Abstract
Objectives:
The Blueprint (BP) Programmed Death Ligand 1 (PD-L1) Immunohistochemistry Comparability Project is a pivotal academic/professional society and industrial collaboration to assess the feasibility of harmonizing the clinical use of five independently developed commercial PD-L1 immunohistochemistry assays. The goal of BP phase 2 (BP2) was to validate the results obtained in BP phase 1 by using real-world clinical lung cancer samples.
Methods:
BP2 were conducted using 81 lung cancer specimens of various histological and sample types, stained with all five trial-validated PD-L1 assays (22C3, 28–8, SP142, SP263, and 73–10); the slides were evaluated by an international panel of pathologists. BP2 also assessed the reliability of PD-L1 scoring by using digital images, and samples prepared for cytological examination. PD-L1 expression was assessed for percentage (tumor proportional score) of tumor cell (TC) and immune cell areas showing PD-L1 staining, with TCs scored continuously or categorically with the cutoffs used in checkpoint inhibitor trials.
Results:
The BP2 results showed highly comparable staining by the 22C3, 28–8 and SP263 assays; less sensitivity with the SP142 assay; and higher sensitivity with the 73–10 assay to detect PD-L1 expression on TCs. Glass slide and digital image scorings were highly concordant (Pearson correlation >0.96). There was very strong reliability among pathologists in TC PD-L1 scoring with all assays (overall intraclass correlation coefficient [ICC] = 0.86–0.93), poor reliability in IC PD-L1 scoring (overall ICC = 0.18–0.19), and good agreement in assessing PD-L1 status on cytological cell block materials (ICC = 0.78–0.85).
Conclusion:
BP2 consolidates the analytical evidence for interchangeability of the 22C3, 28–8, and SP263 assays and lower sensitivity of the SP142 assay for determining tumor proportion score on TCs and demonstrates greater sensitivity of the 73–10 assay compared with that of the other assays.
Keywords: Immunooncology, Checkpoint inhibitors, Companion diagnostics, Complementary diagnostics, Cytology, Pathology
Introduction
Immune checkpoint inhibitor therapies targeting the programmed death 1/programmed death ligand 1 (PD-L1) pathway have become part of the standard of care in oncology.1 At least five inhibitors (nivolumab, pembrolizumab, atezolizumab, durvalumab, and avelumab) have been approved by drug regulatory bodies in one or more countries for the treatment of several tumor types and for various indications. For patients with advanced NSCLC without driver mutations (e.g., EGFR, ALK receptor tyrosine kinase gene [ALK], ROS1, and BRAF) that are treatable by approved targeted therapies, nivolumab, pembrolizumab, and atezolizumab are all available as second-line treatment with (for pembrolizumab) or without (for nivolumab and atezolizumab) biomarker selection. Pembrolizumab is available for rst-line monotherapy but only in patients with high PD-L1 expression,2,3 and in some countries, for use in combination with chemotherapy without any biomarker selection. Importantly, almost all clinical trials involving these inhibitors have demonstrated consistent correlation between their response rates and outcomes and the tumor cell (TC) PD-L1 expression levels, as measured by PD-L1 immunohistochemistry (IHC). Therefore, despite the fact that only pembrolizumab requires a PD-L1 IHC assay as a companion diagnostic to determine patient eligibility for treatment as approved by the U.S. Food and Drug Administration and the European Medicines Agency, PD-L1 IHC has also been established as a complementary diagnostic for nivolumab and atezolizumab to determine NSCLC patient eligibility, respectively.4,5
As each PD-L1 IHC assay was independently developed for specific anti–programmed death 1/PD-L1 therapy using a different PD-L1 diagnostic assays (primary antibody clone plus immunostaining platform/protocol), each assay potentially demonstrates distinct staining properties, which could prohibit the interchangeability of their clinical use. This would pose a significant challenge for pathology laboratories to offer PD-L1 testing, both from laboratory resources and budgetary points of view. Several groups have conducted studies to assess the comparability of the various PD-L1 IHC assays and their potential interchangeability in clinical adoption,6–11 with all the studies demonstrating similar results. The results from the Blueprint (BP) phase 1 (BP1) study demonstrated that three PD-L1 assays (22C3, 28–8, and SP263) showed comparable analytical performance for assessment of PD-L1 expression on TCs, whereas the SP-142 PD-L1 assay appeared to stain fewer TCs compared with the other assays.7 In contrast, all the assays stained tumor-infiltrating immune cells (ICs), but with poor concordance between assays. The BP-1 study had several limitations: (1) samples were obtained from a commercial source and did not necessarily reflect the real-world samples tested clinically, and (2) only three pathologists were involved in the scoring. Since the BP1 study, a fifth PD-L1 assay, which uses the 73–10 clone, has been developed as a potential assay for avelumab.
The goals of BP phase 2 (BP2) were to have a large international panel of clinically active pathologists (1) validate the assay comparability results obtained in BP1 by using real-world clinical lung cancer samples and all five trial-validated PD-L1 assays (22C3, 28–8, SP142, SP263, and 73–10), (2) assess the feasibility of PD-L1 scoring using digital images accessed by a web-based system, and (3) assess the reliability to score PD-L1 expression by using samples prepared for cytologic examination.
Materials and Methods
Materials
Using the respective institutional research ethics board approval, 18 pathologists contributed eight unstained serial sections prepared from paraffin blocks of 81 lung cancer cases that they had collected through their routine clinical practice. The final cases included 39 adenocarcinomas, 26 squamous cell carcinomas, six poorly differentiated non–small cell carcinomas, and 10 small cell carcinomas (Supplementary Table 1). The cases included 21 resections, 20 core needle or bronchial biopsy samples, 18 tumor-positive lymph node excision biopsy or resection samples, and 22 cytological cell blocks.
The PD-L1 IHC 22C3 pharmDx and PD-L1 28–8 pharmDx assays were purchased from Dako (Heverlee, Belgium). The Ventana PD-L1 (SP142) assay and Ventana PD-L1 (SP263) assay were purchased from Ventana (Tucson, AZ). The 73–10 antibody was provided by EMD Serono/Merck KGaA/Pfizer through Dako/Agilent together with its staining protocol (see below).
PD-L1 IHC Staining
Each slide set of 81 cases was stained in a Clinical Laboratory Improvement Amendments–approved IHC laboratory HistoGeneX (Antwerp, Belgium) with use of the U.S. Food and Drug Administration–approved 22C3, 28–8, SP142, and SP263 assays and their respective protocols, which are detailed in the product inserts and autostainers (Dako Autostainer Link48 for the 22C3, 28–8 and 73–10 assays and Ventana BenchMark Ultra for the SP142 and SP263 assays). The PD-L1 73–10 assay was the protocol developed by Dako/Agilent (Santa Clara, CA) for the clinical trials of avelumab and transferred to the HistoGeneX. All immunostained slides and matching hematoxylin and eosin–stained sections were scanned with a Pannoramic 250 Flash III digital scanner (3DHISTECH, Budapest, Hungary) at ×20 magnification, and the scanned images were uploaded and scored on the International Association for the Study of Lung Cancer (IASLC) server in Denver, Colorado. Digital scoring was performed by accessing these images with use of the Pathomation Digital Pathology System (HistoGeneX).
Scoring of PD-L1 Assays
The slides were scored by 24 experienced pulmonary pathologists (IASLC Pathology Committee members) from 15 countries across five continents. Because only some rather than all participants had received company-sponsored assay-specific training, all participants were required to undergo 1.5 days of prestudy group training by two experts from HistoGeneX for the scoring of PD-L1 IHC on TCs and ICs as part of this project. As PD-L1 scoring on TCs is identical for all assays, training on TC scoring was not assay specific. Greater effort was devoted to training for IC scoring using SP-142–stained cases and the SP142 IC scoring algorithm. PD-L1–stained TCs were scored in terms of the tumor proportion score (TPS), which represents the best estimated percentage (0%–100%) of TCs showing partial or complete membranous PD-L1 staining and also into one of seven categories (<1%, 1%–4%, 5%–9%, 10%–24%, 25%–49%, 50%–79%, and 80%–100%). These categories represent cutoffs that have been used in various immune checkpoint inhibitor trials or suggested by the sponsors (e.g., ≥80% for avelumab). All assays were also scored for IC PD-L1 staining on the basis of a pattern scoring method that was developed by HistoGeneX, adapted from the scoring approach described in the Ventana SP142 PD-L1 IHC assay brochure, and detailed in Supplementary Fig. 1. As only one set of glass slides was available for each assay, each IASLC pathologist and one of the trainers were randomly assigned to conduct the scoring of two assays with use of a microscope (glass slide reading) and the scoring of three assays with use of web-based digital images. The trainer’s scores were used as the standard reference score set.
Statistical Analyses
The intraclass correlation coefficient (ICC) was used to assess scoring reliability for continuous TPS scores, and the Fleiss k statistic (FKS) was used for categorical scores after dichotomization based on specified cutoffs. ICCs between 0.75 and 0.9 and those greater than 0.9 were considered to indicate good and excellent reliability, respectively.12 FKS scores of 0.9 or higher were considered near perfect, scores of 0.80 to 0.89 were considered strong, scores of 0.70 to 0.79 were considered moderate, and scores of 0.40 to 0.69 were considered weak.13 The reliability of PD-L1 scoring served as an assessment across all pathologists (excluding the trainer) to compare for each assay and separately for digital and glass image scores. The mean TPS of all the pathologists was used to assess the reliability of their scoring relative to the trainer’s score, and the Pearson correlation and a graphical approach were used to assess the agreement of digital versus glass slide scoring as described by Bland and Altman.14
Results
At the cutoff date for completion of scoring, 114 data sets were available for analyses. These include 50 data sets from glass slide scoring (two assays by 24 IASLC pathologists and one trainer) and 74 data sets from digital image scoring (three assays by 23 IASLC pathologists and five assays by the trainer).
Reliability of Pathologists to Score TC PD-L1 Expression
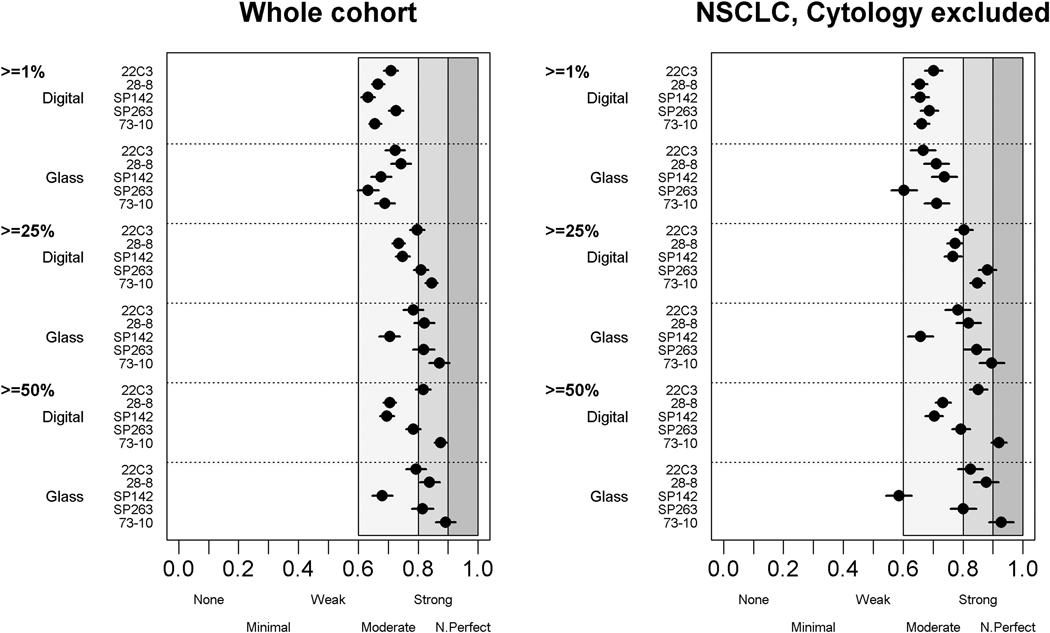
Overall, the ICC among all pathologists for glass slide reading ranged from 0.88 to 0.93, and the ICC for digital image reading ranged from 0.80 to 0.91, demonstrating very good to excellent reliability (Table 1). Comparable or slightly improved scores were obtained when only scores from NSCLC tissue were analyzed after exclusion of scores for small cell and cytological samples. High-level reliability (k > 0.7) was also demonstrated with use of k statistics at the various cutoffs, especially those of at least 5%, 10%, 25%, and 50%, for both glass slide and digital readings (Fig. 1 and Supplementary Fig. 2). Reliability was, however, slightly diminished at the 1% and 80% cutoffs, especially by digital image reading. We further demonstrated (by using the trainer’s scores as a reference) that the pathologists’ scores were strongly comparable (Supplementary Fig. 3).
Table 1.
Reliability (Intraclass Correlation Coefficient) of Scoring PD-L1 Expression on Tumor Cells among All Pathologists (Excluding the Trainer) for All Cases and NSCLC Biopsy Samples/Resected Cases
| Glass Slide Scoring |
Digital Scoring |
|||
|---|---|---|---|---|
| Assay | All Cases | NSCLC Tissue Only | All Cases | NSCLC Tissue Only |
|
| ||||
| 22C3 | 0.89 | 0.88 | 0.91 | 0.91 |
| 28–8 | 0.92 | 0.94 | 0.86 | 0.88 |
| SP-142 | 0.88 | 0.86 | 0.80 | 0.84 |
| SP-263 | 0.89 | 0.92 | 0.90 | 0.93 |
| 73–10 | 0.93 | 0.95 | 0.91 | 0.93 |
| All assays | 0.86 | 0.89 | 0.91 | 0.93 |
PD-L1, programmed death ligand 1.
Figure 1.
Reliability of scoring tumor cells programmed death ligand 1 expression with use of Fleiss k statistics at cutoffs 1%, 25%, and 50% for digital and glass slide readings, respectively, and for all cases (the whole cohort) or NSCLC only, with cytological specimens excluded.
Comparability of TC PD-L1 Scoring by Digital Image versus Glass Slide Readings
Because of logistic and time line challenges, it was not possible to obtain matching digital and glass slide scores from each pathologist. Therefore, to assess the reliability of PD-L1 scoring by digital images compared with that of the standard microscopic assessment on glass slides, we used two statistical methods, with the means of scores by both methods used for each assay. Both Pearson correlation and Bolt and Altman’s methods demonstrated very high correlation and agreement between the two methods of reading PD-L1 IHC results (Supplementary Fig. 4 and 5). These results justify the pooling of all data regardless of scoring method in subsequent analyses.
Comparability of PD-L1 Staining between Five Assays of TC Staining
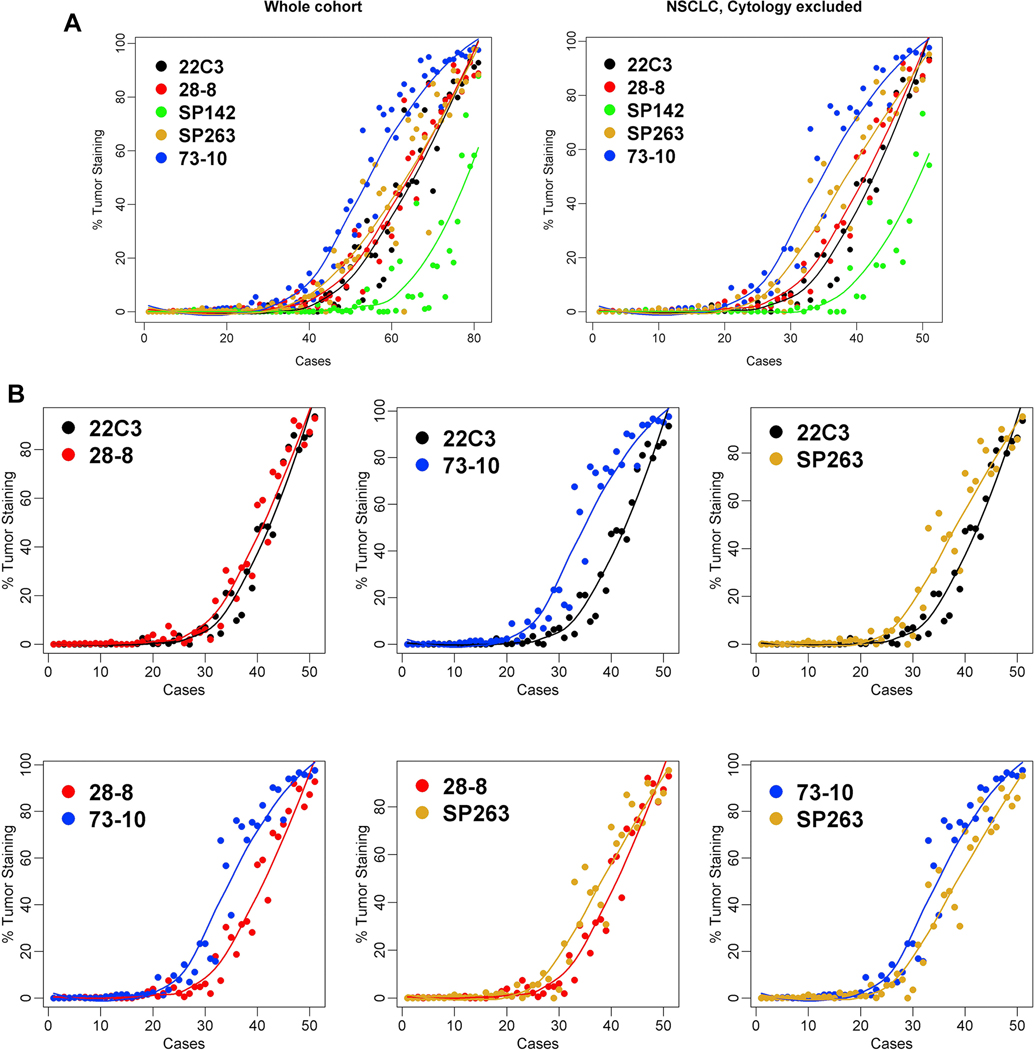
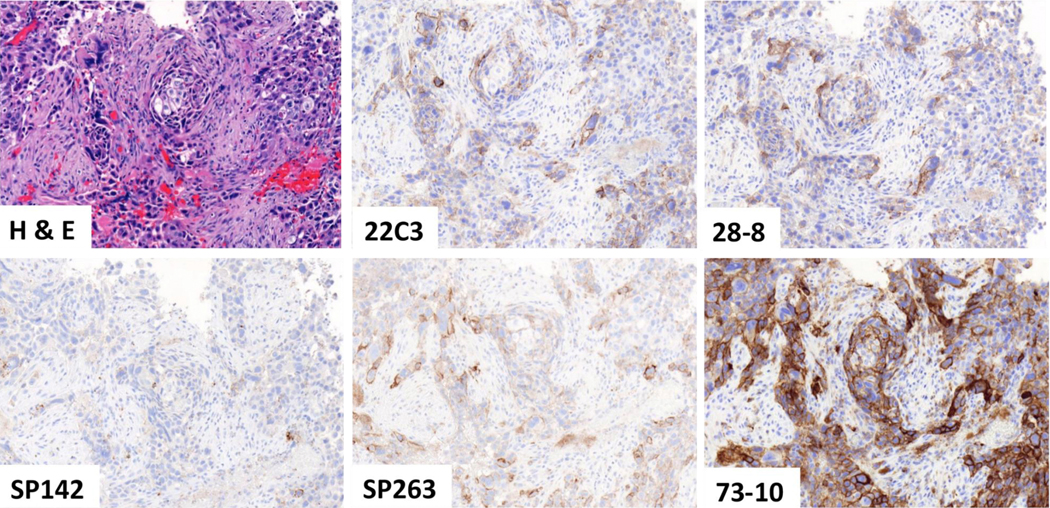
The mean values of the TPS scores across all readers (including that of the trainer) with use of the pooled digital and glass slide scores were derived for each assay and plotted across the samples (Fig. 2A). Three assays (22C3, 28–8 and SP263) showed close approximation between their respective best-fit curves. In contrast, the SP142 assay showed less sensitivity (lower TPS scores) whereas the 73–10 assay showed greater sensitivity (higher TPS scores) to detect PD-L1 expression in the same samples. Comparable results were noted for both the cohort comprising the whole sample and the cohort that excluded small cell carcinoma and cytological samples. Figure 3 shows a representative case that demonstrates the similarities and differences at microscopy levels in staining intensity. Pairwise comparison demonstrated the closest similarity between the 22C3 and 28–8 assays and consistently greater sensitivity for the 73–10 assay versus the 22C3, 28–8, and SP263 assays (Fig. 2B). The SP263 curve and the Bland and Altman plots do infer a slightly greater sensitivity in staining when compared with the 22C3 and 28–8 curves (Fig. 2B and Supplementary Figs. 6 and 7).
Figure 2.
Comparability of programmed death ligand 1 staining on tumor cells among the five assays: overall comparison (A) and pairwise comparisons (B).
Figure 3.
A representative case comparing the programmed death ligand 1 staining on the basis of the five assays.
Reliability of Pathologists to Score IC PD-L1 Expression
As IC scoring for PD-L1 could be done only on tissue sections, we focused the analyses on NSCLC tissue samples only, excluding the cytological aspirate samples. Furthermore, as IC was scored categorically, the FKS was used to assess interpathologist scoring reliability. The FKS among all pathologists for glass slide reading ranged from 0.11 to 0.28, and the FKS for digital image reading ranged from 0.08 to 0.27, demonstrating overall poor agreement for assessment of PD-L1 staining on ICs with use of the system adopted in this study (Supplementary Table 2 and Supplementary Fig. 8). With both approaches, the highest overall reliability was achieved with the SP-142 assay (FKS = 0.27–0.28). There was weak to moderate agreement in scoring IC PD-L1 staining with use of glass slide versus digital scoring, and among pathologists versus the trainer. Interestingly, moderate to strong agreement between pathologists versus the trainer was achieved for SP142 for distinguishing IC0 versus IC1, 2, and 3, but the agreements for higher IC categories were diminished (see Supplementary Fig. 8).
Comparability of PD-L1 Staining between the Five Assays of IC Staining
Among the five assays, the distribution of IC scores among three assays (22C3, 28–8, and SP263) were comparable. In contrast, the 73–10 and SP142 assays showed greater and lesser staining of the IC, respectively, compared with the other three assays (Supplementary Fig. 9).
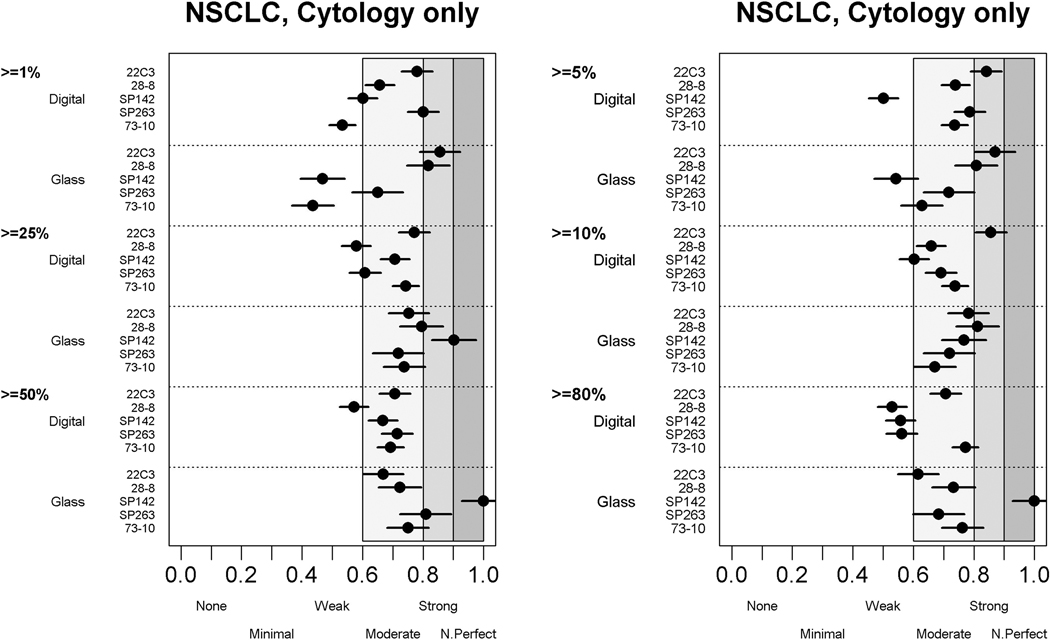
Reliability of PD-L1 Scoring on TCs in Cytological Samples
Overall, the ICCs among all pathologists for reading cytological samples were good, both for glass slide (0.78) and digital (0.85) readings (Supplementary Table 3). However, they were slightly lower than the ICCs for NSCLC tissue–only samples (0.89 and 0.93, respectively) (see Table 1). Comparable moderate levels of agreement (most with k > 0.6) were noted at all cutoff levels (Fig. 4), but overall, these ICCs were lower than those achieved in noncytological NSCLC tissue samples (see Fig. 1 and Supplementary Fig. 2).
Figure 4.
Agreement of scoring tumor cells’ programmed death ligand 1 expression in NSCLC cytological specimens at different cutoffs.
Discussion
This BP2 study using lung cancer diagnostic samples encountered in routine clinical pathology practice has further confirmed that three of the five currently available PD-L1 IHC assays (22C3 DAKO pharmDx, 28–8 DAKO pharmDx, and Ventana SP263) show comparable staining characteristics on TCs, whereas the Ventana SP142 assay shows less sensitivity and the DAKO pharmDx 73–10 IHC assay shows higher sensitivity to detect PD-L1 expression. We have also demonstrated that among a large group of pulmonary pathologists, the overall reliability or agreement in scoring PD-L1 was very strong, especially on NSCLC tissue section samples (overall ICC > 0.89). In contrast, we have shown that despite group training, scoring IC PD-L1 staining levels remains challenging, with low ICCs and poor k scores for all IC groups. Lastly, although the number of cases was limited and the result is considered preliminary, we observed moderate agreement in the pathologists’ assessment of PD-L1 status in needle aspirate cell block specimens.
There have been several studies comparing the analytical performance of the commercially available PD-L1 IHC assays.4 Although the designs, number, and type of samples and the number of pathologists involved in these studies have varied, practically all of them have reported high concordance in TC staining between the 22C3, 28–8, and SP263 assays. Almost all of the published studies, including BP1,7 have used resected NSCLC tissue. To our knowledge, BP2 is the first study to use a mixture of samples that are routinely encountered in clinical practice, including core needle/bronchial and lymph node biopsy, resection, and needle aspirate biopsy samples for cytological evaluation. We also included SCLC samples in our design, as at the time, the prevalence of PD-L1 expression in this type of tumor was unknown. However, of the 10 SCLC samples, only one showed staining for PD-L1, which is consistent with the reported PD-L1 expression in SCLC.15,16 It is important to note that none of the assays are currently approved to assess PD-L1 expression in SCLC. Although this study was not designed to assess the distribution of PD-L1 expression across lung cancer cases, our samples included the full range of PD-L1 expression levels and reflect the frequency of PD-L1–negative cases, especially when lower-stage, surgically resected cases are included (Supplementary Fig. 10).
BP1 results showed that the 22C3, 28–8, and SP263 assays had comparable sensitivity to detect PD-L1 expression on TCs, which is consistent with the results of three other studies,6,9,11 whereas the SP142 assay showed significantly less sensitivity. Furthermore, in both BP1 and a study by Ratcliffe,8 90% or higher levels of agreement were achieved when staining of the same tumor sample was performed by using the SP263 assay versus the 22C3 or 28–8 assays and the scoring was conducted by one pathologist. Perhaps because of these comparisons, the SP263 PD-L1 IHC assay is European Conformity-marked and available as a biomarker of nivolumab and pembrolizumab in Europe. However, our results detected slightly greater sensitivity for the SP263 assay than for the 22C3 and 28–8 assays. It should be noted that both Scheel et al.6 and Hendry et al.10 also detected slightly greater sensitivity of SP263 compared with that of the 22C3 and 28–8 assays. Whether this difference significantly affects the number of PD-L1–positive cases seen at lower cutoffs (e.g., 1%) or has an impact on clinical response rates remains to be determined (Supplementary Fig. 11). In one study, the significant impact in terms of more patients over threshold was seen only for the 1% cutoff.10 Fujimoto et al.17 recently also reported good analytical concordance between the 22C3, 28–8, and SP263 assays (weighted k coefficient = 0.64–0.55) on 40 samples from patients who had been treated with nivolumab; however, they did not observe the higher staining sensitivity of the SP263 assay than with the 22C3 or 28–8 assays, and the three assays demonstrated equivalent predictive performance of response to nivolumab with use of receiver operating characteristic analysis with an area under the curve of 0.75 to 0.82.
BP2 is the first publication to compare the staining characteristics of the 73–10 assay with those of the other four existing PD-L1 assays and show greater sensitivity of the 73–10 assay compared with that of all the other assays. The implications of a significantly more sensitive assay are still to be clarified, but the transferability of such an assay for general use in selecting patients for treatment with agents other than avelumab must be in question, at least with the data currently available. In the EMR100070–005 trial, avelumab was used in first-line treatment of NSCLC patient cohort selected on the basis of being above the 80% cutoff with use of the 73–10 assay. Supplementary Figure 11 shows that although the numbers are small, those patients above this 80% cutoff with use of the 73–10 assay are well matched to the cohort selected by being above the 50% cutoff according to the 22C3 assay. More data are undoubtedly required, but this raises the possibility of alternate biomarker/algorithm selection for avelumab.
In contrast to the other assays’ scoring algorithm, the assessment of PD-L1 staining on ICs in lung cancer is included only in the scoring algorithm of the SP142 PD-L1 IHC assay. The IC score is estimated as a percentage of tumor area (intratumoral and peritumoral desmoplastic stroma) with an IC infiltrate that shows PD-L1 staining regardless of the type of ICs (except macrophages in entrapped lung alveoli). With use of this approach, several studies, including BP1, have previously reported greater variability and poor interpathologist concordance in the scoring of IC PD-L1 expression,6,7,9–11 with ICCs or k values around 0.2. An alternate method (see Supplementary Fig. 1) that is still faithful to the concept of infiltrated tumor area but based on patterns of PD-L1–positive IC infiltrate has been proposed for scoring PD-L1 IC expression. As the SP142 assay often demonstrates a rather unique punctate type of staining not seen in other assays, we tested the feasibility of applying this approach to IC staining to the other assays in the BP2 project. The results demonstrate that reliability when NSCLC tissue sections were used was very poor (FKS = 0.11–0.21) despite extra emphasis during the group training on this scoring system and higher k values for the SP142 assay (0.27–0.28). It is worth noting that the cases used in training included only large sections from resection samples, whereas this study included assessment of ICs in metastatic lymph node and core/bronchial biopsy samples. This could have had an impact on both the effectiveness and relevance of the training received and the reliability of IC scoring, as the assessment methods for ICs are dependent on the spatial distribution of ICs, which is difficult to assess on small samples. The results emphasize the significant challenges in incorporating IC score into routine clinical testing. We also noted that IC staining in the BP2 samples was slightly weaker with the SP142 assay than with the 22C3, 28–8, and SP263 assays, whereas staining with 73–10 was stronger. Although this result was not seen in BP1, it was noted in two other previous studies.9,10 If this observation is confirmed in other studies, the result strongly suggests noncomparability of the other assays for assessing IC staining on the basis of the SP142 scoring algorithm.
Two-thirds of patients present with advanced-stage NSCLC, and a common diagnostic approach for these patients includes cytological assessment of fine-needle aspiration biopsy or endobronchial ultrasound biopsy specimens. In some countries 50% or more of such patients will have only cytological samples available for diagnosis.18 Unfortunately, none of the pivotal immune checkpoint inhibitor trials have included cytological specimens in their biomarker program to develop the companion PD-L1 IHC assays. Thus, strictly speaking, the current clinically used PD-L1 IHC assays have not been validated for all sample types encountered in their clinical application. Consequently, the diagnostic companies that commercialize the various PD-L1 assays have not recommended their use on cytological specimens, and the training offered to pathologists for these assays has not included examples of cytological specimens. Although many pathologists do not consider any difference in their approach to assessment of PD-L1 expression on TCs when using cytological cell block or tissue biopsy/resection sections, a few studies with small sample sizes have reported on the concordance of PD-L1 assessment on cytological versus matching surgical specimens.19–22 In this first part of the BP2 project, our results showed moderately good agreement (ICC = 0.78–0.85 and k = 0.6–0.85) among our group to score TC expression of PD-L1 in cytological samples. Further confirmation of this is awaited in the next phase of BP2, which will compare the concordance of PD-L1 scores from fine-needle aspirate samples versus from core biopsy samples versus from large section samples of the same lung resection specimen.
In conclusion, the BP2 results obtained by using reallife clinical lung cancer samples and scoring by 25 pathologists have further affirmed the results of BP1 and also demonstrate that the new 73–10 assay being developed for avelumab shows greater sensitivity in detecting PD-L1 expression than the other four assays do. We have also confirmed the reliability of PD-L1 scoring by digital images, which were used by a large group of pulmonary pathologists in assessing PD-L1 expression on TCs but not on ICs. Together with other published comparability studies, BP2 consolidates the evidence for interchangeability among three different assays (22C3, 28–8, and SP263) for use in scoring expression of PD-L1 on TCs (on the basis of TPS), allowing a range of cutoffs matched with their respective therapeutic agents to be considered from the assessment of a single PD-L1 IHC test. Studies of the heterogeneity of PD-L1 staining and comparison of the diagnostic values of large specimens versus those of small specimens versus those of cytological specimens from the same tumors are ongoing in Blueprint 2B.
Supplementary Material
Acknowledgments
This study was supported by AstraZeneca, Bristol-Myers Squibb, Hoffmann La Roche/Genentech/Ventana Medical Systema, Merck and Co, Inc., EMD Serono/Merck KGaA/Pfizer, and Agilent Dako. We thank Kristine Brovsky and Chis Rivard in Dr. Fred Hirsch’s laboratory and Dr. Wim Waelput and Dieter Rondas at Histogenex for providing crucial logistic support. Dr. Pelosi wishes to thank Dr. Silvano Bosari from the Division of Anatomic Pathology, Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico, Milan and Department of Pathophysiology and Transplantation, Università degli Studi di Milano, Milan, Italy, for assistance in providing four case materials for this study.
Disclosure: Dr. Tsao reports grants and personal fees from AstraZeneca, Merck, and Pfizer and personal fees from Bristol-Myers Squibb and Ventana/Roche outside the submitted work. Dr. Kerr reports personal fees from Bristol-Myers Squibb, Merck Sharp and Dohme, Merck Serono, Roche, and AstraZeneca outside the submitted work. Dr. Beasley reports pathology consulting work from Genentech, Bristol-Myers Squibb, and Merck outside the submitted work. Dr. Botling reports personal fees from AstraZeneca, Merck Sharp and Dohme, Roche, Pfizer, Bristol-Myers Squibb, Boehringer Ingelheim, and Novartis outside the submitted work. Dr. Bubendorf reports grants and personal fees from Roche and Merck Sharp and Dohme and personal fees from Bristol-Myers Squibb and AstraZeneca during the conduct of the study. Dr. Dacic reports personal fees from Bristol-Myers-Squibb and AstraZeneca outside the submitted work. Dr. Lantuejoul reports grants and personal fees from Bristol-Myers Squibb and personal fees from Merck Sharp and Dohme, Roche, and Novartis outside the submitted work. Dr. Mina-Kenudson reports personal fees from Merrimack Pharmaceuticals, H3 Biomedicine, ACD, and Roche outside the submitted work. Dr. Moreira reports personal fees from Genetech outside the submitted work. Dr. Nicholson reports personal fees from Merck, Boehringer Ingelheim, Novartis, AstraZeneca, Bristol-Myers Squibb, Roche, AstraZeneca, and AbbVie and grants and personal fees from Pfizer outside the submitted work. Dr. Poleri reports personal fees from AstraZeneca, Merck, Roche, and Bristol-Myers Squibb outside the submitted work. Dr. Sauter reports stock ownership in Merck, Thermo Fischer Scientific, and Chemed Corporation outside the submitted work. Dr. Thunnissen reports personal fees from HistoGeneX and Ventana Roche during the conduct of the study. Dr. Wistuba reports grants and personal fees from Genentech/Roche, Bristol-Myers Squibb, Astra Zeneca/Medimmune, Pfizer, HTG Molecular, and Merck; personal fees from Boehringer Ingelheim, Medscape, Asuragen, GlaxoSmithKline, and Bayer; and grants from Oncoplex, DepArray, Adaptive, Adaptimmune, Takeda, Amgen, and EMD Serono outside the submitted work. Dr. Yatabe reports personal fees from Merck Sharp and Dohme, Chugai-pharm, AstraZeneca, Pfizer, and Novartis outside the submitted work. Dr. Hirsch is coinventor of a University of Colorado-owned patent titled: “EGFR IHC and. FISH as Predictive Biomarkers for EGFR Therapy.” Dr. Hirsch has participated in advisory boards for Bristol-Myers Squibb, Genentech/Roche, HTG, Lilly, Merck, Pfizer, and Ventana. Dr. Hirsch’s laboratory has received research grants (through the University of Colorado) from Genentech, Bristol-Myers Squibb, Lilly, Bayer, and Clovis.
Footnotes
Supplementary Data
Note: To access the supplementary material accompanying this article, visit the online version of the Journal of Thoracic Oncology at www.jto.org and at https://doi.org/10.1016/j.jtho.2018.05.013.
The remaining authors declare no conflict of interest.
References
- 1.Gong J, Chehrazi-Raffle A, Reddi S, Salgia R. Development of PD-1 and PD-L1 inhibitors as a form of cancer immunotherapy: a comprehensive review of registration trials and future considerations. J Immunother Cancer. 2018;6:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Melosky B, Chu Q, Juergens R, Leighl N, McLeod D, Hirsch V. Pointed progress in second-line advanced non-small-cell lung cancer: the rapidly evolving field of checkpoint inhibition. J Clin Oncol. 2016;34:1676–1688. [DOI] [PubMed] [Google Scholar]
- 3.Assi HI, Kamphorst AO, Moukalled NM, Ramalingam SS. Immune checkpoint inhibitors in advanced non-small cell lung cancer. Cancer. 2018;124:248–261. [DOI] [PubMed] [Google Scholar]
- 4.Buttner R, Gosney JR, Skov BG, et al. Programmed death-ligand 1 immunohistochemistry testing: a review of analytical assays and clinical implementation in nonsmall-cell lung cancer. J Clin Oncol. 2017;35:3867–3876. [DOI] [PubMed] [Google Scholar]
- 5.Yu H, Boyle TA, Zhou C, Rimm DL, Hirsch FR. PD-L1 expression in lung cancer. J Thorac Oncol. 2016;11: 964–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scheel AH, Dietel M, Heukamp LC, et al. Harmonized PDL1 immunohistochemistry for pulmonary squamous-cell and adenocarcinomas. Mod Pathol. 2016;29:1165–1172. [DOI] [PubMed] [Google Scholar]
- 7.Hirsch FR, McElhinny A, Stanforth D, et al. PD-L1 Immunohistochemistry assays for lung cancer: results from phase 1 of the Blueprint PD-L1 IHC Assay Comparison Project. J Thorac Oncol. 2017;12:208–222. [DOI] [PubMed] [Google Scholar]
- 8.Ratcliffe MJ, Sharpe A, Midha A, et al. Agreement between programmed cell death ligand-1 diagnostic assays across multiple protein expression cutoffs in non-small cell lung cancer. Clin Cancer Res. 2017;23:3585–3591. [DOI] [PubMed] [Google Scholar]
- 9.Rimm DL, Han G, Taube JM, et al. A prospective, multi-institutional, pathologist-based assessment of 4 immunohistochemistry assays forPD-L1expression innon-small cell lung cancer. JAMA Oncol. 2017;3:1051–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hendry S, Byrne DJ, Wright GM, et al. Comparison of four PD-L1 immunohistochemical assays in lung cancer. J Thorac Oncol. 2018;13:367–376. [DOI] [PubMed] [Google Scholar]
- 11.Adam J, Le Stang N, Rouquette I, et al. Multicenter French harmonization study for PD-L1 IHC testing in non-small cell lung cancer. Ann Oncol. 2018;29:953–958. [DOI] [PubMed] [Google Scholar]
- 12.Koo TK, Li MY. A guideline of selecting and reporting intraclass correlation coefficients for reliability research. J Chiropr Med. 2016;15:155–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McHugh ML. Interrater reliability: the kappa statistic. Biochem Med (Zagreb). 2012;22:276–282. [PMC free article] [PubMed] [Google Scholar]
- 14.Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;1:307–310. [PubMed] [Google Scholar]
- 15.Schultheis AM, Scheel AH, Ozretic L, et al. PD-L1 expression in small cell neuroendocrine carcinomas. Eur J Cancer. 2015;51:421–426. [DOI] [PubMed] [Google Scholar]
- 16.Yu H, Batenchuk C, Badzio A, et al. PD-L1 expression by two complementary diagnostic assays and mRNA in situ hybridization in small cell lung cancer. J Thorac Oncol. 2017;12:110–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujimoto D, Sato Y, Uehara K, et al. Predictive performance of four programmed cell death ligand 1 assay systems on nivolumab response in previously treated patients with non-small cell lung cancer. J Thorac Oncol. 2018;13:377–386. [DOI] [PubMed] [Google Scholar]
- 18.Yatabe Y, Kerr KM, Utomo A, et al. EGFR mutation testing practices within the Asia Pacific region: results of a multicenter diagnostic survey. J Thorac Oncol. 2015;10:438–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ilie M, Juco J, Huang L, Hofman V, Khambata-Ford S, Hofman P. Use of the 22C3 anti-programmed death ligand 1 antibody to determine programmed death ligand 1 expression in cytology samples obtained from non-small cell lung cancer patients. Cancer Cytopathol. 2018;126:264–274. [DOI] [PubMed] [Google Scholar]
- 20.Russell-Goldman E, Kravets S, Dahlberg SE, Sholl LM, Vivero M. Cytologic-histologic correlation of programmed death-ligand 1 immunohistochemistry in lung carcinomas. Cancer Cytopathol. 2018;126:253–263. [DOI] [PubMed] [Google Scholar]
- 21.Skov BG, Skov T. Paired comparison of PD-L1 expression on cytologic and histologic specimens from malignancies in the lung assessed with PD-L1 IHC 28–8pharmDx and PDL1 IHC 22C3pharmDx. Appl Immunohistochem Mol Morphol. 2017;25:453–459. [DOI] [PubMed] [Google Scholar]
- 22.Sakakibara R, Inamura K, Tambo Y, et al. EBUS-TBNA as a promising method for the evaluation of tumor PD-L1 expression in lung cancer. Clin Lung Cancer. 2017;18:527–534.e521. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.