Abstract
Recent advances in high throughput single-cell RNA sequencing (scRNA-seq) technology have enabled the simultaneous transcriptomic profiling of thousands of individual cells in a single experiment. To investigate the intrinsic process of retinal development, researchers have leveraged this technology to quantify gene expression in retinal cells across development, in multiple species, and from numerous important models of human disease. In this review, we summarize recent applications of scRNA-seq and discuss how these datasets have complemented and advanced our understanding of retinal progenitor cell competence, cell fate specification, and differentiation. Finally, we also highlight the outstanding questions in the field that advances in single-cell data generation and analysis will soon be able to answer.
Keywords: scRNA-seq, Single-cell rna-sequencing, Retinal development, Neurogenesis, Retinal progenitor cells, Competence, Cell fate specification, Review, Single-cell
1. Introduction
1.1. Achieving cellular diversity in the vertebrate retina
The comprehensive and detailed drawings of individual retinal cells by Santiago Ramón y Cajal served as our initial reference maps of retinal neuron diversity. Though his understanding of the developmental mechanisms influencing neuronal heterogeneity was limited, Cajal correctly postulated the flow of information pertaining to photodetection — from photoreceptors to ganglion cells to the brain. Through these anatomical studies, Cajal reasoned that the information transfer to the brain was shaped by local circuits of interneurons of various shapes and sizes. Cajal said, “[L]ife never succeeded in constructing a machine so subtly devised and so perfectly adapted to an end as the visual apparatus …“ (p. 576 Cajal, 1989). Ever since these initial anatomical observations, we have continued on a quest for understanding how cellular diversity in the retina is achieved and governs visual perception.
Subsequent to these initial characterizations, we have gathered an appreciation for the beauty and detailed structure of the retina and how these diverse cell types contribute to the perception of features including luminance, directed motion, and spectrums of colors. Through comparative anatomical studies dating back to Ramón y Cajal, we have understood how the evolutionarily conserved features and adaptations of retinal structure specify functions, including aspects such as the high acuity vision-promoting structure of the fovea or the presence of oil droplets in bird cone photoreceptors for filtering various wavelengths of light. However, it wasn’t until pioneering studies almost 100 years after the publication of Cajal’s first drawings of the retina, that we began to understand how development sculpts this relationship between form and function. In these first studies, we started to appreciate the developmental relationship of retinal cell types. One individual retinal progenitor cell (RPC) has the capacity to generate all major retinal cell types: six neuronal cell types (retinal ganglion cells [RGCs], horizontal cells, amacrine cells, bipolar cells, and cone and rod photoreceptors) and one glial cell (Müller glia (Holt et al., 1988; Jensen and Raff, 1997; Turner et al., 1990; Turner and Cepko, 1987; Wetts and Fraser, 1988)). This has led to a concentrated focus over the last 30 years, seeking to understand the mechanisms by which a common, multipotent progenitor cell has the ability to generate greater than 50–100 cellular subtypes that comprise the vertebrate retina.
The ability to characterize the biology of individual retinal cells has driven the field for the better part of two decades. Visual analysis of sparsely labelled individual RPCs and their progeny across retinal development by infection with a non-replicating virus led to a pioneering revelation; RPCs are multipotent and give rise to retinal neurons and glia in a sequential but overlapping birth order (Alexiades and Cepko, 1997; Turner et al., 1990; Turner and Cepko, 1987). This discovery, coupled with the advent of the ‘Genomics Age,’ propelled many scientists to pursue the mechanisms of retinal cell type specification by examining the spatial and temporal patterns of gene expression using a multitude of various technologies including in situ hybridization, serial analysis of gene expression (SAGE), microarray technologies, and RNA-sequencing. These studies have led to numerous hypotheses for candidate gene function in the specification and differentiation of individual retinal cell types, catalyzing the identification of transcription factors Atoh7 (Math5), Crx, and Nrl, for RGC, photoreceptor, and rod photoreceptor specification, respectively (Brown et al., 2001; Chen et al., 1997; Mears et al., 2001). Furthermore, lineage tracings of individual progenitors have provided insights into the ‘stochastic’ nature of cell fate decisions (Gomes et al., 2011; He et al., 2012) and correlated cell features with cell division modes (Baye and Link, 2007; Cayouette and Raff, 2003; Clark et al., 2012, 2021; Cohen et al., 2010; Kechad et al., 2012; Lacomme et al., 2016; Malicki, 2004). Additionally, the implementation of genetic reporters (Brzezinski et al., 2011; Katoh et al., 2010; Rowan and Cepko, 2004; Zeng and Sanes, 2017) and gain- and/or loss-of-function studies have integrated our understanding of transcription factor landscapes required during development for lineage restriction and proper cell type specification.
In parallel, morphological and electrophysiological characterizations of innumerable single cells by glass electrode—or more recently multi-electrode arrays—have established the diversity of mature cell (sub) types and their features within the retina. They have contributed a wealth of information detailing the magnitude of cellular function and local circuitry that drive retinal visual processing (Baden et al., 2016; Euler et al., 2014; Helmstaedter et al., 2013; Masland, 2012; Sanes and Masland, 2015; Zeng and Sanes, 2017). Additionally, the relationships of cellular ‘form and function’ and information processing across neural circuits are being integrated with expression of marker genes. With these marker genes, we are able to examine cell subtypes and better understand the molecular features driving the information processing and electrophysiological responses of retinal cells. However, the pairing of molecular (gene expression) identity with both structural and functional cellular diversity at a global level across development has remained a challenge. That is, until recently.
1.2. The dawning of a new age: single-cell profiling of transcriptomes
The first studies to understand the gene expression profiles of individual retinal cells relied on manual isolation of individual cells and transcriptional profiling by qRT-PCR or microarray analysis (Cherry et al., 2009; Kim et al., 2008; Laboissonniere et al., 2017a, 2019; Mizeracka et al., 2013; Mullally et al., 2016; Roesch et al., 2008, 2012, Trimarchi et al., 2007, 2008). These studies provided the first comprehensive insights into the cellular specificity of gene expression. Profiling techniques further advanced towards next-generation sequencing of mRNA (RNA-seq) from individual cells, paired with the electrophysiological and morphological features of individual cells — albeit in limited quantities (Laboissonniere et al., 2017b). However, in 2015 the high-throughput nanoliter droplet-based methods of single cell transcriptomic analysis were published (Klein et al., 2015; Macosko et al., 2015), enabling the profiling of tens of thousands of cells for a reasonable cost. Within two years, the droplet-based method had been commercialized, and the relative ease of unbiased measurements of the transcriptome has gained widespread implementation across numerous studies, including studies of the retina (Fig. 1).
Fig. 1. Publication history of scRNA-seq.
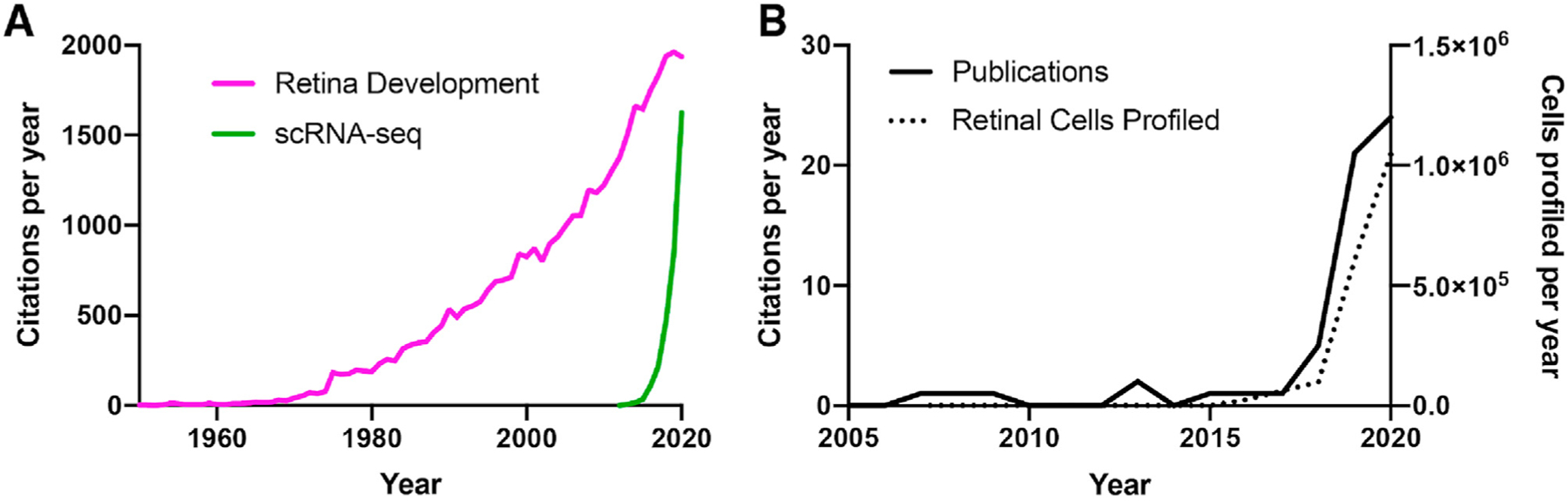
A) Number of publications referencing ‘Retinal Development’ (pink line) and ‘scRNA-seq’ (green line) over time, indicating the rapid utilization of scRNA-seq across the scientific community. B) Number of publications (black line) that have molecularly profiled RNA from individual retinal cells (left axis), and the total number of cells profiled within these studies (dotted line; right axis) per year. With the advancement and increased availability of sequencing technologies, the number of publications and cells profiled is currently increasing at an exponential rate. Data was obtained from a search in pubmed.gov using the following terms: (A) (“scRNA-seq”) OR (“single-cell RNA-sequencing”) OR (“single-cell RNA sequencing) OR (“drop-seq”) versus “Retina Development”; or (B) via a manually curated literature search for papers profiling single-cells within the retina.
1.3. Cell type composition of the retina
While morphological and physiological characteristics of retinal neurons have provided initial guides for the diversity of retinal cell classes and subtypes, single-cell technologies have precisely defined the molecular features of each individual cell (sub)type. Advances in sequencing technologies including Drop-seq (Macosko et al., 2015), InDrop (Klein et al., 2015); 10× Genomics), Smart-seq (Picelli et al., 2014), and others, in addition to widespread advancements in sequencing and data analysis technologies have enabled transcriptional profiling of entire organisms (Cao et al., 2017), organs (Tabula Muris Consortium, 2020), and specialized regions of the nervous system, including the retina (Macosko et al., 2015). Using these technologies, we now have comprehensive maps of retinal cell class and subtype diversity within the mature primate (human and other non-human primates (Liang et al., 2019; Lukowski et al., 2019; Menon et al., 2019; Orozco et al., 2020; Peng et al., 2019; Voigt et al., 2019), and model organisms such as the mouse and chicken (Rheaume et al., 2018; Shekhar et al., 2016; Tran et al., 2019; Yamagata et al., 2021; Yan et al., 2020a). Specifically, within the mouse, we now appreciate that the retina is comprised of at least 129 molecularly distinct retinal neurons and glia within the 7 major classes of retinal cell types: 46 RGCs (Tran et al., 2019), 1 horizontal cell, 2 cone photoreceptors, 63 amacrine cells (Yan et al., 2020a), 1 rod photoreceptor, 15 bipolar cells (Shekhar et al., 2016), and 1 Müller glial cell subtype. These comprehensive maps of cellular diversity, therefore, provide the molecular ‘finish line’ for cell fate specification from multipotent RPCs.
Similar studies have characterized the diversity of cell subtypes in humans (58 cell subtypes; Yan et al., 2020b), macaque (>60 cell subtypes; (Peng et al., 2019), and chicken (135 cell subtypes; Yamagata et al., 2021). While the number of cell subtypes within each species may increase with additional profiling, by identification of rare cell subtypes or distinguishing features of closely related cell subtypes, these initial studies indicate a high degree of variability in the number of retinal cell subtypes between species. Therefore, we must be cognizant that the processes regulating cell fate specification, including cell subtype specification, may vary across evolution.
The age of single-cell RNA-sequencing (scRNA-seq) and single-cell technologies has reshaped the field of developmental biology, eliciting unprecedented detail into the molecular anatomy of individual cells, illuminating novel processes across development, and challenging the definition of a cell type. The application of scRNA-seq over developmental windows, including that of the developing retina, enables unparalleled insight into the transcriptional landscape governing organ development. Here, we review the application of single-cell profiling techniques to the developing vertebrate retina, including early studies identifying markers of individual developing retinal cell types and RPC heterogeneity through the most recent applications characterizing cellular heterogeneity, lineage restriction, and molecular underpinnings of retinal diseases.
2. Retinal progenitor cells—the evolving transcriptome of RPCs across development
How do RPCs confer specification of retinal cell types within discrete temporal windows across retinal development? Heterochronic transplant studies of RPCs into host retinas of different temporal ages indicate that RPCs remain steadfast in ‘developmental age’ from their host tissue (Belliveau et al., 2000; Belliveau and Cepko, 1999; Rapaport et al., 2001), highlighting an autonomous regulation of developmental progression inherent to individual RPCs. The first study interrogating the transcriptomes of individual developing RPCs used microarrays to profile gene expressions of single cells. This study indicated that RPCs exhibit distinct expression profiles that depend on their developmental age (Trimarchi et al., 2008), confirming previous bulk-retina expression profiling using SAGE and RNA in situ hybridization observations (Blackshaw et al., 2004). These studies indicated that murine embryonic and postnatal RPCs can be distinguished by their expression profiles; early RPCs express Sfrp2 while late RPCs express Crym and Car2 (Blackshaw et al., 2004; Trimarchi et al., 2008). Additionally, RPCs from the same time point can express different combinations of transcription factors, with subsets of RPCs expressing neurogenic bHLH factors (Trimarchi et al., 2008).
2.1. Retinal competence model
The heterogeneity in transcript expression within RPCs highlighted intrinsic differences in progenitor populations. As a result, the field has sought to determine the extent to which RPC transcriptome heterogeneity confers both lineage bias and temporally-regulated cell fate specification. Studies examining the expression of the cell-cycle regulators p57Kip2(Cdkn1c) and p27Kip1 (Cdkn1b) within RPCs, as assayed by immunocytochemistry, determined heterogeneity in expression of cell cycle regulators across RPCs at early developmental ages. However, these studies noted that not all heterogeneity amongst RPCs has functional consequences for conferring cell fate (Dyer and Cepko, 2001a,b). The prevailing hypothesis governing retinal cell fate specification has subsequently been refined to a mechanism whereby RPCs progress through a temporally-regulated series of developmental ‘competence windows’, with RPCs making biased, but stochastic cell fate decisions that evolve across developmental time. This model is reminiscent of a Waddington landscape (Waddington, 1957) and those observed during the temporal patterning of multipotent neural progenitors in both the Drosophila embryonic ventral nerve cord and optic lobe medulla and during mammalian corticogenesis (Holguera and Desplan, 2018; Kohwi and Doe, 2013). In the Drosophila ventral nerve cord, neuroblasts utilize a temporal sequence of transcription factors—Hb (Hunchback), Kr (Kruppel), Pdm (POU domain), Cas (Castor), Grh (Grainy head)—to confer cell fate determination across temporal windows (Isshiki et al., 2001; Li et al., 2013a). In the optic lobe medulla, neuroblasts utilize a different sequence—Hth (Homothorax) -Ey (Eyeless), Slp (Sloppy paired), D (Dichaete), and Tll (Tailless) (Li et al., 2013b).
Recent studies have begun examining both the expression and function of the mammalian orthologs of temporal transcription factors within the developing retina. Ikaros (Ikzf1, mouse ortholog of Hb) confers competence in RPCs to generate early born cell types including horizontal cells and amacrine cells during late retinal development (Elliott et al., 2008). Additionally, loss of Ikzf1 expression resulted in a reduced capacity to generate early-born cell types including RGCs, horizontal cells, and amacrine cells (Elliott et al., 2008). Subsequently, expression of Pou2f1 (ortholog of Pdm) within RPCs is sufficient to initiate cone genesis and suppress Casz1 (ortholog of Cas) transcription (Javed et al., 2020). Expression of Casz1 then biases RPCs towards mid-/late-born neuronal fates, but actively inhibits glial specification (Mattar et al., 2015). While these evolutionarily conserved sequences of temporal transcription factors are shown to confer temporal competence, additional mechanisms beyond these simplified models are likely in place. For example, Foxn4 is shown to bias specification of RPCs to generate cones, horizontal cells, amacrine cells, and rods, during the middle period of retinal development by both increasing Casz1 expression and downregulating Ikzf1 expression (Liu et al., 2020). Additionally, the transition of RPCs to generate late-born cell types requires expression of Dicer and the expression of miRNAs let-7, miR-125, and miR-9 (La Torre et al., 2013). While the field has made great strides toward understanding the temporal sequence of retinal competence factor expression and transcriptional mechanisms governing retinal cell fate specification, the finer details that govern competence states within individual RPCs remain elusive.
In order to better appreciate the mechanisms governing temporal cell fate specification, researchers have recently employed large-scale scRNA-seq to profile the in vivo development of retinas in mice, humans, chicken, and zebrafish as well as the in vitro generation of human ES-cell/iPS-cell derived retinal organoids (Clark et al., 2019; Collin et al., 2019; Cowan et al., 2020; Hu et al., 2019; Lu et al., 2020; Raj et al., 2020; Sridhar et al., 2020; Xu et al., 2020; Yamagata et al., 2021). These scRNA-seq studies have answered many questions regarding RPC heterogeneity at individual ages and across development, but have yet to definitively assign competence transitions within the developing vertebrate retina. However, consistent with initial single-cell microarray studies in the retina, more recent analyses within scRNA-seq studies have identified intrinsic heterogeneity within RPCs, classifying RPCs into two broad categories reflective of different RPC transcriptional states: ‘primary’ RPCs—RPCs enriched for cell-cycle phase associated transcripts—and ‘neurogenic RPCs’—RPCs that remain in the cell cycle, but express proneural transcription factors that are indicative of a differentiating mitotic division mode where at least one daughter cell will exit the cell cycle and differentiate as a retinal neuron.
In this section, we examine how scRNA-seq has shaped our understanding of RPC heterogeneity and how changes to the gene expression profiles within RPCs across development may facilitate the temporal specification of retinal cell fates consistent with a retinal competence model.
2.2. Primary RPCs
An initial study of 747 sorted Chx10:GFP(+) cells from the mouse retina profiled RPCs at 3 developmental ages, Embryonic days of gestation (E)14, E18, and Postnatal day (P)2, corresponding to early, mid, and late stages of retina retinal development (Clark et al., 2019). Dimension reduction techniques, implemented to visualize transcriptional similarity across profiled cells, clearly segregated cells into ‘primary’ RPCs, neurogenic RPCs, photoreceptor precursors and a combined total of 24 RGCs and Amacrine Cells. When assessing transcriptional differences amongst RPCs, very little heterogeneity was observed amongst primary RPCs at the same developmental stages. In fact, within primary RPCs of the same developmental age, the most notable transcriptional heterogeneity observed was the expression of transcripts corresponding to distinct cell cycle phases (Clark et al., 2019). Additional studies also examined E14 mouse RPCs, but utilized cell cycle regression in order to uncover transcriptional heterogeneity independent of cell cycle phase. Within these E14 RPCs, further heterogeneity in transcript expression was observed, resembling dorsal-ventral spatial positioning (Vax2os, Bmpr1b) and asymmetries in the temporal progression of RPC maturation (Crym; Lo Giudice et al., 2019), reflective of developmental timing differences across the central to peripheral retinal axis (Hoshino et al., 2017; Young, 1985). However, when examining gene expression signatures of RPCs across developmental ages, global changes in gene expression were observed; results similar to those observed in 42 RPCs profiled by single-cell microarrays (Clark et al., 2019; Trimarchi et al., 2008). These results were further confirmed by examination of ~54,000 primary RPCs from scRNA-seq of dissociated whole mouse retinas across 10 developmental ages of retinal development, from E11-P14 (Clark et al., 2019). Combined, these single-cell transcriptional profiles of RPCs both confirmed and expanded previous lists of transcriptional regulators of RPCs across developmental windows, including Sfrp2, Fgf 15, Foxp1, and Foxp4 in early RPCs and Car2, Crym, Rlbp1, Sox8, Ass1 and the Nfi transcription factors (Nfia, Nfib, and Nfix) in late RPCs (Clark et al., 2019; Trimarchi et al., 2008). Similar observations were made in scRNA-seq experiments profiling RPCs from developing primary human retinas and iPSC-derived human retinal organoids (Hu et al., 2019; Lu et al., 2020; Sridhar et al., 2020).
But how do these developmental transcriptional states correspond to RPC developmental competence windows? Is the transcriptional heterogeneity observed across development indicative of competence state? To date, the answers to these questions still remain largely unresolved. Within the mouse scRNA-seq datasets, primary RPCs seemingly cluster into two distinct transcriptional states across development. These two states broadly reflect early and late developmental windows in which RGCs, horizontal cells, cones and amacrine cells, or amacrine cells, rods, bipolar cells and Müller glia are generated, respectively. However, finer resolution details of the gene expression changes within RPCs traversing discrete competence states is yet to be identified. When examining broad transitions from ‘early’ to ‘late’ RPCs, global transcriptional changes were detected between E16–18 in mice and between 11 and 15 gestational weeks in humans (Clark et al., 2019; Lu et al., 2020).
In contrast to the mammalian species profiled, primary RPCs in embryonic zebrafish do not segregate into distinguishable developmental windows corresponding to generation of early versus late retinal cell types. Instead, researchers identified 7 modules of gene expression profiles within RPCs across 24–48 hours post fertilization (hpf) timepoints of the developing zebrafish (Xu et al., 2020). Of these modules, three displayed transcriptional signatures that were shared by RPCs across each of the time points in which RPCs were profiled (24 hpf, 36 hpf, and 48hpf). Enriched transcripts within these shared modules across zebrafish retinal development include fabp11a and her9 (Module 1), fabp7a and her4.1 (Module 2), and atoh7 and neurod4 (neurogenic module; Module 3). Conversely, four modules of gene expression were only detected within 48hpf RPCs and included expression of transcripts associated with lineage restriction and/or initial fate commitment, including vsx1, otx2, onecut1, nr2e3, and rlbp1a (Xu et al., 2020). Further examinations will be required to determine if zebrafish RPCs can be classified within discrete temporally-defined windows given the consistency of gene expression within ‘primary’ RPC modules across development (Modules 1 and 2) or if the rapid development of the zebrafish retina precludes such distinctions.
Within scRNA-seq datasets of developing retinas from humans and mice, expression of well-established competence factors including Ikaros, Pou2f1, and Casz1 (Elliott et al., 2008; Javed et al., 2020; Mattar et al., 2015) is detectable, albeit to varying degrees. Ikaros, for example, was only weakly detected within mouse datasets (Clark et al., 2019); likely the result of inherent technical limitations of scRNA-seq for capturing transcripts of lowly expressed genes (See ‘Perspectives and Limitations’ Section). However, competence factors including Pou2f1 and Casz1 display expression patterns within mouse RPCs that are consistent with the temporal specification of cones and rods, respectively (Fig. 2A–B; (Clark et al., 2019; Javed et al., 2020; Mattar et al., 2015, 2021). Consistent with a function in specifying mid-development cell types, Foxn4 expression is enriched within mouse RPCs from E14-P2 (Fig. 2A–B; Liu et al., 2020).
Fig. 2.
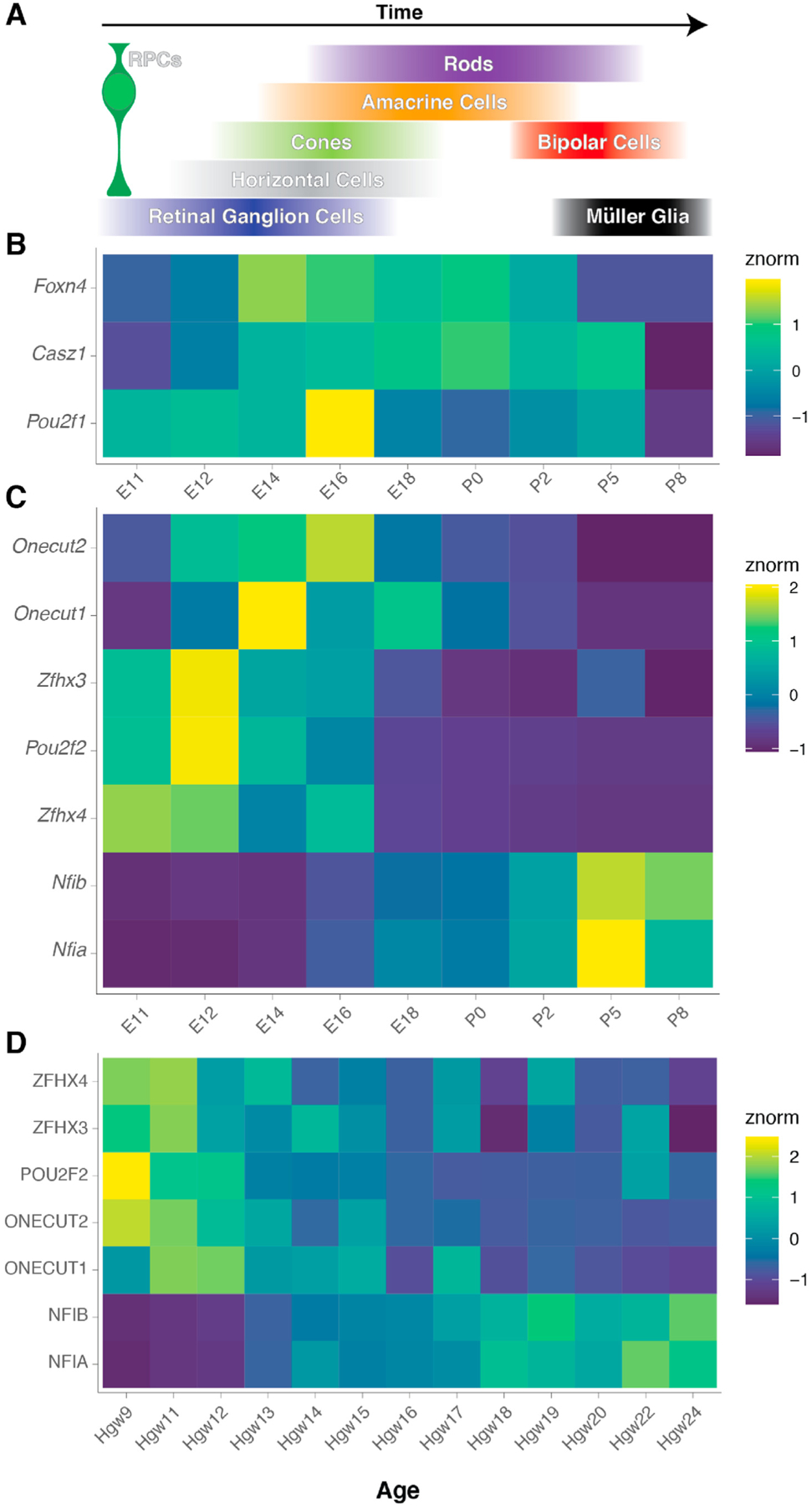
Temporal regulators of cell fate specification.
A) Schematic of the temporal birth windows of retinal cell types from the common multipotent retinal progenitor cell (RPC), highlighting the stereotyped, but overlapping birth order of retinal neurons and glia. B) Heatmap indicating the normalized expression (znorm) of ‘classic’ competence factors — Pou2f1 and Casz1 — and the new retinal competence factor — Foxn4 — within mouse RPCs across scRNA-seq experiments (Clark et al., 2019). Pou2f1 is expressed within early retinal development, followed by expression of Foxn4. Casz1 expression is consistent with the temporal windows in which rod photoreceptors are specified. C-D) Heatmaps indicating the normalized expression (znorm) of temporal neuronal fate regulators (Sagner et al., 2020) across RPCs of the C) developing mouse, and D) developing human retina (Clark et al., 2019; Lu et al., 2020), indicating temporally restricted expression patterns within early (Onecut1/2, Zfhx3/4, Pou2f2) and late (Nfib/Nfia) periods of retinal development.
Additional information regarding temporal specification windows of retinal neurons has also been gleaned from comparisons of retinal development to other regions of the central nervous system, including the forebrain, midbrain, and hindbrain of the developing mouse nervous system. These studies identified the sequential expression of Onecut, Pou domain, Zfhx, and Nfi transcription factors as a common temporal program to specify a diversity of neuronal subtypes within stereotyped birthorders (Sagner et al., 2020). Examination of the temporal expression patterns of these genes in both RPCs of humans and mice suggests conservation of the temporality of this transcription factor code within the developing retina (Fig. 2C–D). However, consistent with the two RPC states defined in RPCs of humans and mice (Clark et al., 2019; Lu et al., 2020), expression of the temporal transcription factors within RPCs, including Onecut1/2, Pou2f2, Zfhx3/4, and Nfia/b, is more reminiscent of ‘early’ and ‘late’ windows of neuronal fate specification than a true sequential temporal progression (Fig. 2C–D).
One possible explanation for lack of a higher resolution competence progression is the inability of current scRNA-seq methods to measure isoform-specific transcript expression. This is an especially important attribute to examine within RPCs as isoforms of the same gene, including competence factor Casz1, specify cell fates to varying degrees (Mattar et al., 2015).
Comparisons of transcriptome profiles of primary RPCs across species have identified multiple species-specific differences in primary RPC gene markers. For example, in mice, Clu displays expression primarily within Müller glia. In human retinas, however, CLU displays widespread expression in both RPCs and Müller glia, amongst other cell types (Lu et al., 2020). Furthermore, Hes1 and Hes5 are expressed in both human and mouse RPCs while zebrafish RPCs express numerous orthologs of these genes (Hes1: her9, her 6, and her 2 and Hes5: her4.4, her4.3, her4.2 and her 15.1). HES4 and its corresponding zebrafish ortholog, her9, are expressed in human and zebrafish RPCs, respectively. However, Hes4 has no ortholog within the mouse genome (Hu et al., 2019; Lu et al., 2020; Raj et al., 2020; Sridhar et al., 2020; Xu et al., 2020). In zebrafish, her9 is required for the differentiation, maintenance, and survival of rods and red/green cones (Coomer et al., 2020). Given that mouse retinas contain only 2 cone subtypes (short-wavelength and green cones), additional studies examining the presence and function of HES4/her9 in retinas of additional cone-dominant species beyond humans and zebrafish will be important for determining the extent to which HES4/her9 expression facilitates expanded color vision.
Unlike the retinas of mammalian species including humans and mice, the fish retina exhibits continuous growth throughout the animal lifespan (Hitchcock and Raymond, 2004). Within the post-embryonic zebrafish, retinal stem cells (RSC) in the ciliary marginal zone (CMZ) asymmetrically divide to generate two daughter cells of distinct fates: an RSC that remains in the peripheral RSC niche and an RPC that populates the retina. These post-embryonic RPCs possess the capacity to generate clones of both size and composition comparable to embryonic RPCs (Wan et al., 2016). Correspondingly, embryonic and post-embryonic zebrafish RPCs display similar gene expression profiles when examined at single-cell resolution (Raj et al., 2020; Xu et al., 2020). Though mice lack the innate ability to undergo retinal regeneration or continuously generate new neurons throughout the lifespan of the animal, studies have demonstrated that Msx1+ progenitor cells derived from the proximal zone of mouse CMZ give rise to both RPCs and non-neural epithelia during embryonic periods of development (Bélanger et al., 2017; Marcucci et al., 2016). These CMZ-derived RPCs produce clones comprising all seven main retinal cell types, though photoreceptors are underrep-resented in comparisons to cellular proportions of the entire retina (Bélanger et al., 2017). In scRNAseq of the developing mouse retina, expression of both Msx1 and Ccnd2 is observed within primary RPCs during early periods of development (Clark et al., 2019). More in-depth profiling of CMZ-derived cells will be required to identify transcriptional and developmental differences from primary RPCs that facilitate cell fate decisions and the early termination of proliferation within CMZ-derived retinal clones.
2.3. Neurogenic RPCs
In order to differentiate as a retinal neuron (or later glial cell), RPCs must undergo both cell-cycle exit and cell fate determination. From initial studies examining marker gene expression, including expression profiles of Atoh7 (also reported as Math5 or Ath5 (Brown et al., 2001; Feng et al., 2010)) or Neurog2 (Ma and Wang, 2006) within the developing retina, we have learned that some RPCs exhibit an increased propensity to divide in a ‘neurogenic’ manner. For example, atoh7:GFP time-lapse imaging in the zebrafish and BrdU experiments in both zebrafish and mouse have shown Atoh7 expression to initiate at S/G2 phase, with Atoh7+ RPCs giving rise to at least one daughter cell that exits the cell cycle and differentiates as a retinal neuron (Brzezinski et al., 2012; Feng et al., 2010; Miesfeld et al., 2018; Poggi et al., 2005). Single-cell profiling studies also highlight such neurogenic RPCs, characterized as RPCs that express different combinations of a host of pro-neural transcription factors including Atoh7, Neurog2 (Ngn2), Neurod1, Ascl1 (Mash1) and Hes6 (Trimarchi et al., 2008). The neurogenic RPCs continue to express genes demarcating cell cycle phase, including G2/M phase (Trimarchi et al., 2008) and neuronal differentiation associated transcripts Dll1 and Btg2 (Bao and Cepko, 1997; Clark et al., 2019; el Ghissassi et al., 2002; Iacopetti et al., 1999; Lu et al., 2020; Sridhar et al., 2020). In combination, these studies support a model that neurogenic RPCs display a transcriptional profile distinct from ‘primary’ RPCs, indicative of an intrinsic bias towards a terminal division where at least one daughter cell exits the cell cycle. Therefore, neurogenic RPCs serve as a transcriptional intermediary state between primary RPCs and terminal cell fate decisions.
Interestingly, neurogenic cells also display a temporally dynamic transcriptome, consistent with primary RPCs and in line with an over-arching progression through developmental competence states. scRNA-seq studies in both mice and humans have determined transcriptional signatures delineating ‘early’ and ‘late’ neurogenic cells, corresponding to neurogenic cells with the capacity to specify neuronal types restricted to early or late windows. For example, many of the genes that take part in RGC genesis—Atoh7 (Brown et al., 2001) and Isl1 (Wu et al., 2015)–are expressed in early neurogenic RPCs. In contrast, Otx2—involved in photoreceptor, horizontal cell and bipolar development (Emerson et al., 2013; Koike et al., 2007; Nishida et al., 2003) is enriched in late neurogenic RPCs.
While many of the transcription factors displaying enriched expression within neurogenic RPCs in scRNA-seq datasets have been previously characterized, scRNA-seq has identified additional transcripts of interest in both humans and mice. These include Pcdh17, Sstr2, Gadd45a, Gadd45g, Btg2, Penk, and Vexin (Mouse—3110035E14Rik; Human—C8orf46). In developing frog retinas, vxn is expressed within the neurogenic RPCs and functions to facilitate proneural transcription factor activity to promote cell cycle exit and neuronal differentiation (Moore et al., 2018). Other genes, including Pcdh17, Gadd45a, Gadd45g, and Btg2, function as tumor suppressors in numerous tissues (el Ghissassi et al., 2002; Hollander and Fornace, 2002; Hu et al., 2013; Hwang et al., 2020; Vairapandi et al., 2002; Yuniati et al., 2019) and therefore, may display enriched expression within neurogenic RPCs in order to promote cell cycle exit. In zebrafish, knock-down of pcdh17 via morpholino injection resulted in smaller eyes with reduced mitotic divisions and reduced cell differentiation, suggesting that pcdh17 may in fact regulate cell cycle exit within RPCs (Chen et al., 2013).
Gadd45a and Gadd45g, two members of the Growth-Associated and DNA Damage protein family, display both enrichment within neurogenic RPCs and complementary, temporally restricted expression patterns. Gadd45a expression is enriched within early neurogenic RPCs in humans and mice, whereas Gadd45g expression is enriched within late neurogenic RPCs (Brodie-Kommit et al., 2021; Clark et al., 2019; Lo Giudice et al., 2019; Lu et al., 2020; Wu et al., 2021). However, it remains to be determined if the temporally regulated expression of transcripts such as Gadd45a or Gadd45g function to drive retinal neurogenesis and/or the temporal specification of retinal cell fates. Other aspects of temporally regulated expression patterns, however, are not conserved across species. For example, Neurod4 is expressed in both early and late neurogenic RPCs in mice but only within late neurogenic RPCs in humans (Clark et al., 2019; Lu et al., 2020).
Numerous bHLH transcription factors display expression enrichment within neurogenic RPCs, including Ascl1, Neurog2, Atoh7, Hes6, Neurod4, and Olig2. However, the extent to which bHLH factors function redundantly or cooperatively in regulating neurogenesis needs to be tested on a case-by-case basis. For example, Neurod4 knockout mouse models displayed minor neurogenic defects within the developing retina. However, when Neurod4 was deleted in combination with other bHLH factors, including Ascl1, Neurog2, or Neurod1, specification of numerous neuronal cell fates was inhibited (Akagi et al., 2004; Inoue et al., 2002; Tomita et al., 2000). In many cases, the loss of neuronal cell types was accompanied by an increase in glial specification, suggesting that Neurod4 works in combination with additional proneural transcription factors to both drive neurogenic competence, inhibit gliogenesis, and specify particular retinal cell fates. Additional details of functional redundancy and/or compensatory mechanisms regulating RPC neurogenesis will be revealed by future scRNA-seq studies examining the consequences of gain/loss-of-function of bHLHs on neurogenic RPC transcriptomes (See “Phenotyping” Section).
2.4. From neurogenesis to restricted lineages
While these large-scale scRNA-seq studies have identified transcriptionally distinct states of RPCs (primary RPCs versus neurogenic RPCs; early RPCs versus late RPCs), to date, scRNA-seq has failed to comprehensively resolve instances of ‘restricted lineage’ progenitors. Years of lineage tracing studies have identified subsets of proliferative cells that display intrinsic biases or limited differentiation potential (Cepko, 2014). For example, Ascl1 lineage tracing indicates that Ascl1+ RPCs generate all retinal cell types except for RGCs (Brzezinski et al., 2011). Similar studies of Olig2+ RPCs suggest lineage restriction of RPCs to generate horizontal cells and cones (Hafler et al., 2012). Additionally, heterogeneity in gene expression within RPCs also biases specification of cellular subtypes. Lineage tracing of Chd6+ RPCs using a chd6-Cre transgenic system in mice identified that chd6+ RPCs give rise to all major retinal cell types. However, expression of Chd6 within progenitors that generate RGCs biases RGC subtype specification towards vertical motion responding, Chd6+ RGCs (De la Huerta et al., 2012).
Within current scRNA-seq studies, evidence for restricted progenitors remains limited (Clark et al., 2019; Lo Giudice et al., 2019; Lu et al., 2020; Sridhar et al., 2020; Xu et al., 2020). One possible explanation is that scRNA-seq analyses depend on the simplification of very high dimensional datasets (dimension reduction), leaving lineage-restricted clones buried within the structure of the dataset. In fact, it has been previously reported that within scRNA-seq data, cells committed to different lineages can “continue to occupy similar states for some time. This causes the early state to appear seemingly multipotent despite the cells within each clone being fate-restricted” (Wagner and Klein, 2020). An additional explanation is that these transient states within neurogenic RPCs have yet to be profiled at sufficient numbers or sequencing depth to parse finer details of fate restriction or lineage bias amongst heterogeneous cells within the restricted lineages. Case in point are scRNA-seq and single-cell microarray studies specifically examining the heterogeneity of lineage-restricted cells within Otx2+ RPCs in the chicken retina and Olig2+ RPCs of the mouse (Ghinia Tegla et al., 2020; Hafler et al., 2012).
Olig2 is expressed within neurogenic RPCs across a broad temporal window during retinal development. Cre-recombinase lineage tracings indicate that Olig2+ RPCs preferentially differentiate as amacrine or horizontal cells and are biased against generating RGCs or Müller glia. However, Olig2 overexpression promotes cell-cycle exit and specification of cone photoreceptors and horizontal cells. The diverse array of cell types generated within the Olig2-restricted lineage is partially explained by heterogeneity in expression of additional fate-promoting transcription factors within Olig2+ RPCs. Single-cell microarray experiments profiling Olig2+ RPCs determined heterogeneous expression of numerous proneural transcription factors across Olig2+ RPCs, including Neurod1, Neurod4, Atoh7, Ptf1a, and Ascl1 (Hafler et al., 2012). Likewise, Otx2 expression within chicken RPCs biases RPCs to become cones and horizontal cells. Despite their restricted potential, these Otx2+ cells express many additional genes enriched within neurogenic cells across development, including Neurog2, Neurod4, Hes5, Dll 4, and Notch1. By performing scRNA-seq on sorted cells from an Otx2 reporter line in parallel with CRISPR-mediated mutation of Otx2, researchers were able to increase the resolution of the scRNA-seq and determine the functional significance of Otx2 in lineage restriction. The authors determined that Otx2 both biases lineage restriction towards cone photoreceptor and horizontal cell fates and also influences specification of cellular subtypes (Ghinia Tegla et al., 2020).
Furthermore, studies in zebrafish - where in vivo, time-lapse recordings of lineage are more readily achievable - highlight the molecular architecture of fate restriction inferences obtainable from scRNA-seq. Detailed analyses of thousands of clones within transgenic embryos have characterized the entire clonal compositions of cell types generated from specific lineages, including those of the atoh7 and vsx1 lineages (Wang et al., 2020). Using the information garnered from lineage trees, the authors were able to re-examine scRNA-seq profiles of 635 neurogenic RPCs from 48hpf zebrafish retinas to examine the gene expression profiles within atoh7 and/or vsx1-positive cells that contribute to lineage restriction (Wang et al., 2020; Xu et al., 2020). First, the authors clustered the 48hpf neurogenic RPCs to examine heterogeneity within this population, identifying both atoh7 positive and negative neurogenic cells. Atoh7 positive clusters of cells could be further subdivided into two additional clusters based on the mutually exclusive presence of vsx1 or oc1 expression. When paired with lineage tracing experiments and additional scRNA-seq of sorted atoh7+ neurogenic RPCs, the profiling studies suggest the following lineage restrictions within 48hpf neurogenic RPCs: atoh7+/vsx1+ RPCs generate amacrine cell/bipolar cell clones; atoh7+ /vsx1-/oc1+ RPCs generate RGC/photoreceptor or amacrine cell/photoreceptor clones; and atoh7-/vsx1+ RPCs generate pre-dominately bipolar cell clones (Wang et al., 2020).
To test their hypotheses that the identified marker genes are contributing to restricted lineages within the oc1 and vsx1 RPCs, the authors examined the consequence of transcription factor overexpression on clonal cell type compositions. For example, overexpression of otx2 caused a significant increase in bipolar cell or photoreceptor cell specification within vsx1+ or onecut1+ RPCs, respectively. Interestingly, overexpression of atoh7 had no significant change on vsx1+ clonal composition but skewed oc1+ RPCs towards RGC fate (Wang et al., 2020).
Given these examples highlighting the use of focused scRNA-seq studies on reporter lines in combination with detailed lineage trees, it seems plausible that additional details of the lineage restriction on individual neurogenic RPCs remain hidden amongst the transcriptional heterogeneity within large-scale datasets. Of note, in studies of 48hpf zebrafish neurogenic RPCs, the authors identified one cluster of neurogenic RPCs that did not display gene expression profiles consistent with cell fate biases, instead remaining seemingly unspecified (Wang et al., 2020). This observation is consistent with the hypothesis that neurogenic RPCs are initially selected to undergo a differentiative division while remaining multipotent, then acquire lineage-restriction prior to terminal mitosis. Further detailed analysis will be required to more comprehensively understand the scope of neurogenic RPC heterogeneity and to integrate the temporal progression through competence phases into lineage restriction models.
Although the presence of material transfer in retinal transplant experiments has raised concerns about the accuracy of developmental lineage tracing studies, detailed examinations of past lineage experiments support the lineage models. For example, studies using in vivo and ex utero retroviral labeling of RPCs with β-galactosidase argue against diffusion of reporters; infection events often contain single clones, and no differences in clone size were observed when comparing clones 4–6 weeks or 1-year post-infection (Turner et al., 1990; Turner and Cepko, 1987; Boudreau-Pinsonneault and Cayouette, 2018). Supporting these results, cultured retinal neuroepithelial cells and retroviral-mediated lineage tracing of RPCs in retinal explants also resulted in clonal sizes and cell type compositions comparable to in vivo studies even though neither experiments relied on reporters to track clonal expansions (Cayouette et al., 2003; Gomes et al., 2011; Boudreau-Pinsonneault and Cayouette, 2018).
If concerns still remain about material transfer in lineage tracing, various recent single-cell lineage tracing tools should be implemented, involving either permanent genomic alterations (scGESTALT [CRISPR in zebrafish; (Raj et al., 2018)]; CLARIN [CRISPR in mouse; (Bowling et al., 2020)]; and Polylox [Cre-recombinase in mouse; (Pei et al., 2017)]) or somatic mutations (Ludwig et al., 2019). Adoption of these tools will help verify previous lineage tracing studies and provide additional information on how cell state affects lineage decisions. One study of zebrafish neurodevelopment has employed scGESTALT to record lineages through barcode editing. However, genome editing had saturated before terminal divisions in the retina, revealing little lineage relationship between the different zebrafish retinal cell types but suggesting that tuning of the system to be active at the onset of retinogenesis could prove to be fruitful for resolving retinal lineages (Raj et al., 2020).
3. Inference of temporal gene expression from static snapshots—pseudotemporal analyses
The stereotypical birth order of retinal cell types is a hallmark of retinal development and has provided an attractive model system for studies of temporal cell fate specification. In fact, the temporal nature of cell fate specification from multipotent RPCs is highly reminiscent of a Waddington landscape architecture (Fig. 3). However, within the developing retina, multiple cell types are born simultaneously over extended and overlapping temporal windows (Fig. 2A; (Cepko et al., 1996; Young, 1985), leading to a more complex model of cell fate specification (Fig. 3B–D). Furthermore, many retinal cell subtypes are also specified within a temporal hierarchy. For example, profiling of amacrine cells from E16 to P8 led to the discovery that GABAergic amacrine cells are generated earlier than glycinergic subtypes (Cherry et al., 2009; Voinescu et al., 2009). Despite this complexity, by implementing pseudotime analyses within scRNA-seq datasets, we can uncover changes in gene expression programs directing RPC specification towards individual cell fates.
Fig. 3. Waddington landscape of the developing mouse retina from scRNA-seq.
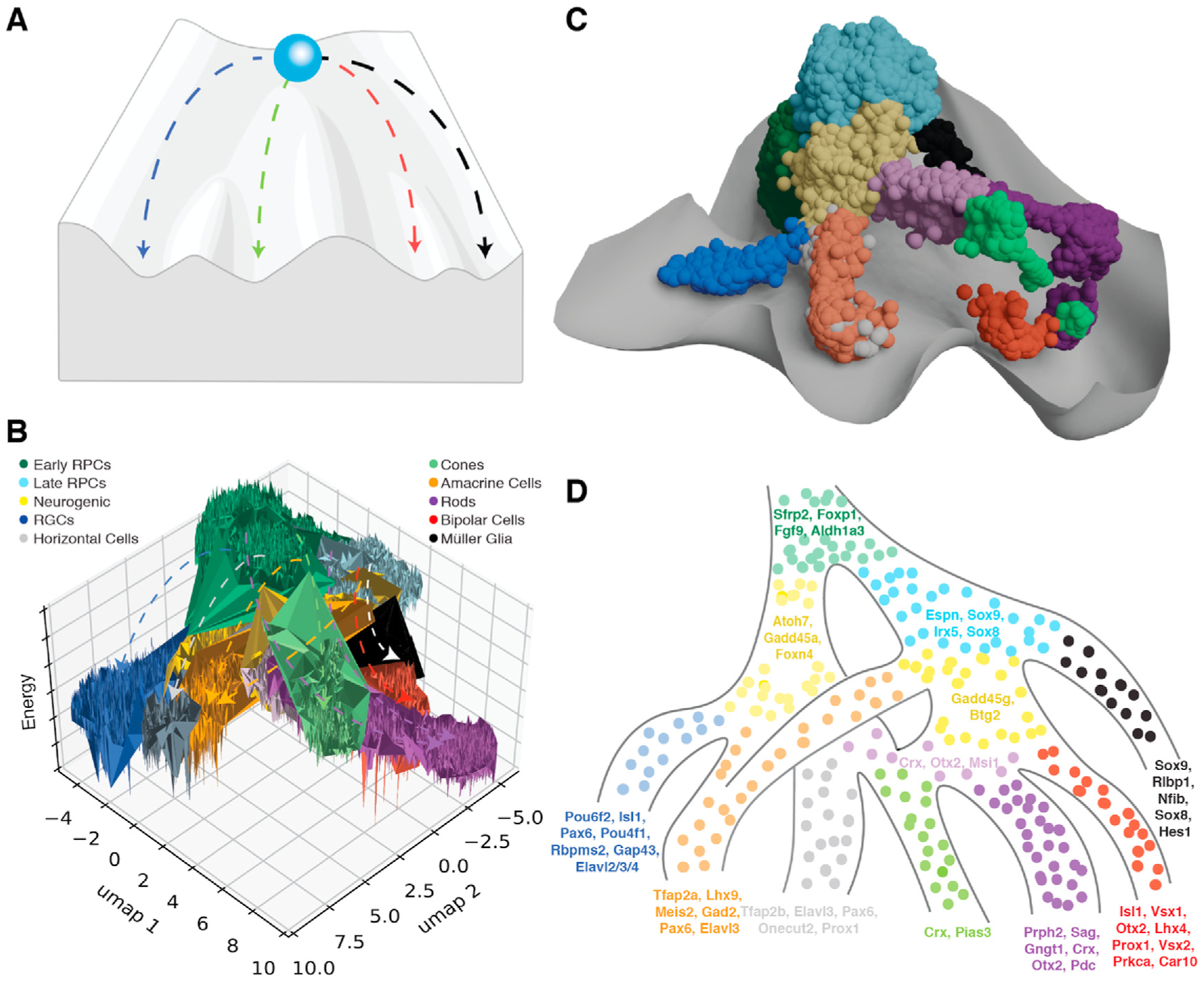
A) Representative Waddington landscape, where a ball - multipotent progenitor -is at the top of a hill. As the ball rolls down the hill - through developmental time -, the peaks and valleys influence where the ball will travel. Ultimately, when the ball reaches the bottom, it is committed to one path, representing a terminal cell fate decision (Waddington, 1957); Waddington landscape generated with Biorender.com. B) Representative potential energy landscape (Soto et al., 2020) as calculated from scRNA-seq results of the developing mouse retina (Clark et al., 2019). Dotted lines represent major cell type trajectories - analogous to valleys within the Waddington model - from early and late RPCs within the developing mouse retina. C) Retinal development waddington landscape derived from mouse single-cell RNA seq data. D) Model of mouse retinal development based on Waddington landscapes. Listed gene transcripts display dynamic RNA velocities - as identified through scVelo (Bergen et al., 2020) — across cellular trajectories for each annotated cell type. Color annotations for B-D are listed in panel B.
First initiated by Monocle (Trapnell et al., 2014) and adapted through additional algorithms (Farrell et al., 2018; Haghverdi et al., 2016), pseudotime analyses of scRNA-seq datasets infer a linear trajectory across cellular states as cells transition through a biological process. As cells exist across a continuum of transcriptional ‘states’ due to asynchronies in progression across development, pseudotime assumes cells of similar gene expression exist at similar transition ‘states.’ Based on this assumption, pseudotime learns the trajectory between cellular states by ordering cells based on similarities in gene expression profiles. The pseudotime order of cells captures the sequence of gene expression changes that occur across biological processes, agnostic to discrete features including ‘true’ developmental time. However, pseudotime trajectories are not necessarily linear. Various trajectory inference techniques can order cells across decision points, where a cell chooses between multiple distinct possible outcomes (decision trees, branchpoints, lineage commitment; Tritschler et al., 2019). The ordering of cells within pseudotime thereby enables assessments of the temporal hierarchy of gene expression during cell state transitions, and therefore, can determine the gene expression programs governing features including cell fate determination. More in-depth discussions and comparisons of pseudotime techniques have previously been discussed at length (Saelens et al., 2019; Tritschler et al., 2019). Implementation of pseudotime trajectory analysis in scRNA-seq studies have determined the gene expression dynamics that govern retinal development, from optic cup formation during zebrafish and xenopus laevis embryogenesis (Briggs et al., 2018; Farrell et al., 2018; Wagner et al., 2018) through terminal cell fate specification, differentiation and maturation stages of all major classes of retinal cell types in mice, humans, and zebrafish (Clark et al., 2019; Lo Giudice et al., 2019; Lu et al., 2020; Raj et al., 2020; Sridhar et al., 2020). Pseudotime techniques and their implementation within retinal development scRNA-seq datasets is highlighted in Fig. 4.
Fig. 4.
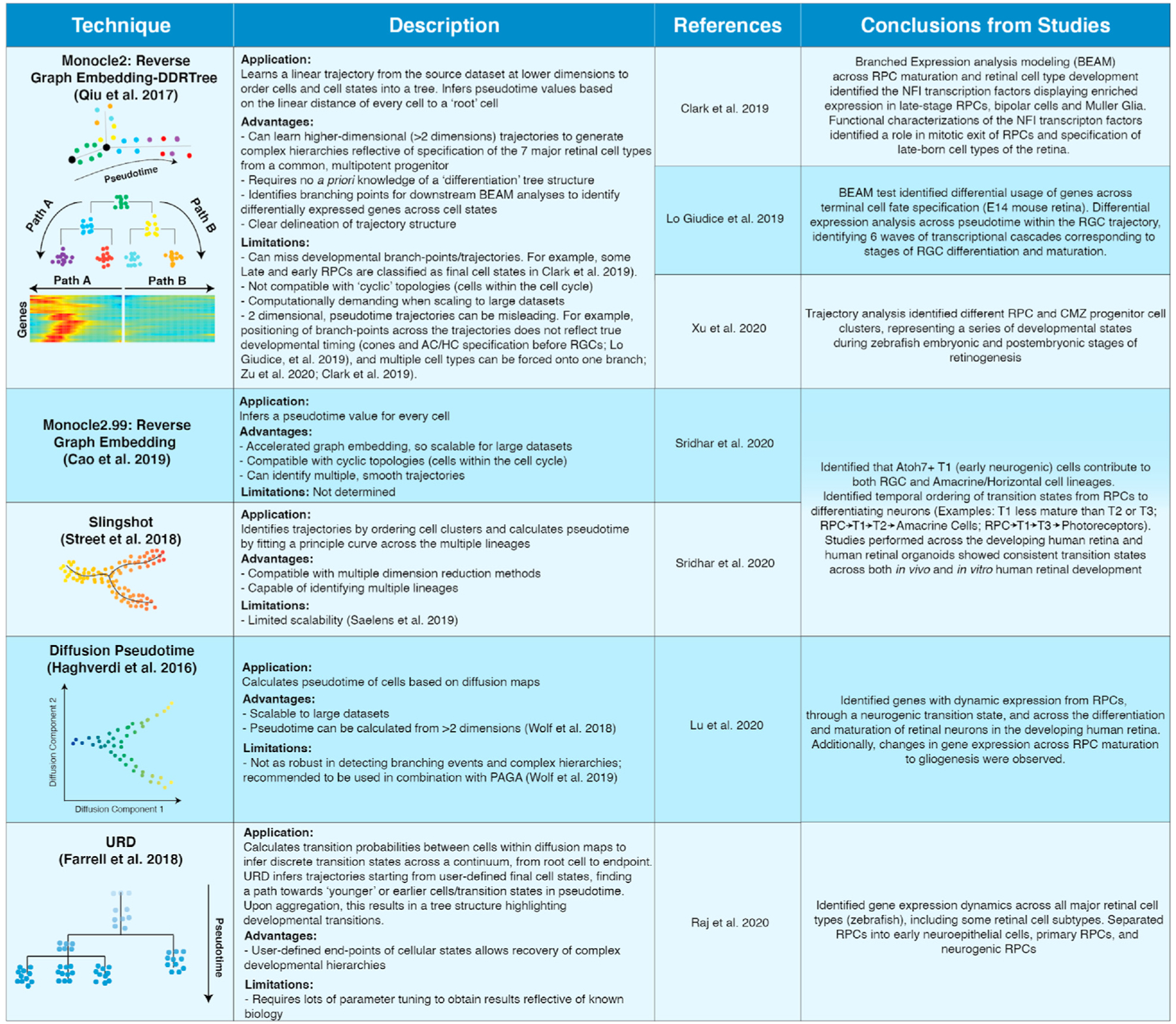
Pseudotime analyses of the developing retina.
Table of pseudotime techniques that have been applied to scRNA-seq datasets of the developing retina, encompassing retinal development of humans, mice, zebrafish, and human retinal organoids.
From these pseudotime trajectory analyses, researchers have identified the prevalence of transcription factor reuse across multiple different cell types. For example, both RGC and amacrine cells express NHLH2 during neuronal differentiation. However, amacrine cells downregulate NHLH2 expression as the cells mature, while RGCs maintain strong expression (Lu et al., 2020). While the functional significance of NHLH2 expression during RGC and amacrine cell differentiation remains to be determined, NHLH2 has been shown to regulate the development of other neural tissues including the hypothalamic-pituitary axis in mice (Good et al., 1997). Additionally, pseudotime analyses indicate expression of MEIS2 within both developing amacrine cells and photoreceptors (Lu et al., 2020). It will be interesting to determine if expression of transcription factors such as MEIS2 across multiple cell fates with similar birth windows is indicative of a restricted lineage precursor, as observed for transcription factors including Otx2 (Emerson and Cepko, 2011). Furthermore, pseudotime analysis highlights Otx2 expression within both photoreceptors and bipolar cells after initial cell fate determination. However, as photoreceptors continue to mature, pseudotime analyses indicate a decrease in Otx2 expression within these cells (Clark et al., 2019). These expression dynamics are consistent with experimental results indicating that high levels of Otx2 expression are required for initial photoreceptor specification, but maintained levels of Otx2 expression facilitates bipolar cell maturation and survival (Yamamoto et al., 2020).
Pseudotime analyses have also revealed coordinated cascades of gene expression changes across the major steps of RPC differentiation towards mature retinal cell fates. For example, pseudotime analysis of primary human RPCs revealed bimodal densities of cells across development, corresponding to early and late RPCs (Lu et al., 2020). Examinations of co-regulated networks of gene expression across the pseudotime trajectory towards RGC fate within the developing mouse retina revealed five discrete transcriptional waves, corresponding to cell cycle transition, neurogenic commitment, cell maturation, synapse assembly, and synaptic transmission (Lo Giudice et al., 2019). These examples highlight the ability of pseudotime analyses to reveal transitional cell states across scRNA-seq datasets, which are populations of cells not readily distinguishable in continuous manifolds of dimension reductions.
On a more global level, integration of pseudotime and trajectory inference methods (Farrell et al., 2018; Qiu et al., 2017) can recreate the hierarchy of retinal cell fate specification, proceeding from the most immature state, RPCs, through intermediate transition states including neurogenic RPCs and cellular precursors, to final cell type classes (Clark et al., 2019; Raj et al., 2020). Performing differential gene analyses both across pseudotime and between different cell type trajectories highlights genes involved in cell fate decisions, differentiation, and maturation. In zebrafish retinas, pseudotime analysis of the bifurcation of photoreceptor precursors into rods and cones identified several known genes required for determination of rod versus cone fates, including six7, nr2f1b, and nr2e3 (Raj et al., 2020; Sotolongo-Lopez et al., 2016; Swaroop et al., 2010). Additionally, these pseudotime analyses identified novel expression patterns of the apelin receptors in photoreceptor progenitors and the apelin ligand in differentiating cones (Raj et al., 2020). As the apelin signaling pathway is implicated in a diverse array of processes including cellular metabolism (reviewed in Chapman et al., 2014), expression of apelin signaling pathway components during photoreceptor specification may coordinate metabolic processes across photoreceptor development. Furthermore, pseudotime analyses on scRNA-seq datasets of human retinas identified transient expression of the transcription factor ATOH7 within photoreceptor precursors and early differentiating cones, implicating ATOH7 in cone photoreceptor specification. Indeed, knockdown of ATOH7 within human retinal explants promoted specification of rod photoreceptors at the expense of cones, suggesting that ATOH7 promotes cone photoreceptor fate during human retinal development (Lu et al., 2020). These cases highlight the ability of pseudotime analyses within scRNA-seq studies to uncover gene expression dynamics across developmental processes, including across the temporal specification of retinal cell fates (Clark et al., 2019; Lo Giudice et al., 2019; Lu et al., 2020; Raj et al., 2020; Xu et al., 2020).
The assumption of pseudotime - that transcription similarities between cells enables linear ordering of cells to recapitulate changes in gene expression across developmental processes can also assist in determining the temporal birth order of cell fates specification from multipotent progenitors. However, users should avoid inferring temporal birth order solely based on pseudotime ordering and in the absence of biological validation. When performed on the developing retina, pseudotime analyses correctly predicted the temporal birth order of retinal cell fates, albeit with one exception - the Müller glia. Years of work have indicated that Müller glia is the last cell type to be specified during retinal development (Bernardos et al., 2007; Jadhav et al., 2009; Ramachandran et al., 2010). However, hierarchical ordering of retinal cell fate specification using pseudotime analyses on scRNA-seq datasets of multiple species position Müller glia early within pseudotime, suggesting specification of Müller glia ‘before’ most retina neurons (Clark et al., 2019; Raj et al., 2020). Such inferences are made agnostic to features such as developmental age of the sampled cells that otherwise indicate specification of Müller glia within later periods of retinal development. In fact, the relative ‘early’ positioning of Müller glia specification within pseudotime is solely based on the transcriptional similarities between Müller glia and late RPCs. scRNA-seq expression profiling indicates that Müller glia express numerous transcripts that are also expressed within late RPCs, including Rlbp1, the Nfi family of transcription factors (Nfia/b/x), and Lhx2 (Clark et al., 2019; Lu et al., 2020). Furthermore, the relatedness of RPCs and Müller glia has been readily established and is highlighted by zebrafish Müller glia, which dedifferentiate to become the source of retinal stem cells during regenerative responses (Bernardos et al., 2007; Jadhav et al., 2009; Ramachandran et al., 2010). Furthermore, Müller glia in other species can be induced through genetic manipulations to generate retinal neurons, suggesting at least partial maintenance of RPC features (Hoang et al., 2020; Jorstad et al., 2017, 2020; Karl et al., 2008; Pollak et al., 2013).
Nonetheless, pseudotime trajectory analyses have uncovered the dynamic nature of gene expression changes underlying retinal cell fate specification and differentiation. Future studies should now focus on the significance of the temporal expression patterns of genes expressed within pseudotime trajectories, assigning function to novel genes expressed during the processes of retinal cell fate specification and differentiation.
4. Phenotyping
4.1. Functional studies
Moving beyond transcriptomic profiling of normal retinal development, single-cell technologies have now extended to phenotypic and molecular characterizations of gene knock-out models, including well established models, such as Atoh7. Previous studies identified Atoh7 as a necessary transcription factor for RGC formation, with Atoh7 mutant models displaying a near complete loss of retinal ganglion cells (Atac et al., 2020; Brown et al., 2001; Ghiasvand et al., 2011; Kay et al., 2001; Prasov et al., 2012). In Atoh7 mutant mouse embryos, few nascent RGCs were present during embryogenesis, suggesting that Atoh7 is necessary for virtually all RGC specification (Brown et al., 2001). However, lineage tracing using an Atoh7:Cre line has suggested that only 55% of RGCs are derived from an Atoh7+ lineage (Brzezinski et al., 2012; Feng et al., 2010; Poggi et al., 2005); a puzzling result given the drastic loss of RGCs in Atoh7 mutant models. Recently, two groups have performed scRNA-seq on the developing mouse retinas of Atoh7 mutant animals to investigate the molecular consequence of Atoh7 loss-of-function on neurogenesis and the specification/differentiation of RGCs. From these studies, we now appreciate that RGCs are specified in Atoh7 mutant retinas, with many RGCs expressing markers of RGC differentiation (Brodie-Kommit et al., 2021; Wu et al., 2021) originally thought to be direct targets of Atoh7, including Pou4f2 and Isl1 (Wu et al., 2015). Additionally, gene module analysis (Brodie-Kommit et al., 2021) and cell cluster analysis (Wu et al., 2021) identified relatively normal transcriptional profiles of neurogenic RPCs, indicating proper selection of RPCs to undergo cell-cycle exit. The scRNA-seq and pseudotime analyses suggest that the specified Atoh7-deficient RGCs don’t progress through normal RGC differentiation, and therefore likely undergo apoptosis as development progresses (Brodie-Kommit et al., 2021; Wu et al., 2021). Confirmation of RGC cell death in Atoh7 mutant retinas was performed using simultaneous deletion of the pro-apoptotic gene Bax. In Atoh7/Bax double mutant retinas, Atoh7-deficient numbers are ‘rescued’ to near normal levels and RGCs persist into adulthood. However, the specified RGCs displayed abnormal characteristics including axon fasciculation and failure to form an optic nerve (Brodie-Kommit et al., 2021). Together, these results suggest that Atoh7 may be dispensable for RGC specification, but may be required both autonomously and non-autonomously for normal RGC differentiation and survival. Further characterizations of Atoh7/Bax double mutant RGCs will be required to determine the necessity of Atoh7 for specification and differentiation of each of the 46 RGC subtypes within the mouse retina.
As previously mentioned, additional studies explored the functional role of OTX2 in lineage restriction of RPCs for production of horizontal cells and cones. scRNA-seq was performed on sorted cells expressing an OTX2-reporter in the presence of either control or OTX2 CRISPR constructs to assess the consequence of OTX2 loss-of-function within the lineage. These studies identified that OTX2 regulates cone photoreceptor specification while inhibiting specification of LHX1+ horizontal cells. Additionally, the authors observed increases in Pax6 expression within OTX2 CRISPR cells, suggesting that OTX2 inhibits Pax6. Cluster analysis also detected the presence of an unusual cell population within the OTX2 mutant cells of the OTX2-reporter lineage, displaying both RGC-like morphology and expression of both horizontal cell and RGC markers. Further investigation will be required to determine if these cells represent a transient RGC state normally present during the course of retinal development or if these cells eventually undergo apoptosis (Ghinia Tegla et al., 2020). However, these studies highlight the utility of scRNA-seq for phenotypic characterizations of mutant phenotypes. Without scRNA-seq studies, the combination of cellular morphology and marker gene expression may have incorrectly categorized these mutant cells as bonafide RGCs, when in fact that is likely not the case.
Additional studies have focused on attributing function to interesting candidate genes from the large-scale scRNA-seq studies. These include examinations of the Nfi transcription factors and the somatostatin receptor Sstr2, that exhibit enriched expression within late RPCs and neurogenic RPCs, respectively (Clark et al., 2019). During retinal development, the Nfi transcription factors Nfia, Nfib, and Nfix are each enriched within primary RPCs during late periods of retinal development (Clark et al., 2019) and display restricted expression patterns within mature retinal cell types including amacrine cells (Keeley and Reese, 2018; Yan et al., 2020a), bipolar cells (Shekhar et al., 2016), and Müller glia (Clark et al., 2019; de Melo et al., 2016). Conditional knockout of Nfia/b/x within the developing retina results in a failure of late RPCs to exit the cell-cycle and differentiate as bipolar cells or Müller glia. Instead, RPCs remain proliferative and continue to undergo neurogenic divisions to specify rod photoreceptors. Using scRNA-seq, the authors were able to confirm these phenotypes and additionally characterize the mutant cells (Clark et al., 2019). Comparisons of scRNA-seq gene expression profiles of ectopic primary and neurogenic RPCs in P14 Nfia/b/x triple knockout retinas to normal retinal development suggest similarities in transcriptomes of Nfia/b/x triple knockout RPCs with late RPCs during development, including expression of late RPC-enriched transcripts Car2, Crym, and Sox8. Furthermore, neurogenic RPCs from Nfia/b/x triple knockout retinas expressed numerous neurogenic RPC-enriched transcripts including Vxn (3110035E14Rik), Otx2, Neurod1, Btg2, and Onecut2, but failed to express other markers of neurogenic RPCs including Olig2, Neurog2, and Dll1 (Clark et al., 2019). These studies highlight the power of scRNA-seq to identify novel processes governing retinal development and provide insight into the mechanisms by which the Nfi transcription factors regulate proliferative quiescence and cell fate specification.
Finally, more recent studies have examined the function of the neurogenic RPC-enriched transcript, Sstr2. In non-retinal tissues, Sstr2 functions as a somatostatin (Sst) receptor, that upon activation, functions to control of cell-cycle exit through accumulation of the downstream effector and cell-cycle inhibitor, p21 (Cdkn1a; Alderton et al., 2001). scRNA-seq of both Sstr2 knockout retinas or Sstr2 agonist-treated retinal explants indicate that Sstr2-activation may function to inhibit neurogenesis and specification of photoreceptors within the developing retina (Weir et al., 2021). In this instance, scRNA-seq studies failed to detect large-scale transcriptional changes after either activation or inhibition of Sstr2 signaling, suggesting an inherent level of functional redundancy in the control of retinal neurogenesis.
4.2. Retinal organoids and in vitro models for development of human disease
Human retinal organoids have emerged as an accessible and manipulatable system for studying retinal development and associated diseases (reviewed in Bell et al., 2020). To establish the authenticity of the organoid system, many studies have tested the extent to which retinal organoid development recapitulates human retinal development, including characterization of cell type composition and global changes in temporal gene expression (Brancati et al., 2020; Collin et al., 2018; Cowan et al., 2020; Kallman et al., 2020; Kim et al., 2019; Lu et al., 2020; Sridhar et al., 2020). To begin to understand the developmental basis of retinal disease, researchers have examined the expression profiles of inherited retinal disease-associated genes across organoid cell types and developmental stages, beginning to elucidate mechanisms by which altered gene function/expression may contribute to disease pathologies (Cowan et al., 2020).
Future scRNA-seq analyses assessing the functional consequence of genetic alterations within human retinal organoids will provide useful tools to evaluate how perturbations, diseases, and organoid differentiation methods affect cell (sub)types, cell states, and gene expression profiles not readily detected via bulk sequencing techniques. For example, recent studies have begun to model NRL mutations on human retinal development. Human patients with mutations in NRL present with either S-cone syndrome, characterized by enhanced blue cone function at the expense of rods, or a more severe phenotype resembling retinitis pigmentosa. scRNA-seq studies of retinal organoids from iPS-derived patient cells harboring a null mutation in the NRL gene determined the existence of two discrete cone populations in NRL mutant organoids that both express short-wavelength opsin. One population of cones resembled S-cones within retinal organoids derived from control patient cells. Conversely, the second S-cone population, which maintained high expression of short-wavelength opsin, displayed an altered transcriptome composed of both cone-specific and rod-specific transcripts (Kallman et al., 2020). These results differed from previous studies that had suggested NRL−/−cells were trans-fated to become S-cones at the expense of rods (Mears et al., 2001).
Additional studies conducted single-cell developmental trajectory and subtype composition analysis of RGCs derived from hiPSCs of primary open angle glaucoma patients with a mutation in SIX6. The SIX6 risk allele impaired proper RGC maturation and led to a deprivation of degeneration-resistant RGC subtypes when compared to control hiPSCs-derived RGCs (Teotia et al., 2017). By modeling human retinal development, these scRNA-seq studies suggest that patients with the SIX6 risk allele exhibit an altered composition of RGC subtypes, which thereby predisposes the patients to increased susceptibility of RGC death (Carnes et al., 2014; Iglesias et al., 2014; Teotia et al., 2017). However, given that enhanced RGC death occurs in most models of retinal organoids and culture systems, further exploration into the significance of SIX6 in RGC subtype survival is required.
5. Perspectives and Limitations
Prior to single-cell genomics technologies, the retinal development field had already discovered many genes involved in the specification and differentiation of retinal cell types, gained insight into the mechanisms regulating neurogenesis and gliogenesis, and identified lineage-restricted populations of RPCs through lineage tracing studies. Given all of this prior knowledge, what advantages have single-cell technologies offered and why should they continue to be applied to the developing retina? First and foremost, scRNA-seq allows us to quantify developmentally regulated gene expression changes and pinpoint them not only to specific cell types, but also to specific developmental cell states across retinogenesis. From these large-scale studies, we can subsequently parse the finer intricacies of gene expression into multiple transcriptional cascades and generate a temporal hierarchy of the gene expression profiles required for specification of individual retinal cell fates. Through profiling of large numbers of genes and cells, we gain the statistical power to examine gene expression patterns across a continuum of dynamic processes, including RPC maturation and cell type differentiation. Identification of these gene expression patterns allows us to move past comparisons of individual gene dynamics and towards evaluation of gene module preservation across multiple datasets (Stein-O’Brien et al., 2019). The extensive profiling of the developing retina across multiple species (mouse, human, zebrafish, and chicken) and in vitro model systems (retinal organoids) offers a treasure trove of new genes and processes to explore. Additionally, given the in-depth characterization of gene expression profiles at the individual cell level, scRNA-seq datasets have provided the information to better identify affected cell type(s) and developmental therapeutic windows in cases of retinal disorders. The advantages of single-cell resolution also extends to hypothesis-driven and phenotyping experiments. Previously, experiments assessing the effects of gene knockouts on cell type specification and differentiation traditionally used marker gene expression to determine changes in cell type proportions after genetic manipulations. Now, using scRNA-seq, we can move beyond discrete cell type classifications based on individual marker genes and instead identify changes in entire transcriptional programs across development and molecularly characterize abnormal or ‘mutant’ cell states that arise as a consequence of altered developmental processes.
Additionally, through integration of gene expression profiles with both cellular morphology and electrophysiology, we are beginning to understand how genetics dictates form and function. Techniques such as Patch-seq have and will continue to further our understanding of the molecular underpinnings of visual processing and behavior (Laboissonniere et al., 2017a; Lipovsek et al., 2021). For example, in zebrafish, we are beginning to connect the transcriptional profiles of RGC subtypes to distinct electrophysiological properties (Kölsch et al., 2021). Using these techniques, we have the potential to correlate alterations in gene expression/function, similar to what may occur in cases of retinal disorders, with altered electrophysiology and morphological consequences.
However, despite the advances in single-cell technologies, obtaining cellular resolution has come at a significant cost in gene expression resolution. Most current technologies profile the 1000–5000 highest expressed genes within individual cells, with many transcripts detected on average at less than one copy per cell. As such, lowly expressed genes displaying significant differential expression by bulk RNA-sequencing methods may be missed by scRNA-seq (Wu et al., 2021). Furthermore, the integration of datasets across experimental conditions poses an additional hurdle, as batch effects are inherent to many commonly used profiling techniques. The development of new technologies to pool experiments without losing experimental condition information is beginning to remedy batch effects Gehring et al., 2020; Luecken and Theis, 2019; McGinnis et al., 2019). Additional focus on combined analysis of multiple datasets within the scRNA-seq field has led to the development of numerous data integration methods aimed at finding shared features across datasets in order to identify and regress the effect of technical noise (Korsunsky et al., 2019; Luecken and Theis, 2019; Stuart et al., 2019; Welch et al., 2019).
As resourceful as these first profiling experiments have been, several limitations of scRNA-seq must be addressed before we can hope to have a truly complete picture of retinal development. First, many of the commonly used technologies to generate scRNA-seq data analysis lack isoform resolution of individual transcripts. This results partially from the decision to use sequence aligners that compress all aligned reads of transcript isoforms to the ‘gene level’ to partially remediate sequence sparsity. As a consequence, isoform information is frequently lost. Importantly, studies have identified that competence factors such as Casz1 generate multiple different isoforms that each bias mid-/late-born neuronal fates differentially (Mattar et al., 2015). However, isoform information in scRNA-seq studies is present within the raw data, only to be uncovered using isoform resolution transcriptome alignment builds and additional technical tricks. For example, the use of long-read sequencing identified novel isoforms of Crb1, including a previously unannotated isoform with unique 5′ and 3′ exons. With these comprehensive isoform maps, one can build a custom transcriptome to specifically analyze isoform usage within the single-cell data. By annotating Crb1 isoforms within alignments of scRNA-seq studies of the developing mouse retina, cell type specificity of Crb1 isoforms is clearly delineated. In the case of Crb1, the canonical Crb1 isoform is expressed in RPCs and Müller glia, whereas the novel Crb1 isoform is expressed in rod photoreceptors (Ray et al., 2020). Further details of isoform usage will be available with continued technical evolution of both mRNA capture efficiency and the ability for sequence aligners to utilize reads across shared exons of transcript isoforms rather than discarding them (Booeshaghi et al., 2020). For example, recent technological advancements in single-cell long-range sequencing could potentially allow us to better characterize differential isoform usage in RPCs (Hagemann-Jensen et al., 2020). When applied to the developing retina, such a technology could further parse the cell type specificity of isoform usage across retinal development. It will be interesting to determine if aspects of retinal development, including RPC competence, can be further explained by heterogeneity in isoform usage across individual cells.
Additionally, while RNA levels are used as a proxy for protein levels, studies indicate that counting RNA transcripts may not be the best predictor of protein expression. For example, the early retinal competence factor, Ikaros, displays low but constant transcript expression across retinal development. However, as development progresses, Ikaros protein levels decrease (Elliott et al., 2008). In neocortex development, progenitor cells also have been shown to accumulate RNA transcripts that encode cell fate determinants; however, these mRNAs are not translated, but function to prime progenitor cells to generate a diverse array of cell types (Li et al., 2020; Zahr et al., 2018). Within the developing retina, future technologies that measure protein expression levels within single cells may be required to assess the relationship of mRNA and protein expression levels within individual cells. Several technologies enable quantification of proteins at single-cell resolution (CITE-seq and REAP-seq), but they both rely on pre-selected antibodies and have been mainly limited to detection and quantification of cell-surface proteins (Peterson et al., 2017; Stoeckius et al., 2017).
Due to the static snapshot and destructive nature of scRNA-seq, we currently cannot trace the steps of individual cells across development in order to decipher the gene expression programs that drove them to their current state. Current techniques, including pseudotime analyses, are predicated on capturing and ordering all transition states across a developmental process. Therefore, pseudotime trajectory and hierarchy analyses predict the average dynamics of cells, overlooking transitions of individual cells. However, the RNA content captured in scRNA-seq includes both spliced and unspliced RNAs. Taking advantage of these features, ‘RNA velocity’ uses the ratio of spliced (previous transcription) to unspliced (ongoing transcription) RNAs to estimate the direction and rate of change for each gene within individual cells to predict a future cell state. Projecting velocity vectors onto lower dimensional embeddings allows tracing of cell states across a developmental process (La Manno et al., 2018).
When applied to the developing retina, RNA velocity predicts that RPCs commit to neurogenesis at G1, well in line with traditional views of time-lapse images and lineage studies that detect Atoh7 protein accumulation and/or reporter expression within RPCs at S/G2-phase (Brzezinski et al., 2012; Le et al., 2006; Miesfeld et al., 2018; Poggi et al., 2005; Yang et al., 2003). RNA velocity analysis recapitulates progression from neurogenic RPCs to terminal cell fates, demonstrating that neurogenic RPCs are an intermediate state prior to commitment to an individual lineage (Lo Giudice et al., 2019). Despite this knowledge and advancements in the computational tools, these tools fail to identify which genes drive neurogenic commitment of G1 RPCs.
A recent update to RNA velocity assumptions and analyses has provided the framework to capture intermediate states and identify the genes/gene networks that demarcate cellular transitions (Bergen et al., 2020). Application of this new algorithm to the developing retina may be able to identify the transcriptional signatures driving RPCs towards neurogenic commitment. However, caution should still be taken when interpreting velocity of cells projected onto lower dimension graphs. Velocities can be miscalculated when data fails to accurately represent the full dynamics of an individual gene. For example, cells can undergo rapid, large-scale gene expression changes that are not fully captured across single-cell profiles (Bergen et al., 2020). Additionally, as pseudotime analyses and RNA velocity are based solely on RNA expression, these techniques do not yet account for the influence of post-translational modifications, asymmetric division modes, and chromatin accessibility on developmental trajectories (Tritschler et al., 2019). With the omission of all these intrinsic factors, neither pseudotime nor RNA velocity can fully delineate the individual molecular events driving lineage choices of an individual cell.
The rapidly developing field of single-cell transcriptomics has offered us an indirect glimpse of the continuous decision-making process of RPCs as they transform from multipotent proliferative cells into the morphologically and functionally divergent cell types that form the intricate architecture of the retina. Several studies have profiled the transcriptomic cascades underlying cell fate specification, but studies addressing cell subtype specification are yet to be completed. Taxonomy datasets of the mature retina highlight the importance of sampling numbers to effectively classify retinal subtypes. For example, fluorescence-activated cell sorting (FACS) enrichment and profiling of 35,699 cells was required to identify all 46 mature RGC subtypes (Tran et al., 2019). Future analysis of the developmental specification of each of these cell subtypes will serve additional challenges. Many of the markers used to enrich cell types of interest by FACS analysis only demarcate mature cell types or display widespread expression across development. For example, Vglut2 is only expressed within the mature RGCs, whereas Vsx2 is widely expressed in RPCs in development before becoming enriched within bipolar cells (Shekhar et al., 2016; Tran et al., 2019). Therefore, to investigate the complete developmental process giving rise to cellular subtype specifications, it is possible that multiple markers/transgenes will have to be employed to mark cell populations of interest across different stages of development.
Despite these limitations of scRNA-seq, a study profiling the developing and adult Drosophila optic lobe at single-cell resolution used a neural network model to assist in classifying cellular subtypes. After training the model on 109,743 annotated cells of the adult optic lobe, the authors recursively annotated the cell types within the optic lobe and retrained the network at five developmental time-points, spanning the developmental processes from neurogenesis through synaptogenesis. Interestingly, the authors observed high transcriptomic heterogeneity amongst cellular subtypes during periods of synaptogenesis, likely driven by differential expression of cell surface molecules mediating circuit formation and dorsal-ventral patterning by WNT signaling gradient. By adulthood, these cellular states converged into transcriptionally stable cellular subtypes (Özel et al., 2021). The similarity between Drosophila optic lobe and mouse retina suggest that large-scale scRNAseq profiling of the retina during periods of synaptogenesis could potentially map out the genetic program underlying circuit formation.
Alternatively, West et al., 2021 leveraged multiplexed RNA FISH technique paired with EdU and BrdU birth-dating to identify birth-windows of the 15 bipolar cell subtypes. The observed birth-dates and genesis locations of each bipolar cell subtype resemble a wave pattern across both developmental time and the dorsal-ventral retinal axis. The subtype birth-dates also occur in an ordered manner and correlate with visual function. Together, this data supports a hierarchical model of subtype specification in the retina and serves as a framework for future studies examining cellular subtype development in the retina (West et al., 2021).
It is also apparent that gene expression is only part of the story of retinal development, and other intrinsic characteristics (epigenetics, post-transcriptional modification, post-translational modification) and external factors (spatial positioning and non-autonomous signaling) play important roles too. For instance, epigenetic profiling of the developing whole retina exemplified how histone modifications correlate with transcriptional changes and identified many cell type and developmental stage specific super enhancers (Aldiri et al., 2017). Examining epigenetic and chromatin landscape changes across retinal development at single-cell resolution will be required to explain the molecular mechanisms that dictate the initiation or repression of cell type specific differentiation and maintenance programs within precursor cells.
Large efforts by the community have made numerous technological advancements for profiling a variety of features within individual cells. These include techniques assaying chromosome organization (Ramani et al., 2017), histone modifications (Grosselin et al., 2019; Kaya-Okur et al., 2019), and DNA accessibility (Buenrostro et al., 2015). Furthermore, techniques identifying transcription factor binding sites within individual cells have also recently been pioneered (Grosselin et al., 2019; Kaya-Okur et al., 2019; Moudgil et al., 2020). With all of these multi-omic datasets, additional efforts have to focus on dataset integration to uncover new biological features. For example, the integration of epigenetic and Hi-C data with scATAC-seq and scRNA-seq datasets from adult retina identified a bipolar cell specific Vsx2 super-enhancer that is required for specification of bipolar cells (Norrie et al., 2019). Future experiments and discoveries will rely on the integration and simultaneous profiling of multiple cell features within individual cells; for example, simultaneous profiling of RNA transcript expression with accessible chromatin is already commercially available.
To date, identification of the mechanisms governing the development of the human macula has provided a formidable challenge. To study this question, one must be able to intersect cellular resolution transcriptomics with spatial positioning. However, the dissociation and lysis steps required for many single-cell techniques lose spatial information. Studies attempting to address macular development have relied on physical separation of the macula and peripheral retina prior to single-cell sequencing library preparation workflows. This technique inherently requires the presence of a defined macula, precluding the measurement of developmental events just prior to macular formation. Recently, several in situ sequencing technologies have emerged to connect spatial and transcriptomic information through cDNA probes targeting pre-selected gene targets or polyT barcoded beads (Larsson et al., 2021; Wang et al., 2018). Though the unbiased mRNA capture efficiency of spatial transcriptomics is only half of the already low capture rate of droplet-based scRNA-seq methods, it is sufficient for the reconstruction of spatial developmental trajectories in the mouse neocortex (Stickels et al., 2020).
Single-molecule fluorescent in situ hybridization (smFISH) can achieve an even higher-resolution of RNA spatial information. Multiplexed smFISH technologies, such as MERFISH (Chen et al., 2015; Moffitt et al., 2018; Xia et al., 2019) and RNA seqFISH+ (Eng et al., 2019), can now image hundreds to thousands of RNA transcripts at single-cell resolution within intact tissues, including samples as complex as the brain. These technologies can help validate the expression of candidate genes within scRNAseq datasets, including their RNA copy number, cellular specificity, and dynamics across developmental time. The convergence of spatial and expression information at cellular resolution has proven useful for deciphering intercellular ligand-receptor interactions and identifying cells that exhibit preferential interactions/connections (Eng et al., 2019). Furthermore, introduction of a barcode editing system to multiplexed FISH has revealed lineage, spatial, and cell state information within the Drosophila brain (Chow et al., 2021).
Within the developing retina, patterning of regions dedicated to high acuity vision is mediated through extrinsic signaling, in particular, that of retinoic acid (da Silva and Cepko, 2017). While scRNA-seq cannot directly detect cell-to-cell signaling, scRNA-seq studies have tried to infer the cell types signaling or receiving retinoic acid through expression of retinoic acid synthesizing and metabolizing enzymes as a proxy for development of human macula (Cowan et al., 2020; Lu et al., 2020). Future studies using spatial transcriptomics to examine transcriptional programs governing spatial asymmetries across the developing retina will offer valuable insights into the mechanisms controlling retinal features including formation of the macula. Additionally, as RPCs and retinal neurons rely on precise molecular programs for migration and formation of the laminar structure of the retina (Amini et al., 2017), spatial transcriptomics may help clarify the interplay between cell state and relative positioning of cells within the developing retina.
The wealth of data obtained through scRNA-seq studies has correlated patterns of gene expressions with biological processes. Future studies must now focus on addressing individual gene necessity and sufficiency across retinal development through expanded functional studies. Given the abundance of novel, developmentally-regulated genes discovered in scRNA-seq studies, it will take years to assay the consequence of perturbations of gene expression levels on a gene-by-gene basis. Instead, several techniques have been developed to assess changes in gene expression using scRNA-seq with pooled CRISPR gRNA libraries that target multiple genes in a single experiment (Datlinger et al., 2017; Dixit et al., 2016; Jaitin et al., 2016). Application of such technologies to the developing mouse cortex revealed gene regulatory networks and cell types affected by mutations in Autism Spectrum Disorder risk genes (Jin et al., 2020). This demonstrates the feasibility of the techniques for studying both gene and gene regulatory element function across the developing retina, and could be readily applied to all genes implicated in the development of retinal disorders (~400 genes).
In conclusion, retinal cell fate decisions are modeled by both stochastic/probabilistic and deterministic (lineage restriction) decisions of RPCs. To date, scRNA-seq studies support both models, whereby the selection of an individual primary RPC to undergo a neurogenic (or later gliogenic) division appears to be largely stochastic, while evidence of lineage restriction is apparent within neurogenic RPCs. Additionally, global changes in the transcriptome of RPCs across developmental time is reminiscent of competence models, supported by both temporally restricted expression patterns of competence factors and temporal fate determinants in scRNA-seq datasets (Fig. 2) and functional studies characterizing individual competence factor functions. However, to more clearly define the roles of stochasticity and lineage restrictions, the field needs to pair either new scRNA-seq based or traditional lineage tracing strategies (reporter lines or time-lapse imaging) with molecular characterizations of gene expression at the individual cell level. With comprehensive lineage restriction maps, one can then proceed backwards in ‘pseudotime’ to begin to uncover the transcriptional hierarchy regulating cell fate specification from individual RPCs. Through these lineage-tracing experiments and pairing of additional multimodal single-cell ‘omics’ technologies, we can strive to achieve a more deterministic model for the sequential gene expression events governing retinal cell fate specification.
Acknowledgements
We thank all members of the Clark lab for critical assessments of the manuscript and H. Halim for helpful discussion on calculating gene regulatory network energy. This work is supported by grants from the NIH (R00EY027844 to BSC), a career development award to BSC from Research to Prevent Blindness, and an unrestricted grant to the Department of Ophthalmology and Visual Sciences from Research to Prevent Blindness in support of PAR and BSC.
References
- Akagi T, Inoue T, Miyoshi G, Bessho Y, Takahashi M, Lee JE, Guillemot F, Kageyama R, 2004. Requirement of multiple basic helix-loop-helix genes for retinal neuronal subtype specification. J. Biol. Chem 279, 28492–28498. 10.1074/jbc.M400871200. [DOI] [PubMed] [Google Scholar]
- Alderton F, Humphrey PP, Sellers LA, 2001. High-intensity p38 kinase activity is critical for p21(cip 1) induction and the antiproliferative function of G(i) protein-coupled receptors. Mol. Pharmacol 59, 1119–1128. 10.1124/mol.59.5.1119. [DOI] [PubMed] [Google Scholar]
- Aldiri I, Xu B, Wang L, Chen X, Hiler D, Griffiths L, Valentine M, Shirinifard A, Thiagarajan S, Sablauer A, Barabas M-E, Zhang J, Johnson D, Frase S, Zhou X, Easton J, Zhang J, Mardis ER, Wilson RK, Downing JR, Dyer MA, St. Jude Children’s Research Hospital—Washington University Pediatric Cancer Genome Project, 2017. The dynamic epigenetic landscape of the retina during development, reprogramming, and tumorigenesis. Neuron 94, 550–568. 10.1016/j.neuron.2017.04.022 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexiades MR, Cepko CL, 1997. Subsets of retinal progenitors display temporally regulated and distinct biases in the fates of their progeny. Development 124, 1119–1131. [DOI] [PubMed] [Google Scholar]
- Amini R, Rocha-Martins M, Norden C, 2017. Neuronal migration and lamination in the vertebrate retina. Front. Neurosci 11, 742. 10.3389/fnins.2017.00742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atac D, Koller S, Hanson JVM, Feil S, Tiwari A, Bahr A, Baehr L, Magyar I, Kottke R, Gerth-Kahlert C, Berger W, 2020. Atonal homolog 7 (ATOH7) loss-of-function mutations in predominant bilateral optic nerve hypoplasia. Hum. Mol. Genet 29, 132–148. 10.1093/hmg/ddz268. [DOI] [PubMed] [Google Scholar]
- Baden T, Berens P, Franke K, Román Rosón M, Bethge M, Euler T, 2016. The functional diversity of retinal ganglion cells in the mouse. Nature 529, 345–350. 10.1038/nature16468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao ZZ, Cepko CL, 1997. The expression and function of Notch pathway genes in the developing rat eye. J. Neurosci 17, 1425–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baye LM, Link BA, 2007. Interkinetic nuclear migration and the selection of neurogenic cell divisions during vertebrate retinogenesis. J. Neurosci 27, 10143–10152. 10.1523/JNEUROSCI.2754-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bélanger M-C, Robert B, Cayouette M, 2017. Msx1-Positive progenitors in the retinal ciliary margin give rise to both neural and non-neural progenies in mammals. Dev. Cell 40, 137–150. 10.1016/j.devcel.2016.11.020. [DOI] [PubMed] [Google Scholar]
- Bell CM, Zack DJ, Berlinicke CA, 2020. Human organoids for the study of retinal development and disease. Annu Rev Vis Sci 6, 91–114. 10.1146/annurev-vision-121219-081855. [DOI] [PubMed] [Google Scholar]
- Belliveau MJ, Cepko CL, 1999. Extrinsic and intrinsic factors control the genesis of amacrine and cone cells in the rat retina. Development 126, 555–566. [DOI] [PubMed] [Google Scholar]
- Belliveau MJ, Young TL, Cepko CL, 2000. Late retinal progenitor cells show intrinsic limitations in the production of cell types and the kinetics of opsin synthesis. J. Neurosci 20, 2247–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergen V, Lange M, Peidli S, Wolf FA, Theis FJ, 2020. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol 38, 1408–1414. 10.1038/s41587-020-0591-3. [DOI] [PubMed] [Google Scholar]
- Bernardos RL, Barthel LK, Meyers JR, Raymond PA, 2007. Late-stage neuronal progenitors in the retina are radial Müller glia that function as retinal stem cells. J. Neurosci 27, 7028–7040. 10.1523/JNEUROSCI.1624-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackshaw S, Harpavat S, Trimarchi J, Cai L, Huang H, Kuo WP, Weber G, Lee K, Fraioli RE, Cho S-H, Yung R, Asch E, Ohno-Machado L, Wong WH, Cepko CL, 2004. Genomic analysis of mouse retinal development. PLoS Biol. 2, E247. 10.1371/journal.pbio.0020247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booeshaghi AS, Sina Booeshaghi A, Yao Z, van Velthoven C, Smith K, Tasic B, Zeng H, Pachter L, 2020. Isoform cell type specificity in the mouse primary motor cortex. bioRxiv. 10.1101/2020.03.05.977991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreau-Pinsonneault C, Cayouette M, 2018. Cell lineage tracing in the retina: could material transfer distort conclusions? Developmental Dynamics. 10.1002/dvdy.24535. [DOI] [PubMed] [Google Scholar]
- Bowling S, Sritharan D, Osorio FG, Nguyen M, Cheung P, Rodriguez-Fraticelli A, Patel S, Yuan W-C, Fujiwara Y, Li BE, Orkin SH, Hormoz S, Camargo FD, 2020. An engineered CRISPR-cas9 mouse line for simultaneous readout of lineage histories and gene expression profiles in single cells. Cell 181, 1693–1694. 10.1016/j.cell.2020.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brancati G, Treutlein B, Camp JG, 2020. Resolving neurodevelopmental and vision disorders using organoid single-cell multi-omics. Neuron 107, 1000–1013. 10.1016/j.neuron.2020.09.001. [DOI] [PubMed] [Google Scholar]
- Briggs JA, Weinreb C, Wagner DE, Megason S, Peshkin L, Kirschner MW, Klein AM, 2018. The dynamics of gene expression in vertebrate embryogenesis at single-cell resolution. Science 360. 10.1126/science.aar5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodie-Kommit J, Clark BS, Shi Q, Shiau F, Kim DW, Langel J, Sheely C, Ruzycki PA, Fries M, Javed A, Cayouette M, Schmidt T, Badea T, Glaser T, Zhao H, Singer J, Blackshaw S, Hattar S, 2021. Atoh7-independent specification of retinal ganglion cell identity. Sci Adv 7. 10.1126/sciadv.abe4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NL, Patel S, Brzezinski J, Glaser T, 2001. Math5 is required for retinal ganglion cell and optic nerve formation. Development 128, 2497–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzezinski JA, Kim EJ, Johnson JE, Reh TA, 2011. Ascl1 expression defines a subpopulation of lineage-restricted progenitors in the mammalian retina. Development 138, 3519–3531. 10.1242/dev.064006, 4th. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzezinski JA, Prasov L, Glaser T, 2012. Math5 defines the ganglion cell competence state in a subpopulation of retinal progenitor cells exiting the cell cycle. Dev. Biol 365, 395–413. 10.1016/j.ydbio.2012.03.006, 4th. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP, Chang HY, Greenleaf WJ, 2015. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–490. 10.1038/nature14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajal S.R.y., 1989. Recollections of my life. 10.7551/mitpress/5817.001.0001. [DOI]
- Cao J, Packer JS, Ramani V, Cusanovich DA, Huynh C, Daza R, Qiu X, Lee C, Furlan SN, Steemers FJ, Adey A, Waterston RH, Trapnell C, Shendure J, 2017. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science 357, 661–667. 10.1126/science.aam8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnes MU, Liu YP, Allingham RR, Whigham BT, Havens S, Garrett ME, Qiao C, , NEIGHBORHOOD Consortium Investigators, Katsanis N, Wiggs JL, Pasquale LR, Ashley-Koch A, Oh EC, Hauser MA, 2014. Discovery and functional annotation of SIX6 variants in primary open-angle glaucoma. PLoS Genet. 10, e1004372. 10.1371/journal.pgen.1004372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayouette M, Barres BA, Raff M, 2003. Importance of intrinsic mechanisms in cell fate decisions in the developing rat retina. Neuron 40, 897–904. 10.1016/s0896-6273(03)00756-6. [DOI] [PubMed] [Google Scholar]
- Cayouette M, Raff M, 2003. The orientation of cell division influences cell-fate choice in the developing mammalian retina. Development 130, 2329–2339. 10.1242/dev.00446. [DOI] [PubMed] [Google Scholar]
- Cepko C, 2014. Intrinsically different retinal progenitor cells produce specific types of progeny. Nat. Rev. Neurosci 15, 615–627. 10.1038/nrn3767. [DOI] [PubMed] [Google Scholar]
- Cepko CL, Austin CP, Yang X, Alexiades M, Ezzeddine D, 1996. Cell fate determination in the vertebrate retina. Proc. Natl. Acad. Sci. U.S.A 93, 589–595. 10.1073/pnas.93.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman NA, Dupré DJ, Rainey JK, 2014. The apelin receptor: physiology, pathology, cell signalling, and ligand modulation of a peptide-activated class A GPCR. Biochem. Cell. Biol 92, 431–440. 10.1139/bcb-2014-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X, 2015. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090. 10.1126/science.aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Wang QL, Nie Z, Sun H, Lennon G, Copeland NG, Gilbert DJ, Jenkins NA, Zack DJ, 1997. Crx, a novel Otx-like paired-homeodomain protein, binds to and transactivates photoreceptor cell-specific genes. Neuron 19, 1017–1030. 10.1016/s0896-6273(00)80394-3. [DOI] [PubMed] [Google Scholar]
- Chen Y, Londraville R, Brickner S, El-Shaar L, Fankhauser K, Dearth C, Fulton L, Sochacka A, Bhattarai S, Marrs JA, Liu Q, 2013. Protocadherin-17 function in Zebrafish retinal development. Dev. Neurobiol 73, 259–273. 10.1002/dneu.22053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry TJ, Trimarchi JM, Stadler MB, Cepko CL, 2009. Development and diversification of retinal amacrine interneurons at single cell resolution. Proc. Natl. Acad. Sci. U.S.A 106, 9495–9500. 10.1073/pnas.0903264106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow K-HK, Budde MW, Granados AA, Cabrera M, Yoon S, Cho S, Huang T-H, Koulena N, Frieda KL, Cai L, Lois C, Elowitz MB, 2021. Imaging cell lineage with a synthetic digital recording system. Science 372. 10.1126/science.abb3099. [DOI] [PubMed] [Google Scholar]
- Clark BS, Cui S, Miesfeld JB, Klezovitch O, Vasioukhin V, Link BA, 2012. Loss of Llgl1 in retinal neuroepithelia reveals links between apical domain size, Notch activity and neurogenesis. Development 139, 1599–1610. 10.1242/dev.078097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark BS, Miesfeld JB, Flinn MA, Collery RF, Link BA, 2021. Dynamic polarization of Rab11a modulates Crb2a localization and impacts signaling to regulate retinal neurogenesis. Front Cell Dev Biol 8, 608112. 10.3389/fcell.2020.608112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark BS, Stein-O’Brien GL, Shiau F, Cannon GH, Davis-Marcisak E, Sherman T, Santiago CP, Hoang TV, Rajaii F, James-Esposito RE, Gronostajski RM, Fertig EJ, Goff LA, Blackshaw S, 2019. Single-cell RNA-seq analysis of retinal development identifies NFI factors as regulating mitotic exit and late-born cell specification. Neuron 102, 1111–1126. 10.1016/j.neuron.2019.04.010 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen AR, Gomes FLAF, Roysam B, Cayouette M, 2010. Computational prediction of neural progenitor cell fates. Nat. Methods 7, 213–218. 10.1038/nmeth.1424. [DOI] [PubMed] [Google Scholar]
- Collin J, Queen R, Mellough CB, Lako M, 2018. Using hESC-derived retinal organoids to investigate the transcriptional profile of emerging photoreceptors. Invest. Ophthalmol. Vis. Sci 59, 570–570. [Google Scholar]
- Collin J, Queen R, Zerti D, Dorgau B, Hussain R, Coxhead J, Cockell S, Lako M, 2019. Deconstructing retinal organoids: single cell RNA-seq reveals the cellular components of human pluripotent stem cell-derived retina. Stem Cell. 37, 593–598. 10.1002/stem.2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coomer CE, Wilson SG, Titialii-Torres KF, Bills JD, Krueger LA, Petersen RA, Turnbaugh EM, Janesch EL, Morris AC, 2020. Her9/Hes4 is required for retinal photoreceptor development, maintenance, and survival. Sci. Rep 10, 11316. 10.1038/s41598-020-68172-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan CS, Renner M, De Gennaro M, Gross-Scherf B, Goldblum D, Hou Y, Munz M, Rodrigues TM, Krol J, Szikra T, Cuttat R, Waldt A, Papasaikas P, Diggelmann R, Patino-Alvarez CP, Galliker P, Spirig SE, Pavlinic D, Gerber-Hollbach N, Schuierer S, Srdanovic A, Balogh M, Panero R, Kusnyerik A, Szabo A, Stadler MB, Orgül S, Picelli S, Hasler PW, Hierlemann A, Scholl HPN, Roma G, Nigsch F, Roska B, 2020. Cell types of the human retina and its organoids at single-cell resolution. Cell 182, 1623–1640. 10.1016/j.cell.2020.08.013 e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva S, Cepko CL, 2017. Fgf 8 expression and degradation of retinoic acid are required for patterning a high-acuity area in the retina. Dev. Cell 42, 68–81. 10.1016/j.devcel.2017.05.024 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datlinger P, Rendeiro AF, Schmidl C, Krausgruber T, Traxler P, Klughammer J, Schuster LC, Kuchler A, Alpar D, Bock C, 2017. Pooled CRISPR screening with single-cell transcriptome readout. Nat. Methods 14, 297–301. 10.1038/nmeth.4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Huerta I, Kim I-J, Voinescu PE, Sanes JR, 2012. Direction-selective retinal ganglion cells arise from molecularly specified multipotential progenitors. Proc. Natl. Acad. Sci. U.S.A 109, 17663–17668. 10.1073/pnas.1215806109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Melo J, Clark BS, Blackshaw S, 2016. Multiple intrinsic factors act in concert with Lhx2 to direct retinal gliogenesis. Sci. Rep 6, 32757. 10.1038/srep32757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit A, Parnas O, Li B, Chen J, Fulco CP, Jerby-Arnon L, Marjanovic ND, Dionne D, Burks T, Raychowdhury R, Adamson B, Norman TM, Lander ES, Weissman JS, Friedman N, Regev A, 2016. Perturb-seq: dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell 167, 1853–1866. 10.1016/j.cell.2016.11.038 e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer MA, Cepko CL, 2001a. p27Kip1 and p57Kip2 regulate proliferation in distinct retinal progenitor cell populations. J. Neurosci 21, 4259–4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer MA, Cepko CL, 2001b. Regulating proliferation during retinal development. Nat. Rev. Neurosci 2, 333–342. 10.1038/35072555. [DOI] [PubMed] [Google Scholar]
- el Ghissassi F, Valsesia-Wittmann S, Falette N, Duriez C, Walden PD, Puisieux A, 2002. BTG2(TIS21/PC3) induces neuronal differentiation and prevents apoptosis of terminally differentiated PC12 cells. Oncogene 21, 6772–6778. 10.1038/sj.onc.1205888. [DOI] [PubMed] [Google Scholar]
- Elliott J, Jolicoeur C, Ramamurthy V, Cayouette M, 2008. Ikaros confers early temporal competence to mouse retinal progenitor cells. Neuron 60, 26–39. 10.1016/j.neuron.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Emerson MM, Cepko CL, 2011. Identification of a retina-specific Otx2 enhancer element active in immature developing photoreceptors. Dev. Biol 360, 241–255. 10.1016/j.ydbio.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson MM, Surzenko N, Goetz JJ, Trimarchi J, Cepko CL, 2013. Otx2 and Onecut1 promote the fates of cone photoreceptors and horizontal cells and repress rod photoreceptors. Dev. Cell 26, 59–72. 10.1016/j.devcel.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng C-HL, Lawson M, Zhu Q, Dries R, Koulena N, Takei Y, Yun J, Cronin C, Karp C, Yuan G-C, Cai L, 2019. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH. Nature 568, 235–239. 10.1038/s41586-019-1049-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euler T, Haverkamp S, Schubert T, Baden T, 2014. Retinal bipolar cells: elementary building blocks of vision. Nat. Rev. Neurosci 15, 507–519. 10.1038/nrn3783. [DOI] [PubMed] [Google Scholar]
- Farrell JA, Wang Y, Riesenfeld SJ, Shekhar K, Regev A, Schier AF, 2018. Single-cell reconstruction of developmental trajectories during zebrafish embryogenesis. Science 360. 10.1126/science.aar3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Xie Z-H, Ding Q, Xie X, Libby RT, Gan L, 2010. MATH5 controls the acquisition of multiple retinal cell fates. Mol. Brain 3, 36. 10.1186/1756-6606-3-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring J, Hwee Park J, Chen S, Thomson M, Pachter L, 2020. Highly multiplexed single-cell RNA-seq by DNA oligonucleotide tagging of cellular proteins. Nat. Biotechnol 38, 35–38. 10.1038/s41587-019-0372-z. [DOI] [PubMed] [Google Scholar]
- Ghiasvand NM, Rudolph DD, Mashayekhi M, Brzezinski JA, Goldman D, Glaser T, 2011. Deletion of a remote enhancer near ATOH7 disrupts retinal neurogenesis, causing NCRNA disease. Nat. Neurosci 14, 578–586. 10.1038/nn.2798, 4th. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghinia Tegla MG, Buenaventura DF, Kim DY, Thakurdin C, Gonzalez KC, Emerson MM, 2020. OTX2 represses sister cell fate choices in the developing retina to promote photoreceptor specification. Elife 9. 10.7554/eLife.54279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes FLAF, Zhang G, Carbonell F, Correa JA, Harris WA, Simons BD, Cayouette M, 2011. Reconstruction of rat retinal progenitor cell lineages in vitro reveals a surprising degree of stochasticity in cell fate decisions. Development 138, 227–235. 10.1242/dev.059683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good DJ, Porter FD, Mahon KA, Parlow AF, Westphal H, Kirsch IR, 1997. Hypogonadism and obesity in mice with a targeted deletion of the Nhlh2 gene. Nat. Genet 15, 397–401. 10.1038/ng0497-397. [DOI] [PubMed] [Google Scholar]
- Grosselin K, Durand A, Marsolier J, Poitou A, Marangoni E, Nemati F, Dahmani A, Lameiras S, Reyal F, Frenoy O, Pousse Y, Reichen M, Woolfe A, Brenan C, Griffiths AD, Vallot C, Gérard A, 2019. High-throughput single-cell ChIP-seq identifies heterogeneity of chromatin states in breast cancer. Nat. Genet 51, 1060–1066. 10.1038/s41588-019-0424-9. [DOI] [PubMed] [Google Scholar]
- Hafler BP, Surzenko N, Beier KT, Punzo C, Trimarchi JM, Kong JH, Cepko CL, 2012. Transcription factor Olig2 defines subpopulations of retinal progenitor cells biased toward specific cell fates. Proc. Natl. Acad. Sci. U.S.A 109, 7882–7887. 10.1073/pnas.1203138109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemann-Jensen M, Ziegenhain C, Chen P, Ramsköld D, Hendriks G-J, Larsson AJM, Faridani OR, Sandberg R, 2020. Single-cell RNA counting at allele and isoform resolution using Smart-seq 3. Nat. Biotechnol 38, 708–714. 10.1038/s41587-020-0497-0. [DOI] [PubMed] [Google Scholar]
- Haghverdi L, Büttner M, Wolf FA, Buettner F, Theis FJ, 2016. Diffusion pseudotime robustly reconstructs lineage branching. Nat. Methods 13, 845–848. 10.1038/nmeth.3971. [DOI] [PubMed] [Google Scholar]
- He J, Zhang G, Almeida AD, Cayouette M, Simons BD, Harris WA, 2012. How variable clones build an invariant retina. Neuron 75, 786–798. 10.1016/j.neuron.2012.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmstaedter M, Briggman KL, Turaga SC, Jain V, Seung HS, Denk W, 2013. Connectomic reconstruction of the inner plexiform layer in the mouse retina. Nature 500, 168–174. 10.1038/nature12346. [DOI] [PubMed] [Google Scholar]
- Hitchcock PF, Raymond PA, 2004. The teleost retina as a model for developmental and regeneration biology. Zebrafish 1, 257–271. 10.1089/zeb.2004.1.257. [DOI] [PubMed] [Google Scholar]
- Hoang T, Wang J, Boyd P, Wang F, Santiago C, Jiang L, Yoo S, Lahne M, Todd LJ, Jia M, Saez C, Keuthan C, Palazzo I, Squires N, Campbell WA, Rajaii F, Parayil T, Trinh V, Kim DW, Wang G, Campbell LJ, Ash J, Fischer AJ, Hyde DR, Qian J, Blackshaw S, 2020. Gene regulatory networks controlling vertebrate retinal regeneration. Science 370. 10.1126/science.abb8598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holguera I, Desplan C, 2018. Neuronal specification in space and time. Science 362, 176–180. 10.1126/science.aas9435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollander MC, Fornace AJ Jr., 2002. Genomic instability, centrosome amplification, cell cycle checkpoints and Gadd45a. Oncogene 21, 6228–6233. 10.1038/sj.onc.1205774. [DOI] [PubMed] [Google Scholar]
- Holt CE, Bertsch TW, Ellis HM, Harris WA, 1988. Cellular determination in the Xenopus retina is independent of lineage and birth date. Neuron 1, 15–26. 10.1016/0896-6273(88)90205-x. [DOI] [PubMed] [Google Scholar]
- Hoshino A, Ratnapriya R, Brooks MJ, Chaitankar V, Wilken MS, Zhang C, Starostik MR, Gieser L, La Torre A, Nishio M, Bates O, Walton A, Bermingham-McDonogh O, Glass IA, Wong ROL, Swaroop A, Reh TA, 2017. Molecular anatomy of the developing human retina. Dev. Cell 43, 763–779. 10.1016/j.devcel.2017.10.029 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Sui X, Li L, Huang X, Rong R, Su X, Shi Q, Mo L, Shu X, Kuang Y, Tao Q, He C, 2013. Protocadherin 17 acts as a tumour suppressor inducing tumour cell apoptosis and autophagy, and is frequently methylated in gastric and colorectal cancers. J. Pathol 229, 62–73. 10.1002/path.4093. [DOI] [PubMed] [Google Scholar]
- Hu Y, Wang X, Hu B, Mao Y, Chen Y, Yan L, Yong J, Dong J, Wei Y, Wang W, Wen L, Qiao J, Tang F, 2019. Dissecting the transcriptome landscape of the human fetal neural retina and retinal pigment epithelium by single-cell RNA-seq analysis. PLoS Biol. 17, e3000365. 10.1371/journal.pbio.3000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang SS, Lim J, Yu Z, Kong P, Sefik E, Xu H, Harman CCD, Kim LK, Lee GR, Li H-B, Flavell RA, 2020. mRNA destabilization by BTG1 and BTG2 maintains T cell quiescence. Science 367, 1255–1260. 10.1126/science.aax0194. [DOI] [PubMed] [Google Scholar]
- Iacopetti P, Michelini M, Stuckmann I, Oback B, Aaku-Saraste E, Huttner WB, 1999. Expression of the antiproliferative gene TIS21 at the onset of neurogenesis identifies single neuroepithelial cells that switch from proliferative to neuron-generating division. Proc. Natl. Acad. Sci. U.S.A 96, 4639–4644. 10.1073/pnas.96.8.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias AI, Springelkamp H, van der Linde H, Severijnen L-A, Amin N, Oostra B, Kockx CEM, van den Hout MCGN, van Ijcken WFJ, Hofman A, Uitterlinden AG, Verdijk RM, Klaver CCW, Willemsen R, van Duijn CM, 2014. Exome sequencing and functional analyses suggest that SIX6 is a gene involved in an altered proliferation-differentiation balance early in life and optic nerve degeneration at old age. Hum. Mol. Genet 23, 1320–1332. 10.1093/hmg/ddt522. [DOI] [PubMed] [Google Scholar]
- Inoue T, Hojo M, Bessho Y, Tano Y, Lee JE, Kageyama R, 2002. Math 3 and NeuroD regulate amacrine cell fate specification in the retina. Development 129, 831–842. [DOI] [PubMed] [Google Scholar]
- Isshiki T, Pearson B, Holbrook S, Doe CQ, 2001. Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell 106, 511–521. 10.1016/s0092-8674(01)00465-2. [DOI] [PubMed] [Google Scholar]
- Jadhav AP, Roesch K, Cepko CL, 2009. Development and neurogenic potential of Müller glial cells in the vertebrate retina. Prog. Retin. Eye Res 28, 249–262. 10.1016/j.preteyeres.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaitin DA, Weiner A, Yofe I, Lara-Astiaso D, Keren-Shaul H, David E, Salame TM, Tanay A, van Oudenaarden A, Amit I, 2016. Dissecting immune circuits by linking CRISPR-pooled screens with single-cell RNA-seq. Cell 167. 10.1016/j.cell.2016.11.039, 1883–1896.e15. [DOI] [PubMed] [Google Scholar]
- Javed A, Mattar P, Lu S, Kruczek K, Kloc M, Gonzalez-Cordero A, Bremner R, Ali RR, Cayouette M, 2020. Pou2f1 and Pou2f2 cooperate to control the timing of cone photoreceptor production in the developing mouse retina. Development 147. 10.1242/dev.188730. [DOI] [PubMed] [Google Scholar]
- Jensen AM, Raff MC, 1997. Continuous observation of multipotential retinal progenitor cells in clonal density culture. Dev. Biol 188, 267–279. 10.1006/dbio.1997.8645. [DOI] [PubMed] [Google Scholar]
- Jin X, Simmons SK, Guo A, Shetty AS, Ko M, Nguyen L, Jokhi V, Robinson E, Oyler P, Curry N, Deangeli G, Lodato S, Levin JZ, Regev A, Zhang F, Arlotta P, 2020. In vivo Perturb-Seq reveals neuronal and glial abnormalities associated with autism risk genes. Science 370. 10.1126/science.aaz6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorstad NL, Wilken MS, Grimes WN, Wohl SG, VandenBosch LS, Yoshimatsu T, Wong RO, Rieke F, Reh TA, 2017. Stimulation of functional neuronal regeneration from Müller glia in adult mice. Nature 548, 103–107. 10.1038/nature23283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorstad NL, Wilken MS, Todd L, Finkbeiner C, Nakamura P, Radulovich N, Hooper MJ, Chitsazan A, Wilkerson BA, Rieke F, Reh TA, 2020. STAT signaling modifies Ascl1 chromatin binding and limits neural regeneration from muller glia in adult mouse retina. Cell Rep. 30, 2195–2208. 10.1016/j.celrep.2020.01.075 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallman A, Capowski EE, Wang J, Kaushik AM, Jansen AD, Edwards KL, Chen L, Berlinicke CA, Joseph Phillips M, Pierce EA, Qian J, Wang T-H, Gamm DM, Zack DJ, 2020. Investigating cone photoreceptor development using patient-derived NRL null retinal organoids. Commun Biol 3, 82. 10.1038/s42003-020-0808-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karl MO, Hayes S, Nelson BR, Tan K, Buckingham B, Reh TA, 2008. Stimulation of neural regeneration in the mouse retina. Proc. Natl. Acad. Sci. U.S.A 105 10.1073/pnas.0807453105, 19508–19513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Omori Y, Onishi A, Sato S, Kondo M, Furukawa T, 2010. Blimp 1 suppresses Chx10 expression in differentiating retinal photoreceptor precursors to ensure proper photoreceptor development. J. Neurosci 30, 6515–6526. 10.1523/JNEUROSCI.0771-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya-Okur HS, Wu SJ, Codomo CA, Pledger ES, Bryson TD, Henikoff JG, Ahmad K, Henikoff S, 2019. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun 10, 1930. 10.1038/s41467-019-09982-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay JN, Finger-Baier KC, Roeser T, Staub W, Baier H, 2001. Retinal ganglion cell genesis requires lakritz, a Zebrafish atonal Homolog. Neuron 30, 725–736. 10.1016/s0896-6273(01)00312-9. [DOI] [PubMed] [Google Scholar]
- Kechad A, Jolicoeur C, Tufford A, Mattar P, Chow RWY, Harris WA, Cayouette M, 2012. Numb is required for the production of terminal asymmetric cell divisions in the developing mouse retina. J. Neurosci 32, 17197–17210. 10.1523/JNEUROSCI.4127-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeley PW, Reese BE, 2018. DNER and NFIA are expressed by developing and mature AII amacrine cells in the mouse retina. J. Comp. Neurol 526, 467–479. 10.1002/cne.24345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DS, Ross SE, Trimarchi JM, Aach J, Greenberg ME, Cepko CL, 2008. Identification of molecular markers of bipolar cells in the murine retina. J. Comp. Neurol 507, 1795–1810. 10.1002/cne.21639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Lowe A, Dharmat R, Lee S, Owen LA, Wang J, Shakoor A, Li Y, Morgan DJ, Hejazi AA, Cvekl A, DeAngelis MM, Zhou ZJ, Chen R, Liu W, 2019. Generation, transcriptome profiling, and functional validation of cone-rich human retinal organoids. Proc. Natl. Acad. Sci. U.S.A 116, 10824–10833. 10.1073/pnas.1901572116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, Kirschner MW, 2015. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161, 1187–1201. 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohwi M, Doe CQ, 2013. Temporal fate specification and neural progenitor competence during development. Nat. Rev. Neurosci 14, 823–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike C, Nishida A, Ueno S, Saito H, Sanuki R, Sato S, Furukawa A, Aizawa S, Matsuo I, Suzuki N, Kondo M, Furukawa T, 2007. Functional roles of Otx2 transcription factor in postnatal mouse retinal development. Mol. Cell Biol 27, 8318–8329. 10.1128/MCB.01209-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kölsch Y, Hahn J, Sappington A, Stemmer M, Fernandes AM, Helmbrecht TO, Lele S, Butrus S, Laurell E, Arnold-Ammer I, Shekhar K, Sanes JR, Baier H, 2021. Molecular classification of zebrafish retinal ganglion cells links genes to cell types to behavior. Neuron 109. 10.1016/j.neuron.2020.12.003, 645–662.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, Baglaenko Y, Brenner M, Loh P-R, Raychaudhuri S, 2019. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296. 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laboissonniere LA, Goetz JJ, Martin GM, Bi R, Lund TJS, Ellson L, Lynch MR, Mooney B, Wickham H, Liu P, Schwartz GW, Trimarchi JM, 2019. Molecular signatures of retinal ganglion cells revealed through single cell profiling. Sci. Rep 9, 15778. 10.1038/s41598-019-52215-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laboissonniere LA, Martin GM, Goetz JJ, Bi R, Pope B, Weinand K, Ellson L, Fru D, Lee M, Wester AK, Liu P, Trimarchi JM, 2017a. Single cell transcriptome profiling of developing chick retinal cells. J. Comp. Neurol 525, 2735–2781. 10.1002/cne.24241. [DOI] [PubMed] [Google Scholar]
- Laboissonniere LA, Sonoda T, Lee SK, Trimarchi JM, Schmidt TM, 2017b. Single-cell RNA-seq of defined subsets of retinal ganglion cells. JoVE. 10.3791/55229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacomme M, Tarchini B, Boudreau-Pinsonneault C, Monat C, Cayouette M, 2016. The LGN protein promotes planar proliferative divisions in the neocortex but apicobasal asymmetric terminal divisions in the retina. Development 143, 575–581. 10.1242/dev.129783. [DOI] [PubMed] [Google Scholar]
- La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, Lidschreiber K, Kastriti ME, Lönnerberg P, Furlan A, Fan J, Borm LE, Liu Z, van Bruggen D, Guo J, He X, Barker R, Sundström E, Castelo-Branco G, Cramer P, Adameyko I, Linnarsson S, Kharchenko PV, 2018. RNA velocity of single cells. Nature 560, 494–498. 10.1038/s41586-018-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson L, Frisén J, Lundeberg J, 2021. Spatially resolved transcriptomics adds a new dimension to genomics. Nat. Methods 18, 15–18. 10.1038/s41592-020-01038-7. [DOI] [PubMed] [Google Scholar]
- La Torre A, Georgi S, Reh TA, 2013. Conserved microRNA pathway regulates developmental timing of retinal neurogenesis. Proc. Natl. Acad. Sci. U.S.A 110, E2362–E2370. 10.1073/pnas.1301837110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le TT, Wroblewski E, Patel S, Riesenberg AN, Brown NL, 2006. Math5 is required for both early retinal neuron differentiation and cell cycle progression. Dev. Biol 295, 764–778. 10.1016/j.ydbio.2006.03.055. [DOI] [PubMed] [Google Scholar]
- Liang Q, Dharmat R, Owen L, Shakoor A, Li Y, Kim S, Vitale A, Kim I, Morgan D, Liang S, Wu N, Chen K, DeAngelis MM, Chen R, 2019. Single-nuclei RNA-seq on human retinal tissue provides improved transcriptome profiling. Nat. Commun 10, 5743. 10.1038/s41467-019-12917-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipovsek M, Bardy C, Cadwell CR, Hadley K, Kobak D, Tripathy SJ, 2021. Patch-seq: past, present, and future. J. Neurosci 41, 937–946. 10.1523/JNEUROSCI.1653-20.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Liu X, Li S, Huang X, Qian H, Jin K, Xiang M, 2020. Foxn4 is a temporal identity factor conferring mid/late-early retinal competence and involved in retinal synaptogenesis. Proc. Natl. Acad. Sci. U.S.A 117, 5016–5027. 10.1073/pnas.1918628117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Chen Z, Desplan C, 2013a. Temporal patterning of neural progenitors in Drosophila. Curr. Top. Dev. Biol 105, 69–96. 10.1016/B978-0-12-396968-2.00003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Erclik T, Bertet C, Chen Z, Voutev R, Venkatesh S, Morante J, Celik A, Desplan C, 2013b. Temporal patterning of Drosophila medulla neuroblasts controls neural fates. Nature 498, 456–462. 10.1038/nature12319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Tyler WA, Zeldich E, Santpere Baró G, Okamoto M, Gao T, Li M, Sestan N, Haydar TF, 2020. Transcriptional priming as a conserved mechanism of lineage diversification in the developing mouse and human neocortex. Sci Adv 6. 10.1126/sciadv.abd2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Giudice Q, Leleu M, La Manno G, Fabre PJ, 2019. Single-cell transcriptional logic of cell-fate specification and axon guidance in early-born retinal neurons. Development 146. 10.1242/dev.178103. [DOI] [PubMed] [Google Scholar]
- Ludwig LS, Lareau CA, Ulirsch JC, Christian E, Muus C, Li LH, Pelka K, Ge W, Oren Y, Brack A, Law T, Rodman C, Chen JH, Boland GM, Hacohen N, Rozenblatt-Rosen O, Aryee MJ, Buenrostro JD, Regev A, Sankaran VG, 2019. Lineage tracing in humans enabled by mitochondrial mutations and single-cell genomics. Cell 176, 1325–1339. 10.1016/j.cell.2019.01.022 e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luecken MD, Theis FJ, 2019. Current best practices in single-cell RNA-seq analysis: a tutorial. Mol. Syst. Biol 15, e8746. 10.15252/msb.20188746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowski SW, Lo CY, Sharov AA, Nguyen Q, Fang L, Hung SS, Zhu L, Zhang T, Grünert U, Nguyen T, Senabouth A, Jabbari JS, Welby E, Sowden JC, Waugh HS, Mackey A, Pollock G, Lamb TD, Wang P-Y, Hewitt AW, Gillies MC, Powell JE, Wong RC, 2019. A single-cell transcriptome atlas of the adult human retina. EMBO J. 38, e100811. 10.15252/embj.2018100811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Shiau F, Yi W, Lu S, Wu Q, Pearson JD, Kallman A, Zhong S, Hoang T, Zuo Z, Zhao F, Zhang M, Tsai N, Zhuo Y, He S, Zhang J, Stein-O’Brien GL, Sherman TD, Duan X, Fertig EJ, Goff LA, Zack DJ, Handa JT, Xue T, Bremner R, Blackshaw S, Wang X, Clark BS, 2020. Single-cell analysis of human retina identifies evolutionarily conserved and species-specific mechanisms controlling development. Dev. Cell 53, 473–491. 10.1016/j.devcel.2020.04.009 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, Trombetta JJ, Weitz DA, Sanes JR, Shalek AK, Regev A, McCarroll SA, 2015. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214. 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malicki J, 2004. Cell fate decisions and patterning in the vertebrate retina: the importance of timing, asymmetry, polarity and waves. Curr. Opin. Neurobiol 14, 15–21. 10.1016/j.conb.2004.01.015. [DOI] [PubMed] [Google Scholar]
- Marcucci F, Murcia-Belmonte V, Wang Q, Coca Y, Ferreiro-Galve S, Kuwajima T, Khalid S, Ross ME, Mason C, Herrera E, 2016. The ciliary margin zone of the mammalian retina generates retinal ganglion cells. Cell Rep. 17, 3153–3164. 10.1016/j.celrep.2016.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masland RH, 2012. The neuronal organization of the retina. Neuron 76, 266–280. 10.1016/j.neuron.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattar P, Ericson J, Blackshaw S, Cayouette M, 2015. A conserved regulatory logic controls temporal identity in mouse neural progenitors. Neuron 85, 497–504. 10.1016/j.neuron.2014.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattar P, Jolicoeur C, Dang T, Shah S, Clark BS, Cayouette M, 2021. A Casz1-NuRD complex regulates temporal identity transitions in neural progenitors. Sci. Rep 11, 3858. 10.1038/s41598-021-83395-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Wang S-Z, 2006. The final fates of neurogenin2-expressing cells include all major neuron types in the mouse retina. Mol. Cell. Neurosci 31, 463–469. 10.1016/j.mcn.2005.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis CS, Patterson DM, Winkler J, Conrad DN, Hein MY, Srivastava V, Hu JL, Murrow LM, Weissman JS, Werb Z, Chow ED, Gartner ZJ, 2019. MULTI-seq: sample multiplexing for single-cell RNA sequencing using lipid-tagged indices. Nat. Methods 16, 619–626. 10.1038/s41592-019-0433-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mears AJ, Kondo M, Swain PK, Takada Y, Bush RA, Saunders TL, Sieving PA, Swaroop A, 2001. Nrl is required for rod photoreceptor development. Nat. Genet 29, 447–452. 10.1038/ng774. [DOI] [PubMed] [Google Scholar]
- Menon M, Mohammadi S, Davila-Velderrain J, Goods BA, Cadwell TD, Xing Y, Stemmer-Rachamimov A, Shalek AK, Love JC, Kellis M, Hafler BP, 2019. Single-cell transcriptomic atlas of the human retina identifies cell types associated with age-related macular degeneration. Nat. Commun 10, 4902. 10.1038/s41467-019-12780-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miesfeld JB, Glaser T, Brown NL, 2018. The dynamics of native Atoh7 protein expression during mouse retinal histogenesis, revealed with a new antibody. Gene Expr. Patterns 27, 114–121. 10.1016/j.gep.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizeracka K, Trimarchi JM, Stadler MB, Cepko CL, 2013. Analysis of gene expression in wild-type and Notch1 mutant retinal cells by single cell profiling. Dev. Dynam 242, 1147–1159. 10.1002/dvdy.24006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt JR, Bambah-Mukku D, Eichhorn SW, Vaughn E, Shekhar K, Perez JD, Rubinstein ND, Hao J, Regev A, Dulac C, Zhuang X, 2018. Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science 362. 10.1126/science.aau5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore KB, Logan MA, Aldiri I, Roberts JM, Steele M, Vetter ML, 2018. C8orf46 homolog encodes a novel protein Vexin that is required for neurogenesis in Xenopus laevis. Dev. Biol 437, 27–40. 10.1016/j.ydbio.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moudgil A, Wilkinson MN, Chen X, He J, Cammack AJ, Vasek MJ, Lagunas T Jr., Qi Z, Lalli MA, Guo C, Morris SA, Dougherty JD, Mitra RD, 2020. Self-reporting transposons enable simultaneous readout of gene expression and transcription factor binding in single cells. Cell 182, 992–1008. 10.1016/j.cell.2020.06.037 e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullally M, Albrecht C, Horton M, Laboissonniere LA, Goetz JJ, Chowdhury R, Manning A, Wester AK, Bose Q, Trimarchi JM, 2016. Expression profiling of developing zebrafish retinal cells. Zebrafish 13, 272–280. 10.1089/zeb.2015.1184. [DOI] [PubMed] [Google Scholar]
- Nishida A, Furukawa A, Koike C, Tano Y, Aizawa S, Matsuo I, Furukawa T, 2003. Otx2 homeobox gene controls retinal photoreceptor cell fate and pineal gland development. Nat. Neurosci 6, 1255–1263. 10.1038/nn1155. [DOI] [PubMed] [Google Scholar]
- Norrie JL, Lupo MS, Xu B, Al Diri I, Valentine M, Putnam D, Griffiths L, Zhang J, Johnson D, Easton J, Shao Y, Honnell V, Frase S, Miller S, Stewart V, Zhou X, Chen X, Dyer MA, 2019. Nucleome dynamics during retinal development. Neuron 104, 512–528. 10.1016/j.neuron.2019.08.002 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco LD, Chen H-H, Cox C, Katschke KJ Jr., Arceo R, Espiritu C, Caplazi P, Nghiem SS, Chen Y-J, Modrusan Z, Dressen A, Goldstein LD, Clarke C, Bhangale T, Yaspan B, Jeanne M, Townsend MJ, van Lookeren Campagne M, Hackney JA, 2020. Integration of eQTL and a single-cell atlas in the human eye identifies causal genes for age-related macular degeneration. Cell Rep. 30, 1246–1259. 10.1016/j.celrep.2019.12.082 e6. [DOI] [PubMed] [Google Scholar]
- Özel MN, Simon F, Jafari S, Holguera I, Chen Y-C, Benhra N, El-Danaf RN, Kapuralin K, Malin JA, Konstantinides N, Desplan C, 2021. Neuronal diversity and convergence in a visual system developmental atlas. Nature 589, 88–95. 10.1038/s41586-020-2879-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei W, Feyerabend TB, Rössler J, Wang X, Postrach D, Busch K, Rode I, Klapproth K, Dietlein N, Quedenau C, Chen W, Sauer S, Wolf S, Höfer T, Rodewald H-R, 2017. Polylox barcoding reveals haematopoietic stem cell fates realized in vivo. Nature 548, 456–460. 10.1038/nature23653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y-R, Shekhar K, Yan W, Herrmann D, Sappington A, Bryman GS, van Zyl T, Do MTH, Regev A, Sanes JR, 2019. Molecular classification and comparative taxonomics of foveal and peripheral cells in primate retina. Cell 176, 1222–1237. 10.1016/j.cell.2019.01.004 e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson VM, Zhang KX, Kumar N, Wong J, Li L, Wilson DC, Moore R, McClanahan TK, Sadekova S, Klappenbach JA, 2017. Multiplexed quantification of proteins and transcripts in single cells. Nat. Biotechnol 35, 936–939. 10.1038/nbt.3973. [DOI] [PubMed] [Google Scholar]
- Picelli S, Faridani OR, Björklund AK, Winberg G, Sagasser S, Sandberg R, 2014. Full-length RNA-seq from single cells using Smart-seq 2. Nat. Protoc 9, 171–181. 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- Poggi L, Vitorino M, Masai I, Harris WA, 2005. Influences on neural lineage and mode of division in the zebrafish retina in vivo. J. Cell Biol 171, 991–999. 10.1083/jcb.200509098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollak J, Wilken MS, Ueki Y, Cox KE, Sullivan JM, Taylor RJ, Levine EM, Reh TA, 2013. ASCL1 reprograms mouse Muller glia into neurogenic retinal progenitors. Development 140, 2619–2631. 10.1242/dev.091355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasov L, Masud T, Khaliq S, Mehdi SQ, Abid A, Oliver ER, Silva ED, Lewanda A, Brodsky MC, Borchert M, Kelberman D, Sowden JC, Dattani MT, Glaser T, 2012. ATOH7 mutations cause autosomal recessive persistent hyperplasia of the primary vitreous. Hum. Mol. Genet 21, 3681–3694. 10.1093/hmg/dds197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, Trapnell C, 2017. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979–982. 10.1038/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj B, Farrell JA, Liu J, El Kholtei J, Carte AN, Navajas Acedo J, Du LY, McKenna A, Relić Đ, Leslie JM, Schier AF, 2020. Emergence of neuronal diversity during vertebrate brain development. Neuron 108, 1058–1074. 10.1016/j.neuron.2020.09.023 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj B, Wagner DE, McKenna A, Pandey S, Klein AM, Shendure J, Gagnon JA, Schier AF, 2018. Simultaneous single-cell profiling of lineages and cell types in the vertebrate brain. Nat. Biotechnol 36, 442–450. 10.1038/nbt.4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R, Fausett BV, Goldman D, 2010. Ascl1a regulates Müller glia dedifferentiation and retinal regeneration through a Lin-28-dependent, let-7 microRNA signalling pathway. Nat. Cell Biol 12, 1101–1107. 10.1038/ncb2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani V, Deng X, Qiu R, Gunderson KL, Steemers FJ, Disteche CM, Noble WS, Duan Z, Shendure J, 2017. Massively multiplex single-cell Hi-C. Nat. Methods 14, 263–266. 10.1038/nmeth.4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport DH, Patheal SL, Harris WA, 2001. Cellular competence plays a role in photoreceptor differentiation in the developing Xenopus retina. J. Neurobiol 49, 129–141. 10.1002/neu.1070. [DOI] [PubMed] [Google Scholar]
- Ray TA, Cochran K, Kozlowski C, Wang J, Alexander G, Cady MA, Spencer WJ, Ruzycki PA, Clark BS, Laeremans A, He M-X, Wang X, Park E, Hao Y, Iannaccone A, Hu G, Fedrigo O, Skiba NP, Arshavsky VY, Kay JN, 2020. Comprehensive identification of mRNA isoforms reveals the diversity of neural cell-surface molecules with roles in retinal development and disease. Nat. Commun 11, 3328. 10.1038/s41467-020-17009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rheaume BA, Jereen A, Bolisetty M, Sajid MS, Yang Y, Renna K, Sun L, Robson P, Trakhtenberg EF, 2018. Single cell transcriptome profiling of retinal ganglion cells identifies cellular subtypes. Nat. Commun 9, 2759. 10.1038/s41467-018-05134-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesch K, Jadhav AP, Trimarchi JM, Stadler MB, Roska B, Sun BB, Cepko CL, 2008. The transcriptome of retinal Müller glial cells. J. Comp. Neurol 509, 225–238. 10.1002/cne.21730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesch K, Stadler MB, Cepko CL, 2012. Gene expression changes within Müller glial cells in retinitis pigmentosa. Mol. Vis 18, 1197–1214. [PMC free article] [PubMed] [Google Scholar]
- Rowan S, Cepko CL, 2004. Genetic analysis of the homeodomain transcription factor Chx10 in the retina using a novel multifunctional BAC transgenic mouse reporter. Dev. Biol 271, 388–402. 10.1016/j.ydbio.2004.03.039. [DOI] [PubMed] [Google Scholar]
- Saelens W, Cannoodt R, Todorov H, Saeys Y, 2019. A comparison of single-cell trajectory inference methods. Nat. Biotechnol 37, 547–554. 10.1038/s41587-019-0071-9. [DOI] [PubMed] [Google Scholar]
- Sagner A, Zhang I, Watson T, Lazaro J, Melchionda M, Briscoe J, 2020. Temporal Patterning of the Central Nervous System by a Shared Transcription Factor Code. bioRxiv. 10.1101/2020.11.10.376491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanes JR, Masland RH, 2015. The types of retinal ganglion cells: current status and implications for neuronal classification. Annu. Rev. Neurosci 38, 221–246. 10.1146/annurev-neuro-071714-034120. [DOI] [PubMed] [Google Scholar]
- Shekhar K, Lapan SW, Whitney IE, Tran NM, Macosko EZ, Kowalczyk M, Adiconis X, Levin JZ, Nemesh J, Goldman M, McCarroll SA, Cepko CL, Regev A, Sanes JR, 2016. Comprehensive classification of retinal bipolar neurons by single-cell transcriptomics. Cell 166, 1308–1323. 10.1016/j.cell.2016.07.054 e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto LM, Bernal-Tamayo JP, Lehmann R, Balsamy S, Martinez-de-Morentin X, Vilas-Zornoza A, San-Martin P, Prosper F, Gomez-Cabrero D, Kiani NA, Tegner J, 2020. scMomentum: Inference of Cell-type-specific Regulatory Networks and Energy Landscapes. Cold Spring Harbor Laboratory. 10.1101/2020.12.30.424887. [DOI] [Google Scholar]
- Sotolongo-Lopez M, Alvarez-Delfin K, Saade CJ, Vera DL, Fadool JM, 2016. Genetic dissection of dual roles for the transcription factor six7 in photoreceptor development and patterning in zebrafish. PLoS Genet. 12, e1005968. 10.1371/journal.pgen.1005968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar A, Hoshino A, Finkbeiner CR, Chitsazan A, Dai L, Haugan AK, Eschenbacher KM, Jackson DL, Trapnell C, Bermingham-McDonogh O, Glass I, Reh TA, 2020. Single-cell transcriptomic comparison of human fetal retina, hPSC-derived retinal organoids, and long-term retinal cultures. Cell Rep. 30, 1644–1659. 10.1016/j.celrep.2020.01.007 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein-O’Brien GL, Clark BS, Sherman T, Zibetti C, Hu Q, Sealfon R, Liu S, Qian J, Colantuoni C, Blackshaw S, Goff LA, Fertig EJ, 2019. Decomposing cell identity for transfer learning across cellular measurements, platforms, tissues, and species. Cell Syst 8, 395–411. 10.1016/j.cels.2019.04.004 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stickels RR, Murray E, Kumar P, Li J, Marshall JL, Di Bella DJ, Arlotta P, Macosko EZ, Chen F, 2020. Highly sensitive spatial transcriptomics at near-cellular resolution with Slide-seqV2. Nat. Biotechnol 10.1038/s41587-020-0739-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Chattopadhyay PK, Swerdlow H, Satija R, Smibert P, 2017. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14, 865–868. 10.1038/nmeth.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, Satija R, 2019. Comprehensive integration of single-cell data. Cell 177. 10.1016/j.cell.2019.05.031, 1888–1902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaroop A, Kim D, Forrest D, 2010. Transcriptional regulation of photoreceptor development and homeostasis in the mammalian retina. Nat. Rev. Neurosci 11, 563–576. 10.1038/nrn2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabula Muris Consortium, 2020. A single-cell transcriptomic atlas characterizes ageing tissues in the mouse. Nature 583, 590–595. 10.1038/s41586-020-2496-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teotia P, Van Hook MJ, Wichman CS, Rand Allingham R, Hauser MA, Ahmad I, 2017. Modeling glaucoma: retinal ganglion cells generated from induced pluripotent stem cells of patients with SIX6 risk allele show developmental abnormalities. Stem Cell. 10.1002/stem.2675. [DOI] [PubMed] [Google Scholar]
- Tomita K, Moriyoshi K, Nakanishi S, Guillemot F, Kageyama R, 2000. Mammalian achaete-scute and atonal homologs regulate neuronal versus glial fate determination in the central nervous system. EMBO J. 19, 5460–5472. 10.1093/emboj/19.20.5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran NM, Shekhar K, Whitney IE, Jacobi A, Benhar I, Hong G, Yan W, Adiconis X, Arnold ME, Lee JM, Levin JZ, Lin D, Wang C, Lieber CM, Regev A, He Z, Sanes JR, 2019. Single-cell profiles of retinal ganglion cells differing in resilience to injury reveal neuroprotective genes. Neuron 104, 1039–1055. 10.1016/j.neuron.2019.11.006 e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL, 2014. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol 32, 381–386. 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimarchi JM, Stadler MB, Cepko CL, 2008. Individual retinal progenitor cells display extensive heterogeneity of gene expression. PloS One 3, e1588. 10.1371/journal.pone.0001588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimarchi JM, Stadler MB, Roska B, Billings N, Sun B, Bartch B, Cepko CL, 2007. Molecular heterogeneity of developing retinal ganglion and amacrine cells revealed through single cell gene expression profiling. J. Comp. Neurol 502, 1047–1065. 10.1002/cne.21368. [DOI] [PubMed] [Google Scholar]
- Tritschler S, Büttner M, Fischer DS, Lange M, Bergen V, Lickert H, Theis FJ, 2019. Concepts and limitations for learning developmental trajectories from single cell genomics. Development 146. 10.1242/dev.170506. [DOI] [PubMed] [Google Scholar]
- Turner DL, Cepko CL, 1987. A common progenitor for neurons and glia persists in rat retina late in development. Nature 328, 131–136. 10.1038/328131a0. [DOI] [PubMed] [Google Scholar]
- Turner DL, Snyder EY, Cepko CL, 1990. Lineage-independent determination of cell type in the embryonic mouse retina. Neuron 4, 833–845. 10.1016/0896-6273(90)90136-4. [DOI] [PubMed] [Google Scholar]
- Vairapandi M, Balliet AG, Hoffman B, Liebermann DA, 2002. GADD45b and GADD45g are cdc 2/cyclinB1 kinase inhibitors with a role in S and G2/M cell cycle checkpoints induced by genotoxic stress. J. Cell. Physiol 192, 327–338. 10.1002/jcp.10140. [DOI] [PubMed] [Google Scholar]
- Voigt AP, Whitmore SS, Flamme-Wiese MJ, Riker MJ, Wiley LA, Tucker BA, Stone EM, Mullins RF, Scheetz TE, 2019. Molecular characterization of foveal versus peripheral human retina by single-cell RNA sequencing. Exp. Eye Res 184, 234–242. 10.1016/j.exer.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinescu PE, Kay JN, Sanes JR, 2009. Birthdays of retinal amacrine cell subtypes are systematically related to their molecular identity and soma position. J. Comp. Neurol 517, 737–750. 10.1002/cne.22200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington CH, 1957. The strategy of the genes, a discussion of some aspects of theoretical biology. In: Waddington CH, et al. (Eds.), With an Appendix [Some Physico-Chemical Aspects of Biological Organisation]. H. Kacser. [Google Scholar]
- Wagner DE, Klein AM, 2020. Lineage tracing meets single-cell omics: opportunities and challenges. Nat. Rev. Genet 21, 410–427. 10.1038/s41576-020-0223-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner DE, Weinreb C, Collins ZM, Briggs JA, Megason SG, Klein AM, 2018. Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science 360, 981–987. 10.1126/science.aar4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Du L, Lee AC, Li Y, Qin H, He J, 2020. Different lineage contexts direct common pro-neural factors to specify distinct retinal cell subtypes. J. Cell Biol 219 10.1083/jcb.202003026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Allen WE, Wright MA, Sylwestrak EL, Samusik N, Vesuna S, Evans K, Liu C, Ramakrishnan C, Liu J, Nolan GP, Bava F-A, Deisseroth K, 2018. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science 361. 10.1126/science.aat5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y, Almeida AD, Rulands S, Chalour N, Muresan L, Wu Y, Simons BD, He J, Harris WA, 2016. The ciliary marginal zone of the zebrafish retina: clonal and time- lapse analysis of a continuously growing tissue. Development 143, 1099–1107. 10.1242/dev.133314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir K, Kim DW, Blackshaw S, 2021. A potential role for somatostatin signaling in regulating retinal neurogenesis. Sci. Rep 11, 10962. 10.1038/s41598-021-90554-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JD, Kozareva V, Ferreira A, Vanderburg C, Martin C, Macosko EZ, 2019. Single-cell multi-omic integration compares and contrasts features of brain cell identity. Cell. 10.1016/j.cell.2019.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West ER, Lapan SW, Lee C, Kajderowicz KM, Li X, Cepko CL, 2021. Spatiotemporal Patterns of Neuronal Subtype Genesis Suggest Hierarchical Development of Retinal Diversity. 10.1101/2021.04.29.442012bioRxiv. [DOI] [PubMed] [Google Scholar]
- Wetts R, Fraser SE, 1988. Multipotent precursors can give rise to all major cell types of the frog retina. Science 239, 1142–1145. 10.1126/science.2449732. [DOI] [PubMed] [Google Scholar]
- Wu F, Bard JE, Kann J, Yergeau D, Sapkota D, Ge Y, Hu Z, Wang J, Liu T, Mu X, 2021. Single cell transcriptomics reveals lineage trajectory of retinal ganglion cells in wild-type and Atoh7-null retinas. Nat. Commun 12, 1465. 10.1038/s41467-021-21704-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Kaczynski TJ, Sethuramanujam S, Li R, Jain V, Slaughter M, Mu X, 2015. Two transcription factors, Pou4f2 and Isl1, are sufficient to specify the retinal ganglion cell fate. Proc. Natl. Acad. Sci. U.S.A 112, E1559–E1568. 10.1073/pnas.1421535112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia C, Fan J, Emanuel G, Hao J, Zhuang X, 2019. Spatial transcriptome profiling by MERFISH reveals subcellular RNA compartmentalization and cell cycle-dependent gene expression. Proc. Natl. Acad. Sci. U.S.A 116 10.1073/pnas.1912459116, 19490–19499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Tang X, Jin M, Zhang H, Du L, Yu S, He J, 2020. Unifying developmental programs for embryonic and postembryonic neurogenesis in the zebrafish retina. Development 147. 10.1242/dev.185660. [DOI] [PubMed] [Google Scholar]
- Yamagata M, Yan W, Sanes JR, 2021. A cell atlas of the chick retina based on single-cell transcriptomics. Elife 10. 10.7554/eLife.63907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Kon T, Omori Y, Furukawa T, 2020. Functional and evolutionary diversification of Otx2 and Crx in vertebrate retinal photoreceptor and bipolar cell development. Cell Rep. 30, 658–671. 10.1016/j.celrep.2019.12.072 e5. [DOI] [PubMed] [Google Scholar]
- Yang Z, Ding K, Pan L, Deng M, Gan L, 2003. Math5 determines the competence state of retinal ganglion cell progenitors. Dev. Biol 264, 240–254. 10.1016/j.ydbio.2003.08.005. [DOI] [PubMed] [Google Scholar]
- Yan W, Laboulaye MA, Tran NM, Whitney IE, Benhar I, Sanes JR, 2020a. Mouse retinal cell atlas: molecular identification of over sixty amacrine cell types. J. Neurosci 40, 5177–5195. 10.1523/JNEUROSCI.0471-20.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W, Peng Y-R, van Zyl T, Regev A, Shekhar K, Juric D, Sanes JR, 2020b. Cell atlas of the human fovea and peripheral retina. Sci. Rep 10, 9802. 10.1038/s41598-020-66092-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RW, 1985. Cell differentiation in the retina of the mouse. Anat. Rec 212, 199–205. 10.1002/ar.1092120215. [DOI] [PubMed] [Google Scholar]
- Yuniati L, Scheijen B, van der Meer LT, van Leeuwen FN, 2019. Tumor suppressors BTG1 and BTG2: beyond growth control. J. Cell. Physiol 234, 5379–5389. 10.1002/jcp.27407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahr SK, Yang G, Kazan H, Borrett MJ, Yuzwa SA, Voronova A, Kaplan DR, Miller FD, 2018. A translational repression complex in developing mammalian neural stem cells that regulates neuronal specification. Neuron 97, 520–537. 10.1016/j.neuron.2017.12.045 e6. [DOI] [PubMed] [Google Scholar]
- Zeng H, Sanes JR, 2017. Neuronal cell-type classification: challenges, opportunities and the path forward. Nat. Rev. Neurosci 18, 530–546. 10.1038/nrn.2017.85. [DOI] [PubMed] [Google Scholar]


