Abstract
Hepatic steatosis increases risk of fatty liver and cardiovascular disease. Perfluorooctanesulfonic acid (PFOS) is a persistent, bio-accumulative pollutant that has been used in industrial and commercial applications. PFOS administration induces hepatic steatosis in rodents and increases lipogenic gene expression signatures in cultured hepatocytes. We hypothesized that PFOS treatment interferes with lipid loss when switching from a high fat diet (HFD) to a standard diet (SD), and augments HFD-induced hepatic steatosis. Male C57BL/6N mice were fed standard chow diet or 60% kCal high-fat diet (HFD) for 4 weeks to increase body weight. Then, some HFD mice were switched to SD and mice were further divided to diet only or diet containing 0.0003% PFOS, for six treatment groups: SD, HFD to SD (H-SD), HFD, SD+PFOS, H-SD + PFOS, or HFD+PFOS. After 10 weeks on study, blood and livers were collected. HFD for 14 weeks increased body weight and hepatic steatosis, whereas H-SD mice returned to SD measures. PFOS administration reduced body weight in mice fed a SD, but not H-SD or HFD. PFOS administration increased liver weight in H-SD+PFOS and HFD+PFOS mice. PFOS increased hepatic steatosis in H-SD and HFD groups. Hepatic mRNA expression and SWATH-MS proteomic analysis revealed that PFOS induced lipid and xenobiotic transporters, as well as metabolism pathways. Overall, the findings herein suggest that PFOS treatment did interfere with lipid loss associated with switch to a SD and similarly augmented hepatic lipid accumulation in mice established on an HFD.
Keywords: Liver, PFOS, perfluorinated compounds, weight loss, NAFLD, PFAS
INTRODUCTION
Non-alcoholic fatty liver disease (NAFLD) affects over 30% of the United States population and its prevalence is expected to increase over the next 15 years (Le et al. 2017; Estes et al. 2018). NAFLD risk factors include obesity, metabolic syndrome, and energy imbalance (Matteoni et al. 1999; Li et al. 2002; Marchesini et al. 2003). Hepatic steatosis, also known as fatty liver, is considered to be the “first hit” in the multiple “hits” that contribute to NAFLD. Hepatic steatosis is defined as accumulation of lipids in hepatocytes at greater than 5% of the total liver weight (Masarone et al. 2014). In the presence of inflammation, fatty liver can progress to nonalcoholic steatohepatitis (NASH). In NASH, lipid peroxidation causes oxidative injury and inflammation that leads to irreversible fibrosis and cirrhosis of liver tissue (George et al. 2003; Kim et al. 2013). Although the fatty liver stage is considered to be relatively benign, it should be noted that it increases risk for cardiovascular disease (Tana et al. 2019). NAFLD is thought to be initiated by the excess of dietary fatty acids within the liver (Petta et al. 2016). When mice are placed on a high fat diet (HFD), a metabolic syndrome-like phenotype is induced that is characterized by elevation of body weight and blood glucose levels as well as hepatic lipid accumulation (Lai et al. 2015). Recently, Cave et al., have suggested that environmental chemicals, including pesticides, solvents, and polychlorinated biphenyls may increase NAFLD risk by modifying lipid metabolism in liver (Al-Eryani et al. 2015).
Per- and poly- fluoroalkyl substances (PFAS) are a complex family of more than 7,000 synthesized chemicals on the market (US EPA, Chemistry Dashboard, 2020). Some PFAS are well characterized for biological effects and toxicokinetics, whereas others are largely unknown (Wang et al. 2017). PFAS are used in fire-fighting foams, household and consumer items, such as heat-, stain-, and water-resistant products. PFAS have been detected in drinking water, air, dust, soil, and sediments (Boulanger et al. 2005; Shoeib et al. 2005; Skutlarek et al. 2006). The carbon-fluorine bonds in PFAS cause these compounds to possess high chemical stability, which causes them to be persistent in the environment and resist biotransformation (Mortensen et al. 2011). Perfluorooctanesulfonic acid (PFOS) is an eight-carbon (C8) perfluoroalkyl that has been detected in human serum (Calafat et al. 2007). PFOS, along with perfluorooctanoic acid (PFOA) another C8 compound, has been phased out of industry in the US and Europe, and the US Environmental Protection Agency (EPA) has issued drinking water lifetime health advisories for PFOA and PFOS at 70 parts per trillion (US EPA 2016 May 5). In humans, PFOS has a long mean serum half-life of approximately 5.4 years (Olsen et al. 2007). In both humans and mice, PFOS distributes mainly to the liver, blood, kidney, and bone (Bogdanska et al. 2011; Pérez et al. 2013; Jian et al. 2018). Through human studies from the National Health and Nutrition Examination Survey (NHANES) and the C8 Health Project, positive associations between PFOS exposure and biomarkers of liver injury have been found (Lin et al. 2010; Gallo et al. 2012; Gleason et al. 2015; Darrow et al. 2016; Bassler et al. 2019), but given the challenges of diagnosing NAFLD due to the lack non-invasive methods, it is still uncertain whether PFOS causes liver steatosis in humans. In adult rodents and monkeys, PFOS administration has been shown to decrease body weights, increase liver weight, lower serum total cholesterol, and cause hepatocellular hypertrophy and lipid vacuolization (Seacat et al. 2002; Seacat et al. 2003; Qazi et al. 2010; Wan et al. 2012). Gene expression studies have suggested that the increased liver weight and lipid vacuolization associated with PFOS treatment in adult rodents is due to peroxisome proliferator-activated receptor alpha (PPAR-α) activation (Takacs and Abbott 2007; Bjork et al. 2011). However, in PPARα knockout mice, Rosen et al. (2010) demonstrated that PFOS treatment for 7 days altered expression of hepatic genes related to lipid metabolism, inflammation, and xenobiotic metabolism. This suggests PPARa-independent effects which may be more relevant to understanding potential human health effects as human have less robust PPARα response (Palmer et al. 1998).
The recommended treatment to reverse hepatic steatosis is diet modification and exercise (Lam and Younossi 2010). Our study seeks to test the hypothesis that due to its capacity to induce steatosis, PFOS has the potential to interfere with the benefits of switching to a healthier diet and may augment hepatic lipid accumulation induced by high fat diet (HFD) feeding. After 4 weeks of HFD or standard diet (SD) alone, PFOS was added to diets at a dose of 0.0003% in food, or ~0.36 mg/kg/day based on average mouse food consumption. The latter dose has been described as a no adverse effect level (NOAEL) dose for PFOS-induced hepatic hypertrophy and vacuolation in monkey and rat (Seacat et al. 2002; Butenhoff et al. 2012; US EPA 2016 May 5). Our data demonstrate that PFOS interferes with the beneficial aspect that dietary modification can provide the fatty liver, and PFOS exacerbates HFD-induced steatosis in mice. Moreover, we also demonstrate robust changes in the hepatic proteome in response to PFOS exposure.
MATERIALS AND METHODS
Chemicals.
All chemicals and solvents were obtained from Sigma Aldrich (St. Louis, MO) or Thermo Fisher Scientific (Waltham, MA) unless specified otherwise.
Animals and treatments.
The animal protocols were reviewed and approved by the University of Rhode Island Institutional Animal Care and Use Committee (IACUC). 5-week-old male C57BL/6N mice weighing approximately 20g were purchased from Charles River Laboratories (Wilmington, MA). The mice were housed under a controlled temperature (20–26°C) with relative humidity (30–70%), and lighting (12 h, light-dark cycles) and acclimated for a week on the standard chow diet. At 6 weeks of age, mice were fed either a standard chow diet (SD; Harlan Teklad Extruded Global Diet, 2020X, n=16) or 60% kCal high fat diet (HFD; Research Diets, D12492, n=32) ad libitum for 4 weeks. The SD contained 3.1 kcal/g of total metabolizable energy, with 18.9% from fat, 16% from protein and 60% from carbohydrates. The HFD contained 5.24 kcal/g of total metabolizable energy, with 60% from fat, 20% from protein, and 20% from carbohydrates. Mice were monitored for weight gain every 2–3 days, and blood glucose changes in tail blood measured by a Bayer Contour glucometer after 3 weeks of HFD feeding (Mishawaka, IN). After 4 weeks, each group was subdivided into 3 diet groups: SD (n=16), mice fed HFD (n=16), or mice switched to SD from HFD (n=16) to mimic weight loss induced improvement of hepatic steatosis (H-SD). They were then further divided into groups that were administered diet alone with or diet with 0.0003% perfluorooctanesulfonic acid (PFOS) potassium salt from Sigma Aldrich (catalog #77282, Lot# BCBH2834V, St. Louis, MO; ~70% linear and ~30% branched isomers based on LC-MS/MS analysis [data not shown]). Based on calculated average food consumption data, daily treatment to PFOS via diet was roughly ~0.36 mg/kg/day. We selected this daily dose because the current EPA health advisory document for PFOS considered 0.3 mg/kg a NOAEL dose for PFOS-induced developmental toxicity in mouse (Wan et al. 2014; US EPA 2016 May 5). PFOS was blended into powdered diet for the initial concentration of starting stock that was 0.03% PFOS (w/w). This stock was further blended into powdered chow for a final concentration of 0.0003% PFOS (w/w). This yielded six treatment groups (n=8) i) SD; ii) SD+PFOS; iii) H-SD; iv) H-SD+PFOS; v) HFD; vi) HFD+PFOS. Mice were monitored for weight gain every 2–3 days, and blood glucose changes for an additional 10 weeks (see supplemental Fig. 2). At necropsy, mice were fasted for 4 h, before decapitation under isoflurane anesthesia. Whole trunk blood was collected in serum separator tubes (BD Microtainer, Franklin Lakes, NJ). Liver, epididymal fat pads, or white adipose tissue (WAT), and both kidneys were removed. Gross liver, kidney, and WAT weights were measured, and ~50 mg liver sections were prepared for histopathology and scoring. The remaining liver tissues and organs were snap frozen with liquid nitrogen and stored at −70°C until analysis.
Histopathology and scoring.
Liver tissue sections were fixed in 10% buffered formalin for a minimum of 24 h, and then processed for paraffin embedding. Paraffin sections (5 μm) were sectioned, H&E stained and examined by a board-certified veterinary pathologist (MG). The sections were scored for lipid accumulation, where the criteria for incidence and semi-quantitative scoring included centrilobular localization of hepatic lipid accumulation, characterized by bridging and/or diffuse expansion of hepatocytes by clear, round, macrovesicular and sometimes microvesicular vacuoles, and inflammation, where the criteria for incidence and semi-quantitative scoring included the number of sinusoidal mononuclear cells (primarily lymphocytes with lesser numbers of macrophages) and/or clustering of inflammatory cells. Sections were sorted into the following categories: 0 (0%), 1 (<10%), 2 (10–25%), 3 (25–40%), 4 (40–50%), and 5 (>50%). Statistical analysis was performed using Kruskal-Wallis test followed by Dunn’s multiple comparison test for multiple comparisons using GraphPad Prism v8.2.0 (La Jolla, CA). Significance was considered to be p ≤ 0.05.
Liver PFOS Extraction.
Liver PFOS extraction method was modified from Chang et al. (2017). Briefly, frozen liver tissues (~100 mg) were homogenized in 2 mL Omni Hard Tissue Homogenizing tubes containing 1.4 mm ceramic beads, with 400 μL cold, deionized water spiked with a fixed amount of a stable isotope-labeled internal standard (13C4-PFOS, Wellington Laboratories, Ontario, Canada, Product code: MPFOS). Using an Omni Bead Ruptor Elite (Omni International, Kennesaw, GA), the mixture was homogenized for 30 sec at 4 m/s. 250 μL of homogenate was then digested overnight at room temperature in 10% 1N KOH. 100 μL of digested homogenate was further treated with 100 μL of 2N HCl, 500 μL 1N formic acid, 500 μL of saturated ammonium sulfate, and 5 mL methyl tert-butyl ether (MTBE). The solution was mixed on a shaker (20–30 min at room temperature). The organic and aqueous layers were separated by centrifugation (2500 x g, 5 min), and an exact volume of MTBE (4.5 mL) was removed from the solution. The top organic layer was subsequently transferred to a new tube and evaporated. The resulting sample was reconstituted with 10 mL of acetonitrile and water (1:1) prior to LC-MS/MS analysis.
Serum PFOS Extraction.
Serum collected at necropsy was prepared according to methods described in Hansen et al. (2001). Briefly, 20 μL of sera, 10 μL of isotope-labeled internal standard (13C4-PFOS, Wellington Laboratories, Ontario, Canada, Product code: MPFOS), 200 μL of 0.5 M tetrabutylammonium bisulfate (TBA; adjusted to pH 10), and 400 μL of 0.25 M sodium carbonate were added to a 15-mL polypropylene tube, and thoroughly mixed. 3 mL of MTBE was added to the solution, and the mixture was placed on a shaker for 20–30 min at room temperature. The organic and aqueous layers were separated by centrifugation (2500 x g, 5 min), and an exact volume of MTBE (2.5 mL) was removed from the solution. The top organic layer was subsequently transferred to a new tube and evaporated overnight. The resulting sample was reconstituted with 1.5 mL of acetonitrile and water (1:1) prior to LC-MS/MS analysis
PFOS Quantification by LC-MS/MS.
Liver and serum samples were vortexed for 30 seconds and passed through a 0.2 μm polyethersulfone membrane syringe filter (MDI Membrane Technologies, Harrisburg, PA) into an autosampler vial. Liquid chromatography was performed on a SHIMADZU Prominence UFLC system (Columbia, MD). Samples and standards were injected (10 μL) on a Waters XBridge C18 column (100 mm X 4.6 mm i.d., 5 μm, Milford, MA) at 40°C. The mobile phase consisted of 0.1% (v/v) formic acid/water (A) and 0.1% (v/v) formic acid/acetonitrile (B) with a gradient elution of 70% of B increased to 90% of B over 8 min with a flow rate of 0.600 mL/min; at 8 min the gradient was reverted to original conditions for column re-equilibration. Analytes were measured on a SCIEX QTRAP 4500 mass spectrometer (MS) with electrospray ionization (ESI) in MRM (Multiple Reaction Monitoring) mode (SCIEX, Framingham, MA). Nitrogen was used for collision-induced dissociation of analytes. MS parameters were optimized as follows: negative ionization, IonSpray voltage, −4500; nebulizer gas, 40; auxiliary heater gas, 45; curtain gas, 20; turbo gas temperature, 400; declustering potential, −60; entrance potential, −10; collision energy, −122; collision cell exit potential, −15. The MRM ion pair used for PFOS quantification was 498.9/79.8 (precursor ion m/z, fragment ion m/z) in conjunction with a matrix matched calibration curve to determine unknown concentration in samples. The data were acquired using Analyst 1.6.3 software and processed using MultiQuant 3.0.1 software (SCIEX, Framingham, MA).
Measurement of cholesterol, triglyceride, and non-esterified fatty acid concentrations.
Whole trunk blood was collected via decapitation in serum separator tubes (BD Microtainer, Franklin Lakes, NJ) at necropsy, and serum was isolated by centrifugation before colorimetric analysis. Liver tissues (~50 mg) were homogenized in 1 mL of phosphate buffer saline (PBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4), and lipids were extracted from 0.2 mL of homogenate with chloroform-methanol (2:1; vol/vol; Folch et al. 1957). The residue was re-suspended in 1% Triton X-100 in 100% ethanol. Liver lipid content was normalized with exact tissue weight. Total cholesterol and triglyceride concentration were measured in duplicates using colorimetric assay kits from Pointe Scientific Inc. (Canton, MI) and non-esterified fatty acid (NEFA) concentration in duplicate was measured using an enzymatic colorimetric assay kit from Wako Diagnostics (Richmond, VA) according to the manufacturer’s protocols.
mRNA Isolation and QuantiGene Plex Assay.
Total RNA was isolated using TRIzol reagent (Invitrogen, Camarillo, CA) from ~50 mg of liver tissue according to the manufacturer’s instructions. RNA was washed with 75% ethanol and resuspended in DEPC-treated water. RNA samples were stored at −80°C until utilized. The purity, concentration and integrity of the extracted RNA was determined by measuring UV absorbance of the samples at 260 nm using a NanoDrop 1000 (ThermoFisher Scientific, Wilmington, DE), and formaldehyde-agarose gel electrophoresis. The quantification of selected mRNA transcripts (supplemental table 1) was performed using the QuantiGene 2.0 Plex Assay kit targeted for 50 individual genes of interest (Invitrogen, Camarillo, CA #QCP139). The assay was conducted according to manufacturer protocols for use with purified RNA with inputs of 150 or 400 ng total RNA. The assay was read on a Bio-Plex® 200 system (Bio-Rad Laboratories, Hercules, CA). The fluorescence intensity (FI; minus background) for each gene was normalized to the housekeeping gene β-actin.
Proteomics sample preparation.
Protein samples were prepared by homogenizing ~50 mg of liver tissue in 1 mL Urea buffer (8 M Urea, 50 mM triethylammoniumbicarbonate, 10 mM dithiothreitol, or DTT). Total protein was quantified using Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, Rockford IL) according to manufacturer’s instructions. Protein samples (~500 μg protein) were spiked with 2 μg bovine serum albumin (BSA), a technical standard to account for variability in sample digestion and denatured with 25 μL DTT (100 mM) at 35°C for 30 min in a shaking water bath (100 rpm). After denaturation, samples were alkylated in the dark with 25 μL iodoacetamide (IAA; 200 mM) for 30 min at room temperature. Samples were subsequently concentrated using the cold water, methanol and chloroform (1:2:1, v/v) precipitation method (centrifugation at 15,000 x g, 5 min at 10°C). The resulting protein pellet was washed with ice-cold methanol and then resuspended in 200 μL of 50 mM ammonium bicarbonate (pH ~8) containing 3% w/v sodium deoxycholate (DOC). 100 μL of the reduced and alkylated protein sample was taken for digestion. Further, TPCK-treated trypsin (10 pg) was added to samples at a ratio of 1:25 (protease:protein) and samples were transferred into digestion tubes (PCT MicroTubes, Pressure Biosciences Inc., Easton, MA). To digest proteins, the barocycler was run at 35°C, for 90 cycles with 60 sec per pressure-cycled (50 sec high pressure - 25000 psi, 10 sec ambient pressure). Further, to 100 μL of digested peptides sample, 10 μL of acetonitrile: water (1:1, v/v containing 5% formic acid) was added to stop the reaction and precipitate DOC (snow white pellet). Samples were spun to remove the pellet and 100 μL supernatant was collected (15,000 x g for 5 min at 10°C). Samples were spun (15,000 x g for 1 min at 10°C) again to remove the remaining precipitate, if any. Subsequently, 20 μL of the resulting peptide solution was injected on the column and samples were analyzed using LC-QTOF/MS. Data dependent acquisition (DDA) and sequential window acquisition of all theoretical mass spectra (SWATH-MS) data acquisition methods were used as described previously with modifications (Jamwal et al. 2017). All experiments were performed on a SCIEX 5600 TripleTOF mass spectrometer equipped with a DuoSpray ion source (SCIEX, Concord, Canada) coupled to Acquity UHPLC HClass system (Waters Corp., Milford, MA, USA). The mass spectrometer was operated in positive electrospray ionization mode for the analysis. Protein database searching was performed against reference UniProt mouse proteome library by ProteinPilot 5.0 (SCIEX, Framingham, MA, USA) using Paragon algorithm (5.0). A comprehensive spectral library of proteins and peptides from DDA runs was prepared. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Vizcaino et al. 2013) partner repository with the dataset identifier PXD01616.
Global Proteomics Analysis.
Raw data was processed on MaxQuant (version 1.6.2.10; Tyanova et al. 2016) and searched against mouse UniProt FASTA database (Swiss-Prot, downloaded Sept 26 2018), with Andromeda search engine at a false discovery rate (FDR) of 0.01. Additional search parameters were as follows, enzyme: trypsin/p, fixed modification: carbamidomethyl, variable modification: oxidation, acetyl, minimum peptide length: 7 amino acids. Resulting label-free quantitation (LFQ) intensities were processed in Perseus (version 1.6.2.3; Tyanova et al. 2016). Here, the data were filtered to remove all protein contaminants, reverse-phase proteins, and those proteins only identified by site - an automated data processing feature of Perseus. The software was then used for imputation, normalization, principal component analysis (PCA), and statistical analysis of the data. Briefly, data for analysis was transformed to a log2 scale and missing values were replaced with values selected from a normal distribution to allow the assignment of the presence or absence of proteins between conditions. PCA was performed on log transformed values. All statistical t-tests, to distinguish proteins differentially expressed between conditions, were performed with a p-value threshold of 0.05. Differentially expressed proteins were further analyzed using Ingenuity® Pathway Analysis (IPA) QIAGEN Bioinformatics (Redwood City, CA, USA) to map statistically significant proteins to the pathways and biological processes in which they were enriched.
Targeted Proteomics Analysis.
Skyline v4.2.0., an open source software, was used to analyze SWATH-MS data in a targeted fashion. Protein sequences of targets of interest was retrieved from UniProt and uploaded onto Skyline. Protein target selection was based on mRNA expression and global pathway analysis. The spectral library generated from DDA files was uploaded in Skyline, and raw SWATH-MS data files were processed using the full scan MS/MS filtering at a resolution of m/z 10,000. One or two peptides were selected per protein, and three fragment ions were selected per peptide. Unique, nonrepetitive peptides were chosen for reproducible fragment ions and verified visually by looking at the peak area, and ratio of the ion across the samples. Wherever necessary, peak boundaries for each selected fragment were adjusted. Areas for each fragment ion were summed for each peptide and averaged for each protein. Resulting protein areas were normalized to BSA area, and the final concentrations of digested protein applied to the column.
Statistical Analysis.
Power analysis using the sample size calculator http://www.jerrydallal.com/LHSP/SIZECALC.HTM was performed with the following assumption that the difference in mean between the test and control group is 50% of the mean and the standard deviation is 25% of the mean values. Based on this analysis, the sample size of n=8 was used. Statistical analysis was performed using GraphPad Prism v8.2.0 (La Jolla, CA). To analyze the body weight time course data, a Two-way ANOVA for repeated measures was conducted for weeks 0–4 to compare the effect of SD vs HFD. A similar two-way ANOVA was conducted for weeks 5–14 and included Fisher’s least significant difference (LSD) post hoc test for multiple comparisons between treatment groups. Other analyses from the conclusion of the study were conducted using one-way ANOVA followed by Fisher’s LSD test for multiple comparisons where appropriate. Significance was considered to be p ≤ 0.05.
RESULTS
Effect of Diet and PFOS on body weight, organ weight, and serum chemistry.
Male C57BL/6N mice were fed either control standard diet (SD) or a 60% kCal high fat diet (HFD) for 4 weeks, which increased body weight, increased fasting blood glucose, and impaired response to a glucose tolerance test (supplemental Fig. 2). At this timepoint, half of the mice fed HFD were switched to a SD (H-SD) to mimic weight loss with switch to a healthier diet. Additionally, mice were further divided into groups that were fed only diet or diet that contained 0.0003% PFOS for an additional 10 weeks. Body weights were recorded weekly (Fig. 1) and clinical parameters associated with response to diet were evaluated (Table 1). HFD feeding for 14 weeks increased body weight by 37%. HFD also increased liver and white adipose tissue weight compared to the SD, by 31% and 68% respectively. HFD feeding also increased serum cholesterol, serum triglycerides and fasting blood glucose, by 75%, 44%, and 28% (Table 1). For the H-SD group, HFD feeding for 4 weeks increased mean body weight (Fig. 1), followed by weight loss with SD feeding. By week 8, the body weight for the H-SD group were similar to SD controls. The organ weights, serum cholesterol, serum triglycerides and fasting blood glucose of the H-SD group were similar to the SD controls, and significantly lower compared to the HFD, except for the serum triglycerides which were not significantly higher than SD, but not significantly lower than the HFD. There was no diet effect on kidney weight.
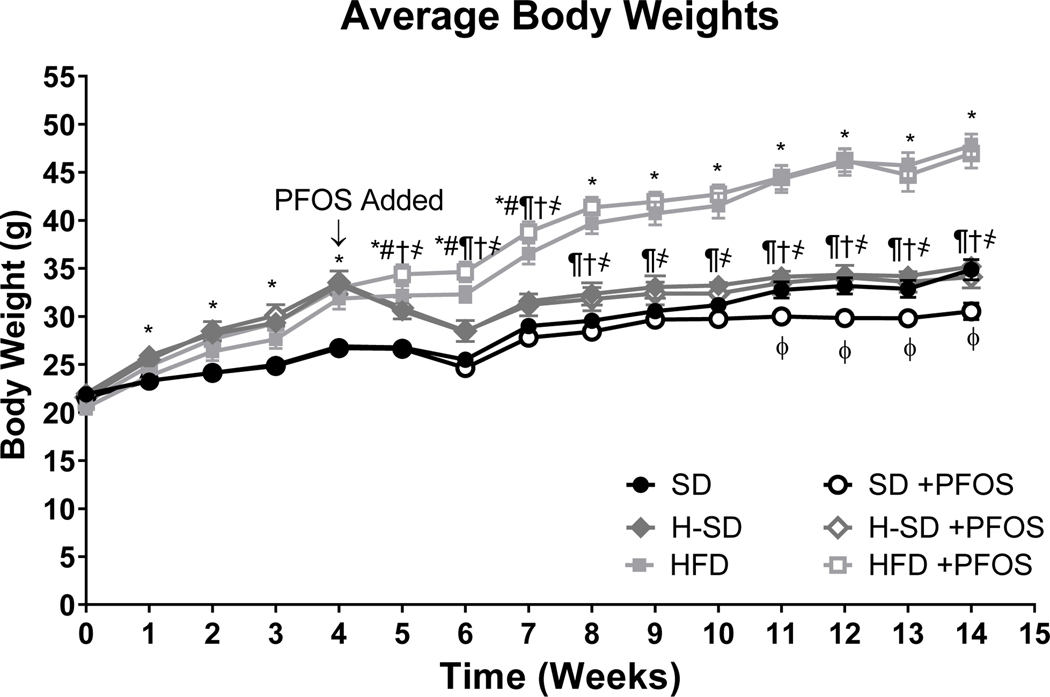
Figure 1. Effect of PFOS on body weight over time.
6-week-old male mice were fed either control standard chow diet (SD) or a 60% kCal high fat diet (HFD), and then after 4 weeks, half of the mice fed HFD were switched to a SD (H-SD) to mimic weight loss. Furthermore, the mice were then exposed to 10 weeks of dietary PFOS treatment (0.0003%). Body weight was measured every week and shown as average body weights ± SEM. HFD feeding for 4 weeks increased body weight. Mice were then switched to a SD lost weight until week 8, and by the end of the end of the study, body weights were similar to SD controls. HFD feeding for an additional 10 weeks increased body weight. For weeks 0–4, calculations were performed using a Two-way ANOVA for repeated measures was conducted to compare the effect SD (N=16) vs HFD (N=27), * indicates p<0.05. For weeks 5–14, calculations were performed using a two-way ANOVA followed by Fisher’s LSD test. (N = 5– 8). * indicates p<0.05 for SD vs HFD, # indicates p<0.05 for SD vs H-SD, ¶ indicates p<0.05 for H-SD vs HFD, † indicates p<0.05 for H-SD+PFOS vs SD+PFOS, ҂indicates p<0.05 for H-SD+PFOS vs HFD+PFOS, and ɸ indicates p<0.05 for SD vs SD+PFOS.
Table 1.
Clinical Parameters in C57BL/6N Mice after PFOS and HFD treatment.
| Parameter | SD | SD +PFOS | H-SD | H-SD +PFOS | HFD | HFD +PFOS |
|---|---|---|---|---|---|---|
|
| ||||||
| Body weight (BW) (g) | 35.4 ± 1.2 | 30.8 ± 0.8# | 36.1 ± 0.9¶ | 36.6 ± 0.5†҂ | 48.6 ± 1.2* | 47.3 ± 1.3* |
| Liver weight (g) | 1.47 ± 0.0 | 1.67 ± 0.1 | 1.49 ± 0.0¶ | 1.86 ± 0.1*#҂ | 1.92 ± 0.1* | 2.71 ± 0.2*# |
| Liver weight/ BW (%BW**) | 4.17 ± 0.1 | 5.38 ± 0.3# | 4.12 ± 0.1 | 5.07 ± 0.3*# | 3.89 ± 0.1 | 5.72 ± 0.4*# |
| White adipose tissue (WAT) (g) | 1.35 ± 0.2 | 0.84 ± 0.1# | 1.57 ± 0.1¶ | 1.37 ± 0.1†҂ | 2.27 ± 0.2* | 1.98 ± 0.2* |
| WAT/BW (%BW**) | 3.75 ± 0.5 | 2.69 ± 0.4# | 4.31 ± 0.2 | 3.75 ± 0.3† | 4.73 ± 0.5 | 4.20 ± 0.5 |
| Kidney weight (g) | 0.34 ± 0.0 | 0.37 ± 0.0 | 0.37 ± 0.0 | 0.38 ± 0.0 | 0.37 ± 0.0 | 0.36 ± 0.0 |
| Serum triglycerides (mg/dL) | 75.8 ± 8.3 | 109.7 ± 9.6# | 90.0 ± 7.9 | 96.8 ± 5.4 | 109 ± 5.1* | 85.3 ± 6.2 |
| Serum total cholesterol (mg/dL) | 190 ± 9.0 | 169 ± 12.6 | 211 ± 12¶ | 184 ± 9.1҂ | 333 ± 14.4* | 253 ± 22.2*# |
| Fasting blood glucose (mg/dL) | 149 ± 14.1 | 130 ± 7.4 | 129 ± 7.2¶ | 121 ± 6.4҂ | 190 ± 21.6* | 196 ± 4.5* |
Male C57BL/6N mice were fed either control standard chow diet (SD) or a 60% kCal high fat diet (HFD), and then after 4 weeks, half of the mice fed HFD were switched to a SD (H-SD) to mimic weight loss. Additionally, mice were then exposed to PFOS in diet for 10 weeks (0.0003%).At 10 weeks, body and organ weights, serum triglyceride and cholesterol concentrations were determined. Calculations were performed using a one-way ANOVA followed by Fisher’s LSD test .All values are means ± SEM; N = 5– 8.
indicates p<0.05 versus control SD
indicates p<0.05 for H-SD versus HFD
indicates p<0.05 versus respective diet controls
indicates p<0.05 for H-SD+PFOS versus SD+PFOS
indicates p<0.05 for H-SD+PFOS versus HFD+PFOS.
percent body weight was calculated as g tissue per g BW multiplied by 100.
Table 1 further illustrates PFOS effects on tissue weights and serum parameters. PFOS treatment in SD mice reduced body weight compared to the control SD mice starting at week 11 (Fig. 1) and at the conclusion of the study the body weight was 13% lower, which has previously been observed in mice at doses 0.3 mg PFOS/kg body weight (Wan et al. 2014). H-SD mice exposed to PFOS had a similar body weights as H-SD mice, and PFOS treatment in HFD mice did not change body weights. Mice consumed 55% less high fat diet (HFD) than standard diet (SD) and PFOS treatment had no effect on food consumption (supplemental Fig. 1). PFOS increased liver weights in the H-SD and HFD groups by 25% and 41%, respectively. There was only a slight increase of liver weight 14% (p=0.0584) in the SD group. Once normalized to body weight, PFOS treatment increased the liver-to-body weight ratio in all treatment groups: SD by 29%, H-SD by 23%, and HFD by 47%. In SD fed mice, PFOS treatment decreased gross WAT weight by 38%, and the WAT to BW ratio by 28%. A similar effect was not seen in the H-SD and HFD groups. PFOS treatment had no effect on kidney weight in all groups. PFOS treatment increased serum triglycerides by 45% in the SD diet. PFOS treatment decreased serum total cholesterol by 24% in the HFD, but not in the H-SD and SD groups.
Liver and Serum PFOS concentrations.
Table 2 depicts serum and liver PFOS concentrations in male C57BL/6N mice exposed to 10 weeks of PFOS at a concentration 0.0003% in diet. Serum concentrations were similar between the SD and the H-SD, and HFD however PFOS concentration the H-SD group was significantly lower than the HFD group, by 18%. Liver PFOS concentrations were 200.2 ± 22.5 μg/ml in the mice fed SD. Interestingly, the PFOS concentrations were reduced by 25% and 32% the livers from H-SD and HFD mice respectively. As there were observed differences in liver weight, the concentrations were normalized to total liver weight;the H-SD mice had 25%. less overall liver PFOS as compared to mice feed the SD, and 23%. less as compared to mice feed the HFD. In HFD mice, the liver-to-serum ratio decreased from 5.2-fold to 3.3-fold. After normalizing PFOS concentrations to the amount of PFOS consumed within the diet, the HFD had much higher concentration of PFOS in the liver and serum, by 67% and 160% respectively.
Table 2.
Serum and Liver PFOS Concentration
| SD | SD +PFOS | H-SD | H-SD +PFOS | HFD | HFD +PFOS | |
|---|---|---|---|---|---|---|
|
| ||||||
| Serum (μg/mL) | <LLOQ | 40.0 ± 6.9 | <LLOQ | 34.8 ± 6.1 ¶ | <LLOQ | 42.4 ± 2.0 |
| Liver PFOS (μg PFOS/g tissue) | <LLOQ | 200.2 ± 22.5 | <LLOQ | 148.0 ± 16.1 * | <LLOQ | 137.0 ± 32.8 * |
| Total Liver PFOS (μg PFOS) | 363.5 ± 20.2 | 272.7 ± 18.0 *¶ | 354.1 ± 38.4 | |||
| Liver/Serum Ratio | 5.2 ± 0.33 | 4.4 ± 0.44 | 3.3 ± 0.34 * | |||
| Serum PFOS (mg PFOS/mL per mg of PFOS consumed) | 2.32 ± 0.14 | 2.19 ± 0.14 ¶ | 6.01 ± 0.13 * | |||
| Liver PFOS (mg PFOS/g tissue per mg of PFOS consumed) | 11.6 ± 0.5 | 9.3 ± 0.4 ¶ | 19.4 ± 2.1 * | |||
Male C57BL/6N mice were fed either control standard chow diet (SD) or a 60% kCal high fat diet (HFD).After 4 weeks, half of HFD mice were switched to a SD (H-SD) to induce weight loss, and mice were further divided with a subset having 0.0003% PFOS in diet. Mice were kept on study for an additional 10 weeks. PFOS was extracted from liver and serum and quantified using LC-MS/MS. All mice that were not dosed with PFOS had concentrations below the lower limit of quantification (LLOQ, 15 ng/mL, 3 μg/g tissue).Total liver PFOS was calculated by multiplying concentration by total liver weights and PFOS concentrations were also normalized to average food consumption within each diet group. The HFD and H-SD mice had a lower liver PFOS concentration, and the H-SD mice had less overall liver PFOS. The HFD mice had a higher liver and serum PFOS concentration relative to the amount of PFOS consumed within the diet. Calculations were performed using a one-way ANOVA followed by Fisher’s LSD test. All values are means ± SD; N=5–8.
indicates p<0.05 versus SD+PFOS
indicates p<0.05 for H-SD+PFOS versus HFD+PFOS.
Diet and PFOS effects on lipid content in mouse liver.
The assessment of liver inflammation showed no significant change in inflammatory cells across any of the treatment groups (supplemental table 2). Assessment of micro and macrovesicular steatosis incidence and severity scores indicated increased lipid accumulation in the HFD group compared to SD group (Table 3). In the HFD control group, 57% of livers had lipid accumulation severity scores of 3 or more (≥3), whereas 100% of the mice in HFD+PFOS group had lipid accumulation severity scores of ≥3. The incidence was steatosis was similar between SD and H-SD mice. There was a slight increase in the incidence of very mild steatosis with PFOS treatment, with only 1 out of 8 mice having some evidence of steatosis, whereas 3 out of 6 H-SD+PFOS mice had some evidence of steatosis, however this trend was not significant.
Table 3.
Effect of PFOS on liver steatosis histopathology
| Scores | SD | SD +PFOS | H-SD | H-SD+PFOS | HFD | HFD +PFOS |
|---|---|---|---|---|---|---|
|
| ||||||
| 0 | 6/8 | 4/5 | 7/8 | 3/6 | 0/7 | 0/6 |
| 1 | 2/8 | 1/5 | 0/8 | 1/6 | 0/7 | 0/6 |
| 2 | 0/8 | 0/5 | 1/8 | 2/6 | 3/7 | 0/6 |
| 3 | 0/8 | 0/5 | 0/8 | 0/6 | 2/7 | 1/6 |
| 4 | 0/8 | 0/5 | 0/8 | 0/6 | 2/7 | 4/6 |
| 5 | 0/8 | 0/5 | 0/8 | 0/6 | 0/7 | 1/6 |
|
| ||||||
| ≥3 | 0/8 (0%) | 0/5 (0%) | 0/8 (0%) ¶ | 0/6 (0%) | 4/7 (57%) * | 6/6 (100%) * |
Formalin fixed hepatic tissue sections were stained with hematoxylin and eosin (Fig. 2A). Livers were scored for lipid accumulation (range from 0 to 5, where 0 is the least and 5 is most severe). Statistical analysis was performed using Kruskal–Wallis test followed by Dunn’s multiple comparison test for multiple comparisons. N=5–8/treatment group
indicates p<0.05 versus SD
indicates p<0.05 for H-SD versus HFD.
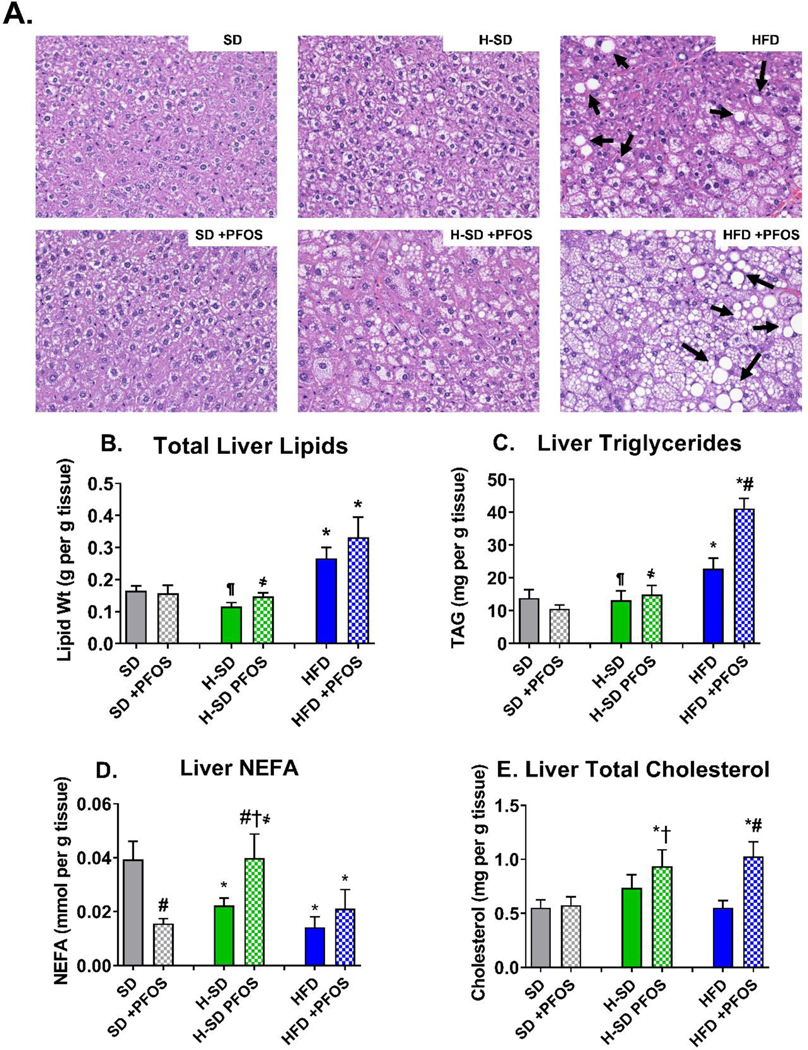
Correlating to the observed pathology, total liver lipid content in the HFD group was 55% higher than SD group (Fig. 2B). In addition, HFD increased liver triglyceride content by 53% (Fig. 2C) and decreased the liver free fatty acid content by 64% (Fig. 2D). H-SD had a similar total liver lipid content as control SD mice. However, the liver free fatty acid concentration was decreased by 43% (Fig. 2D), as weight loss increases β-oxidation (van der Windt et al. 2018). PFOS treatment had no significant effect on total liver lipid content (Fig. 2B). However, PFOS increased the total lipid content in mice fed HFD (Fig. 2B), which matches trends observed in lipid accumulation scores. PFOS treatment in the SD decreased the liver free fatty acid content by 60% (Fig. 2D). In contrast, PFOS treatment in the H-SD weight loss group increased liver free fatty acid content by 79%, which is similar to SD control concentrations of free fatty acids (Fig. 2D). PFOS treatment had no effect on liver free fatty acid in mice fed an HFD. PFOS treatment in mice fed the HFD nearly doubled liver triglyceride content (Fig. 2C) and cholesterol content (Fig. 2E) compared to the HFD controls. PFOS treatment did not affect liver triglycerides and cholesterol in the SD and H-SD groups.
Figure 2. Diet and PFOS effects on liver lipid content.
(A) Liver histopathology representatives for each treatment group viewed at 200X magnification. Hemotoxylin and eosin (H&E) staining of liver tissue illustrates HFD administration increased vacuolization in mice compared to control. Arrows designate lipid vacuoles consistent with histopathology described for hepatic steatosis. (B) Total lipids were isolated and normalized to tissue weight. (C) Triglycerides, (D) non-esterified fatty acids (NEFA), and (E) cholesterol content, and was measured via colorimetric assay and normalized to tissue weight. In the HFD, PFOS treatment caused a slight increase in liver lipids and increased triglycerides and cholesterol. Calculations were done using a one-way ANOVA followed by Fisher’s LSD test. All values are means ± SEM; N = 5–8.* indicates p<0.05 versus control SD, ¶ indicates p<0.05 for H-SD versus HFD, # indicates p<0.05 versus respective diet controls, † indicates p<0.05 for H-SD+PFOS versus SD+PFOS, and ҂ indicates p<0.05 for H-SD+PFOS versus HFD+PFOS.
Diet and PFOS modulate hepatic mRNA expression.
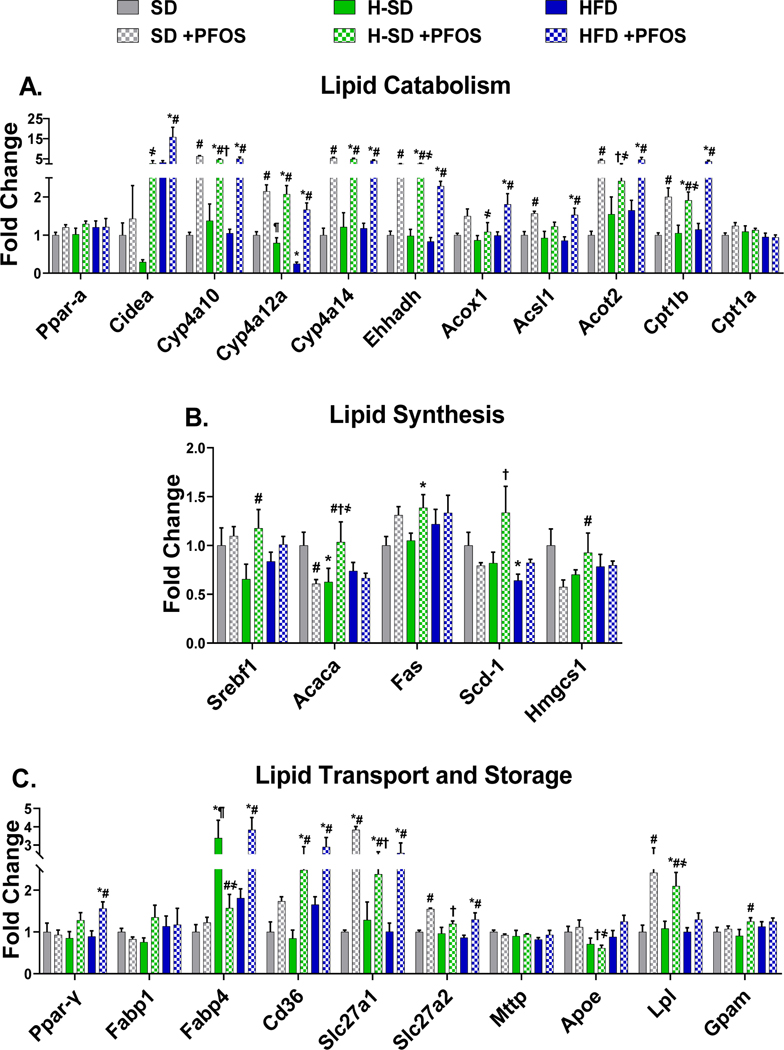
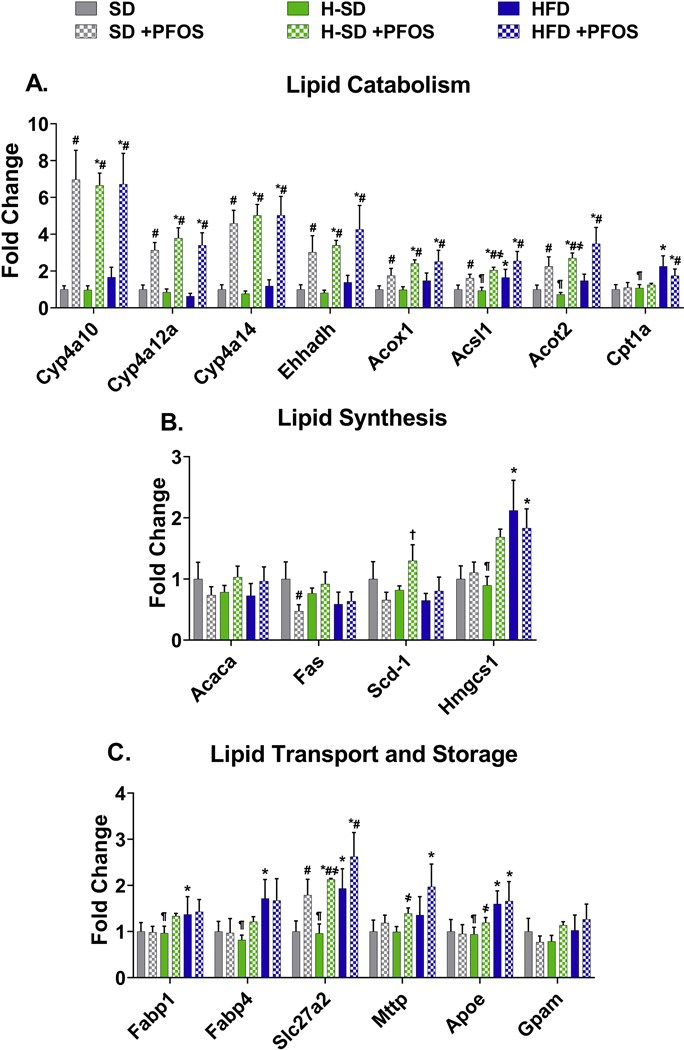
Gene targets were selected based on function related to lipid metabolism, previously reported transcripts modulated by PFAS (Rosen et al. 2008; Tan et al. 2013), and results of untargeted proteomic analysis. Full names and functions of gene targets are listed in supplemental table 1. Ultimately, 50 different targets from lipid catabolism, lipid synthesis, lipid transport, lipid storage, xenobiotic metabolism and transport, detoxification, antioxidant response, and inflammation pathways were analyzed. As described in Fig. 3A, PFOS induced several genes related to lipid catabolism in all three diets, with greater than 5 fold induction of Cyp4a10, Cyp4a14, Ehhadh, Acot2, and Cpt1b. Diet-specific effects on lipid synthesis protein expression were observed - the HFD decreased stearyl-CoA desaturase 1 (Scd-1) expression by 36% and H-SD decreased liver acetyl-CoA carboxylase 1 (Acaca) mRNA expression by 37% (Fig. 3B). Furthermore, in Fig. 3B, PFOS treatment in the SD group repressed Acaca and 3-hydroxy-3-methylglutaryl-CoA synthase 1 (Hmgcs1) by 39% and 43%, whereas PFOS treatment in the H-SD group induced Sterol regulatory element-binding protein 1 (Srebf1) and Acaca by 79% and 65%. H-SD mice had three times higher fatty acid binding protein 4 (Fabp4) levels than SD mice (Fig. 3C). PFOS treatment also induced several genes related lipid transport. In all three diets (SD, H-SD, and HFD), PFOS treatment increased Long-chain fatty acid transport protein 1 (Slc27a1) mRNA expression by 3.8, 2.4, and 2.6-fold compared to the control SD diet. In the SD group, PFOS treatment also increased Slc27a2 and lipoprotein lipase (Lpl) mRNA expression by 1.6 and 2.4-fold. In the H-SD group, PFOS treatment induced fatty acid translocase (Cd36), Lpl, and glycerol-3-phosphate acyltransferase, mitochondrial (Gpam) by 2.9, 1.9, and 1.4-fold compared to H-SD controls. In the HFD group, PFOS treatment increased liver peroxisome proliferator-activated receptor gamma (PparY), Cd36, Fabp4, and Scl27a2 mRNA expression by 1.8, 2.1, 1.8, and 1.5-fold compared to HFD controls.
Figure 3. Hepatic lipid metabolism gene expression.
The quantification of selected liver mRNAs was performed using QuantiGene 2.0 Plex Assay kit with purified RNA. The fluorescence intensity (FI; minus background) for each gene was normalized to the housekeeping gene β-actin. Fold change was calculated between the control SD mice. A) PFOS induces 9 genes related to lipid catabolism. B) PFOS has diet dependent effects on lipid synthesis genes, and C) induces several genes related lipid transport. Calculations were done using a one-way ANOVA followed by Fisher’s LSD test. All values are means ± SEM; N = 5. * indicates p<0.05 versus control SD, ¶ indicates p<0.05 for H-SD versus HFD, # indicates p<0.05 versus respective diet controls, † indicates p<0.05 for H-SD+PFOS versus SD+PFOS, and ҂ indicates p<0.05 for HSD+PFOS versus HFD+PFOS.
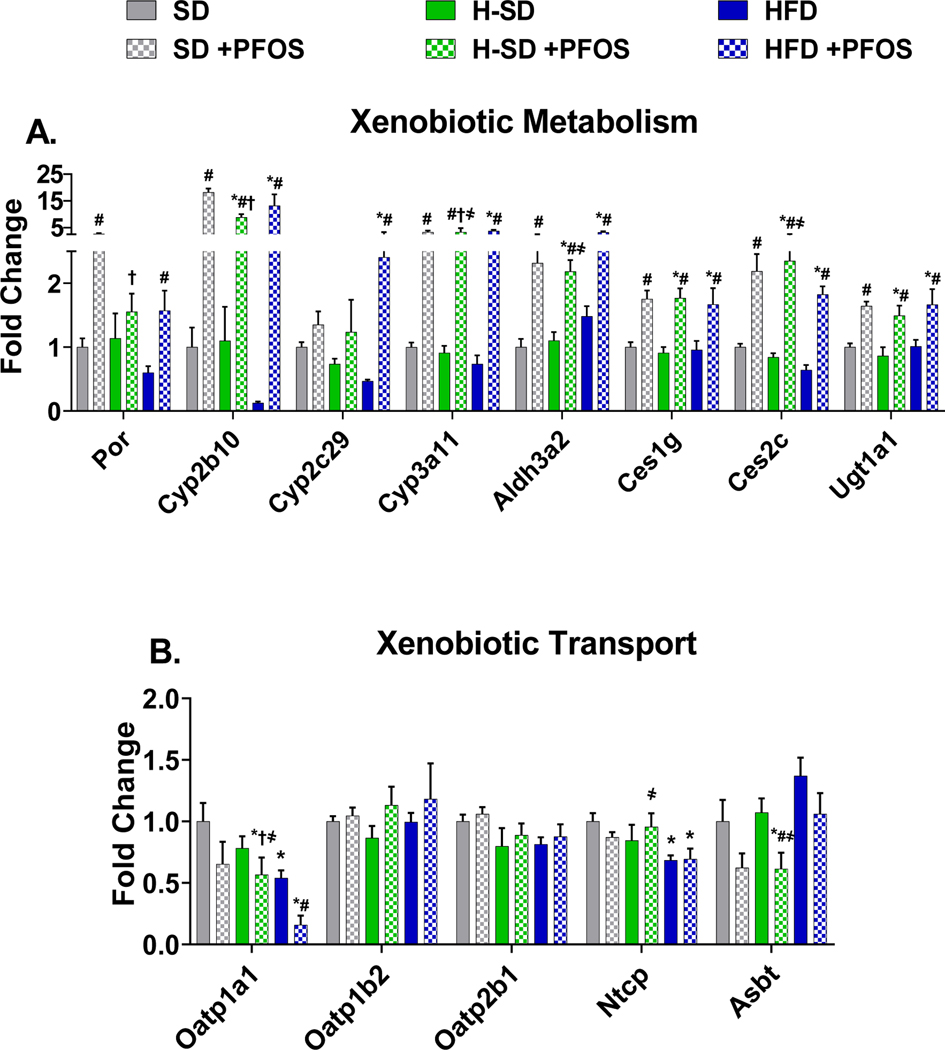
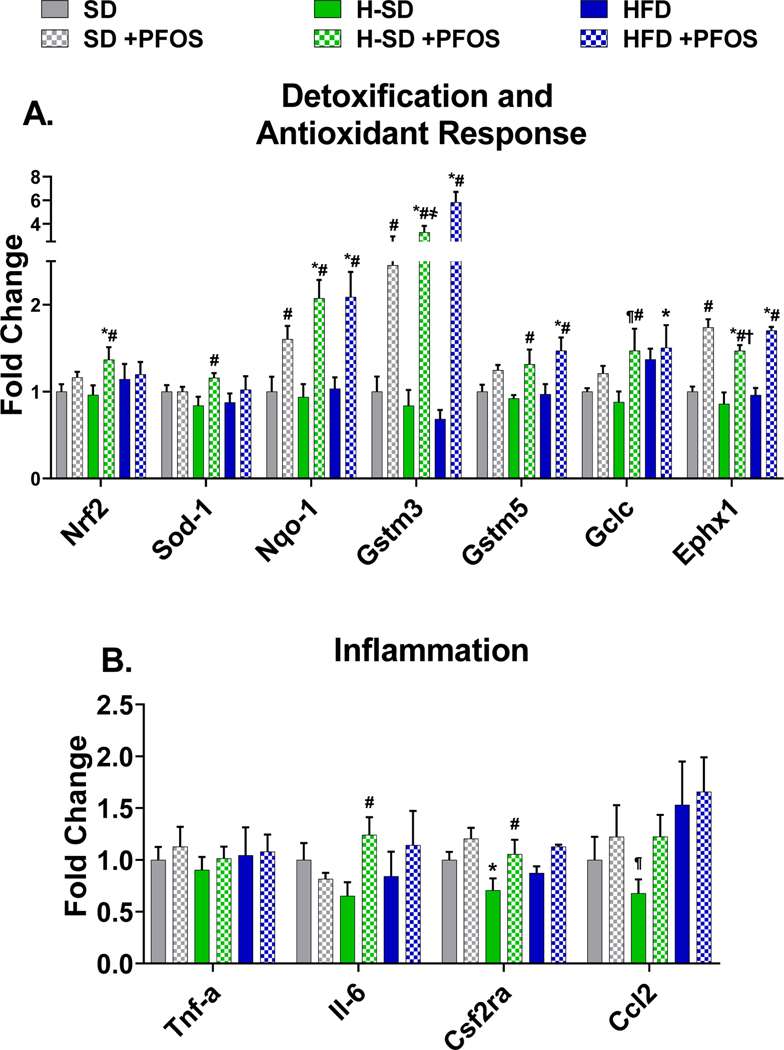
In Fig. 4A, PFOS treatment upregulated 7 out of 8 genes related to xenobiotic metabolism. In all diets PFOS increased Cyp2b10 (~15 fold), Cyp3a11 (~5 fold), Aldh3a2 (~5 fold), Ces1g (~2 fold), Ces2c (~3 fold), and Ugt1a1 (~4 fold). In SD fed mice, PFOS treatment induced Cytochrome P450 oxidoreductase (Por) by 2.7 fold, and in the HFD group, PFOS treatment induced Por, and Cyp2c29 by 2.6 and 5.1 fold compared to HFD control. HFD treatment repressed two xenobiotic transporters, Organic anion transporting protein 1a1 (Oatp1a1) and Sodium-dependent uptake transporter (Ntcp) by 46% and 31% (Fig. 4B). PFOS treatment in the HFD also repressed Oatp1a1 by 70%, as well as Apical sodium dependent bile acid transporter (Asbt) by 43% in the H-SD group. For all diets, PFOS treatment increased the mRNA expression of NAD(P)H quinone dehydrogenase 1 (Nqo1), Glutathione S-transferase mu 3 (Gstm3), and Epoxide hydrolase 1 (Ephx1), which are involved metabolism and the antioxidant response (Fig. 5A). In the -H-SD group, PFOS treatment induced Nuclear factor erythroid 2-related factor 2 (Nrf2), Superoxide dismutase 1 (Sod-1), Glutathione S-transferase mu 5 (Gstm5), and Glutamate-cysteine ligase catalytic subunit (Gclc), and in the HFD group, PFOS treatment induced Gstm5. Livers from H-SD mice had reduced Colony stimulating factor 2 receptor alpha subunit (Csf2ra) expression by 29%, and H-SD administered PFOS induced interleukin 6 (IL-6) and Csf2ra expression by 1.9 and 1.5-fold compared to H-SD control (Fig. 5B).
Figure 4. Hepatic lipid xenobiotic metabolism and transport gene expression.
The quantification of selected liver mRNAs was performed using QuantiGene 2.0 Plex Assay kit with purified RNA. The fluorescence intensity (FI; minus background) for each gene was normalized to the housekeeping gene β-actin. Fold change was calculated between the control SD mice. A) PFOS treatment induced 7 out of 8 transcripts related to xenobiotic metabolism. B) HFD and PFOS treatment represses some transporter gene expression. Calculations were done using a one-way ANOVA followed by Fisher’s LSD test. All values are means ± SEM; N = 5. * indicates p<0.05 versus control SD, ¶ indicates p<0.05 for H-SD versus HFD, # indicates p<0.05 versus respective diet controls, † indicates p<0.05 for H-SD+PFOS versus SD+PFOS, and ҂ indicates p<0.05 for H-SD+PFOS versus HFD+PFOS.
Figure 5. Hepatic lipid oxidative stress gene expression.
The quantification of selected liver mRNAs was performed using QuantiGene 2.0 Plex Assay kit with purified RNA. The fluorescence intensity (FI; minus background) for each gene was normalized to the housekeeping gene β-actin. Fold change was calculated between the control SD mice. A) PFOS treatment induces expression of genes related to detoxification and antioxidant response. B) Only in the H-SD diet, PFOS induces expression of interleukin 6 (Il-6) and colony stimulating factor 2 receptor α (Csf2ra). Calculations were done using a one-way ANOVA followed by Fisher’s LSD test. All values are means ± SEM; N = 5. * indicates p<0.05 versus control SD, ¶ indicates p<0.05 for H-SD versus HFD, # indicates p<0.05 versus respective diet controls, † indicates p<0.05 for H-SD+PFOS versus SD+PFOS, and ҂ indicates p<0.05 for H-SD+PFOS versus HFD+PFOS.
Untargeted Proteomic Analysis.
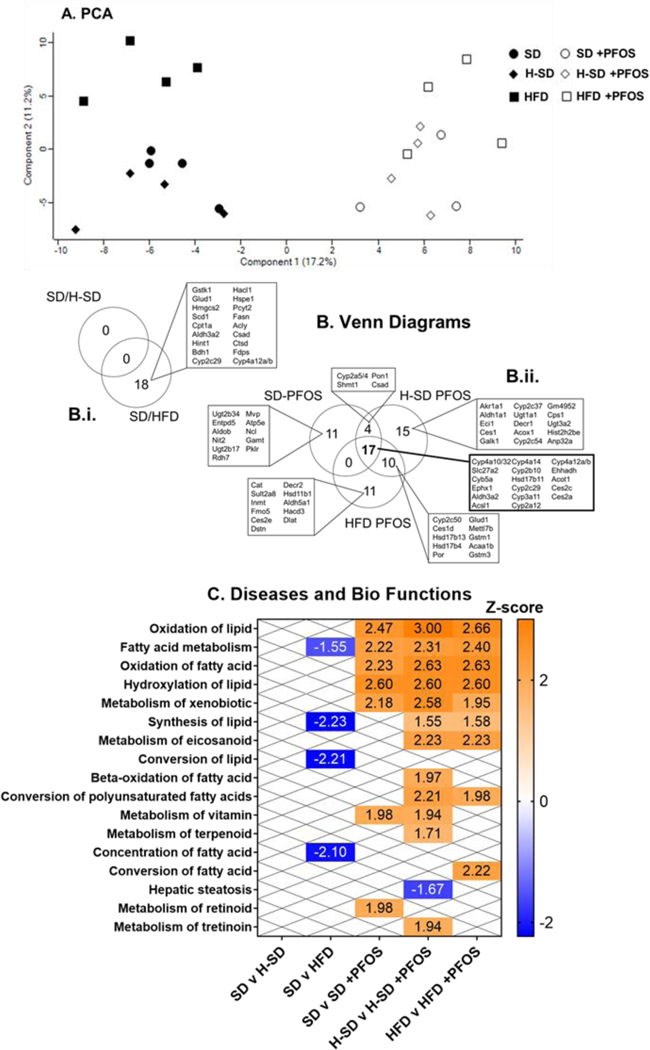
We observed a total of 665 proteins in our untargeted analysis. The data was subjected to PCA and the results are visualized as scatter plots. Fig. 6A illustrates distinct clustering of liver protein expression between the non-treated and PFOS-treated mice, with less clustering occurring with diet. The number of differentially expressed proteins for relevant comparisons (SD/H-SD, SD/HFD, SD/SD+PFOS, H-SD/H-SD+PFOS, and HFD/HFD+PFOS) were calculated and are shown in Fig. 6B. SD and H-SD groups had similar protein expression in liver, whereas livers from mice fed HFD had 18 out of 665 differentially expressed proteins relative to SD (Fig. 6Bi). PFOS treatment altered the liver levels of 32, 38, and 46 proteins for SD, H-SD, and HFD diets, respectively. Of these protein changes, 17 proteins were common among each diet, whereas 11, 15 and 11 proteins showed unique changes in abundance among the SD, H-SD, and HFD, respectively.
Figure 6. Global proteomic analysis.
Protein data was analyzed using Perseus open source software. A) A principle component analysis (PCA) plot is shown. The numbers of significantly increased and decreased proteins for relevant comparisons were calculated. (student’s t-test, p-value ≤ 0.05, false discovery rate (FDR) < 0.05). Comparisons were SD/H-SD, SD/HFD, SD/SD+PFOS, H-SD/H-SD+PFOS, and HFD/HFD+PFOS. B) Venn diagrams illustrating the number of significantly increased and decreased proteins detected and shared between diet and PFOS treatment groups. The H-SD diet had no differentially expressed proteins compared to control and the HFD had 18 differentially expressed proteins compared to the control diet. PFOS treatment in each diet, SD, H-SD, and HFD, induced 32, 38, and 46 significant differentially expressed proteins, respectfully. Of these protein changes, 17 proteins were common among each diet. Differentially expressed proteins among all comparisons were further analyzed using Ingenuity Pathway Analysis (IPA). C) A heat-map describing significant z-scores (≥ 1.5 or ≤ −1.5) of Diseases and Bio Function pathways are shown. Pathways that are expected to be increased are shown in orange and decreased are shown in blue. ―”X” designates comparisons that did not have significant pathway changes.
Differentially expressed proteins among all comparisons were further analyzed using Ingenuity Pathway Analysis (IPA) as illustrated in Fig. 6C and Table 4. The HFD repressed fatty acid metabolism and lipid synthesis related pathways. Upstream analysis indicated that differentially expressed proteins corresponded to repression of Srebf chaperone (Scap) and induction of Por. PFOS treatment, overall, induced proteins involved in lipid utilization and xenobiotic metabolism pathways among all treatment groups (Fig. 6C). Several distinct pathway proteomic changes among the different diets were also observed. The SD-PFOS treatment upregulated metabolism of vitamin and retinoid pathways. PFOS treatment in the H-SD group induced the most pathways, including induction of lipid synthesis, β-oxidation, conversion of polyunsaturated fatty acids, metabolism of vitamin, tretinoin, eicosanoid, and terpenoid pathways, as well as an overall repression of hepatic steatosis. In HFD mice, PFOS treatment induced proteomic changes associated with lipid synthesis, conversion of polyunsaturated fatty acids, metabolism of eicosanoid, and conversion of fatty acids pathways. Upstream analysis (Table 5) shows that PFOS treatment among all three diets, induced Ppar-α, Leptin, Nr1i3 (Car, nuclear receptor subfamily 1 group I member 3), cystic fibrosis transmembrane conductance regulator (Cftr), ATP binding cassette subfamily B member 6 (Abcb6), actin dependent regulator of chromatin, subfamily b, member 1 (Smarcb), and transcription factor 7 like 2 (Tcf7l2) signaling. Additionally, PFOS treatment in the H-SD and HFD groups, induced Nrf2 and Nr1i2 (PXR, nuclear receptor subfamily 1 group I member 2) signaling.
Table 4.
Effect of HFD on upstream regulators of the proteomic response
| Upstream Regulator | Name | Function | Activation Z-score | Target Molecules in Data Set |
|---|---|---|---|---|
|
| ||||
| Scap | sterol regulatory element binding protein (SREBF) chaperone | Unfolded protein response | −2.200 | Acly, Csad, Fasn, Fdps, Hmgcs2, Scd-1 |
|
| ||||
| Por | cytochrome p450 oxidoreductase | donate electrons directly from NADPH to all microsomal P450 enzymes |
2.000 | Acly, Bdh1, Cpt1a, Csad, Cyp2c8, Fdps, Scd-1 |
Protein data were analyzed using Perseus open source software. The numbers of significantly increased and decreased proteins for relevant comparisons were calculated (student’s t-test, p-value ≤ 0.05, false discovery rate (FDR) < 0.05). Diet based comparisons were SD/H-SD and SD/HFD. Differentially expressed proteins were further analyzed using Ingenuity Pathway Analysis (IPA)’s upstream analysis. Significant upstream regulatory molecules are described for SD vs HFD, and an activation z-score was calculated to predict activation of upstream regulators. A positive z-score indicates upregulation and a negative z-score indicates downregulation.
Table 5.
Effect of PFOS on upstream regulators of the proteomic response
| Upstream Regulator | Name | Function | Activation Z-score | Target Molecules in Data Set | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||||
| SD +PFOS | H-SD +PFOS | HFD +PFOS | PFOS treatment | SD +PFOS | SD & H-SD +PFOS | H-SD +PFOS | H-SD & HFD +PFOS | HFD +PFOS | |||
|
| |||||||||||
| Ppar-α | peroxisome proliferator activated receptor alpha | lipid metabolism in the liver | 2.306 | 2.864 | 2.974 | Acot1 Acsl1 Aldh3a2 Cyp2b6 Cyp2c8 Cyp4a11 Cyp4a14 Ehhadh Hsd17b11 Slc27a2 | Aldob Pklr | Csad | Acox1 Cps1 Eci1 | Acaa1b, Cyp2c54, Hsd17b4, Por | Dec1 Cat Decr2 |
|
| |||||||||||
| Lep | leptin | adipokine, pro-inflammatory cytokine | 2.828 | 3.162 | 2.530 | Cyb5a Cyp2a12/Cyp2a22 Cyp2c8 Cyp3a5 Cyp4a11 Cyp4a14 Ehhadh | Aldob | Acox1 Cps1 | Cyp2c54, Gstm3 | Fmo5 | |
|
| |||||||||||
| Nr1i3 | Car, nuclear receptor subfamily 1 group I member 3 | xenobiotic and endobiotic metabolism | 2.583 | 2.946 | 2.771 | Ces2a Ces2c Cyp2b6 Cyp2c8 Cyp3a5 Ephx1 | Ugt2b28 | Cyp2a6 | Aldh1a1 Ugt1a1 | Gstm3, Gstm5, Por | |
|
| |||||||||||
| Cftr | cystic fibrosis transmembrane conductance regulator | involved in AMPK signaling | 2.213 | 2.804 | 2.804 | Cyp2b6 Cyp3a5 Cyp4a11 Hsd17b11 Slc27a2 | Acaa1b, Hsd17b4, Por | ||||
|
| |||||||||||
| Abcb6 | ATP binding cassette subfamily B member 6 | transporter, involved in mitochondrial function | 2.201 | 2.418 | 2.202 | Cyp2a12/Cyp2a22 Cyp2b6 Cyp3a5 Cyp4a11 | Cyp2a6 | Cyp2c54 | |||
|
| |||||||||||
| Smarcb1 | actin dependent regulator of chromatin, subfamily b, member 1 | involved in AMPK signaling | 2.000 | 2.236 | 2.236 | Acsl1 Cyp2c8 Cyp4a11 Cyp4a14 | Cyp2c54 | ||||
|
| |||||||||||
| Tcf7l2 | transcription factor 7 like 2 | Wnt signaling and blood glucose homeostasis | 2.236 | 2.000 | 2.000 | Acsl1 Aldg3a2 Cyp2c8 Ehhadh Slc27a2 | Entpd5 | ||||
|
| |||||||||||
| Nrf2 | nuclear factor, erythroid 2 like 2 | antioxidant and detoxification response | N/A | 2.383 | 2.070 | Akr1a1 Ces1g Ugt1a1 | Cyp2a12/Cyp2a22, Cyp4a14, Cyp4a22, Ephx1, Gstm3, Gstm5 |
Cat Dstn Hacd3 Inmt |
|||
|
| |||||||||||
| Nr1i2 | PXR, nuclear receptor subfamily 1 group I member 2 | xenobiotic and endobiotic metabolism | N/A | 2.218 | 2.218 | Acox1 Ald1a1 Ugt1a1 | Ces2a, Cyp2b6, Cyp3a6, Gstm3, Gstm5, Por |
Cat | |||
Protein data were analyzed using Perseus open source software. The numbers of significantly increased and decreased proteins for relevant comparisons were calculated. (student’s t-test, p-value ≤ 0.05, false discovery rate (FDR) < 0.05). Comparisons were SD/SD+PFOS, H-SD/H-SD+PFOS, and HFD/HFD+PFOS. Differentially expressed proteins among all comparisons were further analyzed using Ingenuity Pathway Analysis (IPA)’s upstream analysis. Significant upstream regulatory molecules are described for PFOS treatment in each diet. For each comparison, an activation z-score was calculated to predict activation of upstream regulators. A positive z-score indicates upregulation and a negative z-score indicates downregulation.
Targeted Proteomic analysis.
After analyzing the total protein changes, peaks for specific targets were analyzed to compare protein expression to mRNA expression changes, as well as evaluate xenobiotic metabolism pathways changes identified in untargeted analysis. In the final analysis, 33 different targets from lipid catabolism, lipid transport and storage, lipid synthesis, redox, and xenobiotic metabolism and transport pathways are presented. SD and H-SD mice had similar protein expression levels (Fig. 7A–C and Fig. 8A–B). HFD upregulated 15 of the 33 proteins in pathways related to lipid metabolism, redox, and xenobiotic metabolism and transport pathways, and downregulated Scd-1, which in an enzyme involved in triglyceride synthesis. Carnitine palmitoyltransferase 1A (Cpt1a) protein expression was approximately doubled in HFD and HFD-PFOS mice compared to SD controls (Fig. 7A). Other proteins that were significantly induced that differed from mRNA expression levels are Hmgcs1, Fabp1, Fabp4, Slc27a2, Apoe, Cyp2b10, Aldh3a2, Oatp1a1, Gclc, and Ephx1.
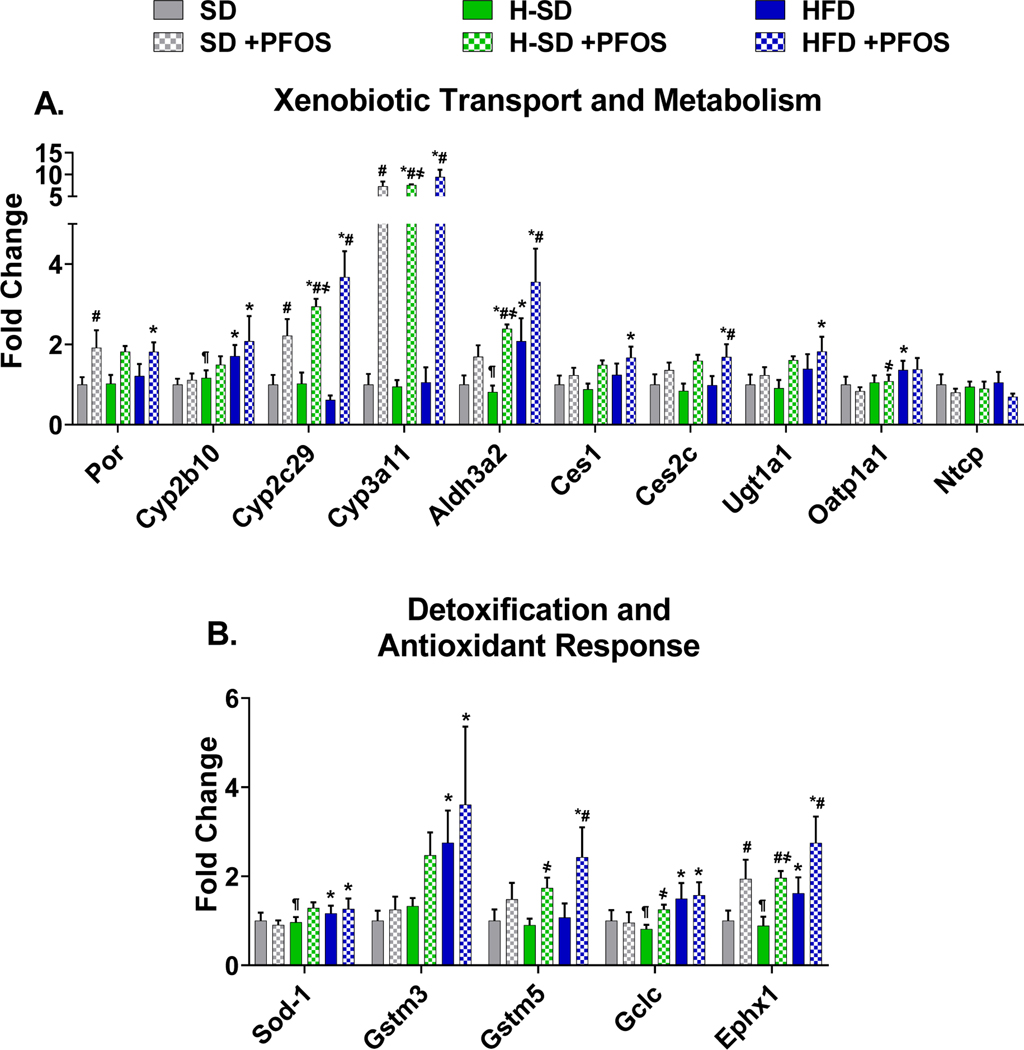
Fig. 7. Targeted proteomics analysis of lipid utilization gene.
Peaks for specific protein targets were analyzed using Skyline open source software. Areas for each fragment ion was summed for each peptide and one or two peptides were averaged for each protein. Areas were normalized to a technical standard for digestion (BSA) and final concentrations of digested proteins was calculated. Fold change was calculated between the control SD. A) PFOS induces several genes related to lipid catabolism genes. B) PFOS significantly reduced fatty acid synthase (Fas) protein expression in the SD diet, and C) HFD induced several genes related lipid transport and PFOS treatment induced very long chain acyl-CoA synthetase member 2 (Slc27a2). Calculations were done using a one-way ANOVA followed by Fisher’s LSD test. All values are means ± SEM; N = 4. * indicates p < 0.05 versus control SD, ¶ indicates p < 0.05 for H-SD versus HFD, # indicates p < 0.05 versus respective diet controls, † indicates p < 0.05 for H-SD + PFOS versus SD + PFOS, and ҂ indicates p < 0.05 for H-SD + PFOS versus HFD + PFOS.
Figure 8. Targeted proteomic analysis of xenobiotic metabolism and transport and oxidative stress genes.
Peaks for specific protein targets were analyzed using Skyline open source software. Areas for each fragment ion was summed for each peptide and one or two peptides were averaged for each protein. Areas were normalized to a technical standard for digestion and final concentrations of digested proteins. Fold change was calculated between the control SD. A) PFOS treatment induced genes related to xenobiotic metabolism. B) PFOS treatment induces expression of genes related to detoxification and antioxidant response. Calculations were done using a one-way ANOVA followed by Fisher’s LSD test. All values are means ± SEM; N = 4. * indicates p<0.05 versus control SD, ¶ indicates p<0.05 for H-SD versus HFD, # indicates p<0.05 versus respective diet controls, † indicates p<0.05 for H-SD+PFOS versus SD+PFOS, and ҂indicates p<0.05 for H-SD+PFOS versus HFD+PFOS.
PFOS treatment increased the expression of proteins involved in lipid metabolism (Fig. 7) and xenobiotic metabolism (Fig. 8) pathways. In Fig. 7A, PFOS increased several proteins (i.e. Cyp4a10, Cyp4a12a, Cyp4a14, Ehhadh, Acox1, Acsl1, and Acot2) related to lipid catabolism in all three diets, which matched observed changes in mRNA expression. SD-PFOS reduced fatty acid synthase (Fas) protein levels (Fig. 7B). In Fig. 7C, the PFOS effect on protein levels in lipid transport and storage pathways followed similar trends as mRNA levels, however fewer changes were significant, and many genes were not detectable in the analysis. PFOS treatment induced Slc27a2 expression the H-SD and HFD groups, by 121% and 36% respectively.
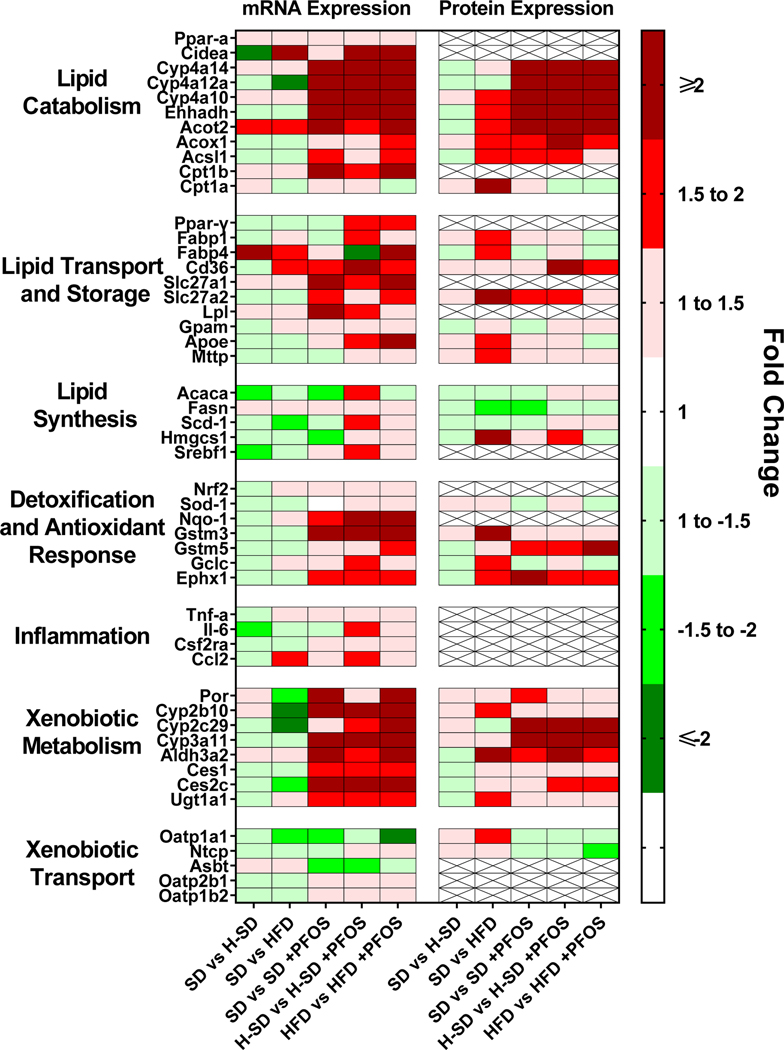
In Fig. 8A, PFOS administration increased liver Cyp3a11 and Cyp2c29 protein expression in all diet groups. SD-PFOS increased Por protein expression by 1.9-fold. Aldh3a2 protein expression in liver was induced in the H-SD and HFD treatment groups by 194% and 71% respectively. PFOS treatment did not significantly alter Cyp2b10, Ces1, Ces2c, and Ugt1a1 protein levels, however there are similar increasing trends with PFOS treatment, as with the mRNA expression level. Although not significant, PFOS treatment, also appears to reduce Oatp1a1 and Ntcp protein expression. As described in Fig. 8B, PFOS treatment in all three diets induced Ephx1 expression, and in the HFD group, PFOS treatment induced Gstm5 by 126%. Fig. 9 is a heat-map comparing gene expression and protein expression changes. Overall, the heat map demonstrates a good concordance between mRNA and protein expression patterns with the platforms used and suggests that many of the protein expression patterns observed are due to effects at the transcriptional level.
Figure 9. Comparison of mRNA and Protein Expression.
The quantification of selected liver mRNAs was performed using QuantiGene 2.0 Plex Assay kit with purified RNA. The fluorescence intensity (FI; minus background) for each gene was normalized to the housekeeping gene β-actin. Liver protein abundance was measured using a method for LC-QTOF/MS for sequential window acquisition of all theoretical mass spectra (SWATH-MS), and peptide peaks for each target were analyzed. Fold change was calculated between the control mice and H-SD and HFD mice to understand the effects of diet on hepatic gene and protein expression and between control mice and PFOS treatment for each diet to understand the effect of PFOS treatment on hepatic gene and protein expression. ―”X‖” indicates proteins that were not detected in proteomic analysis.Cd36 protein expression was included based on a western blot shown in Supplemental Fig. 3.
DISCUSSION
NAFLD is a spectrum disease that begins with the accumulation of lipids within hepatocytes. In the present study, a 60% kCal HFD was administered to induce obesity, glucose intolerance, and fatty liver in mice. After 4 weeks, PFOS was introduced into the diet, and half the mice that received HFD were switched to a standard chow diet to induce weight loss and improve liver steatosis. The rationale for switching to a standard diet is consistent with current AASLD guidance for hepatic steatosis, which recommends dietary modification (Chalasani et al. 2018). As expected, the HFD feeding increased body weight, liver weight, adipose mass, serum lipids, glucose levels, and liver lipids, which is consistent with previous findings (Lai et al. 2015; Sankaralingam et al. 2015). Additionally, the H-SD mice had less weight gain and had similar clinical parameters to the SD mice, suggesting that the dietary intervention was successful in returning HFD fed mice to control clinical parameters.
The purpose of this study was to evaluate whether PFOS administration would interfere with weight-loss induced improvement of NAFLD or augment HFD-induced steatosis. With the diet switch alone compared to the SD, the liver free fatty acid concentration was decreased as weight loss increases β-oxidation (van der Windt et al. 2018). When the H-SD group was combined with PFOS treatment, free fatty acid content was increased compared to the H-SD diet alone and had similar free fatty acids as the SD control livers. This observed increase in the free fatty acids, is supported by the observed upregulation of proteins in the lipid synthesis pathways in the H-SD and HFD with PFOS administration. Our findings suggest that there was resistance to weight-loss induced improvement of NAFLD. When PFOS was combined with HFD, there was an increase in liver triglyceride and cholesterol content, which illustrate worsened HFD-induced steatosis with PFOS treatment. In a study from Jain and Ducatman (2019), NHANES data from 2011 to 2014 was analyzed for associations between PFAS and liver biomarkers in obese and nonobese participants. Jain and Ducatman (2019) found stronger associations of PFAS with liver function biomarkers among obese participants, suggesting an obese population, already susceptible to NAFLD, faces more risk from the exposure to PFAS. The findings herein support this observation in humans and suggest that those predisposed to NAFLD, may have worsened liver lipid outcomes.
PFOS administration induced hepatomegaly among all treatment groups, which is consistent with other studies (Qazi et al. 2010; Wan et al. 2012). Previous work demonstrated that 10 mg/kg PFOS for 7 days in adult male mice increased hepatic lipid content in mice fed a standard diet (Das et al. 2017). Our findings were consistent with findings by Wang et al. (2014), which demonstrated that PFOS administration (5 or 20 mg/kg) for 14 days in combination with HFD induced liver lipid accumulation in 4–5-week-old male Balb/c mice. However, Huck et al. (2018), showed that a low dose (1 mg/kg) of PFOS was protective against HFD-induced hepatic steatosis in 8-week-old male C57BL/6J mice. The latter studies did not measure serum or hepatic PFOS content. Huck et al. (2018) began PFOS administration at the same time as the introduction of HFD, which differed from this work, suggesting the timing of diet treatment may also be critical to determine how PFOS affects the liver. These findings suggest dietary lipid content may affect PFOS uptake or distribution and further studies are warranted to better understand how PFOS enters and affects the liver.
Hepatic gene and protein expression were measured to determine the potential pathways targeted by PFOS within the liver. For the HFD, pathway analysis demonstrated that the 18 differentially expressed proteins corresponded to repression in fatty acid metabolism and lipid synthesis. This finding is consistent with Benard et al. (2016), in which HFD-feeding also suppressed lipid biosynthesis pathways in the mouse proteome. Specifically, the HFD decreased Scd-1 mRNA expression, which was one of the 18 differentially expressed proteins compared to the control diet. Gene expression in the H-SD mice had a significant increase in Fabp4, and decreases in Acaca, and Csf2ra expression, however, the H-SD diet had no detected differentially expressed proteins compared to SD control. The proteomic method used did not include pre-fractionation of the protein samples, which did limit the identification of lower abundant proteins in these samples, and there may be differences in protein targets that were not detected in the proteomic analysis.
Compared to the HFD, PFOS administration modulated the expression of numerous genes and proteins interrogated and including lipid utilization and xenobiotic metabolism genes. PFOS in combination with HFD also induced lipid synthesis. For the hepatic proteome, there were 17 proteins common to PFOS administration among the three diets, with some previously described by Rosen et al. (2013) as upregulated at the mRNA level, such as Cyp4a10, Cyp4a12, Cyp4a14, Ehhadh, Aldh3a2, Acsl1, Slc27a2, Ephx1, Cyp2b10, Cyp3a11, and Ces2c. Within each diet there were also proteins that were uniquely modulated. This suggests that diet may influence how PFOS modulates biological processes in the liver. In the pathway analysis, there were several unique pathway changes among each diet. PFOS administration upregulated lipid synthesis proteins only in the H-SD and HFD, which may explain why PFOS worsened hepatic steatosis in combination with the HFD. The upstream analysis revealed that PFOS treatment in all diets induced Ppar-α and Car signaling, which is consistent with previous studies (Rosen et al. 2008; Rosen et al. 2017). One of the limitations of this study is that rodents were used to model HFD response, and humans are thought to be less sensitive to PPAR-α activation (Bility et al. 2004; Wolf et al. 2008). However, upstream analysis also revealed several non-PPAR-α signals. PFOS treatment induced genes related to leptin signaling in the liver, which may be due to effects on adipokine secretion in the adipose tissue. This finding also may relate to human observations, as there have been an inverse association between PFOS exposure and leptin levels in children (Shelly et al. 2019). Our lab has also previously shown that PFOS can promote adipogenesis in murine-derived preadipocytes and human visceral preadipocytes (Xu et al. 2016). PFOS treatment also induced upstream regulators of mitochondrial function, AMPK, and Wnt signaling pathways. The results herein also reinforce the concept that PFOS induces more than one signaling pathway related to lipid and xenobiotic metabolism.
Although there was no inflammation observed in any of the livers, we observed a 1.2-fold increase in Il-6 mRNA expression with PFOS treatment in the H-SD group as compared to H-SD alone. PFOS has been described to alter inflammatory responses, production of cytokines, and immune responses as reviewed by DeWitt et al. (2012). The typical signaling pathway for Il-6 is based on activation of the NFkB pathway and release of TNF-α in macrophages. PFOS is thought to have anti-inflammatory effects as NFkB is negatively regulated by PPARα agonists like PFOS (Cunard et al. 2002; Andersen et al. 2008; DeWitt et al. 2009). As TNF-α mRNA expression was not induced, it is likely that the observed increase in Il-6 expression maybe due to an TNF-α independent mechanisms, such as the Ca2+/NFAT and glycogen/p38 MAPK pathways as reviewed by Pedersen and Febbraio (2008).
An additional limitation of the study is that mice possess significantly higher clearance mechanisms for PFOS that result in a significantly shorter half-life of weeks compared to years in humans (Olsen et al. 2007; Chang et al. 2012). The consequence of this difference in excretion often increases the PFOS dose used in mice and the NOAEL. For the study herein, we observed serum concentrations ranging from 24.4 to 50.3 μg/mL. The observed PFOS levels in humans are much lower overall with serum levels ranging from 9.8 to 54.6 ng/mL in the US (Calafat et al. 2007). Studies that have evaluated human liver levels of PFOS have found levels of PFOS ranging from 0.375 to 102 ng/g liver (Maestri et al. 2006; Pérez et al. 2013; Yeung et al. 2013). While the serum and liver concentrations we observed were much higher than that detected in human, the daily dose was in line with the reported mouse NOAEL for liver effects (US EPA 2016 May 5). In a study of perinatal PFOS treatment to dams at 0.3 mg/kg/day via oral gavage, dams had serum PFOS concentrations of 15.33 ± 4.62 μg/mL in serum and 40.9 ± 9.88 μg/g liver (Wan et al. 2014). This dose is considered to be a NOAEL dose for liver weight increase and increased insulin resistance of PFOS in mice (US EPA 2016 May 5). In the present study, the observed serum and liver concentrations of PFOS in the SD are higher with concentrations of 40.0 ± 6.9 μg/mL and 200.2 ± 22.5 μg/g liver, which, considering that the latter study used pregnant females that transferred PFOS to pups and the duration of treatment was half as long, the results are consistent with Wan et al. (2014).
An interesting observation was the difference in liver PFOS concentrations between diets. We observed that H-SD mice had much lower liver PFOS concentrations than SD mice, suggesting that diet might impact liver uptake mechanisms. HFD also decreased the serum to liver ratio from 5.2 to 3.3. These results may be explained by the downregulation of uptake transporter expression (i.e. Oatp1a1, 2b1, Ntcp) that has been observed in mouse models of hepatic steatosis (More and Slitt, 2011). Oatps and Ntcp have been described to transport PFOS (Zhao et al. 2017). We observed that HFD decreased Oatp1a1 and Ntcp mRNA expression in liver. We did not measure Bcrp or Abcc2 mRNA or protein expression to assess mechanisms that could enhance PFOS efflux from liver. Overall, the notion that diet or changes to liver with steatosis could impact PFOS uptake by liver has not been explored and warrants further investigation.
Overall, the results of this study demonstrated that PFOS administration had diet dependent effects in mice. This study also demonstrated that PFOS administration can exacerbate hepatic lipid accumulation in mice fed a high fat diet along with robust induction of lipid metabolism pathways at the mRNA and protein level. Liver steatosis and obesity may be an important risk factor PFAS related liver effects, and diet may influence transport of PFAS. Understanding the mechanisms related to diet and PFAS exposure, may help us understand the potential at risk populations from these ubiquitous toxicants and identify ways to intervene in PFAS toxicity.
Supplementary Material
Highlights:
PFOS exposure exacerbates hepatic steatosis in mice fed a high fat diet.
Proteomics reveals modulation of lipid utilization pathways with PFOS exposure
Dietary lipids influence how PFOS affects the liver in mice
Acknowledgements
The authors would like to thank the URI Laboratory Animal Care staff for assistance with monitoring the feed and health of the animals on study, and Cameron Picard, Foquia Mushtaq, Joanna Chang, Miriam Dash, and Kelsey Wright for assistance with blood glucose measurements and tissue collection. Andrew Descoteaux, Tom Dautaj, Maxine Abustan, James Cocozza, Jasmine Ayala, and Brittney Picard for assisting in the isolation of mRNA and colorimetric assays. This work was presented in part at the Gordon Research Conference and Seminar Cellular and Molecular Mechanisms of Toxicity in Summer 2017 (travel award to EM) and Society of Toxicology 2016, 2017, and 2018 annual meetings (travel awards to EM).
Funding
This work was supported by NIH grants R01ES016042, 1R15ES025404–01, and P42ES027706, the URI Council for Research to AS, as well as a URI Undergraduate Research Initiative (UGI) award for Scholarly, Creative and Artistic Projects grant to EM. EM was supported by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number 2 P20 GM103430. This material is based upon work conducted at a Rhode Island NSF EPSCoR research facility, Molecular Characterization Facility, supported in part by the National Science Foundation EPSCoR Cooperative Agreement # OIA-1655221. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations
- PFAS
per- and polyfluoroalkyl substance
- PFOS
Perfluorooctanesulfonaic acid
- NAFLD
nonalcoholic fatty liver disease
- HFD
high fat diet, SD, standard diet
- NASH
nonalcoholic steatohepatitis
- PPAR-α
peroxisome proliferator-activated receptor alpha
- LC-MS/MS
liquid chromatography with tandem mass spectrometry
- SWATH-MS
sequential window acquisition of all theoretical mass spectra
Footnotes
CRediT author statement
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Conflict of Interest
The authors of this manuscript declare no financial conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Al-Eryani L, Wahlang B, Falkner KC, Guardiola JJ, Clair HB, Prough RA, Cave M. 2015. Identification of Environmental Chemicals Associated with the Development of Toxicant-associated Fatty Liver Disease in Rodents. Toxicol Pathol. 43(4):482–497. doi: 10.1177/0192623314549960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen ME, Butenhoff JL, Chang S-C, Farrar DG, Kennedy GL, Lau C, Olsen GW, Seed J, Wallace KB. 2008. Perfluoroalkyl Acids and Related Chemistries—Toxicokinetics and Modes of Action. Toxicol Sci. 102(1):3–14. doi: 10.1093/toxsci/kfm270. [DOI] [PubMed] [Google Scholar]
- Bassler J, Ducatman A, Elliott M, Wen S, Wahlang B, Barnett J, Cave MC. 2019. Environmental perfluoroalkyl acid exposures are associated with liver disease characterized by apoptosis and altered serum adipocytokines. Environ Pollut. 247:1055–1063. doi: 10.1016/j.envpol.2019.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benard O, Lim J, Apontes P, Jing X, Angeletti RH, Chi Y. 2016. Impact of high-fat diet on the proteome of mouse liver. J Nutr Biochem. 31:10–19. doi: 10.1016/j.jnutbio.2015.12.012. [DOI] [PubMed] [Google Scholar]
- Bjork JA, Butenhoff JL, Wallace KB. 2011. Multiplicity of nuclear receptor activation by PFOA and PFOS in primary human and rodent hepatocytes. Toxicology. 288(1–3):8–17. doi: 10.1016/j.tox.2011.06.012. [DOI] [PubMed] [Google Scholar]
- Bogdanska J, Borg D, Sundström M, Bergstrom U, Halldin K, Abedi-Valugerdi M, Bergman Å, Nelson B, DePierre J, Nobel S. 2011. Tissue distribution of 35S-labelled perfluorooctane sulfonate in adult mice after oral exposure to a low environmentally relevant dose or a high experimental dose. Toxicology. 284(1–3):54–62. doi: 10.1016/j.tox.2011.03.014. [DOI] [PubMed] [Google Scholar]
- Boulanger B, Vargo J, Schnoor J, Hornbuckle KC. 2005. Evaluation of perfluorooctane surfactants in a wastewater treatment system and in a commercial surface protection product. Env Sci Technol. 1(39):5524–5530. [DOI] [PubMed] [Google Scholar]
- Butenhoff JL, Chang S-C, Olsen GW, Thomford PJ. 2012. Chronic dietary toxicity and carcinogenicity study with potassium perfluorooctanesulfonate in Sprague Dawley rats. Toxicology. 293(1–3):1–15. doi: 10.1016/j.tox.2012.01.003. [DOI] [PubMed] [Google Scholar]
- Calafat AM, Wong L-Y, Kuklenyik Z, Reidy JA, Needham LL. 2007. Polyfluoroalkyl chemicals in the U.S. population: data from the National Health and Nutrition Examination Survey (NhANES) 2003–2004 and comparisons with NHANES 1999–2000. Environ Health Perspect. 115(11):1596–1602. doi: 10.1289/ehp.10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalasani N, Younossi Z, Lavine JE, Charlton M, Cusi K, Rinella M, Harrison SA, Brunt EM, Sanyal AJ. 2018. The diagnosis and management of nonalcoholic fatty liver disease: Practice guidance from the American Association for the Study of Liver Diseases. Hepatology. 67(1):328–357. doi: 10.1002/hep.29367. [DOI] [PubMed] [Google Scholar]
- Chang S, Allen BC, Andres KL, Ehresman DJ, Falvo R, Provencher A, Olsen GW, Butenhoff JL. 2017. Evaluation of Serum Lipid, Thyroid, and Hepatic Clinical Chemistries in Association With Serum Perfluorooctanesulfonate (PFOS) in Cynomolgus Monkeys After Oral Dosing With Potassium PFOS. Toxicol Sci. 156(2):387–401. doi: 10.1093/toxsci/kfw267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S-C, Noker PE, Gorman GS, Gibson SJ, Hart JA, Ehresman DJ, Butenhoff JL. 2012. Comparative pharmacokinetics of perfluorooctanesulfonate (PFOS) in rats, mice, and monkeys. Reproductive Toxicology. 33(4):428–440. doi: 10.1016/j.reprotox.2011.07.002. [DOI] [PubMed] [Google Scholar]
- Cunard R, DiCampli D, Archer DC, Stevenson JL, Ricote M, Glass CK, Kelly CJ. 2002. WY14,643, a PPARa Ligand, Has Profound Effects on Immune Responses In Vivo. The Journal of Immunology. 169(12):6806–6812. doi: 10.4049/jimmunol.169.12.6806. [DOI] [PubMed] [Google Scholar]
- Darrow LA, Groth AC, Winquist A, Shin H-M, Bartell SM, Steenland K. 2016. Modeled Perfluorooctanoic Acid (PFOA) Exposure and Liver Function in a Mid-Ohio Valley Community. Environ Health Perspect. 124(8):1227–1233. doi: 10.1289/ehp.1510391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das KP, Wood CR, Lin MT, Starkov AA, Lau C, Wallace KB, Corton JC, Abbott BD. 2017. Perfluoroalkyl acids-induced liver steatosis: Effects on genes controlling lipid homeostasis. Toxicology. 378:37–52. doi: 10.1016/j.tox.2016.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWitt JC, Peden-Adams MM, Keller JM, Germolec DR. 2012. Immunotoxicity of Perfluorinated Compounds: Recent Developments. Toxicol Pathol. 40(2):300–311. doi: 10.1177/0192623311428473. [DOI] [PubMed] [Google Scholar]
- DeWitt JC, Shnyra A, Badr MZ, Loveless SE, Hoban D, Frame SR, Cunard R, Anderson SE, Meade BJ, Peden-Adams MM, et al. 2009. Immunotoxicity of perfluorooctanoic acid and perfluorooctane sulfonate and the role of peroxisome proliferator-activated receptor alpha. Crit Rev Toxicol. 39(1):76–94. doi: 10.1080/10408440802209804. [DOI] [PubMed] [Google Scholar]
- Estes C, Razavi H, Loomba R, Younossi Z, Sanyal AJ. 2018. Modeling the epidemic of nonalcoholic fatty liver disease demonstrates an exponential increase in burden of disease. Hepatol Baltim Md. 67(1):123–133. doi: 10.1002/hep.29466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folch J, Lees M, Stanley GHS. 1957. A Simple Method for the Isolation and Purification of Total Lipides from Animal Tissues. J Biol Chem. 226(1):497–509. [PubMed] [Google Scholar]
- Gallo V, Leonardi G, Genser B, Lopez-Espinosa M-J, Frisbee SJ, Karlsson L, Ducatman AM, Fletcher T. 2012. Serum perfluorooctanoate (PFOA) and perfluorooctane sulfonate (PFOS) concentrations and liver function biomarkers in a population with elevated PFOA exposure. Environ Health Perspect. 120(5):655–660. doi: 10.1289/ehp.1104436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George J, Pera N, Phung N, Leclercq I, Yun Hou J, Farrell G. 2003. Lipid peroxidation, stellate cell activation and hepatic fibrogenesis in a rat model of chronic steatohepatitis. J Hepatol. 39(5):756–764. [DOI] [PubMed] [Google Scholar]
- Gleason JA, Post GB, Fagliano JA. 2015. Associations of perfluorinated chemical serum concentrations and biomarkers of liver function and uric acid in the US population (NHANES), 2007–2010. Environ Res. 136:8–14. doi: 10.1016/j.envres.2014.10.004. [DOI] [PubMed] [Google Scholar]
- Hansen KJ, Clemen LA, Ellefson ME, Johnson HO. 2001. Compound-Specific, Quantitative Characterization of Organic Fluorochemicals in Biological Matrices. Environ Sci Technol. 35(4):766–770. doi: 10.1021/es001489z. [DOI] [PubMed] [Google Scholar]
- Huck I, Beggs K, Apte U. 2018. Paradoxical Protective Effect of Perfluorooctanesulfonic Acid Against High-Fat Diet-Induced Hepatic Steatosis in Mice. Int J Toxicol. 37(5):383–392. doi: 10.1177/1091581818790934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain RB, and Ducatman A. (2019). Selective Associations of Recent Low Concentrations of Perfluoroalkyl Substances With Liver Function Biomarkers: NHANES 2011 to 2014 Data on US Adults Aged ≥20 Years. Journal of Occupational and Environmental Medicine 61, 293–302. [DOI] [PubMed] [Google Scholar]
- Jamwal R, Barlock BJ, Adusumalli S, Ogasawara K, Simons BL, Akhlaghi F. 2017. Multiplex and Label-Free Relative Quantification Approach for Studying Protein Abundance of Drug Metabolizing Enzymes in Human Liver Microsomes Using SWATH-MS. J Proteome Res. 16(11 ):4134–4143. doi: 10.1021/acs.jproteome.7b00505. [DOI] [PubMed] [Google Scholar]
- Jian J-M, Chen D, Han F-J, Guo Y, Zeng L, Lu X, Wang F. 2018. A short review on human exposure to and tissue distribution of per- and polyfluoroalkyl substances (PFASs). Science of The Total Environment. 636:1058–1069. doi: 10.1016/j.scitotenv.2018.04.380. [DOI] [PubMed] [Google Scholar]
- Kim D, Kim WR, Kim HJ, Therneau TM. 2013. Association between noninvasive fibrosis markers and mortality among adults with nonalcoholic fatty liver disease in the United States. Hepatol Baltim Md. 57(4):1357–1365. doi: 10.1002/hep.26156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y-S, Chen W-C, Kuo T-C, Ho C-T, Kuo C-H, Tseng YJ, Lu K-H, Lin S-H, Panyod S, Sheen L-Y. 2015. Mass-Spectrometry-Based Serum Metabolomics of a C57BL/6J Mouse Model of High-Fat-Diet-Induced Non-alcoholic Fatty Liver Disease Development. J Agric Food Chem. 63(35):7873–7884. doi: 10.1021/acs.jafc.5b02830. [DOI] [PubMed] [Google Scholar]
- Lam B, Younossi ZM. 2010. Treatment options for nonalcoholic fatty liver disease. Ther Adv Gastroenterol. 3(2):121–137. doi: 10.1177/1756283X09359964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le MH, Devaki P, Ha NB, Jun DW, Te HS, Cheung RC, Nguyen MH. 2017. Prevalence of non-alcoholic fatty liver disease and risk factors for advanced fibrosis and mortality in the United States. PloS One. 12(3):e0173499. doi: 10.1371/journal.pone.0173499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Clark J, Diehl AM. 2002. The liver in obesity and type 2 diabetes mellitus. Clin Liver Dis. 6(4):867–877. doi: 10.1016/S1089-3261(02)00060-0. [DOI] [PubMed] [Google Scholar]
- Lin C-Y, Lin L-Y, Chiang C-K, Wang W-J, Su Y-N, Hung K-Y, Chen P-C. 2010. Investigation of the associations between low-dose serum perfluorinated chemicals and liver enzymes in US adults. Am J Gastroenterol. 105(6):1354–1363. doi: 10.1038/ajg.2009.707. [DOI] [PubMed] [Google Scholar]
- Maestri L, Negri S, Ferrari M, Ghittori S, Fabris F, Danesino P, Imbriani M. 2006. Determination of perfluorooctanoic acid and perfluorooctanesulfonate in human tissues by liquid chromatography/single quadrupole mass spectrometry. Rapid Commun Mass Spectrom. 20(18):2728–2734. doi: 10.1002/rcm.2661. [DOI] [PubMed] [Google Scholar]
- Marchesini G, Bugianesi E, Forlani G, Cerrelli F, Lenzi M, Manini R, Natale S, Vanni E, Villanova N, Melchionda N, et al. 2003. Nonalcoholic fatty liver, steatohepatitis, and the metabolic syndrome. Hepatol Baltim Md. 37(4):917–923. doi: 10.1053/jhep.2003.50161. [DOI] [PubMed] [Google Scholar]
- Masarone M, Federico A, Abenavoli L, Loguercio C, Persico M. 2014. Non alcoholic fatty liver: epidemiology and natural history. Rev Recent Clin Trials. 9(3):126–133. [DOI] [PubMed] [Google Scholar]
- Matteoni CA, Younossi ZM, Gramlich T, Boparai N, Liu YC, McCullough AJ. 1999. Nonalcoholic fatty liver disease: a spectrum of clinical and pathological severity. Gastroenterology. 116(6):1413–1419. [DOI] [PubMed] [Google Scholar]
- More VR, Slitt AL. 2011. Alteration of Hepatic but Not Renal Transporter Expression in Diet-Induced Obese Mice. Drug Metab Dispos. 39(6):992–999. doi: 10.1124/dmd.110.037507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen AS, Letcher RJ, Cangialosi MV, Chu S, Arukwe A. 2011. Tissue bioaccumulation patterns, xenobiotic biotransformation and steroid hormone levels in Atlantic salmon (Salmo salar) fed a diet containing perfluoroactane sulfonic or perfluorooctane carboxylic acids. Chemosphere. 83(8):1035–1044. doi: 10.1016/j.chemosphere.2011.01.067. [DOI] [PubMed] [Google Scholar]
- Nagle CA, Klett EL, Coleman RA. 2009. Hepatic triacylglycerol accumulation and insulin resistance. J Lipid Res. 50(Suppl):S74–S79. doi: 10.1194/jlr.R800053-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen GW, Burris JM, Ehresman DJ, Froehlich JW, Seacat AM, Butenhoff JL, Zobel LR. 2007. Half-Life of Serum Elimination of Perfluorooctanesulfonate, Perfluorohexanesulfonate, and Perfluorooctanoate in Retired Fluorochemical Production Workers. Environ Health Perspect. 115(9):1298–1305. doi: 10.1289/ehp.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer CN, Hsu MH, Griffin KJ, Raucy JL, Johnson EF. 1998. Peroxisome proliferator activated receptor-alpha expression in human liver. Mol Pharmacol. 53(1):14–22. [PubMed] [Google Scholar]
- Pérez F, Nadal M, Navarro-Ortega A, Fabrega F, Domingo JL, Barceló D, Farré M. 2013. Accumulation of perfluoroalkyl substances in human tissues. Environment International. 59:354–362. doi: 10.1016/j.envint.2013.06.004. [DOI] [PubMed] [Google Scholar]
- Pedersen BK, Febbraio MA. 2008. Muscle as an Endocrine Organ: Focus on Muscle-Derived Interleukin-6. Physiological Reviews. 88(4):1379–1406. doi: 10.1152/physrev.90100.2007. [DOI] [PubMed] [Google Scholar]
- Petta S, Gastaldelli A, Rebelos E, Bugianesi E, Messa P, Miele L, Svegliati-Baroni G, Valenti L, Bonino F. 2016. Pathophysiology of Non Alcoholic Fatty Liver Disease. Int J Mol Sci. 17(12). doi: 10.3390/ijms17122082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qazi MR, Abedi MR, Nelson BD, DePierre JW, Abedi-Valugerdi M. 2010. Dietary exposure to perfluorooctanoate or perfluorooctane sulfonate induces hypertrophy in centrilobular hepatocytes and alters the hepatic immune status in mice. Int Immunopharmacol. 10(11):1420–1427. doi: 10.1016/j.intimp.2010.08.009. [DOI] [PubMed] [Google Scholar]
- Rosen MB, Abbott BD, Wolf DC, Corton JC, Wood CR, Schmid JE, Das KP, Zehr RD, Blair ET, Lau C. 2008. Gene Profiling in the Livers of Wild-type and PPARα-Null Mice Exposed to Perfluorooctanoic Acid. Toxicol Pathol. 36(4):592–607. doi: 10.1177/0192623308318208. [DOI] [PubMed] [Google Scholar]
- Rosen MB, Das KP, Rooney J, Abbott B, Lau C, Corton JC. 2017. PPARα-independent transcriptional targets of perfluoroalkyl acids revealed by transcript profiling. Toxicology. 387:95–107. doi: 10.1016/j.tox.2017.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen MB, Das KP, Wood CR, Wolf CJ, Abbott BD, Lau C. 2013Evaluation of Perfluoroalkyl Acid Activity Using Primary Mouse and Human Hepatocytes. Toxicology, 308, 129–137. doi: 10.1016/j.tox.2013.03.011. [DOI] [PubMed] [Google Scholar]
- Rosen MB, Schmid JR, Corton JC, Zehr RD, Das KP, Abbott BD, Lau C. 2010. Gene Expression Profiling in Wild-Type and PPARa-Null Mice Exposed to Perfluorooctane Sulfonate Reveals PPARa-Independent Effects. PPAR Research. doi: 10.1155/2010/794739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaralingam S, Alrob OA, Zhang L, Jaswal JS, Wagg CS, Fukushima A, Padwal RS, Johnstone DE, Sharma AM, Lopaschuk GD. 2015. Lowering Body Weight in Obese Mice With Diastolic Heart Failure Improves Cardiac Insulin Sensitivity and Function: Implications for the Obesity Paradox. Diabetes. 64(5):1643–1657. doi: 10.2337/db14-1050. [DOI] [PubMed] [Google Scholar]
- Seacat AM, Thomford PJ, Hansen KJ, Clemen LA, Eldridge SR, Elcombe CR, Butenhoff JL. 2003. Sub-chronic dietary toxicity of potassium perfluorooctanesulfonate in rats. Toxicology. 183(1–3):117–131. doi: 10.1016/S0300-483X(02)00511-5. [DOI] [PubMed] [Google Scholar]
- Seacat AM, Thomford PJ, Hansen KJ, Olsen GW, Case MT, Butenhoff JL. 2002. Subchronic toxicity studies on perfluorooctanesulfonate potassium salt in cynomolgus monkeys. Toxicol Sci. 68(1):249–264. [DOI] [PubMed] [Google Scholar]
- Shelly C, Grandjean P, Oulhote Y, Plomgaard P, Frikke-Schmidt R, Nielsen F, Zmirou-Navier D, Weihe P, Valvi D. 2019. Early Life Exposures to Perfluoroalkyl Substances in Relation to Adipokine Hormone Levels at Birth and During Childhood. J Clin Endocrinol Metab. 104(11):5338–5348. doi: 10.1210/jc.2019-00385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoeib M, Harner T, Wilford BH, Jones KC, Zhu J. 2005. Perfluorinated sulfonamides in indoor and outdoor air and indoor dust: occurrence, partitioning, and human exposure. Environ Sci Technol. 39(17):6599–6606. [DOI] [PubMed] [Google Scholar]
- Skutlarek D, Exner M, Färber H. 2006. Perfluorinated surfactants in surface and drinking waters. Environ Sci Pollut Res Int. 13(5):299–307. [DOI] [PubMed] [Google Scholar]
- Takacs ML, Abbott BD. 2007. Activation of mouse and human peroxisome proliferator-activated receptors (alpha, beta/delta, gamma) by perfluorooctanoic acid and perfluorooctane sulfonate. Toxicol Sci. 95(1): 108–117. doi: 10.1093/toxsci/kfl135. [DOI] [PubMed] [Google Scholar]
- Tan X, Xie G, Sun Xiuhua, Li Q, Zhong W, Qiao P, Sun Xinguo, Jia W, Zhou Z 2013. High Fat Diet Feeding Exaggerates Perfluorooctanoic Acid-Induced Liver Injury in Mice via Modulating Multiple Metabolic Pathways. PLoS ONE. 8(4) doi: 10.1371/journal.pone.0061409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tana C, Ballestri S, Ricci F, Di Vincenzo A, Ticinesi A, Gallina S, Giamberardino MA, Cipollone F, Sutton R, Vettor R, et al. 2019. Cardiovascular Risk in Non-Alcoholic Fatty Liver Disease: Mechanisms and Therapeutic Implications. Int J Environ Res Public Health. 16(17). doi: 10.3390/ijerph16173104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova S, Temu T, Cox J. 2016. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc. 11 (12):2301–2319. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- United States Environmental Protection Agency, Chemistry Dashboard | PFASMASTER Chemicals. [accessed 2020 Apr 23]. https://comptox.epa.gov/dashboard/chemical_lists/pfasmaster.
- United States Environmental Protection Agency. 2016May5. Drinking Water Health Advisories for PFOA and PFOS. US EPA. [accessed 2019 May 21]. https://www.epa.gov/ground-water-and-drinking-water/drinking-water-health-advisories-pfoa-and-pfos. [Google Scholar]
- van der Windt DJ, Sud V, Zhang H, Tsung A, Huang H. 2018. The Effects of Physical Exercise on Fatty Liver Disease. Gene Expr. 18(2):89–101. doi: 10.3727/105221617X15124844266408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaíno JA, Côté RG, Csordas A, Dianes JA, Fabregat A, Foster JM, Griss J, Alpi E, Birim M, Contell J, et al. 2013. The Proteomics Identifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41(Database issue):D1063–D1069. doi: 10.1093/nar/gks1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan HT, Zhao YG, Leung PY, Wong CKC. 2014. Perinatal Exposure to Perfluorooctane Sulfonate Affects Glucose Metabolism in Adult Offspring. PLOS ONE. 9(1):e87137. doi: 10.1371/journal.pone.0087137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan HT, Zhao YG, Wei X, Hui KY, Giesy JP, Wong CKC. 2012. PFOS-induced hepatic steatosis, the mechanistic actions on p-oxidation and lipid transport. Biochim Biophys Acta. 1820(7):1092–1101. doi: 10.1016/j.bbagen.2012.03.010. [DOI] [PubMed] [Google Scholar]
- Wang L, Wang Y, Liang Y, Li J, Liu Y, Zhang J, Zhang A, Fu J, Jiang G. 2014. PFOS induced lipid metabolism disturbances in BALB/c mice through inhibition of low density lipoproteins excretion. Sci Rep. 4:4582. doi: 10.1038/srep04582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, DeWitt JC, Higgins CP, Cousins IT. 2017. A Never-Ending Story of Per- and Polyfluoroalkyl Substances (PFASs)? Environ Sci Technol. 51(5):2508–2518. doi: 10.1021/acs.est.6b04806. [DOI] [PubMed] [Google Scholar]
- Xu J, Shimpi P, Armstrong L, Salter D, Slitt AL. 2016. PFOS induces adipogenesis and glucose uptake in association with activation of Nrf2 signaling pathway. Toxicol Appl Pharmacol. 290:21–30. doi: 10.1016/j.taap.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung LWY, Guruge KS, Taniyasu S, Yamashita N, Angus PW, Herath CB. 2013. Profiles of perfluoroalkyl substances in the liver and serum of patients with liver cancer and cirrhosis in Australia. Ecotoxicology and Environmental Safety. 96:139–146. doi: 10.1016/j.ecoenv.2013.06.006. [DOI] [PubMed] [Google Scholar]
- Zhao W, Zitzow JD, Weaver Y, Ehresman DJ, Chang S- C, Butenhoff JL, Hagenbuch B. 2017. Organic Anion Transporting Polypeptides Contribute to the Disposition of Perfluoroalkyl Acids in Humans and Rats. Toxicol Sci. 156(1):84–95. doi: 10.1093/toxsci/kfw236. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.