Abstract
Members of the ATP-binding cassette (ABC) transporter superfamily translocate a broad spectrum of chemically diverse substrates. While their eponymous ATP-binding cassette in the nucleotide-binding domains (NBDs) is highly conserved, their transmembrane domains (TMDs) forming the translocation pathway exhibit distinct folds and topologies, suggesting that during evolution the ancient motor domains were combined with different transmembrane mechanical systems to orchestrate a variety of cellular processes. In recent years, it has become increasingly evident that the distinct TMD folds are best suited to categorize the multitude of ABC transporters. We therefore propose a new ABC transporter classification that is based on structural homology in the TMDs.
Keywords: ABC transporters, ATPases, cryo-EM, membrane proteins, molecular machines, phylogeny, primary active transporters, sequence alignment, structural biology, X-ray crystallography
We suggest a new classification of the ABC transporter superfamily that is based on the TMD fold. Historically, first hints of the ABC protein superfamily came from sequence alignments of bacterial proteins that revealed highly conserved motifs in their ATPase domains [1]. The superfamily of ABC proteins was subsequently divided into three main classes [2-4]: exporters, nontransporter ABC proteins, and a third class consisting primarily of importers. The mammalian ABC systems, in particular, were grouped into seven subfamilies (ABCA to ABCG), based on NBD and TMD sequence homology, gene structure, and domain order [5-7]. It should be noted that ABCE and ABCF are not transporters, but exist as twin-NBDs without TMDs and are involved in mRNA translation control [8]. Detailed membrane topology and sequence analyses of exporters uncovered that, in contrast to the NBDs, the TMDs are polyphyletic and can serve as references to categorize ABC transporters into three distinct types (ABC1-3) [9,10]. According to this classification, the cystic fibrosis transmembrane conductance regulator (CFTR), the transporter associated with antigen processing (TAP), and the drug efflux pump P-glycoprotein (P-gp) belong to the ABC1 transporters; ABCG2 and ABCG5/G8 are members of the ABC2 group, which also comprises importers; and the macrolide translocator MacB is categorized as an ABC3 system. Yet, another classification scheme currently in use differentiates between the three types of importers predominantly found in prokaryotes [11-14] and two types of exporters, exemplified by Sav1866 [15] and ABCG5/8 [16], in addition to the LptB2FG-type [17,18] and MacB-type [19-22] transporters.
Our motivation for proposing a revised nomenclature stems from the recent wealth of ABC transporter structures determined by X-ray crystallography and single-particle cryo-electron microscopy, which has unveiled a remarkable diversity of TMD folds and evolutionary relationships between bacterial and eukaryotic/mammalian transporters [16-21,23-26]. This affluence of structural information provides the opportunity to introduce a universal nomenclature that combines previous phylogenetic analyses with the new findings coming from high-resolution structures. The nomenclature groups ABC transporters into distinct types, I–VII, based on their TMD fold (Fig. 1, Tables 1 and 2). This classification is supported by quantitative analyses using TM-scores based on pairwise structural alignment of TMDs (Tables S1-S6, Fig. S1). The classification focuses on the transporter-forming TMDs and does not consider additional membrane integrated domains, as for example observed in TAP1/TAP2 [27,28].
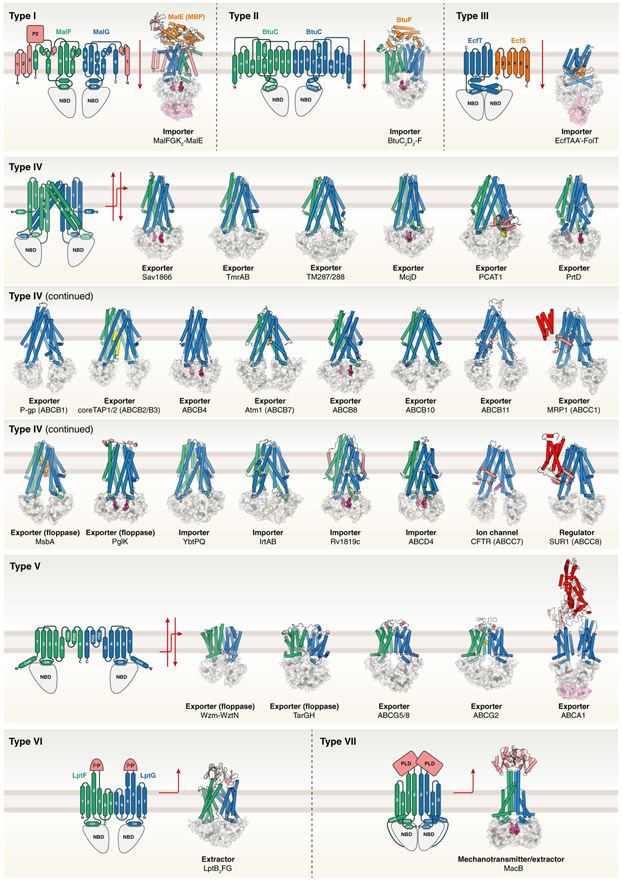
Fig. 1.
The different types within the ABC transporter superfamily. Members of the superfamily of ABC transporters can be grouped into distinct types based on their TMD fold. The TMDs of representative experimentally determined structures are depicted as cartoons, and their NBDs are shown in surface representation. The TMD architecture of the first structure of each type is illustrated by a topology diagram. The number of structures shown for each transporter type does not necessarily reflect the abundance or importance of the respective type, but highlights the common scaffold and functional diversity of the transporters. The two TMDs of each transporter are shown in green and blue, respectively, except for cases where the TMDs are part of the same polypeptide chain (uniform blue color). Please note that the type V ABC transporters also include the retina-specific importer ABCA4 and importers in plants. Substrate-binding components of type I-III folds are illustrated in orange, and auxiliary domains and additional (TM) helices are shown in red, salmon, and violet, respectively. Bound (occluded) nucleotides and Mg2+ ions in the NBDs are shown as dark pink spheres. Transported substrates and inhibitors are shown in yellow (carbon) and in CPK colors (remaining atoms in small-molecule compounds), respectively. The directions of substrate transport are indicated by solid and dashed red arrows. The structures have the following Protein Data Bank (PDB) accession codes: MalFGK2-MalE: 2R6G [12]; BtuC2D2-BtuF: 4FI3 [50]; EcfTAA′-FolT: 4HUQ [14]; Sav1866: 2HYD [15]; TmrAB: 5MKK [51]; TM287/288: 4Q4H [52]; McjD: 4PL0 [53]; PCAT1: 6V9Z [54]; Atm1: 4MYH [55]; MRP1: 5UJA [56]; PrtD: 5L22 [57]; P-gp: 4M1M [58]; TAP1/2: 5U1D [59]; ABCB4: 6S7P [60]; ABCB8: 5OCH; ABCB10: 3ZDQ [61]; ABCB11: 6LR0 [62]; MsbA: 5TV4 [63]; PglK: 6HRC [64]; YbtPQ: 6P6J [31]; IrtAB: 6TEJ [32]; Rv1819c: 6TQF [33]; ABCD4: 6JBJ [30]; CFTR: 5UAK [65]; SUR1: 6BAA [66]; Wzm-WztN: 6OIH [25]; TarGH: 6JBH [26]; ABCG5/8: 5DO7 [16]; ABCG2: 6HCO [67]; ABCA1: 5XJY [23]; LptB2FG: 5X5Y [17]; MacB: 5LJ7 [21]. ABC, ATP-binding cassette; β-jr, β-jellyroll-like domain; C, C terminus; CH, coupling helix; CoH, connecting helix; EH, elbow helix; N, N terminus; NBD, nucleotide-binding domain; P2, extracytoplasmic loop; PG, periplasmic gate helix; PLD, periplasmic domain; TMD, transmembrane domain.
Table 1.
Prokaryotic ABC transporters classified according to their TMD folds.
| TMD fold | TM helix organization |
Experimentally determined structures |
PDB codesa | Function |
|---|---|---|---|---|
| Type I | (5-6) + (5-6/8)b | MalFGK2(-E) | 2R6G, 3FH6, 3PUV, 3PUW, 3PUX, 3RLF, 4JBW | Maltose import |
| ModB2C2(-A) | 2ONK, 3D31 | Molybdate import | ||
| MetNI(-Q) | 3DHW, 3TUI, 3TUJ, 3TUZ, 6CVL | Methionine import | ||
| Art(QN)2 | 4YMS, 4YMT, 4YMU, 4YMV, 4YMW | Amino acid import | ||
| AlgM1M2SS-Q2 | 4TQU | Alginate import | ||
| Type II | 10 + 10 | BtuC2D2(-F) | 1L7V, 2QI9, 4DBL, 4FI3, 4R9U | Cobalamin import |
| MolBC | 2NQ2 | Import of molybdate and tungstate | ||
| HmuUV | 4G1U | Heme import | ||
| BhuUV(-T) | 5B57, 5B58 | Heme import | ||
| Type III | 4-8 (T) + 6-7 (S) | EcfTAA′-Fo1T | 4HUQ, 5D3M, 5JSZ | Folate import |
| EcfTAA′-PdxU2 | 4HZU | Pyridoxine import | ||
| LbECF-PanT | 4RFS | Pantothenate import | ||
| CbiMQO | 5X3X, 5X41 | Co2+ import | ||
| ECF-CbrT | 6FNP | Cobalamin import | ||
| Type IV | 6 + 6 Homodimer Heterodimer Single chain |
Sav1866 | 2HYD, 2ONJ | Multidrug export |
| MsbA | 3B60, 3B5Y, 3B5Z, 5TV4, 6BPL, 6BPP, 6BL6, 6O30, 6UZ2, 6UZL | Lipid A/LPS flopping | ||
| NaAtm1 | 4MRR, 4MRS, 4MRV, 4MRN, 4MRP | Export of GSH, GSH-related compounds, and metal-GSH complexes | ||
| TM287/288 | 4Q4A, 4Q4H, 4Q4J, 6QUZ, 6QV0, 6QV1, 6QV2 | Daunorubicin export | ||
| McjD | 4PL0, 5EG1, 5OFR | Antimicrobial peptide export | ||
| PCAT1 | 4RY2, 6V9Z | Polypeptide export | ||
| PglK | 5C76, 5C78, 5NBD, 6HRC | Export (flopping) of lipid-linked oligosaccharides | ||
| TmrAB | 5MKK, 6RAF, 6RAG, 6RAH, 6RAI, 6RAJ, 6RAK, 6RAL, 6RAM, 6RAN | Peptide export | ||
| PrtD | 5L22 | Polypeptide type-1 secretion system | ||
| YbtPQ | 6P6I, 6P6J | Metal–siderophore import | ||
| Rv1819c | 6TQE, 6TQF | Import of cobalamin and bleomycin | ||
| IrtAB | 6TEJ | Iron–siderophore import | ||
| Type V | 6 + 6 Homodimer Heterodimer Single chain |
Wzm-WztN TarGH |
6OIH, 6M96 | O-antigen export (flopping) |
| TarGH | 6JBH | Export (flopping) of wall teichoic acid | ||
| Type VI | 6 + 6 Heterodimer |
LptB2FG(C) | 5X5Y, 5L75, 6MIT, 6MJP, 6MHU, 6MHZ, 6MI7, 6MI8, 6S8G, 6S8H, 6S8N |
LPS extraction |
| Type VII | 4 + 4 | MacB | 5GKO, 5WS4, 5LIL, 5LJ6, 5LJ7, 5XU1 | Export of macrolides and polypeptide virulence factors |
GSH, glutathione; LPS, lipopolysaccharide.
Only PDB codes of structures with an overall resolution equal to or better than 4.5 Å were included.
Conserved TMs in bold.
Table 2.
Eukaryotic ABC transporters classified according to their TMD foldsa.
| TMD fold | TM helix organization |
Experimentally determined structures |
PDB codesb | Function |
|---|---|---|---|---|
| Type IV | 6 + 6 Homodimer Heterodimer Single chain |
ABCB subfamily | ||
| P-gp (ABCB1) | 4F4C, 4M1M, 4M2S, 4M2T, 4Q9H, 4Q9I, 4Q9J, 4Q9K, 4Q9L, 4XWK, 5KPD, 5KPI, 5KPJ, 5KO2, 5KOY, 6C0V | Multidrug export | ||
| CmABCB1 | 3WME, 3WMF, 3WMG, 6A6M, 6A6N | Multidrug export | ||
| ScAtm1 (ABCB7) | 4MYC, 4MYH | Unknown substrate for Fe/S protein biogenesis | ||
| TAP1/2 (ABCB2/3) | 5U1D | Peptide export | ||
| ABCB4 | 6S7P | Lipid export | ||
| ABCB8 | 5OCH | Unknown | ||
| ABCB10 | 3ZDQ, 4AYT, 4AYW, 4AYX | Unknown | ||
| ABCB11 | 6LR0 | Bile salt export | ||
| ABCC subfamily | ||||
| MRP1 (ABCC1) | 5UJA, 5UJ9, 6BHU, 6UY0 | Leukotriene, sphingolipid, and multidrug export | ||
| CFTR (ABCC7) | 5UAR, 5UAK, 5W81, 6D3R, 6MSM, 6O1V, 6O2P | Chloride channel | ||
| SUR1 (ABCC8) | 6BAA, 6C3O, 5YKE, 5YKF, 5YWC, 5YWD, 5YW7, 5YW8, 6JB1, 6JB3, 6PZ9,6PZA, 6PZC, 6PZI | Regulatory module of KATP channel | ||
| ABCD subfamily | ||||
| ABCD4 | 6JBJ | Cobalamin import | ||
| Type V | 6 + 6 Homodimer Heterodimer Single chain |
ABCA subfamily | ||
| ABCA1 | 5XJY | Phospholipid/cholesterol export | ||
| ABCG subfamily | ||||
| ABCG5/8 | 5DO7 | Sterol export | ||
| ABCG2 | 5NJG, 5NJ3, 6ETI, 6FEQ, 6FFC, 6HIJ, 6HCO, 6HBU, 6HZM, 6VXF, 6VXH, 6VXI, 6VXJ | Multidrug export |
Excluding ABC proteins of the ABCH and ABCI subfamilies, which most likely can be classified as type V and type III systems, respectively.
Only PDB codes of structures with an overall resolution equal to or better than 4.5 Å were included.
As before, types I-III of the new nomenclature cover the three different importer architectures (Fig. 1, Table 1, Tables S2 and S3; TM-score for pairwise structural alignment between the type III systems CbiQ (PDB code 5X3X) and EcfT from Lactobacillus brevis (PDB code 4HUQ): 0.736). It is noteworthy that prokaryotic importers typically operate with periplasmic, extracellular, or membrane-embedded substrate-binding proteins whose structural features correlate with the type of TMD fold [29].
Based on the characteristic structure of the founding member Sav1866, which includes a domain-swapped TMD arrangement, type IV members of the new nomenclature have previously been classified as type I ABC exporters [15]. However, a significant and growing number of these ABC proteins have nonexporter functions, i.e., the gated chloride channel CFTR, the regulatory KATP channel modules SUR1/2, the lysosomal cobalamin (vitamin B12) transporter ABCD4 [30], the bacterial siderophore importers YbtPQ and IrtAB, and the cobalamin/antimicrobial peptide importer Rv1819c [31-33], as well as several type IV systems with importer functions in plants [34-39]. This striking functional diversity mediated by the same structural framework (Fig. 1, Tables 1 and 2, Tables S4 and S5) makes the type IV ABC transporters stand out and is also the main reason why we suggest the more universal taxonomy based on structural principles.
According to the new classification, type V systems are ABC transporters of the ABCG/ABCA/Wzm type (Fig. 1, Tables 1 and 2, Table S6). They include channel-forming biopolymer secretion systems in bacteria [25,26]. Remarkably, although many type V systems are exporters, this type also comprises transporters with import function, including the retina-specific importer (flippase) ABCA4 (rim protein) [40,41] and importers in plants [42-44].
Finally, LptB2FG and MacB are the founding members of type VI and type VII ABC transporters, respectively. We are aware that LptF and LptG have TMD folds that resemble type V members, and the TMD of MacB is reminiscent of type V systems and LptF/G. Yet, they exhibit distinct features that warrant classifications into separate groups. These include the lack of an amphipathic N-terminal ‘elbow helix’ and no extracellular reentrant helices between TM5 and TM6. In addition, MacB contains only four proper TM helices as well as an additional coupling helix, thereby defining a separate transporter architecture. In accordance with differences in TMD topologies, the LptFG and MacB transporters also display diverging dimerization interfaces. Thus, we have chosen to assign LptFG and MacB to separate types. This notion is corroborated by the TM-score-based quantitative analysis (Table S6 and Fig. S1). Of note, at the time of writing, publicly available, yet unpublished structures of the lipid transporter complex MlaFEDB of Gram-negative bacteria reveal some resemblance of MlaE to LptF/G and MacB. However, the number of TM helices differs between LptFG (six TM helices), MlaE (five TM helices), and MacB (four TM helices) [45-48] (Table S6 and Fig. S1).
We would like to point out that the classification of the mammalian ABC transporters into the ABCA-G subfamilies can be maintained as subcategories of type IV (subfamilies B–D) and type V (subfamilies A and G) within the new nomenclature (Table 2). We are also not proposing any changes to gene symbols. Most importantly, the new nomenclature based on TMD architecture can be universally applied to ABC transporters beyond their particular physiological functions and across the three domains of life. Hence, it allows any newly discovered transporter fold to be compared with the existing types and seamlessly incorporated into the classification scheme, possibly as a new type. Since the new nomenclature depends on TMD architecture, it requires structural information in order to classify new transporter systems. At the same time, we regard the nomenclature as a dynamic platform that can be upgraded, adjusted, or refined whenever necessary due to novel insights that add extra dimensions to our understanding of ABC systems.
The recent advances in structural mapping of the diverse superfamily of ABC transporters have revealed a vast area of mechanistically uncharted territory. One key objective of future research should be to fully comprehend how type IV systems perform so many different functions, i.e., as importer, exporter, lipid floppase, ion channel, and regulator, by employing a single structural scaffold. However, we do not exclude that other types might turn out to be as functionally diverse as type IV systems. Exploring the different modes of operation and accompanying conformational landscapes [49] and the dynamics of the multifarious ABC systems will require integrative experimental approaches that include electron paramagnetic resonance (EPR), nuclear magnetic resonance (NMR), single-molecule techniques, and single-turnover experiments. We are confident that future studies of such kind will provide major new insights into the mechanisms of these fascinating molecular machines.
Supplementary Material
Table S1. TM-scores based on pairwise structural alignment of representatives of the different TMD types.
Table S2. TM-scores based on pairwise structural alignment of type I TMDs.
Table S3. TM-scores based on pairwise structural alignment of type II TMDs.
Table S4. TM-scores based on pairwise structural alignment of type IV TMDs in inward-facing conformations.
Table S5. TM-scores based on pairwise structural alignment of type IV TMDs in (semi-) occluded/outward-facing conformations.
Table S6. TM-scores based on pairwise structural alignment of type V, VI, and VII TMDsa.
Acknowledgements
K.B. acknowledges support by a grant of the Medical Research Council (MR/N020103/1). M.D. is supported in part by the Intramural Program of the NIH. V.K. acknowledges support by the Medical Research Council (MR/N000994/1) and Wellcome Trust (101828/Z/13/Z). R.L. acknowledges generous financial support from German Research Foundation (LI 415/5). D.P.T. is supported in part by the Canada Research Chairs program. This work was supported by the German Research Foundation (SFB 807 and TA157/12-1 (Reinhart Koselleck Award Program) to R.T.).
Abbreviations
- ABC
ATP-binding cassette
- cryo-EM
cryogenic electron microscopy
- NBD
nucleotide-binding domain
- TMD
transmembrane domain
Footnotes
Supporting information
Additional supporting information may be found online in the Supporting Information section at the end of the article.
References
- 1.Higgins CF, Hiles ID, Salmond GPC, Gill DR, Downie JA, Evans IJ, Holland IB, Gray L, Buckel SD, Bell AW et al. (1986) A family of related ATP-binding subunits coupled to many distinct biological processes in bacteria. Nature 323, 448–450. [DOI] [PubMed] [Google Scholar]
- 2.Dassa E and Bouige P (2001) The ABC of ABCS: a phylogenetic and functional classification of ABC systems in living organisms. Res Microbiol 152, 211–229. [DOI] [PubMed] [Google Scholar]
- 3.Bouige P, Laurent D, Piloyan L and Dassa E (2002) Phylogenetic and functional classification of ATP-binding cassette (ABC) systems. Curr Protein Pept Sci 3, 541–559. [DOI] [PubMed] [Google Scholar]
- 4.Saurin W, Hofnung M and Dassa E (1999) Getting in or out: early segregation between importers and exporters in the evolution of ATP-binding cassette (ABC) transporters. J Mol Evol 48, 22–41. [DOI] [PubMed] [Google Scholar]
- 5.Dean M, Rzhetsky A and Allikmets R (2001) The human ATP-binding cassette (ABC) transporter superfamily. Genome Res 11, 1156–1166. [DOI] [PubMed] [Google Scholar]
- 6.Klein I, Sarkadi B and Varadi A (1999) An inventory of the human ABC proteins. Biochim Biophys Acta 1461, 237–262. [DOI] [PubMed] [Google Scholar]
- 7.Tusnady GE, Sarkadi B, Simon I and Varadi A (2006) Membrane topology of human ABC proteins. FEBS Lett 580, 1017–1022. [DOI] [PubMed] [Google Scholar]
- 8.Gerovac M and Tampé R (2019) Control of mRNA translation by versatile ATP-driven machines. Trends Biochem Sci 44, 167–180. [DOI] [PubMed] [Google Scholar]
- 9.Khwaja M, Ma Q and Saier MH Jr (2005) Topological analysis of integral membrane constituents of prokaryotic ABC efflux systems. Res Microbiol 156, 270–277. [DOI] [PubMed] [Google Scholar]
- 10.Wang B, Dukarevich M, Sun EI, Yen MR and Saier MH Jr (2009) Membrane porters of ATP-binding cassette transport systems are polyphyletic. J Membr Biol 231, 1–10. [DOI] [PubMed] [Google Scholar]
- 11.Locher KP, Lee AT and Rees DC (2002) The E. coli BtuCD structure: a framework for ABC transporter architecture and mechanism. Science 296, 1091–1098. [DOI] [PubMed] [Google Scholar]
- 12.Oldham ML, Khare D, Quiocho FA, Davidson AL and Chen J (2007) Crystal structure of a catalytic intermediate of the maltose transporter. Nature 450, 515–521. [DOI] [PubMed] [Google Scholar]
- 13.Wang T, Fu G, Pan X, Wu J, Gong X, Wang J and Shi Y (2013) Structure of a bacterial energy-coupling factor transporter. Nature 497, 272–276. [DOI] [PubMed] [Google Scholar]
- 14.Xu K, Zhang M, Zhao Q, Yu F, Guo H, Wang C, He F, Ding J and Zhang P (2013) Crystal structure of a folate energy-coupling factor transporter from Lactobacillus brevis. Nature 497, 268–271. [DOI] [PubMed] [Google Scholar]
- 15.Dawson RJ and Locher KP (2006) Structure of a bacterial multidrug ABC transporter. Nature 443, 180–185. [DOI] [PubMed] [Google Scholar]
- 16.Lee J-Y, Kinch LN, Borek DM, Wang J, Wang J, Urbatsch IL, Xie X-S, Grishin NV, Cohen JC, Otwinowski Z et al. (2016) Crystal structure of the human sterol transporter ABCG5/ABCG8. Nature 533, 561–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo Q, Yang X, Yu S, Shi H, Wang K, Xiao L, Zhu G, Sun C, Li T, Li D et al. (2017) Structural basis for lipopolysaccharide extraction by ABC transporter LptB2FG. Nat Struct Mol Biol 24, 469–474. [DOI] [PubMed] [Google Scholar]
- 18.Dong H, Zhang Z, Tang X, Paterson NG and Dong C (2017) Structural and functional insights into the lipopolysaccharide ABC transporter LptB2FG. Nat Commun 8, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fitzpatrick AWP, Llabrés S, Neuberger A, Blaza JN, Bai X-C, Okada U, Murakami S, van Veen HW, Zachariae U, Scheres SHW et al. (2017) Structure of the MacAB-TolC ABC-type tripartite multidrug efflux pump. Nat Microbiol 2, 17070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okada U, Yamashita E, Neuberger A, Morimoto M, van Veen HW and Murakami S (2017) Crystal structure of tripartite-type ABC transporter MacB from Acinetobacter baumannii. Nat Commun 8, 1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crow A, Greene NP, Kaplan E and Koronakis V (2017) Structure and mechanotransmission mechanism of the MacB ABC transporter superfamily. Proc Natl Acad Sci USA 114, 12572–12577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang HB, Hou WT, Cheng MT, Jiang YL, Chen Y and Zhou CZ (2018) Structure of a MacAB-like efflux pump from Streptococcus pneumoniae. Nat Commun 9, 196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qian H, Zhao X, Cao P, Lei J, Yan N and Gong X (2017) Structure of the human lipid exporter ABCA1. Cell 169, 1228–1239.e10. [DOI] [PubMed] [Google Scholar]
- 24.Taylor NMI, Manolaridis I, Jackson SM, Kowal J, Stahlberg H and Locher KP (2017) Structure of the human multidrug transporter ABCG2. Nature 546, 504–509. [DOI] [PubMed] [Google Scholar]
- 25.Bi Y, Mann E, Whitfield C and Zimmer J (2018) Architecture of a channel-forming O-antigen polysaccharide ABC transporter. Nature 553, 361–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen L, Hou W-T, Fan T, Liu B, Pan T, Li Y-H, Jiang Y-L, Wen W, Chen Z-P, Sun L et al. (2020) Cryo-electron microscopy structure and transport mechanism of a wall teichoic acid ABC transporter. MBio 11, e02749–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koch J, Guntrum R, Heintke S, Kyritsis C and Tampé R (2004) Functional dissection of the transmembrane domains of the transporter associated with antigen processing (TAP). J Biol Chem 279, 10142–10147. [DOI] [PubMed] [Google Scholar]
- 28.Thomas C and Tampé R (2020) Structural and mechanistic principles of ABC transporters. Annu Rev Biochem 89, 605–636. [DOI] [PubMed] [Google Scholar]
- 29.Scheepers GH, Lycklama ANJA and Poolman B (2016) An updated structural classification of substrate-binding proteins. FEBS Lett 590, 4393–4401. [DOI] [PubMed] [Google Scholar]
- 30.Xu D, Feng Z, Hou WT, Jiang YL, Wang L, Sun L, Zhou CZ and Chen Y (2019) Cryo-EM structure of human lysosomal cobalamin exporter ABCD4. Cell Res 29, 1039–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Z, Hu W and Zheng H (2020) Pathogenic siderophore ABC importer YbtPQ adopts a surprising fold of exporter. Sci Adv 6, eaay7997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arnold FM, Weber MS, Gonda I, Gallenito MJ, Adenau S, Egloff P, Zimmermann I, Hutter CAJ, Hürlimann LM, Peters EE et al. (2020) The ABC exporter IrtAB imports and reduces mycobacterial siderophores. Nature 580, 413–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rempel S, Gati C, Nijland M, Thangaratnarajah C, Karyolaimos A, de Gier JW, Guskov A and Slotboom DJ (2020) A mycobacterial ABC transporter mediates the uptake of hydrophilic compounds. Nature 580, 409–412. [DOI] [PubMed] [Google Scholar]
- 34.Shitan N, Bazin I, Dan K, Obata K, Kigawa K, Ueda K, Sato F, Forestier C and Yazaki K (2003) Involvement of CjMDR1, a plant multidrug-resistance-type ATP-binding cassette protein, in alkaloid transport in Coptis japonica. Proc Natl Acad Sci USA 100, 751–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Terasaka K, Blakeslee JJ, Titapiwatanakun B, Peer WA, Bandyopadhyay A, Makam SN, Lee OR, Richards EL, Murphy AS, Sato F et al. (2005) PGP4, an ATP binding cassette P-glycoprotein, catalyzes auxin transport in Arabidopsis thaliana roots. Plant Cell 17, 2922–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee M, Choi Y, Burla B, Kim Y-Y, Jeon B, Maeshima M, Yoo J-Y, Martinoia E and Lee Y (2008) The ABC transporter AtABCB14 is a malate importer and modulates stomatal response to CO2. Nat Cell Biol 10, 1217–1223. [DOI] [PubMed] [Google Scholar]
- 37.Yang H and Murphy AS (2009) Functional expression and characterization of Arabidopsis ABCB, AUX 1 and PIN auxin transporters in Schizosaccharomyces pombe. Plant J 59, 179–191. [DOI] [PubMed] [Google Scholar]
- 38.Kamimoto Y, Terasaka K, Hamamoto M, Takanashi K, Fukuda S, Shitan N, Sugiyama A, Suzuki H, Shibata D, Wang B et al. (2012) Arabidopsis ABCB21 is a facultative auxin importer/exporter regulated by cytoplasmic auxin concentration. Plant Cell Physiol 53, 2090–2100. [DOI] [PubMed] [Google Scholar]
- 39.Shitan N, Dalmas F, Dan K, Kato N, Ueda K, Sato F, Forestier C and Yazaki K (2013) Characterization of Coptis japonica CjABCB2, an ATP-binding cassette protein involved in alkaloid transport. Phytochemistry 91, 109–116. [DOI] [PubMed] [Google Scholar]
- 40.Allikmets R, Shroyer NF, Singh N, Seddon JM, Lewis RA, Bernstein PS, Peiffer A, Zabriskie NA, Li Y, Hutchinson A et al. (1997) Mutation of the Stargardt disease gene (ABCR) in age-related macular degeneration. Science 277, 1805–1807. [DOI] [PubMed] [Google Scholar]
- 41.Quazi F, Lenevich S and Molday RS (2012) ABCA4 is an N-retinylidene-phosphatidylethanolamine and phosphatidylethanolamine importer. Nat Commun 3, 925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kang J, Hwang JU, Lee M, Kim YY, Assmann SM, Martinoia E and Lee Y (2010) PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid. Proc Natl Acad Sci USA 107, 2355–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xi J, Xu P and Xiang CB (2012) Loss of AtPDR11, a plasma membrane-localized ABC transporter, confers paraquat tolerance in Arabidopsis thaliana. Plant J 69, 782–791. [DOI] [PubMed] [Google Scholar]
- 44.Kang J, Yim S, Choi H, Kim A, Lee KP, Lopez-Molina L, Martinoia E and Lee Y (2015) Abscisic acid transporters cooperate to control seed germination. Nat Commun 6, 8113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coudray N, Isom GL, MacRae MR, Saiduddin MN, Bhabha G and Ekiert DC (2020) Structure of MlaFEDB lipid transporter reveals an ABC exporter fold and two bound phospholipids. bioRxiv 10.1101/2020.06.02.129247 [DOI] [Google Scholar]
- 46.Mann D, Fan J, Farrell DP, Somboon K, Andrew Muenks S, Tzokov S, Khalid F, Dimaio SM and Bergeron JRC (2020) Stuctural basis for lipid transpot by the MLA complex. bioRxiv 10.1101/2020.05.30.125013 [DOI] [Google Scholar]
- 47.Tang X, Chang S, Qiao W, Luo Q, Chen Y, Jia Z, Coleman J, Zhang K, Wang T, Zhang Z et al. (2020) Structural insight into outer membrane asymmetry maintenance of Gram-negative bacteria by the phospholipid transporter MlaFEDB. bioRxiv 10.1101/2020.06.04.133611 [DOI] [PubMed] [Google Scholar]
- 48.Chi X, Fan Q, Zhang Y, Liang K, Wan L, Zhou Q and Li Y (2020) Structural mechanism of phospholipids translocation by MlaFEDB complex. Cell Res. 10.1038/s41422-020-00404-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hofmann S, Januliene D, Mehdipour AR, Thomas C, Stefan E, Brüchert S, Kuhn BT, Geertsma ER, Hummer G, Tampé R et al. (2019) Conformation space of a heterodimeric ABC exporter under turnover conditions. Nature 571, 580–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Korkhov VM, Mireku SA and Locher KP (2012) Structure of AMP-PNP-bound vitamin B12 transporter BtuCD-F. Nature 490, 367–372. [DOI] [PubMed] [Google Scholar]
- 51.Nöll A, Thomas C, Herbring V, Zollmann T, Barth K, Mehdipour AR, Tomasiak TM, Brüchert S, Joseph B, Abele R et al. (2017) Crystal structure and mechanistic basis of a functional homolog of the antigen transporter TAP. Proc Natl Acad Sci USA 114, E438–E447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hohl M, Hurlimann LM, Bohm S, Schoppe J, Grutter MG, Bordignon E and Seeger MA (2014) Structural basis for allosteric cross-talk between the asymmetric nucleotide binding sites of a heterodimeric ABC exporter. Proc Natl Acad Sci USA 111, 11025–11030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Choudhury HG, Tong Z, Mathavan I, Li Y, Iwata S, Zirah S, Rebuffat S, van Veen HW and Beis K (2014) Structure of an antibacterial peptide ATP-binding cassette transporter in a novel outward occluded state. Proc Natl Acad Sci USA 111, 9145–9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kieuvongngam V, Olinares PDB, Palillo A, Oldham ML, Chait BT and Chen J (2020) Structural basis of substrate recognition by a polypeptide processing and secretion transporter. Elife 9, e51492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Srinivasan V, Pierik AJ and Lill R (2014) Crystal structures of nucleotide-free and glutathione-bound mitochondrial ABC transporter Atm1. Science 343, 1137–1140. [DOI] [PubMed] [Google Scholar]
- 56.Johnson ZL and Chen J (2017) Structural basis of substrate recognition by the multidrug resistance protein MRP1. Cell 168, 1075–1085.e9. [DOI] [PubMed] [Google Scholar]
- 57.Morgan JLW, Acheson JF and Zimmer J (2017) Structure of a type-1 secretion system ABC transporter. Structure 25, 522–529. [DOI] [PubMed] [Google Scholar]
- 58.Li J, Jaimes KF and Aller SG (2014) Refined structures of mouse P-glycoprotein. Protein Sci 23, 34–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oldham ML, Grigorieff N and Chen J (2016) Structure of the transporter associated with antigen processing trapped by herpes simplex virus. eLife 5, e21829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Olsen JA, Alam A, Kowal J, Stieger B and Locher KP (2020) Structure of the human lipid exporter ABCB4 in a lipid environment. Nat Struct Mol Biol 27, 62–70. [DOI] [PubMed] [Google Scholar]
- 61.Shintre CA, Pike ACW, Li Q, Kim J-I, Barr AJ, Goubin S, Shrestha L, Yang J, Berridge G, Ross J et al. (2013) Structures of ABCB10, a human ATP-binding cassette transporter in apo- and nucleotide-bound states. Proc Natl Acad Sci USA 110, 9710–9715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang L, Hou WT, Chen L, Jiang YL, Xu D, Sun L, Zhou CZ and Chen Y (2020) Cryo-EM structure of human bile salts exporter ABCB11. Cell Res 30, 623–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mi W, Li Y, Yoon SH, Ernst RK, Walz T and Liao M (2017) Structural basis of MsbA-mediated lipopolysaccharide transport. Nature 549, 233–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Perez C, Mehdipour AR, Hummer G and Locher KP (2019) Structure of outward-facing PglK and molecular dynamics of lipid-linked oligosaccharide recognition and translocation. Structure 27, 669–678.e5. [DOI] [PubMed] [Google Scholar]
- 65.Liu F, Zhang Z, Csanady L, Gadsby DC and Chen J (2017) Molecular structure of the human CFTR ion channel. Cell 169, 85–95.e8. [DOI] [PubMed] [Google Scholar]
- 66.Martin GM, Yoshioka C, Rex EA, Fay JF, Xie Q, Whorton MR, Chen JZ and Shyng SL (2017) Cryo-EM structure of the ATP-sensitive potassium channel illuminates mechanisms of assembly and gating. Elife 6, e24149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Manolaridis I, Jackson SM, Taylor NMI, Kowal J, Stahlberg H and Locher KP (2018) Cryo-EM structures of a human ABCG2 mutant trapped in ATP-bound and substrate-bound states. Nature 563, 426–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. TM-scores based on pairwise structural alignment of representatives of the different TMD types.
Table S2. TM-scores based on pairwise structural alignment of type I TMDs.
Table S3. TM-scores based on pairwise structural alignment of type II TMDs.
Table S4. TM-scores based on pairwise structural alignment of type IV TMDs in inward-facing conformations.
Table S5. TM-scores based on pairwise structural alignment of type IV TMDs in (semi-) occluded/outward-facing conformations.
Table S6. TM-scores based on pairwise structural alignment of type V, VI, and VII TMDsa.