Abstract
The gut microbiota may play a role in breast cancer etiology by regulating hormonal, metabolic, and immunologic pathways. We investigated associations of fecal bacteria with breast cancer and non-malignant breast disease in a case-control study conducted in Ghana, a country with rising breast cancer incidence and mortality. To do this, we sequenced the V4 region of the 16S rRNA gene to characterize bacteria in fecal samples collected at the time of breast biopsy (N=379 breast cancer cases, N=102 non-malignant breast disease cases, N=414 population-based controls). We estimated associations of alpha diversity (observed amplicon sequence variants [ASVs], Shannon index, and Faith’s phylogenetic diversity), beta diversity (Bray Curtis and unweighted/weighted UniFrac distance), and presence and relative abundance of select taxa with breast cancer and non-malignant breast disease using multivariable unconditional polytomous logistic regression. All alpha diversity metrics were strongly, inversely associated with odds of breast cancer and for those in the highest vs. lowest tertile of observed ASVs, the odds ratio (95% confidence interval) was 0.21 (0.13–0.36; Ptrend<0.001). Alpha diversity associations were similar for non-malignant breast disease and breast cancer grade/molecular subtype. All beta diversity distance matrices and multiple taxa with possible estrogen-conjugating and immune-related functions were strongly associated with breast cancer (all P’s<0.001). There were no statistically significant differences between breast cancer and non-malignant breast disease cases in any microbiota metric. In conclusion, fecal bacteria characteristics were strongly and similarly associated with breast cancer and non-malignant breast disease. Our findings provide novel insight into potential microbially-mediated mechanisms of breast disease.
Keywords: microbiome, breast cancer, non-malignant breast diseases
Introduction
Breast cancer incidence and mortality are rising in sub-Saharan Africa (1). Women of African-ancestry tend to be diagnosed with breast cancer at younger ages, at a later stage, and with more aggressive subtypes than women of non-African ancestry (1,2). Increasing breast cancer incidence in sub-Saharan Africa is likely due to the adoption of Westernized lifestyles, changes in reproductive factors, and population aging (due to increased life expectancy) (2–4). However, breast cancer risk factors remain understudied among African populations.
It is likely that some established breast cancer risk factors (e.g., obesity) (5,6), in addition to other unknown risk factors, may influence the composition and function of the gut microbiota, including trillions of bacteria. In turn, the gut microbiota influences multiple pathways that are mechanistically linked to the initiation and growth of breast neoplasms (7–10). For example, accumulating evidence supports the role of the gut microbiota in regulating endogenous estrogens (7,11) and systemic inflammation (12,13).
The role of the gut microbiota in breast cancer risk remains unclear. Two previous studies investigated the associations of the fecal microbiota with breast cancer, but they had small sample sizes and conflicting findings, perhaps due to differences in study populations and sequencing technologies (14,15). No studies have been conducted in an African population or of the gut microbiota in association with non-malignant breast disease. Herein, we report an investigation of the associations of fecal bacteria with breast cancer and non-malignant breast disease in a population-based case-control study conducted in Ghana.
Methods
Study design and population
In the Ghana Breast Health study, described previously (16–18), 2,218 breast cancer cases or non-malignant breast disease cases and 2,352 controls were recruited at three hospitals in Ghana, including Korle Bu Teaching Hospital in Accra and Komfo Anokye Teaching Hospital and Peace and Love Hospital in Kumasi.
Eligible cases included women aged 18–74 years residing in the defined catchment areas surrounding the two cities for ≥one year who were diagnosed, referred for biopsy due to suspicion of breast cancer, or treated at breast clinics in the three hospitals. As the majority of women were recruited at biopsy, both breast cancer and non-malignant breast disease cases were included. Diagnoses were based on pathologic review of core biopsies by pathologists in Ghana and the NCI. Population controls were identified using a household census of randomly selected enumeration areas that gave rise to the cases.
To select participants for the microbiota study, we selected all breast cancer cases (N=415) with ≥ 1 stool sample available. We then selected controls frequency-matched by city (N=447), prioritizing stool samples collected in the clinic over home-collected samples. Next, we randomly selected enough city frequency-matched non-malignant breast disease cases to reach 972 total samples total (N=110). Of the 972 study samples, we excluded 10 samples that failed sequencing, 62 samples with < 6,250 reads after rarefaction (see below; 28 malignant cases, 7 non-malignant cases, and 27 controls), and five cases that were diagnosed with a cancer other than breast. The final sample size for this study was N=895.
Tumor characteristics
Prior to undergoing treatment, 4–8 core-needle biopsies (14-gauge) were fixed in 10% neutral buffered formalin for 24–72 hours. Then, they were processed into formalin-fixed paraffin embedded blocks for diagnosis using standardized protocols (16). Blocks not required for diagnosis were shipped to the NCI for additional pathological review (80% of the 1,126 breast cancer cases in the original study). Since organized mammography screening is not standard practice in Ghana, 96% of tumors presented clinically as > 2 cm (18).
Information was obtained on key immunohistochemical estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2) markers from pathology departments in Ghana for 69% of the cases. If ≥10% of tumor cells stained positive, tumors were considered ER and PR status positive. For HER2, tumors were considered positive if they had a homogeneous, dark pattern of staining in ≥10% of the tumor cells. Indeterminate and negative cases were combined and considered negative for HER2. Agreement of the immunohistochemical assays was compared between pathology departments in Ghana and an NCI laboratory for 87 cases using two tumor tissue samples from the same patient. Agreement for the assays was: 79% for ER (P < 0. 0001), 78% for HER2 (P < 0.0001), and 65% for PR (P = 0.002).
Data collection
The original interview response rate was 99.2% and 91.9% for non-malignant and malignant cases and controls. Participants completed standardized questionnaires assessing breast cancer risk factors, such as socioeconomic status, age at menarche and menopause, number of births, age at first birth, breastfeeding history, family history of breast cancer, body size, alcohol consumption, physical activity levels, occupational exposures, and screening history. Weight using the Seca 869 Physician Scale and height measurements were taken by study coordinators and recorded on standardized forms.
Stool collection
Stool was collected among a subset of the Ghana Breast Health Study cases (prior to undergoing treatment) and controls. When possible, case and control stool samples were collected at the initial clinic study visit. Participants were provided a collection cup and two empty pre-labeled stool collection Falcon tubes, and were instructed to add a single scoop of the stool into each vial. At the study visit, if controls were unable to provide stool they were provided the stool collection materials to take home and be collected and transported immediately to the laboratory at a later date by study personnel. Upon receipt in the laboratory, one Sarstedt vial was snap frozen at −80° C and one vial had RNAlater added and was then frozen at −80° C. Both home- and clinic-collected stool samples were processed using the same protocol. The samples were shipped using liquid nitrogen every 3–4 months for storage at Fisher Biorepository (Frederick, Maryland).
Of 3,066 subjects approached, 58.1% of both case groups and 46.7% of controls provided stool samples. On average, compared to those who did not provide stool, those who provided stool were formally educated (p = 0.25) and had a similar distribution of family history of breast cancer (p = 0.68). On average, they were similar ages (p = 0.94), breastfed for similar lengths of time (p = 0.12), and had similar BMI (p = 0.25). Stool providers were more likely to have had ≥ 1 live birth (p < 0.001). Breast cancer cases had similar distributions in cancer grades (p = 0.29). Of non-stool provider cases, 23.8% had triple-negative breast cancer compared with 31% among stool provider cases (p = 0.02). In KBTH, 22% provided stool; in KATH, 41% provided stool; in PLH, 27% provided stool (p < 0.001).
DNA extraction and sequencing
Stool samples were sent to the Knight Laboratory (University of California, San Diego) on dry ice. Samples were thawed at 4°C and kept on ice during plating. Stool specimens were sampled using a swab (Puritan Cotton Tipped Applicators – Puritan Medical Products), which was then used for the DNA extraction. Within each DNA extraction batch, two artificial community and two blank quality control (QC) samples were included (19). DNA extraction and 16S rRNA amplicon sequencing were done using Earth Microbiome Project (EMP) standard protocols (http://www.earthmicrobiome.org/protocols-and-standards/16s). DNA was extracted using the MO-BIO PowerMag Soil DNA Isolation Kit with beadbeating. Amplicon PCR was performed on the V4 region of the 16S rRNA gene using primer pair 515f/806r with Golay error-correcting barcodes on the reverse primer (20,21). Negative controls included no-template controls for DNA extraction and PCR amplification. Amplicons were barcoded and pooled in equal concentrations for sequencing. The amplicon pool was purified with the Qiagen UltraClean PCR cleanup kit and sequenced on the Illumina MiSeq sequencing platform with paired end 150 cycle chemistry. Sequence data were demultiplexed and minimally quality filtered using the QIIME 1.9.1 script split_libraries_fastq.py, with a Phred quality threshold of 3 and default parameters to generate FASTA sequence files.
Bioinformatics
Using the DADA2 pipeline 1.2.1 (22), sequence variant tables and phylogenetic trees were generated based on pair-end sequence reads. For quality filtering, the first ten bases were trimmed from forward and reverse reads. Forward/reverse reads were truncated at 140 bases. Then, the reads were merged using the default ‘mergePairs’ DADA2 function. After merging and error correction, amplicon sequence variants (ASVs) (i.e., 100% OTUs) were identified. After removal of chimeras, using the ‘removeBimeraDenovo’ function, 85% of the sequence reads were retained. Taxonomy was assigned to the resulting ASVs using the SILVA v123 database. Six sequences aligning to human mitochondria were filtered.
Observed ASVs, Shannon Index, Faith’s Phylogenetic Diversity (PD) was computed using QIIME 1.9.1 (q2‐diversity). Beta diversity measures were calculated based on Bray-Curtis, weighted UniFrac, and unweighted UniFrac distance matrices. For relative abundance analyses, we restricted our analyses to taxa present in 50% of the population at a mean relative abundance of >0.01% (56 taxa); for presence/absence analyses, we restricted to those taxa present in 5 to 95% of the population (159 taxa). Based on rarefaction curves for alpha diversity (Supplemental Figure 1), we rarefied the alpha and beta diversity metrics to 6,250 reads. All participants excluded from alpha/beta diversity analyses were similarly excluded from the relative abundance and presence/absence analyses. From the 895 samples included in our analysis, 4,206 sequence variants were identified comprising 34 phyla, 58 classes, 103 orders, 156 families, and 414 genera. A median of 19,782 reads were generated per sample.
For quality control analysis, the taxonomic composition of the artificial community was compared to the known composition and was similar based on visual inspection. For artificial community samples placed in separate batches, average inter-batch alpha diversity coefficients of variation were 13.1%, 7.5%, and 10.9% for observed ASVs, Shannon index, and Faith’s PD, respectively. In the QC blanks, the median number of reads was 231 and out of 22 blanks only two remained after rarefaction.
Statistical analysis
We summarized and compared the characteristics of the study participants by case/control status using chi-square tests for categorical variables and ANOVA for continuous variables. We used multivariable polytomous logistic regression to estimate associations of the microbiota parameters (alpha diversity, beta diversity, and taxa relative abundance or presence/absence) with breast cancer and non-malignant breast disease. We also estimated associations of alpha and beta diversity with breast cancer grade (grade 1 and 2 or grade 3) and subtype, focusing our analyses on the most common subtypes (estrogen receptor [ER] −/+, triple-negative (ER−, PR−, and HER2−), and luminal-like A (ER+ or PR+ and HER2−) (4)) and tested for heterogeneity by grade/subtype using a case-only multivariable logistic regression analysis with grade/subtype as the dependent variable. For all alpha diversity analyses, we categorized participants into tertiles of the alpha diversity metrics based on the distribution among the controls. To test for trend, we assigned each participant the median value of their tertile and modeled the value continuously in the regression model.
For beta diversity analyses, we visually evaluated principle coordinate analysis (PCoA) plots and estimated associations of each distance metric’s first six standardized (using the controls’ standard deviation) principal coordinate axes with breast cancer and non-malignant breast disease. The first six vectors of the principal coordinates explained 48%, 36%, and 86% of the variability in Bray Curtis, unweighted UniFrac, and weighted UniFrac distance, respectively. We also conducted the microbiome regression-based kernel association test (23) (MiRKAT), using exact methods, to calculate p-values for overall differences in microbiome composition by non-malignant breast disease or breast cancer status based on kernel similarity matrices for Bray Curtis and unweighted and weighted UniFrac distance, individually and overall. For associations with P-values < 0.05, we repeated the corresponding MiRKAT models with 10,000 permutations to check that the P-value remained statistically significant under the empirical null distribution of the test statistic. For the alpha diversity, beta diversity, and taxonomic analyses, we estimated pairwise associations between alpha diversity metrics, principal coordinates axes, and taxa presence/relative abundance using general linear regression. We assessed the sensitivity of the above associations to excluding participants with stool collected at home and excluding those who took antibiotics within the last 30 days.
We considered covariates in the above-described regression models based on biological plausibility and previous literature. We also considered associations of the variable with alpha and beta diversity among controls. As described below, observed ASVs were strongly, inversely associated with odds of breast disease, and therefore may serve as a confounder or mediator of the associations of beta diversity and taxa presence with disease. Based on our conclusions from causal diagrams and observed changes in the magnitude of the associations when including observed ASVs in the model, we evaluated the impact of adjustment for observed ASVs in the beta diversity and taxa presence models. Other possible covariates included age, study center (KATH, KBTH, or PLH), stool collection method (home or hospital), body mass index (BMI) (kg/m2), education (junior secondary school or lower, senior secondary school/some college or technical school or more, other or unknown), family history of cancer (yes, no, unknown), antibiotic use (missing, ≤ 30 days ago, and >30 days ago, this year, or never), number of full-term pregnancies (0, 1–2, 3–4, 5+ pregnancies), history of breastfeeding, age at menarche, age at menopause, and current hormonal contraceptive use. Final covariates are listed in table footnotes. We assessed potential effect measure modification by age (≥50 or <50), menopausal status, and body mass index (BMI) category by comparing stratum specific estimates and calculating P-interactions using the likelihood ratio test.
All statistical analyses were conducted using R, version 3.5.2. Two-sided P-values <0.05 or 95% confidence intervals (CI) that excluded 1.0 were considered statistically significant. To account for multiple comparisons in each analyses, we used Bonferroni correction of P-values by the number of tests conducted (indicated in table footnotes).
Results
Selected characteristics of the study participants are presented in Table 1. Compared to breast cancer cases and controls, non-malignant cases were younger and more likely to be premenopausal, nulliparous, and formally educated. Antibiotic use in the previous 30 days was more common among breast cancer cases than in non-malignant breast disease cases and controls. The three groups did not differ in average BMI or use of hormonal contraception. All three fecal microbiota alpha diversity estimates were lower in both case groups, compared to controls. Among the breast cancer cases, 31.4% had triple-negative breast cancer and 71% had grade 3 histology.
Table 1.
Characteristics of study participants in the Ghana Breast Health case-control study of breast cancer and non-malignant breast disease (N=895)
| Characteristics | Breast cancer cases (N = 379) | Non-malignant cases (N = 102) | Controls (N = 414) | ||||
|---|---|---|---|---|---|---|---|
| Mean (SD) | % | Mean (SD) | % | Mean (SD) | % | P-valuea | |
| Participation information | |||||||
| Stool Collected at the Clinic | 100 | 100 | 44.9 | <0.001 | |||
| Study center | <0.001 | ||||||
| KATH | 30.6 | 23.5 | 59.2 | ||||
| KBTH | 16.6 | 23.5 | 15.2 | ||||
| PLH | 52.8 | 52.9 | 25.6 | ||||
| Demographics | |||||||
| Age, y | 50.8 (12.3) | 38.8 (12.8) | 46.9 (12.9) | <0.001 | |||
| Married/living with partner | 54.1 | 52.9 | 60.4 | 0.01 | |||
| Senior secondary or higher education | 31.1 | 53.9 | 24.2 | <0.001 | |||
| Premenopausal | 43.8 | 74.5 | 57.2 | <0.001 | |||
| Medical history | |||||||
| Family history of breast cancer | 7.1 | 6.9 | 2.9 | 0.06 | |||
| Had no full-term pregnancies | 8.7 | 29.4 | 10.6 | <0.001 | |||
| Took antibiotics within the last 30 days | 27.2 | 20.6 | 19.3 | 0.03 | |||
| Age at menarche, y | 15.6 (2.6) | 14.9 (1.7) | 15.2 (2.0) | 0.02 | |||
| Ever breastfed >1 month (parous only) | 86.5 | 69.6 | 88.4 | <0.001 | |||
| Currently using hormonal contraceptive | 30.3 | 32.4 | 30.9 | 0.93 | |||
| Lifestyle characteristics | |||||||
| Never tobacco user | 94.7 | 99.0 | 99.0 | 0.004 | |||
| BMI, kg/m2 | 27.1 (5.9) | 27.7 (6.5) | 28.0 (7.9) | 0.45 | |||
| Microbiota metrics | |||||||
| Shannon index | 4.2 (1.2) | 4.2 (1.1) | 4.6 (0.9) | <0.001 | |||
| Observed ASVs | 92.6 (52.8) | 90.9 (46.9) | 120 (44.5) | <0.001 | |||
| Faith’s PD | 20.2 (9.2) | 20.1 (8.6) | 25.6 (7.8) | <0.001 | |||
P-values calculated using χ2 test for categorical variables and ANOVA for continuous variables
Abbreviations: ASVs, amplicon sequence variants; BMI, body mass index; KATH, Komfo Anokye Teaching Hospital; KBTH, Korle Bu Teaching Hospital; PLH, Peace and Love Hospital; PD, phylogenetic diversity; SD, standard deviation
All alpha diversity metrics were strongly, inversely associated with odds of breast cancer and non-malignant breast disease (Table 2). Compared to controls, the odds of breast cancer were incrementally lower with each higher tertile of alpha diversity (all Ptrends < 0.001). For example, women in the highest compared to the lowest tertile of observed ASVs had a statistically significant 79% lower odds (95% CI: 64%−87%) of breast cancer. The estimated alpha diversity associations were similar for non-malignant cases compared to controls. Alpha diversity estimates did not statistically significantly differ between breast cancer and non-malignant cases. In our analysis of associations of taxa presence or abundance with alpha diversity, multiple taxa were strongly, statistically significantly associated with the alpha diversity metrics (Supplemental Table 1), most notably presence of taxa in Family Ruminococcaceae and Lachnospiraceae.
Table 2.
Multivariable associations of alpha diversity estimates with breast cancer and non-malignant breast disease in the Ghana Breast Health study (N = 102 non-malignant breast disease cases, 379 malignant cases, and 414 controls)
| Alpha Diversity Metric | No. breast cancer cases/non-malignant breast disease cases/controls | Breast cancer cases vs. controls | Non-malignant cases vs. controls | Breast cancer cases vs. non-malignant cases |
|---|---|---|---|---|
| OR (95% CI)a | OR (95% CI)a | OR (95% CI)a | ||
| Shannon | ||||
| Continuous | 0.62 (0.51, 0.75) | 0.65 (0.50, 0.84) | 0.98 (0.77, 1.23) | |
| Tertile 1 (1.91 – 4.11) | 185/53/137 | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) |
| Tertile 2 (4.12 – 5.02) | 107/21/139 | 0.57 (0.36, 0.92) | 0.43 (0.22, 0.85) | 1.28 (0.69, 2.42) |
| Tertile 3 (5.03 – 7.40) | 87/28/138 | 0.34 (0.21, 0.55) | 0.43 (0.23, 0.80) | 0.80 (0.43, 1.48) |
| Ptrend | <0.001 | 0.004 | 0.66 | |
| Observed ASVs | ||||
| Continuous | 0.99 (0.98, 0.99) | 0.99 (0.98, 0.99) | 1.00 (1.00, 1.01) | |
| Tertile 1 (26 – 94) | 214/53/136 | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) |
| Tertile 2 (95 – 139) | 99/34/140 | 0.41 (0.26, 0.67) | 0.63 (0.34, 1.17) | 0.72 (0.40, 1.28) |
| Tertile 3 (140 – 338) | 66/15/138 | 0.21 (0.13, 0.36) | 0.22 (0.10, 0.46) | 1.27 (0.61, 2.76) |
| Ptrend | <0.001 | <0.001 | 0.79 | |
| Faith’s PD | ||||
| Continuous | 0.91 (0.89, 0.94) | 0.91 (0.88, 0.94) | 1.01 (0.98, 1.04) | |
| Tertile 1 (8.13 – 21.56) | 219/56/137 | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) |
| Tertile 2 (21.66 – 28.90) | 93/30/139 | 0.35 (0.22, 0.55) | 0.42 (0.23, 0.79) | 0.86 (0.48, 1.55) |
| Tertile 3 (28.97 – 86.36) | 67/16/138 | 0.20 (0.12, 0.33) | 0.19 (0.09, 0.40) | 1.17 (0.57, 2.50) |
| Ptrend | <0.001 | <0.001 | 0.86 |
Covariates for unconditional polytomous logistic regression models included: age (continuous), study center (Komfo Anoyke Teaching Hospital, Korle Bu Teaching Hospital, or Peace and Love Hospital), collection method (home or clinic), body mass index (kg/m2), education (junior secondary school or lower, senior secondary school/some college or technical school or more, other or unknown), family history of cancer (yes, no, unknown), antibiotic use (>30 days ago, this year, never, or missing/<30 days ago), number of full-term pregnancies (0, 1–2, 3–4, 5+ pregnancies), and current hormonal contraceptive use
Abbreviations: ASVs, amplicon sequence variants; CI, confidence interval; OR, odds ratio; PD, Phylogenetic Diversity
Beta diversity was strongly and similarly associated with breast cancer and non-malignant breast disease. Based on multivariable-adjusted MiRKAT tests, compared to controls, Bray Curtis and weighted and unweighted UniFrac distance matrices were strongly associated with breast cancer and non-malignant breast disease (all P-values ≤ 0.001), but comparing breast cancer to non-malignant cases the corresponding P-values were > 0.05 (Supplemental Table 2). As shown in Figure 1, based on principal coordinates plots of the three distance matrices, both case groups visually clustered together and they clustered separately from controls. Multiple principal coordinate axes of each distance metric were statistically significantly associated with breast cancer and non-malignant breast disease, with slightly fewer remaining statistically significant after adjusting for observed ASVs (Supplemental Table 3). For example, after observed ASVs adjustment, there was an 86% (95% CI: 52%−227%) higher odds of breast cancer per one standard deviation increase in the first Bray Curtis principle coordinate axis. Breast cancer cases did not differ from non-malignant cases based on any principal coordinate axis. Presence of multiple taxa, including Faecalibacterium, Ruminococcaceae UCG-002/UCG-005, and Christensenellaceae R-7 group, were strongly associated with the principal coordinate axes that were significantly associated with breast cancer (Supplemental Table 4).
Figure 1.
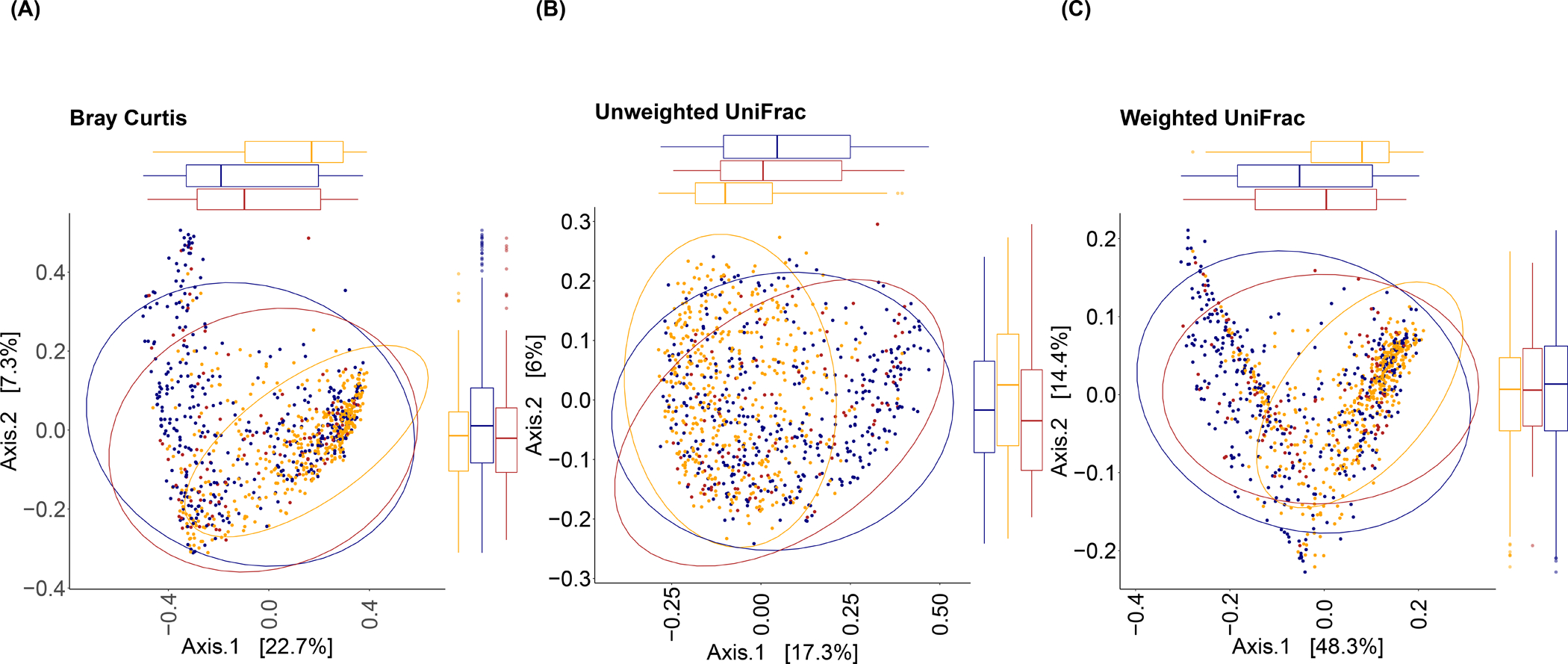
Principal coordinates plots based on (A) Bray Curtis, (B) Unweighted UniFrac, and (C) Weighted UniFrac distance in fecal samples collected in the Ghana Breast Health study (N=102 non-malignant breast disease cases, 379 malignant cases, and 414 controls). Ellipses were calculated using the ‘stat_ellipse’ function to compute normal confidence ellipses in R package ggplot2 (44).
Multivariable associations of taxa relative abundance with breast cancer and non-malignant breast disease are shown in Figure 2A–C (all mean relative abundances, ORs, and 95% CIs are presented in Supplemental Table 5). Bacteroides was most strongly, positively associated with breast cancer and for every one-percentage increase in its relative abundance there was a statistically significant 5% higher odds (95% CI: 3%−6%) of breast cancer. Romboutsia and Coprococcus 2 were most strongly inversely associated with breast cancer, and for every one-percentage increase in their relative abundance there was a statistically significant 91% (95% CI: 76%−97%) and 55% (95% CI: 40%−67%) lower odds of breast cancer, respectively. Relative abundance associations were in similar directions but slightly weaker for non-malignant breast disease and no taxon was statistically significantly different comparing the two case groups.
Figure 2.
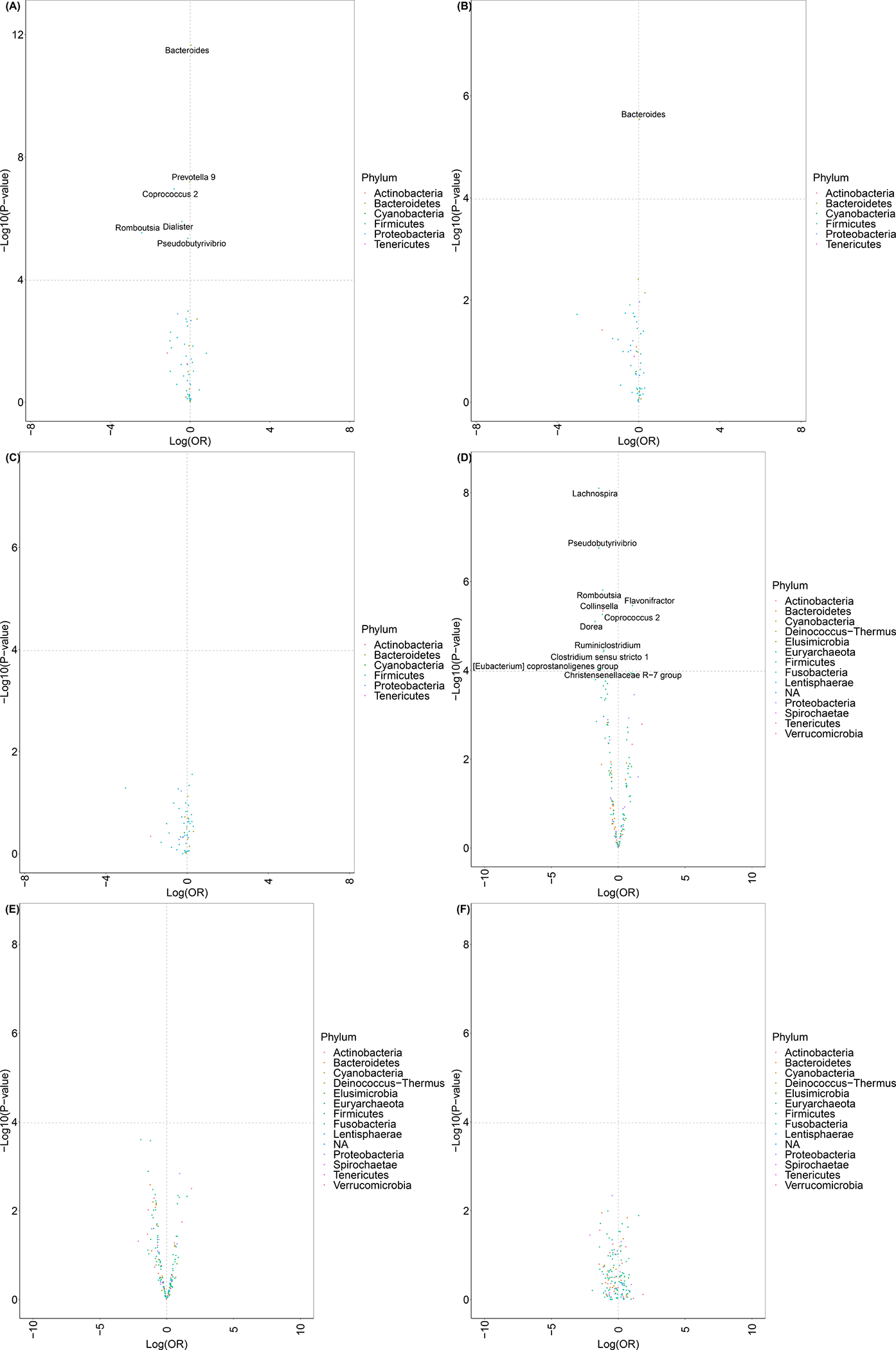
Multivariable associations of genus-level relative abundance comparing: (A) breast cancer cases vs. controls, (B) non-malignant breast disease cases vs. controls, and (C) breast cancer cases vs. non-malignant breast disease cases for the most abundant genera in the Ghana Breast Health Study; multivariable associations of genus-level presence/absence comparing (D) breast cancer cases vs. controls, (E) non-malignant breast disease cases vs. controls, and (F) breast cancer cases vs. non-malignant breast disease cases among the most genera prevalent in 5% to 95% of the Ghana Breast Health Study (N=102 non-malignant breast disease cases, 379 malignant cases, and 414 controls)
Multivariable associations of presence of taxa with breast cancer and non-malignant breast disease are presented in Figure 2D–F (% presence, ORs, and 95% CIs are presented in Supplemental Table 6). Prior to observed ASV adjustment, 53 taxa were statistically significantly associated with breast cancer; whereas, after observed ASV adjustment, 12 taxa were statistically significantly associated with breast cancer (Supplemental Table 6). Significant taxa included multiple genera in family Ruminococcaceae, Lachnospiraceae, and Prevotellaceae. Faecalibacterium, which was present in almost 100% of controls and only 90% of both case groups, was most strongly, inversely associated with odds of breast cancer and non-malignant breast disease. Christensenellaceae R-7 group, Dorea, [Eubacterium] coprostanoligenes group, Pseudobutyrivibrio, and Lachnospira were also inversely associated with odds of breast cancer; whereas, Flavonifractor and Family Ruminococcaceae were positively associated with breast cancer. Similar to our relative abundance findings, associations were generally slightly weaker for non-malignant breast disease and no taxon statistically significantly differed between the two case groups.
Alpha diversity associations by breast cancer grade and molecular subtype are shown in Table 3 and by participant characteristics in Table 4. Compared to controls, the inverse alpha diversity associations were similar for all grades/subtypes of breast cancer but slightly stronger for Grade 3 breast cancers (Pheterogeneity = 0.05 for Shannon and Faith’s PD). Beta diversity, as assessed using a MiRKAT test, statistically significantly differed between each grade/subtype of breast cancer compared to controls (all P’s <0.001) but did not significantly differ when comparing the grades/subtypes to non-malignant cases (data not shown). Alpha diversity associations were slightly stronger among those who were ≥ 50 years old or post-menopausal.
Table 3.
Multivariable associationsa of alpha diversity estimates with breast cancer grade and molecular subtype and in the Ghana Breast Health study (N=379 malignant cases and 414 controls)
| Grade | ER Status | Molecular Subtypeb | ||||
|---|---|---|---|---|---|---|
| 1 or 2 | 3 | ER− | ER+ | Triple-negative | Luminal A | |
| N cases | 81 | 198 | 135 | 129 | 82 | 105 |
| Shannon Index | ||||||
| Tertile 1 (1.91 – 4.11) | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) |
| Tertile 2 (4.12 – 5.02) | 0.73 (0.37, 1.47) | 0.56 (0.32, 0.98) | 0.61 (0.33, 1.11) | 0.85 (0.47, 1.55) | 0.68 (0.32, 1.44) | 0.82 (0.43, 1.59) |
| Tertile 3 (5.03 – 7.40) | 0.58 (0.29, 1.14) | 0.30 (0.17, 0.53) | 0.35 (0.19, 0.66) | 0.55 (0.30, 1.01) | 0.55 (0.26, 1.16) | 0.52 (0.26, 1.02) |
| Ptrend | 0.11 | <0.001 | 0.001 | 0.06 | 0.11 | 0.06 |
| Pheterogeneity | 0.05 | 0.11 | 0.95 | |||
| Observed sequence variants | ||||||
| Tertile 1 (26 – 94) | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) |
| Tertile 2 (95 – 139) | 0.55 (0.28, 1.07) | 0.38 (0.22, 0.66) | 0.35 (0.19, 0.65) | 0.59 (0.33, 1.06) | 0.44 (0.21, 0.91) | 0.47 (0.24, 0.89) |
| Tertile 3 (140 – 338) | 0.32 (0.15, 0.67) | 0.17 (0.09, 0.31) | 0.21 (0.11, 0.41) | 0.27 (0.14, 0.53) | 0.29 (0.13, 0.65) | 0.23 (0.11, 0.48) |
| Ptrend | 0.003 | <0.001 | <0.001 | <0.001 | 0.002 | <0.001 |
| Pheterogeneity | 0.06 | 0.19 | 0.97 | |||
| Faith’s PD | ||||||
| Tertile 1 (8.13 – 21.56) | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) |
| Tertile 2 (21.66 – 28.90) | 0.50 (0.25, 0.97) | 0.32 (0.18, 0.55) | 0.30 (0.16, 0.56) | 0.54 (0.30, 0.97) | 0.38 (0.18, 0.80) | 0.46 (0.24, 0.87) |
| Tertile 3 (28.97 – 86.36) | 0.27 (0.13, 0.56) | 0.17 (0.09, 0.31) | 0.16 (0.08, 0.33) | 0.25 (0.13, 0.49) | 0.23 (0.10, 0.51) | 0.22 (0.10, 0.45) |
| Ptrend | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 |
| Pheterogeneity | 0.05 | 0.24 | 0.76 | |||
Abbreviations: ER, estrogen receptor
Fully adjusted unconditional polytomous logistic regression models include age (continuous), study center (Komfo Anoyke Teaching Hospital, Korle Bu Teaching Hospital, or Peace and Love Hospital), and collection method (home or hospital), body mass index (kg/m2), education (junior secondary school or lower, senior secondary school/Some college or technical school or more, other or unknown), family history of cancer (yes, no, unknown), antibiotic use (missing, ≤ 30 days ago, and >30 days ago, this year, or never), number of full-term pregnancies (0, 1–2, 3–4, 5+ pregnancies), and current hormonal contraceptive use
Triple-negative breast cancers were ER−, PR−, and HER2−; luminal-like A breast cancers were ER+ or PR+ and HER2−
Table 4.
Multivariable associationsa of alpha diversity estimates with breast cancer by participant characteristics in the Ghana Breast Health study (N=379 malignant cases and 414 controls)
| Age | Menopausal status | BMIb | |||||
|---|---|---|---|---|---|---|---|
| < 50 | ≥ 50 | Pre-menopausal | Post-menopausal | Normal | Overweight | Obese | |
| N cases | 184 | 195 | 213 | 166 | 118 | 119 | 109 |
| Shannon Index | |||||||
| Tertile 1 (1.91 – 4.11) | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) |
| Tertile 2 (4.12 – 5.02) | 0.57 (0.30, 1.10) | 0.67 (0.31, 1.43) | 0.71 (0.37, 1.36) | 0.50 (0.23, 1.07) | 0.53 (0.22, 1.22) | 0.43 (0.15, 1.14) | 0.88 (0.34, 2.30) |
| Tertile 3 (5.03 – 7.40) | 0.46 (0.23, 0.91) | 0.25 (0.11, 0.53) | 0.51 (0.26, 0.98) | 0.26 (0.11, 0.58) | 0.39 (0.17, 0.88) | 0.26 (0.10, 0.69) | 0.28 (0.10, 0.75) |
| Ptrend | 0.02 | 0.001 | 0.04 | 0.00 | 0.02 | 0.01 | 0.02 |
| P-interaction | 0.26 | 0.28 | 0.94 | ||||
| Observed sequence variants | |||||||
| Tertile 1 (26 – 94) | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) |
| Tertile 2 (95 – 139) | 0.53 (0.27, 1.04) | 0.25 (0.11, 0.54) | 0.52 (0.26, 1.02) | 0.22 (0.09, 0.48) | 0.34 (0.14, 0.81) | 0.70 (0.26, 1.88) | 0.45 (0.17, 1.16) |
| Tertile 3 (140 – 338) | 0.31 (0.15, 0.63) | 0.12 (0.05, 0.28) | 0.34 (0.17, 0.68) | 0.12 (0.05, 0.28) | 0.17 (0.07, 0.42) | 0.19 (0.06, 0.52) | 0.20 (0.07, 0.58) |
| Ptrend | 0.001 | <0.001 | 0.002 | <0.001 | <0.001 | 0.002 | 0.004 |
| P-interaction | 0.05 | 0.06 | 0.56 | ||||
| Faith’s PD | |||||||
| Tertile 1 (8.13 – 21.56) | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) | 1.00 (referent) |
| Tertile 2 (21.66 – 28.90) | 0.43 (0.22, 0.85) | 0.23 (0.10, 0.49) | 0.47 (0.24, 0.91) | 0.22 (0.10, 0.48) | 0.35 (0.14, 0.84) | 0.40 (0.16, 1.00) | 0.59 (0.22, 1.55) |
| Tertile 3 (28.97 – 86.36) | 0.23 (0.11, 0.47) | 0.15 (0.06, 0.34) | 0.25 (0.12, 0.49) | 0.15 (0.06, 0.35) | 0.19 (0.07, 0.44) | 0.28 (0.09, 0.82) | 0.06 (0.02, 0.22) |
| Ptrend | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | 0.01 | <0.001 |
| P-interaction | 0.17 | 0.27 | 0.27 | ||||
Abbreviations: BMI, body mass index
Fully adjusted unconditional logistic regression models include age (continuous), study center (Komfo Anoyke Teaching Hospital, Korle Bu Teaching Hospital, or Peace and Love Hospital), and collection method (home or hospital), body mass index (kg/m2), education (junior secondary school or lower, senior secondary school/Some college or technical school or more, other or unknown), family history of cancer (yes, no, unknown), antibiotic use (missing, ≤ 30 days ago, and >30 days ago, this year, or never), number of full-term pregnancies (0, 1–2, 3–4, 5+ pregnancies), and current hormonal contraceptive use
Normal BMI: 18.5 −24.99 kg/m2; Overweight BMI: 25–29.99 kg/m2; Obese: 30+ kg/m2
When excluding control stool samples collected at home or excluding those who used antibiotics within the previous 30 days, alpha diversity (Supplemental Table 7), beta diversity, relative abundance, and presence/absence associations were minimally affected (data not shown).
Discussion
In this population-based case control study of breast cancer and non-malignant breast disease in Ghana, we found that 1) breast cancer and non-malignant breast disease cases had similar fecal microbial profiles, but different profiles from controls; 2) alpha diversity was strongly, inversely associated with breast cancer and non-malignant breast disease; 3) multiple taxa with multifunctional roles in estrogen metabolism and immune homeostasis, as described below, were associated with breast cancer and non-malignant breast disease; and, 4) alpha diversity was similarly and strongly inversely associated with breast cancer by stage and molecular subtype, including ER −/+ and triple-negative breast cancers. These findings potentially support that the gut microbiota may have multifunctional roles in development of breast disease.
We found that alpha diversity was strongly, inversely associated with breast cancer and non-malignant breast disease. Similar to our findings, in a case-control investigation of the association of 16S rRNA gene sequenced fecal bacteria with breast cancer (N=48 postmenopausal breast cancer cases, N=48 postmenopausal controls), alpha diversity was inversely associated with breast cancer (15). In contrast, in another study using shotgun metagenomics to characterize the fecal microbiota of 62 breast cancer cases and 71 controls, compared to their control counterparts, the mean observed species was higher among post-menopausal breast cancer cases and the mean Shannon index was higher among pre-menopausal cases; however, only unadjusted analyses were presented (14).
Collectively, the gut microbiota has a well-documented role in regulating systemic estrogens (7,11,24–26), which are in turn mechanistically linked to the development of hormone-receptor positive breast cancers (27–29). Among postmenopausal women, alpha diversity was previously suggested to be negatively associated with estrogen concentrations in feces and, in urine, positively associated with both estrogen concentrations (11) and the ratio of estrogen metabolites to parent estrogens (25). Lower alpha diversity was also previously positively associated with multiple metabolic markers–including markers of adiposity, inflammation, and dyslipidemia (30))–which may be associated with breast cancer risk (8–10).
We found strong taxa–breast disease associations for multiple genera. Some of our taxonomic findings were similarly noted in previous 16S rRNA studies in the US (15); however, of note, it is possible taxa-disease associations may differ by geographic location (31). Intriguingly, some gut taxa associated with breast disease in our study were previously present at higher abundances in breast tumor tissue compared to normal tissue (e.g., taxa in Family Prevotellaceae and Family Ruminococcaceae) (32,33).
It is plausible that bacteria may be involved in breast disease through multiple pathways including those related and unrelated to hormonal regulation. Taxa with estrogen-deconjugating enzymatic activity, characterized by presence of genes for β-glucuronidase or β-galactosidase (7) are hypothesized to have enzymatic activities that promote reabsorption of unconjugated estrogens into circulation, contributing to estrogenic burden (7). We found that genus previously noted to contain β-galactosidase were both positively associated (Ruminococcaceae [Family] and Bacteroides) and inversely associated (Eubacterium coprostanoligenes group, Coprococcus, Dorea, Collinsella, Faecalibacterium, and Prevotella) (7) with breast disease. Additionally, genera previously noted to contain β-glucuronidase (Collinsella and Faecalibacterium) were inversely associated with breast disease. We also found that bacteria previously suggested to be associated with markers of systemic inflammation (34) (e.g., Faecalibacterium, Prevotella, and Family Ruminococcaceae) were associated with breast cancer and non-malignant breast disease. Furthermore, some species within our breast disease-associated taxa (e.g., Bacteroides, Dialister, Coprococcus, Faecalibacterium, Pseudobutyrivibrio, and Romboutsia) are thought to be involved in short chain fatty acid metabolism, which in turn affects gut barrier integrity and systemic inflammation (35–37). In the metagenomic case-control study described above, the pathway for the short chain fatty acid, butyrate, was suggested to be inversely associated with postmenopausal breast cancer (14). Higher resolution sequencing approaches are required to provide further evidence of the role taxa functionality in breast disease.
There are multiple strengths of our study. There are no published studies investigating microbiota-breast disease associations in an African population, which is generally underrepresented in the microbiota and disease literature. We had a well-defined population, large sample size, and population-based controls. Also, our findings were robust across multiple sensitivity analyses.
There were also some limitations. About half of the stool samples among controls were collected at home, whereas all case samples were collected in the clinic. Despite this potential bias, we found that excluding the home-collected samples had minimal impact on our observed associations. Our findings are cross-sectional, so we cannot determine the temporality of the associations, such that the presence of breast lesions or associated behavioral differences could be driving the observed associations. We did not collection information on certain potential confounders, such as diet and probiotic use, but future studies should evaluate whether these variables are important confounders of microbiome-breast disease associations. Finally, we used 16S rRNA gene sequencing which cannot ascertain species-, gene-, and function-level detail of the bacteria.
Since this study was not originally designed to assess non-malignant breast diseases, an additional limitation of this study was that we did not have detailed pathological information on non-malignant breast disease diagnoses. Thus, this case group comprises those with a variety of suspicious breast lesions that may not be representative of those in the general population. Bearing this in mind, we found robust similarities in fecal microbiota characteristics between breast cancer and non-malignant breast disease cases similar to other studies that found similar associations of breast cancer risk factors (e.g., parity and childhood body size) with both breast cancer and non-malignant breast diseases such as fibroadenoma and proliferative benign breast disease (38–40). Since women with certain types of non-malignant breast diseases may have higher breast cancer risk (38,41–43), future studies should address this issue in detail.
In conclusion, our findings suggest that alpha diversity, overall microbiota composition, and taxa with hypothesized estrogen-conjugation and immune-related functions may be associated with breast diseases. Our findings support further study of the gut microbiota’s role in breast disease etiology and should be followed with prospective studies and studies with gene- and function-level of the gut microbiota among diverse populations.
Supplementary Material
Supplemental Figure 1. Rarefaction curves for observed amplicon sequence variants in fecal samples collected in the Ghana Breast Health study by (A) disease status, (B) stool collection location, and (C) history of antibiotic use (N=102 non-malignant breast disease cases, 379 malignant cases, and 414 controls)
Supplemental Figure 2. Boxplots of alpha diversity by disease status for (A) observed amplicon sequence variants, (B) the Shannon index, and (C) Faith’s phylogenetic diversity in the Ghana Breast Health study (N=102 non-malignant breast disease cases, 379 malignant cases, and 414 controls)
Novelty and impact:
Our study is the largest study to investigate associations of the fecal microbiota with breast cancer to date, and the first to investigate these associations in Sub-Saharan Africa, a population with rising breast cancer incidence and mortality. Our findings set up intriguing hypotheses whereby the gut microbiota may be associated with breast disease and motivate continued study of gut microbiota-breast disease/breast cancer associations in diverse study populations.
Acknowledgements:
We would like to thank Greg Humphreys for his help with sample handling and processing and sequencing of the 16S rRNA gene. The success of this investigation would not have been possible without exceptional teamwork and the diligence of the field staff who oversaw the recruitment, interviews and collection of data from study subjects. Special thanks are due to the following individuals: Korle Bu Teaching Hospital, Accra—Dr Adu-Aryee, Obed Ekpedzor,Angela Kenu, Victoria Okyne, Naomi Oyoe Ohene Oti, Evelyn Tay; Komfo Anoyke Teaching Hospital, Kumasi—Marion Alcpaloo, Bernard Arhin, Emmanuel Asiamah, Isaac Boakye, Samuel Ka-chungu and; Peace and Love Hospital, Kumasi—Samuel Amanama, Emma Abaidoo, Prince Agyapong, Thomas Agyei, Debora Boateng-Ansong, Margaret Frempong, Bridget Nortey Mensah, Richard Opoku, and Kofi Owusu Gyimah. The study was further enhanced by surgical expertise provided by Dr Lisa Newman of the University of Michigan and by pathological expertise provided by Drs. Stephen Hewitt and Petra Lenz of the National Cancer Institute and Dr. Maire A. Duggan from the Cumming School of Medicine, University of Calgary, Canada. Study management assistance was received from Ricardo Diaz, Shelley Niwa, Usha Singh, Ann Truelove, and Michelle Brotzman at Westat, Inc., at NCI. Appreciation is also expressed to the many women who agreed to participate in the study and to provide information and biospecimens in hopes of preventing and improving outcomes of breast cancer in Ghana.
Funding:
This work was supported by the Intramural Research Program in the Division of Cancer Epidemiology and Genetics, the US National Institutes of Health (NIH), National Cancer Institute (NCI).
Abbreviations:
- ASV
amplicon sequence variants
- BMI
body mass index
- CI
confidence interval
- ER
estrogen receptor
- HER2
human epidermal growth factor receptor
- KATH
Komfo Anokye Teaching Hospital
- KBTH
Korle Bu Teaching Hospital
- MiRKAT
microbiome regression-based kernel association test
- NCI
National Cancer Institute
- PLH
Peace and Love Hospital
- PD
phylogenetic diversity
- PR
progesterone receptor
- QC
quality control
- SD
standard deviation
Footnotes
Conflicts of Interest: The authors declare no potential conflicts of interest.
Ethics Statement: All participants provided written informed consent. This study was approved by the Special Studies Institutional Review Board of the National Cancer Institute (NCI; Rockville, MD, USA; FWA #: 00005897 and IORG #: 00010), the Ghana Heath Service Ethical Review Committee, and Institutional Review Boards at the University of Ghana Noguchi Memorial Institute for Medical Research (Accra, Ghana; FWA #: 00001824 and IORG #: 0000908), the Kwame Nkrumah University of Science and Technology (Kumasi, Ghana), the School of Medical Sciences at Komfo Anokye Teaching Hospital (Kumasi, Ghana), and Westat (Rockville, MD, USA).
Data availability statement:
The sequencing data that support the findings of this study are openly in the in the National Center for Biotechnology (NCBI) Sequence Read Archive (http://www.ncbi.nlm.nih.gov/bioproject/658160; bioproject ID PRJNA658160). The patient data generated in the current study are not publicly available due to data privacy of patients, but are available from the corresponding author on reasonable request.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. [DOI] [PubMed] [Google Scholar]
- 2.Akarolo-Anthony SN, Ogundiran TO, Adebamowo CA. Emerging breast cancer epidemic: Evidence from Africa. Breast Cancer Res. 2010;12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brinton LA, Figueroa JD, Awuah B, Yarney J, Wiafe S, Wood S, et al. Breast Cancer in Sub-Saharan Africa: Opportunities for Prevention. Breast Cancer Res Treat. 2014;144:467–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Figueroa JD, Davis Lynn BC, Edusei L, Titiloye N, Adjei E, Clegg‐Lamptey J, et al. Reproductive Factors and Risk of Breast Cancer by Tumor Subtypes among Ghanaian Women: A Population‐based Case–control Study. Int J Cancer. 2020;44:ijc.32929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tett A, Huang KD, Asnicar F, Fehlner-Peach H, Pasolli E, Karcher N, et al. The Prevotella copri Complex Comprises Four Distinct Clades Underrepresented in Westernized Populations. Cell Host Microbe. 2019;26:666–679. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sonnenburg JL, Bäckhed F. Diet-microbiota interactions as moderators of human metabolism. Nature. 2016;535:56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwa M, Plottel CS, Blaser MJ, Adams S. The Intestinal Microbiome and Estrogen Receptor–Positive Female Breast Cancer. JNCI J Natl Cancer Inst. 2016;108:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tobias DK, Akinkuolie AO, Chandler PD, Lawler PR, Manson JE, Buring JE, et al. Markers of Inflammation and Incident Breast Cancer Risk in the Women’s Health Study. Am J Epidemiol. 2018;187:705–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clarke CA, Canchola AJ, Moy LM, Neuhausen SL, Chung NT, Lacey JV., et al. Regular and low-dose aspirin, other non-steroidal anti-inflammatory medications and prospective risk of HER2-defined breast cancer: The California Teachers Study. Breast Cancer Res. 2017;19:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia-Estevez L, Moreno-Bueno G. Updating the role of obesity and cholesterol in breast cancer. Breast Cancer Res. 2019;21:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flores R, Shi J, Fuhrman B, Xu X, Veenstra TD, Gail MH, et al. Fecal microbial determinants of fecal and systemic estrogens and estrogen metabolites: A cross-sectional study. J Transl Med. 2012;10:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maynard CL, Elson CO, Hatton RD, Weaver CT. Reciprocal interactions of the intestinal microbiota and immune system. Nature. 2012;489:231–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu J, Liao M, Yao Z, Liang W, Li Q, Liu J, et al. Breast cancer in postmenopausal women is associated with an altered gut metagenome. Microbiome. 2018;6:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goedert JJ, Jones G, Hua X, Xu X, Yu G, Flores R, et al. Investigation of the Association Between the Fecal Microbiota and Breast Cancer in Postmenopausal Women: A Population-Based Case-Control Pilot Study. J Natl Cancer Inst. 2015;107:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brinton LA, Awuah B, Nat Clegg-Lamptey J, Wiafe-Addai B, Ansong D, Nyarko KM, et al. Design considerations for identifying breast cancer risk factors in a population-based study in Africa. Int J Cancer. 2017;140:2667–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brinton LA, Figueroa JD, Ansong D, Nyarko KM, Wiafe S, Yarney J, et al. Skin lighteners and hair relaxers as risk factors for breast cancer: Results from the Ghana breast health study. Carcinogenesis. 2018;39:571–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brinton L, Figueroa J, Adjei E, Ansong D, Biritwum R, Edusei L, et al. Factors contributing to delays in diagnosis of breast cancers in Ghana, West Africa. Breast Cancer Res Treat. 2017;162:105–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sinha R, Abu-Ali G, Vogtmann E, Fodor A. Assessment of variation in microbial community amplicon sequencing by the Microbiome Quality Control (MBQC) project consortium. Nat Biotechnol2. 2017;35:1077–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walters W, Hyde ER, Berg-Lyons D, Ackermann G, Humphrey G, Parada A, et al. Improved Bacterial 16S rRNA Gene (V4 and V4–5) and Fungal Internal Transcribed Spacer Marker Gene Primers for Microbial Community Surveys. mSystems. 2016; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao N, Chen J, Carroll IM, Ringel-Kulka T, Epstein MP, Zhou H, et al. Testing in microbiome-profiling studies with MiRKAT, the microbiome regression-based kernel association test. Am J Hum Genet. 2015;96:797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parida S, Sharma D. The Microbiome–Estrogen Connection and Breast Cancer Risk. Cells. 2019;8:1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fuhrman BJ, Feigelson HS, Flores R, Gail MH, Xu X, Ravel J, et al. Associations of the fecal microbiome with urinary estrogens and estrogen metabolites in postmenopausal women. J Clin Endocrinol Metab. 2014;99:4632–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parida P, Sharma S. Microbial Alterations and Risk Factors of Breast Cancer: Connections and Mechanistic Insights. Cells. 2020; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harbeck N, Penault-Llorca F, Cortes J, Gnant M, Houssami N, Poortmans P, et al. Breast cancer. Nat Rev Dis Prim. 2019. [DOI] [PubMed] [Google Scholar]
- 28.Colditz GA. Relationship between estrogen levels, use of hormone replacement therapy, and breast cancer. J Natl Cancer Inst. 1998;90:814–23. [DOI] [PubMed] [Google Scholar]
- 29.Rosenberg L, Bethea TN, Viscidi E, Hong CC, Troester MA, Bandera EV., et al. Postmenopausal female hormone use and estrogen receptor-positive and -negative breast cancer in african American women. J Natl Cancer Inst. 2016;108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–6. [DOI] [PubMed] [Google Scholar]
- 31.He Y, Wu W, Zheng HM, Li P, McDonald D, Sheng HF, et al. Regional variation limits applications of healthy gut microbiome reference ranges and disease models. Nat Med. 2018;24:1532–5. [DOI] [PubMed] [Google Scholar]
- 32.Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science (80-). 2020;368:973–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klann E, Williamson JM, Tagliamonte MS, Ukhanova M, Asirvatham JR, Chim H, et al. Microbiota composition in bilateral healthy breast tissue and breast tumors. Cancer Causes Control. 2020;31:1027–38. [DOI] [PubMed] [Google Scholar]
- 34.van den Munckhof ICL, Kurilshikov A, ter Horst R, Riksen NP, Joosten LAB, Zhernakova A, et al. Role of gut microbiota in chronic low-grade inflammation as potential driver for atherosclerotic cardiovascular disease: a systematic review of human studies. Obes Rev. 2018;19:1719–34. [DOI] [PubMed] [Google Scholar]
- 35.Zhang LS, Davies SS. Microbial metabolism of dietary components to bioactive metabolites: Opportunities for new therapeutic interventions. Genome Med. 2016;8:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, et al. Human genetics shape the gut microbiome. Cell. 2014;159:789–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li J-W, Fang B, Pang G-F, Zhang M, Ren F-Z. Age- and diet-specific effects of chronic exposure to chlorpyrifos on hormones, inflammation and gut microbiota in rats. Pestic Biochem Physiol. 2019;159:68–79. [DOI] [PubMed] [Google Scholar]
- 38.Li J, Humphreys K, Ho PJ, Eriksson M, Darai-Ramqvist E, Lindström LS, et al. Family History, Reproductive, and Lifestyle Risk Factors for Fibroadenoma and Breast Cancer. JNCI Cancer Spectr. 2018;2:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goldberg M, Cohn BA, Houghton LC, Flom JD, Wei Y, Cirillo P, et al. Early-Life Growth and Benign Breast Disease. Am J Epidemiol. 2019;188:1646–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baer HJ, Schnitt SJ, Connolly JL, Byrne C, Willett WC, Rosner B, et al. Early life factors and incidence of proliferative benign breast disease. Cancer Epidemiol Biomarkers Prev. 2005;14:2889–97. [DOI] [PubMed] [Google Scholar]
- 41.Visscher DW, Frank RD, Carter JM, Vierkant RA, Winham SJ, Heinzen EP, et al. Breast Cancer Risk and Progressive Histology in Serial Benign Biopsies. J Natl Cancer Inst. 2017;109:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schnitt SJ, Morrow M, Tung NM. Refining Risk Assessment in Women With Benign Breast Disease: An Ongoing Dilemma. J Natl Cancer Inst. 2017;109:1–2. [DOI] [PubMed] [Google Scholar]
- 43.Kabat GC, Jones JG, Olson N, Negassa A, Duggan C, Ginsberg M, et al. A multi-center prospective cohort study of benign breast disease and risk of subsequent breast cancer. Cancer Causes Control. 2010;21:821–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wickham H ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag. 2016;New York. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Rarefaction curves for observed amplicon sequence variants in fecal samples collected in the Ghana Breast Health study by (A) disease status, (B) stool collection location, and (C) history of antibiotic use (N=102 non-malignant breast disease cases, 379 malignant cases, and 414 controls)
Supplemental Figure 2. Boxplots of alpha diversity by disease status for (A) observed amplicon sequence variants, (B) the Shannon index, and (C) Faith’s phylogenetic diversity in the Ghana Breast Health study (N=102 non-malignant breast disease cases, 379 malignant cases, and 414 controls)
Data Availability Statement
The sequencing data that support the findings of this study are openly in the in the National Center for Biotechnology (NCBI) Sequence Read Archive (http://www.ncbi.nlm.nih.gov/bioproject/658160; bioproject ID PRJNA658160). The patient data generated in the current study are not publicly available due to data privacy of patients, but are available from the corresponding author on reasonable request.


