Abstract
Members of the MEF2 family of transcription factors bind as homo- and heterodimers to the MEF2 site found in the promoter regions of numerous muscle-specific, growth- or stress-induced genes. We showed previously that the transactivation activity of MEF2C is stimulated by p38 mitogen-activated protein (MAP) kinase. In this study, we examined the potential role of the p38 MAP kinase pathway in regulating the other MEF2 family members. We found that MEF2A, but not MEF2B or MEF2D, is a substrate for p38. Among the four p38 group members, p38 is the most potent kinase for MEF2A. Threonines 312 and 319 within the transcription activation domain of MEF2A are the regulatory sites phosphorylated by p38. Phosphorylation of MEF2A in a MEF2A-MEF2D heterodimer enhances MEF2-dependent gene expression. These results demonstrate that the MAP kinase signaling pathway can discriminate between different MEF2 isoforms and can regulate MEF2-dependent genes through posttranslational activation of preexisting MEF2 protein.
The transactivation activity of many transcription factors is regulated by phosphorylation (2). The mitogen-activated protein (MAP) kinase family of serine/threonine kinases has been shown to play important roles in regulating gene expression via transcription factor phosphorylation (5, 10, 16, 38, 40, 42). Unique structural features, specific activation pathways, and different substrate specificities provide evidence to support the contention that different MAP kinases are independently regulated and control different cellular responses to extracellular stimuli (7, 38, 40, 44).
p38 MAP kinase was first identified in studies designed to explore how bacterial endotoxin induces cytokine expression (11, 13, 23). Following the initial description of p38 (p38α), three additional isoforms of this MAP kinase group have been cloned and characterized: p38β (18), p38γ (also termed ERK6 or SAPK3) (22, 24, 30), and p38δ (also termed SAPK4) (4, 17, 41). p38α and p38β are sensitive to pyridinyl imidazole derivatives, whereas p38γ and p38δ are not (4). In mammalian cells, these closely related p38 isoforms are activated coordinately by a broad panel of stimuli which include physical-chemical stresses and proinflammatory cytokines (17, 36). Two MAP kinase kinases (MKK), MKK3 and MKK6, are the upstream activators of the p38 group MAP kinases (6, 12, 14, 37). Several proteins including transcription factors such as CHOP 10 (GADD153) (42), Sap1 (16), MEF2C (10), enzymes such as cPLA2 (20), and the protein kinases MAPKAPK2/3 (27, 29, 39), MNK1/2 (8, 45), and p38-regulated/activated protein kinase (33) have been shown by us and others to be substrates of p38.
We showed that MEF2C, a member of the MEF2 family of transcription factors, is phosphorylated by p38 and that this event regulates the transactivation activity of MEF2C (10). Our studies showed that p38 specifically phosphorylates serine 387 and threonines 293 and 300 within the MEF2C transactivation domain (10). MEF2C phosphorylation by p38 was shown to play an important role in regulation of c-Jun expression in monocytic cells (10). Recently, Kato et al. have shown that MEF2C is also a substrate for BMK1/ERK5 and, interestingly, that MEF2C can be regulated by p38 and ERK5 signaling pathways via somewhat different phosphorylation patterns (19). Serine 59 of MEF2C is also constitutively phosphorylated in vivo, probably by casein kinase II, and this phosphorylation enhances DNA binding activity (32).
There are four members of the MEF2 family (28), MEF2A to D, that bind as homo- and heterodimers to the DNA consensus sequence CTA(A/T)4TA(G/A) (2). This sequence is found in the promoter regions of numerous muscle-specific, growth factor- and stress-induced genes (10, 15, 19, 46). The MEF2 isoforms share homology in an amino-terminal 56-amino-acid MADS domain and an adjacent 29-amino-acid MEF2 domain, which together mediate DNA binding and dimerization (34). The sequences outside these two domains are relatively divergent. The constitutively phosphorylated serine residue of MEF2C is located between the MADS and MEF2 domains (32), and the regulatory phosphorylation sites are located outside the highly conserved amino-terminal portion of MEF2C (32).
Here we describe studies carried out to further evaluate the role of the p38 MAP kinase pathway in the regulation of all MEF2 family members. We show that MEF2A, but not MEF2B or MEF2D, is a substrate for p38. Threonines 312 and 319 are the key regulatory phosphorylation sites by p38 in MEF2A. Phosphorylation at these sites enhances transcriptional activity of MEF2A. Our results also show that MEF2A dimerizes with MEF2D and that the phosphorylation of MEF2A in this complex plays a positive role in regulating MEF2-dependent gene expression.
MATERIALS AND METHODS
Cell culture.
CHO, 293, and HeLa cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 2 mM glutamine, 50 U of penicillin and 50 mg of streptomycin per ml, and 1% nonessential amino acids.
cDNA constructs and expression plasmids.
Expression plasmids for the GAL4 DNA binding domain [GAL4(DBD)] fused with the transactivation domains of human MEF2A (amino acids 87 to 505) [plasmid GAL4-MEF2A(wt)] mouse MEF2B (87 to 349), human MEF2C (87 to 442), mouse MEF2D1b (87 to 506), MEF2A(T312, 319A), MEF2A(S355A), MEF2A(S453A), and MEF2A(S479A) were constructed as described elsewhere (10). The oligonucleotide sequences used for PCR mutagenesis are available upon request. Bacterial expression constructs of His-tagged MKK6b(E) and p38α were described previously (17). The His-tagged full-length MEF2A, MEF2B, MEF2C, MEF2D, MEF2A(T312, 319A), MEF2A(S355A), MEF2A(S453A), and MEF2A(S479A) were cloned into the bacterial expression vector pETM1 by a PCR-recombination method as described elsewhere (18).
Preparation of recombinant proteins.
Escherichia coli BL21(DE3) was transformed with the vector pETM1 containing cDNAs encoding MEF2A, MEF2B, MEF2C, MEF2D, MEF2A(T312, 319A), MEF2A(S355A), MEF2A(S453A), and MEF2A(S479A). The transformed bacteria were grown at 37°C in LB broth until the A600 was 0.5, at which time isopropyl-β-d-thiogalactopyranoside at a final concentration of 1 mM was added for 5 h. Cells were collected by centrifugation at 8,000 × g for 10 min, and the bacterial pellet was resuspended in 10 ml of buffer A (30 mM NaCl, 10 mM EDTA, 20 mM Tris-HCl, 2 mM phenylmethylsulfonyl fluoride) for every 100 ml of original bacterial culture. The cell suspension was sonicated, and cellular debris was removed by centrifugation at 10,000 × g for 30 min. Recombinant proteins were purified from the cleared lysate by using a Ni-nitrilotriacetic acid purification system (Qiagen). Recombinant p38 isoforms and MKKs were prepared by the same method. Full activation of recombinant p38α, p38β, p38γ, or p38δ in vitro was achieved by incubation with recombinant MKK6(E) at a 5:1 molar ratio at 37°C for 15 min in the presence of ATP. Full activation of recombinant ERK2 and JNK2 was achieved by incubation with MEK1(E) and MKK7(D), respectively. Different amount of MKKs and times of incubation were tested to optimize the conditions for full activation of these recombinant MAP kinases (data not shown).
Protein kinase assays.
In vitro kinase assays were carried out at 37°C for 30 min, using 0.2 μg of recombinant kinase, 5 μg of kinase substrate, 250 μM ATP, and 10 μCi of [γ-32P]ATP in 20 μl of kinase reaction buffer as described previously (18). Reactions were terminated by the addition of Laemmli sample buffer. Reaction products were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 12% gel. Phosphorylated proteins were visualized by autoradiography.
Preparation of cell lysates and Western blotting.
These experiments were performed as described previously (13). Briefly, cells were rapidly chilled on ice, washed twice with ice-cold washing buffer (10 mM Tris-HCl, 150 mM NaCl, 1 mM Na3VO4 [pH 7.5]), and then lysed in 250 μl (per 106 cells) lysis buffer (20 mM Tris-HCl, 120 mM NaCl, 10% glycerol, 1 mM Na3VO4, 2 mM EDTA, 1% Triton X-100, 1 mM phenylmethaylsulfonyl fluoride [pH 7.5]). The proteins were separated by SDS-PAGE and transferred to a nitrocellulose membrane. Monoclonal anti-GAL4(DBD) antibody RK5C1 (Santa Cruz Biotechnology) was used to detect GAL4-MEF2 fusion proteins.
Phosphoamino acid analysis and phosphopeptide mapping.
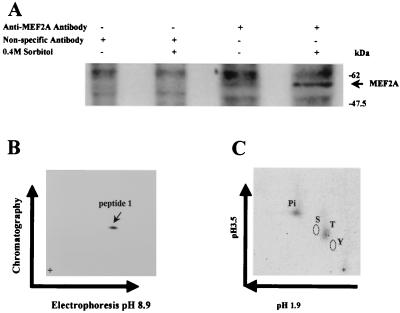
The experiments were performed as described by Boyle et al. (3). Recombinant MEF2A and MEF2A mutants were phosphorylated in vitro by p38 as described above. Phosphorylated MEF2A proteins were separated by SDS-PAGE, transferred to nitrocellulose membranes, visualized by autoradiography, and excised. Eluted proteins were digested with trypsin, and peptides were analyzed by thin-layer electrophoresis and thin-layer chromatography as described elsewhere (3). In some cases, phosphopeptides were recovered from thin-layer chromatography plates and phosphoamino acid analysis was performed as described elsewhere (3). Phosphopeptides were visualized by autoradiography or phosphorimaging and quantitated by phosphorimaging. MEF2A protein from 32Pi-labeled 293 cells treated with or without 0.4 M sorbitol was immunoprecipitated with anti-MEF2A antibody (Santa Cruz Biotechnology), using 100 μg of antibody per 5 × 106 cells, and subjected to peptide mapping and phosphoamino acid analysis as described above.
Electrophoretic mobility shift assay (EMSA).
Nuclear extracts of 293 cells and HeLa cells were incubated with a double-stranded, 32P-labeled oligonucleotide containing a MEF2 binding site as a probe. Antisera specific for MEF2A (Santa Cruz Biotechnology), MEF2B, MEF2C, and MEF2D (gift from R. Prywes, Columbia University) and nonspecific antiserum were used to detect binding of each of the four isoforms of MEF2 as described elsewhere (21).
Reporter gene expression.
The GAL4-responsive plasmid pG5E1bLuc contains five GAL4 sites cloned upstream of a minimal promoter driving a luciferase gene (9). Plasmids encoding a luciferase gene driven by the wild-type c-Jun promoter (pJluc) were kindly provided by R. Prywes (15). The reporter plasmid pG5E1bLuc was cotransfected into cells along with a construct encoding the GAL4(DBD) fused to MEF2A, MEF2B, MEF2C, MEF2D, MEF2A(T312, 319A), MEF2A(S355A), MEF2A(S453A), and MEF2A(S479A) along with the expression vector encoding constitutively active forms of MKKs, termed MKK6b(E), MKK1(E), MKK7(D), and MEK5(D). Cells were grown on 35-mm-diameter multiwell plates (Nunc, Naperville, Ill.) and transiently transfected with 1 μg of total plasmid DNA, using Lipefectamine reagent (Gibco BRL, Gaithersburg, Md.). A β-galactosidase expression plasmid (pCMV-β-gal; Clontech, Palo Alto, Calif.) was used to control for transfection efficiency. The total amount of DNA for each transfection was kept constant by using the empty vector pcDNA3. After 24 h, the medium was changed to serum-free Dulbecco’s modified Eagle’s medium supplemented with 2 mM glutamine and nonessential amino acids; 48 h after transfection, cell extracts were prepared and the activities of β-galactosidase and luciferase were measured as described elsewhere (10, 18). In some experiments, different concentrations of SB203580 (stock solution is 20 mM in dimethyl sulfoxide) were included in the medium 30 min before transfection and kept thereafter.
RESULTS
MEF2A is a preferred substrate for p38 in vitro.
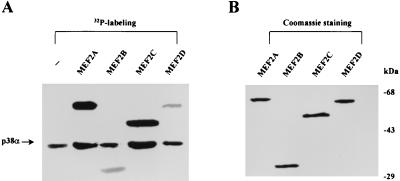
Our previous studies using a yeast two-hybrid system identified MEF2C as a physiological substrate for p38 (10). This screen failed to identify other members of the MEF2 family, possibly because some low-abundance clones were missed during the screen. Indeed, we have determined that MEF2C is more abundant than the other MEF2 isoforms in the brain library used for the two-hybrid screen. To determine whether other MEF2 family members are substrates of p38, we performed in vitro kinase assays using recombinant forms of each of the MEF2 family members. As shown in Fig. 1A, MEF2A and MEF2C are preferred substrates for p38 compared with MEF2B and MEF2D. Equal amounts of each MEF2 isoform were used in these assays, as determined in Fig. 1B. Although the same amount of p38 was used in each kinase reaction, the degrees of autophosphorylation of p38 appear different, possibly because of kinase-substrate interaction. The enhancement of p38 autophosphorylation has also been observed when other substrate proteins such as ATF2 are present (data not shown).
FIG. 1.
Phosphorylation of bacterially expressed MEF2 proteins by p38 in vitro. (A) His-tagged MEF2A, MEF2B, MEF2C, and MEF2D proteins were phosphorylated in vitro by purified p38 (p38α) as described in Materials and Methods. Autophosphorylated p38 is indicated by an arrow. (B) Coomassie blue staining of MEF2 proteins used in the kinase assay.
Transactivation activity of MEF2A is up-regulated by activation of the p38 pathway.
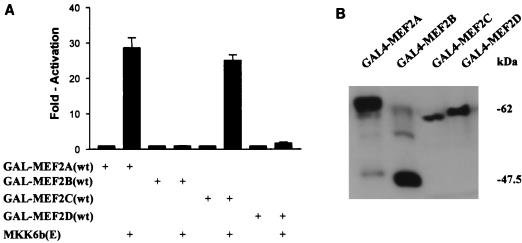
Previous studies showed that p38 augments the transcriptional activity of MEF2C by phosphorylation of the transactivation domain. To determine whether p38 had a similar effect on MEF2A, we used fusion proteins containing the transactivation domain of each of the four MEF2 family members fused to GAL4(DBD) (see Materials and Methods). To assess the transcriptional activity of the GAL4-MEF2 proteins, the human embryonic kidney cell line 293 was cotransfected with a luciferase reporter gene containing five copies of a GAL4 binding site upstream of a minimal promoter and GAL4-MEF2 expression plasmids. Activation of the p38 pathway was achieved by cotransfection of the constitutively active p38 activator, MKK6b(E). As reported previously, activation of the p38 pathway enhanced MEF2C-dependent reporter gene expression (Fig. 2A) (10). In addition, activation of p38 also dramatically enhanced MEF2A-dependent, but not MEF2B- or MEF2D-dependent, reporter gene expression (Fig. 2A). This enhancement correlated well with the in vitro phosphorylation of these proteins by p38 (Fig. 1). The inability of MKK6b(E) to activate MEF2B and MEF2D is not due to lack of the expression of GAL4-MEF2B or GAL4-MEF2D, since these fusion proteins were expressed similarly, as detected by Western blotting with an anti-GAL4(DBD) monoclonal antibody (Fig. 2B).
FIG. 2.
Activation of the p38 pathway by MKK6b(E) up-regulates MEF2A and MEF2C activity. 293 cells were cultured in six-well plates and cotransfected with β-galactosidase expression vector pCMV-β-gal (0.2 μg), the reporter plasmid pG5ElbLuc (0.2 μg), GAL4-MEF2A(wt), and expression vectors for GAL4(DBD) fused to various MEF2 isoforms (0.3 μg), the MKK6b(E) expression plasmid (0.3 μg), or the control empty vector pcDNA3. Empty vector pcDNA3 was used to normalize the total DNA to 1 μg per transfection. Cell extracts were prepared 48 h following transfection. The ratio of luciferase activity to β-galactosidase activity is presented as the mean ± standard deviation (n = 3) (A). Three experiments were performed with comparable results. The results of one experiment are shown. The expression of GAL4 fusion proteins in extracts of transfected cells was determined by Western blotting using anti-GAL4(DBD) monoclonal antibody RK5C1 (B).
Identification of p38-catalyzed phosphorylation sites in MEF2A.
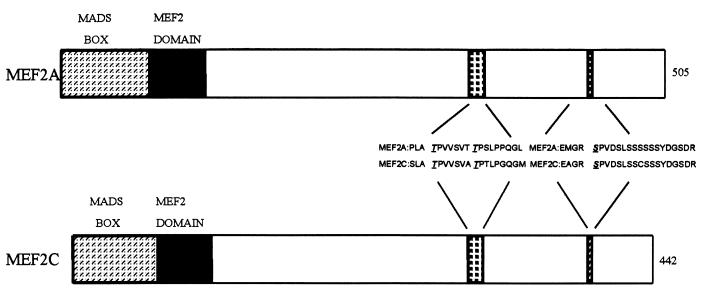
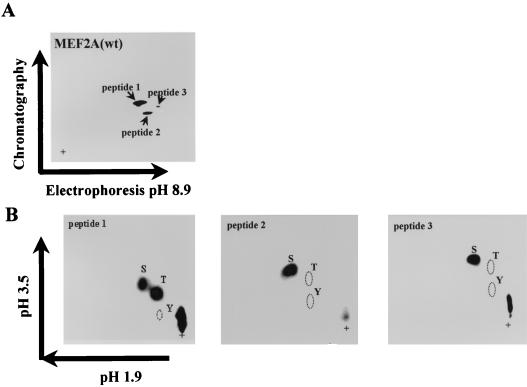
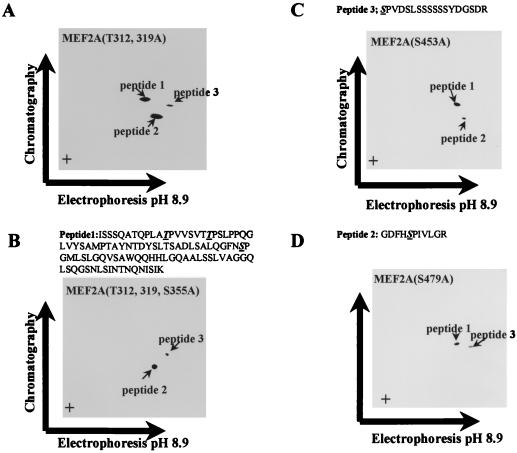
Sequence alignment of MEF2 family members showed that Thr-293, Thr-300, and Ser-387 of MEF2C and surrounding sequences were conserved in MEF2A (Thr-312, Thr-319, and Ser-453) (Fig. 3). These three residues have been shown to be phosphorylated by p38 (10). We sought to determine if the corresponding residues were the phosphorylation sites for p38 in MEF2A. To identify the residues of MEF2A phosphorylated by p38 in vitro, we used a phosphopeptide mapping approach. This analysis revealed two major phosphorylated peptides and one weakly phosphorylated peptide (Fig. 4A). The relative phospho-intensity of these three peptides is 8.1:2.4:1, based on phosphorimaging analysis. Phosphoamino acid analysis showed that peptide 1 contained both phosphoserine and phosphothreonine, whereas peptides 2 and 3 contained only phosphoserine (Fig. 4B).
FIG. 3.
Schematic representations of MEF2A and MEF2C. The number of amino acids in each protein is indicated at the right. MEF2A and MEF2C without the last alternatively spliced exon are shown. The amino acid sequence in the regions containing p38 phosphorylation sites of MEF2C and the corresponding sequence in MEF2A are shown. Thr-293, Thr-300, and Ser-387 in MEF2C and Thr-312, Thr-319, and Ser-453 in MEF2A are underlined.
FIG. 4.
Tryptic phosphopeptide mapping and phosphoamino acid analysis of wild-type MEF2A [MEF2A(wt)] phosphorylated by p38. (A) Two-dimensional tryptic peptide map obtained from 32P-labeled MEF2A; (B) phosphoamino acid analysis of each phosphorylated peptide from panel A. There are three phosphorylated peptides. Peptide 1 contains phosphoserine and phosphothreonine, while peptides 2 and 3 contain only phosphoserine. The positions of phosphoserine (S), phosphothreonine (T), and phosphotyrosine (Y) standards are indicated; + designates the origin.
We predicted the peptide 1 sequence as shown above Fig. 5B because only this tryptic peptide of MEF2A contains both Ser-Pro and Thr-Pro, which are consensus phosphorylation sites for MAP kinases. Since the intensity of phosphothreonine is about two times higher than that of phosphoserine when quantitated by phosphorimaging, it appeared that both threonines, followed by prolines in this peptide, were phosphorylated by p38. This prediction was confirmed by point mutations. Mutation of Thr-312 and Thr-319 to alanines reduced the intensity of the phosphorylation of peptide 1 by about two-thirds (the ratio of the phosphorylation of peptides is 2.7:2.8:1 [Fig. 5A], compared to 8.1:2.4:1 for the wild-type protein [Fig. 4A]), which also supports our contention that two threonines are phosphorylated. An additional mutation of Ser-355 completely abolished the phosphorylation of this peptide (Fig. 5B). The totality of our data support the conclusion that the peptide 1 sequence is as shown in Fig. 5B. Thus, threonines 312 and 319 in MEF2A, corresponding to Thr-293 and Thr-300 of MEF2C, are phosphorylation sites for p38 (Fig. 3). When we mutated serine 453 to alanine, we observed that phosphorylation of peptide 3 by p38 was abolished, suggesting that Ser-453 is a site of p38 phosphorylation (Fig. 5C). To determine the phosphorylated serine residues in peptide 2, we analyzed the electrical charge and mass of tryptic peptides of MEF2A and predicted that peptide 2 was the peptide containing Ser-479. Mutation of Ser-479 to alanine [MEF2A(S479A)] abolished the phosphorylation of peptide 2 (Fig. 5D), which suggests that Ser-479 represents an additional phosphorylation site of MEF2A. The sequence of phosphorylated peptides identified in the preceding analyses are shown above the chromatographs.
FIG. 5.
In vitro phosphorylation sites of MEF2A by p38. (A) Phosphopeptide mapping of MEF2A(T312, 319A) phosphorylated by p38 in vitro; (B) phosphopeptide mapping of MEF2A(T312, 319, S355A) phosphorylated by p38 in vitro; (C) phosphopeptide mapping of MEF2A(S479A) phosphorylated by p38 in vitro; (D) phosphopeptide mapping of MEF2A(S453A) phosphorylated by p38 in vitro. The sequences of predicted phosphopeptides are shown above panels B to D, with phosphorylated residues underlined.
To investigate if any of these sites in MEF2A are phosphorylated after p38 activation in intact cells, 293 cells were labeled with 32Pi. Endogenous MEF2A was immunoprecipitated after the cells were treated with or without 0.4 M sorbitol to activate p38. Increased phosphorylation of MEF2A protein was detected in cells stimulated with sorbitol (Fig. 6A). Peptide mapping revealed that phosphorylation occurred on a peptide with an Rf identical to that of p38 phosphorylated peptide 1 (0.46) (Fig. 6B and 4A). The mobility (mr) of this peptide in the electrophoresis dimension was also similar to that of peptide 1 (Fig. 6B and 4A). Phosphoamino acid analysis of this phosphopeptide showed that the phosphorylation occurred on threonine residues (Fig. 6C). These data suggested that Thr-312 and/or Thr-319 are the in vivo phosphorylation sites of MEF2A. This predication is consistent with the analysis of electrical charge, mass, and hydrophobicity of the tryptic peptides. For example, peptide 1 phosphorylated by p38 in vitro had a calculated Rf of 0.447 and mr of 9.0 × 10−4, while the Rf and mr of peptide 1 with phosphorylation on two threonine residues were 0.446 and 7.1 × 10−4, respectively. Since the only other tryptic peptide of MEF2A that could be phosphorylated by MAP kinase on threonine should migrate quite differently (Rf = 0.375 and mr = 4.0 × 10−4) than the one we detected, the only peptide that is likely to be phosphorylated in vivo is peptide 1. Thus, we concluded that the Thr-312 and/or 319 residues located in peptide 1 of MEF2A are the in vivo phosphorylation sites.
FIG. 6.
In vivo phosphorylation of MEF2A. (A) MEF2A was immunoprecipitated from sorbitol-stimulated or nonstimulated 293 cells that were metabolically labeled with 32P. Only the region of the gel containing MEF2A is shown. (B) Phosphopeptide mapping of MEF2A isolated by SDS-PAGE shown in panel A. The only 32P-labeled peptide obtained had Rf and mr values very similar to those of phosphopeptide 1 of p38-phosphorylated MEF2A in vitro (Fig. 4). (C) Phosphoamino acid analysis of the peptide shown in panel B.
Thr-312 and Thr-319 are essential regulatory sites in MEF2A.
The sites in MEF2A phosphorylated by p38 in vitro have been determined to be Thr-312, Thr-319, Ser-355, Ser-453, and Ser-479. Mutants of the GAL4-MEF2A fusion protein were used to determine if these phosphorylation sites play a role in p38-mediated MEF2A activation in vivo. MEF2A mutations with changes of Thr-312 and Thr-319 to alanine [GAL-MEF2A(T312, 319A)], Ser-355 to alanine [GAL-MEF2A(S355A)], Ser-453 to alanine [GAL-MEF2A(S453A)], and Ser-479 to alanine [GAL-MEF2A(S479A)] were constructed. Mutation of Thr-312 and Thr-319 in MEF2A completely abolished the ability of p38 to enhance activation of the GAL4-driven luciferase reporter in 293 cells (Fig. 7). In contrast, the mutations in GAL4-MEF2A(S387A), GAL4-MEF2A(S453A), and GAL4-MEF2A(S479A) had no effect on p38-mediated MEF2A activation (Fig. 7). All GAL4 fusion proteins were expressed at comparable levels, as demonstrated by Western blotting (Fig. 7A). We observed similar results when CHO and HeLa cells were used (data not shown). These data are consistent with the in vivo phosphorylation sites analysis (Fig. 6) and support the contention that Thr-312 and Thr-319 are the key phosphorylation sites in MEF2A that are regulated by p38.
FIG. 7.
Enhancement of the MEF2A transactivation activity by p38 pathway is dependent on phosphorylation of Thr-312 and Thr-319. 293 cells were cotransfected with pCMV-β-gal, pG5ElbLuc, MKK6b(E), GAL-MEF2A(wt), and plasmids expressing GAL4(DBD) fused to various MEF2A isoforms, and reporter gene expression was assayed as described for Fig. 2. The expression of GAL4–MEF2A and its mutants was determined by Western blotting using anti-GAL4(DBD) monoclonal antibody RK5C1 (top). Two experiments were performed, and the results of one experiment are shown.
MEF2A activation is specifically mediated by p38.
To examine if the other MAP kinase pathways can regulate MEF2A activation, the GAL4-driven luciferase reporter system described above was used. As expected, activation of the p38 pathway by dominant active MKK6 dramatically enhanced MEF2A-dependent reporter gene expression (Fig. 8A). In contrast, activation of the ERK, JNK, and ERK5/BMK pathways by dominant active MEK1, MKK7, and MEK5, respectively, did not lead to an increase in the reporter gene expression (Fig. 8A). As positive controls, we tested the responsiveness of GAL4-ATF2 and GAL4-ELK1 fusion proteins to the above kinases. Expression of MEK1(E) enhanced ELK-1-dependent gene expression, MKK7(D) enhanced ATF2-dependent gene expression, and MEK5(D) weakly enhanced MEF2C-dependent gene expression (Fig. 8B).
FIG. 8.
MEF2A activation is specifically mediated by the p38, but not ERK, JNK, or ERK5/BMK, pathway. 293 cells were cotransfected with pCMV-β-gal, pG5ElbLuc, and GAL4-MEF2A(wt). The p38 pathway, ERK pathway, JNK pathway, or ERK5/BMK pathway was activated by expression of MKK6b(E), MEK1(E), MKK7(D), or MEK5(D), respectively. (A) Reporter gene expression determined as described for Fig. 2; (B) positive control of ERK, JNK, or ERK5/BMK activation. Comparable results were obtained in two experiments.
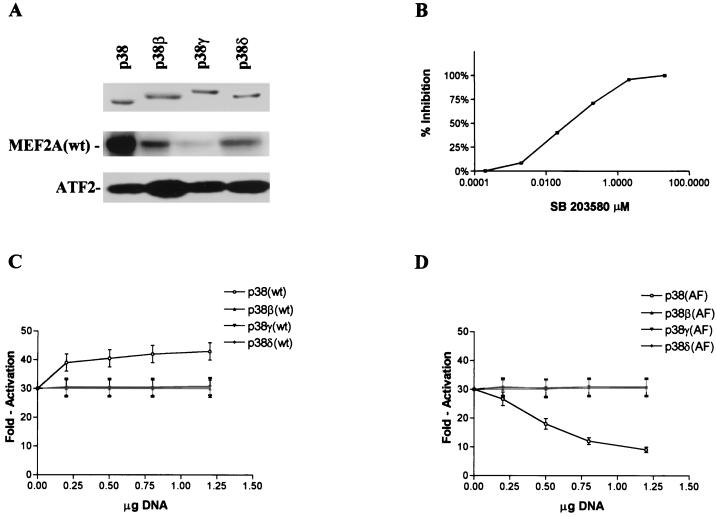
There are four members of the p38 group of kinases that can be activated by MKK6. To examine which p38 isoform is the principal kinase which mediated MKK6-induced MEF2A activation, we did an in vitro kinase assay using recombinant p38 isoforms that were activated by MKK6(E) (see Materials and Methods) as kinase and MEF2A protein as substrate. As shown in Fig. 9A, p38 was the most potent kinase for MEF2A when equal amounts of kinase were used. A glutathione S-transferase fusion protein of the ATF2 N-terminal portion (amino acid 1 to 109), which can be phosphorylated by all p38 isoforms in vitro, was included as positive control for the kinase assays (Fig. 9A, bottom). We then used SB203580, a specific inhibitor of p38 and p38β, to test if p38 or p38β is involved in MKK6-mediated MEF2A activation in vivo. As shown in Fig. 9B, SB203580 dose dependently blocked MKK6-enhanced MEF2A-dependent gene expression with a 50% inhibitory concentration of 0.8 μM, which is similar to that of the p38 substrate p38-regulated/activated protein kinase (33). This finding suggested that either p38 or p38β is the kinase downstream of MKK6 that mediates MEF2A activation. To further examine the contribution of different p38 isoforms to MKK6-induced MEF2A activation, we coexpressed p38, p38β, p38γ, and p38δ and their inactive mutants with the reporter gene. As shown in Fig. 9C, overexpression of p38 enhanced reporter gene expression, while the expression of the other p38 isoforms had little or no effect. Expression of the inactive mutant of p38 [termed p38(AF)] dose dependently reduced reporter gene expression (Fig. 9D). In contrast, expression of inactive mutants of other p38 isoforms had no effect on reporter gene expression. These data support the idea that p38 is the enzyme downstream of MKK6 in vivo that mediates MKK6-induced MEF2A activation.
FIG. 9.
Activation of MEF2A by MKK6b(E) is mediated by p38 in 293 cells. (A) In vitro phosphorylation of MEF2A by different p38 isoforms. Glutathione S-transferase–ATF2(1–109) protein, which is efficiently phosphorylated by all p38 isoforms, was included as a positive control. (B) 293 cells pretreated for 30 min with different doses of SB203580 were cotransfected with pCMV-β-gal, pG5ElbLuc, MKK6b(E), GAL-MEF2A(wt), MKK6b(E), or empty vector as described for Fig. 2. MKK6b(E)-induced reporter gene expression was inhibited dose dependently by SB203580. Comparable results were obtained in two experiments. (C) 293 cells were cotransfected with pCMV-β-gal (0.1 μg), pG5ElbLuc (0.1 μg), MKK6b(E) (0.1 μg), GAL-MEF2A(wt) (0.1 μg), and p38, p38β, p38γ, or p38δ in the amounts of DNA indicated. Empty vector pcDNA3 was used to normalize the total DNA to 1.6 μg per transfection. (D) 293 cells were cotransfected with pCMV-β-gal, pG5ElbLuc, MKK6b(E), GAL-MEF2A(wt), and kinase-inactive mutants of four p38 isoforms as indicated.
Characterization of MEF2 binding activities in 293 cells.
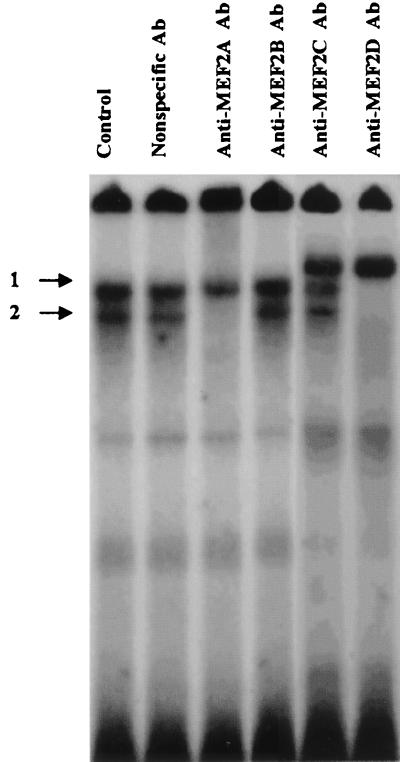
MEF2 family members bind DNA as homo- or heterodimers in vivo (35). To investigate the composition of MEF2 dimers in the cells used for these studies, an EMSA in combination with supershift analyses using antibodies specific to MEF2A, MEF2B, MEF2C, and MEF2D were performed. The MEF2 site binding activities can be detected in 293 cells as two major DNA-protein complexes (numbered 1 and 2 in Fig. 10). In 293 cells, anti-MEF2A antibody completely eliminated complex 2, suggesting that this DNA-protein complex contains MEF2A. No significant change of the gel shift pattern was seen when MEF2B antibody was used. A shift of complex 1 was induced by MEF2C antibody, suggesting MEF2C is present in this complex. MEF2D antibody eliminated both bands. Based on these data, we conclude that all dimers in complex 1 are comprised of MEF2D-MEF2C heterodimers and MEF2D-MEF2D homodimers, whereas the major component of complex 2 is a MEF2A-MEF2D heterodimer.
FIG. 10.
Compositions of MEF2 dimers in 293 cells. Nuclear protein extracts from 293 cells were used in EMSA. Incubation of extracts and probe with specific MEF2A, MEF2B, MEF2C, and MEF2D immune sera or an anti-p50 antiserum as a nonspecific antibody (Ab) was carried out to test for the presence of MEF2 isoforms in DNA-protein complex. Complex 2 appears to represent MEF2A-MEF2D heterodimers; complex 1 contains MEF2C-MEF2D and MEF2D-MEF2D dimers.
Phosphorylation of MEF2A in MEF2A-MEF2D heterodimers enhances its transactivation activity.
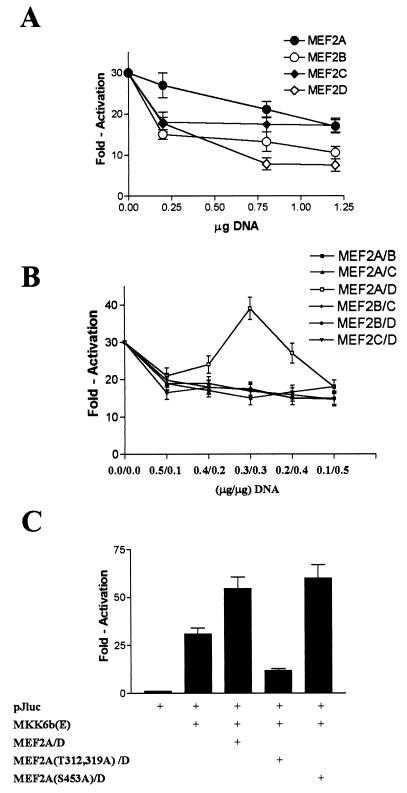
To determine which dimers play a positive role in regulating MEF2-dependent gene expression, we used the reporter construct pJluc, in which the luciferase gene is driven by the c-Jun promoter (−225 to +150 bp). Previously, a MEF2 site has been shown to play a positive regulatory role in activation of this promoter (15). As previously reported for MEF2C (10), we observed that overexpression of MEF2A, MEF2B, or MEF2D decreased MKK6b(E)-mediated pJluc reporter gene expression (Fig. 11A). Overexpression of each MEF2 isoform should lead to an increase in a given homodimer concentration inside cells. The inhibitory effect associated with this increased homodimer concentration can be interpreted as following: transcriptional activities of various homodimers are not regulated by p38 phosphorylation; and increased homodimers may compete with a p38-regulatable heterodimer by occupying MEF2 sites and thereby inhibit the up-regulation of MEF2-dependent gene expression by MKK6b(E). Next we investigated the impact of phosphorylation of MEF2 various heterodimers in 293 cells. The concentration of a given heterodimer 293 cells can be increased by coexpression of two MEF2 isoforms. Since the level of transient expression of proteins can be different even with the same expression vector, we cannot predict under which condition the equal expression of two isoforms can be achieved. Therefore, we cotransfected cells with different ratios of two expression vectors to cover the range in which the expression of two MEF2 isoforms would be equal. As shown in Fig. 11B, we found that coexpression of MEF2A and MEF2D in 293 cells led to an up-regulation in pJluc reporter expression. Maximal enhancement of MKK6b(E)-induced pJluc expression was achieved when the ratio of MEF2A and MEF2D expression vector was 1:1, which may be due to similar levels of expression of these two proteins. All other combinations produced a negative effect on reporter gene expression. Although enhancement of pJluc expression is small, the negative effect of all other combinations supports the conclusion that the MEF2A-MEF2D heterodimer is a specific activator. We think that the relatively small effect of overexpressing MEF2A-MEF2D reflects the presence of sufficient endogenous MEF2A-MEF2D inside cells. Nonetheless, these data suggest that the MEF2A-MEF2D heterodimer, a major component of complex 2 (Fig. 10), is a downstream element of p38 in MKK6-induced pJluc expression. Coexpression of phosphorylation-defective MEF2A with MEF2D reduced the expression of the pJluc reporter gene, supporting the contention that phosphorylation plays an important role in regulating MEF2A activity under physiological conditions (Fig. 11C). Taken together, these data suggested that in 293 cells, the MEF2A-MEF2D heterodimer is a regulator of MEF2-dependent gene expression, and p38-catalyzed phosphorylation of MEF2A in the complex plays a positive role in gene expression.
FIG. 11.
Effect of overexpression of MEF2 homo- or heterodimers on MEF2-dependent reporter gene expression in 293 cells. 293 cells were cotransfected with pCMV-β-gal (0.1 μg), pJluc (0.1 μg), MKK6b(E) (0.1 μg), and MEF2A, MEF2B, MEF2C, or MEF2D. Empty vector pcDNA3 was used to normalize the total DNA to 1.5 μg per transfection (A). 293 cells were cotransfected with pCMV-β-gal (0.1 μg), pJluc (0.1 μg), MKK6b(E) (0.1 μg), and different combinations of MEF2 isoforms with different DNA ratio as indicated (B) and with MEF2A or its mutants together with MEF2D at a 1:1 ratio (C). Reporter gene pJluc expression is shown.
DISCUSSION
MEF2 was originally described as a muscle-specific DNA binding protein that recognizes an A/T-rich element within the promoter regions of many muscle-specific genes (46). Recently, the MEF2 element has been shown to play a role in growth factor- and stress stimulus-induced early gene responses (15). Thus, the regulation of MEF2-mediated gene expression may be controlled at multiple levels: by tissue specificity and by signal transduction pathways activated by growth or stress stimuli. Tissue-specific expression of certain MEF2 family members and tissue-specific splicing have been shown to play an important role in the control of MEF2-regulated genes (28, 46). Phosphorylation of MEF2C has also been recently reported to play an important role in regulation of MEF2-dependent gene expression (10, 19). We now extend our previous observations by showing that MEF2A is regulated by the p38 pathway. We found that in 293 cells, MEF2A forms a heterodimer with MEF2D. Phosphorylation of MEF2A in the MEF2A-MEF2D dimer by p38 enhanced MEF2-dependent gene expression. These data extend the possible regulatory pathways that depend on p38-mediated phosphorylation of members of the MEF2 family of transcription factors.
Our in vitro studies demonstrated that MEF2A can be phosphorylated by p38 at two threonines and two serines. Importantly, however, we found that only the two threonines, which are phosphorylated in stress-activated cells, play a regulatory role in p38-mediated MEF2A activation in 293 cells; differences between in vitro and in vivo phosphorylation patterns were also found in our studies with MEF2C. Although three phosphorylation sites in MEF2C have been identified, cell studies using either CHO or 293 cells revealed that two threonines comprised the essential regulatory sites (10, 19). Nonetheless, the regulatory phosphorylation sites appear to be cell type dependent because MEF2C was phosphorylated at all three sites in macrophages in response to lipopolysaccharide stimulation (10). By analogy with MEF2C, it is possible that the other in vitro phosphorylation sites in MEF2A are of importance in cell types other than those examined in this study. How cell-type-specific phosphorylation of MEF2 isoforms occurs remains to be determined. There may be cell-type-specific cofactors involved in the regulation of MEF2 activity. Other explanations may include differences in composition of dimers in different cell types or the possibility that tissue-specific alternatively spliced isoforms are differentially phosphorylated.
Our data support the contention that phosphorylation of one component of the MEF2A-MEF2D heterodimer is sufficient to augment transactivation activity. Since multiple MEF2 family members can be phosphorylated by one or more kinases, it would be interesting to know if there is phosphorylation of both components in vivo and the consequences of such phosphorylation. Moreover, phosphorylation of a component in a dimer can be carried out by different kinases, which can phosphorylate MEF2 family members through different phosphorylation sites. For example, p38 and ERK5 have been shown to regulate MEF2C via different phosphorylation sites (19). Whether the transactivation activity of MEF2A is regulated by different kinases awaits further investigation. In this regard, Kato et al. (19a) also examined phosphorylation of four MEF2 isoforms by ERK5 and found that MEF2D is the preferred substrate for ERK5. Therefore, studies to determine if the p38 and ERK5 pathways are integrated in controlling MEF2A-MEF2D activity are needed. Although MEF2 family members can be targeted by MAP kinases of two different groups, within the p38 group, only a single isoform appears to influence MEF2A activity. Thus, while there is complexity at the level of kinases, there also appears to be a high degree of selectivity at the enzyme-substrate level.
Targeted disruption of mef2c in mice and mef2 in Drosophila provides evidence for a crucial role of MEF2 family transcriptional factors in both skeletal and cardiac muscle development (25, 26). It has been shown recently that sole activation of the p38 pathway can lead to hypertrophy of cultured cardiomyocytes (43, 47). Because of the pivotal role of MEF2C protein in cardiac development (26), it will be of interest to address whether phosphorylation of MEF2 family members by p38 has a role in cardiomyocyte hypertrophy. MEF2 proteins do not function by themselves but rather cooperate with other transcription factors, such as basic helix-loop-helix (bHLH) transcription factors (1, 31). Although the interaction between bHLH transcriptional factors and MEF2 proteins is mediated by the DNA binding and dimerization domains (1), the effect of phosphorylation within the MEF2 transactivation domain on the synergistic effect of these transcriptional factors could be important. Since cooperation of myogenic bHLH proteins with members of MEF2 group protein plays an essential role in the establishment of skeletal muscle lineages (31), it is reasonable to conclude that regulation MEF2 protein activity by p38 may play a role in the process of myocyte differentiation. Indeed, Puri et al. (35a) observed that activation of the p38 pathway by transient expression of dominant active MKK6 drove C2C12 cell differentiation to myotubes. The investigation of the potential role of the p38 pathway in the initiation of myogenesis is now a priority.
In summary, we have examined the regulatory effect of p38 pathway on four different MEF2 family members. We found that MEF2A and MEF2C can be phosphorylated and regulated by p38 and that regulation occurs when MEF2 is present as a dimer. In the future, it will be of interest to determine tissue- and/or cell-type-specific regulation of these transcription factors as well as to investigate how several MAP kinase pathways are coordinated in regulating MEF2 family member activities. Since MEF2 factors play pivotal roles in differentiation of skeletal and cardiac muscle cells, it will be especially interesting to determine whether MAP kinase signaling affects muscle gene expression through modulation of MEF2 activity.
ACKNOWLEDGMENTS
We thank R. Prywes for the MEF2D antibody and B. Chastain for secretarial assistance.
This work was supported by grants from the National Institutes of Health (to J.H., R.J.U., and E.N.O.), American Heart Association (to J.H.), Muscular Dystrophy Association (to E.N.O.), Robert A. Welch Foundation (to E.N.O.), and Human Frontier Science Foundation (to E.N.O.). J.H. is an Established Investigator of the American Heart Association.
Footnotes
Publication no. 11506-IMM from the Department of Immunology of The Scripps Research Institute.
REFERENCES
- 1.Black B L, Molkentin J D, Olson E N. Multiple roles for the MyoD basic region in transmission of transcriptional activation signals and interaction with MEF2. Mol Cell Biol. 1998;18:69–77. doi: 10.1128/mcb.18.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Black, B., and E. Olson. Transcription control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu. Rev. Cell Dev. Biol., in press. [DOI] [PubMed]
- 3.Boyle W J, VanDerGeer P, Hunter T. Phosphopeptide mapping and phosphoamino acid analysis by two dimensional separation on thin-layer cellulose plates. Methods Enzymol. 1991;201:110–148. doi: 10.1016/0076-6879(91)01013-r. [DOI] [PubMed] [Google Scholar]
- 4.Cuenda A, Cohen P, Buee-Scherrer V, Goedert M. Activation of stress-activated protein kinase-3 (SAPK3) by cytokines and cellular stresses is mediated via SAPKK3 (MKK6); comparison of the specificities of SAPK3 and SAPK2 (RK/p38) EMBO J. 1997;16:295–305. doi: 10.1093/emboj/16.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis R J. MAPKs: new JNK expands the group. Trends Biochem Sci. 1994;19:470–473. doi: 10.1016/0968-0004(94)90132-5. [DOI] [PubMed] [Google Scholar]
- 6.Derijard B, Raingeaud J, Barrett T, Wu I, Han J, Ulevitch R J, Davis R J. Independent human MAP kinase signal transduction pathways defined by MEK and MKK isoforms. Science. 1995;267:682–685. doi: 10.1126/science.7839144. [DOI] [PubMed] [Google Scholar]
- 7.Fanger G R, Gerwins P, Widmann C, Jarpe M B, Johnson G L. MEKKs, GCKs, MLKs, PAKs, TAKs, and Tpls: upstream regulators of the c-Jun amino-terminal kinases? Curr Opin Genet Dev. 1997;7:67–74. doi: 10.1016/s0959-437x(97)80111-6. [DOI] [PubMed] [Google Scholar]
- 8.Fukunaga R, Hunter T. MNK1, a new MAP kinase-activated protein kinase, isolated by a novel expression screening method for identifying protein kinase substrates. EMBO J. 1997;16:1921–1933. doi: 10.1093/emboj/16.8.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta S, Campbell D, Derijard B, Davis R J. Transcription Factor ATF2 regulation by the JNK signal transduction pathway. Science. 1995;267:389–393. doi: 10.1126/science.7824938. [DOI] [PubMed] [Google Scholar]
- 10.Han J, Jiang Y, Li Z, Kravchenko V V, Ulevitch R T. Activation of the transcription factor MEF2C by the MAP kinase p38 in inflammation. Nature. 1997;386:296–299. doi: 10.1038/386296a0. [DOI] [PubMed] [Google Scholar]
- 11.Han J, Lee J-D, Bibbs L, Ulevitch R J. A MAP kinase targeted by endotoxin and hyperosmolarity in mammalian cells. Science. 1994;265:808–811. doi: 10.1126/science.7914033. [DOI] [PubMed] [Google Scholar]
- 12.Han J, Lee J-D, Jiang Y, Li Z, Feng L, Ulevitch R J. Characterization of the structure and function of a novel MAP kinase kinase (MKK6) J Biol Chem. 1996;271:2886–2891. doi: 10.1074/jbc.271.6.2886. [DOI] [PubMed] [Google Scholar]
- 13.Han J, Lee J D, Tobias P S, Ulevitch R J. Endotoxin induces rapid protein tyrosine phosphorylation in 70Z/3 cells expressing CD14. J Biol Chem. 1993;268:25009–25014. [PubMed] [Google Scholar]
- 14.Han J, Wang X, Jiang Y, Ulevitch R J, Lin S. Identification and characterization of a predominant isoform of human MKK3. FEBS Lett. 1996;403:19–22. doi: 10.1016/s0014-5793(97)00021-5. [DOI] [PubMed] [Google Scholar]
- 15.Han T-H, Prywes R. Regulatory role of MEF2D in serum induction of the c-jun promoter. Mol Cell Biol. 1995;15:2907–2915. doi: 10.1128/mcb.15.6.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janknecht R, Hunter T. Convergence of MAP kinase pathways on the ternary complex factor Sap-1a. EMBO J. 1997;16:1620–1627. doi: 10.1093/emboj/16.7.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang Y, Gram H, Zhao M, New L, Gu J, Feng L, Padova F, Ulevitch R, Han J. Characterization of the structure and function of the fourth member of p38 group MAP kinase-p38δ. J Biol Chem. 1997;272:30122–30128. doi: 10.1074/jbc.272.48.30122. [DOI] [PubMed] [Google Scholar]
- 18.Jiang Y, Chen C, Li Z, Guo W, Gegner J A, Lin S, Han J. Characterization of the structure and function of a new mitogen-activated protein kinase (p38β) J Biol Chem. 1996;271:17920–17926. doi: 10.1074/jbc.271.30.17920. [DOI] [PubMed] [Google Scholar]
- 19.Kato Y, Kravchenko V, Tapping R, Han J, Ulevitch R, Lee J. BMK1/ERK5 regulates serum-induced early gene expression through transcription factor MEF2C. EMBO J. 1997;16:7054–7066. doi: 10.1093/emboj/16.23.7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19a.Kato, Y., et al. Unpublished data.
- 20.Kramer R M, Roberts E F, Um S L, Borsch-Haubold A G, Watson S P, Fisher M J, Jakubowski J A. p38 mitogen-activated protein kinase phosphorylates cytosolic phospholipase A2 (cPLA2) in thrombin-stimulated platelets. Evidence that proline-directed phosphorylation is not required for mobilization of arachidonic acid by cPLA2. J Biol Chem. 1996;271:27723–27729. doi: 10.1074/jbc.271.44.27723. [DOI] [PubMed] [Google Scholar]
- 21.Kravchenko V V, Pan Z, Han J, Herbert J-M, Ulevitch R J, Ye R D. Platelet-activating factor induces NF-κB activation through a G protein-coupled pathway. J Biol Chem. 1995;270:14928–14934. doi: 10.1074/jbc.270.25.14928. [DOI] [PubMed] [Google Scholar]
- 22.Lechner C, Zahalka M A, Giot J-F, Moler N P H, Ullrich A. ERK6, a mitogen-activated protein kinase involved in C2C12 myoblast differentiation. Proc Natl Acad Sci USA. 1996;93:4355–4359. doi: 10.1073/pnas.93.9.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee J C, Laydon J T, McDonnell P C, Gallagher T F, Kumar S, Green D, McNulty D, Blumenthal M J, Heyes R J, Landvatter S W, Strickler J E, McLaughlin M M, Siemens I, Fisher S, Livi G P, White J R, Adams J L, Young P R. Identification and characterization of a novel protein kinase involved in the regulation of inflammatory cytokine biosynthesis. Nature. 1994;372:739–746. doi: 10.1038/372739a0. [DOI] [PubMed] [Google Scholar]
- 24.Li Z, Jiang Y, Ulevitch R J, Han J. The primary structure of p38gamma: a new member of p38 group of MAP kinase. Biochem Biophys Res Commun. 1996;228:334–340. doi: 10.1006/bbrc.1996.1662. [DOI] [PubMed] [Google Scholar]
- 25.Lilly B, Zhao B, Ranganayakulu G, Paterson B M, Schulz R A, Olson E N. Requirement of MADS domain transcription factor D-MEF2 for muscle formation in Drosophila. Science. 1995;267:688–693. doi: 10.1126/science.7839146. [DOI] [PubMed] [Google Scholar]
- 26.Lin Q, Schwarz J, Bucana C, Olson E N. Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science. 1997;276:1404–1407. doi: 10.1126/science.276.5317.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ludwig S, Engel K, Hoffmeyer A, Sithanandam G, Neufeld B, Palm D, Gaestel M, Rapp U R. 3pK, a novel mitogen-activated protein (MAP) kinase-activated protein kinase, is targeted by three MAP kinase pathways. Mol Cell Biol. 1996;16:6687–6697. doi: 10.1128/mcb.16.12.6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin J F, Miano J M, Hustad C M, Copeland N G, Jenkins N A, Olson E N. A Mef2 gene that generates a muscle-specific isoform via alternative mRNA splicing. Mol Cell Biol. 1994;14:1647–1656. doi: 10.1128/mcb.14.3.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McLaughlin M M, Kumar S, McDonnell P C, Van Horn S, Lee J C, Livi G P, Young P R. Identification of mitogen-activated protein (MAP) kinase-activated protein kinase-3, a novel substrate of CSBP p38 MAP kinase. J Biol Chem. 1996;271:8488–8492. doi: 10.1074/jbc.271.14.8488. [DOI] [PubMed] [Google Scholar]
- 30.Mertens S, Craxton M, Goedert M. SAP kinase-3, a new member of the family of mammalian stress-activated protein kinases. FEBS Lett. 1996;383:273–276. doi: 10.1016/0014-5793(96)00255-4. [DOI] [PubMed] [Google Scholar]
- 31.Molkentin J D, Black B L, Martin J F, Olson E N. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell. 1995;83:1125–1136. doi: 10.1016/0092-8674(95)90139-6. [DOI] [PubMed] [Google Scholar]
- 32.Molkentin J D, Li L, Olson E N. Phosphorylation of the MADS-box transcription factor MEF2C enhances its DNA binding activity. J Biol Chem. 1996;271:17199–17204. doi: 10.1074/jbc.271.29.17199. [DOI] [PubMed] [Google Scholar]
- 33.New L, Jiang Y, Zhao M, Liu K, Zhu W, Flood L J, Kato Y, Parry G C, Han J. PRAK, a novel protein kinase regulated by the p38 MAP kinase. EMBO J. 1998;17:3372–3384. doi: 10.1093/emboj/17.12.3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olson E N, Perry M, Schulz R A. Regulation of muscle differentiation by the MEF2 family of MADS box transcription factors. Dev Biol. 1995;172:2–14. doi: 10.1006/dbio.1995.0002. [DOI] [PubMed] [Google Scholar]
- 35.Ornatsky O I, McDermott J C. MEF2 protein expression, DNA binding specificity and complex composition, and transcriptional activity in muscle and non-muscle cells. J Biol Chem. 1996;271:24927–24933. doi: 10.1074/jbc.271.40.24927. [DOI] [PubMed] [Google Scholar]
- 35a.Puri, P. L., et al. Personal communication.
- 36.Raingeaud J, Gupta S, Rogers J S, Dickens M, Han J, Ulevitch R J, David R J. Pro-inflammatory cytokines and environmental stress cause p38 MAP kinase activation by dual phosphorylation on tyrosine and threonine. J Biol Chem. 1995;270:7420–7426. doi: 10.1074/jbc.270.13.7420. [DOI] [PubMed] [Google Scholar]
- 37.Raingeaud J, Whitmarsh A J, Barrett T, Derijard B, Davis R J. MKK3- and MKK6-regulated gene expression is mediated by the p38 mitogen-activated protein kinase signal transduction pathway. Mol Cell Biol. 1996;16:1247–1255. doi: 10.1128/mcb.16.3.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robinson M J, Cobb M H. Mitogen-activated protein kinase pathways. Curr Opin Cell Biol. 1997;9:180–186. doi: 10.1016/s0955-0674(97)80061-0. [DOI] [PubMed] [Google Scholar]
- 39.Rouse J, Cohen P, Trigon S, Morange M, Alonso-Llamazares A, Zamanillo D, Hunt T, Nebreda A R. A novel kinase cascade triggered by stress and heat shock that stimulates MAPKAP kinase-2 and phosphorylation of the small heat shock proteins. Cell. 1994;78:1027–1037. doi: 10.1016/0092-8674(94)90277-1. [DOI] [PubMed] [Google Scholar]
- 40.Su B, Karin M. Mitogen-activated protein kinase cascades and regulation of gene expression. Curr Opin Immunol. 1996;8:402–411. doi: 10.1016/s0952-7915(96)80131-2. [DOI] [PubMed] [Google Scholar]
- 41.Wang X S, Diener K, Manthey C L, Wang S, Rosenzweig B, Bray J, Delaney J, Cole C N, Chan-Hui P Y, Mantlo N, Lichenstein H S, Zukowski M, Yao Z. Molecular cloning and characterization of a novel p38 mitogen-activated protein kinase. J Biol Chem. 1997;272:23668–23674. doi: 10.1074/jbc.272.38.23668. [DOI] [PubMed] [Google Scholar]
- 42.Wang X-Z, Ron D. Stress-induced phosphorylation and activation of the transcription factor CHOP (GADD153) by p38 MAP kinase. Science. 1996;272:1347–1349. doi: 10.1126/science.272.5266.1347. [DOI] [PubMed] [Google Scholar]
- 43.Wang Y, Huang S, Sah V, Ross J, Brown J, Han J, Chien K. Cardiac muscle cell hypertrophy and apoptosis induced by distinct members of p38 mitogen-activated protein kinase family. J Biol Chem. 1998;273:2161–2168. doi: 10.1074/jbc.273.4.2161. [DOI] [PubMed] [Google Scholar]
- 44.Waskiewicz A J, Cooper J A. Mitogen and stress response pathways: MAP kinase cascades and phosphatase regulation in mammals and yeast. Curr Opin Cell Biol. 1995;7:798–805. doi: 10.1016/0955-0674(95)80063-8. [DOI] [PubMed] [Google Scholar]
- 45.Waskiewicz A J, Flynn A, Proud C G, Cooper J A. Mitogen-activated protein kinases activate the serine/threonine kinases Mnk1 and Mnk2. EMBO J. 1997;16:1909–1920. doi: 10.1093/emboj/16.8.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu Y-T, Breitbart R E, Smoot L B, Lee Y, Mahdavi V, Nadal-Ginard B. Human myocyte-specific enhancer factor 2 comprises a group of tissue-restricted MADS box transcription factors. Genes Dev. 1992;6:1783–1798. doi: 10.1101/gad.6.9.1783. [DOI] [PubMed] [Google Scholar]
- 47.Zechner D, Thuerauf D J, Hanford D S, McDonough P M, Glembotski C C. A role for the p38 mitogen-activated protein kinase pathway in myocardial cell growth, sarcomeric organization, and cardiac-specific gene expression. J Cell Biol. 1997;139:115–127. doi: 10.1083/jcb.139.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]