3−/− and WT control littermates to a series of two-bottle choice experiments in which all mice were offered the choice between water and a range of nutritive and non-nutritive tastants (17). We found that obesity-resistant Irx3−/− mice display a reduced preference for sucrose, but not lipid or protein, compared to WT animals (Fig. 2B and fig. S7C). Altered sweet preference has not been shown as a phenotype in humans harboring risk alleles in the FTO obesity-associated region. To test this, we obtained GWAS summary statistics from 118,950 genotyped individuals responding to a sweet preference questionnaire from 23andMe (Table S2). A GWAS of these data indicated that SNPs within the FTO obesity-associated region represent the second strongest association with sweet preference in humans, with the C allele of rs1421085 associated with sweet food preference over salty (3.6X10−23, OR=1.1) (Fig. 2C). Taken together, our in vivo mouse data establish a central nervous system role of Irx3 in the regulation of metabolism and feeding behavior analogous to phenotypes associated with allelic variants of obesity-associated SNPs within FTO in humans, including alterations in consummatory behavior. Previous work has described reciprocal counterregulatory mechanisms between peripheral energy expenditure and energy intake, with perturbations in diet and nutritional status inducing long-term changes in hypothalamic neurocircuit development (18). Future work should determine whether the alterations in feeding behavior in Irx3−/− mice result from primary, autonomous dysfunction of regulatory circuits within the central nervous system, including the hypothalamus, or are secondary to peripheral effects, through the intersection of neuro-hormonal cues from adipose and other peripheral tissues.
Fig. 2. Irx3 acts in the brain to regulate metabolism and changes in macronutrient selection.
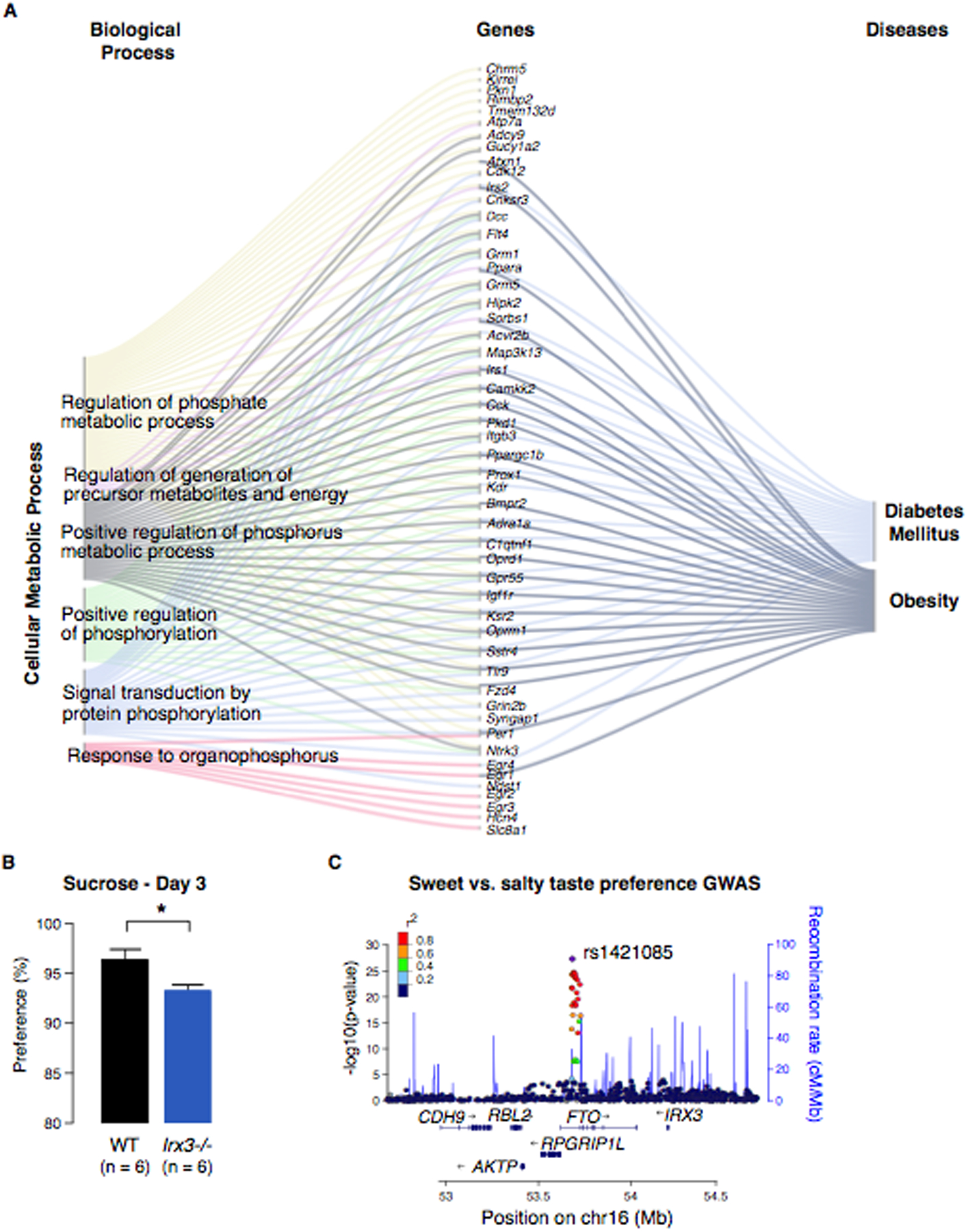
(A) Expression analysis of differentially expressed genes between hypothalami of Irx3−/− and WT mice using Gene Ontology (GO) annotations. Sankey flow diagram showing all genes upregulated in the hypothalami from Irx3−/− animals with high enrichment for Cellular Metabolic Processes and the top ranked diseases related to them. Gene symbols are shown. (B) Two-bottle choice experiment comparing Irx3−/− and WT mice. Data are expressed as mean ± SEM. *P < 0.05 compared to WT group. Error bars represent standard deviations. (C) A regional association plot of the FTO locus. LocusZoom was used to plot the negative log10 p-value of every SNP within +/− 700 kb of rs1421085, the lead SNP in the locus.
Having uncovered a central nervous system role of Irx3 in metabolism and feeding behavior, we next sought to characterize the regulatory potential of obesity-associated SNPs within FTO. To functionally classify regulatory variants in neurons and adipocytes, thought to represent tissues that participate in the genetics of obesity in humans (19), we used orthogonal computational and experimental approaches. For computational regulatory variant predictions, we derived multiple variant features from sequence-based methods which harness cross-species functional sequence conservation and sequence-based regulatory evidence (20). Experimentally, we used a Massively Parallel Reporter Assay (MPRA) to identify variants located in enhancers in hippocampal (HT22) and preadipocyte (3T3-L1) mouse cell lines. We tested all 87 common (MAF>=5%) variants in strong linkage disequilibrium (r2>0.8) with the lead obesity GWAS associated SNP rs1558902 (19). We found 21 SNPs in 3T3-L1 preadipocytes and 18 SNPs in HT22 neuronal cells located in enhancers in at least three replicates tested in each cell line (Table S3). Of these, 5 SNPs displayed allelic-specific enhancer activities in preadipocyte and/or neuronal cells. Each was located in independent enhancers spread over 31 kb (Fig. 3A, fig. S8 and Tables S4 and S5). Using a luciferase reporter assay, we confirmed allele-specific enhancer properties and directional effects of 4 variants in preadipocytes, 2 of which changed regulatory activity in neuronal cells as well (Fig. 3B). Of note, 3 of the 4 SNPs map within accessible chromatin regions in human adipose and brain tissues, assayed by the Roadmap Epigenomics Consortium (Fig. 3A). In addition, we confirmed that all accessible variants score highest across multiple, orthogonal sequence-based computational metrics, including high functional conservation scores for the variant flanking 120bp regions, as evaluated with PMCA (21) (Table S6), sequence-based predicted functional significance scores < 0.01, as evaluated with DeepSEA (22) (Table S7), and all four SNPs showed remarkably consistent allele-specific chromatin accessibility with the Basset model when comparing the experimentally-derived allelic activity in pre-adipocytes and hypothalamic neurons (Table S8). All 4 SNPs are co-inherited as one common haplotype, with each allele in the obesity-risk haplotype associated with increased enhancer activity (Fig. 3C), suggesting that they may coordinately regulate target gene expression in the same direction (LDhap tool: https://ldlink.nci.nih.gov). Our data suggest that multiple genetic variants in this locus may regulate gene expression in both adipose and neuronal tissues. This supports a model in which GWAS signals may result from a complex genetic architecture whereby allelic heterogeneity of multiple regulatory variants in distinct regulatory elements imparts shared effects across tissues, regulating the quantitative and spatial expression of multiple genes (23). We next determined the impact of these enhancers on gene expression. Because all four regulatory regions with allele-specific enhancer properties map within the 20 kb region that we deleted in the mouse genome (fig. S2B), we used mmFtoΔ20 mice to evaluate the impact, in vivo, of this deletion on the expression of neighboring genes in adipose and brain tissues. We initially assayed the expression of genes in the Fto-Irxb cluster during adipocyte differentiation. We isolated primary preadipocytes from mmFtoΔ20 and WT mice and observed a decreased expression of Irx3 and Irx5 in mmFtoΔ20, but not of other genes in the locus (Fig. 4A). The impact of deleting these enhancers on the expression of Irx3 and Irx5 was restricted to preadipocytes, with no effect on expression in mature adipocytes, as previously described (8).
Fig. 3. Functional variants within the FTO association locus modulate enhancer activity in brain and adipose.
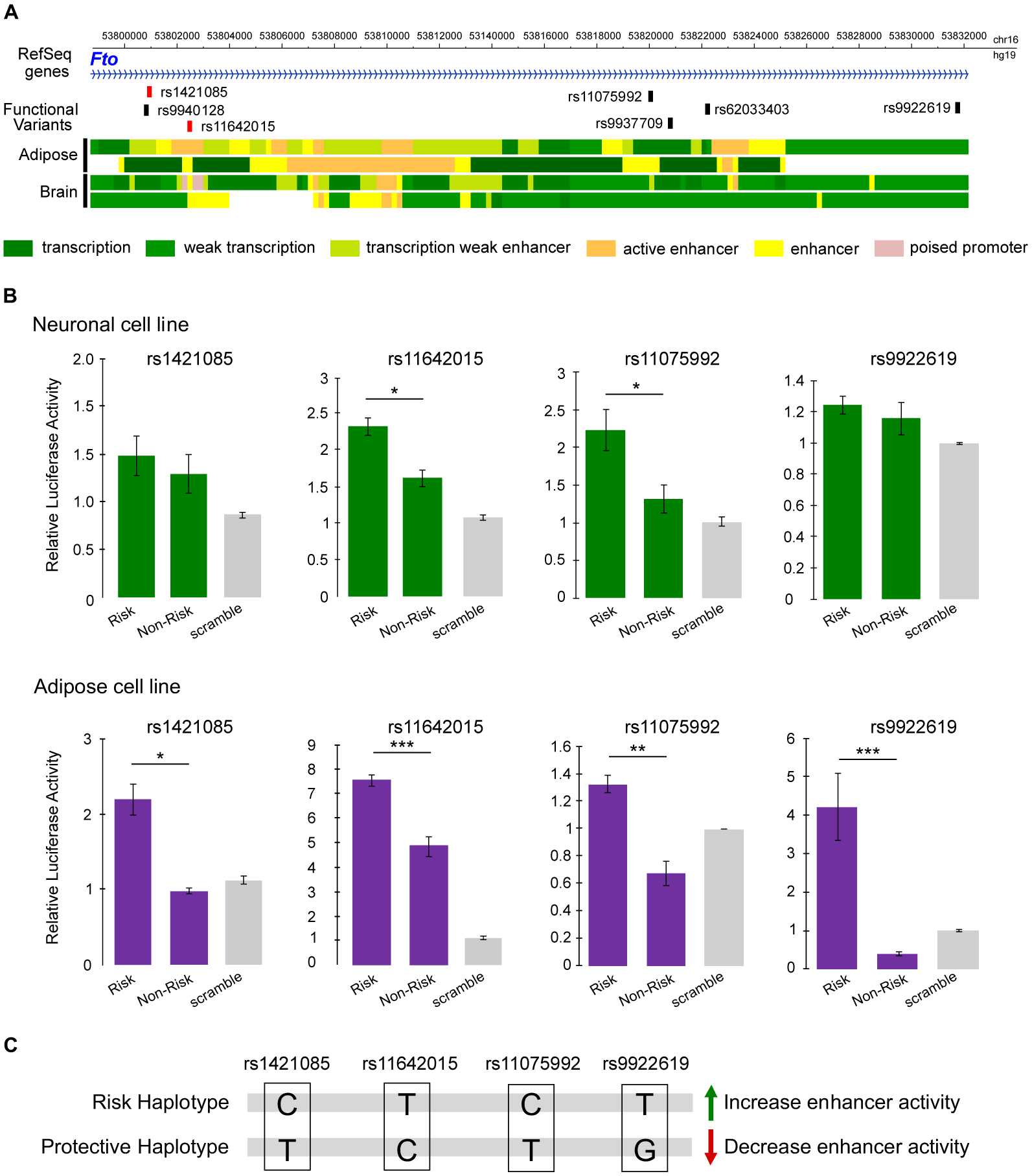
(A) Functional variants that showed allele-specific activity using MPRA (black boxes) and PMCA (red boxes). Colored bars indicate the chromatin state annotations from Roadmap Epigenomics Project. Tissues: adipose-derived mesenchymal stem cell cultured imputed (E025) and adipose nuclei imputed (E063); brain hippocampus middle (E071) and fetal brain male (E081). (B) Comparison of allele-specific activity of four variants in the FTO obesity-associated interval using luciferase reporter assay. The plots show the mean ± SEM from five triplicate experiments. *P < 0.05, **P < 0.01, and ***P < 0.001. (C) Segregation of alleles by risk or non-risk haplotype and effect on enhancer activity.
Fig. 4. Evaluation of enhancer activity in the FTO obesity-associated locus in neuronal and adipose tissues.
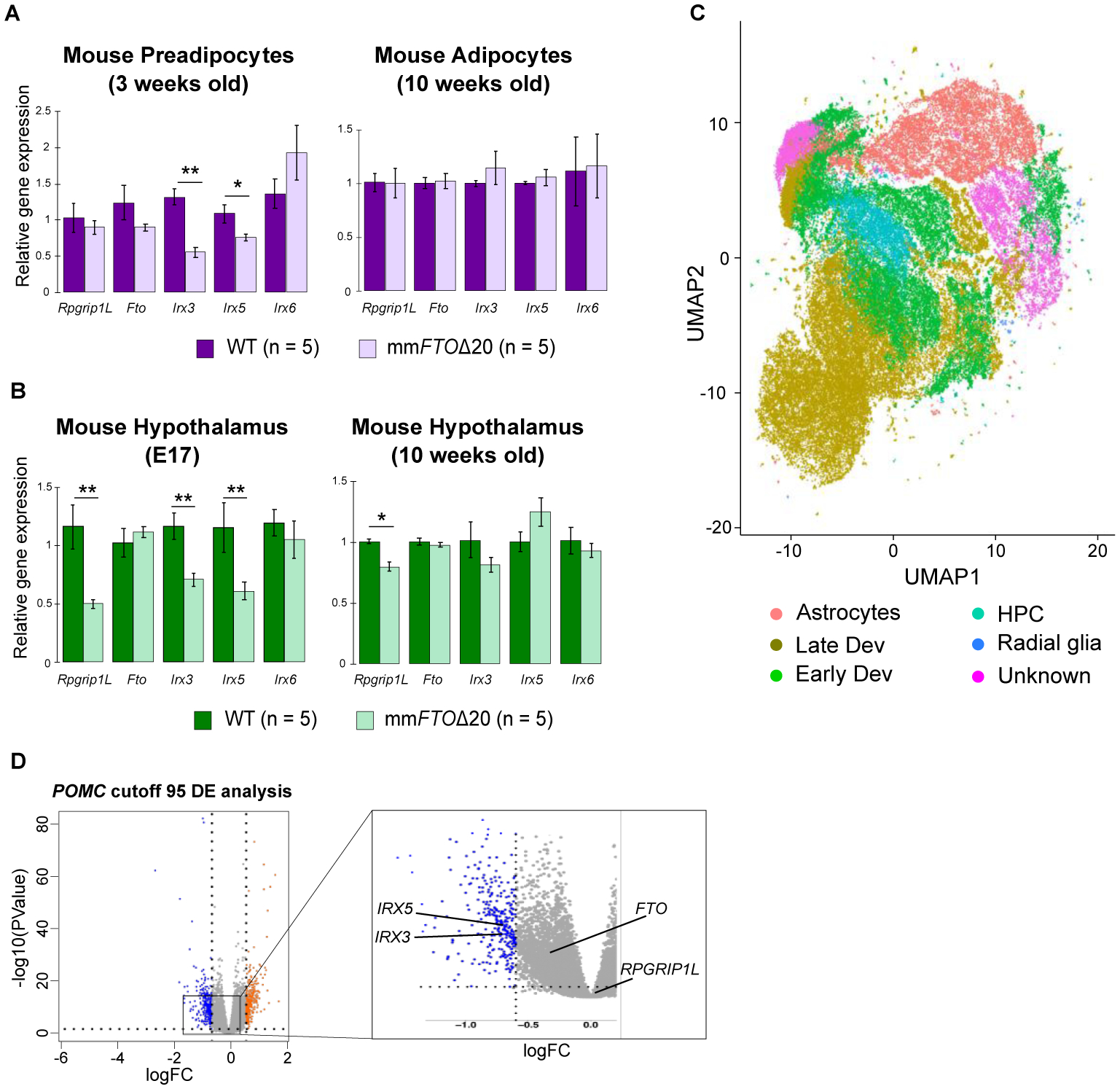
Relative expression of Rpgrip1L, Fto, Irx3, Ixr5, and Irx6 genes in (A) mouse preadipocyte cells, adipose tissue, and (B) hypothalamus. (C) UMAP plot showing the different cell populations identified using single cell sequencing. (D) Volcano plot of the differential gene expression (DE) analysis between WT and hsFTOΔ36 hypothalamic precursor cells with POMC cutoff 95 (counts) and KNN K=11. Gray dots represent genes not significantly changed. Blue and orange dots are genes significantly down and up-regulated, respectively. The log fold change (logFC) is shown on the x axis and the negative log10 of the adjusted P value is shown on the y axis (logFC cutoff > 0.6 or < −0.6, and adjusted P value < 0.05 as significantly differentially expressed). IRX3 and IRX5 are significantly differentially expressed across two conditions with KNN K ranging from 10 to 13 (IRX3), from 11 to 13 (IRX5), and cutoff value above 80 or 85 (counts). The abbreviations are: HPC (hypothalamic progenitor cells); Early Dev (hypothalamic neurons at early development time point); Late Dev (hypothalamic neurons at late development time point). For qPCR analysis data are expressed as mean ± SEM. *P < 0.05 and **P < 0.01 compared to WT.
We next assayed the impact of the 20 kb deletion on gene expression in mouse hypothalamus during embryonic development (E17) as well as in adult mice (10 weeks). At E17, the 20 kb deletion leads to downregulation of Irx3 and Irx5, with no impact on the expression of Fto and Irx6 (Fig. 4B). Similar to adipose, this effect was restricted to embryonic development, with no alterations in Irx3 and Irx5 expression in adult hypothalami. To further explore the temporally-restricted expression of Irx3 and Irx5 in the developing hypothalamus, we assessed single-cell gene expression across windows of mouse hypothalamic development in mice (24), and determined that the expression of Irx3 and Irx5 is highest at mid-gestation and decreases steadily afterwards, being barely detectable in adult neurons (fig. S9). The expression of Rpgrip1L was also decreased in hypothalami of mmFtoΔ20 mice (Fig. 4B), raising the possibility that regulation of Rpgrip1l in the brain may also contribute to obesity risk, as previously suggested (10).
Our data suggest that variants in multiple enhancers within the FTO obesity-associated region regulate the expression of multiple genes in at least two major obesity-relevant tissues, adipose and brain, in mice. Next, we tested the impact of the obesity-associated region on gene expression in human hypothalamic neuronal precursors. We first assayed the dynamic expression of IRX3 and IRX5 during differentiation of human iPSCs into hypothalamic neurons and observed that IRX3 and IRX5 expression is highly correlated and peaks at an early stage of hypothalamic neuronal differentiation, decreasing at later developmental stages, paralleling the observations in mice (fig. S10A and B). These data further support the possibility that some of the allelic effects of obesity-associated SNPs on gene expression may involve developmental phenotypes restricted to specific temporal windows and not detected in differentiated, adult tissues. A recent report uncovered evidence that the FTO locus variants have effects on BMI in early childhood (25), further raising the prospect that the association with BMI may involve a combination of developmental and growth phenotypes.
To test the effect of modulating these enhancers in a model of human hypothalamic neurons, we subsequently generated in human iPSCs a genomic deletion of a 36,100 bp segment which encompasses the FTO obesity-associated locus and corresponds to the deletion engineered in mmFtoΔ20 mice. We differentiated WT and 36,100 bp deletion (hsFTOΔ36) iPSCs into hypothalamic arcuate-like neurons (fig. S10C and D) (26–28). We performed single cell RNA sequencing (scRNA-seq) in 91,825 cells at the neuron progenitor stage to assess transcriptome differences between WT and hsFTOΔ36 cells. Single-cell transcriptomic profiling identified distinct cell populations within the hypothalamic neuron precursor stage, grouped into distinct subtypes. We defined different developmental stages and cell types based on the expression of known neuronal markers (29, 30). Cell subtypes were designated as (1) hypothalamic neurons at late development time point (Late Dev), (2) hypothalamic neurons at an early developmental time point (Early Dev), (3) hypothalamic progenitor cells (HPC), and (4) radial glia, with all four subtypes together constituting the neurogenic lineage (Fig. 4C and Table S9). We found IRX3 and IRX5 expressed in all hypothalamic cell subtypes. To assay for alterations in gene expression in cellular sub-groups, we clustered cells based on the expression of 8 major neural and hypothalamic markers, including ARNT2, NES, NEUROD1, NHLH2, NKX2–1, NPY, OTP, and POMC (fig. S11). We found that only in cells expressing POMC, which is critical in regulating normal feeding behavior and energy homeostasis, the deletion of the 36 kb resulted in reduced expression of IRX3 and IRX5 compared to WT cells, supporting our findings in mouse hypothalami (Fig. 4D). No other gene in the locus was differentially expressed between the two groups in any other cell type cluster. While we performed our analysis in hypothalamic cells, there currently is no clear delineation of the precise brain cell populations in which the expression of IRX3 and IRX5 is regulated by enhancers and allelic variants within these enhancers in the obesity-associated region. Future work tackling this outstanding question will be critical to demarcate the molecular, cellular, and organismal phenotypes involved in obesity susceptibility in this locus.
Taken together, our data highlight the complexities that arise during the functional dissection of disease-associated loci in humans. Recent work has suggested extensive pleiotropy of loci, SNPs, and gene sets underlying associations with polygenic traits in humans (4). Also, GTEx has shown that the regulatory effects of eQTLs tend to be highly shared across tissues (31). Furthermore, the impact of regulatory variants on molecular phenotypes is often dependent on developmental context, with changes in gene expression restricted to specific temporal windows (32). Our findings support all these observations, demonstrating how the collective effects of regulatory variants are integrated across tissues and developmental stages and result in a convergence of phenotypes reminiscent of homeostatic mechanisms governing complex physiological traits in vivo, such as body weight regulation.
There are important limitations to our study. The choice of immortalized cell lines for the reporter assays may mask allelic effects of SNPs that would be seen in primary cells. Also, the manipulation of candidate genes in mice may result in organismal phenotypes that are quantitatively and qualitatively different than the small effect phenotypes elicited by allelic variants of SNPs associated with the human trait. Finally, the congruent macronutrient preference phenotypes we describe between Irx3−/− mice and humans represent but a subset of the feeding behavior phenotypes associated with this locus in humans. This may reflect species differences in the function of these genes, but also that there are other functions associated with IRX3, IRX5, or other genes in the locus (RPGRIP1L or FTO) that contribute to the BMI association in humans.
Our work suggests that the genetic architecture of a disease-associated locus may include allelic heterogeneity, with multiple variants modifying the regulatory properties of distinct enhancers with broad tissue-specificity and regulating multiple genes in limited temporal windows. These insights provide a mechanistic framework to explain the genetic and functional architecture of GWAS loci, predicting that it will often encompass multiple phenotypic mechanisms that ultimately converge to modulate disease susceptibility.
Supplementary Material
Acknowledgements:
We thank Romain Barrès, Michelle C. Ward and Xiaochang Zhang for comments and valuable suggestions. We thank Juan Tena and Jose Luis Gómez-Skarmeta for help with the 4C-seq analyses; and Sebastian Pott for assistance with the single cell RNA-seq analysis; We would also like to thank Martin Wabitsch for his generous gift of the SGBS human preadipocyte cells and David Schubert for Murine HT22 hippocampal neuronal cell line. We thank the customers of 23andMe. Inc. for answering surveys and participating in this research.
Funding:
This research was supported by a Novo Nordisk Foundation Challenge Grant (NNF18OC0033754) to M.A.N.; grants from the National Institutes of Health, R01HL128075, R01HL119577, 5P30DK020595, and R01DK114661 to M.A.N., and RO1HL085197 to C.O. IW and WAB are supported by MRC University Unit programme grant MC_UU_00007/2.
Footnotes
Data and materials availability:
RNA-seq (E-MTAB-10186), scRNA-seq (E-MTAB-10201), PCHi-C sequencing (E-MTAB-10200), ATAC-seq (E-MTAB-10257) and 4C-seq data (E-MTAB-10195) deposited at https://www.ebi.ac.uk/arrayexpress/.
References and Notes
- 1.Timpson NJ, Greenwood CMT, Soranzo N, Lawson DJ, Richards JB, Genetic architecture: The shape of the genetic contribution to human traits and disease. Nat. Rev. Genet (2018),, doi: 10.1038/nrg.2017.101. [DOI] [PubMed] [Google Scholar]
- 2.Visscher PM, Wray NR, Zhang Q, Sklar P, McCarthy MI, Brown MA, Yang J, 10 Years of GWAS Discovery: Biology, Function, and Translation. Am. J. Hum. Genet (2017),, doi: 10.1016/j.ajhg.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science (2020), doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Watanabe K, Stringer S, Frei O, Umićević Mirkov M, de Leeuw C, Polderman TJC, van der Sluis S, Andreassen OA, Neale BM, Posthuma D, A global overview of pleiotropy and genetic architecture in complex traits. Nat. Genet (2019), doi: 10.1038/s41588-019-0481-0. [DOI] [PubMed] [Google Scholar]
- 5.Cecil JE, Tavendale R, Watt P, Hetherington MM, Palmer CNA, An Obesity-Associated FTO Gene Variant and Increased Energy Intake in Children. N. Engl. J. Med 359, 2558–2566 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B, Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature (2012), doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smemo S, Tena JJ, Kim K-H, Gamazon ER, Sakabe NJ, Gómez-Marín C, Aneas I, Credidio FL, Sobreira DR, Wasserman NF, Lee JH, Puviindran V, Tam D, Shen M, Son JE, Vakili NA, Sung H-K, Naranjo S, Acemel RD, Manzanares M, Nagy A, Cox NJ, Hui C-C, Gomez-Skarmeta JL, a Nóbrega M, Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 507, 371–5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Claussnitzer M, Dankel SN, Kim K-H, Quon G, Meuleman W, Haugen C, Glunk V, Sousa IS, Beaudry JL, Puviindran V, Abdennur NA, Liu J, Svensson P-A, Hsu Y-H, Drucker DJ, Mellgren G, Hui C-C, Hauner H, Kellis M, FTO Obesity Variant Circuitry and Adipocyte Browning in Humans. N. Engl. J. Med 373, 895–907 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fischer J, Koch L, Emmerling C, Vierkotten J, Peters T, Brüning JC, Rüther U, Inactivation of the Fto gene protects from obesity. Nature (2009), doi: 10.1038/nature07848. [DOI] [PubMed] [Google Scholar]
- 10.Stratigopoulos G, Burnett LC, Rausch R, Gill R, Penn DB, Skowronski AA, LeDuc CA, Lanzano AJ, Zhang P, Storm DR, Egli D, Leibel RL, Hypomorphism of Fto and Rpgrip1l causes obesity in mice. J. Clin. Invest (2016), doi: 10.1172/JCI85526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wardle J, Llewellyn C, Sanderson S, Plomin R, The FTO gene and measured food intake in children. Int. J. Obes 33, 42–45 (2009). [DOI] [PubMed] [Google Scholar]
- 12.Madsen JGS, Madsen MS, Rauch A, Traynor S, Van Hauwaert EL, Haakonsson AK, Javierre BM, Hyldahl M, Fraser P, Mandrup S, Highly interconnected enhancer communities control lineage-determining genes in human mesenchymal stem cells. Nat. Genet (2020), doi: 10.1038/s41588-020-0709-z. [DOI] [PubMed] [Google Scholar]
- 13.Ranzenhofer LM, Mayer LES, Davis HA, Mielke-Maday HK, McInerney H, Korn R, Gupta N, Brown AJ, Schebendach J, Tanofsky-Kraff M, Thaker V, Chung WK, Leibel RL, Walsh BT, Rosenbaum M, The FTO Gene and Measured Food Intake in 5- to 10-Year-Old Children Without Obesity. Obesity (2019), doi: 10.1002/oby.22464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gömez-Skarmeta JL, Del Corral RD, De La Calle-Mustienes E, Ferrés-Marcó D, Modolell J, Araucan and Caupolican, two members of the novel iroquois complex, encode homeoproteins that control proneural and vein-forming genes. Cell (1996), doi: 10.1016/S0092-8674(00)81085-5. [DOI] [PubMed] [Google Scholar]
- 15.Bellefroid EJ, Kobbe A, Gruss P, Pieler T, Gurdon JB, Papalopulu N, Xiro3 encodes a xenopus homolog of the Drosophila Iroquois genes and functions in neural specification. EMBO J. (1998), doi: 10.1093/emboj/17.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anselme I, Laclef C, Lanaud M, Rüther U, Schneider-Maunoury S, Defects in brain patterning and head morphogenesis in the mouse mutant Fused toes. Dev. Biol (2007), doi: 10.1016/j.ydbio.2006.12.025. [DOI] [PubMed] [Google Scholar]
- 17.Von Holstein-Rathlou S, Bondurant LD, Peltekian L, Naber MC, Yin TC, Claflin KE, Urizar AI, Madsen AN, Ratner C, Holst B, Karstoft K, Vandenbeuch A, Anderson CB, Cassell MD, Thompson AP, Solomon TP, Rahmouni K, Kinnamon SC, Pieper AA, Gillum MP, Potthoff MJ, FGF21 mediates endocrine control of simple sugar intake and sweet taste preference by the liver. Cell Metab. (2016), doi: 10.1016/j.cmet.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vogt MC, Paeger L, Hess S, Steculorum SM, Awazawa M, Hampel B, Neupert S, Nicholls HT, Mauer J, Hausen AC, Predel R, Kloppenburg P, Horvath TL, Brüning JC, Neonatal insulin action impairs hypothalamic neurocircuit formation in response to maternal high-fat feeding. Cell (2014), doi: 10.1016/j.cell.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, Powell C, Vedantam S, Buchkovich ML, Yang J, Croteau-Chonka DC, Esko T, Fall T, Ferreira T, Gustafsson S, Kutalik Z, Luan J, Mägi R, Randall JC, Winkler TW, Wood AR, Workalemahu T, Faul JD, Smith JA, Zhao JH, Zhao W, Chen J, Fehrmann R, Hedman ÅK, Karjalainen J, Schmidt EM, Absher D, Amin N, Anderson D, Beekman M, Bolton JL, Bragg-Gresham JL, Buyske S, Demirkan A, Deng G, Ehret GB, Feenstra B, Feitosa MF, Fischer K, Goel A, Gong J, Jackson AU, Kanoni S, Kleber ME, Kristiansson K, Lim U, Lotay V, Mangino M, Leach IM, Medina-Gomez C, Medland SE, Nalls MA, Palmer CD, Pasko D, Pechlivanis S, Peters MJ, Prokopenko I, Shungin D, Stančáková A, Strawbridge RJ, Sung YJ, Tanaka T, Teumer A, Trompet S, van der Laan SW, van Setten J, V Van Vliet-Ostaptchouk J, Wang Z, Yengo L, Zhang W, Isaacs A, Albrecht E, Ärnlöv J, Arscott GM, Attwood AP, Bandinelli S, Barrett A, Bas IN, Bellis C, Bennett AJ, Berne C, Blagieva R, Blüher M, Böhringer S, Bonnycastle LL, Böttcher Y, Boyd HA, Bruinenberg M, Caspersen IH, Chen Y-DI, Clarke R, Daw EW, de Craen AJM, Delgado G, Dimitriou M, Doney ASF, Eklund N, Estrada K, Eury E, Folkersen L, Fraser RM, Garcia ME, Geller F, Giedraitis V, Gigante B, Go AS, Golay A, Goodall AH, Gordon SD, Gorski M, Grabe H-J, Grallert H, Grammer TB, Gräßler J, Grönberg H, Groves CJ, Gusto G, Haessler J, Hall P, Haller T, Hallmans G, Hartman CA, Hassinen M, Hayward C, Heard-Costa NL, Helmer Q, Hengstenberg C, Holmen O, Hottenga J-J, James AL, Jeff JM, Johansson Å, Jolley J, Juliusdottir T, Kinnunen L, Koenig W, Koskenvuo M, Kratzer W, Laitinen J, Lamina C, Leander K, Lee NR, Lichtner P, Lind L, Lindström J, Lo KS, Lobbens S, Lorbeer R, Lu Y, Mach F, Magnusson PKE, Mahajan A, McArdle WL, McLachlan S, Menni C, Merger S, Mihailov E, Milani L, Moayyeri A, Monda KL, Morken MA, Mulas A, Müller G, Müller-Nurasyid M, Musk AW, Nagaraja R, Nöthen MM, Nolte IM, Pilz S, Rayner NW, Renstrom F, Rettig R, Ried JS, Ripke S, Robertson NR, Rose LM, Sanna S, Scharnagl H, Scholtens S, Schumacher FR, Scott WR, Seufferlein T, Shi J, Smith AV, Smolonska J, V Stanton A, Steinthorsdottir V, Stirrups K, Stringham HM, Sundström J, Swertz MA, Swift AJ, Syvänen A-C, Tan S-T, Tayo BO, Thorand B, Thorleifsson G, Tyrer JP, Uh H-W, Vandenput L, Verhulst FC, Vermeulen SH, Verweij N, Vonk JM, Waite LL, Warren HR, Waterworth D, Weedon MN, Wilkens LR, Willenborg C, Wilsgaard T, Wojczynski MK, Wong A, Wright AF, Zhang Q, T. L. C. LifeLines Cohort Study, Brennan EP, Choi M, Dastani Z, Drong AW, Eriksson P, Franco-Cereceda A, Gådin JR, Gharavi AG, Goddard ME, Handsaker RE, Huang J, Karpe F, Kathiresan S, Keildson S, Kiryluk K, Kubo M, Lee J-Y, Liang L, Lifton RP, Ma B, McCarroll SA, McKnight AJ, Min JL, Moffatt MF, Montgomery GW, Murabito JM, Nicholson G, Nyholt DR, Okada Y, Perry JRB, Dorajoo R, Reinmaa E, Salem RM, Sandholm N, Scott RA, Stolk L, Takahashi A, Tanaka T, van ‘t Hooft FM, Vinkhuyzen AAE, Westra H-J, Zheng W, Zondervan KT, Adipog T. ADIPOGen Consortium, T. A.-B. W. AGEN-BMI Working Group, T. Cardiogram. CARDIOGRAMplusC4D Consortium, T. Ckdg. CKDGen Consortium, T. GLGC, T. ICBP, T. M. MAGIC Investigators, T. M. MuTHER Consortium, T. Mig. MIGen Consortium, T. P. PAGE Consortium, T. R. ReproGen Consortium, T. G. GENIE Consortium, T. I. E. International Endogene Consortium, Heath AC, Arveiler D, Bakker SJL, Beilby J, Bergman RN, Blangero J, Bovet P, Campbell H, Caulfield MJ, Cesana G, Chakravarti A, Chasman DI, Chines PS, Collins FS, Crawford DC, Cupples LA, Cusi D, Danesh J, de Faire U, den Ruijter HM, Dominiczak AF, Erbel R, Erdmann J, Eriksson JG, Farrall M, Felix SB, Ferrannini E, Ferrières J, Ford I, Forouhi NG, Forrester T, Franco OH, Gansevoort RT, V Gejman P, Gieger C, Gottesman O, Gudnason V, Gyllensten U, Hall AS, Harris TB, Hattersley AT, Hicks AA, Hindorff LA, Hingorani AD, Hofman A, Homuth G, Hovingh GK, Humphries SE, Hunt SC, Hyppönen E, Illig T, Jacobs KB, Jarvelin M-R, Jöckel K-H, Johansen B, Jousilahti P, Jukema JW, Jula AM, Kaprio J, Kastelein JJP, Keinanen-Kiukaanniemi SM, Kiemeney LA, Knekt P, Kooner JS, Kooperberg C, Kovacs P, Kraja AT, Kumari M, Kuusisto J, Lakka TA, Langenberg C, Le Marchand L, Lehtimäki T, Lyssenko V, Männistö S, Marette A, Matise TC, McKenzie CA, McKnight B, Moll FL, Morris AD, Morris AP, Murray JC, Nelis M, Ohlsson C, Oldehinkel AJ, Ong KK, Madden PAF, Pasterkamp G, Peden JF, Peters A, Postma DS, Pramstaller PP, Price JF, Qi L, Raitakari OT, Rankinen T, Rao DC, Rice TK, Ridker PM, Rioux JD, Ritchie MD, Rudan I, Salomaa V, Samani NJ, Saramies J, Sarzynski MA, Schunkert H, Schwarz PEH, Sever P, Shuldiner AR, Sinisalo J, Stolk RP, Strauch K, Tönjes A, Trégouët D-A, Tremblay A, Tremoli E, Virtamo J, Vohl M-C, Völker U, Waeber G, Willemsen G, Witteman JC, Zillikens MC, Adair LS, Amouyel P, Asselbergs FW, Assimes TL, Bochud M, Boehm BO, Boerwinkle E, Bornstein SR, Bottinger EP, Bouchard C, Cauchi S, Chambers JC, Chanock SJ, Cooper RS, de Bakker PIW, Dedoussis G, Ferrucci L, Franks PW, Froguel P, Groop LC, Haiman CA, Hamsten A, Hui J, Hunter DJ, Hveem K, Kaplan RC, Kivimaki M, Kuh D, Laakso M, Liu Y, Martin NG, März W, Melbye M, Metspalu A, Moebus S, Munroe PB, Njølstad I, Oostra BA, Palmer CNA, Pedersen NL, Perola M, Pérusse L, Peters U, Power C, Quertermous T, Rauramaa R, Rivadeneira F, Saaristo TE, Saleheen D, Sattar N, Schadt EE, Schlessinger D, Slagboom PE, Snieder H, Spector TD, Thorsteinsdottir U, Stumvoll M, Tuomilehto J, Uitterlinden AG, Uusitupa M, van der Harst P, Walker M, Wallaschofski H, Wareham NJ, Watkins H, Weir DR, Wichmann H-E, Wilson JF, Zanen P, Borecki IB, Deloukas P, Fox CS, Heid IM, O’Connell JR, Strachan DP, Stefansson K, van Duijn CM, Abecasis GR, Franke L, Frayling TM, McCarthy MI, Visscher PM, Scherag A, Willer CJ, Boehnke M, Mohlke KL, Lindgren CM, Beckmann JS, Barroso I, North KE, Ingelsson E, Hirschhorn JN, Loos RJF, Speliotes EK, Genetic studies of body mass index yield new insights for obesity biology. Nature. 518, 197–206 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sinnott-Armstrong N, Sousa IS, Laber S, Rendina-Ruedy E, Nitter Dankel SE, Ferreira T, Mellgren G, Karasik D, Rivas M, Pritchard J, Guntur AR, Cox RD, Lindgren CM, Hauner H, Sallari R, Rosen CJ, Hsu Y-H, Lander ES, Kiel DP, Claussnitzer M, A regulatory variant at 3q21.1 confers an increased pleiotropic risk for hyperglycemia and altered bone mineral density. Cell Metab. (2021), doi: 10.1016/j.cmet.2021.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Claussnitzer M, Dankel SN, Klocke B, Grallert H, Glunk V, Berulava T, Lee H, Oskolkov N, Fadista J, Ehlers K, Wahl S, Hoffmann C, Qian K, Rönn T, Riess H, Müller-Nurasyid M, Bretschneider N, Schroeder T, Skurk T, Horsthemke B, Spieler D, Klingenspor M, Seifert M, Kern MJ, Mejhert N, Dahlman I, Hansson O, Hauck SM, Blüher M, Arner P, Groop L, Illig T, Suhre K, Hsu YH, Mellgren G, Hauner H, Laumen H, Leveraging cross-species transcription factor binding site patterns: From diabetes risk loci to disease mechanisms. Cell (2014), doi: 10.1016/j.cell.2013.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou J, Troyanskaya OG, Predicting effects of noncoding variants with deep learning-based sequence model. Nat. Methods 12 (2015), doi: 10.1038/nmeth.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Corradin O, Saiakhova A, Akhtar-Zaidi B, Myeroff L, Willis J, Cowper-Sallari R, Lupien M, Markowitz S, Scacheri PC, Combinatorial effects of multiple enhancer variants in linkage disequilibrium dictate levels of gene expression to confer susceptibility to common traits. Genome Res. 24 (2014), doi: 10.1101/gr.164079.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Romanov RA, Tretiakov EO, Kastriti ME, Zupancic M, Häring M, Korchynska S, Popadin K, Benevento M, Rebernik P, Lallemend F, Nishimori K, Clotman F, Andrews WD, Parnavelas JG, Farlik M, Bock C, Adameyko I, Hökfelt T, Keimpema E, Harkany T, Molecular design of hypothalamus development. Nature (2020), doi: 10.1038/s41586-020-2266-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Helgeland Ø, Vaudel M, Juliusson PB, Lingaas Holmen O, Juodakis J, Bacelis J, Jacobsson B, Lindekleiv H, Hveem K, Lie RT, Knudsen GP, Stoltenberg C, Magnus P, Sagen JV, Molven A, Johansson S, Njølstad PR, Genome-wide association study reveals dynamic role of genetic variation in infant and early childhood growth. Nat. Commun (2019), doi: 10.1038/s41467-019-12308-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yao L, Liu Y, Qiu Z, Kumar S, Curran JE, Blangero J, Chen Y, Lehman DM, Molecular Profiling of Human Induced Pluripotent Stem Cell-Derived Hypothalamic Neurones Provides Developmental Insights into Genetic Loci for Body Weight Regulation. J. Neuroendocrinol (2017), doi: 10.1111/jne.12455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merkle FT, Maroof A, Wataya T, Sasai Y, Studer L, Eggan K, Schier AF, Generation of neuropeptidergic hypothalamic neurons from human pluripotent stem cells. Dev. (2015), doi: 10.1242/dev.117978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang L, Meece K, Williams DJ, Lo KA, Zimmer M, Heinrich G, Carli JM, Leduc CA, Sun L, Zeltser LM, Freeby M, Goland R, Tsang SH, Wardlaw SL, Egli D, Leibel RL, Differentiation of hypothalamic-like neurons from human pluripotent stem cells. J. Clin. Invest (2015), doi: 10.1172/JCI79220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shin J, Berg DA, Zhu Y, Shin JY, Song J, Bonaguidi MA, Enikolopov G, Nauen DW, Christian KM, Ming GL, Song H, Single-Cell RNA-Seq with Waterfall Reveals Molecular Cascades underlying Adult Neurogenesis. Cell Stem Cell (2015), doi: 10.1016/j.stem.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Artegiani B, Lyubimova A, Muraro M, van Es JH, van Oudenaarden A, Clevers H, A Single-Cell RNA Sequencing Study Reveals Cellular and Molecular Dynamics of the Hippocampal Neurogenic Niche. Cell Rep. (2017), doi: 10.1016/j.celrep.2017.11.050. [DOI] [PubMed] [Google Scholar]
- 31.Aguet F, Barbeira AN, Bonazzola R, Brown A, Castel SE, Jo B, Kasela S, Kim-Hellmuth S, Liang Y, Oliva M, Parsana PE, Flynn E, Fresard L, Gaamzon ER, Hamel AR, He Y, Hormozdiari F, Mohammadi P, Muñoz-Aguirre M, Park Y, Saha A, V Segrć A, Strober BJ, Wen X, Wucher V, Das S, Garrido-Martín D, Gay NR, Handsaker RE, Hoffman PJ, Kashin S, Kwong A, Li X, MacArthur D, Rouhana JM, Stephens M, Todres E, Viñuela A, Wang G, Zou Y, T. Gte. Consortium, Brown CD, Cox N, Dermitzakis E, Engelhardt BE, Getz G, Guigo R, Montgomery SB, Stranger BE, Im HK, Battle A, Ardlie KG, Lappalainen T, The GTEx Consortium atlas of genetic regulatory effects across human tissues. bioRxiv (2019), doi: 10.1101/787903. [DOI] [Google Scholar]
- 32.Strober BJ, Elorbany R, Rhodes K, Krishnan N, Tayeb K, Battle A, Gilad Y, Dynamic genetic regulation of gene expression during cellular differentiation. Science (80-.) 364, 1287–1290 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wabitsch M, Brenner RE, Melzner I, Braun M, Möller P, Heinze E, Debatin KM, Hauner H, Characterization of a human preadipocyte cell strain with high capacity for adipose differentiation. Int. J. Obes (2001), doi: 10.1038/sj.ijo.0801520. [DOI] [PubMed] [Google Scholar]
- 34.Fischer-Posovszky P, Newell FS, Wabitsch M, Tornqvist HE, Human SGBS cells - A unique tool for studies of human fat cell biology. Obes. Facts (2008),, doi: 10.1159/000145784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Y, Maher P, Schubert D, A role for 12-lipoxygenase in nerve cell death caused by glutathione depletion. Neuron (1997), doi: 10.1016/S0896-6273(00)80953-8. [DOI] [PubMed] [Google Scholar]
- 36.Suo Z, Wu M, Citron BA, Palazzo RE, Festoff BW, Rapid tau aggregation and delayed hippocampal neuronal death induced by persistent thrombin signaling. J. Biol. Chem (2003), doi: 10.1074/jbc.M301406200. [DOI] [PubMed] [Google Scholar]
- 37.Knowles DA, Burrows CK, Blischak JD, Patterson KM, Serie DJ, Norton N, Ober C, Pritchard JK, Gilad Y, Determining the genetic basis of anthracycline-cardiotoxicity by molecular response QTL mapping in induced cardiomyocytes. Elife (2018), doi: 10.7554/elife.33480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Melnikov A, Murugan A, Zhang X, Tesileanu T, Wang L, Rogov P, Feizi S, Gnirke A, C. G. C. Jr, Kinney JB, Kellis M, Lander ES, Mikkelsen TS, Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat. Biotechnol 30, 271–277 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maurano MT, Haugen E, Sandstrom R, Vierstra J, Shafer A, Kaul R, Stamatoyannopoulos JA, Large-scale identification of sequence variants influencing human transcription factor occupancy in vivo. Nat. Genet (2015), doi: 10.1038/ng.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Korhonen J, Martinmäki P, Pizzi C, Rastas P, Ukkonen E, MOODS: Fast search for position weight matrix matches in DNA sequences. Bioinformatics (2009), doi: 10.1093/bioinformatics/btp554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nasser J, Bergman DT, Fulco CP, Guckelberger P, Doughty BR, Patwardhan TA, Jones TR, Nguyen TH, Ulirsch JC, Natri HM, Weeks EM, Munson G, Kane M, Kang HY, Cui A, Ray JP, Eisenhaure TM, Mualim K, Collins RL, Dey K, Price AL, Epstein CB, Kundaje A, Xavier RJ, Daly MJ, Huang H, Finucane HK, Hacohen N, Lander ES, Engreitz JM, Genome-wide maps of enhancer regulation connect risk variants to disease genes. bioRxiv (2020),, doi: 10.1101/2020.09.01.278093. [DOI] [Google Scholar]
- 42.Montefiori LE, Sobreira DR, Sakabe NJ, Aneas I, Joslin AC, Hansen GT, Bozek G, Moskowitz IP, McNally EM, Nóbrega MA, A promoter interaction map for cardiovascular disease genetics. Elife (2018), doi: 10.7554/eLife.35788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Williamson I, Eskeland R, Lettice LA, Hill AE, Boyle S, Grimes GR, Hill RE, Bickmore WA, Anterior-posterior differences in HoxD chromatin topology in limb development. Dev. (2012), doi: 10.1242/dev.081174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morey C, Da Silva NR, Perry P, Bickmore WA, Nuclear reorganisation and chromatin decondensation are conserved, but distinct, mechanisms linked to Hox gene activation. Development. 134, 909–19 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, Boehnke M, Abecasis GR, Willer CJ, Frishman D, in Bioinformatics (2011). [DOI] [PMC free article] [PubMed]
- 46.Hinds DA, McMahon G, Kiefer AK, Do CB, Eriksson N, Evans DM, St Pourcain B, Ring SM, Mountain JL, Francke U, Davey-Smith G, Timpson NJ, Tung JY, A genome-wide association meta-analysis of self-reported allergy identifies shared and allergy-specific susceptibility loci. Nat. Genet (2013), doi: 10.1038/ng.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gupta RM, Hadaya J, Trehan A, Zekavat SM, Roselli C, Klarin D, Emdin CA, Hilvering CRE, Bianchi V, Mueller C, Khera AV, Ryan RJH, Engreitz JM, Issner R, Shoresh N, Epstein CB, de Laat W, Brown JD, Schnabel RB, Bernstein BE, Kathiresan S, A Genetic Variant Associated with Five Vascular Diseases Is a Distal Regulator of Endothelin-1 Gene Expression. Cell (2017), doi: 10.1016/j.cell.2017.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler a. D., The Human Genome Browser at UCSC. Genome Res. (2002), doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frankish A, Diekhans M, Ferreira AM, Johnson R, Jungreis I, Loveland J, Mudge JM, Sisu C, Wright J, Armstrong J, Barnes I, Berry A, Bignell A, Carbonell Sala S, Chrast J, Cunningham F, Di Domenico T, Donaldson S, Fiddes IT, García Girón C, Gonzalez JM, Grego T, Hardy M, Hourlier T, Hunt T, Izuogu OG, Lagarde J, Martin FJ, Martínez L, Mohanan S, Muir P, Navarro FCP, Parker A, Pei B, Pozo F, Ruffier M, Schmitt BM, Stapleton E, Suner MM, Sycheva I, Uszczynska-Ratajczak B, Xu J, Yates A, Zerbino D, Zhang Y, Aken B, Choudhary JS, Gerstein M, Guigó R, Hubbard TJP, Kellis M, Paten B, Reymond A, Tress ML, Flicek P, GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. (2019), doi: 10.1093/nar/gky955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR, STAR: Ultrafast universal RNA-seq aligner. Bioinformatics (2013), doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wagner F, Yan Y, Yanai I, K-nearest neighbor smoothing for high-throughput single-cell RNA-Seq data. bioRxiv (2017), doi: 10.1101/217737. [DOI] [Google Scholar]
- 52.Lun A, Overcoming systematic errors caused by log-transformation of normalized single-cell RNA sequencing data. bioRxiv (2018), doi: 10.1101/404962. [DOI] [Google Scholar]
- 53.Luecken MD, Theis FJ, Current best practices in single‐cell RNA‐seq analysis: a tutorial. Mol. Syst. Biol (2019), doi: 10.15252/msb.20188746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robinson MD, McCarthy DJ, Smyth GK, edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics (2009), doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Soneson C, Robinson MD, Bias, robustness and scalability in single-cell differential expression analysis. Nat. Methods (2018), doi: 10.1038/nmeth.4612. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq (E-MTAB-10186), scRNA-seq (E-MTAB-10201), PCHi-C sequencing (E-MTAB-10200), ATAC-seq (E-MTAB-10257) and 4C-seq data (E-MTAB-10195) deposited at https://www.ebi.ac.uk/arrayexpress/.


