Abstract
Hox genes code for transcription factors and are evolutionarily conserved. They regulate a plethora of downstream targets to define the anterior-posterior (AP) body axis of a developing bilaterian embryo. Early work suggested a possible role of clustering and ordering of Hox to regulate their expression in a spatially restricted manner along the AP axis. However, the recent availability of many genome assemblies for different organisms uncovered several examples that defy this constraint. With recent advancements in genomics, the current review discusses the arrangement of Hox in various organisms. Further, we revisit their discovery and regulation in Drosophila melanogaster. We also review their regulation in different arthropods and vertebrates, with a significant focus on Hox expression in the crustacean Parahyale hawaiensis. It is noteworthy that subtle changes in the levels of Hox gene expression can contribute to the development of novel features in an organism. We, therefore, delve into the distinct regulation of these genes during primary axis formation, segment identity, and extra-embryonic roles such as in the formation of hair follicles or misregulation leading to cancer. Toward the end of each section, we emphasize the possibilities of several experiments involving various organisms, owing to the advancements in the field of genomics and CRISPR-based genome engineering. Overall, we present a holistic view of the functioning of Hox in the animal world.
Keywords: hox, evolution, patterning, gene regulation, bithorax complex, vertebrate hox, modularity and adaptability, homeotic transformation
Introduction
The development of an animal progresses three-dimensionally across anterior-posterior (AP), dorsal-ventral (DV), and left-right (LR) axes. A combination of various transcription factors, epigenetic regulators, cell receptors, and signaling molecules are involved in the overall development of an organism (François et al., 1994; Beddington and Robertson, 1999; Levin, 2005; Peel et al., 2005; Dequéant and Pourquié, 2008; Basson, 2012; Perrimon et al., 2012; Coutelis et al., 2013; Berenguer et al., 2020). Homeotic genes or Hox are one of the significant contributors to bilaterian development and are evolutionarily conserved. They are often present in clusters and code for transcription factors (HOX) that act on the downstream genes to provide identity to developing segments along the AP axis of a bilaterian embryo (Akam et al., 1988; Akam, 1998; Lewis, 1998).
A series of genetic crossings and recombinations lead to the discovery of Hox in Drosophila melanogaster. Interestingly, all the genes were mapped to the right arm of the third chromosome in the fruit fly. Further, the genes were clustered together in two complexes of ∼300 Kb each – Antennapedia complex (ANT-C) and bithorax complex (BX-C), named after the initial phenotypes obtained for different alleles in both complexes. Strikingly, the arrangement of these genes on the chromosome followed an order that was collinear to the segments affected in mutants from anterior to posterior direction. Thus, in the fly, the concept of spatial collinearity was established for Hox expression (Lewis, 1978, 1998; Kaufman et al., 1990). This expression is under the control of several cis-regulatory elements (CREs) that cluster together to form cis-regulatory modules (CRMs) (Peifer and Bender, 1986; Peifer et al., 1987; Celniker et al., 1989; Martin et al., 1995; Maeda, 2009; Chopra, 2011; Bekiaris et al., 2018). Series of discoveries toward the turn of the 20th century showed the presence of Hox in all bilaterians and even in cnidarians (Ferrier et al., 2000; Kourakis and Martindale, 2001; Ferrier and Minguillón, 2003; Ikuta et al., 2004; Duboule, 2007; Mooi and David, 2008; Mallo et al., 2010; Ikuta, 2011; Janssen et al., 2014; Fritsch et al., 2015; Schiemann et al., 2017; Wanninger and Wollesen, 2019; Nong et al., 2020). The transcription factors coded by these genes have a conserved helix-turn-helix motif-containing DNA binding domain. The domain binds to DNA in a sequence-specific manner and is called the homeodomain due to its discovery in the factors coded by Hox. Many transcription factors in addition to HOX across animals, plants, and fungi have the homeodomain (McGinnis et al., 1984b, a; Scott and Weiner, 1984; Suzuki and Yagi, 1994; Williams, 1998; Holland, 2001; Holland et al., 2007; Son et al., 2020). Therefore, all Hox genes are homeobox genes, but all homeobox genes are not Hox genes. The complex interplay of HOX proteins with other players in the system contributes toward diversity in the animal kingdom (Akam et al., 1988; Akam, 1998; Lewis, 2007; Holland, 2015; Rogers, 2020).
In the current review, we discuss the arrangement and copies of Hox genes in different organisms. We then revisit their discovery and regulation in D. melanogaster, subsequently commenting upon their cis-regulation in vertebrates. Further, the review highlights the presence of these genes in other arthropods and their expansion in vertebrates, with a significant focus on Hox expression in Parhyale hawaiensis. The crustacean is an emerging model organism with established gene-editing techniques such as CRISPR-Cas9 to decipher the role of Hox, adding them to the league of other classical models, including fruit fly, zebrafish, or mouse (Martin et al., 2016; Sun and Patel, 2019). Notably, the function of these genes is not limited to segment identity and homeotic transformations (Castelli-Gair Hombría and Lovegrove, 2003). We also underline many upcoming reports that describe their role in tissue homeostasis, embryonic cell fate determination, organogenesis including abdominal epithelium in flies or hair follicles in mammals, maintenance of stem cells niche, and misregulation leading to cancer (Lewis, 2000; Awgulewitsch, 2003; Shah and Sukumar, 2010; Estacio-Gómez and Díaz-Benjumea, 2014; Singh and Mishra, 2014; Domsch et al., 2019). Toward the end of each section, we emphasize the possibilities of novel experiments to understand the regulation and functioning of Hox genes in different organisms. This largely owes to the recent advances in genomics and genome editing technologies, including CRISPR-Cas9. We thus present a bird’ eye view of the Hox field and prospective investigations required to understand their role in various organisms.
Hox Clusters: Arrangement, Positioning, and Duplications
The animal kingdom has diverse body forms, symmetries, and developmental axes. Hox are one of the key contributors to this diversity as they provide identity to different segments during embryonic development, are involved in tissue homeostasis and organ positioning, and help in maintaining cellular identities post-embryonic development (Lewis, 2000; Castelli-Gair Hombría and Lovegrove, 2003; Lovegrove et al., 2006; Mallo et al., 2010; Sánchez-Herrero, 2013; Papagiannouli and Lohmann, 2015; Hrycaj and Wellik, 2016; Domsch et al., 2020). They are present in cnidarians with ancestral elements of the anterior and posterior Hox genes (Chourrout et al., 2006; Ikuta, 2011; Gaunt, 2018; Rentzsch and Holstein, 2018; Nong et al., 2020). During evolution, bilaterians acquired another set of central Hox genes and formed a complete set of genes responsible for the animal development across the anterior-posterior body axis (Chourrout et al., 2006; Hrycaj and Wellik, 2016). Usually, Hox genes are present in a cluster and exhibit spatial collinearity; the genes present in one end of the cluster are expressed in the anterior-most regions (or segments) of the developing embryo. At the same time, the genes present on the opposite end are responsible for posterior development (Gaunt, 2015). However, this is not universally true.
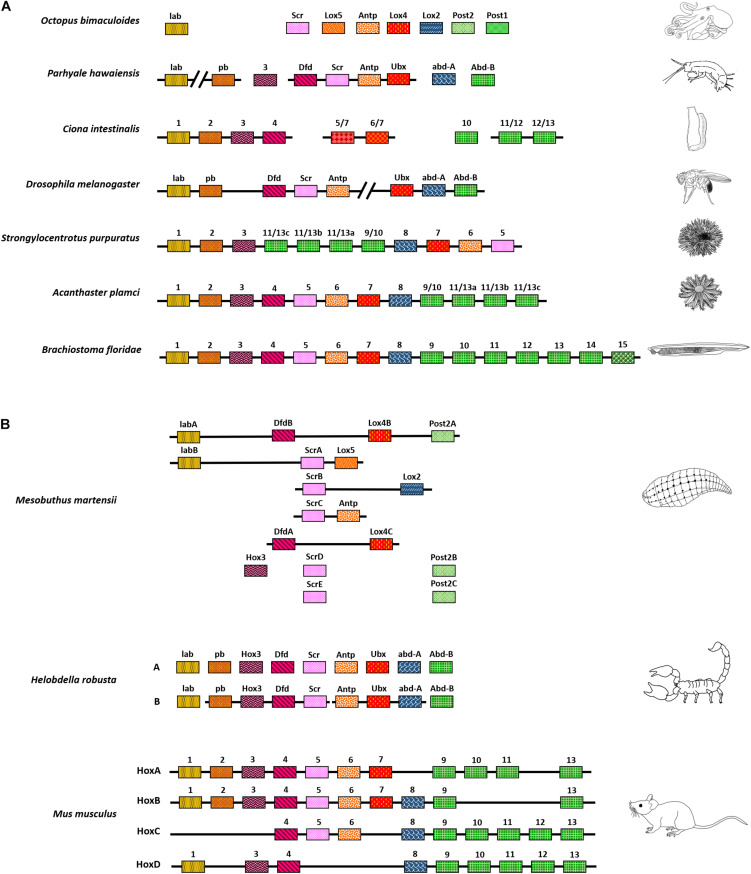
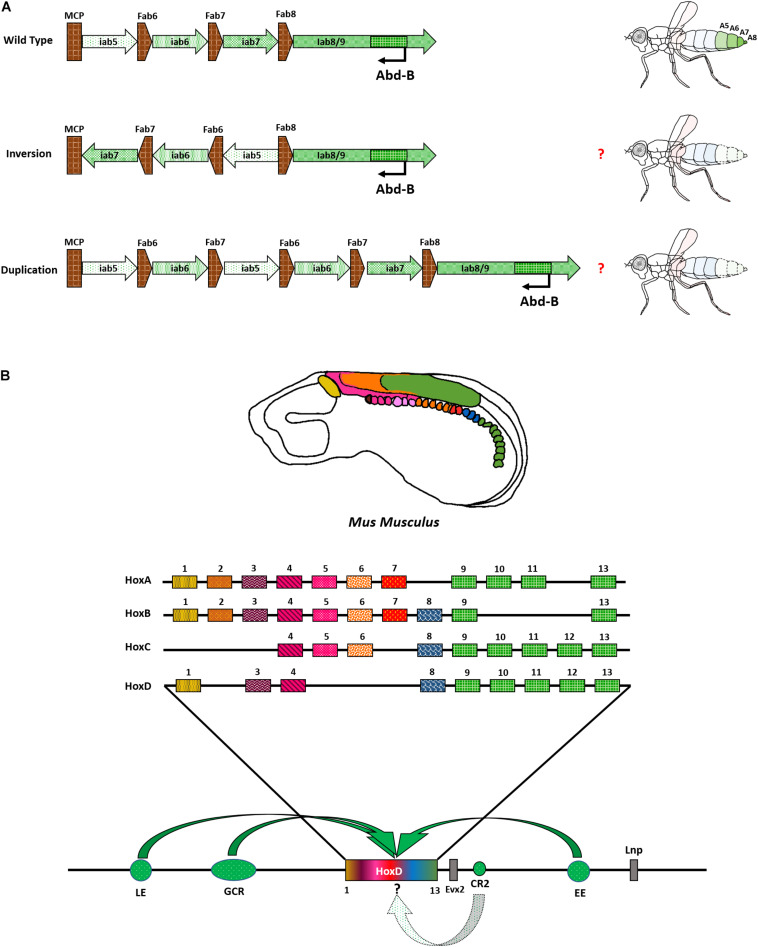
The Hox genes of California two-eyed octopus, Octopus bimaculoides, are completely dispersed across the genome (Albertin et al., 2015). Other than the octopus, most other bilaterians show clustering of at least two Hox genes in cis-. For example, in Parhyale hawaiensis, a crustacean, some of the Hox are arranged in clusters of two and four genes. However, the detailed arrangement of all Hox genes in Parhyale remains elusive due to the absence of long contigs (Serano et al., 2016). Even in a marine chordate, Ciona intestinalis, Hox appear to be present in an exceptionally dispersed cluster, or they could even be disseminated across the genome (Spagnuolo et al., 2003; Ikuta et al., 2004). D. melanogaster has a partially contiguous arrangement of Hox. As mentioned earlier, the Hox genes cluster of Drosophila is split into two complexes with 5 and 3 Hox in them. Both complexes are around 300 Kb in length and are separated by a distance of ∼9 Mb (Dessain and McGinnis, 1993; Rogers, 2020). Other than Drosophila, the red fluor beetle, Tribolium castaneum, has been a subject of extensive studies for patterning and evolution in insects. Both the insects have similar expressions of Hox orthologs in anterior-posterior segments. However, their arrangement is quite different in the genome. T. castaneum Hox are organized in a single tight cluster as opposed to the split found in Drosophila (Beeman, 1987; Telford, 2000; Brown et al., 2002; Shippy et al., 2008). In other organisms such as the starfish, Acanthaster planci, and sea urchin Strongylocentrotus purpuratus, Hox are present in a cluster. Still, either their orientation is altered, or they have re-ordered arrangement when compared to the majority of bilaterians that follow collinearity. In S. purpuratus, posterior Hox (Hox11/13) have relocated to positions analogous to central Hox and vice-versa (Howard-Ashby et al., 2006; Baughman et al., 2014). In contrast, the Hox genes of cephalochordate amphioxus, Branchiostoma floridae, are present as a single intact cluster in the order of their evolutionary homologs, along the anterior-posterior body axis. It is the most cohesive cluster of Hox discovered in the animal kingdom from Hox1 to Hox14. Later, analysis of the region between Hox14 and EvxA – EvxB led to the finding of another paralogous posterior Hox gene called Hox15. Thus, the cluster of Hox genes in amphioxus is by far the largest intact cluster in terms of the number of Hox genes and spans 470 Kb (Holland et al., 2008). The above examples suggest that animals have varied arrangements of Hox genes as they underwent multiple combinations of convergent and divergent evolutionary processes throughout the tree of life (Figure 1A).
FIGURE 1.
Chromosomal arrangement of Hox genes. (A) Variety of Hox arrangement observed in different animals, from a completely scattered arrangement in octopus to a fully intact Hox cluster in Amphioxus. (B) Hox have also undergone duplication in several organisms. They have fragmented arrangements in animals such as the freshwater leech or are present in an intact cluster, as seen in vertebrates. The depiction here is independent of phylogeny and represents the order of clustering. Also, the organisms shown here do not represent their respective phyla.
In several organisms, Hox genes are present in multiple copies of paralogous genes. For instance, the cluster of Hox genes in the annelid Helobdella robusta appear fragmented along with varying copies of different homeotic genes. This is especially true for anterior and central Hox orthologs such as Deformed (Dfd) and Sex combs reduced (Scr) present in two and five copies, respectively. The Leech homeobox gene (Lox4) is also present in two copies, whereas orthologs like Proboscipedia (Pb), Ultrabithorax (Ubx), and abdominal-A (abd-A) appear completely absent. The posterior Hox gene, Post2, is also present in three copies (Kourakis and Martindale, 2001).
Similarly, the Chinese scorpion, Mesobuthus martensii, has two sets of Hox genes, with one of the sets being more clustered than the other. Interestingly, the duplication of Hox genes in scorpions is associated with variation in and extension of the posterior-most segments of the animal, including telson (Di et al., 2015). Vertebrates have at least four different paralogous complexes of Hox genes clusters. Each complex has a different number of Hox homologs arranged in a tight cluster of ∼100 Kb. The clustering also follows spatial collinearity like its invertebrate counterparts. In addition to that, vertebrate Hox genes are also expressed in a temporally collinear manner. The genes present in one end of the complex are expressed earlier during embryonic development and vice-versa. The different complexes work independently, as well as in concert, to fine-tune the growth of a developing embryo (Figure 1B; Burke et al., 1995; Medina-Martínez et al., 2000; Suemori and Noguchi, 2000; Spitz et al., 2001; Kmita et al., 2005; Tschopp et al., 2009; Yamada et al., 2021).
In conclusion, the Hox genes are present in different positions and numbers across the genome, from an atomized and dispersed manner in octopus to cleanly clustered complexes in vertebrates (Figures 1A,B). It, therefore, becomes important to understand the significance of clustering and ordering of Hox genes in different organisms. Many of the available genome sequences still lack chromosome level assemblies. With the advancement of long-read nanopore sequencing and the use of proximity ligation assays like Hi-C, it is possible to achieve chromosome level assemblies (Wang et al., 2014; Kadota et al., 2020). The ongoing earth biogenome project shall benefit from these techniques, and analysis of Hox genes arrangement in different animals will help us better understand their clustering and ordering throughout the tree of life (Lewin et al., 2018). An in-depth overview of known Hox genes clusters and their arrangement across different organisms is nicely covered in a review by Stephen Gaunt (2018).
Drosophila Hox Complex: A Split That Unified the Field
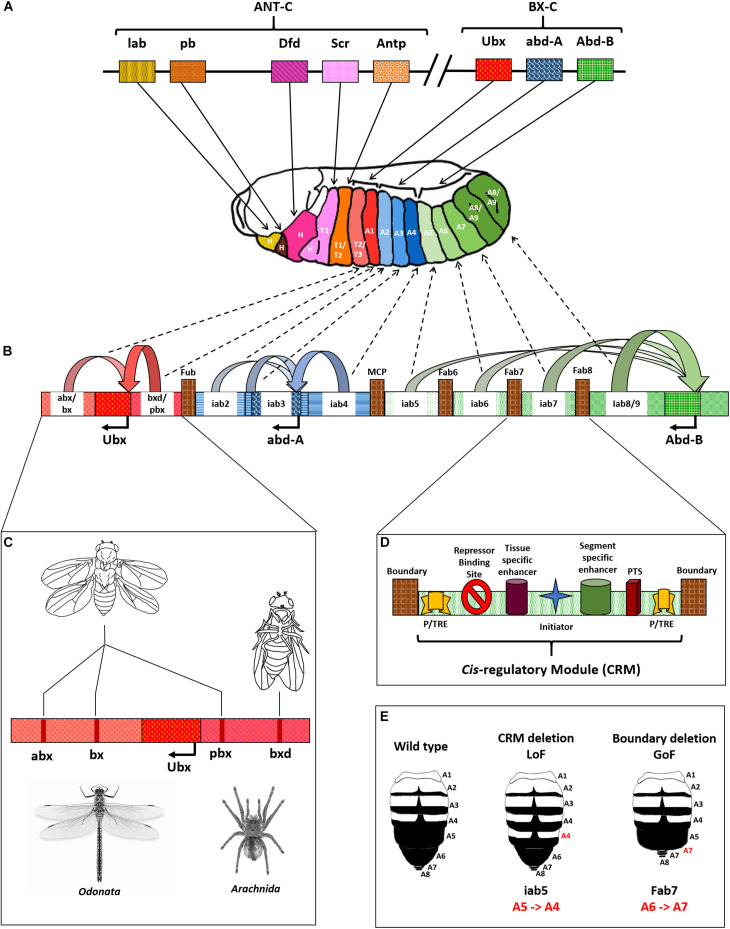
Homeotic genes were discovered by Ed Lewis in D. melanogaster in the latter half of the 20th century (Lewis, 1978). There are two clusters of these genes in the fruit fly, the Antennapedia complex (ANT-C) and the bithorax complex (BX-C). The ANT-C is responsible for the identity of anterior segments of the fly from the head through thoracic segment 2 (T2) and has five Hox genes. In the proximo-distal arrangement concerning centromere-telomere, these genes are ordered as labial (lab), Proboscipedia (Pb), Deformed (Dfd), Sex comb reduced (Scr), and Antennapedia (Antp). The BX-C has three genes in the centromeric proximo-distal order of Ultrabithorax (Ubx), abdominal-A (abd-A), and Abdominal-B (Abd-B). These genes provide identity to the posterior two-thirds of the fly’s body axis from T3 to abdominal segment 8/9 (A8/9), which is the terminal segment in the fly (Figure 2A; Lewis, 1978, 1998; Kaufman et al., 1990; Dessain and McGinnis, 1993). It is noteworthy that there are various CRMs for each Hox gene in the fly. These CRMs consist of numerous regulatory elements, including enhancers, initiators, insulators or boundary elements (BE), Polycomb/Trithorax response elements (P/TRE), and promoter tethering sequences (PTS), that together orchestrate the segment-specific expression of these genes (Figures 2B,D; Celniker et al., 1989, 1990; Simon et al., 1990; Sánchez-Herrero, 1991; Castelli-gair et al., 1992; Muller and Bienz, 1992; Mishra and Karch, 1999; Bender and Hudson, 2000; Calhoun and Levine, 2003; Lin et al., 2003; Mihaly et al., 2006; Iampietro et al., 2010; Li et al., 2015).
FIGURE 2.
Drosophila Hox complex and regulation of BX-C. (A) Drosophila Hox genes are split into two complexes, Antennapedia complex (ANT-C), and bithorax complex (BX-C), as shown. Each gene is responsible for providing identity to a specific segment, as indicated by bold arrows. (B) Cis-regulatory modules (CRMs) of the BX-C cause differential expression of Hox genes in a segment-specific manner. The genes influenced by their CRMs are shown as curved arrows with respective colors, and the dotted arrows indicate the segments they influence. (C) Representation of cis-regulatory module with different elements including boundary/insulator, Polycomb/Trithorax Response Elements (P/TRE), and promoter tethering sequences (PTS). (D) Deletion mutations in CRMs of Ubx leading to phenotypes that look similar to an odonate, like dragonfly or an arachnid, like spider. (E) Loss and gain of function mutations in the abdominal region of Drosophila due to deletions of CRMs or BEs.
There are nine CRMs of the three BX-C genes in the order anterobithorax/bithorax (abx/bx) and bithoraxoid/postbithorax (bxd/pbx) for Ubx, infra-abdominal2 (iab2), iab3 and iab4 for abd-A, and iab5, iab6, iab7, and iab8/9 for Abd-B. Each of the CRM drives segment-specific expression of the associated gene in embryonic parasegment 5 (PS5) through PS14, corresponding to segments T3 through A8/9 in the adult fly. Deletions of CRMs cause loss of function (LoF) phenotypes for the associated Hox genes and lead to anteriorization of respective segments. For example, deletion of iab5 causes homeotic transformation of A5 to A4. The mutant has two copies of A4 after A3 that follow the normal occurrence of A6, A7, and genitalia (A8/9) (Figure 2E; Martinez-Arias and Lawrence, 1985; Peifer and Bender, 1986; Turner and Kaufman, 1987; Galloni et al., 1993; Casares and Sanchez-Herrero, 1995; Hendrickson and Sakonju, 1995; Martin et al., 1995; Bender and Hudson, 2000; Bae et al., 2002; Estrada et al., 2002; Deutsch, 2004; Mihaly et al., 2006; Starr et al., 2011). Further, chromatin domain boundaries separate the CRMs of the BX-C. These include Front-ultraabdominal (Fub) that separates bxd/pbx from iab2, Mis-cadastral pigmentation (MCP) separating iab4 and iab5, Frontabdominal6 (Fab6) between iab5 and iab6, (Fab7) demarcating the domains of iab6 and iab7, followed by (Fab8), which is present between iab7 and iab8/9 (Figure 2B). These BEs maintain the autonomous domains of functioning for different CRMs and genes. In contrast to the LoF phenotypes of CRM deletions, the deletions of BEs cause gain of function phenotypes for the associated Hox genes leading to posteriorization of the related segments. This phenotype is due to the ectopic activation of posterior CRM and its prevalence over the anterior one. For instance, deletion of the chromatin domain boundary, Fab7 leads to the homeotic transformation of A6 to A7 as depicted in Figure 2E (Simon et al., 1990; Karch et al., 1994; Hagstrom et al., 1996; Zhou et al., 1996; Mihaly et al., 1998; Mishra and Karch, 1999; Muller et al., 1999; Barges et al., 2000; Schweinsberg et al., 2004; Bender and Lucas, 2013; Postika et al., 2018, 2021). Furthermore, multiple P/TREs adjacent to the BEs and inside CRMs maintain the repressed or activated states of associated CRMs. A combination of boundaries and PREs maintain the distinct autonomy of CRMs wherein the PREs are known to function via DNA kissing (Simon et al., 1993; Chan et al., 1994; Mishra et al., 2001; Vazquez et al., 2006; Lanzuolo et al., 2007; Bantignies et al., 2011; Négre et al., 2011; Singh and Mishra, 2015). The CRMs of BX-C are also present in a spatially collinear manner in tune with their associated genes (Lewis, 1978, 1998; Maeda, 2006). Figure 2 summarizes the arrangement of D. melanogaster Hox and the elements of the bithorax complex. Notably, the significance of positioning of CRMs in a collinear manner is still elusive. A significant merit could be the sequential regulation of the Hox genes by upstream regulators as proposed in the open for business model of BX-C regulation (see next section) (Maeda and Karch, 2007, 2015; Kyrchanova et al., 2015).
Since Hox genes provide identities to a developing segment, altering the levels of these genes can tip the scale in favor of distinct traits gained or lost during evolution, albeit they are not the sole drivers of the process (Ho et al., 2009; Holland, 2015). For example, mutations in the CRMs of the D.mel Ubx gene manifest fascinating phenotypes. A triple deletion mutant for abx, bx, and pbx causes homeotic transformation of T3 into a copy of T2. The T3 of flies possesses a pair of rudimentary wings called halteres that help maintain balance during flight (Lewis, 1978; Dickinson et al., 1999; Yarger and Fox, 2016). In the triple mutant, the halteres get completely transformed into wings, and the posterior thorax attains the morphology of the anterior one resulting in a fly with four wings, compared to a pair of wings and halteres in normal conditions (Figure 2C). Since the CRMs maintain required levels of Ubx in T3, their absence leads to a lack of expression of the gene. This loss of function causes T3 to anteriorize into a copy of T2 (Little et al., 1990; Martínez-Laborda et al., 1996). It was a remarkable achievement for two reasons – (1) All three mutations were within a span of 100 kb of each other and were therefore extremely difficult to obtain in cis- through the genetic crossing. (2) The fruit fly, a dipteran, looks strikingly similar to an odonate like dragonfly or damselfly with four distinct wings (Lewis, 1978). A combination of three inter-genic mutations led to the development of body morphology that diverged almost 500 million years before the arrival of dipterans (Figure 2C; Misof et al., 2014). Similarly, flies hemizygous for bxd have a partial transformation of A1 into a copy of T3, resulting in a fly with four pairs of legs instead of three. This feature is similar to an arachnid that includes spiders, scorpions, and ticks (Figure 2C; Shultz, 1989).
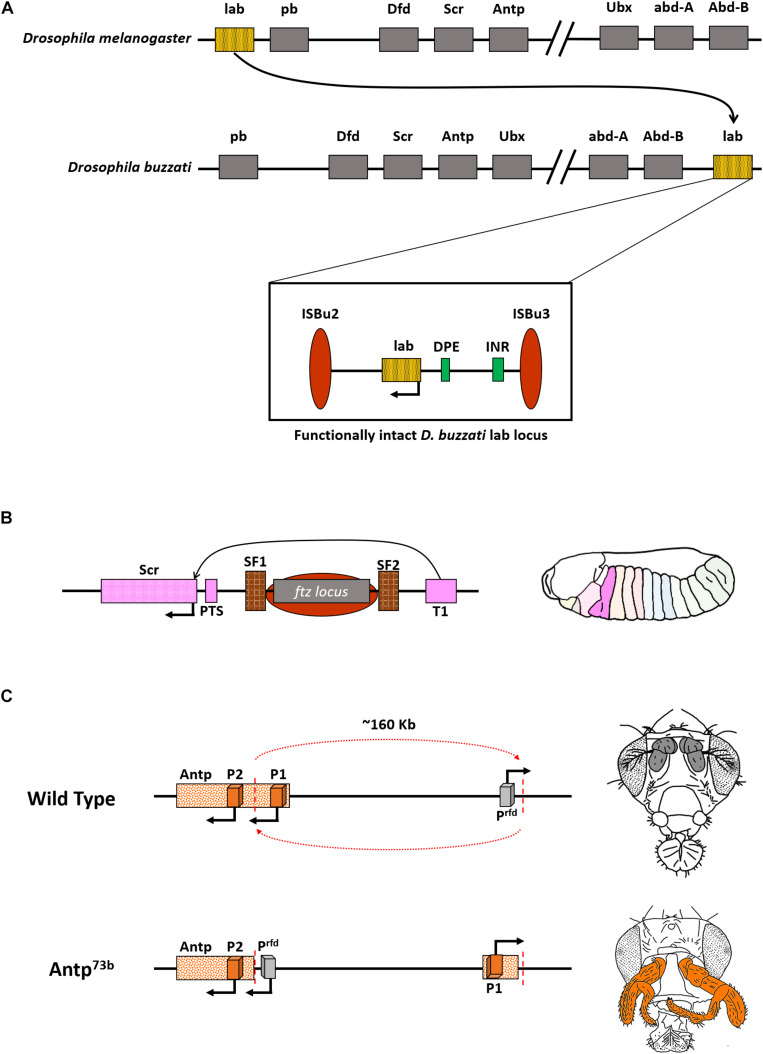
The presence of intact CRMs juxtaposed with genes would ensure that they provide coordinated expression during embryonic development. This is evident from the case of Drosophila buzzati, where the gene labial (lab), an anterior gene, is relocated to a position nearer to abd-A and Abd-B, the genes that define the posterior development of the fly. Nevertheless, the expression pattern for all Hox genes remains similar to D. melanogaster. The rearrangement of the lab locus was attributed to the presence of two transposable elements, ISBu2 and ISBu3, that stabilized over generations. These transposons together flank the gene lab and its associated non-coding elements. So, the overall arrangement of transposons, associated non-coding elements, and absence of any other coding gene indicate the functional intactness of the locus (Negre et al., 2003). The D. buzzati lab, hence, still expresses in the anterior part of the body, unlike its neighbors abd-A and Abd-B (Figure 3A).
FIGURE 3.
Functional CRMs can accommodate changes in gene order. (A) Representation of Hox in D. melanogaster and D. buzzati. Note that the gene labial (lab) is present next to Abdominal-B (Abd-B), and the split is between Ubx and abd-A, wherein Ubx is now a part of ANT-C. Inset: Functional intactness of lab locus despite rearrangement owing to two transposons, ISBu2/3, that flank the regulatory cassette of the gene. (B) Illustration depicting regulation of Scr gene bypassing ftz locus. The ftz gene and regulators are flanked by boundary elements SF1 and SF2. T1 is the enhancer for Scr. Right: Domain expression of Scr in a normal condition. (C) Inversion involving Antp (Antp73b) spanning ∼160 Kb juxtaposes the gene to an ectopic promoter that drives its expression in antennae and arista of the flies (colored bold gray in wild type fly). This leads to the homeotic transformation of antennae to legs (highlighted in orange).
The process of cis-regulation can be effectively carried out even in the presence of a non-related DNA element in between. For example, few of the cis-regulators of the Dmel Scr gene are present after a non-homeotic gene, ftz (Gindhart et al., 1995; Calhoun and Levine, 2003). The ftz gene is, however, flanked by two strong boundary elements SF1 and SF2, that presumably loop out the gene and its associated regulators, thus, facilitating proper interaction of Scr enhancer, T1, with the Scr gene (Figure 3B; Nègre et al., 2010; Li et al., 2015). This process is similar to insulator bypass events, observed in BX-C in the presence of boundary elements like MCP, Fab-7, or Fab-8 (Sipos et al., 1998; Mishra and Karch, 1999; Kyrchanova et al., 2011, 2015, 2019).
Though the genes and CRMs can together relocate to various positions across the genome or be reorganized by chromatin domain boundaries, an arbitrary split in the middle of CRMs is deleterious. This is apparent from the famous Antennapedia mutant, Antp73b. The Dmel Antp gene has two promoters, P1 and P2. A breakpoint of 45 Kb upstream of P2 separates it from P1 and results in an inversion that repositioned P2 around 160 Kb away from its original locus. The inversion also leads to repositioning a non-specific promoter of an uncharacterized gene, responsible for dominant phenotype (rfd), in the Antp locus. This promoter (Prfd) causes ectopic expression of Antp, leading to a gain of function phenotype, characterized by the homeotic transformation of antennae and arista in the fly into a pair of legs (Figure 3C). Embryos homozygous for Antp73b die early during development. These findings support the theory that ectopic promoters can drive the expression of nearby genes in a non-specific manner (Laughon et al., 1986; Schneuwly et al., 1987). Along with gaining insights into the regulation of BX-C, Scr locus, and Antp associated dominant phenotype, the understanding of the Hox complex in Drosophila was pivotal for dissecting the embryonic development of an organism and also led to a better understanding of crucial facets of gene regulation (Figure 3). A plethora of subsequent studies in the following decades after the discovery of Hox revealed their existence in all bilaterians as well as cnidarians (Burke et al., 1995; Brooke et al., 1998; Peterson et al., 2000; Ferrier and Holland, 2001; Samadi and Steiner, 2010; Ikuta, 2011; Gaunt, 2018). Deciphering the functioning of the Drosophila Hox genes complex, in particular, the BX-C, led to a better understanding of embryonic development, molecular biology, patterning, and evolution. Welcome Bender rightly proposed that the regulation of BX-C should enter textbooks at par with lac operon, phage transcription, and yeast mating-type (Bender, 2020).
Clustering, Cis-Regulation, and Remote Controls of Hox Expression
Segment-specific activation and expression of Hox genes are important for segment identity. Transcription factor coding genes including Gap, Pair-rule, and segment polarity genes act upstream of Hox genes and regulate their expression via associated CRMs in insects (Capdevila and Garcia-bellido, 1981; Reinitz and Levine, 1990; Kornberg and Tabata, 1993; Casares and Sanchez-Herrero, 1995; Drewell et al., 2014). As mentioned earlier, there are nine CRMs in the BX-C that direct expression levels and patterns of Ubx, abd-A, and Abd-B in a segment-specific manner.
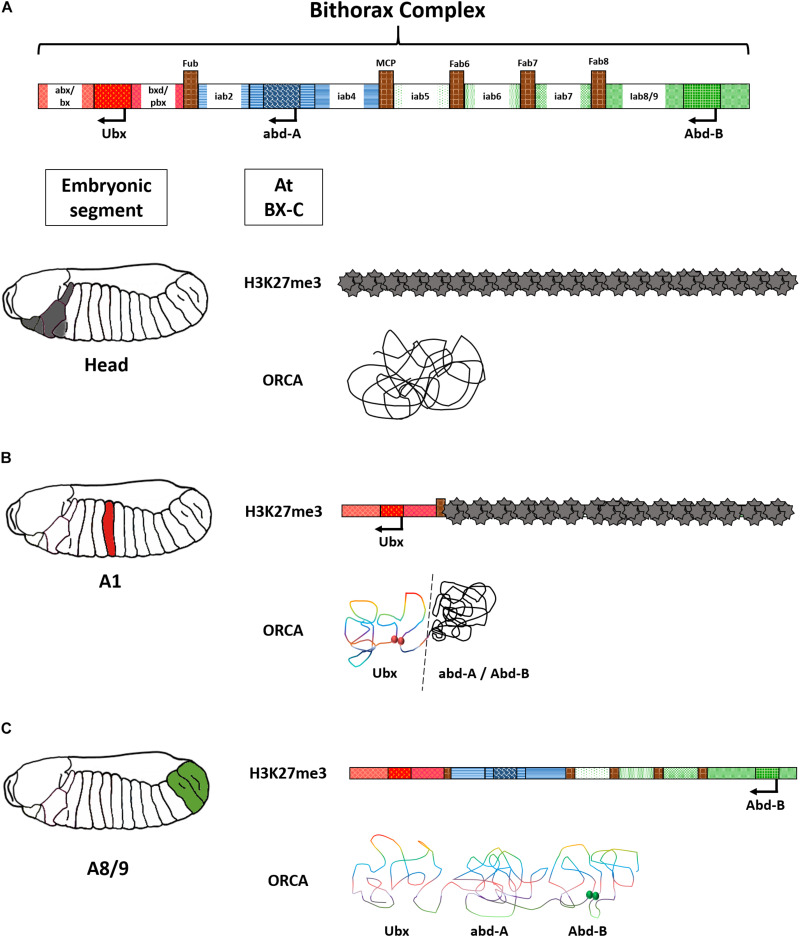
These regions are tightly regulated. Probing the chromatin landscapes of Hox locus has shed some light on their mode of regulation. Segment-specific ChIP-seq for H3K27me3 repressive marks on Drosophila BX-C has pinpointed regions that were sequentially de-methylated from anterior to posterior segment in the fly embryo. For instance, in the head, the BX-C is marked with H3K27me3, coinciding with the absence of expression of all the three genes in the complex. While in A1, the Ubx domain lacked H3K27me3 marks corroborating with the expression status of Ubx in the segment. However, the other two genes of the same complex, abd-A, and Abd-B, were still carrying the repressive marks (Figures 4A–C). This indicated a segment-specific “opening” of BX-C CRMs as one would move from the anterior to the posterior regions in the fly axis and was aptly called the open for business model of the bithorax complex (Bowman et al., 2014; Maeda and Karch, 2015). This model was later reinforced by visualization of chromatin landscape of the BX-C using the optical reconstruction of chromatin architecture (ORCA) technique. It deploys sequential probing of the region of interest on a chromosome, which in this case was ∼320 Kb of the BX-C, by fluorescent probes. The probes are hybridized and washed in a series. They are then coupled with continuous imaging using two customized microscope platforms optimized for HiLo illumination (Mateo et al., 2019). ORCA is advantageous over conventional confocal microscopy due to the single-molecule resolution possible using the said platform. Like conventional imaging, samples are uniformly illuminated, but a high pass filter rejects the illuminated regions that are outside focus. The extracted data is fused with low-frequency in-focus illumination to render a spatially resolved, sharp image (Ford et al., 2012). ORCA of BX-C revealed interactions of segment-specific enhancers with the associated promoters in an in vivo context. Regions devoid of repressive marks were forming a distinct loop, while the ones that remained repressed were forming another closed loop domain (Figures 4A–D; Mateo et al., 2019). The clustering of the CRMs and genes in a relatively short region can be an efficient way to moderate Hox levels. In the unusual cases of Hox arrangement like octopus, sea star, or sea urchin, deciphering 3D genome architecture would provide crucial insights into their functioning.
FIGURE 4.
Open for business model of the bithorax complex. (A) H3K27me3 marks on the bithorax complex in the head reveal complete repression of the locus. The same is corroborated by a representation of the ORCA image that shows clustering of the entire BX-C in one domain. (B) In A1, only the Ubx domain is derepressed and forms a distinct loop. Red spheres indicate contact points of Ubx enhancers with its promoter. (C) In A8/9, the entire BX-C is de-repressed and forms multiple loop domains. Green spheres indicate contact points of Abd-B enhancers with its regulators. The above image is a conceptual representation of data published by Bowman et al. (2014) and Mateo et al. (2019).
The range of available model organisms limits our current knowledge. Nevertheless, in silico and synthetic biology approaches can help in designing experiments of physiological and evolutionary relevance. Crocker et al. (2017) modeled functional enhancers based on the binding sites of various transcription factors across different species of Drosophila. They produced several synthetic enhancers which could be validated in vivo in a developing fly embryo. However, only a limited number of the predicted enhancers could emulate the expression ability of the native ones (Crocker et al., 2017). This could be because of additional factors like insulators and Polycomb/Trithorax response elements (P/TREs) that contribute to regulatory aspects of the genome. Toward this, Srinivasan et al. developed in silico tools to predict chromatin domain boundaries and P/TREs in Drosophila and other insects (Srinivasan and Mishra, 2012, 2020). With the ever-expanding availability of genome sequences, such tools can be extended to model regulation of genes, including Hox, in a diverse set of organisms (Lewin et al., 2018).
Notably, many of the regulatory elements of the genome, like enhancers and insulators, are known to interact with regions that are several Mbs apart (Long et al., 2016). Despite that, the clustering of CRMs and Hox genes in complex organisms suggests a very strong functional consequence. It is speculated that the order of genes within the Hox complex is important for proper body axis development. However, it may be the order of CRMs that might be equally important.
An intriguing region to understand the significance of relative positioning of CRMs can be the Abd-B locus in the BX-C. Each of the iabs (CRM) in the region is demarcated by chromatin domain boundaries (BEs) (Figures 2B,D,E). For example, iab5 specifies PS10 (A5) identity and is followed by a BE, Fab6. The BE separates iab5 from the next CRM iab6, which specifies PS11 (A6) of the fly embryo, thus ensuring autonomous domains of the two CRMs (Galloni et al., 1993; Lewis, 2007; Bender et al., 2011; Postika et al., 2021). Together, the four iabs of the Abd-B locus provide identities to four abdominal segments in the fly from A5 to A8 (terminalia). Hence, the number of these CRMs and their relative positioning in the genome is collinear with the segment they provide identity (Maeda, 2006; Lewis, 2007; Kyrchanova et al., 2015; Maeda and Karch, 2015). Furthermore, many of the BEs are known to function in an orientation-dependent manner. However, most of these studies are done in a transgenic context or a narrow region within the BX-C (Galloni et al., 1993; Martin et al., 1995; Bender and Hudson, 2000; Kyrchanova et al., 2016, 2019). In principle, the iterative arrangement of CRMs and BEs in the Abd-B locus is a compelling case to decipher their role in complex systems. An interesting experiment would be to generate targeted inversions and duplications of CRMs in the BX-C and examine the resulting novel phenotypes. The re-arrangements should be developed in a manner that does not affect binding sites for transcription factors, repressors, or chromatin remodelers obtained from existing ChIP data in the modENCODE consortium (Celniker et al., 2009; Nègre et al., 2010; Négre et al., 2011). Moreover, these re-arrangements should not fuse the domains of two nearby genes or known regulators, as indicated in Figure 5A. One could harness the potential of Cre-LoxP or FLP-FRT systems to bring about these changes. The recombinase recognition sequences can be knocked in at specific sites using CRISPR/Cas9 (Li et al., 2020). For instance, a reorganized locus with MCP followed by iab7, iab6, and iab5 will offer a new playground for cis-/trans- factors to regulate Abd-B. The rearrangement would render iab7 flanked by MCP and Fab7 in opposite directions, whereas Fab6 and Fab8 boundaries will flank iab5. Although the relative positioning of iab6 would remain the same, but, according to the open for business model of BX-C regulation, either iab5 and iab8 will become accessible to Abd-B promoter, or iab7 will be accessible irrespective of re-ordering in PS11 (prospective A6). Such an experiment can unfold the aspects of directionality, ordering, and relative positioning of CRMs within the particular Hox gene locus. Similarly, generating duplications of CRMs like iab5 and iab6 will provide a better understanding of the significance of the number of CRMs required to specify a segment (Figure 5A). Inversions in several cis-regulators in vertebrates have revealed the significance of positioning distal enhancers concerning Hox genes (Kmita et al., 2000; Zakany et al., 2004). Site-specific rearrangements and deletions of vertebrate cis-regulators revealed modularity associated with their arrangements and caused changes in the topologically associated domains (TADs) in which they reside. This leads to the ectopic expression of Hox genes in non-specific regions of the limb, thereby suggesting a significant role of the positions of CREs (Fabre et al., 2017). Since BX-C has a spatially collinear arrangement of the CRMs with a clear understanding of their components, the re-engineered locus will provide a deeper understanding of the evolution of CRM positioning and copy number variations (CNVs).
FIGURE 5.
Re-arrangement of Abd-B locus and vertebrate Hox complex with CRMs. (A) Representation of proposed experiment to re-arrange the CRMs of Abd-B locus in the BX-C. (B) Hox genes are expressed as indicated in Mus musculus. HoxD locus is shown as a representation of regulatory modules associated with Hox complexes. Bold, curved arrows indicate their approximate presence and interaction with Hox complex (not to scale and point precisely on a particular Hox). The role of CR2 in modulating Hox is still unknown and is represented by a dotted arrow.
Overall, the situation is perhaps a bit more complex in vertebrates. They have a minimum of 4 Hox complexes distributed across different chromosomes. Each complex has its own set of regulators. Their embryonic expression follows spatio-temporal collinearity. This means that the genes present toward one end of the cluster are expressed earlier in the anterior regions. The genes present toward the other end of the cluster are expressed later in time in the posterior regions. So, there is an added temporal aspect of regulation in addition to the pre-existing spatial one. Furthermore, the clustering of Hox is more pronounced in vertebrates, with no non-homeotic genes present in the complex. The intergenic distance is also drastically reduced and the entire Hox complex resides within a span of ∼100 Kb. In contrast, both Hox complexes in Drosophila are larger than 300 Kb. The reduction in the size of the complex can be attributed to the positioning of CRMs of vertebrate Hox, outside the cluster on either end, several Kbs away. These regions constitute the global control regions (GCRs), early enhancers (EE), late enhancers (LE), and many other uncharacterized regulatory elements (Soshnikova, 2014). The tight clustering of Hox in vertebrates might also help in robust regulation during secondary axis formation in the limbs when the collinearity is replayed (Soshnikova and Duboule, 2009; Mallo et al., 2010; Mallo and Alonso, 2013; Soshnikova, 2014). Some studies have shown several regions that are ultra-conserved near the HoxD/Evx locus of vertebrates. One of these regions, called conserved region 2 (CR2), was shown to have an early enhancer but late repressor activity in a transgenic context in zebrafish, Danio rerio (Sabarinadh et al., 2004; Kushawah and Mishra, 2017). The exact mechanism and mode of function of these elements are still unknown. It is also not known whether these regions have an impact on Hox genes. Deletions of these regions in several combinations can help us dissect their significance (Figure 5B).
The spatio-temporal regulation of Hox genes in vertebrates has some fascinating offshoots. Marsupials like Tammar wallaby, Macropus eugenii, have delayed expression of posterior Hox genes, HoxA13 and HoxD13, attributed to weaker hind limbs in newborn animals. The forelimbs are relatively stronger, which helps them to climb the brood pouch of their parent. The delayed expression of the posterior Hox is yet another example of modularity and differential expression, possibly due to differences in clustering and accessibility of CRMs which can be accessed via the genome sequence available for marsupials (Chew et al., 2012; Deakin, 2012).
Similar variations of spatio-temporal regulation can be observed in simpler chordates like amphioxus. Despite being in a tight cluster, the spatially collinear expression of Hox genes is perturbed in Branchiostoma floridae. Hox6, a central Hox gene, expresses almost ubiquitously across the neural tube, posterior to the cerebral vesicle. While Hox14, a posterior Hox gene, is expressed in the most anterior cerebral vesicle. Furthermore, Hox14 mRNA is also detected in the pharyngeal endoderm. Interestingly, levels of Hox6 vary greatly in closely related species. Unlike B. floridae Hox6, which shows a uniform expression throughout the neural tube, the B. lanceolatum homolog expresses in a spatially restricted manner (Pascual-Anaya et al., 2012). This indicates subtle modulations of HOX levels in closely related species and is similar to changes observed in invertebrates. Deep sequencing of flanking regions of Hox loci in multiple organisms along with a Bag-of-Motif analysis to understand protein-DNA interactions can shed light on putative regulatory mechanisms associated with the clustering of CRMs.
In simpler organisms like annelids or mollusks, the arrangement of Hox thus seems to be dispensable, but with the evolution of complexity, clustering becomes a necessity for co-regulation.
Modulating Hox in Arthropods
The property of a system to separate and re-integrate its components to form a viable system is called modularity. Subtle changes in Hox expression can quickly orchestrate the evolutionary modularity. The studies are not limited to fruit flies. In an amphipod crustacean, Parhyale hawaiensis, the interplay between various Hox genes and their ability to act independently was comprehended by a series of sophisticated experiments involving manipulation of Hox levels (Liubicich et al., 2009; Martin et al., 2016; Sun and Patel, 2019).
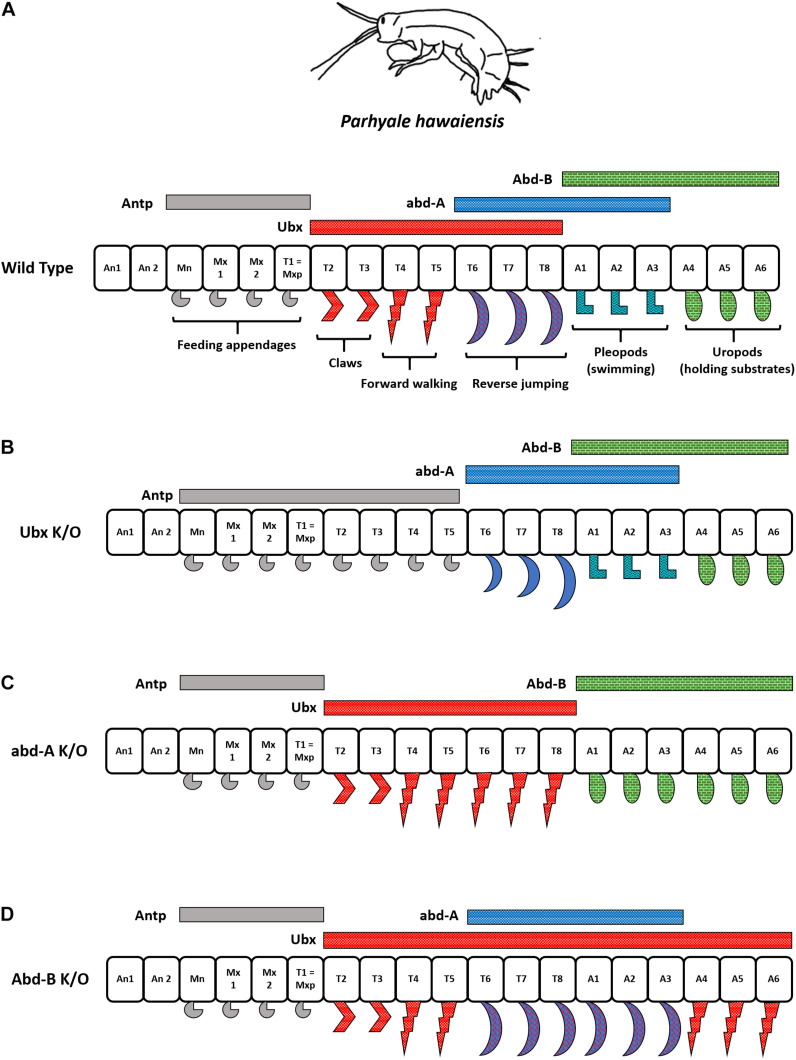
The amphipod is bilaterally symmetrical and has multiple segments with specialized appendages. A group of metameric segments evolved to perform a common function is called tagma, and the associated evolutionary process is called tagmatization (Abzhanov and Kaufman, 2000). The arrangement of appendages in the order of their occurrence from anterior to posterior segments in Parhyale is as follows – feeding appendages (mandible, Mn and maxillipeds Mx, or, gnathopods), claws (T2–T3), forward (T4–T5), and reverse (T6–T8) walking legs (pereopods), swimming appendages (pleopods or swimmerets) in the segments A1 to A3, and appendages for holding substrates (uropods) formed in A4–A6. A simple representation of P. hawaiensis tagmatization is depicted in Figure 6A.
FIGURE 6.
Interplay of different Hox genes in Parahyale hawaiensis. (A) Hox expression in Parhyale hawaiensis. Note that abd-A has overlapping regions of functioning with Ubx as well as Abd-B. (B) Ubx knock-out animals show anteriorization of forward walking legs. (C) abd-A knock-out animals show anteriorization of reverse walking legs and posteriorization of swimming appendages. (D) Abd-B knock-out animals show anteriorization of swimming appendages into reverse walking legs and uropods in forward walking legs.
Recent advances in CRISPR-based gene editing have allowed researchers to perform knock-out experiments in P. hawaiensis. Martin et al. (2016) knocked out Ph Ubx by CRISPR/Cas9 and observe that the locomotor appendages acquire the identity of feeding appendages (T2–T5 → Mn/Mxp). Further, knocking out a posterior Hox gene Ph abd-A, which would otherwise be responsible for forming reverse walking legs in the crustacean, now has them transformed into a copy of forward walking legs (T6–T8 → T4); Figure 6B. This was as expected from previous studies in Drosophila that indicate the anteriorization of segments in the absence of posterior Hox; a property called the posterior prevalence of Hox genes. However, another class of legs called pleopods or swimming appendages was transformed into a copy of posterior appendages, uropods (A1–A3 → A4), in the Ph abd-A knock-out animals. This was an apparent effect of the additive function of Ph abd-A to regulate segment identity in either direction along the AP axis. In the anterior regions, Ph abd-A works with Ph Ubx to develop segments with reverse walking legs (T6–T8). In the posterior regions, Ph abd-A functions with Ph Abd-B to develop swimming appendages (A1–A3) as depicted in Figure 6C. Strikingly, knock-out of an even more posterior gene Ph Abd-B displayed a spectacular non-linear transformation of uropods into copies of forward walking legs but not swimmerets (A4–A6 → T5; not A3); Figure 6D. This suggested that the ABD-B represses Ph Ubx in the posterior segments of the Parhyale from A1 to A6, whereas Ph abd-A expression is independent of the ABD-B levels. Ph Abd-B knock-out animals had derepression of Ph Ubx in the posterior segments, leading to extreme transformation into forward walking legs. The segment with overlapping domains of Ph Ubx and Ph abd-A developed reverse walking legs in Ph Abd-B knock-outs while swimmerets were altogether absent from the organism (A1–A3 → T8); Figures 6A,D. The studies from the crustacean suggest that alongside collinear expression of Hox, the co-regulation, inter-regulation, and cross-talk between different HOX cause varying phenotypes. The interplay between these genes brings about diversity in the animal kingdom (Martin et al., 2016).
In addition to the AP axis, a handful of studies also show the role of Hox genes in LR and DV axis determination (Thickett and Morgan, 2002; Mohit et al., 2006; Coutelis et al., 2013). An exemplar is the Dmel Abd-B regulating MyosinID (MyoID), a protein responsible for complete dextral (clockwise 360°) rotation of spermiduct around hindgut during metamorphosis. Abd-B knocked down flies show partial sinistral (anti-clockwise) rotation to varying degrees that causes male sterility due to rotation of external genitalia (Spéder et al., 2006; Coutelis et al., 2013). Crustaceans such as P. hawaiensis develop symmetrically along the LR axis, and early knock-down of Ph Ubx in one of the sides causes asymmetrical homeotic transformation of segments, including appendage formation. This was done by injecting morpholinos for Ph Ubx knockdown in one of two-celled stage embryo cells. Each cell follows its fate separately across the LR axis of development. Although the system was utilized to compare wild type versus knockdown phenotypes in the same organism (Browne et al., 2005; Liubicich et al., 2009; Pavlopoulos et al., 2009), the study also implies asymmetrical differentiation of body segments upon differential expression of Hox genes. In tune with this, in the Xenopus embryo, HoxC8 expresses asymmetrically along the left-right axis of development in the lateral plate mesoderm (Thickett and Morgan, 2002). One interesting organism worth probing for Hox genes regulation and determination of the LR axis is the fiddler crab. It is a natural example of left-right asymmetry in appendage formation. The female fiddler crabs have similar-sized left and right feeding appendages. In comparison, males have one of their feeding appendages extraordinarily enlarged. They use this appendage to fight competitors during mating and display handedness (Pardo et al., 2020). A detailed understanding of Hox expression in these organisms can shed light upon the formation of segments in AP and LR axes of development.
Another example of a modified and rather intriguing appendage is the scorpion’s tail, including the terminal telson. Scorpions have undergone duplications of Hox genes, which are correlated to the heteronomy of the posterior segments (Sharma et al., 2014). Arizona bark scorpion, Centruroides sculpturatus, has 19 Hox genes instead of 10 in its sister groups. The dual copies are expressed in varying degrees from antero-central to telson. These include Antp, Ubx, abd-A, and Abd-B. In C. sculpturatus, extended-expression of the two copies of Cs Antp and Cs Ubx is corroborated with enlarged telson in a distinct shape for an appendage. Notedly, the telson is formed posterior to terminalia (anus). It would be interesting to delete one or multiple copies of each of these Hox genes and observe the changes in body patterning. The tagmatization could be affected to the extent that the body form might become less elongated, as is the case with Opiliones, harvestmen, or instigated to form a telson-less scorpion (Sharma et al., 2012). The opposite spectrum of body formation is seen in Tardigrades, in which deletion of several Hox genes correlates with their compact body plan with simpler, repetitive, and less (four) number of segments (Smith et al., 2016).
Other than the levels of HOX, structural modifications in the transcription factors can help in diverse functions. Recent experiments with flies provided evidence of functional conservation of mouse Hox genes. Singh et al. (2020) replaced labial, the anterior-most gene in Drosophila Hox complex, with Hox1 from Mus musculus. Interestingly, out of the three copies of Hox1 in the form of HoxA1, HoxB1, and HoxD1, only HoxA1 could rescue the labial knock-out phenotype completely. They also developed animals with chimeric HOX proteins and discovered a six-amino acid C-terminal motif in HoxA1 essential for its interaction with PBX. The ortholog-specific interaction leads to differential occupancy of HoxA1 across the genome. This study strongly supports the notion of evolutionary modularity in Hox complex by causing structural changes in HOX that lead to similar yet functionally divergent protein products (Singh et al., 2020).
An ordered arrangement of Hox could have played an important role in their sequential co-regulation along the AP axis, as indicated by our understanding of BX-C regulation. One can consider Hox genes as switches to control different electrical equipment at home. They can be present anywhere across the house and can still function, as is the case of an octopus. But clustering on a switchboard gives quick, precise, and perhaps, robust control over the spatio-temporal regulation of Hox genes. This modularity could have been one reason for arthropods to surpass mollusks as the richest bio-diverse species on our planet (Benton, 2010). Many genes are co-regulated in different organisms (Snel et al., 2004). Overall, clustering is more abundant in vertebrates than invertebrates (Elizondo et al., 2009; Ferrier, 2016). Nevertheless, in addition to clustering, the ordering is an important property of Hox complexes that need to be pondered upon. The past decade has witnessed rapid advancements in our understanding of epigenetic factors, inter-genic regulators, and chromatin organization (Narlikar and Ovcharenko, 2009; Hübner et al., 2013; Allis and Jenuwein, 2016; Hug and Vaquerizas, 2018). Understanding them in the context of gene clusters, including Hox complexes, will be riveting. The Hox genes have a tremendous potential to modulate diversity by teaming up with multiple partners and setting a stage for downstream players in various axes. Different combinations of cis- and trans- regulators together bring about manifold changes that can drive evolution.
Hox Genes: Master Regulators Beyond Embryogenesis and Homeosis
Classically, mutations in Hox genes are associated with the homeotic transformation of one body segment into another, a process called homeosis (Lewis, 1994). These mutations transformed embryonic segments, and therefore the Hox genes were established as the regulators during embryonic development (Pradel and White, 1998). However, even during embryonic development, Hox genes can still play a non-homeotic role by specifically affecting tissue homeostasis and organogenesis (Castelli-Gair Hombría and Lovegrove, 2003).
Recent studies opened new horizons to understand the role of Hox genes in an organism. A rising number of articles suggest their role beyond homeotic functions and embryonic development (Wang et al., 2009; Estacio-Gómez and Díaz-Benjumea, 2014; Gummalla et al., 2014; Rux and Wellik, 2017). In D. melanogaster, prolonged expression of Hox genes beyond embryogenesis is observed in developing larva and pupa (Wang et al., 2009). The three genes of the bithorax complex, Ubx, abd-A, and Abd-B, have defined anterior limits of expression in Drosophila larvae. The larva undergoes metamorphosis during pupal stages of development, ultimately eclosing as adults. One key event during this process is autophagy of most of the larval tissues, including the fat body, salivary glands, and trachea. This is further coupled with the differentiation of adult tissues that goes on till eclosion. Interestingly, all the three genes of BX-C, Ubx, abd-A, and Abd-B are expressed in the larval fat body (Marchetti et al., 2003). Down regulation of Ubx is accompanied by developmental and starvation-induced autophagy, whereas sustained expression of the Hox gene inhibits autophagy and delays metamorphosis (Banreti et al., 2014).
Like the larval fat body, larval epithelial cells (LECs) also undergo apoptosis during metamorphosis. Further, another group of cells called histoblast nest cells (HNCs) differentiates to form adult abdominal epithelial cells during pupation. Posterior BX-C genes abd-A and Abd-B have overlapping expressions in the LECs. Loss of abd-A impairs the apoptotic pathway in LECs and cannot be rescued by Abd-B alone. Moreover, HNC proliferation is hindered by abd-A down regulations, and the cells fail to form a complete epithelium in abd-A knocked down pupae. Thus ABD-A is required for both, apoptosis of LECs as well as the proliferation of HNCs to form mature abdominal epithelium in adults. The study showed that ABD-A was present in the LECs and contributed toward development together with the posterior Hox gene product ABD-B, therefore defying the property of posterior prevalence (Singh and Mishra, 2014). The study also contributed to our understanding of Hox genes’ modular capacity in an extra-homeotic and extra-embryonic manner.
Similar reports for Abd-B were observed in testis development, where it remains active in pre-meiotic spermatocytes. Tissue-specific knockdown of Abd-B in adult testes leads to a loss of maintenance of the stem cell niche required to produce normal sperms. This is because ABD-B has direct binding sites on src42A and sec63, members of Boss signaling involved in testes formation and sperm differentiation. Abd-B also has an extended effect on the orientation of centrosomes and the division rates of germline stem cells (Papagiannouli and Lohmann, 2015).
Obtaining tissue-specific cells for further studies of Hox was a Herculean task a couple of years back, as one had to do neck-breaking dissections to get ounces of desirable material. Although now, endogenous tagging of Hox genes has solved a lot of such problems. Cell sorting of fluorescently labeled HOX expressing tissues followed by multi-omics experiments can help us understand the genome-wide effects of HOX in adult tissues. Domsch et al. (2019) reported an endogenously tagged line for Ubx with GFP at the N-terminal. They utilized this resource to establish the role of Ubx as a major repressor of factors involved in alternate fate development in mesodermal cells. Sorting GFP expressing cells followed by ChIP and Co-IP experiments helped in a deeper understanding of modalities of Ubx functioning. This revealed UBX’s ability to cause repression by constantly interacting with a member of Polycomb Repressive Complex protein Pleiohomeotic (PHO) (Domsch et al., 2019). In their recent work, Paul et al. (2021) showed that not only the presence of HOX but also their dosage determines the formation of appendages – in their case, wing appendages.
The extraembryonic roles of Hox are more distinct in vertebrates. As early as 2003, it was evident that Hox genes play a role in non-homeotic fashion owing to the near-complete loss of hair formation in mice deficient for HoxC13. Although the mouse also had patterning defects, hair growth was uniformly reduced across the body (Awgulewitsch, 2003).
Recent reports showed several HoxC genes in the dermal papilla and associated it with regional follicle variation. In a mutant mouse line called Koala mutant, a 1 Mb inversion encompassed disintegration of HoxC4 to HoxC13 from the main complex leading to their misexpression. CTCF ChiP-seq revealed changes in levels of CTCF binding within the HoxC complex and perturbation of topologically associated domains (TADs) (Millar, 2018). Similar deletion studies have identified the role of HoxA genes in mammary gland formation during specific transition developmental periods (Lewis, 2000).
Owing to their multifaceted roles during and after development, levels of Hox proteins need to be tightly regulated. Misexpression of these genes has been observed in various cancers like breast cancer, melanoma, bone cancer, blood cancer, and colorectal cancer (Shah and Sukumar, 2010). Central and posterior Hox genes, HoxA5 and HoxD9, have been implicated in esophageal squamous cell carcinoma. Strikingly, they were found to localize more in the cytoplasm of the mucosa cells in esophageal cancer than in the nucleus in normal cellular conditions (Takahashi et al., 2007). Similarly, ectopic expression of HoxC6, HoxC11, HoxD1, and HoxD8 are observed in different cases of neuroblastoma (Manohar et al., 1996; Zhang et al., 2007). Overexpression of posterior Hox genes, particularly HoxA9-11, HoxB13, and HoxC10, is linked to the onset and tumor progression of ovarian, cervical, and prostate cancers (Jung et al., 2004; Cheng et al., 2005; Miao et al., 2007; Zhai et al., 2007). Misexpression of HoxA9, HoxA10, and HoxC6 was also reported in cases of Leukemia caused by translocations of mixed-lineage leukemia Mll gene. MLL is the vertebrate homolog of Drosophila Trithorax (TRX) protein and helps maintain an active state of Hox expression in required domains (Armstrong et al., 2002; Ono et al., 2005). Hox-associated cancer is not limited to genetic mutations. Rauch et al. (2007) showed increased methylation of HoxA7 and HoxA9 associated CpG islands. The study highlighted epigenetic misregulation as a putative cause for Hox-related lung tumors. Likewise, promoter methylation of HoxA5 and downregulation of HoxA10 are associated with progressive breast carcinoma. The disease can also be caused by overexpression of HoxB7 and HoxB13 in these tissues (Raman et al., 2000; Chu et al., 2004; Chen et al., 2008; Jerevall et al., 2008). Misexpression studies in Drosophila confirmed the causal effect and established flies as a model to study Hox-associated oncogenesis. The outcome of the study was the ability of Dfd, Ubx, and abd-A genes to be leukemogenic when overexpressed in fat body and lamellocytes (Ponrathnam et al., 2021).
Detailed understanding of Hox genes expression and interaction during embryogenesis, tissue formation, organogenesis, and cellular homeostasis is required to delineate their functional modalities. Due to their overarching involvement in multiple processes of body formation, patterning, and evolution, Hox genes occupy a prime position in our quest toward understanding these processes in depth.
Concluding Remarks
A long-debated topic in the field of Hox genes was their presence in the form of clusters and the property of spatio-temporal collinearity. Some recent developments also demonstrated the functioning of Hox independent of clustering. However, coordinated functioning is better when they are clustered together, as implied by the open for business model of the bithorax complex. Alterations of CRMs throughout the Hox led to a myriad of homeotic transformations. Similar genomic alterations across evolution might have experimented with Hox modules and their expression to bring about the enormous diversity we see today. Individual notes are pleasant to hear, but it’s the symphony that conveys the melody. Hox come together to set up the primary and secondary axes and provide constant inputs in different tissues, therefore orchestrating the developmental design sublimely. In vivo experiments with the latest genome editing tools and a better understanding of non-coding DNA become important for comprehending the conductors of this symphony.
Author Contributions
RKM and NH conceived the design of the article and edited the manuscript. NH wrote the manuscript with inputs from RKM, and conceptualized and drew illustrations upon discussion with RKM. Both authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors acknowledge the members of the RM group, especially, Shubhanshu Pandey, Ashish Bihani, M. S. Soujanya, Sonu Yadav, and Lorraine Teron for carefully reviewing the manuscript. The authors would also like to extend gratitude toward Surbhi Lambhate for continuous discussions and suggestions while preparing the manuscript.
Footnotes
Funding. NH is a fellow of the Department of Biotechnology, Govt. of India, and thanks the agency for providing timely financial support. RKM is recipient of JC Bose National fellowship, India. The authors would also like to thank CSIR, India, JC Bose National Fellowship, India and Tata Institute for Genetics and Society, India for financial support at various levels.
References
- Abzhanov A., Kaufman T. C. (2000). Crustacean (malacostracan) Hox genes and the evolution of the arthropod trunk. Development 127 2239–2249. 10.1242/dev.127.11.2239 [DOI] [PubMed] [Google Scholar]
- Akam M. (1998). Hox genes, homeosis and the evolution of segment identity: No need for hopeless monsters. Int. J. Dev. Biol. 42 445–451. 10.1387/ijdb.9654030 [DOI] [PubMed] [Google Scholar]
- Akam M., Dawson I., Tear G. (1988). Homeotic genes and the control of segment diversity. Development 133 123–133. [Google Scholar]
- Albertin C. B., Simakov O., Mitros T., Wang Z. Y., Pungor J. R., Edsinger-Gonzales E., et al. (2015). The octopus genome and the evolution of cephalopod neural and morphological novelties. Nature 524 220–224. 10.1038/nature14668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allis C. D., Jenuwein T. (2016). The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 17 487–500. 10.1038/nrg.2016.59 [DOI] [PubMed] [Google Scholar]
- Armstrong S. A., Staunton J. E., Silverman L. B., Pieters R., Den Boer M. L., Minden M. D., et al. (2002). MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat. Genet. 30 41–47. 10.1038/ng765 [DOI] [PubMed] [Google Scholar]
- Awgulewitsch A. (2003). Hox in hair growth and development. Naturwissenschaften 90 193–211. 10.1007/s00114-003-0417-4 [DOI] [PubMed] [Google Scholar]
- Bae E., Calhoun V. C., Levine M., Lewis E. B., Drewell R. A. (2002). Characterization of the intergenic RNA profile at abdominal-A and abdominal-B in the Drosophila bithorax complex. Proc. Natl. Acad. Sci. U. S. A. 99 16847–16852. 10.1073/pnas.222671299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banreti A., Hudry B., Sass M., Saurin A. J., Graba Y. (2014). Hox Proteins Mediate Developmental and Environmental Control of Autophagy. Dev. Cell 28 56–69. 10.1016/j.devcel.2013.11.024 [DOI] [PubMed] [Google Scholar]
- Bantignies F., Roure V., Comet I., Leblanc B., Schuettengruber B., Bonnet J., et al. (2011). Polycomb-dependent regulatory contacts between distant hox loci in drosophila. Cell 144 214–226. 10.1016/j.cell.2010.12.026 [DOI] [PubMed] [Google Scholar]
- Barges S., Mihaly J., Galloni M., Hagstrom K., Müller M., Shanower G., et al. (2000). The Fab-8 boundary defines the distal limit of the bithorax complex iab-7 domain and insulates iab-7 from initiation elements and a PRE in the adjacent iab-8 domain. Development 127 779–790. [DOI] [PubMed] [Google Scholar]
- Basson M. A. (2012). Signaling in Cell Differentiation and Morphogenesis. Cold Spring Harb. Perspect. Biol. 4 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baughman K. W., Mcdougall C., Cummins S. F., Hall M., Degnan B. M., Satoh N., et al. (2014). Genomic organization of Hox and ParaHox clusters in the echinoderm, Acanthaster planci. Genesis 52 952–958. 10.1002/dvg.22840 [DOI] [PubMed] [Google Scholar]
- Beddington R. S. P., Robertson E. J. (1999). Axis development and early asymmetry in mammals. Cell 96 195–209. 10.1016/S0092-8674(00)80560-7 [DOI] [PubMed] [Google Scholar]
- Bekiaris P. S., Tekath T., Staiger D., Danisman S. (2018). Computational exploration of cis -regulatory modules in rhythmic expression data using the “Exploration of Distinctive CREs and CRMs” (EDCC) and “CRM Network Generator” (CNG) programs. PLoS One 13:e0190421. 10.1371/journal.pone.0190421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeman R. W. (1987). A homoeotic gene cluster in the red flour beetle. Nature 327, 3–5.3574463 [Google Scholar]
- Bender W., Akam M., Karch F., Beachy P. A., Peifer M., Spierer P., et al. (2011). Molecular Genetics of the Bithorax Complex in Drosophila melanogaster. Sciecne 221 23–29. [DOI] [PubMed] [Google Scholar]
- Bender W., Hudson A. (2000). P element homing to the Drosophila bithorax complex. Development 127 3981–3992. [DOI] [PubMed] [Google Scholar]
- Bender W., Lucas M. (2013). The Border Between the Ultrabithorax and abdominal-A Regulatory Domains. Genetics 193 1135–1147. 10.1534/genetics.112.146340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender W. W. (2020). Molecular Lessons from the Drosophila Bithorax Complex. Genetics 216 613–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton M. J. (2010). The origins of modern biodiversity on land. Philos. Trans. R. Soc. B Biol. Sci. 365 3667–3679. 10.1098/rstb.2010.0269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berenguer M., Meyer K. F., Yin J., Duester G. (2020). Discovery of genes required for body axis and limb formation by global identification of retinoic acid-regulated epigenetic marks. PLoS Biol. 18:e3000719. 10.1371/journal.pbio.3000719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman S. K., Deaton A. M., Domingues H., Wang P. I., Sadreyev R. I., Kingston R. E., et al. (2014). H3K27 modifications define segmental regulatory domains in the Drosophila bithorax complex. Elife 3 1–13. 10.7554/eLife.02833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooke N. M., Garcia-Fernàndez J., Holland P. W. H. (1998). The ParaHox gene cluster is an evolutionary sister of the Hox gene cluster. Nature 392 920–922. 10.1038/31933 [DOI] [PubMed] [Google Scholar]
- Brown S. J., Fellers J. P., Shippy T. D., Richardson E. A., Maxwell M., Stuart J. J., et al. (2002). Sequence of the Tribolium castaneum homeotic complex: the region corresponding to the Drosophila melanogaster antennapedia complex. Genetics 1074, 1067–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne W. E., Price A. L., Gerberding M., Patel N. H. (2005). Stages of embryonic development in the amphipod crustacean, Parhyale hawaiensis. Genesis 42 124–149. 10.1002/gene.20145 [DOI] [PubMed] [Google Scholar]
- Burke A. C., Nelson C. E., Morgan B. A., Tabin C. (1995). Hox genes and the evolution of vertebrate axial morphology. Development 121 333–346. 10.1242/dev.121.2.333 [DOI] [PubMed] [Google Scholar]
- Calhoun V. C., Levine M. (2003). Long-range enhancer-promoter interactions in the Scr-Antp interval of the Drosophila Antennapedia complex. Proc. Natl. Acad. Sci. U. S. A. 100 9878–9883. 10.1073/pnas.1233791100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capdevila M., Garcia-bellido A. (1981). Genes Involved in the Activation of the Bithorax Complex in Drosophila. Wilehm Roux Arch. Dev. Biol. 190 339–350. [DOI] [PubMed] [Google Scholar]
- Casares F., Sanchez-Herrero E. (1995). Regulation of the infraabdominal regions of the bithorax complex of Drosophila by gap genes. Development 121 1855–1866. 10.1242/dev.121.6.1855 [DOI] [PubMed] [Google Scholar]
- Castelli-gair J. E., Capdevila M., Micol J., Garcia-bellido A. (1992). Positive and negative cis-regulatory elements in the bithoraxoid region of the Drosophila Ultrabithorax gene. Mol. Genet. Genomics 234 177–184. [DOI] [PubMed] [Google Scholar]
- Castelli-Gair Hombría J., Lovegrove B. (2003). Beyond homeosis - HOX function in morphogenesis and organogenesis. Differentiation 71 461–476. 10.1046/j.1432-0436.2003.7108004.x [DOI] [PubMed] [Google Scholar]
- Celniker S. E., Dillon L. A. L., Gerstein M. B., Gunsalus K. C., Henikoff S., Karpen G. H., et al. (2009). Unlocking the secrets of the genome. Nature 459 927–930. 10.1038/459927a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker S. E., Keelan D. J., Lewis E. B. (1989). The molecular genetics of the bithorax complex of Drosophila: characterization of the products of the Abdominal-B domain. Genes Dev. 3 1424–1436. 10.1101/gad.3.9.1424 [DOI] [PubMed] [Google Scholar]
- Celniker S. E., Sharma S., Keelan D. J., Lewis E. B. (1990). The molecular genetics of the bithorax complex of Drosophila: cis-regulation in the Abdominal-B domain. EMBO J. 9 4277–4286. 10.1002/j.1460-2075.1990.tb07876.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C. S., Rastelli L., Pirrotia V. (1994). A Polycomb response element in the Ubx gene that determines an epigenetically inherited state of repression. EMBO J. 13 2553–2564. 10.1002/j.1460-2075.1994.tb06545.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Ji S. L., Liang X., Zhang H., Zhu T., Zhang Z., et al. (2008). Hoxb7 inhibits transgenic HER-2/neu-induced mouse mammary tumor onset but promotes progression and lung metastasis. Cancer Res. 68 3637–3644. 10.1158/0008-5472.CAN-07-2926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W., Liu J., Yoshida H., Rosen D., Naora H. (2005). Lineage infidelity of epithelial ovarian cancers is controlled by HOX genes that specify regional identity in the reproductive tract. Nat. Med. 11 531–537. 10.1038/nm1230 [DOI] [PubMed] [Google Scholar]
- Chew K. Y., Yu H., Pask A. J., Shaw G., Renfree M. B. (2012). HOXA13 and HOXD13 expression during development of the syndactylous digits in the marsupial Macropus eugenii. BMC Dev. Biol. 12:2. 10.1186/1471-213X-12-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra V. S. (2011). Chromosomal organization at the level of gene complexes. Cell. Mol. Life Sci. 68 977–990. 10.1007/s00018-010-0585-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chourrout D., Delsuc F., Chourrout P., Edvardsen R. B., Rentzsch F., Renfer E., et al. (2006). Minimal ProtoHox cluster inferred from bilaterian and cnidarian Hox complements. Nature 442 684–687. 10.1038/nature04863 [DOI] [PubMed] [Google Scholar]
- Chu M. C., Selam F. B., Taylor H. S. (2004). HOXA10 regulates p53 expression and matrigel invasion in human breast cancer cells. Cancer Biol. Ther. 3 568–572. 10.4161/cbt.3.6.848 [DOI] [PubMed] [Google Scholar]
- Coutelis J. B., Géminard C., Spéder P., Suzanne M., Petzoldt A. G., Noselli S. (2013). Drosophila Left/Right Asymmetry Establishment Is Controlled by the Hox Gene Abdominal-B. Dev. Cell 24 89–97. 10.1016/j.devcel.2012.11.013 [DOI] [PubMed] [Google Scholar]
- Crocker J., Tsai A., Stern D. L. (2017). A Fully Synthetic Transcriptional Platform for a Multicellular Eukaryote. Cell Rep. 18 287–296. 10.1016/j.celrep.2016.12.025 [DOI] [PubMed] [Google Scholar]
- Deakin J. E. (2012). Marsupial genome sequences: providing insight into evolution and disease. Scientifica 2012:543176. 10.6064/2012/543176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dequéant M. L., Pourquié O. (2008). Segmental patterning of the vertebrate embryonic axis. Nat. Rev. Genet. 9 370–382. 10.1038/nrg2320 [DOI] [PubMed] [Google Scholar]
- Dessain S., McGinnis W. (1993). Drosophila Homeobox Genes. Adv. Dev. Biochem. 2 1–55. 10.1016/s1064-2722(08)60035-3 [DOI] [Google Scholar]
- Deutsch J. S. (2004). Segments and parasegments in arthropods: a functional perspective. BioEssays 26 1117–1125. 10.1002/bies.20111 [DOI] [PubMed] [Google Scholar]
- Di Z., Yu Y., Wu Y., Hao P., He Y., Zhao H., et al. (2015). Genome-wide analysis of homeobox genes from Mesobuthus martensii reveals Hox gene duplication in scorpions. Insect Biochem. Mol. Biol. 61 25–33. 10.1016/j.ibmb.2015.04.002 [DOI] [PubMed] [Google Scholar]
- Dickinson M. H., Trans P., Lond R. S., Dickinson M. H. (1999). Haltere - mediated equilibrium reflexes of the fruit fly, Drosophila melanogaster. Philos. Trans. R Soc. Lond. B Biol. Sci. 354 903–916. 10.1098/rstb.1999.0442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domsch K., Carnesecchi J., Disela V., Friedrich J., Trost N., Ermakova O., et al. (2019). The Hox Transcription Factor Ubx stabilizes Lineage Commitment by Suppressing Cellular Plasticity. Elife 8:e42675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domsch K., Schröder J., Janeschik M., Schaub C., Lohmann I. (2020). The Hox transcription factor Ubx ensures somatic myogenesis by suppressing the mesodermal master regulator Twist. BioRxiv 1–29. 10.1101/2020.02.24.963231 [DOI] [PubMed] [Google Scholar]
- Drewell R. A., Nevarez M. J., Kurata J. S., Winkler L. N., Li L., Dresch J. M. (2014). Deciphering the combinatorial architecture of a Drosophila homeotic gene enhancer. Mech. Dev. 131 68–77. 10.1016/j.mod.2013.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duboule D. (2007). The rise and fall of Hox gene clusters. Development 134 2549–2560. 10.1242/dev.001065 [DOI] [PubMed] [Google Scholar]
- Elizondo L., Jafar-Nejad P., Clewing J., Boerkoel C. (2009). Gene Clusters, Molecular Evolution and Disease: a Speculation. Curr. Genomics 10 64–75. 10.2174/138920209787581271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estacio-Gómez A., Díaz-Benjumea F. J. (2014). Roles of Hox genes in the patterning of the central nervous system of Drosophila. Fly 8 26–32. 10.4161/fly.27424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada B., Casares F., Busturia A., Sánchez-herrero E. (2002). Genetic and molecular characterization of a novel iab-8 regulatory domain in the Abdominal-B gene of Drosophila melanogaster. Development 5204 5195–5204. [DOI] [PubMed] [Google Scholar]
- Fabre P. J., Leleu M., Mormann B. H., Lopez-Delisle L., Noordermeer D., Beccari L., et al. (2017). Large scale genomic reorganization of topological domains at the HoxD locus. Genome Biol. 18 1–15. 10.1186/s13059-017-1278-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrier D. E. K. (2016). Evolution of homeobox gene clusters in animals: the Giga-cluster and Primary vs. secondary clustering. Front. Ecol. Evol. 4:36. 10.3389/fevo.2016.00036 [DOI] [Google Scholar]
- Ferrier D. E. K., Holland P. W. H. (2001). Ancient Origin of the Hox Gene Cluster. Nat. Rev. Genet. 2 33–8. [DOI] [PubMed] [Google Scholar]
- Ferrier D. E. K., Minguillón C. (2003). Evolution of the Hox/ParaHox gene clusters. Int. J. Dev. Biol. 47 605–611. [PubMed] [Google Scholar]
- Ferrier D. E. K., Minguillón C., Holland P. W. H., Garcia-Fernàndez J. (2000). The amphioxus Hox cluster: deuterostome posterior flexibility and Hox14. Evol. Dev. 2 284–293. 10.1046/j.1525-142X.2000.00070.x [DOI] [PubMed] [Google Scholar]
- Ford T. N., Lim D., Mertz J. (2012). Fast optically sectioned fluorescence HiLo endomicroscopy. J. Biomed. Opt. 17:021105. 10.1117/1.jbo.17.2.021105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- François V., Solloway M., O’Neill J. W., Emery J., Bier E. (1994). Dorsal-ventral patterning of the Drosophila embryo depends on a putative negative growth factor encoded by the short gastrulation gene. Genes Dev. 8 2602–2616. 10.1101/gad.8.21.2602 [DOI] [PubMed] [Google Scholar]
- Fritsch M., Wollesen T., De Oliveira A. L., Wanninger A. (2015). Unexpected co-linearity of Hox gene expression in an aculiferan mollusk. BMC Evol. Biol. 15:151. 10.1186/s12862-015-0414-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloni M., Gyurkovics H., Schedl P., Karch F. (1993). The bluetail transposon: evidence for independent cis-regulatory domains and domain boundaries in the bithorax complex. EMBO J. 12 1087–1097. 10.1002/j.1460-2075.1993.tb05750.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaunt S. J. (2015). The significance of Hox gene collinearity. Int. J. Dev. Biol. 59, 159–170. 10.1387/ijdb.150223sg [DOI] [PubMed] [Google Scholar]
- Gaunt S. J. (2018). Hox cluster genes and collinearities throughout the tree of animal life. Int. J. Dev. Biol. 62 673–683. 10.1387/ijdb.180162sg [DOI] [PubMed] [Google Scholar]
- Gindhart J. G., King A. N., Kaufman T. C. (1995). Characterization of the cis-Regulatory Region of the Drosophila Homeotic Gene Sex combs reduced. Genetics 139 781–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gummalla M., Galetti S., Maeda R. K., Karch F. (2014). Hox gene regulation in the central nervous system of Drosophila. Front. Cell. Neurosci. 8:96. 10.3389/fncel.2014.00096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagstrom K., Muller M., Schedl P. (1996). Fab-7 functions as a chromatin domain boundary to ensure proper segment specification by the Drosophila bithorax complex. Genes Dev. 10 3202–3215. 10.1101/gad.10.24.3202 [DOI] [PubMed] [Google Scholar]
- Hendrickson J. E., Sakonju S. (1995). Cis and trans interactions between the iab regulatory regions and abdominal-A and abdominal-B in Drosophila melanogaster. Genetics 139 835–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho M. C. W., Johnsen H., Goetz S. E., Schiller B. J., Bae E., Tran D. A., et al. (2009). Functional evolution of cis-regulatory modules at a homeotic gene in Drosophila. PLoS Genet. 5:e1000709. 10.1371/journal.pgen.1000709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland L. Z., Albalat R., Azumi K., Benito-Gutiérrez È, Blow M. J., Bronner-Fraser M., et al. (2008). The amphioxus genome illuminates vertebrate origins and cephalochordate biology. Genome Res. 18 1100–1111. 10.1101/gr.073676.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland P. W. H. (2001). Beyond the Hox: how widespread is homeobox gene clustering?. J. Anat. 199 13–23. 10.1017/S0021878201008275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland P. W. H. (2015). Did homeobox gene duplications contribute to the Cambrian explosion?. Zool. Lett. 1:1. 10.1186/s40851-014-0004-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland P. W. H., Booth H. A. F., Bruford E. A. (2007). Classification and nomenclature of all human homeobox genes. BMC Biol. 5:47. 10.1186/1741-7007-5-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard-Ashby M., Materna S. C., Brown C. T., Chen L., Cameron R. A., Davidson E. H. (2006). Identification and characterization of homeobox transcription factor genes in Strongylocentrotus purpuratus, and their expression in embryonic development. Dev. Biol. 300 74–89. 10.1016/j.ydbio.2006.08.039 [DOI] [PubMed] [Google Scholar]
- Hrycaj S. M., Wellik D. M. (2016). Hox genes and evolution. F1000Res. 5:859. 10.12688/f1000research.7663.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hübner M. R., Eckersley-Maslin M. A., Spector D. L. (2013). Chromatin organization and transcriptional regulation. Curr. Opin. Genet. Dev. 23 89–95. 10.1016/j.gde.2012.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug C. B., Vaquerizas J. M. (2018). The Birth of the 3D Genome during Early Embryonic Development. Trends Genet. 34 903–914. 10.1016/j.tig.2018.09.002 [DOI] [PubMed] [Google Scholar]
- Iampietro C., Gummalla M., Mutero A., Maeda R. K. (2010). Initiator Elements Function to Determine the Activity State of BX-C Enhancers. PLoS Genet. 6:e1001260. 10.1371/journal.pgen.1001260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikuta T. (2011). Evolution of invertebrate deuterostomes and Hox/ParaHox genes. Genom. Proteom. Bioinf. 9 77–96. 10.1016/S1672-0229(11)60011-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikuta T., Yoshida N., Satoh N., Saiga H. (2004). Ciona intestinalis Hox gene cluster: its dispersed structure and residual colinear expression in development. Proc. Natl. Acad. Sci. U. S. A. 101 15118–15123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen R., Eriksson B., Tait N. N., Budd G. E. (2014). Onychophoran Hox genes and the evolution of arthropod Hox gene expression. Front. Zool. 11:22. 10.1186/1742-9994-11-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerevall P. L., Brommesson S., Strand C., Gruvberger-Saal S., Malmström P., Nordenskjöld B., et al. (2008). Exploring the two-gene ratio in breast cancer-independent roles for HOXB13 and IL17BR in prediction of clinical outcome. Breast Cancer Res. Treat. 107 225–234. 10.1007/s10549-007-9541-8 [DOI] [PubMed] [Google Scholar]
- Jung C., Kim R. S., Zhang H. J., Lee S. J., Jeng M. H. (2004). HOXB13 induces growth suppression of prostate cancer cells as a repressor of hormone-activated androgen receptor signaling. Cancer Res. 64 9185–9192. 10.1158/0008-5472.CAN-04-1330 [DOI] [PubMed] [Google Scholar]
- Kadota M., Nishimura O., Miura H., Tanaka K., Hiratani I., Kuraku S. (2020). Multifaceted Hi-C benchmarking: what makes a difference in chromosome-scale genome scaffolding?. Gigascience 9:giz158. 10.1093/gigascience/giz158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karch F., Galloni M., Sipos L., Gausz J., Gyurkovics H., Schedl P. (1994). Mcp and Fab-7: molecular analysis of putative boundaries of cis-regulatory domains in the bithorax complex of Drosophila melanogaster. Nucleic Acids Res. 22 3138–3146. 10.1093/nar/22.15.3138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman T. C., Seeger M. A., Olsen G. (1990). Molecular and Genetic Organization of the Antennapedia Gene Complex of Drosophila melanogaster. Adv. Genet. 27 309–362. [DOI] [PubMed] [Google Scholar]
- Kmita M., Kondo T., Duboule D. (2000). Targeted inversion of a polar silencer within the HoxD complex re-allocates domains of enhancer sharing. Nat. Genet. 26 451–454. 10.1038/82593 [DOI] [PubMed] [Google Scholar]
- Kmita M., Tarchini B., Zàkàny J., Logan M., Tabin C. J., Duboule D. (2005). Early developmental arrest of mammalian limbs lacking HoxA/HoxD gene function. Nature 435 1113–1116. 10.1038/nature03648 [DOI] [PubMed] [Google Scholar]
- Kornberg T. B., Tabata T. (1993). Segmentation of the Drosophila embryo. Curr. Opin. Genet. Dev. 3 585–593. 10.1016/0959-437X(93)90094-6 [DOI] [PubMed] [Google Scholar]
- Kourakis M. J., Martindale M. Q. (2001). Hox gene duplication and deployment in the annelid leech Helobdella. Evol. Dev. 3 145–153. 10.1046/j.1525-142X.2001.003003145.x [DOI] [PubMed] [Google Scholar]
- Kushawah G., Mishra R. K. (2017). Ultraconserved sequences associated with HoxD cluster have strong repression activity. Genome Biol. Evol. 9 2134–2139. 10.1093/gbe/evx148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrchanova O., Ivlieva T., Toshchakov S., Parshikov A., Maksimenko O., Georgiev P. (2011). Selective interactions of boundaries with upstream region of Abd-B promoter in Drosophila bithorax complex and role of dCTCF in this process. Nucleic Acids Res. 39 3042–3052. 10.1093/nar/gkq1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrchanova O., Mogila V., Wolle D., Deshpande G. (2016). Functional Dissection of the Blocking and Bypass Activities of the Fab-8 Boundary in the Drosophila Bithorax Complex. PLoS Genet. 12:e1006188. 10.1371/journal.pgen.1006188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrchanova O., Mogila V., Wolle D., Paolo J., White R., Georgiev P., et al. (2015). The boundary paradox in the Bithorax complex. Mech. Dev. 138 122–132. 10.1016/j.mod.2015.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrchanova O., Sabirov M., Mogila V., Kurbidaeva A., Postika N., Maksimenko O., et al. (2019). Complete reconstitution of bypass and blocking functions in a minimal artificial Fab-7 insulator from Drosophila bithorax complex. Proc. Natl. Acad. Sci. U. S. A. 116 13462–13467. 10.1073/pnas.1907190116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzuolo C., Roure V., Dekker J., Bantignies F., Orlando V. (2007). Polycomb response elements mediate the formation of chromosome higher-order structures in the bithorax complex. Nat. Cell Biol. 9 1167–1174. 10.1038/ncb1637 [DOI] [PubMed] [Google Scholar]
- Laughon A., Boulet A. M., Bermingham J. R., Laymon R. A., Scott M. P. (1986). Structure of transcripts from the homeotic Antennapedia gene of Drosophila melanogaster: two promoters control the major protein-coding region. Mol. Cell. Biol. 6 4676–4689. 10.1128/mcb.6.12.4676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin M. (2005). Left-right asymmetry in embryonic development: a comprehensive review. Mech. Dev. 122 3–25. 10.1016/j.mod.2004.08.006 [DOI] [PubMed] [Google Scholar]
- Lewin H. A., Robinson G. E., Kress W. J., Baker W. J., Coddington J., Crandall K. A., et al. (2018). Earth BioGenome Project: sequencing life for the future of life. Proc. Natl. Acad. Sci. U. S. A. 115 4325–4333. 10.1073/pnas.1720115115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis E. B. (1978). A gene complex controlling segmentation in Drosophila. Nature 276 565–570. [DOI] [PubMed] [Google Scholar]
- Lewis E. B. (1994). Homeosis: the first 100 years. Trends Genet. 10 341–343. [DOI] [PubMed] [Google Scholar]
- Lewis E. B. (1998). The bithorax complex: the first fifty years. Int. J. Biomed. Sci. 42 403–415. 10.1007/978-1-4020-6345-9_32 [DOI] [PubMed] [Google Scholar]
- Lewis E. B. (2007). “A gene complex controlling segmentation in Drosophila,” in Genes, Development, and Cancer, ed. Lipshitz H. D. (Dordrecht: Springer; ), 229–242. 10.1007/978-1-4020-6345-9_10 [DOI] [Google Scholar]
- Lewis M. T. (2000). Homeobox genes in mammary gland development and neoplasia. Breast Cancer Res. 2 158–169. 10.1186/bcr49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Li Y., Pawlik K. M., Napierala J. S., Napierala M. (2020). A CRISPR-Cas9, Crelox, and Flp-FRT Cascade Strategy for the Precise and Efficient Integration of Exogenous DNA into Cellular Genomes. CRISPR J. 3 470–486. 10.1089/crispr.2020.0042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Ma Z., Liu J. K., Roy S., Patel S. K., Lane D. C., et al. (2015). An Organizational Hub of Developmentally Regulated Chromatin Loops in the Drosophila Antennapedia Complex. Mol. Cell. Biol. 35 4018–4029. 10.1128/MCB.00663-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q., Wu D., Zhou J. (2003). The promoter targeting sequence facilitates and restricts a distant enhancer to a single promoter in the Drosophila embryo. Development 130 519–526. 10.1242/dev.00227 [DOI] [PubMed] [Google Scholar]
- Little J. W., Byrd C. A., Brower D. L. (1990). Effect of abx, bx and pbx mutations on expression of homeotic genes in Drosophila larvae. Genetics 124 899–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liubicich D. M., Serano J. M., Pavlopoulos A., Kontarakis Z., Protas M. E., Kwan E., et al. (2009). Knockdown of Parhyale Ultrabithorax recapitulates evolutionary changes in crustacean appendage morphology. Proc. Natl. Acad. Sci. U. S. A. 106 13892–13896. 10.1073/pnas.0903105106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long H. K., Prescott S. L., Wysocka J. (2016). Ever-Changing Landscapes: transcriptional Enhancers in Development and Evolution. Cell 167 1170–1187. 10.1016/j.cell.2016.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovegrove B., Simões S., Rivas M. L., Sotillos S., Johnson K., Knust E., et al. (2006). Coordinated Control of Cell Adhesion, Polarity, and Cytoskeleton Underlies Hox-Induced Organogenesis in Drosophila. Curr. Biol. 16 2206–2216. 10.1016/j.cub.2006.09.029 [DOI] [PubMed] [Google Scholar]
- Maeda R. K. (2006). The ABC of the BX-C: the bithorax complex explained. Development 133 1413–1422. 10.1242/dev.02323 [DOI] [PubMed] [Google Scholar]
- Maeda R. K. (2009). The Bithorax Complex of Drosophila: an Exceptional Hox Cluster. Curr. Top. Dev. Biol. 88 1–33. 10.1016/S0070-2153(09)88001-0 [DOI] [PubMed] [Google Scholar]
- Maeda R. K., Karch F. (2007). Making connections: boundaries and insulators in Drosophila. Curr. Opin. Genet. Dev. 17 394–399. 10.1016/j.gde.2007.08.002 [DOI] [PubMed] [Google Scholar]
- Maeda R. K., Karch F. (2015). The open for business model of the bithorax complex in Drosophila. Chromosoma 124 293–307. 10.1007/s00412-015-0522-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallo M., Alonso C. R. (2013). The regulation of Hox gene expression during animal development. Development 140 3951–3963. 10.1242/dev.068346 [DOI] [PubMed] [Google Scholar]
- Mallo M., Wellik D. M., Deschamps J. (2010). Hox genes and regional patterning of the vertebrate body plan. Dev. Biol. 344 7–15. 10.1016/j.ydbio.2010.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manohar C. F., Salwen H. R., Furtado M. R., Cohn S. L. (1996). Up-Regulation of HOXC6, HOXD1 and HOXD8 Homeobox Gene Expression in Human Neuroblastoma Cells following Chemical Induction of Differentiation. Tumor Biol. 17 34–47. [DOI] [PubMed] [Google Scholar]
- Marchetti M., Fanti L., Berloco M., Pimpinelli S. (2003). Differential expression of the Drosophila BX-C in polytene chromosomes in cells of larval fat bodies: a cytological approach to identifying in vivo targets of the homeotic Ubx, Abd-A and Abd-B proteins. Development 130 3683–3689. 10.1242/dev.00587 [DOI] [PubMed] [Google Scholar]
- Martin A., Serano J. M., Jarvis E., Bruce H. S., Wang J., Ray S., et al. (2016). CRISPR/Cas9 Mutagenesis Reveals Versatile Roles of Hox Genes in Crustacean Limb Specification and Evolution. Curr. Biol. 26 14–26. 10.1016/j.cub.2015.11.021 [DOI] [PubMed] [Google Scholar]
- Martin C. H., Mayeda C. A., Davis C. A., Ericsson C. L., Knafels J. D., Mathog D. R., et al. (1995). Complete sequence of the bithorax complex of Drosophila. Proc. Natl. Acad. Sci. U. S. A. 92 8398–8402. 10.1073/pnas.92.18.8398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Arias A., Lawrence P. A. (1985). Parasegments and compartments in the Drosophila embryo. Nature 313 639–642. 10.1038/313639a0 [DOI] [PubMed] [Google Scholar]
- Martínez-Laborda A., Serrano-Cartagena J., Micol J. L. (1996). A genetic analysis of bx bxd cis double mutants in the Drosophila Ultrabithorax gene. Mol. Gen. Genet. 250 540–546. 10.1007/s004380050107 [DOI] [PubMed] [Google Scholar]
- Mateo L. J., Murphy S. E., Hafner A., Cinquini I. S., Walker C. A., Boettiger A. N. (2019). Visualizing DNA folding and RNA in embryos at single-cell resolution. Nature 568 49–54. 10.1038/s41586-019-1035-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis W., Garber R. L., Wirz J., Kuroiwa A., Gehring W. J. (1984a). A homologous protein-coding sequence in drosophila homeotic genes and its conservation in other metazoans. Cell 37 403–408. 10.1016/0092-8674(84)90370-2 [DOI] [PubMed] [Google Scholar]
- McGinnis W., Levine M. S., Hafen E., Kuroiwa A., Gehring W. J. (1984b). A conserved DNA sequence in homoeotic genes of the Drosophila Antennapedia and bithorax complexes. Nature 308 428–433. 10.1038/308428a0 [DOI] [PubMed] [Google Scholar]
- Medina-Martínez O., Bradley A., Ramírez-Solis R. (2000). A large targeted deletion of Hoxb1-Hoxb9 produces a series of single- segment anterior homeotic transformations. Dev. Biol. 222 71–83. 10.1006/dbio.2000.9683 [DOI] [PubMed] [Google Scholar]
- Miao J., Wang Z., Provencher H., Muir B., Dahiya S., Carney E., et al. (2007). HOXB13 promotes ovarian cancer progression. Proc. Natl. Acad. Sci. U. S. A. 104 17093–17098. 10.1073/pnas.0707938104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaly J., Barges S., Sipos L., Maeda R., Cléard F., Hogga I., et al. (2006). Dissecting the regulatory landscape of the Abd-B gene of the bithorax complex. Development 133 2983–2993. 10.1242/dev.02451 [DOI] [PubMed] [Google Scholar]
- Mihaly J., Hogga I., Barges S., Galloni M., Mishra R. K., Hagstrom K., et al. (1998). Chromatin domain boundaries in the Bithorax complex. Cell. Mol. Life Sci. 54 60–70. 10.1007/s000180050125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar S. E. (2018). Hox in the Niche Controls Hairy-geneity. Cell Stem Cell 23 457–458. 10.1016/j.stem.2018.09.012 [DOI] [PubMed] [Google Scholar]
- Mishra R. K., Karch F. (1999). Boundaries that demarcate structural and functional domains of chromatin. J. Biosci. 24 377–399. 10.1007/bf02941252 [DOI] [Google Scholar]
- Mishra R. K., Mihaly J., Barges S., Spierer A., Karch F., Hagstrom K., et al. (2001). The iab-7 polycomb response element maps to a nucleosome-free region of chromatin and requires both GAGA and pleiohomeotic for silencing activity. Mol. Cell. Biol. 21 1311–1318. 10.1128/MCB.21.4.1311-1318.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misof B., Jun Tong K., Duchêne S., Ho S. Y. W., Lo N. (2014). Phylogenomics resolves the timing and pattern of insect evolution. Science 346:763. 10.1126/science.aaa5460 [DOI] [PubMed] [Google Scholar]
- Mohit P., Makhijani K., Madhavi M. B., Bharathi V., Lal A., Sirdesai G., et al. (2006). Modulation of AP and DV signaling pathways by the homeotic gene Ultrabithorax during haltere development in Drosophila. Dev. Biol. 291 356–367. 10.1016/j.ydbio.2005.12.022 [DOI] [PubMed] [Google Scholar]
- Mooi R., David B. (2008). Radial Symmetry, the Anterior / Posterior Axis, and Echinoderm Hox Genes. Annu. Rev. Ecol. Evol. Syst. 39 43–62. 10.1146/annurev.ecolsys.39.110707.173521 [DOI] [Google Scholar]
- Muller J., Bienz M. (1992). Sharp anterior boundary of homeotic gene expression conferred by the fushi tarazu protein. EMBO J. 11 3653–3661. 10.1002/j.1460-2075.1992.tb05450.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M., Hagstrom K., Gyurkovics H., Pirrotta V., Schedl P. (1999). The Mcp Element From the Drosophila melanogaster Bithorax Complex Mediates Long-Distance Regulatory Interactions. Genetics 153 1333–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narlikar L., Ovcharenko I. (2009). Identifying regulatory elements in eukaryotic genomes. Brief. Funct. Genomics Proteomics 8 215–230. 10.1093/bfgp/elp014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negre B., Ranz J. M., Casals F., Cáceres M., Ruiz A. (2003). A New Split of the Hox Gene Complex in Drosophila: relocation and Evolution of the Gene labial. Mol. Biol. Evol. 20 2042–2054. 10.1093/molbev/msg238 [DOI] [PubMed] [Google Scholar]
- Négre N., Brown C. D., Ma L., Bristow C. A., Miller S. W., Wagner U., et al. (2011). A cis-regulatory map of the Drosophila genome. Nature 471 527–531. 10.1038/nature09990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nègre N., Brown C. D., Shah P. K., Kheradpour P., Morrison C. A., Henikoff J. G., et al. (2010). A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet. 6:e1000814. 10.1371/journal.pgen.1000814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nong W., Cao J., Li Y., Qu Z., Sun J., Swale T., et al. (2020). Jellyfish genomes reveal distinct homeobox gene clusters and conservation of small RNA processing. Nat. Commun. 11:3051. 10.1038/s41467-020-16801-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono R., Nosaka T., Hayashi Y. (2005). Roles of a trithorax group gene. MLL, in hematopoiesis. Int. J. Hematol. 81 288–293. 10.1532/IJH97.04196 [DOI] [PubMed] [Google Scholar]
- Papagiannouli F., Lohmann I. (2015). Stage-specific control of stem cell niche architecture in the Drosophila testis by the posterior Hox gene Abd-B. Comput. Struct. Biotechnol. J. 13 122–130. 10.1016/j.csbj.2015.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo J. C. F., Stefanelli-Silva G., Christy J. H., Costa T. M. (2020). Fiddler crabs and their above-ground sedimentary structures: a review. J. Ethol. 38 137–154. 10.1007/s10164-020-00647-1 [DOI] [Google Scholar]
- Pascual-Anaya J., Adachi N., Álvarez S., Kuratani S., D’Aniello S., Garcia-Fernàndez J. (2012). Broken colinearity of the amphioxus Hox cluster. Evodevo 3:28. 10.1186/2041-9139-3-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul R., Giraud G., Domsch K., Duffraisse M., Marmigère F., Khan S., et al. (2021). Hox dosage contributes to flight appendage morphology in Drosophila. Nat. Commun. 12:2892. 10.1038/s41467-021-23293-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlopoulos A., Kontarakis Z., Liubicich D. M., Serano J. M., Akam M., Patel N. H., et al. (2009). Probing the evolution of appendage specialization by Hox gene misexpression in an emerging model crustacean. Proc. Natl. Acad. Sci. U. S. A. 106 13897–13902. 10.1073/pnas.0902804106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peel A. D., Chipman A. D., Akam M. (2005). Arthropod segmentation: beyond the Drosophila paradigm. Nat. Rev. Genet. 6 905–916. 10.1038/nrg1724 [DOI] [PubMed] [Google Scholar]
- Peifer M., Bender W. (1986). The anterobithorax and bithorax mutations of the bithorax complex. EMBO J. 5 2293–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peifer M., Karch F. F., Bender W. (1987). The bithorax complex: control of segment identity. Genes Dev. 1 891–898. [DOI] [PubMed] [Google Scholar]
- Perrimon N., Pitsouli C., Shilo B. Z. (2012). Signaling mechanisms controlling cell fate and embryonic patterning. Cold Spring Harb. Perspect. Biol. 4:a005975. 10.1101/cshperspect.a005975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson K. J., Cameron R. A., Davidson E. H. (2000). Bilaterian Origins: significance of New Experimental Observations. Dev. Biol. 17 1–17. 10.1006/dbio.1999.9475 [DOI] [PubMed] [Google Scholar]
- Ponrathnam T., Saini R., Banu S., Mishra R. K. (2021). Drosophila Hox genes induce melanized pseudo-tumors when misexpressed in hemocytes. Sci. Rep. 11:1838. 10.1038/s41598-021-81472-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postika N., Metzler M., Affolter M., Müller M., Schedl P., Georgiev P., et al. (2018). Boundaries mediate long-distance interactions between enhancers and promoters in the Drosophila Bithorax complex. PLoS Genet. 14:e1007702. 10.1371/journal.pgen.1007702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postika N., Schedl P., Georgiev P., Kyrchanova O. (2021). Mapping of functional elements of the Fab-6 boundary involved in the regulation of the Abd-B hox gene in Drosophila melanogaster. Sci. Rep. 11:4156. 10.1038/s41598-021-83734-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradel J., White R. A. H. (1998). From selectors to realizators. Int. J. Dev. Biol. 421 417–421. [PubMed] [Google Scholar]
- Raman V., Martenser S. A., Reisman D., Evron E., Odenwald W. F., Jaffee E., et al. (2000). Compromised HOXA5 function can limit p53 expression in human breast tumours. Nature 405 974–978. 10.1038/35016125 [DOI] [PubMed] [Google Scholar]
- Rauch T., Wang Z., Zhang X., Zhong X., Wu X., Lau S. K., et al. (2007). Homeobox gene methylation in lung cancer studied by genome-wide analysis with a microarray-based methylated CpG island recovery assay. Proc. Natl. Acad. Sci. U. S. A. 104 5527–5532. 10.1073/pnas.0701059104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinitz J., Levine M. (1990). Control of the initiation of homeotic gene expression by the gap genes giant and tailless in Drosophila. Dev. Biol. 140 57–72. 10.1016/0012-1606(90)90053-L [DOI] [PubMed] [Google Scholar]
- Rentzsch F., Holstein T. W. (2018). Making head or tail of cnidarian hox gene function. Nat. Commun. 9 9–11. 10.1038/s41467-018-04585-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers B. T. (2020). Drosophila Embryo: Homeotic Genes in Specification of the Anterior–Posterior Axis. Hoboken: Wiley Online Library. [Google Scholar]
- Rux D. R., Wellik D. M. (2017). Hox genes in the adult skeleton: novel functions beyond embryonic development. Dev. Dyn. 246 310–317. 10.1002/dvdy.24482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabarinadh C., Subramanian S., Tripathi A., Mishra R. K. (2004). Extreme conservation of noncoding DNA near HoxD complex of vertebrates. BMC Genomics 5:75. 10.1186/1471-2164-5-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samadi L., Steiner G. (2010). Expression of Hox genes during the larval development of the snail, Gibbula varia (L.)-further evidence of non-colinearity in molluscs. Dev. Genes Evol. 220 161–172. 10.1007/s00427-010-0338-0 [DOI] [PubMed] [Google Scholar]
- Sánchez-Herrero E. (1991). Control of the expression of the bithorax complex genes abdominal-A and abdominal-B by cis-regulatory regions in Drosophila embryos. Development 111 437–449. [DOI] [PubMed] [Google Scholar]
- Sánchez-Herrero E. (2013). Hox Targets and Cellular Functions. Scientifica 2013:738257. 10.1155/2013/738257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiemann S. M., Martín-durán J. M., Aina B., Vellutini B. C., Passamaneck Y. J. (2017). Clustered brachiopod Hox genes are not expressed collinearly and are associated with lophotrochozoan novelties. Proc. Natl. Acad. Sci. U. S. A. 7 1913–1922. 10.1073/pnas.1614501114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneuwly S., Kuroiwa A., Gehring W. J. (1987). Molecular analysis of the dominant homeotic Antennapedia phenotype. EMBO J. 6 201–206. 10.1002/j.1460-2075.1987.tb04739.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweinsberg S., Hagstrom K., Gohl D., Schedl P., Kumar R. P., Mishra R., et al. (2004). The enhancer-blocking activity of the Fab-7 boundary from the Drosophila bithorax complex requires GAGA-factor-binding sites. Genetics 168 1371–1384. 10.1534/genetics.104.029561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott M. P., Weiner A. J. (1984). Structural relationships among genes that control development: sequence homology between the antennapedia, ultrabithorax, and fushi tarazu loci of Drosophila. Proc. Natl. Acad. Sci. U. S. A. 81 4115–4119. 10.1073/pnas.81.13.4115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serano J. M., Martin A., Liubicich D. M., Jarvis E., Bruce H. S., La K., et al. (2016). Comprehensive analysis of Hox gene expression in the amphipod crustacean Parhyale hawaiensis. Dev. Biol. 409 297–309. 10.1016/j.ydbio.2015.10.029 [DOI] [PubMed] [Google Scholar]
- Shah N., Sukumar S. (2010). The Hox genes and their roles in oncogenesis. Nat. Rev. Cancer 10 361–371. 10.1038/nrc2826 [DOI] [PubMed] [Google Scholar]
- Sharma P. P., Schwager E. E., Extavour C. G., Giribet G. (2012). Hox gene expression in the harvestman Phalangium opilio reveals divergent patterning of the chelicerate opisthosoma. Evol. Dev. 14 450–463. 10.1111/j.1525-142X.2012.00565.x [DOI] [PubMed] [Google Scholar]
- Sharma P. P., Schwager E. E., Extavour C. G., Wheeler W. C. (2014). Hox gene duplications correlate with posterior heteronomy in scorpions. Proc. Biol. Sci. 281:20140661. 10.1098/rspb.2014.0661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shippy T. D., Ronshaugen M., Cande J., He J., Beeman R. W., Levine M., et al. (2008). Analysis of the Tribolium homeotic complex: insights into mechanisms constraining insect Hox clusters. Dev. Genes Evol. 218, 127–139. 10.1007/s00427-008-0213-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultz J. W. (1989). Morphology of locomotor appendages in Arachnida: evolutionary trends and phylogenetic implications. Zool. J. Linn. Soc. 97 1–55. 10.1111/j.1096-3642.1989.tb00552.x [DOI] [Google Scholar]
- Simon J., Chiang A., Bender W., Shimell M. J., Connor M. O. (1993). Elements of the Drosophila Bithorax Complex That Mediate Repression by Polycomb Group Products. Dev. Biol. 158 131–144. 10.1006/dbio.1993.1174 [DOI] [PubMed] [Google Scholar]
- Simon J., Peifer M., Bender W., O’Connor M. (1990). Regulatory elements of the bithorax complex that control expression along the anterior-posterior axis. EMBO J. 9 3945–3956. 10.1002/j.1460-2075.1990.tb07615.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N. P., de Kumar B., Paulson A., Parrish M. E., Zhang Y., Florens L., et al. (2020). A six-amino-acid motif is a major determinant in functional evolution of HOX1 proteins. Genes Dev. 34 1680–1696. 10.1101/gad.342329.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N. P., Mishra R. K. (2014). Role of abd-A and Abd-B in Development of Abdominal Epithelia Breaks Posterior Prevalence Rule. PLoS Genet. 10:e1004717. 10.1371/journal.pgen.1004717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N. P., Mishra R. K. (2015). Specific combinations of boundary element and Polycomb response element are required for the regulation of the Hox genes in Drosophila melanogaster. Mech. Dev. 138 141–150. 10.1016/j.mod.2015.07.016 [DOI] [PubMed] [Google Scholar]
- Sipos L., Mihály J., Karch F., Schedl P., Gausz J., Gyurkovics H. (1998). Transvection in the Drosophila Abd-B domain: extensive upstream sequences are involved in anchoring distant cis-regulatory regions to the promoter. Genetics 149 1031–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith F. W., Boothby T. C., Giovannini I., Rebecchi L., Jockusch E. L., Goldstein B. (2016). The Compact Body Plan of Tardigrades Evolved by the Loss of a Large Body Region. Curr. Biol. 26 224–229. 10.1016/j.cub.2015.11.059 [DOI] [PubMed] [Google Scholar]
- Snel B., van Noort V., Huynen M. A. (2004). Gene co-regulation is highly conserved in the evolution of eukaryotes and prokaryotes. Nucleic Acids Res. 32 4725–4731. 10.1093/nar/gkh815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son S. H., Son Y. E., Cho H. J., Chen W., Lee M. K., Kim L. H., et al. (2020). Homeobox proteins are essential for fungal differentiation and secondary metabolism in Aspergillus nidulans. Sci. Rep. 10:6094. 10.1038/s41598-020-63300-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soshnikova N. (2014). Hox genes regulation in vertebrates. Dev. Dyn. 243 49–58. 10.1002/dvdy.24014 [DOI] [PubMed] [Google Scholar]
- Soshnikova N., Duboule D. (2009). Epigenetic temporal control of mouse hox genes in vivo. Science 324 1321–1323. 10.1126/science.1171468 [DOI] [PubMed] [Google Scholar]
- Spagnuolo A., Ristoratore F., Di Gregorio A., Aniello F., Branno M., Di Lauro R. (2003). Unusual number and genomic organization of Hox genes in the tunicate Ciona intestinalis. Gene 309 71–79. 10.1016/S0378-1119(03)00488-8 [DOI] [PubMed] [Google Scholar]
- Spéder P., Ádám G., Noselli S. (2006). Type ID unconventional myosin controls left-right asymmetry in Drosophila. Nature 440 803–807. 10.1038/nature04623 [DOI] [PubMed] [Google Scholar]
- Spitz F., Gonzalez F., Peichel C., Vogt T. F., Duboule D., Zákány J. (2001). Large scale transgenic and cluster deletion analysis of the HoxD complex separate an ancestral regulatory module from evolutionary innovations. Genes Dev. 15 2209–2214. 10.1101/gad.205701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan A., Mishra R. K. (2012). Chromatin domain boundary element search tool for Drosophila. Nucleic Acids Res. 40 4385–4395. 10.1093/nar/gks045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan A., Mishra R. K. (2020). Genomic organization of Polycomb Response Elements and its functional implication in Drosophila and other insects. J. Biosci. 45:12. 10.1007/s12038-019-9975-2 [DOI] [PubMed] [Google Scholar]
- Starr M. O., Ho M. C. W., Gunther E. J. M., Tu Y. K., Shur A. S., Goetz S. E., et al. (2011). Molecular dissection of cis-regulatory modules at the Drosophila bithorax complex reveals critical transcription factor signature motifs. Dev. Biol. 359 176–189. 10.1016/j.ydbio.2011.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suemori H., Noguchi S. (2000). Hox C cluster genes are dispensable for overall body plan of mouse embryonic development. Dev. Biol. 220 333–342. 10.1006/dbio.2000.9651 [DOI] [PubMed] [Google Scholar]
- Sun D. A., Patel N. H. (2019). The amphipod crustacean Parhyale hawaiensis: an emerging comparative model of arthropod development, evolution, and regeneration. Wiley Interdiscip. Rev. Dev. Biol. 8:e355. 10.1002/wdev.355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M., Yagi N. (1994). DNA recognition code of transcription factors in the helix-turn-helix, probe helix, hormone receptor, and zinc finger families. Proc. Natl. Acad. Sci. U. S. A. 91 12357–12361. 10.1073/pnas.91.26.12357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi O., Hamada J., Abe M., Hata S., Asano T., Takahashi Y., et al. (2007). Dysregulated expression of HOX and ParaHOX genes in human esophageal squamous cell carcinoma. Oncol. Rep. 17 753–760. [PubMed] [Google Scholar]
- Telford M. J. (2000). Evidence for the derivation of the Drosophila fushi tarazu gene from a Hox gene orthologous to lophotrochozoan Lox5. Curr. Biol. 10, 349–352. 10.1016/S0960-9822(00)00387-0 [DOI] [PubMed] [Google Scholar]
- Thickett C., Morgan R. (2002). Hoxc-8 expression shows left-right asymmetry in the posterior lateral plate mesoderm. Gene Expr. Patterns 2 5–6. 10.1016/S0925-4773(02)00353-2 [DOI] [PubMed] [Google Scholar]
- Tschopp P., Tarchini B., Spitz F., Zakany J., Duboule D. (2009). Uncoupling time and space in the collinear regulation of Hox genes. PLoS Genet. 5:e1000398. 10.1371/journal.pgen.1000398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner F. R., Kaufman T. C. (1987). A Developmental Genetic Analysis of the Gene Regulator of postbithorax in Drosophila melanogaster. Dev. Biol. 432 418–432. [DOI] [PubMed] [Google Scholar]
- Vazquez J., Muller M., Pirrotta V., Sedat J. W. (2006). The Mcp Element Mediates Stable Long-Range Chromosome–Chromosome Interactions in Drosophila. Mol. Biol. Cell 17 2158–2165. 10.1091/mbc.E06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K. C., Helms J. A., Chang H. Y. (2009). Regeneration, repair and remembering identity: the three Rs of Hox gene expression. Trends Cell Biol. 19 268–275. 10.1016/j.tcb.2009.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Yang Q., Wang Z. (2014). The evolution of nanopore sequencing. Front. Genet. 5:449. 10.3389/fgene.2014.00449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanninger A., Wollesen T. (2019). The evolution of molluscs. Biol. Rev. 94 102–115. 10.1111/brv.12439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R. W. (1998). Plant homeobox genes: many functions stem from a common motif. BioEssays 20 280–282. [DOI] [PubMed] [Google Scholar]
- Yamada K., Maeno A., Araki S., Kikuchi M., Suzuki M., Ishizaka M., et al. (2021). An atlas of seven zebrafish hox cluster mutants provides insights into sub/neofunctionalization of vertebrate hox clusters. Development 148:dev198325. 10.1242/DEV.198325 [DOI] [PubMed] [Google Scholar]
- Yarger A. M., Fox J. L. (2016). Dipteran Halteres: perspectives on Function and Integration for a Unique Sensory Organ. Integr. Comp. Biol. 56 865–876. 10.1093/icb/icw086 [DOI] [PubMed] [Google Scholar]
- Zakany J., Kmita M., Duboule D. (2004). A Dual Role for Hox Genes in Limb Anterior-Posterior. Science 51 1669–1672. [DOI] [PubMed] [Google Scholar]
- Zhai Y., Kuick R., Nan B., Ota I., Weiss S. J., Trimble C. L., et al. (2007). Gene expression analysis of preinvasive and invasive cervical squamous cell carcinomas identifies HOXC10 as a key mediator of invasion. Cancer Res. 67 10163–10172. 10.1158/0008-5472.CAN-07-2056 [DOI] [PubMed] [Google Scholar]
- Zhang X., Hamada J. I., Nishimoto A., Takahashi Y., Murai T., Tada M., et al. (2007). HOXC6 and HOXC11 increase transcription of S100β gene in BrdU-induced in vitro differentiation of GOTO neuroblastoma cells into Schwannian cells. J. Cell. Mol. Med. 11 299–306. 10.1111/j.1582-4934.2007.00020.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Barolo S., Szymanski P., Levine M. (1996). The Fab-7 element of the bithorax complex attenuates enhancer-promoter interactions in the Drosophila embryo. Genes Dev. 10 3195–3201. 10.1101/gad.10.24.3195 [DOI] [PubMed] [Google Scholar]