Abstract
Many human genes are transcribed from both strands and produce sense-antisense gene pairs. Sense-antisense (SAS) chimeric transcripts are produced upon the coalescing of exons/introns from both sense and antisense transcripts of the same gene. SAS chimera was first reported in prostate cancer cells. Subsequently, numerous SAS chimeras have been reported in the ChiTaRS-2.1 database. However, the landscape of their expression in human cells and functional aspects are still unknown. We found that longer palindromic sequences are a unique feature of SAS chimeras. Structural analysis indicates that a long hairpin-like structure formed by many consecutive Watson-Crick base pairs appears because of these long palindromic sequences, which possibly play a similar role as double-stranded RNA (dsRNA), interfering with gene expression. RNA–RNA interaction analysis suggested that SAS chimeras could significantly interact with their parental mRNAs, indicating their potential regulatory features. Here, 267 SAS chimeras were mapped in RNA-seq data from 16 healthy human tissues, revealing their expression in normal cells. Evolutionary analysis suggested the positive selection favoring sense-antisense fusions that significantly impacted the evolution of their function and structure. Overall, our study provides detailed insight into the expression landscape of SAS chimeras in human cells and identifies potential regulatory features.
INTRODUCTION
The fusion of exons or introns from two different genes can lead to the production of chimeric transcripts (1,2). Numerous studies have addressed the functional roles of various chimera in cancer progression, neurological disorders and other genetic abnormalities (3–10). Abundant levels of chimeric transcripts are also found in normal cells (11,12). Chimeric transcripts can be produced by several mechanisms, including cis-splicing of adjacent genes (13,14), trans-splicing (15,16), chromosomal translocation (17) and transcriptional slippage (11,18). Trans-splicing is frequently observed in lower eukaryotes, where it plays a vital role in generating functional diversity and regulating the expression of genes involved in cell viability and growth (19). Trans-splicing is also believed to be an important reason for the production of fusion transcripts in normal cells (20,21). However, trans-splicing occurs at a very low frequency in higher vertebrates, and its underlying mechanisms remain unknown. Increasing evidence has shown that trans-splicing frequently occurs in physiological and pathological processes (11,19,21), although these results have been questioned due to the possibility of chimeric artifacts appearing as a result of reverse transcription (22). Furthermore, detecting several recurrent chimeric transcripts in high throughput RNA-seq data supports the claim that chimeric transcripts could be produced via trans-splicing mechanisms (12). Both splicing and gene fusions are common phenomena that appear in a substantial proportion of normal tissues, as well as in cancer cells (21,23,24).
The exponential increase in next-generation sequencing data has helped identify over 150,000 chimeric transcripts collected in various databases and public datasets (25). One popular chimeric RNA resource is ChiTaRS (Chimeric Transcripts and RNA-Seq data), originally released in 2012 and subsequently updated (26–29). In ChiTaRS 2.1, a new sub-class of 6044 gene fusions, sense-antisense (SAS) chimeras, was reported (27). These chimeric RNAs are generated by the fusion of exons and/or introns from the sense and antisense strands of the same gene. A significant number of mammalian genes were reported to be transcribed from both the strands of the same gene and producing sense-antisense gene pairs (30). The antisense transcripts of the genes are generally long non-coding RNA transcripts and served as a regulator of sense transcripts (31). Co-expression of both sense and antisense transcripts in the cells is crucial for maintaining normal functional processes (32). SAS chimeras were first reported in prostate cancer cells, where antisense transcripts of the KLK4 gene form chimeras with the KLK4 sense transcript (18). Subsequent RNA-seq analysis of >300 samples showed a significant proportion of recurrent chimeric SAS transcripts in different cancer cell lines (27–29). However, the underlying mechanism of SAS chimera formations and their functional significance have not yet been explored.
To explore the potential roles of SAS chimeric transcripts in humans, the present study aimed to investigate their structural and functional features. We detected long palindromic sequences present at or near the junction sites of most SAS chimeras. We also predicted that >85% of SAS chimeric transcripts are long non-coding RNA (lncRNA). Analysis of RNA secondary structure often plays a significant role in determining the function of lncRNA transcripts (33,34). Many regulatory RNA functions depend on secondary structure, which can be altered in response to a diverse array of cellular conditions (35). Hence, we were interested in studying structural aspects of these SAS chimeras, particularly the functional role of the palindromic sequences. We found that SAS chimeras form long hairpin-like structures along the length of the palindromic regions, indicative of their possible function as double-stranded RNA (dsRNA) that can serve to inhibit gene expression. Furthermore, RNA-RNA interaction analysis by free energy minimization uncovered the potential interaction of SAS chimeras with their parental mRNAs. This result indicates that this long hairpin-like structure could enable SAS chimeras to interact with their parental mRNA transcripts and regulate their expression in response to different cellular conditions. Next, we mapped several SAS chimeras in different healthy human tissues and detected potential orthologs in mice. Our study thus uncovered essential regulatory features of SAS chimeras and highlighted their expression landscape in human cells.
MATERIALS AND METHODS
Mining SAS chimeras and mapping them in RNA-seq data
From 66,243 chimeric RNAs available in the ChiTaRS database (29), we isolated a specific set of chimeric fusions that occurred between the sense and antisense strands of the same gene. We thus collected 5180 human SAS chimeras. First, we defined the junction region of each chimera using the formula: Junction region = overlapping sequence +15 bp upstream sequence +15 bp downstream sequence. Pre-processing of human chimeras was performed to filter-out low-quality chimeras. In this process, low-quality chimeras were filtered out based on the criteria: (i) a junction sequence of a chimera identical to a human genome or transcriptome sequence (identity 95% and query coverage 100%) was removed, (ii) chimeras with duplicate junction sequences were excluded, (iii) chimeras with high percentages of A, T and N nucleotides at junction sequences were removed because of minimization of the possibilities of poly-A tails or ambiguous nucleotides, (iv) chimeras with overlaps 40 bp-long were excluded and (v) chimeras with short–length parental genes of <50 bp were removed. Accordingly, we trimmed the number of human SAS chimeras with a unique junction to 2896 based on the above stringent criteria.
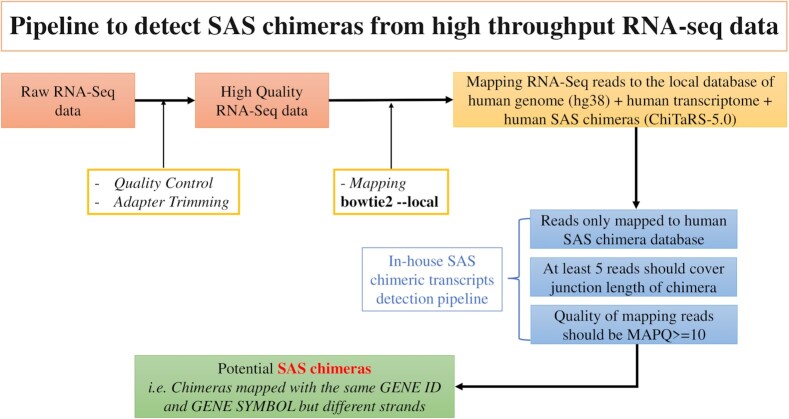
Next, to map the SAS chimeras in RNA-seq data, we employed the pipeline depicted in Figure 1. First, we built a local database consisting of the human genome (hg38), human transcriptome and human SAS chimeras (ChiTaRS-5.0). Then, we used Bowtie2 (36) for mapping RNA-seq reads to this database. The potential SAS chimeras were identified from the RNA-seq data if they satisfied the following three criteria: (i) Reads only mapped with human SAS chimeras and not with the human genome or transcriptome, (ii) at least five reads covered the junction length of the chimera and (iii) the quality of mapping reads was MAPQ> = 10.
Figure 1.
The in-house pipeline used to detect SAS chimeras in RNA-seq data.
Predicting the protein-coding and non-coding nature of SAS chimeric transcripts
To predict the protein-coding abilities of SAS chimeric transcripts, we used CNIT (http://cnit.noncode.org/CNIT/) (37), CPAT (http://lilab.research.bcm.edu/cpat/) (38) and LncFinder (39). CPAT (38) can accurately distinguish between coding and non-coding mammalian transcripts (40). CNIT (37) is a recently published tool for more accurately identifying the coding ability of RNA transcripts based on intrinsic features of the sequences. CNIT predicts the protein-coding ability of human transcripts with 98% accuracy. LncFinder (39) can predict novel lncRNAs using machine-learning approaches based on features extracted from sequence-intrinsic composition, secondary structure and physicochemical properties. We annotated the SAS chimeras as protein-coding or as lncRNA when the output of these three tools agreed.
Prediction of RNA secondary structure
The secondary structures of selected SAS chimeras were predicted using the RNAfold tool from the ViennaRNA Package (41). To predict RNA secondary structure, the tool minimizes free energy (MFE structure) using the Zuker algorithm (42). The energy model is loop-based, and free energy of RNA secondary structure is calculated as the sum of the free energies of all loops. The structures were colored according to base-pairing probabilities, as calculated with a partition function that sums all Boltzmann-weighted free energies of all possible secondary structures (43). The nucleic acid sequence of chimeras was provided as input to the command line tool with default parameters. For each SAS chimera, we predicted structural accessibility, reflecting nucleotides as being paired or unpaired using Raccess (44). CentroidFold (45) was used to compute the base-pairing probability plot. LncTar (46) was used to detect interactions between the lncRNA (SAS chimeric transcript) with the mRNA sequence of the parental gene. Information regarding free energy of binding and duplex formation between the mRNAs (parental genes) and the SAS chimeras was obtained using the RNAfold web server (41) of ViennaRNA. The RNAup server was used to predict RNA–RNA interactions. RNAup (47) first assesses the energy needed to open the structure for every stretch of bases up to a certain length for both RNA molecules. Then, the interaction-free energy is computed and combined with the opening energy for the entire micro-state range to obtain the total binding energy. IntaRNA (48) was also used to predict RNA–RNA interactions and for visualization.
Gene Ontology analysis and analysis in cancers
Gene ontology and pathway enrichment analyses were performed using ShinyGO v0.61(49) and Panther (49). To investigate associations of genes forming SAS chimeras in cancer, we used TumorPortal (50) and CancerMine (51), and performed thorough data mining of PubMed resources. We downloaded RNA-seq data of breast tumors from the Pan-Cancer Analysis of Whole Genomes (PCAWG) project (52,53) and also downloaded RNA-seq of the breast cancer cell line MCF-7 from the Cancer Cell Line Encyclopedia (CCLE) (54).
RESULTS
Characterization of SAS chimeric transcripts based on their predicted coding potentialities
Predicting the protein-coding abilities of transcripts is important for understanding their functions. Therefore, we collected 5180 human SAS chimeras (Supplementary Table S1) found in the ChiTaRS 5.0 database (29) and predicted their coding abilities. SAS chimeras were thus classified as protein-coding or long non-coding RNA (lncRNA) transcripts. It is, however, important to note that computational prediction of a transcript as non-coding does not always mean that that transcript acts as a functional lncRNA. Indeed, recent studies have demonstrated that few annotated lncRNAs can be translated into micro-peptides (55,56). Therefore, we used the three most widely used tools for evaluating coding potentiality (see Materials and Methods) and only annotated SAS chimera coding abilities when the output from all three tools was the same. We thus found that only 5.97% of the transcripts possess protein-coding ability and that 85.1% of SAS chimeric transcripts cannot be translated into proteins (Supplementary Table S2). We could not annotate the coding abilities of 8.93% of the SAS chimeras because the three tools used for the predicting coding ability did not agree. These results highlight that SAS chimeras are less likely to generate fusion proteins and may instead act as lncRNAs.
Sequence feature analysis reveals that SAS chimeras contain longer palindromic sequences
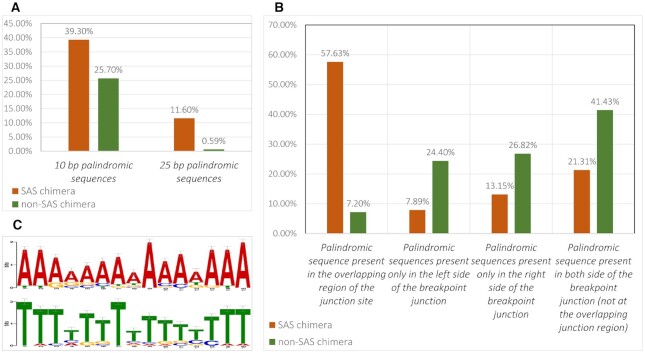
We consulted the ChiTaRS 5.0 database (29) and downloaded FASTA sequences and associated details for 5180 SAS chimeras identified in humans. We trimmed this number to the 2896 SAS chimeras that matched the stringent criteria delineated in the Materials and Methods. We analyzed sequence features of the SAS chimeras and found that a significant number included palindromic sequences. We explicitly checked for patterns of palindromic sequences present in both SAS and non-SAS chimera groups. For the purposes of this study, the minimum length of a palindrome was fixed to 10 bp, and the number of permitted mismatches was set at zero to avoid false positives. Among the 2896 SAS chimeras, palindromic sequences were present in 1140 chimeras (39.3%), with a median length of 17 bp. To assess the significance of the longer palindromic sequences noted in SAS chimeras, we checked the status of palindromic sequences in all other non-SAS chimeras that have been identified in humans. We thus extracted a total of 66,243 human chimeric RNAs from the ChiTaRS 5.0 database (29) and reduced this list to the 16,706 that matched the strict criteria for high-quality chimeras using our in-house detection pipeline. As with the SAS chimeras, the minimum palindrome length was fixed at 10 bp, and the number of permitted mismatches was set at zero. Among these 16,706 non-SAS chimeras, 4298 chimeric RNAs contain palindromic sequences (25.7% of all non-SAS chimera), with a median length of 11 bp (Figure 2A). We further compared the SAS and non-SAS chimera groups based on the presence of at least 25 bp-long palindromic sequences. We found 336 SAS chimeras (11.6%) and 100 non-SAS chimeras (0.59%) with palindromic sequences >25 bp. This indicates that the presence of longer palindromic regions is a unique feature of SAS chimeras.
Figure 2.
(A) Comparison of 10 bp- and 25 bp-long palindromic sequences in SAS and non-SAS chimeras. (B) Location of palindromic sequences in SAS and non-SAS chimeras. (C) The most frequently observed palindromic sequence motifs present at the junction regions of SAS chimeras.
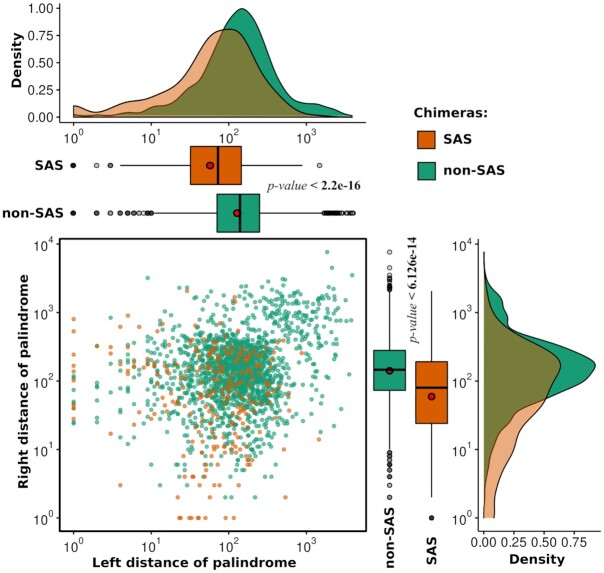
Next, we addressed the specific locations of the palindromic sequences within chimeras from both the SAS and non-SAS groups. We observed that palindromic sequences are located within the overlapping region of the junction site of 57.63% SAS chimeras and 7.2% non-SAS chimeras (Figure 2B). Since several palindromes appeared at the breakpoint junction site of SAS chimeras, we represented these regions in the form of a motif (Supplementary Table S3, Sheet 2) using the MEME motif discovery tool (57). The most frequently observed palindromic sequence at junction regions (E-value 2.6E–004) is represented in Figure 2C. In 21.31% SAS chimeras and 41.43% non-SAS chimeras, we found palindromic sequences that were not exactly located in the overlapping regions of the junction, but rather are present on both sides of the junction region. For this group of SAS and non-SAS chimeras where palindromic sequences are located on both sides of the junction, we calculated the distance of the palindromic sequences from the breakpoint junction regions. We found this distance to be significantly higher in non-SAS chimeras than in SAS chimeras (Figure 3). Moreover, we observed that the presence of palindromic sequences on only one side of the breakpoint junction site was lower in the case of SAS chimeras, as compared to non-SAS chimeras (Figure 2B). This result indicates that the longer palindromic sequences within or near the junction regions of the SAS chimeras might play significant roles in chimera functionality. Therefore, from this list of SAS chimeras, we selected the top 100 SAS chimeric RNAs with palindromic regions 41 bp-long or above for subsequent in-depth analysis. We further sought to identify the functional role and repercussions of such lengthy palindromes in the human genome.
Figure 3.
Distances of the palindromic sequences from the breakpoint junction regions for SAS and non-SAS chimeras where palindromic sequences are located on both sides of the junction.
Structural analysis of SAS chimeras with long palindromic sequences
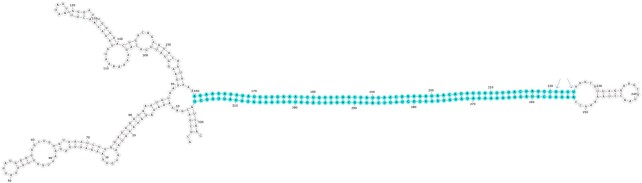
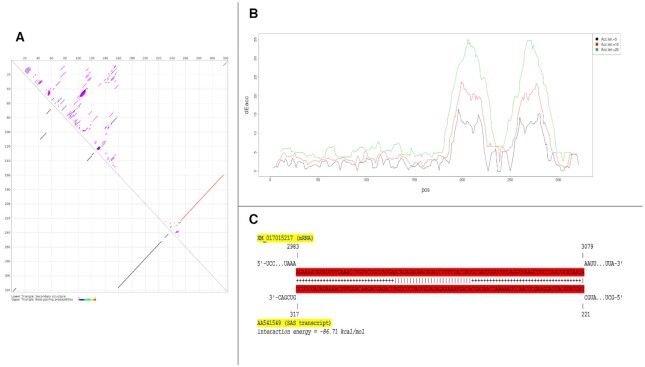
The study of RNA secondary structure is critical to understanding the function and regulation of RNA transcripts (58–61). Hence, we predicted the secondary structure of the top 100 SAS chimeras with long palindromic sequences using the RNAfold tool of the ViennaRNA package (41). Of the 100 chimeras analyzed, 91 formed long paired structures within the palindromic regions across the two strands. These strands projected outwards, forming a hairpin-like structure. Nine other chimeras also formed similar structures, although a few nucleotides were missing toward the pairing edge. This result strengthened our hypothesis that the palindromic regions play a significant role in forming the SAS chimeras considered here. From the top 100 SAS chimeras, identified on the basis of palindromic sequence length, a representative image of chimera ID AA541549 is depicted in Figure 4, where the highlighted base-pairing portion represents the palindromic region. From the base-pairing probability dot plot (62) generated by CentroidFold (45) of this SAS chimera (Figure 5A), it can be seen that the palindromic repeat region is present at the junction site of the chimera, where overlapping nucleotides from both the sense (region 160–224) and anti-sense strands (region 253–317) of the gene are seen, and which could give rise to a hairpin-like structure. We assume that such structures enable the formation of SAS chimeras (which are generally single-stranded) that can function as dsRNA.
Figure 4.
Minimum free energy (MFE) secondary structure of a SAS chimeric transcript (Chimera ID: AA541549). Highlighted base-pairing in the figure represents the palindromic region. The arrow in the figure represents the overlapping junction region.
Figure 5.
(A) Base-pairing probability plot of an SAS chimeric transcript (Chimera ID: AA541549). (B) Structural accessibility of the SAS chimera AA541549. Raccess computed the accessibility of segment [a, b] = [x, x+l-1] in the transcript for all positions x with a fixed length of l (Acc.len) = 5, 10, 20. In the figure, access energy ([a, b]) is plotted at (x + l/2). (C) Interaction of SAS chimera AA541549 with its parental mRNA (Gene ID: XM_017015217). The highlighted base-pairs for a given pair of sequences represent the minimal energy of the RNA–RNA interaction that can be formed between the two RNAs at each sequence position.
Several natural dsRNAs are found in human cells which participate in major biological processes (63). Recent studies have shown that dsRNAs can give rise to small interfering RNA (siRNA), upon degradation (64). Only organisms possessing RNA-dependent RNA polymerase (RdRP) activity are known to show gene regulation due to endogenous siRNA (65). However, since humans do not possess RdRP, the nature of endogenous siRNA activity and the related gene regulation is still unclear (65). We hypothesized that SAS chimeras could function as dsRNAs in humans through a similar mechanism as seen in RdRP-presenting species. Accessibility across the target sites has been shown to influence the efficacy of siRNA activity (66). Hence, to test our assumption that SAS chimeric transcripts are involving in regulating their parental genes, we first calculated the structural accessibility of SAS chimeras using Raccess software (44). We observed that the structural accessibility of SAS chimeras is maximal across the length of palindromic sequences, which indicate these regions could potentially be involved in interactions with target sites. For chimera ID AA541549, one can see that the palindromic regions span base pairs 160–224 and 253–317 (Figure 4), and how these can base-pair (Figure 5A), while Figure 5B, revealing that structural accessibility is highest in these regions. We further analyzed the palindromic sequences of 100 SAS chimeras using LncTar (46) to test if they potentially interact with their parental mRNAs. We identified 77 SAS chimeras that can potentially interact with their parental genes (above a cutoff of normalized deltaG = -0.1) (Supplementary Table S4). For detailed understanding, we represented the interaction of chimera ID AA541549 with its parental mRNA XM_017015217.2 using IntaRNA (48) (Figure 5C), which revealed that the region spanning base pairs 221–317 interacts with its parental mRNA (normalized deltaG for LncTar analysis = -0.7371). This portion of the SAS chimera is found within the palindromic regions and also shows the highest structural accessibility (as determined from Figure 5A ,B). Finally, RNAup (47) was used to predict that position in the sequence which could generate the optimal secondary structure upon interaction. Such analysis revealed that 229 to 252 positions, corresponding to the intermediate location of the palindromic regions (160–224 and 253–317) are crucial for generating the optimal structure upon interaction. Therefore, we speculate that SAS chimeras with longer palindromic sequences are important for generating a dsRNA structure, which has significance in functional regulation.
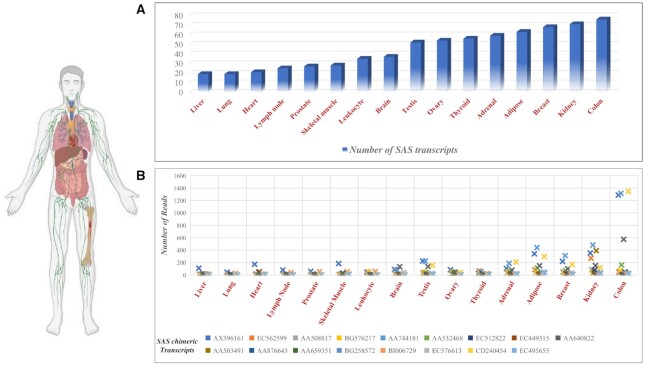
Analysis of SAS chimeras in RNA-seq data of the Human Body Map project
In the ChiTaRS database, SAS chimeras were mapped in RNA-seq data collected from the different cancer cell lines, with some being expressed in cancers (27–29). However, their expressions in different human tissues under normal physiological conditions are not known. Hence, we analyzed RNA-seq data collected from 16 human tissues as part of the Illumina Human Body Map project 2.0 (GEO accession: GSE30611). We were able to map 267 SAS chimeras in 16 different tissues. Of these, 119 were mapped to more than one tissue (Figure 6A and Supplementary Table S5). We observed that tissue-specific SAS chimeras have relatively lower mapping reads than SAS chimeras which are expressed in multiple tissues. Seventeen SAS chimeras were mapped in at least 10 different tissues (Figure 6B). Among these, 11 are produced from mitochondrial genes, including ATP6, COX3, CYTB, ND2, ND3, ND4 and ND6. Recent transcriptome profiling of human mitochondria revealed that mitochondrial genes are transcribed from both strands and can generate sense-antisense pairs (67). Therefore, there could be possibilities that the fusion of the sense and antisense transcripts of the same mitochondrial gene might be necessary for diversifying cellular functions.
Figure 6.
(A) The number of SAS chimeras per tissue. (B) The number of reads of SAS chimeras expressed in at least 10 tissues.
To understand the potential functional associations of SAS chimeras mapped in the RNA-seq data of 16 human tissues, we analyzed functional characteristics of their parental genes. For this, we performed gene ontology and pathway enrichment analyses using ShinyGO v0.61 with a P-value cutoff of 0.05 (68). The results indicated the involvement of many parental genes in various cellular functions. The most enriched GO biological processes were response to organic and inorganic substances, regulation of multicellular organismal processes, cellular localization and cell death. Molecular function analysis indicated that parental genes are involved in binding (GO:0005488)-related processes, including enzyme binding, protein-containing complex binding, ubiquitin-protein ligase binding, RNA binding, cell adhesion molecule binding, protein kinase binding and signaling receptor binding. This implies susceptibility of the genes to selective, non-covalent interactions with other molecules. In assessing cellular component GO terms, extracellular region and cell junction were associated with a majority of the enriched genes. Analysis of gene enrichment against the curated Reactome database (69) highlighted the potential role of these genes in disease, immune systems, cellular responses to stress and metabolism of proteins. Taken together, these findings highlight that SAS chimeras could be important for regulating various cellular functions. Results of the enrichment analysis are provided in Supplementary Table S6.
Evolutionary trends in sense-antisense fusion
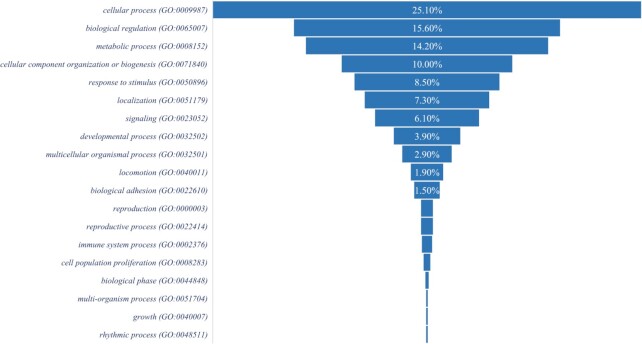
Most of the recurrent fusion transcripts which are found in the normal cells could be essential for physiological functions specific to an organism (12). The mechanism of gene fusion depends on an organism’s genome complexity. One can check whether such fusion events are beneficial through the evolutionary selection across species. However, it is difficult to analyze interspecies divergence at the transcript level due to several reasons: (i) fusion breakpoints within a gene can vary between species, (ii) to define a true fusion transcript, we considered the junctions of fusions as being unique. Therefore, sequence conservation at the fusion junction might be low across different species; (iii) there may be a lack of orthologous expression, and (iv) if fusion transcripts are expressed at specific developmental stages or under stress conditions in an organism, there is a minimal chance of analyzing conservation across species. Therefore, to enable direct comparison of sense-antisense fusion events, we identified common genes that can produce SAS chimeric transcripts across species. We extracted the list of SAS chimeras and their sequences in different organisms from the ChiTaRS database (29). We observed that Homo sapiens and Mus musculus (mouse) conserved 429 parental genes that can generate multiple SAS chimeras (Supplementary Table S7). Similarly, we observed 14 conserved parental genes between Homo sapiens and Sus scrofa (wild boar) and 16 conserved parental genes between Homo sapiens and Drosophila melanogaster (fruit fly) that are involved in the appearance of multiple SAS chimeras in these organisms. To understand the functions associated with a large number of conserved parental genes of SAS chimeras in humans and mice, we performed gene enrichment analysis and found enrichment for cellular processes, biological regulation and metabolic processes (Figure 7). We speculated that the formation of these SAS chimeras has functional importance for both species. Thus, they underwent evolutionary selection. Further availability of SAS chimeras from different organisms and transcriptome profiling of various organisms could provide additional detailed evolutionary insight into the orthologous expression of SAS chimeras.
Figure 7.
Gene enrichment analysis for biological processes of parental genes found to generate multiple SAS chimeras in humans and mice.
Is the appearance of SAS chimeric transcripts a consequence of trans-splicing or splicing errors?
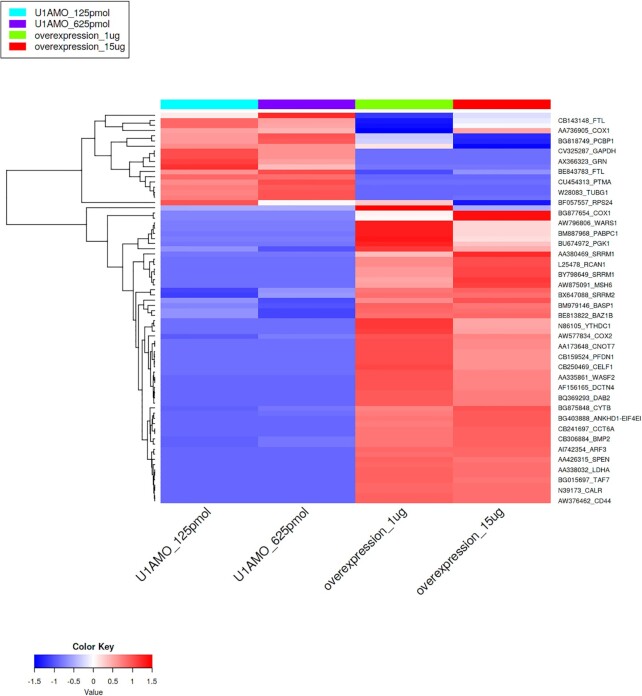
Trans-splicing of RNAs could generate the chimeric transcripts in cancer, as well as in normal cells (21,70). Recent studies have shown that trans-splicing employs splicing machinery similar to that of alternative splicing (19). Evidence from recent studies suggested that the U1 snRNP spliceosome complex promotes trans-splicing in Drosophila (71) and trypanosomes (72). U1 snRNP has been recognized as an important player for the regulation of alternative splicing in mammals (73,74). The binding of U1snRNP to the 5′ splice site promotes initial splicing complex formation and regulates subsequent spliceosome assembly (75). Therefore, we tested the hypothesis that U1snRNP regulates trans-splicing in humans, which promotes the generation of SAS transcripts. To assess whether knockdown and overexpression of U1 snRNP modulates the differential expression of SAS transcripts in cells, we downloaded RNA-seq dataset from Bioproject PRJNA590153 (76). To generate this dataset, knockdown and over-expression of U1 snRNP in the HeLa cell line were performed and total RNA-seq data were collected. These data were downloaded (GSE140543) and mapped to the SAS chimeric transcripts. We detected 421 SAS chimeras found in at least in 50% samples. Among these, 141 SAS chimeras were detected in all samples from the U1 snRNP knockdown and overexpression dataset. Next, we performed differential analysis of SAS chimeras using DEseq2 (77) with a cutoff P value <0.05 and fold-change >2. We observed 58 significantly up-regulated SAS chimeras, and 18 significantly down-regulated SAS chimeras in the over-expression dataset (Figure 8 and Supplementary Table S8). The large number of up-regulated SAS chimeras in the over-expressed U1 snRNP dataset indicates that U1 snRNP could be associated with trans-splicing in human cells and play a vital role in the production of SAS chimeric transcripts.
Figure 8.
Hierarchical clustering with Euclidean distances of differentially expressed SAS chimeras in U1 snRNP knockdown and overexpressed samples in the HeLa cell line.
Alternation of splicing factors is responsible for switching transcript variants and for the generation of several oncogenic chimeric transcripts, which can generate cancer (15,21,78,79). Human Far Upstream Element Binding Protein 1 (FUBP1) is a key regulator of transcription, translation and RNA splicing and plays crucial roles in regulating multiple cellular processes (80). FUBP1 regulates the landscape of alternative splicing of tumor suppressors and oncogenes to control neoplastic transformation (81). To understand whether alternation of FUBP1 expression could lead to the formation of SAS chimeric transcripts in cancer, it is important to correlate dysregulation of the complex splicing processes with SAS chimera levels in cancer. Mutations in the splicing factor FUBP1 are frequently observed in low-grade glioma (82, 83). To investigate the impacts of FUBP1 mutations on aberrant splicing and gene expression, we downloaded the RNA-seq dataset from Bioproject: PRJNA392042 (84). To generate this dataset, the authors used U87MG, a glioblastoma cell line, in which they knocked-down FUBP1 (splicing factor) levels using targeted siRNA and then performed RNA-seq. We downloaded the RNA-seq data for U87MG cell line samples, including three treated with siRNA against FUBP1 and three treated with a non-specific siRNA, and performed SAS chimera analysis. We considered those SAS chimeras found in at least 50% of the samples. We performed the differential expression analysis but did not observe any significant variations in SAS chimeric transcript formation between FUBP1 knocked down and control cells. We detected 52 SAS chimeras (Supplementary Table S9) from all samples, 41 of which overlapped with the RNA-seq data from 16 human tissues in the Illumina Human Body Map project, indicating that these SAS chimeras are part of normal cellular processes. Of the remaining 11 SAS chimeras, we did not find any unique transcripts in either the siRNA-treated FUBP1 samples or in non-specific siRNA-treated samples. This result suggests that unlike other chimeric transcripts, dysregulation of alternative splicing is less likely to generate SAS chimeric transcripts. However, a more detailed analysis of different cancer-related datasets with a larger sample cohort could provide detailed insight into this question.
Characterization of the parental genes of SAS chimeric transcripts in cancer
Chimeric RNAs are characteristic cytogenetic signatures of many cancers and have been successfully used as biomarkers and therapeutic targets (85,86). Several SAS chimeras available in the ChiTaRS database were mapped to different cancer cell lines (29). To understand potential functional implications in cancer, we characterized the functional roles of the parental genes in cancer. Information about the oncogenic and tumor suppressor activity of the parental genes was retrieved from the TumorPortal (50) and CancerMine (51) databases, as well as from PubMed references. From the 5180 SAS chimeras, we detected 3262 unique parental genes. Among these, 471 (14.43%) were identified as oncogenes, 236 (7.23%) were characterized as tumor suppressor genes, and 347 (10.63%) had been reported to act as both oncogenes and tumor suppressors, depending upon the context. In total, 271 genes are identified as cancer-driver genes. The classification of genes as oncogenes or tumor suppressors, or both, is detailed in Supplementary Table S10. Although mutations in oncogenes or tumor suppressor genes could lead to tumorigenesis, fusions involving such genes also correspond to an essential class of cancer-driving events (87). In consulting our dataset of SAS chimera parental genes (Supplementary Table S10), we found MYC, EGFR and TP53 to be the top three genes that can act as both oncogenes and tumor suppressor genes, as well as cancer-driver genes. In the ChiTaRS database, MYC was found to generate one SAS chimera, while EGFR can generate two, and TP53 can generate three such chimera. Several natural antisense transcripts (NATs) were identified for MYC (88,89), EGFR (90,91) and TP53 (92,93), which are involved in the antisense‐mediated regulation of these genes and play pivotal roles in cancer (94). Therefore, it is possible that trans-splicing could promote the fusion of the sense and antisense transcripts of the same gene and produce sense-antisense transcripts. However, it remains unknown if SAS chimeras originating from these tumor suppressors and oncogenes are associated with cancers.
Analysis of SAS chimeric transcripts in PCAWG (ICGC/TCGA) breast cancer samples and the MCF-7 breast cancer cell line
To understand if there are potential associations of SAS chimeras with cancer, we analyzed the case study of breast cancer. For this, we downloaded RNA-seq data of eight tumors from breast cancer patients attained by the Pan-Cancer Analysis of Whole Genomes (PCAWG) project (52,53). We also downloaded RNA-seq data from the MCF-7 breast cancer cell line collected by CCLE (54). We thus identified 91 SAS chimeric transcripts in the RNA-seq data from the eight different breast cancer patient samples and 81 SAS chimeric transcripts from the MCF-7 breast cancer cell line (Supplementary Table S11). Next, to assess whether there are SAS chimeras presenting breast cancer-specific expression, we compared normal SAS chimeras from the human body map project and the MCF12A normal breast cell line (Bioproject: PRJNA491862) and excluded those SAS chimeras identified in normal samples from the cancer-specific cell line and tumor data. Using this criterion, we eliminated 11 SAS chimeras from the MCF-7 cell line and 15 SAS chimeras from the tumor data. The number of mapped SAS chimeras depends on the quality of the RNA-seq data, such as read lengths and total reads per sample. As SAS chimera expression levels are low, it is possible that these could not be mapped in the RNA-seq data when shorter read lengths and lower numbers of total reads were involved.
To define breast cancer-specific SAS chimeras, we first selected SAS chimeras found to be expressed in both patient samples and in the MCF-7 breast cancer cell line. In this manner, we were able to identify 30 SAS chimeras that are present in both patient samples and MCF-7 cell-line. Although these SAS chimeras were not detected in normal samples, we nonetheless employed a more stringent criterion to further rule out the possibility of these chimeras being involved in normal physiological functions. For this, we excluded SAS chimeras whose parental genes were found to generate other SAS isoforms in normal samples. Using this criterion, we found only five SAS chimeras uniquely expressed in the MCF-7 breast cancer cell line and in tumors from breast cancer patients. The parental genes of these five SAS chimeras are EEF1A1P5, RPL30, SLC39A6, PPIA and ND5. Previous studies demonstrated that Zinc transporter LIV-1 (SLC39A6) is present in higher amounts in estrogen receptor-positive breast cancers, and it is positively associated with metastasis to regional lymph nodes (95). We found the expression of a SAS chimera (Chimera ID: BE166679) produced from the SLC39A6 gene in the MCF-7 breast cancer cell line and one breast cancer patient sample but not in normal breast tissue or the MCF12A normal breast cell line. Therefore, the expression of this SAS chimera could be associated with breast cancer. Detailed analysis of SAS chimeras in different cancers is needed to understand their potential regulatory roles in this disease.
Mapping of SAS chimeras in nanopore direct RNA-sequencing data argues against these chimeras being RT-PCR artifacts
The recently developed technique of nanopore direct RNA-sequencing is a revolutionary approach by which native RNA can be sequenced directly, without the need for PCR amplification (96,97). The main advantage of this approach is that it eliminates any RT-PCR-based bias, which could introduce errors in the sequencing data generated by conventional RNA-seq methods (98). Therefore, the detecting of SAS chimeras in the direct RNA-sequencing data further supports the claim that these are not artifacts or sequencing errors. Still, the nanopore sequencing data is plagued by a high error rate (∼14%) (99). Therefore, there remains the chance that several chimeras could be missed due to the higher sequence identity and high mapping quality in the alignment of chimeric junction sequences required for chimeric transcript identification. Nonetheless, there is still high confidence that those chimeras detected in the nanopore direct RNA-seq data with higher sequence identity and high mapping quality are true chimeric sequences. With the aim of detecting only high-quality SAS chimeras, we downloaded direct RNA-seq data for the human GM12878 cell line (100). We then modified our pipeline used for SAS chimera detection in the nanopore long-read RNA-seq data. For this, we used minimap2 (101) instead of bowtie2, as the minimap2 algorithm is efficient for aligning long-read sequences. As expected, we detected only five SAS chimeras by this process (Supplementary Figure S1). Of these, two (Chimera_ID: BP417791 and HY093779) presently significant high read counts in the junction sequence. Both of these SAS chimeras are generated from the IGK (Immunoglobulin Kappa Locus) gene with distinct junction sequences. To further confirm these SAS chimeras, we conducted a BLASTn search of the junction sequences of these two SAS chimeras against the nanopore FASTQ reads. The individual reads in which the junctions of these two SAS chimeras aligned with >90% sequence identity were then extracted for downstream analysis. We found 17 reads for SAS chimera BP417791 and three reads for HY093779. Next, a BLASTn search was performed with the extracted FASTQ reads against the human transcriptome database to further confirm that these reads do not belong to any known human transcript. Finally, UCSC-BLAT (102) analysis was performed to confirm that the reads are involved in chimera organization in the sense and antisense directions (Supplementary Figure S1). For these two SAS chimeras, we observed the reads to be mapped in both the sense and antisense directions. These findings argue that sense-antisense chimeras are not RT-PCR-derived artifacts.
DISCUSSION
Bidirectional transcription has become recognized as more common in mammalian genomes than was initially thought (103). Evidence from global transcriptome analysis demonstrated that a large proportion of the genome could produce transcripts from both strands (104). The antisense transcript of a bi-directionally transcribed gene was shown to be involved in the epigenetic regulation of gene expression via degradation of the corresponding sense transcripts (105). Therefore, bidirectional transcription can regulate both transcriptional gene activation and suppression in response to various intrinsic and extrinsic cellular signals. However, the mechanism of regulation needed to maintain the expression of both sense and antisense transcripts remained poorly defined. Fusion of the sense-antisense strands of the same gene was first observed in prostate cancer cells, where transcripts from both strands of the KLK4 gene formed a chimera (18). Subsequently, with the release of the ChiTaRS-2.1 database (27), several sense-antisense chimeras were identified as a novel subset of fusion transcripts. Sense-antisense fusion is an evolutionarily conserved phenomenon, and numerous SAS chimeras have been detected in humans, mice and fruit flies (27). However, the expression of SAS chimeras in human cells and their potential roles remain uninvestigated.
In the current study, we analyzed the latest dataset of curated human SAS chimeras from ChiTaRS 5.0 (29) and found that SAS chimeras contain longer palindromic sequences compared to non-SAS chimeras. Predictions of the secondary structure of these chimeric SAS RNAs revealed the presence of a hairpin-like structure along the length of the palindromic region, which could be essential for converting the single-stranded chimeric RNAs into dsRNAs. The existence of natural dsRNAs, which are potentially involved in regulatory functions, such as post-mitotic changes, apoptotic alterations and antiviral signaling, has already been described in humans (63,106). Furthermore, predictions of protein-coding abilities supported that >85% of SAS chimeras are lncRNA transcripts. LncRNAs that form extended intramolecular hairpins might be processed into siRNAs and participate in siRNA-mediated gene regulation (107,108). We hypothesize that the palindromic sequences of SAS long non-coding RNA transcripts are important for generating the hairpin structure, leading to the formation of dsRNAs, which might play a similar role as siRNA-mediated gene regulation. Further, we assume that as SAS chimeric transcripts are generated from the fusion of sense and antisense strands of the same gene, such transcripts could be involved in regulating the expression of parental genes in response to different cellular conditions. To test this assumption, we analyzed RNA–RNA interactions between SAS long non-coding transcripts and their parental mRNAs, based on free energy minimization. We found that 77 of our list of top 100 SAS chimeras presenting longer palindromic sequences significantly interacted with their parental mRNA (Supplementary Table S4). Hence, our study suggests that SAS chimeras are important for maintaining the co-expression of sense and antisense transcripts in the same cell and potentially involved in repression and activation of gene expression in a manner that determines cellular responses.
Analysis of RNA-seq data from 16 different healthy human tissues revealed the expression of several SAS chimeric transcripts in more than one tissue. This finding indicates putative association of SAS chimeras in normal physiological processes. In addition, evolutionary analysis of the sense-antisense fusion across species identified several common genes that can produce SAS chimeric transcripts in humans, mice, fruit flies and pigs. This result indicates that the formation of SAS chimeras from certain genes could be evolutionarily beneficial as they underwent evolutionary selection in those organisms to generate functional diversity in response to various cellular conditions. Several evolutionary conserved sense-antisense gene pairs between human and mouse were reported in the earlier study and suggested that bidirectional transcription have a significant effect on vertebrate genome evolution (30). For each gene in the sense-antisense pair, its evolution is restricted not only by sequence features but also by encoded functional features. Therefore, selection has favored sense-antisense fusion as an important mechanism to regulate the co-expression of the sense-antisense gene pairs. Sense-antisense fusions are evolutionary dynamic, and this process could allow cells to get the most from limited genetic information in adapting to various physiological conditions.
Next, to verify that the generation of SAS chimeras were not RT-PCR-based artifacts, we mapped these chimeras in the publicly available nanopore direct RNA-sequencing data from the GM12878 cell line. We detected two highly expressed SAS chimeras that can be mapped to several unmapped FASTQ reads with very high quality and high sequence identity. This finding suggests that the chimeras are not the result of RT-PCR-derived artifacts or sequencing errors. Furthermore, we confirmed that bias was not introduced when the SAS chimeras mapped in the various RNA-seq data were analyzed using our in-house pipeline. For this, we downloaded RNA-seq data of the LN-229 cell line from the NCBI-GEO database (SRR10342173) (109) and mapped SAS chimeras using both STAR aligner (110) and our in-house pipeline. We thus detected 53 SAS chimeras using STAR and 43 SAS chimeras using our in-house pipeline (Supplementary Table S12). We found 28 common SAS chimeras identified by both STAR and our pipeline. Most of the highly expressed SAS chimeras were detected by using our in-house pipeline were also detected by STAR. It would thus appear that SAS chimera identification is not due to bias introduced by the mapping algorithms used.
In summary, the findings reported here elucidate several interesting aspects of SAS chimeric transcripts. Moreover, our results support that U1 snRNP could be the potential splicing factor behind the trans-splicing in humans as with their knockdown and overexpression, SAS chimeras are found to be differentially expressed. Our study also raised several interesting questions that can provide new insight into biological regulation. One of the most important questions is whether SAS chimeric transcripts reflect normal or aberrant gene transcription. For instance, one can ask if SAS chimeric transcripts are products of dysregulation of normal transcriptional and/or splicing processes. If so, understanding the underlying mechanisms that mediate the formation of SAS chimeras is needed. It is also important to know why specific genes produce SAS chimeric transcripts and why some are tissue-specific. Definitive answers to these questions are still not available, with more advanced experimental studies being needed.
CONCLUSION
Our study provides detailed insight into the expressions of SAS chimeras in human cells and identified potential regulatory features. We found that most of the SAS chimeras were annotated as lncRNAs and proposed that such transcripts could potentially involve in regulating gene expression. Palindromic sequences are located within the junction sites of most SAS chimeras, where they could generate a hairpin-like structure and lead to the formation of dsRNA, which raises the possibility of their interfering with gene expression. Additionally, significant interactions of SAS chimeras with their parental mRNAs were found, which supports their potential regulatory role. Finally, the expression analysis of various SAS chimeras in different human tissues and expression of their orthologs in mice highlights their importance in the functional regulation of normal physiological processes. Further functional validation based on our findings could open novel directions for understanding SAS chimera-mediated gene regulation in various physiological and pathological conditions.
DATA AVAILABILITY
The detailed protocol for identifying SAS chimeras from RNA-seq data is provided in the GitHub repository (https://github.com/Rajesh-Detroja/SAS-Chimeras).
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr. Gideon Baum for his guidance regarding nanopore direct RNA sequencing data analysis.
Contributor Information
Sumit Mukherjee, Cancer Genomics and BioComputing of Complex Diseases Lab, Azrieli Faculty of Medicine, Bar-Ilan University, Safed 1311502, Israel.
Rajesh Detroja, Cancer Genomics and BioComputing of Complex Diseases Lab, Azrieli Faculty of Medicine, Bar-Ilan University, Safed 1311502, Israel.
Deepak Balamurali, Cancer Genomics and BioComputing of Complex Diseases Lab, Azrieli Faculty of Medicine, Bar-Ilan University, Safed 1311502, Israel.
Elena Matveishina, Department of Bioengineering and Bioinformatics, Lomonosov Moscow State University, Moscow 119234, Russian Federation; Institute of Bioengineering, Research Centre of Biotechnology, Russian Academy of Sciences, Moscow 117312, Russian Federation.
Yulia A Medvedeva, Institute of Bioengineering, Research Centre of Biotechnology, Russian Academy of Sciences, Moscow 117312, Russian Federation; Department of Biomedical Physics, Moscow Institute of Technology, Dolgoprudny 141701, Russian Federation.
Alfonso Valencia, Barcelona Supercomputing Center (BSC), C/ Jordi Girona 29, 08034, Barcelona, Spain; ICREA, Pg. Lluís Companys 23, 08010 Barcelona, Spain.
Alessandro Gorohovski, Cancer Genomics and BioComputing of Complex Diseases Lab, Azrieli Faculty of Medicine, Bar-Ilan University, Safed 1311502, Israel.
Milana Frenkel-Morgenstern, Cancer Genomics and BioComputing of Complex Diseases Lab, Azrieli Faculty of Medicine, Bar-Ilan University, Safed 1311502, Israel.
SUPPLEMENTARY DATA
Supplementary Data are available at NARGAB Online.
FUNDING
Israeli Council for Higher Education [PBC Fellowship for Outstanding Post-Doctoral Fellows, 2019-2021 to S.M.]; Israel Innovation Authority [66824, 2019–2021 to M.F-M.]; RSF [18–14-00240 to Y.A.M. (in part)].
Conflict of interest statement. None declared.
REFERENCES
- 1.Romani A., Guerra E., Trerotola M., Alberti S.. Detection and analysis of spliced chimeric mRNAs in sequence databanks. Nucleic Acids Res. 2003; 31:e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akiva P., Toporik A., Edelheit S., Peretz Y., Diber A., Shemesh R., Novik A., Sorek R.. Transcription-mediated gene fusion in the human genome. Genome Res. 2006; 16:30–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mitelman F., Johansson B., Mertens F.. The impact of translocations and gene fusions on cancer causation. Nat. Rev. Cancer. 2007; 7:233–245. [DOI] [PubMed] [Google Scholar]
- 4.Kannan K., Wang L., Wang J., Ittmann M.M., Li W., Yen L.. Recurrent chimeric RNAs enriched in human prostate cancer identified by deep sequencing. Proc. Natl. Acad. Sci. USA. 2011; 108:9172–9177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frenkel-Morgenstern M., Lacroix V., Ezkurdia I., Levin Y., Gabashvili A., Prilusky J., Del Pozo A., Tress M., Johnson R., Guigo R.et al.. Chimeras taking shape: potential functions of proteins encoded by chimeric RNA transcripts. Genome Res. 2012; 22:1231–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asmann Y.W., Necela B.M., Kalari K.R., Hossain A., Baker T.R., Carr J.M., Davis C., Getz J.E., Hostetter G., Li X.et al.. Detection of redundant fusion transcripts as biomarkers or disease-specific therapeutic targets in breast cancer. Cancer Res. 2012; 72:1921–1928. [DOI] [PubMed] [Google Scholar]
- 7.Suzuki M., Makinoshima H., Matsumoto S., Suzuki A., Mimaki S., Matsushima K., Yoh K., Goto K., Suzuki Y., Ishii G.et al.. Identification of a lung adenocarcinoma cell line with CCDC6-RET fusion gene and the effect of RET inhibitors in vitro and in vivo. Cancer Sci. 2013; 104:896–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu C.S., Yu C.Y., Chuang C.Y., Hsiao M., Kao C.F., Kuo H.C., Chuang T.J.. Integrative transcriptome sequencing identifies trans-splicing events with important roles in human embryonic stem cell pluripotency. Genome Res. 2014; 24:25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nome T., Thomassen G.O.S., Bruun J., Ahlquist T., Bakken A.C., Hoff A.M., Rognum T., Nesbakken A., Lorenz S., Sun J.et al.. Common fusion transcripts identified in colorectal cancer cell lines by high-throughput RNA sequencing. Transl. Oncol. 2013; 6:546–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Latysheva N.S., Babu M.M.. Molecular signatures of fusion proteins in cancer. ACS Pharmacol. Transl. Sci. 2019; 2:122–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chwalenia K., Facemire L., Li H.. Chimeric RNAs in cancer and normal physiology. Wiley Interdiscip. Rev. RNA. 2017; 8:e1427. [DOI] [PubMed] [Google Scholar]
- 12.Singh S., Qin F., Kumar S., Elfman J., Lin E., Pham L.P., Yang A., Li H.. The landscape of chimeric RNAs in non-diseased tissues and cells. Nucleic Acids Res. 2020; 48:1764–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y., Gong M., Yuan H., Park H.G., Frierson H.F., Li H.. Chimeric transcript generated by cis- splicing of adjacent genes regulates prostate cancer cell proliferation. Cancer Discov. 2012; 2:598–607. [DOI] [PubMed] [Google Scholar]
- 14.Kumar-Sinha C., Kalyana-Sundaram S., Chinnaiyan A.M.. SLC45A3-ELK4 chimera in prostate cancer: spotlight on cis-splicing. Cancer Discov. 2012; 2:582–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jia Y., Xie Z., Li H.. Intergenically spliced chimeric RNAs in cancer. Trends Cancer. 2016; 2:475–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McManus C.J., Duff M.O., Eipper-Mains J., Graveley B.R.. Global analysis of trans-splicing in drosophila. Proc. Natl. Acad. Sci. USA. 2010; 107:12975–12979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mori H., Colman S.M., Xiao Z., Ford A.M., Healy L.E., Donaldson C., Hows J.M., Navarrete C., Greaves M.. Chromosome translocations and covert leukemic clones are generated during normal fetal development. Proc. Natl. Acad. Sci. USA. 2002; 99:8242–8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lai J., Lehman M.L., Dinger M.E., Hendy S.C., Mercer T.R., Seim I., Lawrence M.G., Mattick J.S., Clements J.A., Nelson C.C.. A variant of the KLK4 gene is expressed as a cis sense-antisense chimeric transcript in prostate cancer cells. RNA. 2010; 16:1156–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lei Q., Li C., Zuo Z., Huang C., Cheng H., Zhou R.. Evolutionary insights into RNA trans-Splicing in vertebrates. Genome Biol. Evol. 2016; 8:562–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jividen K., Li H.. Chimeric RNAs generated by intergenic splicing in normal and cancer cells. Genes Chromosom. Cancer. 2014; 53:963–971. [DOI] [PubMed] [Google Scholar]
- 21.Li H., Wang J., Ma X., Sklar J.. Gene fusions and RNA trans-splicing in normal and neoplastic human cells. Cell Cycle. 2009; 8:218–222. [DOI] [PubMed] [Google Scholar]
- 22.Djebali S., Lagarde J., Kapranov P., Lacroix V., Borel C., Mudge J.M., Howald C., Foissac S., Ucla C., Chrast J.et al.. Evidence for transcript networks composed of chimeric rnas in human cells. PLoS One. 2012; 7:e28213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Babiceanu M., Qin F., Xie Z., Jia Y., Lopez K., Janus N., Facemire L., Kumar S., Pang Y., Qi Y.et al.. Recurrent chimeric fusion RNAs in non-cancer tissues and cells. Nucleic Acids Res. 2016; 44:2859–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gupta S.K., Luo L., Yen L.. RNA-mediated gene fusion in mammalian cells. Proc. Natl. Acad. Sci. USA. 2018; 115:E12295–E12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Panigrahi P., Jere A., Anamika K.. FusionHub: a unified web platform for annotation and visualization of gene fusion events in human cancer. PLoS One. 2018; 13:e0196588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frenkel-Morgenstern M., Gorohovski A., Lacroix V., Rogers M., Ibanez K., Boullosa C., Leon E.A., Ben-Hur A., Valencia A.. ChiTaRS: a database of human, mouse and fruit fly chimeric transcripts and RNA-sequencing data. Nucleic Acids Res. 2013; 41:D142–D151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frenkel-Morgenstern M., Gorohovski A., Vucenovic D., Maestre L., Valencia A.. ChiTaRS 2.1-an improved database of the chimeric transcripts and RNA-seq data with novel sense-antisense chimeric RNA transcripts. Nucleic Acids Res. 2015; 43:D68–D75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gorohovski A., Tagore S., Palande V., Malka A., Raviv-Shay D., Frenkel-Morgenstern M.. ChiTaRS-3.1-the enhanced chimeric transcripts and RNA-seq database matched with protein-protein interactions. Nucleic Acids Res. 2017; 45:D790–D795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Balamurali D., Gorohovski A., Detroja R., Palande V., Raviv-Shay D., Frenkel-Morgenstern M.. ChiTaRS 5.0: the comprehensive database of chimeric transcripts matched with druggable fusions and 3D chromatin maps. Nucleic Acids Res. 2019; 48:D825–D834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galante P.A.F., Vidal D.O., de Souza J.E., Camargo A.A., de Souza S.J.. Sense-antisense pairs in mammals: functional and evolutionary considerations. Genome Biol. 2007; 8:R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murray S.C., Haenni S., Howe F.S., Fischl H., Chocian K., Nair A., Mellor J.. Sense and antisense transcription are associated with distinct chromatin architectures across genes. Nucleic Acids Res. 2015; 43:7823–7837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pelechano V., Steinmetz L.M.. Gene regulation by antisense transcription. Nat. Rev. Genet. 2013; 14:880–893. [DOI] [PubMed] [Google Scholar]
- 33.Zampetaki A., Albrecht A., Steinhofel K.. Long non-coding RNA structure and function: is there a link. Front. Physiol. 2018; 9:1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chillón I., Marcia M.. The molecular structure of long non-coding RNAs: emerging patterns and functional implications. Crit. Rev. Biochem. Mol. Biol. 2020; 55:662–690. [DOI] [PubMed] [Google Scholar]
- 35.Ganser L.R., Kelly M.L., Herschlag D., Al-Hashimi H.M.. The roles of structural dynamics in the cellular functions of RNAs. Nat. Rev. Mol. Cell Biol. 2019; 20:474–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langmead B., Salzberg S.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2013; 94:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo J.C., Fang S.S., Wu Y., Zhang J.H., Chen Y., Liu J., Wu B., Wu J.R., Li E.M., Xu L.Y.et al.. CNIT: a fast and accurate web tool for identifying protein-coding and long non-coding transcripts based on intrinsic sequence composition. Nucleic Acids Res. 2019; 47:W516–W522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang L., Park H.J., Dasari S., Wang S., Kocher J.P., Li W.. CPAT: Coding-potential assessment tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013; 41:e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han S., Liang Y., Ma Q., Xu Y., Zhang Y., Du W., Wang C., Li Y.. LncFinder: an integrated platform for long non-coding RNA identification utilizing sequence intrinsic composition, structural information and physicochemical property. Brief. Bioinform. 2019; 20:2009–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Antonov I.V., Mazurov E., Borodovsky M., Medvedeva Y.A.. Prediction of lncRNAs and their interactions with nucleic acids: benchmarking bioinformatics tools. Brief. Bioinform. 2019; 20:551–564. [DOI] [PubMed] [Google Scholar]
- 41.Lorenz R., Bernhart S.H., Höner Zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L.. ViennaRNA package 2.0. Algorithms Mol. Biol. 2011; 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zuker M., Stiegler P.. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981; 9:133–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCaskill J.S.The equilibrium partition function and base pair binding probabilities for RNA secondary structure. Biopolymers. 1990; 29:1105–1119. [DOI] [PubMed] [Google Scholar]
- 44.Kiryu H., Terai G., Imamura O., Yoneyama H., Suzuki K., Asai K.. A detailed investigation of accessibilities around target sites of sirnas and mirnas. Bioinformatics. 2011; 27:1788–1797. [DOI] [PubMed] [Google Scholar]
- 45.Hamada M., Kiryu H., Sato K., Mituyama T., Asai K.. Prediction of RNA secondary structure using generalized centroid estimators. Bioinformatics. 2009; 25:465–473. [DOI] [PubMed] [Google Scholar]
- 46.Li J., Ma W., Zeng P., Wang J., Geng B., Yang J., Cui Q.. LncTar: a tool for predicting the RNA targets of long noncoding RNAs. Brief. Bioinform. 2014; 16:806–812. [DOI] [PubMed] [Google Scholar]
- 47.Mückstein U., Tafer H., Hackermüller J., Bernhart S.H., Stadler P.F., Hofacker I.L.. Thermodynamics of RNA-RNA binding. Bioinformatics. 2006; 22:1177–1182. [DOI] [PubMed] [Google Scholar]
- 48.Mann M., Wright P.R., Backofen R.. IntaRNA 2.0: enhanced and customizable prediction of RNA-RNA interactions. Nucleic Acids Res. 2017; 45:W435–W439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mi H., Muruganujan A., Ebert D., Huang X., Thomas P.D.. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019; 47:D419–D426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lawrence M.S., Stojanov P., Mermel C.H., Robinson J.T., Garraway L.A., Golub T.R., Meyerson M., Gabriel S.B., Lander E.S., Getz G.. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014; 505:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lever J., Zhao E.Y., Grewal J., Jones M.R., Jones S.J.M.. CancerMine: a literature-mined resource for drivers, oncogenes and tumor suppressors in cancer. Nat. Methods. 2019; 16:505–507. [DOI] [PubMed] [Google Scholar]
- 52.Campbell P.J., Getz G., Korbel J.O., Stuart J.M., Jennings J.L., Stein L.D., Perry M.D., Nahal-Bose H.K., Ouellette B.F.F., Li C.H.et al.. Pan-cancer analysis of whole genomes. Nature. 2020; 578:82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Calabrese C., Davidson N.R., Demircioglu D., Fonseca N.A., He Y., Kahles A., Lehmann K.V, Liu F., Shiraishi Y., Soulette C.M.et al.. Genomic basis for RNA alterations in cancer. Nature. 2020; 578:129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ghandi M., Huang F.W., Jané-Valbuena J., Kryukov G.V., Lo C.C., McDonald E.R., Barretina J., Gelfand E.T., Bielski C.M., Li H.et al.. Next-generation characterization of the cancer cell line encyclopedia. Nature. 2019; 569:503–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Choi S.W., Kim H.W., Nam J.W.. The small peptide world in long noncoding RNAs. Brief. Bioinform. 2019; 20:1853–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ruiz-Orera J., Albà M.M.. Conserved regions in long non-coding RNAs contain abundant translation and protein–RNA interaction signatures. NAR Genomics Bioinform. 2019; 1:e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bailey T.L., Johnson J., Grant C.E., Noble W.S.. The MEME suite. Nucleic Acids Res. 2015; 43:W39–W49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Piao M., Sun L., Zhang Q.C.. RNA regulations and functions decoded by Transcriptome-wide RNA structure probing insights from probing RNA structuromes. Genomics, Proteomics Bioinforma. 2017; 15:267–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mukherjee S., Barash D., Sengupta S.. Comparative genomics and phylogenomic analyses of lysine riboswitch distributions in bacteria. PLoS One. 2017; 12:e0184314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mukherjee S., Das Mandal S., Gupta N., Drory-Retwitzer M., Barash D., Sengupta S.. RiboD: a comprehensive database for prokaryotic riboswitches. Bioinformatics. 2019; 35:3541–3543. [DOI] [PubMed] [Google Scholar]
- 61.Andrzejewska A., Zawadzka M., Pachulska-Wieczorek K.. On the way to understanding the interplay between the rna structure and functions in cells: a genome-wide perspective. Int. J. Mol. Sci. 2020; 21:6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kiryu H., Kin T., Asai K.. Robust prediction of consensus secondary structures using averaged base pairing probability matrices. Bioinformatics. 2007; 23:434–441. [DOI] [PubMed] [Google Scholar]
- 63.Portal M.M., Pavet V., Erb C., Gronemeyer H.. Human cells contain natural double-stranded RNAs with potential regulatory functions. Nat. Struct. Mol. Biol. 2015; 22:89–97. [DOI] [PubMed] [Google Scholar]
- 64.Lipardi C., Wei Q., Paterson B.M.. RNAi as random degradative PCR: siRNA primers convert mRNA into dsRNAs that are degraded to generate new siRNAs. Cell. 2001; 107:297–307. [DOI] [PubMed] [Google Scholar]
- 65.Watanabe T., Totoki Y., Toyoda A., Kaneda M., Kuramochi-Miyagawa S., Obata Y., Chiba H., Kohara Y., Kono T., Nakano T.et al.. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008; 453:539–543. [DOI] [PubMed] [Google Scholar]
- 66.Hofacker I.L., Tafer H.. Designing optimal siRNA based on target site accessibility. Methods Mol. Biol. 2010; 623:137–154. [DOI] [PubMed] [Google Scholar]
- 67.Mercer T.R., Neph S., Dinger M.E., Crawford J., Smith M.A., Shearwood A.M.J., Haugen E., Bracken C.P., Rackham O., Stamatoyannopoulos J.A.et al.. The human mitochondrial transcriptome. Cell. 2011; 146:645–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ge S.X., Jung D., Jung D., Yao R.. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics. 2020; 36:2628–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fabregat A., Jupe S., Matthews L., Sidiropoulos K., Gillespie M., Garapati P., Haw R., Jassal B., Korninger F., May B.et al.. The reactome pathway knowledgebase. Nucleic Acids Res. 2018; 48:D498–D503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li H., Wang J., Mor G., Sklar J.. A neoplastic gene fusion mimics trans-splicing of RNAs in normal human cells. Science (80-.). 2008; 321:1357–1361. [DOI] [PubMed] [Google Scholar]
- 71.Gao J.L., Fan Y.J., Wang X.Y., Zhang Y., Pu J., Li L., Shao W., Zhan S., Hao J., Xu Y.Z.. A conserved intronic U1 snRNP-binding sequence promotes trans-splicing in drosophila. Genes Dev. 2015; 29:760–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Preußer C., Rossbach O., Hung L.H., Li D., Bindereif A.. Genome-wide RNA-binding analysis of the trypanosome U1 snRNP proteins U1C and U1-70K reveals cis/trans-spliceosomal network. Nucleic Acids Res. 2014; 42:6603–6615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fukumura K., Inoue K.. Role and mechanism of U1-independent pre-mRNA splicing in the regulation of alternative splicing. RNA Biol. 2009; 6:395–398. [DOI] [PubMed] [Google Scholar]
- 74.Buratti E., Baralle D.. Novel roles of U1 snRNP in alternative splicing regulation. RNA Biol. 2010; 7:412–419. [DOI] [PubMed] [Google Scholar]
- 75.Charenton C., Wilkinson M.E., Nagai K.. Mechanism of 5′ splice site transfer for human spliceosome activation. Science (80-.). 2019; 364:362–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Oh J.M., Venters C.C., Di C., Pinto A.M., Wan L., Younis I., Cai Z., Arai C., So B.R., Duan J.et al.. U1 snRNP regulates cancer cell migration and invasion in vitro. Nat. Commun. 2020; 11:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Love M.I., Anders S., Huber W.. Differential analysis of count data - the DESeq2 package. Genome Biol. 2014; 15:10–1186. [Google Scholar]
- 78.Anczukow O., Krainer A.R.. Splicing-factor alterations in cancers. RNA. 2016; 22:1285–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Neckles C., Sundara Rajan S., Caplen N.J.. Fusion transcripts: unexploited vulnerabilities in cancer. Wiley Interdiscip. Rev. RNA. 2020; 11:e1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Debaize L., Troadec M.B.. The master regulator FUBP1: its emerging role in normal cell function and malignant development. Cell. Mol. Life Sci. 2019; 76:259–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Elman J.S., Ni T.K., Mengwasser K.E., Jin D., Wronski A., Elledge S.J., Kuperwasser C.. Identification of FUBP1 as a long tail cancer driver and widespread regulator of tumor suppressor and oncogene alternative splicing. Cell Rep. 2019; 28:3435–3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jacob A.G., Singh R.K., Mohammad F., Bebee T.W., Chandler D.S.. The splicing factor FUBP1 is required for the efficient splicing of oncogene MDM2 pre-mRNA. J. Biol. Chem. 2014; 289:17350–17364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Baumgarten P., Harter P.N., Tönjes M., Capper D., Blank A.E., Sahm F., von Deimling A., Kolluru V., Schwamb B., Rabenhorst U.et al.. Loss of FUBP1 expression in gliomas predicts FUBP1 mutation and is associated with oligodendroglial differentiation, IDH1 mutation and 1p/19q loss of heterozygosity. Neuropathol. Appl. Neurobiol. 2014; 40:205–216. [DOI] [PubMed] [Google Scholar]
- 84.Seiler M., Peng S., Agrawal A.A., Palacino J., Teng T., Zhu P., Smith P.G., Caesar-Johnson S.J., Demchok J.A., Felau I.et al.. Somatic mutational landscape of splicing factor genes and their functional consequences across 33 cancer types. Cell Rep. 2018; 23:282–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wu H., Li X., Li H.. Gene fusions and chimeric RNAs, and their implications in cancer. Genes Dis. 2019; 6:385–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li Z., Qin F., Li H.. Chimeric RNAs and their implications in cancer. Curr. Opin. Genet. Dev. 2018; 48:36–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gao Q., Liang W.W., Foltz S.M., Mutharasu G., Jayasinghe R.G., Cao S., Liao W.W., Reynolds S.M., Wyczalkowski M.A., Yao L.et al.. Driver fusions and their implications in the development and treatment of human cancers. Cell Rep. 2018; 23:227–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kim V.N., Han J., Siomi M.C.. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009; 10:126–139. [DOI] [PubMed] [Google Scholar]
- 89.Napoli S., Piccinelli V., Mapelli S.N., Pisignano G., Catapano C.V.. Natural antisense transcripts drive a regulatory cascade controlling c-MYC transcription. RNA Biol. 2017; 14:1742–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fan W.H., Lu Y.L., Deng F., Ge X.M., Liu S., Tang P.H.. EGFR antisense RNA blocks expression of the epidermal growth factor receptor and partially reverse the malignant phenotype of human breast cancer MDA-MB-231 cells. Cell Res. 1998; 8:63–71. [DOI] [PubMed] [Google Scholar]
- 91.Wang A., Bao Y., Wu Z., Zhao T., Wang D., Shi J., Liu B., Sun S., Yang F., Wang L.et al.. Long noncoding RNA EGFR-AS1 promotes cell growth and metastasis via affecting HuR mediated mRNA stability of EGFR in renal cancer. Cell Death Dis. 2019; 9:e49658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Farnebo M.Wrap53, a novel regulator of p53. Cell Cycle. 2009; 8:2343–2346. [DOI] [PubMed] [Google Scholar]
- 93.Mahmoudi S., Henriksson S., Corcoran M., Méndez-Vidal C., Wiman K.G., Farnebo M.. Wrap53, a natural p53 antisense transcript required for p53 induction upon DNA damage. Mol. Cell. 2009; 33:462–471. [DOI] [PubMed] [Google Scholar]
- 94.Zhao S., Zhang X., Chen S., Zhang S.. Natural antisense transcripts in the biological hallmarks of cancer: powerful regulators hidden in the dark. J. Exp. Clin. Cancer Res. 2020; 39:187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Taylor K.M., Morgan H.E., Smart K., Zahari N.M., Pumford S., Ellis I.O., Robertson J.F.R., Nicholson R.I.. The emerging role of the LIV-1 subfamily of zinc transporters in breast cancer. Mol. Med. 2007; 13:396–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Soneson C., Yao Y., Bratus-Neuenschwander A., Patrignani A., Robinson M.D., Hussain S.. A comprehensive examination of nanopore native RNA sequencing for characterization of complex transcriptomes. Nat. Commun. 2019; 10:3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Parker M.T., Knop K., Sherwood A.V., Schurch N.J., Mackinnon K., Gould P.D., Hall A.J.W., Barton G.J., Simpson G.G.. Nanopore direct RNA sequencing maps the complexity of arabidopsis mRNA processing and m6A modification. Elife. 2020; 9:e49658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhao L., Zhang H., Kohnen M.V., Prasad K.V.S.K., Gu L., Reddy A.S.N.. Analysis of transcriptome and epitranscriptome in plants using pacbio iso-seq and nanopore-based direct RNA sequencing. Front. Genet. 2019; 10:253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cozzuto L., Liu H., Pryszcz L.P., Pulido T.H., Delgado-Tejedor A., Ponomarenko J., Novoa E.M.. MasterOfPores: a workflow for the analysis of oxford nanopore direct RNA sequencing datasets. Front. Genet. 2020; 11:211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Workman R.E., Tang A.D., Tang P.S., Jain M., Tyson J.R., Razaghi R., Zuzarte P.C., Gilpatrick T., Payne A., Quick J.et al.. Nanopore native RNA sequencing of a human poly(A) transcriptome. Nat. Methods. 2019; 16:1297–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li H.Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018; 34:3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kent W.J.BLAT - The BLAST-like alignment tool. Genome Res. 2002; 12:656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Orekhova A.S., Rubtsov P.M.. Bidirectional promoters in the transcription of mammalian genomes. Biochem. 2013; 78:335–341. [DOI] [PubMed] [Google Scholar]
- 104.Katayama S., Tomaru Y., Kasukawa T., Waki K., Nakanishi M., Nakamura M., Nishida H., Yap C.C., Suzuki M., Kawai J.et al.. Antisense transcription in the mammalian transcriptome. Science (80-.). 2005; 309:1564–1566. [DOI] [PubMed] [Google Scholar]
- 105.Morris K.V., Santoso S., Turner A.M., Pastori C., Hawkins P.G.. Bidirectional transcription directs both transcriptional gene activation and suppression in human cells. PLoS Genet. 2008; 4:e1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dhir A., Dhir S., Borowski L.S., Jimenez L., Teitell M., Rötig A., Crow Y.J., Rice G.I., Duffy D., Tamby C.et al.. Mitochondrial double-stranded RNA triggers antiviral signalling in humans. Nature. 2018; 560:238–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Golden D.E., Gerbasi V.R., Sontheimer E.J.. An inside job for siRNAs. Mol. Cell. 2008; 31:309–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Czech B., Malone C.D., Zhou R., Stark A., Schlingeheyde C., Dus M., Perrimon N., Kellis M., Wohlschlegel J.A., Sachidanandam R.et al.. An endogenous small interfering RNA pathway in drosophila. Nature. 2008; 453:798–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Clough E., Barrett T.. The gene expression omnibus database. Statistical genomics. 2016; NY: Humana Press; 93–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The detailed protocol for identifying SAS chimeras from RNA-seq data is provided in the GitHub repository (https://github.com/Rajesh-Detroja/SAS-Chimeras).