Abstract
COVID-19 has emerged as one of the deadliest pandemics that has ever crept on humanity. Screening tests are currently the most reliable and accurate steps in detecting severe acute respiratory syndrome coronavirus in a patient, and the most used is RT-PCR testing. Various researchers and early studies implied that visual indicators (abnormalities) in a patient's Chest X-Ray (CXR) or computed tomography (CT) imaging were a valuable characteristic of a COVID-19 patient that can be leveraged to find out virus in a vast population. Motivated by various contributions to open-source community to tackle COVID-19 pandemic, we introduce SARS-Net, a CADx system combining Graph Convolutional Networks and Convolutional Neural Networks for detecting abnormalities in a patient's CXR images for presence of COVID-19 infection in a patient.
In this paper, we introduce and evaluate the performance of a custom-made deep learning architecture SARS-Net, to classify and detect the Chest X-ray images for COVID-19 diagnosis. Quantitative analysis shows that the proposed model achieves more accuracy than previously mentioned state-of-the-art methods. It was found that our proposed model achieved an accuracy of 97.60% and a sensitivity of 92.90% on the validation set.
Keywords: Convolutional neural network, Graph convolutional network, COVID-19 detection, Chest X-ray, Deep learning
1. Introduction
COVID-19, also known as the coronavirus disease 2019, is an infectious disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), known as 2019 novel coronavirus (2019-nCoV) previously. The cases started spreading worldwide, mainly from Wuhan, China, where first cases were reported in late December 2019 [40].
The severe acute respiratory syndrome coronavirus 2 has a comparatively more significant risk on humanity due to its high reproductive number than SARS coronavirus (mean R0 for SARS coronavirus 2 is 3.28) [38]. The mean serial interval was calculated as 3.96 days, considerably shorter than mean serial interval calculated for SARS (8.4 days) or MERS (14.6 days) [8]. Various measures have been taken to suppress its exponential Reproductive Number (R0) and Series Interval. It is crucial to immediately isolate the infected person from uninfected population if there is an absence of specific therapeutic drugs or vaccines for COVID-19. While screening tests have been very useful in finding infection in a patient, the patient gets quick treatment and is isolated to stop the virus spread. RT-PCR testing has been instrumental during initial stage of virus spread and is considered gold standard. (Bell et al.) However, it has some flaws that might emerge once the most affected countries enter community transmission phase.
As the virus enters community transmission phase, there'll be a need for finding an optimal tradeoff between testing accuracy (performance) and time taken for testing to be done. With many cases emerging every day, it will be harmful to the human body to wait for a few days before screening tests. Therefore, some feasible alternative method is very much needed. One such method is COVID-19 detection through radiography images. Chest radiography imaging includes chest X-ray (CXR) or Computed Tomography (CT) imaging, that is conducted by some radiologists who further look for visual indicators (abnormalities) associated with SARS COVID-19 infection.
Chest CT and CXR imaging modalities have been used to diagnose Pneumonia for a long time. [26] They are currently the best method available because of their less complexity and high availability, which aids in the faster diagnosis of a patient. They are the most commonly used imaging modality for diagnosing and screening various chest-related diseases and play a vital role in clinical and epidemiological studies. Recent studies have shed light on Chest CT, demonstrating typical radiographic features of most covid infected patients, including ground-glass opacity, multifocal patchy consolidation, and interstitial changes with a peripheral distribution. As per a study conducted by (Daniel J Bell et al., 2020) chest radiography is less sensitive than chest CT; it is typically the first-line imaging modality used for patients suspected of COVID-19. Of the Chest radiographs of patients with covid requiring hospitalization, 69% (Wong et al., 2019) of total patients had an abnormal chest radiograph when admitted. After admission, approximately 80% had abnormalities at any time during hospitalization. The above findings were most extensive about 10-12 days after symptoms.
Radiography examinations are usually done for any respiratory-related ailments and therefore it is readily available. Any symptomatic/asymptomatic COVID-19 patient will show respiratory ailments as a symptom. Therefore, when radiography examinations are performed over a patient, the radiologist can look for abnormalities in CXR images and detect the presence of the SARS-CoV-2 virus. As the COVID-19 pandemic progresses, there will be more reliance on radiography techniques than RT-PCR testing because of its advantages. The biggest challenge in detecting COVID-19 through radiography is the unavailability of experienced radiologists who can readily spot any abnormality in a patient's CXR image. As Pneumonia and COVID-19 have similar symptoms, a radiologist should correctly interpret the radiographic images as the visual indicators can be very minute. This challenge can be tackled using Computer-aided Diagnostic (CADx) systems, that can help a radiologist spot the visual indicators more efficiently and accurately. The pressure on an individual radiologist or a doctor also increases resulting in human error, which is not the case in machine-aided detection tailored to detect COVID-19 accurately.
CADx tools can lower the clinical burden by facilitating Classification and Interpretation using Machine Learning and AI. Deep Learning (DL) plays a vital role in medical image analysis due to its excellent feature extraction ability. DL models complete tasks by automatically analyzing multi-modal medical images. Some examples of the application of DL include the diagnosis of diabetic retinopathy (P. [3]), cancer detection and classification (B. [11]), polyp detection during colonoscopy (R. [35]), and multi-classification of multi-modality skin lesions (L. [2]),
Many articles provide systematic reviews of machine learning techniques in detecting COVID-19 and the potential of Neural Networks and DL for the task [1,6]. [17] developed an on-device COVID-19 screening system that uses CXR images to identify COVID-19 infections. Afshar et al. developed a capsule-based network framework to identify COVID-19 infection in CXR images. [15] have worked on developing a semi-supervised technique for the identification of COVID-19 infection.
Computer Vision refers to using a specific set of algorithms for processing visual data and how a computer might gain information and understanding from this process. Convolutional Neural Network (CNN), a class of DL algorithm, has been proven very useful in image recognition & classification, video analysis, medical image analysis & other vision-related tasks. Generally, due to the limited availability of data (especially in the case of medical data), pre-trained CNN models trained on large datasets like ImageNet are used to capitalize on the knowledge of generic features from the images for a target application. Advanced computer-aided diagnosis schemes are primarily based on state-of-the-art methods.
[18] employed learning of relation-aware representation (RAR) along with image-level representation. When evaluating each data point individually, RAR is a method that uses the relationships between data points collected as a whole to improve decisions unbiasedly. Taking inspiration from their work, we used RAR over an entire cohort of X-ray images to assess the associations between them. We determined chest-related diseases in each image unbiasedly. The above association can be effectively modeled if we treat each image as a 'node.' [10] proposed a new AI framework, a graph convolutional network (GCN), that can learn RARs of nodes by studying graph structure and node features. This work aims to learn image-level features using a conventional CNN, and relation-aware representation (RAR) features using a Graph Convolutional Network. Through extensive experiments, we find the combination could outperform any network operating alone. This research aims to present a CADx system that uses a combination of CNN and GCN to make more accurate diagnoses for detecting COVID-19 from CXR images.
The rest of paper's organization is described as follows: Section 2 describes the Literature survey required for the study. The proposed Methodology is depicted and described in detail in Section 3. The experiments, quantitative comparison, and analysis of results are discussed in Section 4. Finally, concluding remarks have been made in Section 5.
2. Literature survey
Since COVID-19 has become widespread, many literature works have described the application of computer vision and DL in the diagnosis of the disease based on medical images and have achieved promising results. Chest CT and X-ray images have proven successful in detecting acute Pneumonia through various DL methods described in the literature [13,21]. [34] developed a DL-based CNN prediction model to classify COVID-19 Pneumonia and influenza with an accuracy of 86.7%. CT images were used as an imaging modality by [33], who utilized a modified Inception TL model and obtained an accuracy of 89.5%. [22] used a DL algorithm to characterize COVID-19 Pneumonia in chest CT images. A methodology based on DeTrac CNNs was presented for the classification of images into COVID-19 and non-COVID-19. The results were promising, with an accuracy of above 90%. (Abbas et al., 2020)
(Luz et al., 2020) employed TL technique to use a class of CNNs known as Efficient-Nets to detect the pattern of COVID-19 in CXR images, that are famous for their high accuracy and low computation. They also used a hierarchical classifier for the task, making it suitable to embed medical equipment or mobile phones. [9] use the TL strategy to fine-tune a ResNet-50 model in the classification of CXR images for detecting COVID-19 along with bacterial and viral Pneumonia. [20] also used TL to develop Deep-Covid to predict COVID-19 from CXR images. They curated a dataset of 5000 CXR images by board-certified radiologists and used a subset of those images to train four CNNs, namely ResNet-18, ResNet-50, Squeeze-Net, and DenseNet-121. [31] used Squeeze-Net with a Bayesian optimization additive for COVID-19 diagnosis. They report better performance and obtain higher COVID-19 diagnosis accuracy on their proposed network due to fine-tuned hyperparameters and augmented datasets. [23] used a combination of SVM and CNNs to detect COVID-19 from CXR images. [25] presented a weakly labeled data augmentation method on COVID-19 CXR images.
[30] give insights into an extensive set of statistical results obtained from the currently available public datasets, besides providing results on a medium-size COVID CXR dataset. [14] propose Coro-Net, a CNN-based model to automate the detection of COVID-19 infection from CXR images. They use TL to train Xception CNN architecture, pretrained on the ImageNet dataset, and employ a 4-fold cross-validation method for evaluation. [24] used Inception-V3 to extract features from CXR images for COVID-19 and classifiers such as KNNs, Decision Trees, Random Forests, SVMs for their classification. They reported an F-1 score of 0.89 for the COVID-19 class, using hierarchical analysis for the task of COVID-19 detection. The authors of [12] analyzed X-ray images and combined the texture and morphological features to classify them into COVID-19, bacterial Pneumonia, and non COVID-19 viral Pneumonia.
3. Methodology
The contributions of this research entail the following points: (1) First, we designed a base network SARS-Net CNN (Net-1), a CNN consisting of convolution layers and Inception blocks. (2) Next, we added various improvement techniques in the base network, Net-1, namely Coordinate-Convolutions (CC), anti-aliased (AA) CNNs, and rank-based stochastic pooling (RSP) to obtain Net-2, Net-3, and Net-4. The names of Net-2, Net-3, and Net-4 are SARS-Net CC, SARS-Net AA, and SARS-Net RSP, respectively. (3) Last, we developed SARS-Net (Net-5), where we combined Net-1 with our proposed 2-layer graph convolutional network (GCN). We prefer to use the Net-i naming convention for ease of understanding where i = 1 to 5. Further experiments showed that Net-5 gives the best results amongst all proposed five networks. In addition, Net-5 was superior to state-of-the-art approaches.
3.1. Dataset
The dataset used for training and validation is COVIDx, introduced in the COVID-NET paper [32]. It consists of 13,975 CXR images spreading across 13,870 patient cases. So far, the COVIDx dataset is the most prominent openly available CXR dataset in terms of the number of COVID-19 positive patient cases.
The distribution of data samples from the COVIDx dataset used for training and testing is depicted in Table 2. We use 90% of the data samples for training and validation, and the rest 10% is used for testing. To generate the COVIDx dataset, the CXR images were collected from various publicly available dataset repositories, namely:
-
a
COVID-19 Image Data Collection [5]
-
b
COVID-19 Chest X-ray Dataset Initiative [4]
-
c
ActualMed COVID-19 Chest X-ray Dataset Initiative [4]
-
d
COVID-19 radiography database [COVID-19 radiography database]
-
e
RSNA Pneumonia Detection Challenge dataset [RSNA pneumonia detection challenge]
Table 2.
Data split of COVIDx into train and test split with the number of samples belonging to each Patient case.
| Cases | Train-Split | Test-Split |
|---|---|---|
| Normal | 7966 | 885 |
| Pneumonia | 5451 | 594 |
| COVID-19 | 258 | 100 |
| Total | 13675 | 1579 |
For generating the final dataset for training and evaluation of SARS-Net, the following type of patient cases were used from each of the repositories:
| COVID-19 | Pneumonia | No pneumonia and non-COVID-19 |
| (a), (b), (c), (d) | (a) | (e) |
3.2. Data augmentation methods (DA)
All the variants of SARS-Net architecture were trained on the COVIDx dataset. First, as a pre-processing step, the CXR images were Centre cropped before training to remove the commonly-found embedded textual patient information. Then, various DA methods were tried out to decide the optimal methods which enhance the performance of SARS-Net and prevent overfitting. We tried several combinations of DA strategies and proceeded with the ones that gave the optimal model performance during training and evaluation. Initially, we relied on the augmentation techniques that are effective for CXR imaging modality [32] and subsequently added other DA methods in the pipeline. DA helps us increase the number of samples and introduce variability in the dataset without collecting new samples. Furthermore, to train the architectures, DA was leveraged with the following augmentation types: Elastic, Translation, CutBlur, Rotation, Random Horizontal flip, Zoom, MotionBlur, Intensity shift, and CutNoise. Finally, the input images in each batch were normalized.
3.3. SARS-Net CNN architecture
Most of the methods used in the literature so far for COVID-19 detection have used transfer learning techniques (Luz et al., [9], Minaee et al., Periera et al., [14]). DL models previously trained on ImageNet dataset are initialized and fine-tuned according to requirements. The underlying principle in Transfer Learning (TL) lies in the fact that DL models transfer the weights learned while capturing generic features from ImageNet datasets for aforementioned tasks.
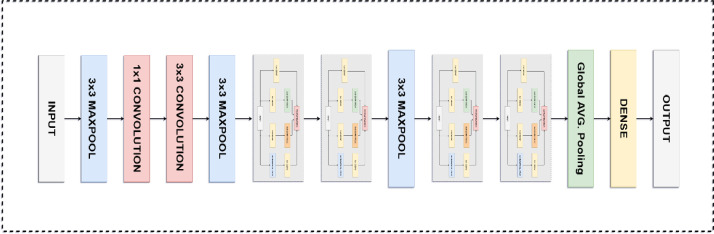
The proposed SARS-Net CNN network architecture is shown in Fig. 3, that employs parallel concatenation in a single block known as Inception Block, which was introduced by (Szegedy et al., 2020).
Fig. 3.
Illustration of Architecture of SARS-Net consisting of Inception Blocks.
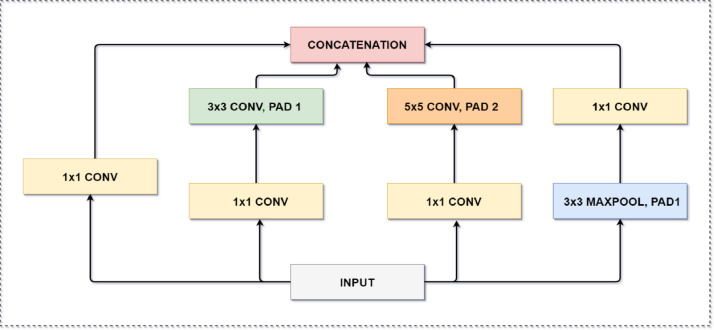
As shown in Fig. 2, each Inception block comprises four parallel paths leading to concatenation. Different spatial sizes are extracted by first three paths, which use filter sizes of 1 × 1, 3 × 3, and 5 × 5. We use a convolution of 1 × 1 in two middle paths to reduce the number of input channels. It also helps in reducing the model complexity. The fourth and last path uses a max-pooling of 3 × 3, followed by a 1 × 1 convolution, to change the channels. All paths use padding to maintain same height and width of the block's input and output. The outputs along each path are concatenated along the channel to constitute the Inception block output.
Fig. 2.
Illustration of An Inception Block.
Mathematically speaking, for a given image I and filter K, the convolution operation can be expressed as:
| (1) |
| (2) |
| (3) |
Where is the floor function of
There are also some special types of Convolution:
-
•
Valid convolution: p=0
-
•
Same convolution: output size = input size
-
•
convolution: , it might be helpful to in some cases to shrink the number of channels without changing the other dimensions . In the example below, we filled the filter with numbers for the sake of illustration. In a CNN, the Filter parameters are learned through backpropagation.
Our proposed SARS-Net CNN consists of a stack of four Inception blocks and a global average pooling to predict the final output. The dimensionality is reduced by using max-pooling layers in between the Inception blocks. The input is passed through convolutional blocks having a kernel size of 7 × 7, 3 × 3, and 1 × 1 with max-pooling between them. ReLU is used as an activation function for the convolutional blocks.
The Pooling operation can be expressed as:
| (4) |
| (5) |
Pooling Layer:
-
•
Input: with size being the image input
-
•
Padding: (rarely used), stride:
-
•
Size of pooling filter:
-
•
Pooling function:
-
•
Output: with size
We can assert that:
| (6) |
| (7) |
With
| (8) |
| (9) |
| (10) |
3.4. Proposed model variants of SARS-Net CNN: Net-2, Net-3, and Net-4
In this section, we discuss various improvement techniques that we adopted over the base network, SARS-Net CNN, to formulate and design the other three nets, namely SARS-Net CC (Net-2), SARS-Net AA (Net-3), and SARS-Net RSP (Net-4).
3.4.1. SARS-Net CC (Net-2)
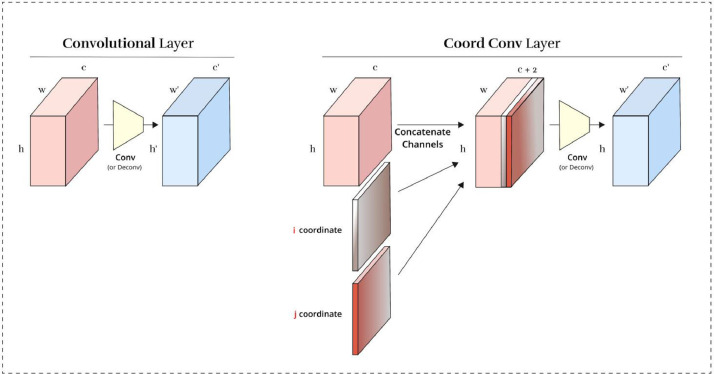
For designing SARS-Net CC (Net-2), we add an extra module of CoordConv [19] that allows SARS-Net to learn either varying degrees of translational dependence or complete translational invariance without compromising computational and parametric efficiency of ordinary convolution. The proposed CoordConv layer is a simple addition to the standard convolutional layer. It works by giving convolution access to its input coordinates using extra coordinate channels. Fig. 4 depicts the operation where two coordinates, i and j, are added. The i-coordinate channel is an h × w rank-1 matrix with its first row filled with 0′s, its second row with 1′s, its third with 2′s, and j coordinate channel is similar but with columns filled in with constant values instead of rows.
Fig. 4.
Illustration of a convolutional and CoordConv layers. (Left) A standard convolutional layer maps from a representation block with shape h × w × c to a new representation of shape h' × w' × c'. (Right) A CoordConv layer has the same functional signature, but accomplishes the mapping by first concatenating extra channels to the incoming representation.
The CoordConv layer is implemented as a simple extension of standard convolution. We first instantiate extra channels and fill them with coordinate information (constant, untrained). Once we have them ready, they are concatenated channel-wise to the input representation and a standard convolutional layer is applied. We apply a final linear scaling of both i and j coordinate values in all experiments to make them fall in the range [−1, 1].
3.4.2. SARS-Net AA
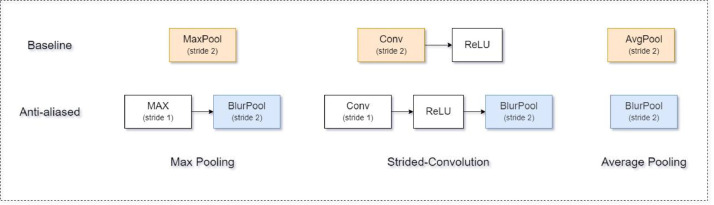
SARS-Net AA (Net-3) consists of an extra module of Anti-aliasing layers [36] that allows the SARS-Net to improve shift-equivariance without compromising the computational and parametric efficiency of ordinary convolution. The Anti-aliasing layer is a simple modification of the standard downsampling and Strided convolutional layer. It works by integrating low-pass filters before downsampling to achieve anti-alias, a common signal processing technique.
Modern CNNs are not shift-invariant, as small input shifts or translations can cause drastic changes in the output. Commonly used downsampling methods, such as max-pooling, strided-convolution, and average pooling, ignore the sampling theorem. The well-known signal processing fix is anti-aliasing by low-pass filtering before downsampling. Conventional methods for reducing spatial resolution – max-pooling, average pooling, and Strided convolution – all break shift-equivariance. We use improvements, shown in Fig. 5.
Fig. 5.
Block Diagram of common Anti-aliasing downsampling layers. (Top) Max-pooling, Strided-convolution, and average-pooling can each be better anti-aliased (bottom) with the shown architectural modification.
Anti-aliasing to improve shift-equivariance:
-
•
MaxPool → MaxBlurPool
-
•
StridedConv → ConvBlurPool
Strided-convolutions suffer from the same issue, and the same method applies.(12)
-
•
AveragePool → BlurPool
Blurred downsampling with a box filter is the same as average pooling. Replacing it with a more robust filter provides better shift-equivariance.(13)
3.4.3. SARS-Net RSP
For SARS-Net RSP (Net-4), we replace the traditional module of max pooling with rank-based stochastic pooling (RSP) [29] that allows the SARS-Net to improve its performance without compromising its computational and parametric efficiency. Rank-based pooling is calculated based on the matrix ranks rather than their absolute values, as in the case of max and average pooling.
The output of the conv layer, feature maps, are usually overly sizable in terms of their length, width, and the number of channels that can cause substantial computational burdens during training. The problem is solved by the pooling layers, a process of nonlinear downsampling. Pooling layers also help the CNN to be translation invariant. Pooling layer (PL) is a procedure of nonlinear downsampling (NLDS) to solve the above problem. Additionally, PL could provide invariance-to-translation characteristics to those AMs. Given a region with size of 2 × 2, let the pixels of = {}, (m= 1, 2, n= 1, 2) are
| (14) |
We added a constant 1/, here denotes the size of region . = 4 for a 2 × 2 NLDS pooling.
The two commonly used Downsampling procedures are average pooling (AP) and max pooling (MP). AP computes the mean value in as
| (15) |
MP works on and chooses its maximum value:
| (16) |
Rank-based pooling (RP) is another type of pooling method. Three typical algorithms are rank-based average pooling (RAP), rank-based weighted pooling (RWP), and rank-based stochastic pooling (RSP). All pooling operations in RP are calculated based on the ranks other than the realistic values. First, the 2 × 2 region is vectorized, and the rank matrix (RM) is calculated via the values of every entry , k (1, 1, 1, 2, 2, 1, 2, 2), usually lower ranks R are assigned to higher values () as
| (17) |
Providing tied values ( = ), a constraint is added to Eq. (17).
| (18) |
RAP used the greatest activations
| (19) |
= 2 is defined in this work. RWP and RSP are calculated on the exponential rank (ER) vector E = {}, which is defined as
| (20) |
where α is a hyper-parameter, here α = 0.5.
At this setting, Eq. (20) can be updated as = 0.5 × = . RWP is defined as the summation of and as below
| (21) |
Suppose is an outcome from a binary discrete random variable = { ,…, }, then RSP is defined as
| (22) |
3.5. SARS-Net: SARS-Net CNN + graph convolutional network (GCN)
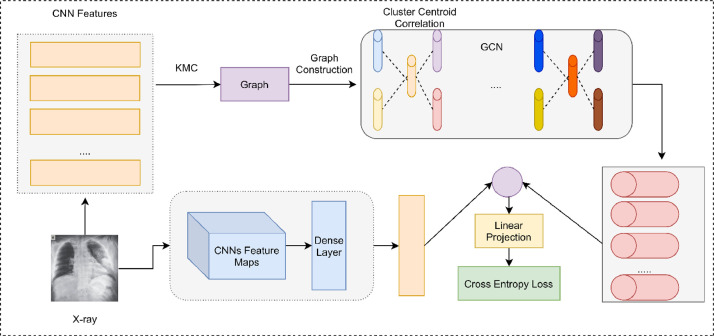
As an improvement over our base model, we also use Graph Convolutional Network (GCN) to learn the Relation Aware Representation (RAR) from the CXR images to improve the performance of SARS-Net. The CNN prioritizes the local characteristics of the image and objects, such as edges, corners and interest points when the kernel performs the convolution over the visual feature maps; thus, a lot of global information is lost. GCN helps in generalizing the regular convolution operation to graph convolution and works well with non-Euclidean data [27]. When we represent data patterns in a non-Euclidean way, we are giving it an inductive bias. We can prioritize and reward the model into learning certain patterns in data by changing its structure, given data of an arbitrary type, format and size. Generally, the inductive bias that is used is relational, in majority of current research pursuits and literature. We define an Adjacency Matrix (ADM) A ∈ RN × N for studying the relationship of the nodes {vi} of a graph G = (V, E), where there are N nodes vi ∈ V, i = 1,⋯, N and related links (vi, vj) ∈ E.
Graph Convolutional Network is used to represent a graph G with the help of a Neural network (X, A) in which X ∈ RN × D, where D represents the feature dimension of every node, N represents the number of nodes, and AX represents the sum of features of all neighboring nodes. Thus, GCN can successfully learn the Relation Aware Representation (RAR) feature [7].
A multi-layer GCN uses the layer-wise rule to update all the nodes' feature representation:
| (23) |
Where ∈ RN × N denotes the normalized form of the adjacency matrix A, and denotes the ReLU activation function. H ( l ) ∈ RN × dl represents the feature representation of the l-th layer. For finding the normalization matrix of A, i.e., A ↦ , following steps are followed:
Firstly, the degree matrix dm ∈ RN × N that is a diagonal matrix is computed as:
| (24) |
Afterward, is deduced via degree matrix dm and ADM A [16].
Each image can be represented by its image-level features and its neighbor features by combining CNN and the two layer-GCN [28]. We use a two-layer Graph Convolutional Network:
Here X = H (0), so we have
| (25) |
| (26) |
Where ∈ Rd 0× dC, and ∈ RdC × d 2 are two trainable weight matrixes.
We follow the same method as implemented by [37]. For the COVID-19 detection task, the SARS-Net CNN consisting of Inception blocks was initially used to give an image-level presentation of CXR images. As the CNNs do not consider the inter-image dependencies, so their RARs are learned by GCN. The GCN was combined with SARS-Net CNN. The Fully connected layer represented as Dense in Fig. 3 is utilized as the individual image-level representation I ∈ RD where D = 100 in this work.
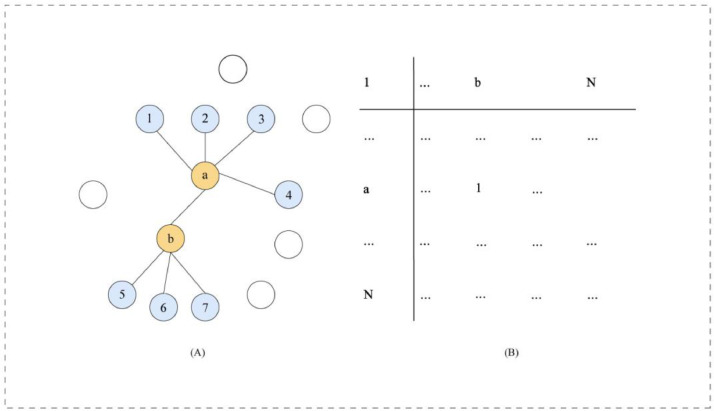
Once we obtain the individual image-level representations, we make them pass through k-means clustering (KMC) to obtain N cluster centroids (CCs) X ∈ RN × D. The Cluster Centroid correlation is used to display the relationships of the images. Then the adjacency matrix A ∈ RN × N is deduced as
| (27) |
KNN represents the cosine similarity (CS)-based kNN, its neighbor number is set to kKNN.
In the above example, where the four CS-based nearest neighbors of nodes a & b are KNN(Xb) = (5, 6, 7, a), KNN(Xa) = (1, 2, 3, 4). So, we have Xa ∈ KNN(Xb), Xb KNN(Xa).
Using the 'or' operation, we can conclude Aij = 1.
The node features X, and the adjacency matrix A is then passed to a two-layered- GCN, and we obtained H(2) ∈ RN×D if we set d2 = D = 100. The H(2) is then combined with I via dot product
| (28) |
Through a linear projection with trainable weights W(2) ∈ RN×NC, in which NC denotes the number of classes, we can define
| (29) |
Where z ∈ RNc, and Wb represent the bias. Here, NC = 3 due to this tertiary classification problem, i.e., Normal, Non-COVID-19 or COVID-19. Hence, we only need to train (W(0), W(1), W(2)) and their related biases for the two-layer Graph Convolutional Network.
Fig. 8 illustrates the basic building block of SARS-Net, which is a combination of GCN along with SARS-Net CNN. CNN representations are obtained during the inference stage, along with their corresponding GCN representations using a qualified 2L-GCN and a pre-built graph.
Fig. 8.
Basic building blocks of SARS-Net. The top rows indicate the complete GCN pipeline, where the CNN features and GCN features are combined, while the bottom row shows the SARS-Net CNN pipeline.
3.6. Implementation details
The proposed SARS-Net is trained and validated on the COVIDx dataset. The division of the COVIDx dataset for training and testing is depicted in Table 2, where we split 90% of the data for training and validation (train split), and the rest 10% is the test split. Out of the 90% train split, we further divide 10% of the data for validation and use the rest for training. A scheduling LR policy is used to update the learning rate where the learning rate is decreased by a small amount when learning remains the same for an extended period. Adam optimizer is used for optimizing the SARS-Net.
The hyperparameters used for training were:
-
•
LR =1e-3
-
•
Epochs=100
-
•
Batch size=32
Various data augmentation techniques were used to prevent the overfitting of SARS-Net model on the training dataset, namely rotation, zoom, horizontal flip, and translation.
The SARS-Net model was built and evaluated using PyTorch, a DL library. The convolution operation used in the architecture can be summarized as a series of convolutional products using various kernels of filters applied to an input image. Finally, the output is passed through an activation function. In the end, we have fully connected layers for the classification of the convoluted feature outputs.
More specifically, at any lth layer of a CNN we have,
-
•
Input: with size being the image input
-
•
Padding: stride:
-
•
Number of filters: where each has dimension:
-
•
Bias of the convolution:
-
•
Activation function:
-
•
Output: with size
And we have:
| (30) |
Thus:
| (31) |
| (32) |
With:
| (33) |
| (34) |
The learned parameters at the layer are:
-
•
Filter with parameters
-
•
Bias with parameters
The operation of the Fully connected layer:
In general, considering the node as layer, we have the following equations:
| (35) |
The input the output of a convolution or pooling layer with the dimensions .
We reshape the tensor to a 1D vector to plug it into the fully connected layer, having the dimension: , thus:
| (36) |
The learned parameters at the layer is:
-
•
Weightswith parameters
-
•
Bias with parameters
The algorithm for training the SARS-Net:
-
•
Initialization of the model parameters and weights.
-
•For i =1,2,…N, (N=Total epochs):
-
1Forward propagation:
-
iCompute the predicted value of through the DL model: .
-
iiEvaluate the function: where m is total number of input images, stands for the model parameters, and is the cost function.
-
i
-
2Perform Backpropagation
-
iApply a gradient descent method to update the parameters:
-
i
-
1
3.7. Dealing with the class imbalance problem
We use Cross-Entropy as the loss function/criterion for training our SARS-Net because it minimizes the distance between two probability distributions, i.e., the actual and the predicted. This section shows how class imbalance can cause problems while training DL models by creating a bias against the majority class. We prove the above statements by using the formula for cross-entropy loss, and finally, we use class-specific weight factors to handle the data imbalance.
The cross-entropy loss contribution from the ith training data case is:
| (37) |
Where xi and yi are the input features and labels, and f(xi) is the SARS-Net output, the probability that it is positive. Suppose we use a normal cross-entropy loss function with a highly unbalanced dataset. In that case, the algorithm will be trained to prioritize the majority class (i.e., Non-COVID cases) since it contributes more to the loss. For any case, i.e., (Yi = 0) or (1- Yi = 0), so only one of these terms contributes to the loss (the other term is multiplied by zero and becomes zero).
We can rewrite the overall average cross-entropy loss over the entire training set D of size N as follows:
| (38) |
Using this formulation, we can see that if there is a large imbalance with very few positive training cases, the negative class dominates the loss. Summing the contribution over all the training cases for each class (i.e., pathological condition), we see that the contribution of each class (i.e., positive or negative) is:
| (39) |
| (40) |
As we see in our case, positive COVID-19 contributions are significantly lower than negative ones. However, we want contributions to be equal. One way of doing this is by multiplying each example from each class by a class-specific weight factor Wpos, and Wneg, so that the overall contribution of each class is the same.
To implement this, we want:
| (41) |
which can be implemented by:
| (42) |
| (43) |
This way, we balance the contribution of positive and negative labels. After computing the weights, our final weighted loss for each training case is:
| (44) |
4. Experiments and results
4.1. Quantitative analysis
To evaluate the efficiency and demonstrate the usefulness of the proposed SARS-Net, we carry out extensive quantitative analysis to better understand its detection performance. We calculate the test Accuracy, Sensitivity, and PPV (Positive Predictive Value) for each case (Normal, Pneumonia, COVID-19) on the COVIDx dataset.
The Positive Predictive Value of each of the individual disease cases on various CNN architectures is shown in Table 3. PPV is defined as the probability of patients who have a positive test result actually having the disease. It is commonly used in medical testing where a "positive" result means that a patient actually has the disease.
Table 3.
Performance of tested CNN architectures on the COVIDx test dataset in terms of Positive Predictive Value (PPV).
| Architecture | PPV (%) | ||
|---|---|---|---|
| Normal | Non COVID-19 | COVID-19 | |
| VGG-16 | 87.30 | 81.10 | 94.40 |
| VGG-19 | 89.60 | 81.30 | 94.30 |
| ResNet-50 | 92.20 | 86.80 | 95.60 |
| SARS-Net CNN | 91.50 | 91.30 | 97.90 |
| SARS-Net | 92.80 | 92.50 | 98.60 |
The Accuracy and Sensitivity of each patient case on various CNN architectures during testing are shown in Table 4. From the table, it can be observed that SARS-Net achieves good accuracy by achieving a test accuracy of 97.60%. The average inference time on CPU and GPU and the model input size is mentioned in Table 5. It can be noted that the proposed SARS-Net model has a relatively shorter inference time of 0.035 milliseconds and 0.015 milliseconds on CPU and GPU, respectively. Finally, the comparisons of five network model variants of SARS-Net are shown in Table 6. Getting a quick test result is crucial in scenarios like COVID-19 detection, where time is essential in testing pathology and hospitals.
Table 4.
Performance of tested CNN architectures on the COVIDx test dataset in terms of Sensitivity (%) and Accuracy (%).
| Architecture | Sensitivity (%) | Accuracy (%) | ||
|---|---|---|---|---|
| Normal | Non COVID-19 | COVID-19 | ||
| VGG-16 | 92.00 | 87.00 | 56.50 | 91.32 |
| VGG-19 | 93.67 | 89.00 | 61.50 | 92.07 |
| ResNet-50 | 95.00 | 91.40 | 79.80 | 94.67 |
| SARS-Net CNN | 94.00 | 93.65 | 90.50 | 95.38 |
| SARS-Net | 94.80 | 95.67 | 92.90 | 97.60 |
Table 5.
Performance of tested CNN architectures on the COVIDx test dataset on inference time on CPU and GPUs.
| Architecture | Input Size(Ch, H, W) | Average Inference time on CPU (in MS) | Average Inference time on GPU (in MS) |
|---|---|---|---|
| SARS-Net | 3*224*224 | 0.035 | 0.015 |
| SARS-Net CNN | 3*224*224 | 0.030 | 0.009 |
| ResNet18 | 3*224*224 | 0.234 | 0.015 |
| ResNet50 | 3*224*224 | 0.587 | 0.040 |
| Alex Net | 3*224*224 | 0.670 | 0.010 |
| VGG16 | 3*224*224 | 0.782 | 0.050 |
| VGG19 | 3*224*224 | 0.901 | 0.067 |
Table 6.
Comparison of five network model variants of SARS-Net.
| Architecture | Sensitivity (%) | Accuracy (%) | ||
|---|---|---|---|---|
| Normal | Non COVID-19 | COVID-19 | ||
| SARS-Net CNN | 94.00 | 93.65 | 90.50 | 95.38 |
| SARS-Net CC | 94.48 | 93.23 | 90.73 | 95.56 |
| SARS-Net AA | 94.86 | 94.06 | 92.60 | 96.40 |
| SARS-Net RSP | 94.59 | 94.91 | 92.78 | 97.08 |
| SARS-Net | 94.80 | 95.67 | 92.90 | 97.60 |
Evaluation Metrics used:
-
iAccuracy
(45) -
iiSensitivity
(46) -
iiiPositive Predictive Value (PPV)
(47)
As mentioned in the Inception blocks, SARS-Net uses parallel concatenation to extract information in parallel, employing different kernel sizes ranging from 1 × 1 to 11 × 11. Besides, it has long-range connectivity and uses repeated blocks to learn complex features of the COVID-19 infected CXR images and produce good results compared to other CNN architectures. It is also advantageous to use various sized kernels in DL models to extract complex features.
As can be inferred from the above values, the addition of Graph Convolutional Network (GCN) enhances the performance of the proposed SARS-Net as GCNs can quickly learn the RARs among the test set. Therefore, classifiers with GCNs provide more precise results than those without GCNs.
4.2. Comparison to state of the art approaches
We compare the performance of SARS-Net with various other SOTA model literature that adopt a similar methodology and report the results in terms of Accuracy and Sensitivity for COVID-19 detection. Table 7 shows the results in Accuracy and Sensitivity for detecting COVID-19 of our proposed SARS-Net model compared to other State of the art literature studies. As is shown, SARS-Net yielded the following performance in terms of Sensitivity = 92.90 and Accuracy = 97.60.
Table 7.
Performance of State of the Art (SOTA) CNN architectures reproduced on the prepared COVIDx test dataset in terms of Accuracy and Sensitivity for COVID-19 Cases.
| Architecture | Accuracy (%) | Sensitivity (%) |
|---|---|---|
| Afshar et al. | 95.15 | 90.00 |
| Luz et al. | 93.90 | 90.80 |
| Wang et al. | 93.30 | 90.00 |
| Ozturk et al. | 87.02 | 85.35 |
| Rahimzadeh et al. | 91.40 | 80.53 |
| Wang Z et al. | 93.65 | 90.92 |
| SARS-Net CNN | 95.38 | 90.50 |
| SARS-Net | 97.60 | 92.90 |
4.3. Model interpretation
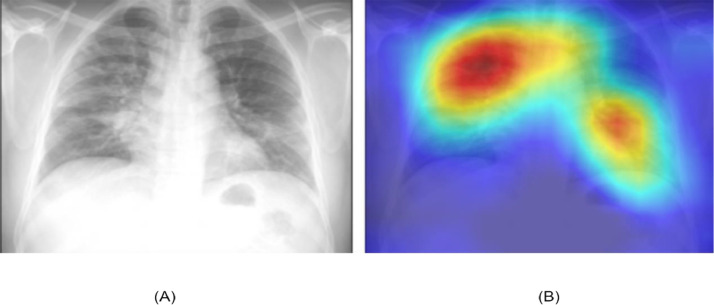
To get a better insight into how DL models make their decision, we analyze the output of GRAD-CAM, which highlights the areas of the input image in the form of heat maps that impact SARS-Net decisions. To interpret SARS-Net predictions, we produce the heatmaps to highlight the image areas most indicative of the disease using the Class Activation maps [39], generated by a method known as GRAD-CAM. The left portion of Fig. 11 (A) shows an input image fed into the SARS-Net, and the right portion (B) showcases a heatmap pointing out the areas of the image that indicate COVID-19.
Fig. 11.
Output of GRADCAM for the COVID-19 Detection.
The fully trained convolutional network, SARS-Net, is fed with an image to generate the Class Activation Mappings, and the feature maps are extracted from the output of the last convolutional layer. Suppose W c.k is the last classification layer's weight for feature map k belonging to class c and be the feature map. To generate a mapping of the most significant salient features of an input image belonging to a class c, we calculate the weighted sum of the feature maps using the weights connected to them. Specifically, a mapping is calculated as:
| (48) |
In the last step to obtain the Class activation mapping to visualize the most critical features of an input image, we upscale the mapping according to the dimensions of the image, followed by overlaying the image.
5. Conclusion
In this paper, we introduce SARS-Net, a CADx system combining Graph Convolutional Network and Convolutional Neural Network for COVID-19 detection from CXR images. Extensive experiments showed that the proposed SARS-Net model attains the best results among all other proposed networks and attains superior performances to state-of-the-art methods. The SARS-Net is a combination of the SARS-Net CNN model and a 2L-GCN model. Here, SARS-Net CNN aids to extract image-level features, whereas GCN helps to extract relation-awareness features. The combination of both two networks helps increase the performance of our proposed SARS-Net model. Medical image processing has been gaining much attention recently due to the emergence of deeper and high-accuracy networks that can compete against humans and speed up medical research to a great extent. COVID-19 detection has always been a crucial step for diagnosis in the present pandemic scenario, and recently many computer-aided analysis approaches are being used for quick and more reliable analysis. In this paper, the proposed SARS-Net model incorporates the latest profound DL advancements to diagnose COVID-19 from CXR images accurately. Our SARS-Net achieves an accuracy of 97.60% on the test set. The application of our method in hospitals is promising. Carrying out automated COVID-19 detection can improve the recovery rate and enable faster cures, thus improving people's overall health and quality of life. By leveraging automation by experts, CAD-X systems can aid healthcare services and reduce the burden of radiologists and doctors in developing countries where healthcare services are limited. Future scope includes works to improve SARS-Net's generalization and Sensitivity as more and more COVID-19 infected CXR images are available. The future research directions contain the following steps: (i) We also hope to develop a variant of SARS-Net trained on CT imaging modality when sufficient datasets are released, making our prediction model robust to different imaging modalities. (ii) Test out other combination mechanics of CNN and GCN to improve the classification results. (iii) Test other recent DA methods.
Figs. 1,6,7,9,10 and Tables 1 and 8
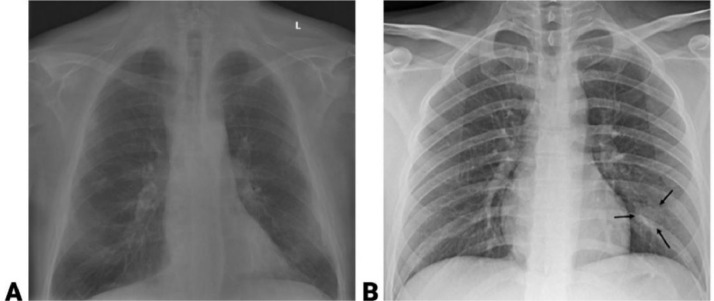
Fig. 1.
Example of two CXR images from the COVID x dataset showing (A) non-COVID infection and (B) COVID-19 infection.
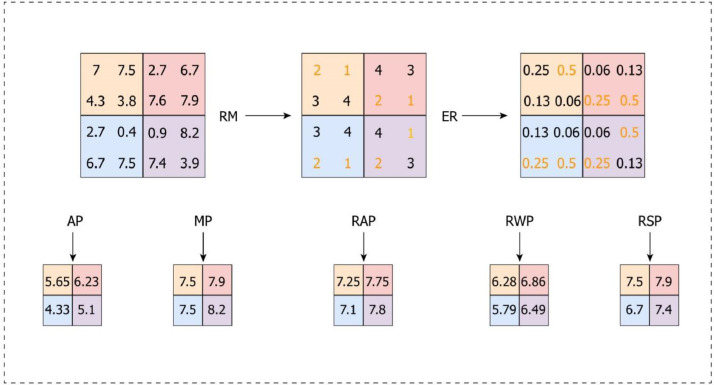
Fig. 6.
A toy example of various pooling technologies. AP: average pooling, MP: max pooling, RM: rank matrix, ER: exponential rank, RAP: rank-based average pooling, RWP: rank-based weighted pooling, RSP: rank-based stochastic pooling, RM: rank matrix, ER: exponential rank.
Fig. 7.
Illustration of a cosine-similarity k-nearest neighbor (KNN) based adjacency matrix ADM. (A) denotes the Graph and (B) denotes its corresponding Adjacency matrix generated by cosine similarity (CS)-based kNN.
Fig. 9.
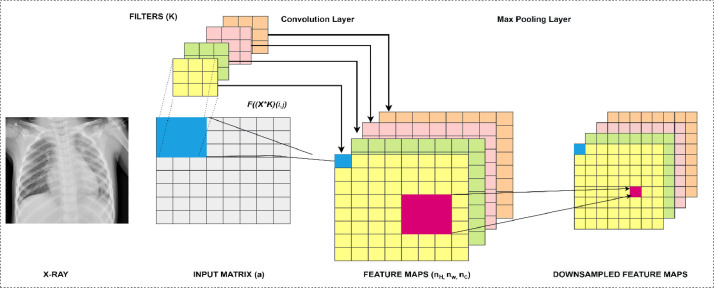
Pipeline of Convolution and Max-pooling layer operations.
Fig. 10.
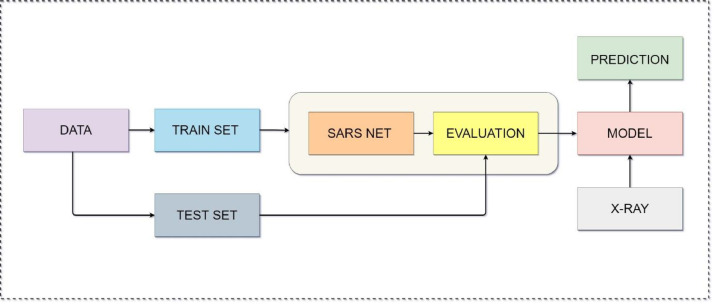
The training and testing workflow of the SARS-Net CADx system is depicted in the following flow chart.
Table 1.
The five proposed networks used in our study.
| Index | Modules | Network Name | Description |
|---|---|---|---|
| Net-1 | Base Network | SARS-Net CNN | CNN consisting of conv. layers and Inception blocks |
| Net-2 | ←Net-1 + CC | SARS-Net CC | Add coordinate-convolutions to Net-1 |
| Net-3 | ←Net-1 + AA | SARS-Net AA | Add anti-aliased CNNs to Net-1 |
| Net-4 | ←Net-1 + RSP | SARS-Net RSP | Use RSP to replace Max-Pooling in Net-1 |
| Net-5 | ←Net-1 + GCN | SARS-Net | Add 2-layer GCN to Net-1 |
Table 8.
Pseudocode for SARS-Net Training (Algorithm 1).
| Algorithm 1 Training procedure for SARS-Net |
| Input: image is the training data split of COVID-19 X-rays; class label is the labels assigned to the images; K is the number of epochs |
| Output: the trained model m; training time T |
| 1: (X) ← (generate augmented CXR image from all data repositories) |
| 2: (Y) ← (class label) |
| 3: (trainX, trainY), (valX, valY) ← split((X,Y), split size=0.1) |
| 4: for each epoch e in Range K do |
| 5: mt ← modelTrain(Adam,(trainX,trainY)) |
| 6: me ← modelEvaluate(mt,(valX,valY)) |
| 7: if earlyStopping(me) is TRUE then |
| 8: break |
| 9: end if |
| 10: end for |
| 11: mbest ← save bestModel{(mt, me) It = 1,2,. . .,K} |
Declaration of Competing Interest
We all authors declare that we have no conflict of Interest.
Biographies
Aayush Kumar: He is in currently doing his under graduate study in Computer Science Engineering at KIIT Deemed to be University, Bhubaneswear, India. He has keen interest in Machine Learning and Deep Learning for Healthcare services.
Ayush R Tripathi: He is in currently doing his under graduate study in Computer Science Engineering at KIIT Deemed to be University, Bhubaneswear, India. He has shown interest for AI in Healthcare.
Dr Suresh Chandra Satapathy is a Ph.D in Computer Science Engineering, currently working as Professor of School of Computer Engg and Dean- Research at KIIT (Deemed to be University), Bhubaneshwar, Odisha, India. He is quite active in research in the areas of Swarm Intelligence, Machine Learning, Data Mining and Cognitive Sciences. He has developed two new optimization algorithms known as Social Group Optimization (SGO) published in Springer Journal and SELO(Social Evolution and Learning Algorithm) published in Elsevier. He has more than 150 publications in reputed journals and conf proceedings.
Prof. Yu-Dong Zhang (M’08, SM’15) received his BE in Information Sciences in 2004, and MPhil in Communication and Information Engineering in 2007, from Nanjing University of Aeronautics and Astronautics. He received the PhD degree in Signal and Information Processing from Southeast University in 2010. He worked as a postdoc from 2010 to 2012 with Columbia University; and from 2012 to 2013 with Research Foundation of Mental Hygiene. He served as a Full Professor from 2013 to 2017 with Nanjing Normal University. Now he serves as Professor with University of Leicester
References
- 1.Albahri A.S., Hamid R.A., Alwan J.K., Al-qays Z.T., Zaidan A.A., Zaidan B.B., et al. Role of biological data mining and machine learning techniques in detecting and diagnosing the novel coronavirus (COVID-19): a systematic review. J. Med. Syst. 2020;44(11) doi: 10.1007/s10916-020-01582-x. Article ID: 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bi L., Feng D.D.F., Fulham M., Kim J. Multi-label classification of multi-modality skin lesion via hyper-connected convolutional neural network. Pattern Recognit. 2020;107 [Google Scholar]
- 3.Chudzik P., Majumdar S., Caliva F., Al-Diri B., Hunter A. Medical Imaging 2018: Image Processing, 10574. SPIE; 2018. Exudate segmentation using fully convolutional neural networks and inception modules; pp. 785–792. [Google Scholar]
- 4.Chung, A. Actualmed COVID-19 chest x-ray data initiative. https://github.com/agchung/Actualmed-COVID-chestxray-dataset (2020).
- 5.Cohen, J.P., Morrison, P., & Dao, L. COVID-19 image data collection. arXiv:2003.11597 (2020). Chung, A. Figure 1 COVID-19 chest x-ray data initiative. https://github.com/agchung/Figure1-COVID-chestxray-dataset (2020).
- 6.De Felice F., Polimeni A. Coronavirus disease (COVID-19): a machine learning bibliometric analysis. In Vivo. 2020;34:1613–1617. doi: 10.21873/invivo.11951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Derr Ty.ler, et al. Proceedings of the 13th International Conference on Web Search and Data Mining. 2020. Epidemic graph convolutional network. [Google Scholar]
- 8.Du Z., et al. Serial interval of COVID-19 among publicly reported confirmed cases. Emerg. Infect. Dis. 2020 doi: 10.3201/eid2606.200357. Mar 19[e-pub] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.M. Farooq, A. Hafeez, 2020 Covid-resnet: a deep learning framework for screening of covid19 from radiographs, arXiv:2003.14395.
- 10.Fukunaga Itsu.ki, et al. Prediction of the health effects of food peptides and elucidation of the mode-of-action using multi-task graph convolutional neural network. Mol. Inf. 2020;39(1–2) doi: 10.1002/minf.201900134. [DOI] [PubMed] [Google Scholar]
- 11.Gecer B., Aksoy S., Mercan E., Shapiro L., Weaver D., Elmore J. Detection and classification of cancer in whole slide breast histopathology images using deep convolutional networks. Pattern Recognit. 2018;84 doi: 10.1016/j.patcog.2018.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hussain La.l, et al. Machine-learning classification of texture features of portable chest X-ray accurately classifies COVID-19 lung infection. BioMedical Engineering OnLine. 2020;19(1):1–18. doi: 10.1186/s12938-020-00831-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jacobi Adam, B. A., Chung Mic.hael, Eber C. Portable chest x-ray in coronavirus disease-19 (covid-19): a pictorial review. Clin. Imaging. 2020 doi: 10.1016/j.clinimag.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khan A.I., Shah J.L., Bhat M.M. Coronet: a deep neural network for detection and diagnosis of covid-19 from chest x-ray images. Comput. Methods Programs Biomed. 2020;196 doi: 10.1016/j.cmpb.2020.105581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khobahi, S., Agarwal, C. & Soltanalian, M. Coronet: a deep network architecture for semi-supervised task-based identification of covid-19 from chest x-ray images. medRxiv doi: 10.1101/2020.04.14.20065722 (2020). [DOI]
- 16.Kipf T.N., Welling M. presented at the International Conference on Learning Representations (ICLR) 2017. Semi-supervised classification with graph convolutional networks. [Google Scholar]
- 17.Li, X., Li, C. & Zhu, D. Covid-mobile expert: on-device covid-19 screening using snapshots of a chest x-ray. arXiv:2004.03042 (2020).
- 18.Li Y., Luo Y., Huang Z. Australasian Database Conference. 2020. Graph-based relation-aware representation learning for clothing matching; pp. 189–197. [Google Scholar]
- 19.Liu Ros.anne, Lehman Jo.el, Molino Pier.o, Such Fe.lipe Petr.oski, Frank Er.ic, Sergeev Ale.x, Yosinski Jas.on. An intriguing failing of convolutional neural networks and the coordconv solution. Adv. Neural Inf. Process. Syst. 2018:9628–9639. [Google Scholar]
- 20.Minaee Sherv.in, et al. Deep-covid: predicting covid-19 from chest x-ray images using deep transfer learning. Med. Image Anal. 2020;65 doi: 10.1016/j.media.2020.101794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nair A., et al. A British Society of Thoracic Imaging statement: considerations in designing local imaging diagnostic algorithms for the COVID-19 pandemic. Clin. Radiol. 2020 doi: 10.1016/j.crad.2020.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ni Q.Q., et al. A deep learning approach to characterize 2019 coronavirus disease (COVID-19) pneumonia in chest CT images. Eur. Radiol. 2020:11. doi: 10.1007/s00330-020-07044-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Novitasari D.C.R., et al. detection of covid-19 chest x-ray using support vector machine and convolutional neural network. Commun. Math Biol Neurosci. 2020:19. Article ID. 42. [Google Scholar]
- 24.Pereira R.M., Bertolini D., Teixeira L.O., Silla Jr C.N., Costa Y.M. Covid-19 identification in chest x-ray images on flat and hierarchical classification scenarios. Comput. Methods Programs Biomed. 2020 doi: 10.1016/j.cmpb.2020.105532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rajaraman S., et al. Weakly labeled data augmentation for deep learning: a study on COVID-19 detection in chest x-rays. Diagnostics. 2020;10(6):17. doi: 10.3390/diagnostics10060358. Article ID. 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raoof Suh.ail, et al. Interpretation of plain chest roentgenogram. Chest. 2012;141(2):545–558. doi: 10.1378/chest.10-1302. [DOI] [PubMed] [Google Scholar]
- 27.Seti X., Wumaier A., Yibulayin T., Paerhati D., Wang L., Saimaiti A. Named-entity recognition in sports field based on a character-level graph convolutional network. Information. 2020;11(1):30. [Google Scholar]
- 28.Shi J., Wang R., Zheng Y., Jiang Z., Yu L. Second MICCAI Workshop on Computational Pathology (COMPAT) 2019. Graph convolutional networks for cervical cell classification. [Google Scholar]
- 29.Shi Z.L., Ye Y.D., Wu Y.P. Rank-based pooling for deep convolutional neural networks. Neural Netw. 2016;83:21–31. doi: 10.1016/j.neunet.2016.07.003. Nov. [DOI] [PubMed] [Google Scholar]
- 30.Tartaglione E., et al. Unveiling covid-19 from chest x-ray with deep learning: a hurdles race with small data. Int. J. Environ. Res. Public Health. 2020;17(18):6933. doi: 10.3390/ijerph17186933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ucar F., Korkmaz D. COVIDiagnosis-net: deep Bayes-squeeze net-based diagnosis of the coronavirus disease 2019 (COVID-19) from x-ray images. Med. Hypotheses. 2020 doi: 10.1016/j.mehy.2020.109761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Lind.a, Lin Zh.ong Q.iu, Wong Alexa.nder. Covid-net: a tailored deep convolutional neural network design for detection of covid-19 cases from chest x-ray images. Sci. Rep. 2020;10(1):1–12. doi: 10.1038/s41598-020-76550-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong Ho Yu.en Fra.nk, et al. Frequency and distribution of chest radiographic findings in patients positive for COVID-19. Radiology. 2020;296(2):E72–E78. doi: 10.1148/radiol.2020201160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu Xia.owei, et al. A deep learning system to screen novel coronavirus disease 2019 pneumonia. Engineering. 2020;6(10):1122–1129. doi: 10.1016/j.eng.2020.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang R., Zheng Y., Poon C.C., Shen D., Lau J.Y. Polyp detection during colonoscopy using a regression-based convolutional neural network with a tracker. Pattern Recognit. 2018;83:209–219. doi: 10.1016/j.patcog.2018.05.026. 298594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Richard. International Conference on Machine Learning. PMLR; 2019. Making convolutional networks shift-invariant again; pp. 7324–7334. [Google Scholar]
- 37.Zhang Y.D., Satapathy S.C., Guttery D.S., Górriz J.M., Wang S.H. Improved breast cancer classification through combining graph convolutional network and convolutional neural network. Inf. Process. Manag. 2021;58(2) [Google Scholar]
- 38.Zhao S., Ran J., Musa S.S. et al. Preliminary estimation of the basic reproduction number of novel coronavirus (2019-nCoV) in China, from 2019 to 2020: a data-driven analysis in the early phase of the outbreak. 2020 bioRxiv 2020. doi: 10.1101/2020.01.23.916395. [DOI] [PMC free article] [PubMed]
- 39.Zhou Bol.ei, et al. Proceedings of the IEEE conference on computer vision and pattern recognition. 2016. Learning deep features for discriminative localization. [Google Scholar]
- 40.Zhu N., Zhang D., Wang W., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]