Abstract
Saccharomyces cerevisiae telomeres consist of a continuous 325 ± 75-bp tract of the heterogeneous repeat TG1-3 which contains irregularly spaced, high-affinity sites for the protein Rap1p. Yeast cells monitor or count the number of telomeric Rap1p molecules in a negative feedback mechanism which modulates telomere length. To investigate the mechanism by which Rap1p molecules are counted, the continuous telomeric TG1-3 sequences were divided into internal TG1-3 sequences and a terminal tract separated by nontelomeric spacers of different lengths. While all of the internal sequences were counted as part of the terminal tract across a 38-bp spacer, a 138-bp disruption completely prevented the internal TG1-3 sequences from being considered part of the telomere and defined the terminal tract as a discrete entity separate from the subtelomeric sequences. We also used regularly spaced arrays of six Rap1p sites internal to the terminal TG1-3 repeats to show that each Rap1p molecule was counted as about 19 bp of TG1-3 in vivo and that cells could count Rap1p molecules with different spacings between tandem sites. As previous in vitro experiments had shown that telomeric Rap1p sites occur about once every 18 bp, all Rap1p molecules at the junction of telomeric and nontelomeric chromatin (the telomere-nontelomere junction) must participate in telomere length measurement. The conserved arrangement of these six Rap1p molecules at the telomere-nontelomere junction in independent transformants also caused the elongated TG1-3 tracts to be maintained at nearly identical lengths, showing that sequences at the telomere-nontelomere junction had an effect on length regulation. These results can be explained by a model in which telomeres beyond a threshold length form a folded structure that links the chromosome terminus to the telomere-nontelomere junction and prevents telomere elongation.
Telomeres are the nucleoprotein complexes that make up the ends of eukaryotic chromosomes. In the majority of organisms examined, telomeres consist of short repeated DNA sequences and their associated sequence-specific binding proteins (reviewed in reference 11). In the yeast Saccharomyces cerevisiae, telomeres consist of the heterogeneous repeat (TG)1-6TG2-3, abbreviated as TG1-3, which contains irregularly spaced binding sites for the major yeast telomere binding protein Rap1p (reviewed in reference 54). The length of the TG1-3 repeats is regulated, being maintained within a small range of 325 ± 75 bp (40). Telomeres can be formed on linear yeast plasmids by transformation followed by the addition of TG1-3 repeats. Sequencing of these linear plasmid repeats has shown that the precise TG1-3 sequences of independently formed telomeres are different (48). When individual telomeres derived from the same formation event are isolated from different cells, they have different TG1-3 sequences for the half of the TG1-3 tract nearest the chromosome end. This sequence heterogeneity is thought to arise from telomeres going through cycles of telomere shortening followed by the addition of TG1-3 repeats with a different sequence (48). This shortening and lengthening by the addition of different TG1-3 sequences probably also occurs with chromosomal telomeres because the lengths of individual telomeres vary between different cell lineages (43). A direct consequence of this sequence heterogeneity is that the spacing between individual Rap1p molecules bound to these repeats varies between different telomeres, and so the heterogeneity of the telomere DNA sequences causes heterogeneity in telomere chromatin structure.
The fact that telomere length varies as cells grow but still remains within a specific length range has led to the hypothesis that telomere length is regulated by balancing lengthening and shortening mechanisms (11). Presumably, telomere length is somehow measured, and depending on the result, telomere lengthening or telomere shortening occurs. Telomere lengthening occurs primarily via telomerase, a cellular reverse transcriptase with its own RNA template (11, 24), and can also occur through recombination-dependent mechanisms (36). Telomere shortening occurs by incomplete replication of the 5′ chromosome end, owing to removal of the RNA primer for DNA synthesis (reviewed in reference 54), and by active degradation (6, 49) and deletion mechanisms (23). Similar processes occur in other organisms, including humans (11, 28, 32). How telomere length is measured and how these processes are controlled are not yet understood.
Telomere binding proteins play an important role in regulating telomere length. The major yeast telomere binding protein Rap1p can be divided into at least two domains, a large DNA binding domain (amino acids 361 to 596 [16]) and a large C-terminal domain (amino acids 600 to 827) which contains multiple subdomains involved in telomere length control, transcriptional silencing, and transcriptional activation (5, 12, 20, 26, 45). Rap1p is known to play a negative regulatory role by inhibiting telomere elongation. Missense or deletion mutations in the Rap1p C terminus cause yeast telomere length to increase (20, 21, 25, 45). These rap1 mutations eliminate interactions between Rap1p and other proteins important for telomere length control (13, 52). Overproduction of the Rap1p C-terminal domain in vivo causes chromosomal telomeres to lengthen (5, 12, 41, 51), presumably because Rap1p-interacting factors are titrated away from telomeres. Experiments with the Rap1p homolog of Kluyveromyces lactis indicate that the C termini of Rap1p molecules bound to the very ends of the chromosome interact with proteins that inhibit telomere elongation, presumably by inhibiting the access of telomerase to the chromosome terminus (19). These data have led to the hypothesis that the Rap1p C terminus and proteins bound to it inhibit telomere lengthening (11).
The Rap1p C terminus can also play a positive role in telomere elongation. The Rap1p C terminus can stimulate the formation of chromosomal telomeres in a transformation assay, a model system for the elongation of short telomeres (38). As with the inhibition of telomere lengthening, the ability of the Rap1p C terminus to stimulate telomere elongation most likely involves other proteins that bind to Rap1p.
Several factors that interact with the Rap1p C terminus and regulate telomere length and transcriptional silencing have been identified. These factors include Rif1p, Rif2p, and Sir3p (13, 33, 52). Sir3p forms a complex with Sir2p, Sir4p, and chromatin to silence transcription of nearby genes, a phenomenon known as telomere position effect (14, 15, 27, 39, 44). Overproduction of Rif1p and Rif2p causes telomeres to shorten, deletion of either gene causes telomeres to lengthen by ∼200 bp, and simultaneous deletion of both genes causes telomere length to increase from ∼325 bp to 2 to 4 kb of TG1-3 (52). The rif1 rif2 double-mutant phenotype is identical to that of rap1t mutants which lack the Rap1p C terminus (20). The 2- to 4-kb TG1-3 repeats in rap1t and rif1Δ rif2Δ cells are thought to arise by telomere elongation occurring in an unregulated fashion (20, 21). Thus, Rif1p and Rif2p are important components for telomere length regulation. Recent work has shown that yeast normally measure telomere length by counting Rap1p molecules (29), and Rif1p and Rif2p probably play essential roles in this process. However, the mechanism by which Rap1p molecules are counted is unknown.
In measuring telomere length, the cell must determine where the telomere starts, where it ends, and how many Rap1p molecules are present in between. How the beginning and the end of a telomeric TG1-3 tract are demarcated is unknown. While in vitro studies show that Rap1p binds to sites present at an average of about 1 per 18 bp, site spacing is irregular and some sites overlap (7). This variable placement of Rap1p binding sites means that these telomeric Rap1p molecules are not presented to the cell in a uniform manner. Thus, it is unknown whether all of these Rap1p molecules are used to measure telomere length in vivo.
To investigate the mechanism of telomere length regulation, we constructed a series of synthetic telomeres with additional Rap1p molecules internal to the terminal TG1-3 repeats and determined their effects on telomere length. The goal of this approach was to mimic a portion of the internal telomere nucleoprotein complex and determine how changes in this structure alter length regulation. If the internal sequences are detected by the cell as part of the telomere, the terminal TG1-3 tract will be elongated to a length less than that of a natural telomere. We constructed synthetic telomeres that separated adjacent TG1-3 tracts with different lengths of nontelomeric DNA or that contained regularly spaced arrays of Rap1p sites internal to the terminal TG1-3 tract. As the separation between adjacent TG1-3 tracts increased in size from 38 to 50 bp, cells counted smaller portions of the internal TG1-3 tract as part of the TG1-3 tract. Cells could not accommodate a 138-bp disruption; the internal sequences were not counted, and a new telomere-nontelomere junction was established. Placing tandem arrays of regularly spaced Rap1p binding sites just internal to the TG1-3 repeats showed that yeast cells can accommodate different spacings of 13 to 35 bp between consecutive Rap1p sites while measuring telomere length and that cells equated one Rap1p molecule with about 19 bp of TG1-3 DNA in vivo. These results indicate that all of the Rap1p molecules at the telomere-nontelomere junction participate in telomere length regulation. The internal tandem Rap1p sites had the unexpected effect of eliminating the variation in average telomere lengths normally seen between independently formed telomeres. These data suggest that the boundaries of the telomere, i.e., the chromosome terminus and the junction between the Rap1p-containing and non-Rap1p-containing chromatin (the telomere-nontelomere junction), communicate with one another when telomere length is measured. We therefore propose that telomere length measurement involves forming a folded chromatin structure that is dependent on the presence of a threshold number of telomeric Rap1p molecules. This folded structure links the chromosome terminus and the telomere-nontelomere junction and inhibits telomere lengthening.
MATERIALS AND METHODS
Strains.
Recombinant DNA manipulations were in Escherichia coli MC1066 (r− m+ pyrF::Tn5 trpC leuB). S. cerevisiae strains used were KR36-6L (MATa ade2-1 or 101 ade8-18 ura3-52 trp1Δ1 leu2-ΔRC his3Δ) (40) and the gal4Δ strain YM708 (MATα ade2-101 ura3-52 trp1Δ-901 his3-200 lys2-801 LEU2 canR gal4-542; from Mark Johnston).
Plasmids.
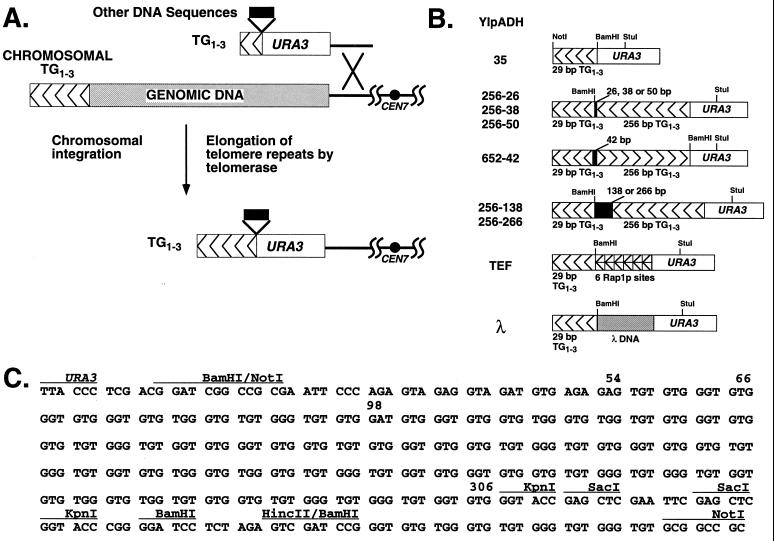
Plasmid YIpADH35 contains ADH4-URA3-TG1-3 sequences where the 29-bp segments of TG1-3 sequences consist of the oligonucleotides GATCCGGGTGTGTGGGTGTGTGGGTGTGGGTGTGC and GGCCGCACACCCACACCCACACACCCACACACCCG in pBR322 as described elsewhere (38) (boldfaced and underlined bases denote Rap1p binding sites). The HincII-NotI fragment bearing the 256 bp of TG1-3 was isolated from pCT300 (256 bp of TG1-3 in pVZ-1; from K. Runge, R. Wellinger, J. Wright, and V. Zakian), filled in, and then cloned into the filled-in BamHI site of YIpADH35 to give YIpADH256-50 with the orientation of the TG1-3 repeats the same as the terminal 29-bp TG1-3 repeats (see Fig. 1B and C). This plasmid was previously called YIpADH-275 (38). This 256-bp tract was derived from a ∼350-bp tract of TG1-3 sequences (as determined by Southern blotting) cloned by circularization of a yeast linear plasmid that was directly transformed into yeast (40). Upon rescuing the plasmid into E. coli, some of the TG1-3 sequences were deleted. Sequencing of multiple YIpADH256-XX constructions gave the same sequence for the 256-bp insert (Fig. 1C), showing it was stable in bacteria. YIpADH256-50 has a 50-bp polylinker spacer between the terminal 29 bp of TG1-3 and 256 bp of TG1-3. YIpADH652-42 has the 256 bp of TG1-3 repeats in the opposite orientation to telomere with a 42-bp polylinker spacer between the terminal 29-bp TG1-3 and 256-bp TG1-3.
FIG. 1.
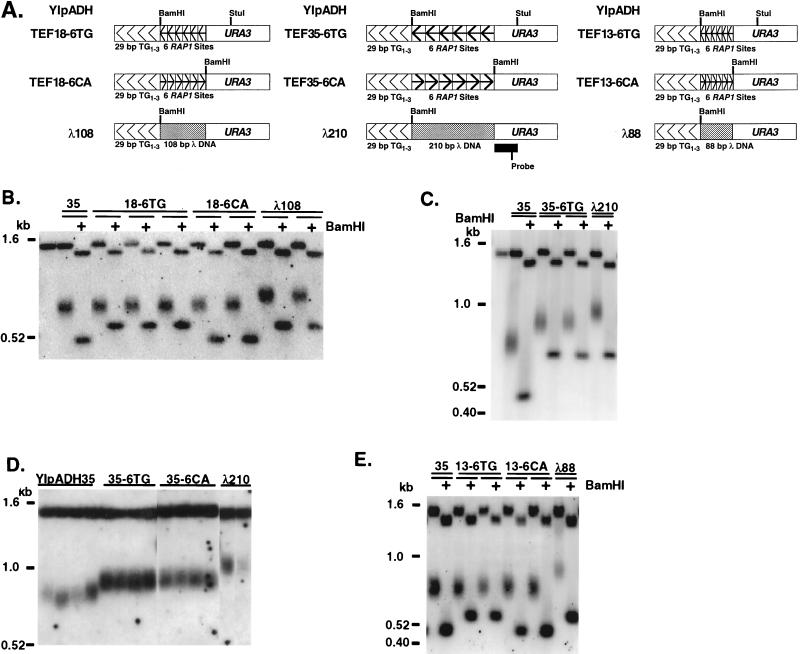
Introduction of synthetic telomeres into yeast. (A) Replacement of the left telomere of chromosome VII by homologous integration at the ADH4 locus, using a synthetic telomere adjacent to URA3. Different inserts, shown in panel B, were cloned between URA3 and the TG1-3 sequences to attempt to alter telomere length regulation. (B) The synthetic telomeres used in this work. The nontelomeric spacers between the TG1-3 tracts are described in Materials and Methods. The orientation of the TG1-3 repeats is indicated by arrowheads. YIpADH652-42 has the internal 256-bp TG1-3 tract in the reverse orientation relative to the terminal 29-bp tract. The specific YIpADHTEF and YIpADHλ telomeres are presented in Fig. 5. (C) Sequences of the TG1-3 repeats and spacer in the YIpADH256-50 construction. The sites used to construct the plasmids in panel B are indicated. The TG1-3 repeats are from positions 54 to 309 in the 5′ KpnI site, and the single A in the TG1-3 repeats is at position 98.
To reduce the polylinker spacer, YIpADH256-50 was separately digested with KpnI or SacI (two sites each are present in the polylinker [Fig. 1C]) and self-ligated to generate, respectively, the 26- and 38-bp polylinkers between the 29 and 256 bp of TG1-3 to form YIpADH256-26 and YIpADH256-38. To increase the length of the polylinker spacer, we cloned 210-bp (bp 10101 to 10310) and 88-bp (bp 10101 to 10188) PCR fragments of lambda phage DNA (described below) into the BamHI site of YIpADH256-50 to form YIpADH256-266 and YIpADH256-138.
The single BamHI site between URA3 and the TG1-3 repeats of YIpADH35 was the site of insertion of the TEF18-6 oligonucleotide, which contains six tandem repeats of the same sequence with one Rap1p binding site every 18 bp, and the 78-bp TEF13-6 oligonucleotide with six tandem repeats bearing one Rap1p site every 13 bp (Table 1). The renatured, kinase-treated oligonucleotides were cloned into the BamHI site of YIpADH35 to form YIpADHTEF18-6TG and YIpADHTEF13-6TG with the BamHI site closest to the NotI site. Plasmids with the six TEF oligonucleotides in opposite orientation (YIpADHTEF18-6CA and YIpADHTEF13-6CA) were also constructed. All constructions were verified by DNA sequencing. The negative control plasmid YIpADHλ210, YIpADH35 containing a 210-bp insert of lambda DNA, has already been described (38). Lambda PCR fragments were also cloned into YIpADH35, with the BamHI site closest to the NotI site, to generate YIpADHλ88 and YIpADHλ108. To form the PCR fragments for the λ210, λ108, and λ88 constructions, PCR primers λ-10101 and, respectively, λ-10310r, λ-10208r, and λ-10188r (Table 1) were used with 10 ng of λ DNA (New England Biolabs) and 35 cycles of 94°C-1 min, 55°C-45 s, and 72°C-1 min in a Perkin-Elmer Cetus 9600 PCR machine.
TABLE 1.
Oligonucleotides and PCR primers used to construct synthetic telomeres
| Name | Sequencea |
|---|---|
| Oligonucleotides | |
| TEF18-6 A | GAT CTA ATG TGT GGG TGC AAC TAA ATG TGT GGG TGC AAC TAA ATG TGT GGG TGC AAC TAA ATG TGT GGG TGC AAC TAA ATG TGT GGG TGC AAC TAA ATG TGT GGG TGC G |
| TEF18-6 B | GAT CCG CAC CCA CAC ATT TAG TTG CAC CCA CAC ATT TAG TTG CAC CCA CAC ATT TAG TTG CAC CCA CAC ATT TAG TTG CAC CCA CAC ATT TAG TTG CAC CCA CAC ATT A |
| TEF13-6 A | GAT CTA ATG TGT GGG TGCAAT GTG TGG GTG CAA TGT GTG GGT GCA ATG TGT GGG TGCAAT GTG TGG GTG CAA TGT GTG GGT GCG |
| TEF13-6 B | GAT CCG CAC CCA CAC ATTGCA CCC ACA CAT TGC ACC CAC ACA TTG CAC CCA CAC ATTGCA CCC ACA CAT TGC ACC CAC ACA TTA |
| TEF35 A | GAT CTG GTC TAA ATG TGT GGG TGC AAC ATG AAT GG |
| TEF35 B | GAT CCC ATT CAT GTT GCA CCC ACA CAT TTA GAC CA |
| PCR primers | |
| λ-10101 | GGC CGG ATC CGT TTC TGC GGG AAA GTG T |
| λ-10188r | GGC CGG AGA TCT GCA CGC CAG TCG GGT CGC C |
| λ-10208r | GGC CGG AGA TCT CCG TGG ATG ACA TCC CGG C |
| λ-10310r | GGC CAG ATC TAA AAC AGG CTG AGC ACG G |
| 5′ URA3 StuI | GGC CTT TTG ATG TTA GCA GAA TTG |
| 3′ polylinker | GGA TCG ACT CTA GAG GAT CCC CGG GTA CC |
| Primer 1 | AGA GGA GCT CGG TAC CCA CAC C |
Rap1p sites are indicated by boldface type and by underlines. The Rap1p site was derived from the TEF2 UAS (4).
Plasmids bearing six tandem repeats of a 35-bp oligonucleotide bearing a single Rap1p site in the same (YIpADHTEF35-6TG) or opposite (YIpADHTEF35-6CA) orientation as the terminal 29-bp TG1-3 tract have been described (YIpADHTEF35-6TG is the same as YIpADHTEF-35 [38]). The tandem repeats were constructed in pBRA8, a pBR322 plasmid with the 2.0-kb BamHI-EcoRI ADE8 fragment cloned into the AvaI site of pBR322, both as blunt-end fragments. The plasmid was cleaved with BamHI (in the Tetr gene) and BglII (in ADE8), and the TEF35 A-B oligonucleotide pair was cloned into this vector. A clone (pBRA-TEF35-2TG) that contained both BglII and BamHI sites was selected and sequenced. This clone has two tandem repeats of the TEF35 oligonucleotide. An EcoRI-BglII fragment from pBRA-TEF35-2TG was cloned into the same plasmid cut with EcoRI-BamHI to convert the double repeat to four head-to-tail repeats. The four head-to-tail repeats were cloned into pBRA-TEF35-2TG in the same manner to generate six head-to-tail repeats in pBRA-TEF-6TG. Inserts were verified by sequencing at each step of the oligomerization process. For unknown reasons, these repeats could not be oligomerized in vitro and subsequently cloned into high-copy-number bacterial pUC-based vectors, and these repeats were stable in E. coli MC1066 but not in Sure or DH10B cells. The six-repeat BamHI-BglII fragment was then cloned into the BamHI site of YIpADH35 to generate YIpADHTEF35-6TG and YIpADHTEF35-6CA.
Telomere formation and integration.
Digestion of all of the YIpADH plasmids with SalI and NotI releases a SalI-ADH4-URA3-insert-TG1-3-NotI fragment which can replace the left telomere of chromosome VII. Transformations using 5 μg of SalI- and NotI-digested plasmid into S. cerevisiae KR36-6L or YM708 were performed as described previously (38). The relative efficiencies of telomere formation previously noted (38) were confirmed in these experiments. In most cases, 100 to 200 transformants of each type were picked to a master plate lacking uracil, grown overnight at 30°C, replica plated to a plate containing uracil, grown overnight, and assayed for telomere position effect by testing for growth on YC-Ura and 5-fluoro-orotic acid plates. Subsequently, several transformants of each type predicted to have a URA3 telomere were then used for Southern analysis using a PCR fragment equivalent to the StuI-NsiI 3′ fragment of URA3 as the probe. Integration of YIpADH256-50 at internal loci (URA3) does not give detectable levels of telomere position effect (data not shown).
DNA isolation and Southern blotting.
Genomic yeast DNA was isolated according to the protocol of V. Schulz (42a). Briefly, stationary-phase cells (5 ml) were washed with water and resuspended in 250 μl of lysis buffer (100 mM Tris-HCl [pH 8.0], 50 mM EDTA, 1% sodium dodecyl sulfate [SDS]), and acid-washed glass beads (Sigma type V; diameter of 400 to 500 μm) were added to the 0.5-ml mark. The tube was shaken for 10 min in a Vortex-Genie (VWR Scientific) with the shaker attachment at top speed, and 150 μl of 7.5 M ammonium acetate was then added. The tube was heated at 65°C for 15 min and placed on ice for 15 min, and then 500 μl of CHCl3 was added. After mixing, the tube was spun for 8 min at top speed in a microcentrifuge. The aqueous phase was added to a 1.5-ml tube with 0.5 ml of room temperature isopropanol and spun at room temperature for 5 min. The DNA pellet was washed with 70% ethanol and resuspended in 50 μl of TE (10 mM Tris-HCl [pH 8.0], 1 mM EDTA). To this preparation, 1.5 μl of RNase A (5 mg/ml) was added, and the mixture was incubated at 37°C for 15 min; this was followed by the addition of 1.5 μl of proteinase K (10 mg/ml) and incubation at 37°C for another 15 min. The solution was brought to 300 μl with water, extracted once with an equal volume of phenol-CHCl3 (4:1), and extracted once with CHCl3. The DNA was precipitated by adding 150 μl of 7.5 M ammonium acetate (pH 7) and 1 ml of 95% ethanol at −20°C, spun, and resuspended in 50 μl of TE. Five microliters was digested for each gel lane. Southern analysis was done as described in reference 42. Prehybridization solution contained 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 5× Denhardt’s solution, 0.5% SDS, and 100 μg of denatured, fragmented salmon sperm DNA per ml, and prehybridization was at 68°C for 2 to 3 h. Denatured probe was added to the filter in hybridization solution (6× SSC, 0.5% SDS, 100 μg of denatured salmon sperm DNA per ml), and the filter was hybridized overnight in a hybridization oven at 68°C. The filters were washed as described elsewhere (42).
Telomere length measurement.
In the Southern blots used for these analyses, the molecular size standards at 1.636 and 0.517 kb were used to construct the standard curve for migration versus molecular weight and then checked against the migration of the 1.018-kb standard. In all cases, the derived molecular size of the 1.018-kb standard was within 8 bp of its actual molecular size. In addition, the molecular size of the chromosomal URA3 StuI band was within ±15 bp in these blots. The amount of TG1-3 equivalent to the TEF35-6, TEF18-6, or TEF13-6 tandem arrays was determined by subtracting the size of the YIpADHTEFXX-6 StuI fragment from the YIpADHλ control with the same insert size (e.g., YIpADHλ210 size − YIpADHTEF35-6 size). For example, since YIpADHλ210 contains an insert which is not counted as TG1-3 whereas YIpADHTEF35-6 contains an insert that is equivalent to TG1-3, this difference gives the “equivalent TG1-3 length” of the TEF35-6 tandem array. Two advantages of this approach were that (i) the minor differences in migration for the YIpADHTEF insert also occurred for the YIpADHλ insert on the same blot, and so deviations in gel migration for these terminal restriction fragments on an individual blot are similar, and (ii) only one comparison is performed, and so errors inherent in each measurement are added together only once. In addition, each telomere was also cleaved with StuI plus BamHI to verify that the inserts were still present in vivo and of the correct size. For YIpADHTEF13-6, the YIpADHλ88 telomere was used as a control. Corrections for the 4- to 6-bp differences between the insert sizes of the YIpADHλ and YIpADHTEF telomeres were included in these calculations. For Fig. 3C, length measurements were from the blots in Fig. 3A and B and two other blots similar to that in Fig. 2B that ran more uniformly (not shown).
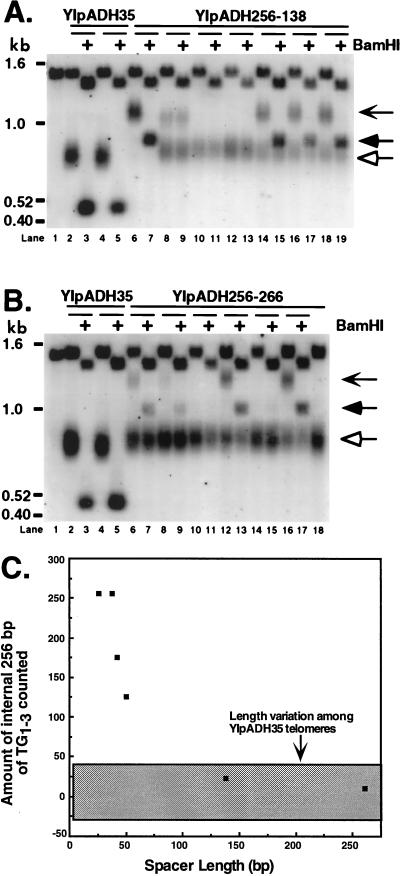
FIG. 3.
A 138-bp nontelomeric spacer establishes a new telomere-nontelomere junction. (A) Individual YM708 transformants bearing the YIpADH256-138 telomere were analyzed as for Fig. 2. Lanes 6 and 7, a YIpADH256-138 transformant with long telomeres; lanes 10 to 13, transformants with short telomeres; lanes 8 and 9 and 14 to 19, transformants with short and long telomeres. The transformant in lanes 8 and 9 had lost the BamHI site in the long telomeres and was not used for the analysis in panel C. The chevron-headed arrow indicates the long telomeres, the filled arrowhead indicates the StuI-BamHI long telomere fragment, and the hollow arrowhead indicates the short telomeres. Lane 1 contains DNA from the untransformed strain. (B) Individual transformants bearing the YIpADH256-266 telomere were analyzed as for Fig. 2. Lanes 10, 11, 14, 15, and 18 show transformants with short telomeres, while lanes 6 to 9, 12, 13, 16, and 17 show transformants with short and long telomeres. Arrows are used as in panel A. Lane 1 contains DNA from the untransformed strain. (C) The amount of the internal 256-bp tract counted as part of the elongated TG1-3 tract plotted against the length of the nontelomeric spacer. The gray box represents the length heterogeneity due to normal variation and was calculated by using seven independent YIpADH35 telomeres. The amount of internal TG1-3 counted was determined by subtracting the average length of the terminal TG1-3 tract distal to the BamHI site for different nontelomeric spacer telomeres (the ? in Fig. 2A) from the average modal length of the TG1-3 tract for the control YIpADH35 telomere (Fig. 2A). The standard error of the telomere length measurements for each spacer telomere was ±10% except for YIpADH652-42 (±53%).
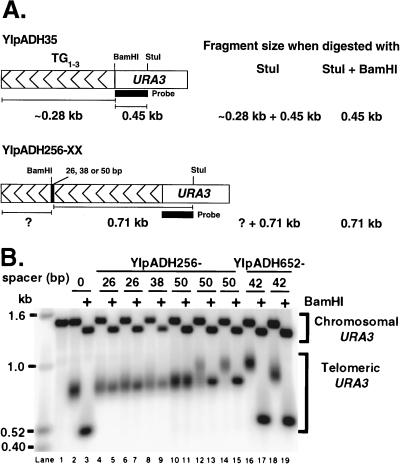
FIG. 2.
Some of the internal TG1-3 sequences are considered part of the telomere across a 50-bp nontelomeric spacer. (A) Predicted sizes of the telomere restriction fragments on Southern blots. Telomere length, i.e., the length of the elongated terminal TG1-3 tract, was determined by subtracting the StuI-BamHI fragment length from the StuI fragment length measured on the same blot. Analysis on the same blot was important for giving reproducible and accurate length determination for different control telomeres. With lanes 14 and 15 of B as an example, the length of the elongated terminal TG1-3 sequences (the ? in the diagram) is 150 bp. (B) Representative genomic DNAs from cells bearing the YIpADH256-26, -38, and -50 telomeres and the YIpADH652-42 telomere were cleaved with StuI or StuI plus BamHI and analyzed by Southern blotting using the URA3 fragment in panel A as probe. All telomeres were formed in yeast strain YM708. The 0 spacer is YIpADH35. Each pair of lanes (indicated by a bar under the number) represents an individual transformant. Lane 1, YM708 with no synthetic telomere; lanes 10 and 11, a YIpADH256-50 transformant with short telomeres; lanes 14 and 15, a YIpADH256-50 transformant with long telomeres; lanes 12 and 13, a YIpADH256-50 transformant with short and long telomeres (mixed telomeres). Note that the YIpADH652-42 construction places the BamHI site closer to URA3 (Fig. 1B). Telomere fragment sizes were determined as described in Materials and Methods.
PCR.
The terminal portions of the 26- and 38-bp spacers between the 256- and 29-bp TG1-3 sequences were detected by a hot-start PCR method with the 5′ URA3 StuI and 3′ polylinker primers (Table 1). For this purpose, the primer mix (50 pmol of both primers in 50 μl of H2O) was heated at 94°C for 10 min, and the remaining components (∼2 μg of genomic DNA plus deoxynucleoside triphosphates, buffer components and Taq DNA polymerase in 50 μl) were separately heated at 94°C for 2 min. After these two reaction mixtures were combined, the mixture was topped with 50 μl of mineral oil and PCR was performed by 5′ denaturation at 94°C for 1 cycle, then 5 cycles of 94°C-1 min, 60°C-1 min, and 72°C-1 min, followed by 30 cycles of 94°C-1 min, 55°C-1 min, and 72°C-1 min, and ending with 72°C-10 min.
The most internal 12 bp of the 50- and 138-bp spacers were detected with the 5′ URA3 StuI primer and primer 1 (Table 1), using 5 to 15 μl of a 1:1,000 dilution of yeast genomic DNA (20 to 45 ng). DNA was denatured in a partial PCR for 5 min at 94°C in a heating block, and then each primer, preincubated at 94°C for 5 min, and Taq polymerase were sequentially added, keeping the reaction at 94°C between each addition. The reaction was topped with 50 μl of mineral oil and then transferred to a PCR machine heated to 94°C. The PCR program was 1 cycle for 5 min at 94°C, then 5 cycles of 94°C-1 min, 68°C-1 min, and 72°C-1 min, followed by 30 cycles of 94°C-1 min, 60°C-1 min, and 72°C-1 min, and ending with 72°C-10 min. The correct 760-bp PCR product was confirmed by NsiI digestion. Of the 13 long and mixed telomeres and 14 short telomeres tested, most are shown in Fig. 2 (lanes 2, 10, 12, and 14), 3A (lanes 6, 8, 10, 12, 14, and 18), 4B (lanes 4, 5, 6, 8, 9, and 13), and 4C (lanes 6, 8, 10, 11, 13, 16, and 17). All of the long telomeres from Fig. 4B and C in this sample retained the BamHI site, showing that these subclones retained the entire spacer.
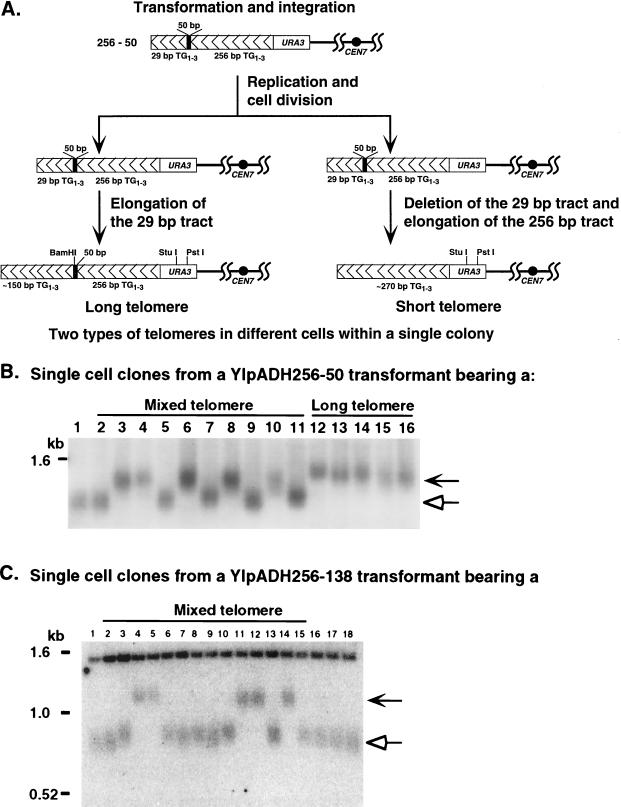
FIG. 4.
The transformants with mixed telomeres are colonies of cells with short or long telomeres. (A) Hypothesis for the formation of single transformant colonies bearing mixed telomeres and the predicted outcomes. After integration of the construction, the cell replicates the telomere and divides prior to either elongating the terminal 29 bp TG1-3 tract or deleting the terminal 29-bp tract and elongating the internal 256-bp tract. After these events, the telomeres are stably maintained in separate cells within the same colony. Subclones derived from single cells from these individual transformants bearing two types of telomeres should have either short or long telomeres but never both. (B) Genomic DNAs from single colonies derived from a single YIpADH256-50 transformant bearing either mixed telomeres (lanes 2 to 11) or only long telomeres (lanes 12 to 16). DNAs were cleaved with PstI, which cleaves in URA3 (see panel A) and analyzed as for Fig. 2. Lane 1 is DNA from a cell transformed with YIpADH35. (C) Genomic DNAs from a single colony derived from a single YIpADH256-138 transformant bearing long and short telomeres (Fig. 3A, lane 14) cleaved with StuI and analyzed as for Fig. 2 (lanes 2 to 15). Lane 1 is DNA from a cell transformed with YIpADH35; lanes 16 to 18 are single-cell subclones of the short telomere transformant in Fig. 3A, lane 12. In panels B and C, the chevron-headed arrows indicate long telomeres that retain the nontelomeric spacer and hollow arrows indicate short telomeres that have lost the spacer. All transformants were in strain YM708.
RESULTS
Model systems for altering telomere length regulation.
Two classes of synthetic telomeres were constructed to investigate the mechanism of telomere length regulation. The first class involved separating an internal 256-bp tract of TG1-3 repeats from a terminal 29-bp TG1-3 tract with a nontelomeric spacer of various lengths (Fig. 1, the YIpADH256-XX and YIpADH652-42 telomeres). These nontelomeric spacers contained no Rap1p binding sites. After transformation into yeast and conversion of the synthetic telomere to a chromosomal telomere (Fig. 1A), the 29-bp TG1-3 tract will be elongated to 325 ± 75 bp of TG1-3 (40) if the internal tract is not seen as part of the telomere (i.e., not counted as telomeric TG1-3). However, if the internal tract is seen as part of the telomere, the 29-bp TG1-3 tract will not be elongated as much. These spacer telomeres allowed us to determine how a disruption in the continuous TG1-3 tract altered length measurement. These telomeres also tested the minimum size of the disruption that functionally separated the internal and terminal TG1-3 sequences to form a new internal boundary for the telomere. The orientation dependence of the internal TG1-3 tract on telomere length measurement was also examined (Fig. 1, YIpADH652-42).
The second class of synthetic telomeres, the YIpADHTEF telomeres (Fig. 1B), consisted of internal tandem arrays of high-affinity, nontelomeric Rap1p sites derived from the upstream activation sequence (UAS) of the yeast TEF2 gene (4) followed by a short 29-bp TG1-3 tract. As with the first class of telomeres, the 29-bp TG1-3 tract will be elongated to a length that depends on whether the internal nontelomeric Rap1p sites are counted as part of the telomere. These telomeres were used to define how Rap1p molecules are counted to measure telomere length. Because Rap1p sites in natural telomeres are irregularly spaced and overlapping, it was unknown if all of the bound Rap1p molecules participate in telomere length control. The YIpADHTEF telomeres provided a more defined system because the internal Rap1p sites were regularly spaced at three different intervals and none of the sites overlapped.
To measure the length of the terminal TG1-3 tract after conversion of each of these telomeres to chromosomal telomeres (Fig. 1A), genomic DNAs were digested with either StuI, which cuts in URA3, or StuI plus BamHI, which cuts between URA3 and the elongated TG1-3 tract (Fig. 2A). The difference between the lengths of the StuI and StuI-BamHI fragments gave the length of the terminal TG1-3 tract and showed how much elongation occurred in vivo (the question mark in Fig. 2A). The terminal restriction fragment of a single telomere is heterogeneous, giving rise to a disperse band (Fig. 2B, lanes 2, 4, and 6), because it represents a population of molecules which have different TG1-3 sequences at the very end of the chromosome (48). Therefore, the size of each telomere was measured at the most intense point of hybridization in the disperse band, which is the mode (and usually the average) of the telomere lengths in the band being analyzed. We shall refer to this telomere size as the modal telomere length.
An internal 256-bp TG1-3 tract can be partially counted as part of the telomere in both orientations and across a 50-bp spacer.
As described above, the first class of synthetic telomeres contained an internal 256-bp tract of TG1-3 sequences (Fig. 1B and C). The synthetic telomeres containing this sequence were named YIpADH256-XX, where XX is the 26-, 38-, or 50-bp nontelomeric spacer between the 256-bp tract and the terminal 29-bp tract (Fig. 1B). In these constructs, the terminal and internal TG1-3 tracts were in the same orientation. In addition, a telomere with the 256-bp tract in the opposite orientation followed by a 42-bp spacer and the terminal 29 bp TG1-3 tract, YIpADH652-42 (Fig. 1B), was also constructed.
Several transformants bearing either the 26- or 38-bp spacer telomeres were examined for telomere length measurement (representative transformants are shown in Fig. 2B). The terminal StuI restriction fragment was indistinguishable in length from the control YIpADH35 telomeres containing no disruption in the continuous TG1-3 tract (Fig. 2B, lane 2 versus lanes 4, 6, and 8). Digestion with BamHI collapsed the heterogeneous band to a sharper one; therefore, the BamHI site was still present (lane 6 versus lane 7). In addition, the presence of the BamHI site in the 26- and 38-bp spacer telomeres was also confirmed by PCR (see Materials and Methods). The StuI-BamHI fragments of the 26- and 38-bp spacer telomeres were slightly more diffuse than expected for a discrete fragment (lanes 5, 7, and 9). One possible explanation for this result is that since these spacers were very close to the chromosome end, the random lengthening and shortening reactions that occur during cell growth (48) may have eliminated the spacer and BamHI site in a small subpopulation of cells. However, since the heterogeneity of the band was reduced by BamHI digestion, the majority of cells retained the spacer. Thus, the 26- and 38-bp nontelomeric spacers did not prevent the cell from counting all of the internal 256 bp of TG1-3 sequences as part of the elongated TG1-3 tract.
The 50-bp spacer telomere gave rise to three types of telomeres when digested with StuI: short telomeres that were identical in length to the YIpADH35 telomere, long telomeres that were ∼150 bp longer than the control YIpADH35 telomeres, and telomeres that contained a mixture of these two lengths (mixed telomeres) (Fig. 2B and data not shown). Digestion with StuI and BamHI had no effect on the short telomeres, caused the long telomeres to collapse to a compact band, and caused the mixed telomeres to form a compacted band superimposed on the short telomere band. Thus, the long telomeres contained the BamHI site and were the only ones that retained the entire 50-bp spacer (verified below). Analysis of the long telomeres showed that the 50-bp spacer allowed ∼130 bp of the internal 256-bp TG1-3 tract to be counted since the terminal tract was extended to only ∼150 bp, versus the 280-bp extension for the YIpADH35 telomere on this blot (Fig. 2B). By PCR assay and Southern blotting, the short telomeres were shown to lack the 50-bp spacer (data not shown; see Materials and Methods). We show below that the mixed telomeres were from individual transformants that formed colonies with single cells bearing either a short or a long telomere. These results show that the internal TG1-3 tract was counted across a 50-bp spacer but not as efficiently as across the shorter 26- and 38-bp spacers.
DNA from cells bearing the telomere containing the 256-bp tract in the opposite orientation with respect to the elongated TG1-3 sequences, YIpADH652-42 (Fig. 1B), gave StuI restriction fragments that were slightly longer than the YIpADH35 telomere (Fig. 2B). Of the four transformants examined, three were ∼70 bp longer and one was ∼170 bp longer than the YIpADH35 telomere with no spacer (Fig. 2B; isolates that are ∼70 [lane 18] and ∼170 [lane 16] bp longer are shown). The average length of the StuI fragment of the four YIpADH652-42 telomeres was thus 95 bp greater than that of the StuI fragment of the YIpADH35 telomere. Since the elongated TG1-3 tract of the YIpADH652-42 telomeres were not as long as the elongated TG1-3 of the YIpADH35 telomere, some of the internal TG1-3 sequences were counted as part of the elongated TG1-3 tract. The length of the YIpADH35 telomere averaged ∼270 bp on blots used for these measurements (Materials and Methods and data not shown). We therefore concluded that ∼175 bp of the internal TG1-3 sequences in the opposite orientation were counted as part of the elongated TG1-3 tract. Results with the 50-bp spacer telomere and with the YIpADHTEF telomeres (described below) suggested that the partial counting of the internal TG1-3 tract was due to the length of the 42-bp spacer and not the reverse orientation of the 256-bp tract. Thus, internal TG1-3 sequences in the reverse orientation were counted as part of the telomere.
The internal 256-bp TG1-3 tract is not counted as part of the telomere across a 138-bp spacer.
Because the 50-bp spacer allowed only ∼130 bp of the internal 256 bp TG1-3 tract to be counted as part of the telomere, larger spacers were constructed to determine how large a disruption was required to abrogate counting of the internal TG1-3 sequence as part of the TG1-3 tract and establish a new internal boundary for the telomere (i.e., a telomere-nontelomere junction). Nontelomeric DNA spacers of 138 and 266 bp, which did not contain Rap1p sites, were constructed (Fig. 1B and C) and then introduced into yeast.
The majority of transformants bearing the 138- or 266-bp spacer telomeres contained both short and long telomeres (Fig. 3A and B). As with the 50-bp spacer, only the long telomeres retained the entire spacer and BamHI site (Fig. 3; see also below). Therefore, only the long telomeres that retained the BamHI site were used to determine the effects of 138- and 266-bp spacers on length measurement. Measurement of the long telomeres bearing the 138- or 266-bp spacers (marked with chevron arrows in Fig. 3A and B) revealed that the elongated TG1-3 tract was only 20 and 10 bp shorter, respectively, than the average TG1-3 tract of the YIpADH35 control telomeres bearing a continuous TG1-3 tract. The modal lengths of seven independent YIpADH35 telomeres (measured at the point of the most intense hybridization of the telomeric band) were determined and found to vary with a range of ±30 bp. This range (the gray box in Fig. 3C) encompassed the small modal length differences between the elongated TG1-3 tracts of the long 138- and 266-bp spacer telomeres and the TG1-3 tract of the YIpADH35 telomere (Fig. 3C). Thus, the elongated tracts for the 138- and 266-bp spacer telomeres were indistinguishable in length from control telomeres with no internal 256-bp TG1-3 tract. We therefore concluded that the 138-bp spacer prevented the internal TG1-3 tract from being counted as part of the telomere and so established a new telomere-nontelomere junction and functionally separated the telomere from the subtelomeric sequences.
Telomere processing may not occur immediately after integration of the synthetic telomere.
Because some single transformants contained mixed telomeres (Fig. 2B, 3A, and 3B), it appeared that a single transformation event had given rise to a colony containing individual cells with either short or long telomeres. One hypothesis to explain these data is that integration of the synthetic telomere construct and its conversion to a chromosomal telomere by the cellular machinery did not occur in the same cell cycle (Fig. 4A). Yeast telomeres as short as ∼30 bp are sufficient for chromosomal maintenance since tel1Δ hdf1Δ cells have telomeres in this size range and are viable (37). After the synthetic telomere was integrated, the cell may have divided to produce two progeny before altering the length of the terminal 29-bp TG1-3 tract. In these two progeny cells, one cell may have elongated the 29-bp tract to give rise to the long telomeres, while the other cell may have degraded the terminal TG1-3 tract (and thus the BamHI site in the spacer) and formed a telomere by elongating the internal 256-bp TG1-3 tract to give rise to the short telomeres. In this model, the short and long telomeres exist in separate cells within the colony and are stable once formed after the first few cell divisions. A second, alternative model is that the telomere is immediately formed after integration and long telomeres undergo deletion events during colony growth to form the short telomeres. In the first model, single-cell subclones derived from the mixed telomere colonies should have either short telomeres or long telomeres but not both. Long telomeres should be stable to subsequent growth, and so transformants with long telomeres should yield subclones with only long telomeres (Fig. 4A). In the second model, where the long telomeres undergo deletion events, cells with long telomeres should frequently give rise to subclones that have mixed telomeres.
Twenty single-cell subclones (ten each isolated from two YIpADH256-50 colonies that contained mixed telomeres similar to the transformant in Fig. 2B, lane 12) were examined for telomere length. The clonal lines derived from the mixed telomere colonies were of two types: some had only short telomeres, while others had only long telomeres (10 clones derived from a single colony are shown in Fig. 4B, lanes 2 to 11). No clonal line derived from a single cell had either mixed telomeres or telomeres of intermediate length. In addition, we examined five single-cell clones derived from a colony with only short telomeres and five single-cell clones derived from a colony with only long telomeres. The clonal lines derived from the short-telomere colony contained only short telomeres (not shown), and the lines derived from long-telomere colonies contained only long telomeres (Fig. 4B, lanes 12 to 16). Subclones from cells bearing long telomeres grown for 40 generations gave rise to cells that had only long telomeres (data not shown and reference 38), indicating that the long telomeres were stably maintained. Of 15 long-telomere YIpADH256-50 clones examined, none contained mixed telomeres (Fig. 4B and data not shown). Therefore, the short telomeres in the mixed-telomere colonies did not arise from deletion events that shorten the long telomeres during colony growth. These data agreed with the first hypothesis (Fig. 4A) and strongly suggested that telomere formation occurred after integration of the transforming DNA and cell division.
Formation of mixed telomere colonies also occurred with the larger 138- and 266-bp spacers (Fig. 3A and B). Four of the seven transformants bearing the 266-bp spacer contained mixed telomeres, as did four of the seven YIpADH256-138 transformants. The mixed telomeres in these transformants were also due to single cells containing either short or long telomeres (shown for YIpADH256-138 in Fig. 4C). No single cells bearing a mixed telomere or telomere of intermediate length were isolated. Interestingly, one of the 138-bp spacer telomere mixed transformants was not cleaved by BamHI (Fig. 3A, lanes 8 and 9) but did retain the spacer (shown below). This unusual transformant deleted the BamHI site but not the rest of the spacer before forming the long telomere.
The short telomeres in the mixed transformants could have arisen by complete deletion of the spacer (Fig. 4A) or deletion of all but ≤38 bp of the spacer, which would not interfere with counting the internal TG1-3 sequences (Fig. 2B). To determine if a portion of the spacer was retained in some of the short telomeres, a PCR method using a URA3 primer and primer 1, which hybridizes to the junction of the spacer and the internal 256 bp TG1-3 tract (bases 301 to 318 of Fig. 1C), was developed to test for the presence of the most internal 12 bp of the spacer (Materials and Methods). As expected, all eight long telomeres tested in this assay (seven bearing the 50-bp spacer and one bearing the 138-bp spacer) and five mixed telomeres (two bearing the 50-bp spacer and three bearing the 138-bp spacer, including the transformant in Fig. 3A, lane 8) retained these 12 bp of the spacer. In contrast, none of the 16 short telomeres tested (2 YIpADH35 telomeres, 6 short telomeres derived from the 50-bp spacer, and 8 derived from the 138 bp-spacer constructs) retained the internal 12 bp of the spacer. These short telomeres included eight short telomere subclones derived from three original transformants bearing mixed telomeres, indicating that the short telomere in these mixed telomere colonies lacked the entire spacer. Given that all of the short telomeres lacking the BamHI site in Fig. 2 and 3 were in the same size range, the most likely possibility is that all of them had lost the entire spacer.
The hypothesis that explains the mixed telomeres in the YIpADH256-50, -138, and -266 transformants (Fig. 4A) predicts that the 26- and 38-bp spacers should also have formed colonies with mixed telomeres. Of the eight colonies bearing the 26- or 38-bp spacer telomeres examined, two of the four 26-bp spacer and three of the four 38-bp spacer telomere transformants had telomeric restriction fragments that did not become less heterogeneous after digestion with StuI plus BamHI (data not shown), indicating that the majority of cells in these four transformants lacked the BamHI site. However, some cells in each of these transformants had the BamHI site as assayed by PCR. Therefore, the 26- and 38-bp spacer telomeres also gave rise to mixed colonies in the same way as the 50-, 138-, and 266-bp spacer telomeres did.
Mixed colonies were not observed in YIpADH652-42 transformants bearing the 42-bp spacer telomere with the 256-bp tract in the opposite orientation (data not shown). This result may reflect the observation that telomere sequences in the opposite orientation are poor substrates for telomere formation (36), and so degradation past the spacer would not allow telomere formation and would not yield a viable transformant.
Cells measure one Rap1p molecule as 19 bp of TG1-3 sequences in vivo.
The irregular spacing of Rap1p sites within yeast telomere sequences makes it unclear whether all Rap1p sites are counted to monitor telomere length, and no experiments have yet determined whether all Rap1p molecules are counted in vivo. This information is particularly important because a model of yeast telomere length regulation predicts that several Rap1p molecules at the telomere-nontelomere junction do not participate in length regulation (3, 8). To determine how many Rap1p molecules are counted to measure telomere length, we varied the spacing between six tandem Rap1p sites at the telomere-nontelomere junction and determined the effect on telomere length. The fact that a 26- or 38-bp nontelomeric spacer does not disrupt counting (Fig. 2 and 3) indicates that the cell can accommodate variable spacing between Rap1p sites. This consideration indicates that if six Rap1p molecules are arranged in regular arrays with different spacing, then they should be counted as a constant length of TG1-3 sequences even though the total DNA length of the Rap1p binding site array varies.
Synthetic telomeres containing arrays of six nontelomeric Rap1p sites spaced once every 35, 18, or 13 bp were constructed (YIpADHTEF telomeres [Fig. 5A]). Each repeat in the array contained a high-affinity, 13-bp Rap1p site from the TEF2 UAS (4) (Materials and Methods). The 1-site-per-35-bp spacing was chosen to match the frequency of exact matches to the Rap1p consensus in TG1-3 sequences (48), the 1-site-per-18-bp spacing was chosen to match the frequency of in vitro Rap1p binding to TG1-3 sequences (7), and the 1-site-per-13-bp spacing was chosen because molecular model building based on the crystal structure of the Rap1p DNA binding domain suggests that Rap1p molecules can bind as closely as once every 11 to 12 bp (17). The spacing between the 3′ end and 5′ start of each Rap1p binding site in the phased arrays was 22, 5, or 0 bp for the sites spaced every 35, 18, or 13 bp, respectively. Since these spacings were shorter than the 26-bp spacer that did not affect the counting of internal TG1-3 sequences (Fig. 2B), the nontelomeric DNA between the Rap1p sites should not affect counting of Rap1p molecules. Cells bearing control telomeres with no insert (the YIpADH35 telomeres [Fig. 1B]) or telomeres containing lambda DNA inserts the same size as the TEF inserts, (the YIpADHλ telomeres [Fig. 5A]) were also constructed.
FIG. 5.
Cells count one Rap1p molecule as ∼19 bp of TG1-3. (A) Telomere constructions contained either phased arrays of Rap1p sites internal to the 29 bp TG1-3 tract or the same-size fragment of λ DNA. The orientation of the Rap1p site array is indicated by the arrowheads as in Fig. 1. The locations of the BamHI site and the URA3 fragment used to probe the Southern blots are shown. (B) Genomic Southern analysis of telomeres bearing six Rap1p sites spaced one site every 18 bp in strain KR36-6L. DNAs were digested with StuI or StuI plus BamHI and analyzed by Southern blotting using the URA3 probe shown in panel A. Two YIpADHλ108 telomeres were included as negative controls for telomeres bearing inserts of equal size with no Rap1p sites. The leftmost lane of B and C is DNA from the untransformed KR36-6L strain; 35 denotes YIpADH35 (Fig. 1B). (C) Genomic Southern analysis of telomeres bearing six Rap1p sites spaced one site every 35 bp in strain KR36-6L analyzed as for panel B (reprinted from reference 38 with permission). A YIpADHλ210 telomere was included as a negative control for an insert of equal size with no Rap1p sites. (D) Genomic Southern analysis of telomeres in strain KR36-6L bearing six Rap1p sites spaced one site every 35 bp. All DNAs were digested with StuI. All lanes are from the same gel. (E) Genomic Southern analysis of telomeres in strain KR36-6L bearing six Rap1p sites spaced one site every 13 bp. A YIpADHλ88 telomere was included as a negative control for an insert of nearly equal size with no Rap1p sites.
If the cell considers the TEF insert as part of the telomere and the Rap1p site spacing in the TEF insert is close to the spacing of the counted Rap1p molecules in the TG1-3 repeats, then the sizes of the elongated YIpADH35 and YIpADHTEF telomeric StuI fragments will be the same. If Rap1p molecules alone are insufficient for the cell to consider the TEF inserts as part of the telomere, then the sizes of the elongated YIpADHTEF and elongated YIpADHλ StuI fragments will be the same.
These synthetic telomeres (Fig. 5A) were transformed into yeast, and Southern blot analysis of at least two independent transformants of each type was performed to determine telomere length. In all cases, the YIpADHTEF StuI fragments were shorter than the control YIpADHλ StuI fragments (Fig. 5B to E). Thus, the array of nontelomeric Rap1p sites reduced the length of the elongated TG1-3 tract when present in either orientation, and so Rap1p molecules bound to non-TG1-3 sequences were counted in both orientations as part of the telomere. Therefore, Rap1p molecules, and not TG1-3 sequences, served as the metric for telomere length regulation.
The StuI terminal restriction fragments of YIpADH35 and YIpADHTEF18-6TG and YIpADHTEF18-6CA were nearly identical in length (Fig. 5B). Therefore, the 108-bp array of six nontelomeric Rap1p sites spaced one every 18 bp functionally replaced the same length of TG1-3 sequences. The length (in base pairs) of TG1-3 that the cell equated with the array of Rap1p molecules could be determined by comparing the modal telomere length of the elongated TEF telomere to the modal length of the λ negative control telomere. The lengths of the TEF and λ telomeres were determined by measuring the point of most intense hybridization. The difference between the average length of the YIpADHTEF18-6 StuI fragments (which contained an insert counted by the cell as TG1-3 repeats) and the average length of the YIpADHλ108 StuI fragments (which contained an insert not counted by the cell as TG1-3) revealed the length in base pairs of TG1-3 that the cell equated with the six Rap1p molecules. The Rap1p arrays were counted as 114 bp (YIpADHTEF18-6TG) or 90 bp (YIpADHTEF18-6CA) of TG1-3 (Table 2). Thus, one Rap1p molecule was counted as either 19.0 or 15.0 bp of TG1-3 DNA for these telomeres.
TABLE 2.
Differently spaced, tandem Rap1p sites are counted as similar lengths of TG1-3 sequences in the YIpADHTEF telomeres
| TEF repeat(s) | na | bp/Rap1p site | Length of tandem sites (bp) | Equivalent bp of TG1-3b | bp of TG1-3/Rap1pc |
|---|---|---|---|---|---|
| 35-6TG | 4 | 35 | 210 | 124 (9) | 20.7 |
| 35-6CA | 4 | 35 | 210 | 128 (0) | 21.3 |
| 18-6TG | 3 | 18 | 108 | 114 (5) | 19.0 |
| 18-6CA | 2 | 18 | 108 | 90 (0) | 15.0 |
| 13-6TG | 2 | 13 | 78 | 106 (5) | 17.7 |
| 13-6CA | 2 | 13 | 78 | 103 (0) | 17.2 |
| All TGd | 9 | NAe | NA | 115 | 19.2 |
| All CAd | 8 | NA | NA | 107 | 17.8 |
| All repeatsd | 17 | NA | NA | 111 | 18.5 |
Number of independent telomeres measured for each construction.
Determined by subtracting the average modal YIpADHTEFXX-6 StuI fragment length from the appropriate average modal YIpADHλXX StuI fragment length. Standard deviations of length measurements for the YIpADHTEF StuI fragments in base pairs are given in parentheses.
Value from the fifth column divided by 6 (number of Rap1p sites per TEF repeat).
Average of telomeres with different spacing between Rap1p sites described above.
NA, not applicable, as multiple constructs are being considered.
The YIpADHTEF35-6TG and -CA telomeres also possessed elongated, terminal TG1-3 tracts that were shorter than the control YIpADH35 terminal TG1-3 tract (Fig. 5C and D). In these cases, the length of the array of Rap1p sites was 210 bp but was counted as 124 bp (TG orientation) or 128 bp (CA orientation) of TG1-3 in comparison to the YIpADHλ210 telomere (Fig. 5C and D; Table 2). These values indicate that one Rap1p molecule was counted as 20.7 or 21.3 bp of TG1-3 in the TEF35-6 telomeres. These data show that when the cell measures telomere length, it can efficiently compensate for variations in spacing between Rap1p sites that exceed the equivalent number of base pairs of TG1-3 per Rap1p molecule.
The arrays of six Rap1p molecules spaced one every 13 bp in the YIpADHTEF13-6TG and -CA telomeres were counted as either 106 bp (TG orientation) or 103 bp (CA orientation) of TG1-3 (Fig. 5E; Table 2). These results suggested that one Rap1p was equivalent to 17.7 or 17.2 bp of TG1-3 DNA in these telomeres. In this case, even though the molecules were more closely spaced than the average spacing of Rap1p binding sites in TG1-3 determined in vitro (7), the cell counted the six Rap1p molecules as nearly the same length of TG1-3 DNA as when the Rap1p sites were spaced one every 18 or 35 bp.
Taken together, these data show that the cell can accommodate a variable spacing of Rap1p molecules to measure a given length of telomeric DNA. In all cases, the elongated tract of TG1-3 sequences was shorter by 106 to 124 bp when the TEF array was in the same orientation as the TG1-3 repeats and 90 to 128 bp when the TEF array was in the opposite orientation (Table 2), even though the size of the TEF array varied (from 78 to 210 bp). The average number of bp of TG1-3 the cell equated with one Rap1p molecule was 19.2 bp for sites in the correct orientation, 17.8 bp for sites in the opposite orientation, and 18.5 bp for all sites tested here. These data indicate that for the six Rap1p molecules at the telomere-nontelomere junction, each Rap1p molecule was counted as ∼19 bp of TG1-3.
Regularly spaced arrays of internal Rap1p sites eliminate telomere length heterogeneity between individual transformants.
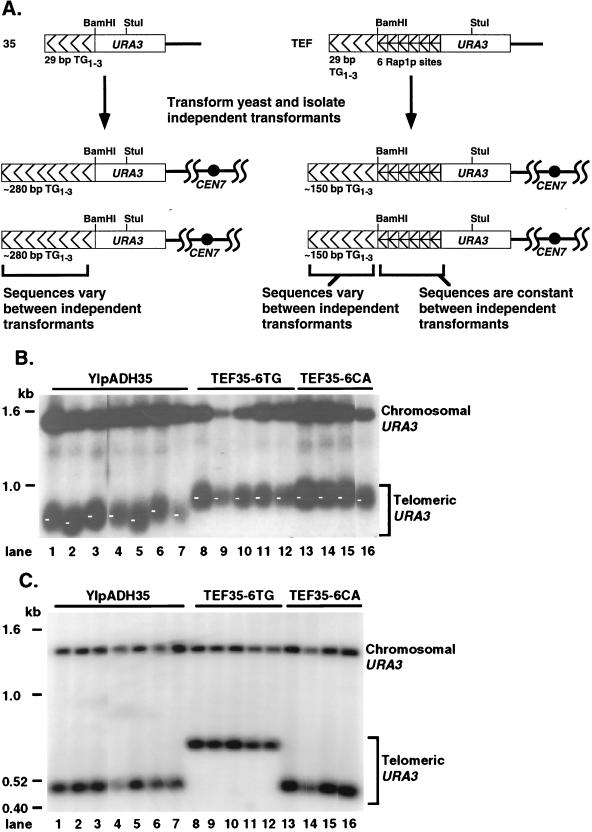
The terminal chromosome restriction fragment gives a dispersed band on Southern blots because the length of a specific telomere fragment in individual cells differs. An internal restriction fragment gives a sharp band because it is the same length in all cells (e.g., the telomeric StuI restriction fragments versus the StuI-BamHI fragments in Fig. 5). This terminal restriction fragment length heterogeneity is due to sequence differences between the terminal TG1-3 tracts of different telomeres (48). An additional heterogeneity is observed between telomeres formed in independent transformants, because when new synthetic telomeres are formed, the sequence of all but the first 11 bp of TG1-3 added is random (18, 48) (Fig. 6A).
FIG. 6.
Phased arrays of Rap1p sites eliminate the length variation between transformants. (A) Structures of steady-state telomeres in independent transformants derived from the YIpADH35 (35) and YIpADHTEF (TEF) constructions. When telomeres are formed with YIpADH35, 90% of the telomere consists of newly synthesized TG1-3 repeats. Since the TG1-3 sequences added after the first 11 bp are random (18), each of the independent YIpADH35 transformants will have a different telomere sequence near the telomere-nontelomere junction. In contrast, all of the YIpADHTEF telomeres retain the same nontelomeric Rap1p sites at the junction. The terminal 120 to 150 bp of TG1-3 sequences for both the YIpADH35 and YIpADHTEF telomeres are randomized during cell growth (48), and so the only differences between the YIpADH35 and YIpADHTEF telomeres are the sequences near their telomere-nontelomere junctions. (B) Genomic DNAs digested with StuI from seven independent YIpADH35 transformants, five independent YIpADHTEF35-6TG transformants, and four independent YIpADHTEF35-6CA transformants (all in strain KR36-6L) were analyzed by Southern blotting as for Fig. 5. All lanes are from the same gel. The length of each telomere was measured at the point indicated by the small white bar in each lane, which is the point of most intense hybridization (the modal telomere length). The lengths of the YIpADH35 telomeres varied over a ∼60-bp range, while the lengths of most of the YIpADHTEF35-6 telomeres were within a ∼10-bp range (see text). (C) The same DNAs as in panel B were digested with StuI and BamHI to show that the length heterogeneity between the independently formed telomeres was due to different lengths of the terminal TG1-3 sequences.
To examine modal length variation in telomeres where almost all of the TG1-3 sequences came from new synthesis, telomeres from seven independent YIpADH35 transformants were compared on the same blot. The size of the most intensely hybridizing portion of these terminal restriction fragments, i.e., the modal length, varied over a ∼60-bp range (Fig. 6B; similar length differences can also be observed in Fig. 5D). This heterogeneity was due to variation in the length of the TG1-3 sequences because the length of the internal URA3 StuI-BamHI fragment was the same in all YIpADH35 transformants (Fig. 6C).
In contrast to the YIpADH35 results, the array of six internal Rap1p molecules eliminated much of the variation in modal telomere lengths between individual transformants. All five of the YIpADHTEF35-6TG telomeres and three of four YIpADHTEF35-6CA telomeres had lengths that only varied over an ∼10-bp range (Fig. 6B). The length of the fourth YIpADHTEF35-6CA telomere (Fig. 6B, lane 16) was within ∼20 bp of the average TEF35-6 StuI fragment length. This elimination of modal telomere length variation was significant given that the elongated TG1-3 repeats on each of the independently formed TEF telomeres should have a different terminal TG1-3 sequence, because the sequence of the added TG1-3 repeats should have been random (18) and changed as the colony grew (48) while the internal sequences were maintained (Fig. 6C). A similar lack of modal telomere length heterogeneity was also observed among eight individual YIpADHTEF18-6 transformants and among eight individual YIpADHTEF13-6 transformants (Fig. 5B and E and data not shown). Thus, the modal telomere length heterogeneity between individual YIpADHTEF transformants, each bearing the same sequences at the telomere-nontelomere junction, was significantly reduced compared to the length heterogeneity of YIpADH35 telomeres, each bearing different TG1-3 sequences at the junction. These results showed that the conserved spacing of six Rap1p molecules at the telomere-nontelomere junction in different transformants had a significant effect on steady-state telomere length. These results indicate that the change in modal telomere length was due to TG1-3 sequences near the telomere-nontelomere junction whereas the disperse nature of the terminal restriction fragment was due to sequence variation in the terminal TG1-3 repeats.
DISCUSSION
The placement of TG1-3 sequences or nontelomeric Rap1p sites adjacent to the telomeric TG1-3 tract caused yeast cells to maintain the TG1-3 tract at a shorter equilibrium telomere length. These results indicated that yeast measure telomere length by counting Rap1p molecules instead of TG1-3 sequences. Yeast detected internal TG1-3 sequences as part of the telomere, or counted the internal sequences, across a 50-bp spacer. However, only part of the internal TG1-3 tract was counted across a spacer larger than 38 bp, and a 138-bp nontelomeric spacer was sufficient to establish a new internal boundary and separate the telomere from the subtelomeric sequences. Cells counted each of the six telomeric Rap1p molecules in the TEF telomeres as ∼19 bp of TG1-3 in vivo. Interestingly, the conserved spacing of Rap1p molecules near the telomerenontelomere junction in different transformants eliminated the modal length heterogeneity commonly observed between independent telomere formation events.
Cells transformed with the YIpADH256-XX telomere formed colonies containing individual cells bearing either short or long telomeres (Fig. 2 to 4), and these telomere lengths were stably maintained over many divisions (Fig. 4; data not shown; reference 38). The mixed transformants occurred at high frequency and with all nontelomeric spacers, suggesting that the events that gave rise to them are normal cellular processes. A hypothesis consistent with these findings is that after integration, the original telomere construct was transiently stable for one or more cell divisions before either the terminal 29-bp TG1-3 tract was elongated or the terminal 29-bp TG1-3 tract was deleted and the internal 256-bp tract was lengthened (see Results and Fig. 4A). The high frequency of mixed transformants in the YIpADH256-138 and -266 telomeres (Fig. 3A and B) suggests that most integration events can give rise to a mixed transformant. Therefore, these results strongly suggest that telomere formation was not completed in the first cell cycle after integration.
The phased array of Rap1p sites at the telomere-nontelomere junction in the YIpADHTEF telomeres showed that the spacing between the six internal Rap1p molecules could be varied widely and still be counted as nearly the same length of TG1-3 sequences, such that 78 to 210 bp of nontelomeric Rap1p sites was counted as 90 to 128 bp of TG1-3. The results presented here indicate that yeast equated each Rap1p molecule with ∼19 bp of TG1-3 (Table 2). In vitro experiments by others have shown that Rap1p bound to cloned telomeric sequences at a frequency of slightly greater than one molecule per 18 bp of TG1-3 (7). The fact that the frequency of Rap1p binding sites in vitro and the equivalence of Rap1p for a given amount of TG1-3 sequences in vivo is so close indicates all of the Rap1p molecules near the telomere DNA-nontelomere DNA junction participate in telomere length measurement.
All three arrays eliminated the modal length heterogeneity between individual transformants (Fig. 5 and 6). Thus, all three arrays had similar overall effects on the cellular telomere length measurement apparatus in that the most internal Rap1p molecules can affect the lengthening and shortening reactions that occur at the chromosome end. These data indicate that the chromosome end and telomere-nontelomere junction must somehow communicate when telomere length is measured.
While this work was in progress, Marcand et al. showed that internal TG1-3 sequences or fusion proteins containing the Gal4p DNA binding domain and the Rap1p C terminus tethered internal to the terminal TG1-3 tract could cause telomeres to be maintained at a shorter equilibrium length (29). They concluded that telomere length is regulated by a negative feedback mechanism that counts Rap1p molecules. Their data and ours are consistent with a wide variety of previous genetic experiments that showed that titration of telomere components from chromosome ends altered telomere length regulation (40) and that the Rap1p C terminus tethers these components to telomeres (5, 13, 19, 20, 25, 51, 52). The use of the Gal4p-Rap1p fusion by Marcand et al. required that only low levels of the protein be produced because overexpression of the Rap1p C terminus causes telomere lengthening (5, 12), most likely by titrating away negative telomere length regulatory components (13, 52). Otherwise, telomere lengthening caused by overproduction of the Rap1p C terminus would mask the telomere shortening caused by tethering Rap1p C termini to telomeric sites. This low level of fusion protein raised the possibility that the Gal4p binding sites were not completely occupied (footnote 18 in reference 29), preventing quantitative analysis of the number of bp of TG1-3 that the cell equates with one Rap1p. In contrast, our approach was quantitative because the nontelomeric Rap1p sites in this work did not perturb the cellular levels of telomere length regulatory components. Thus, the length change of these telomeres (Fig. 5) was due only to the arrangement of Rap1p molecules at the telomere-nontelomere junction. In addition, comparing the modal lengths of the YIpADHTEF telomeres with the control YIpADHλ telomeres allowed us to determine that the cell equates one Rap1p molecule with ∼19 bp of TG1-3 in vivo (Table 2). Since the spacing of Rap1p sites is one per 18 bp of TG1-3 in vitro (7), our results indicate that all six of the arrayed Rap1p molecules at the telomere-nontelomere junction participated in telomere length measurement.
Models for telomere length measurement in yeast need to account for the partial counting of the internal 256-bp TG1-3 tract across the 42- and 50-bp spacers (Fig. 2B), for a 138-bp nontelomeric spacer establishing a new internal telomere-nontelomere junction (Fig. 3A and C), for all Rap1p molecules near the telomere-nontelomere junction participating in telomere length measurement, and for the effect of these Rap1p molecules on telomere length (Fig. 5 and 6). In addition, a wide variety of studies in S. cerevisiae and K. lactis indicate that the Rap1p C terminus at the ends of established telomeres plays an important role in limiting telomere elongation (5, 12, 19, 20, 25). Finally, the Rap1p C terminus at the telomere-nontelomere junction plays a role in the transcriptional silencing of genes near the telomere by nucleating a complex of proteins that then spreads internally by coating chromatin with Sir3p (15, 27, 39, 44). Thus, a model for yeast telomere length regulation must accommodate these different functions.
One current model for telomere structure suggests that the telomere is divided into counted and uncounted regions where the uncounted region consists of Rap1p molecules near the telomere-nontelomere junction bound by Sir3p and Sir4p (3, 8). This model is inconsistent with our data showing that all Rap1p molecules at the telomere-nontelomere junction are counted by the cell. The recent data of Marcand et al. (29) are also inconsistent with this model. Their studies showed that tethering just two Rap1p C termini at the telomere-nontelomere junction can cause the elongated TG1-3 tract to be maintained at a length ∼30 bp shorter than that of control telomeres (29), indicating that at least one or possibly both of these Rap1p C termini participate in telomere length measurement. Given our results that each Rap1p molecule at the telomere-nontelomere junction is counted as ∼19 bp of TG1-3 (Table 2), both of these Gal4-Rap1p fusions must have participated in telomere length measurement. However, data from several labs indicate that the Sir3p-Rap1p interaction is required to establish silencing of genes near telomeres (15, 27, 33, 44). A simple explanation to accommodate these two distinct Rap1p functions is to propose that they are temporally separated, occurring at different times in the cell cycle.
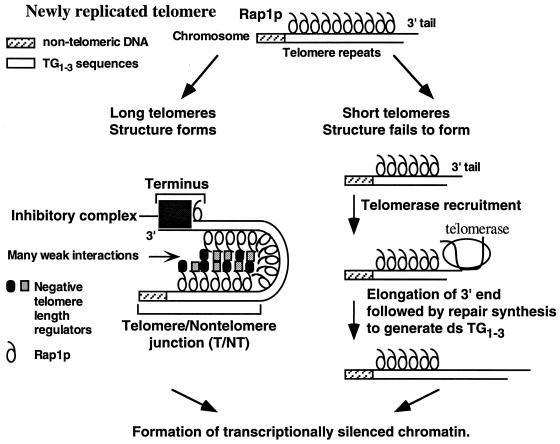
Our working model for telomere length measurement is that after telomeres have been replicated, Rap1p molecules are counted by the formation of a folded telomere structure that is dependent on many weak interactions between Rap1p and negative regulators of telomere length (Fig. 7). This model of the yeast telomere as a highly folded structure is consistent with previous studies showing that the chromatin that includes yeast TG1-3 repeats is a single unit as defined by micrococcal nuclease digestion (53). In addition, Rap1p binding causes a 90° to 100° bend in DNA in vitro (7), and ∼18 Rap1p molecules should be present in a 325-bp telomere, giving rise to a highly folded structure. We hypothesize that many interactions between Rap1p and other proteins constrain the folding of the TG1-3 sequences so that the chromosome end is brought close to the telomere-nontelomere junction (Fig. 7). Formation of this structure then blocks telomere elongation because the structure itself blocks telomerase access to the chromosome end, or the structure recruits an inhibitory complex that blocks telomerase access. If the structure is not formed, telomere elongation by telomerase may occur. We propose that subsequent to structure formation and elongation of short telomeres (Fig. 7), the yeast Ku proteins may rebind to the chromosome terminus (10) and recruit Sir proteins (46) to form the heterochromatin complex of Sir proteins and Rap1p involved in telomeric silencing. Once silencing is established, genes would then become refractory to transcriptional activation by transactivators.
FIG. 7.
A working model for telomere length regulation. Telomere length is monitored by counting Rap1p molecules to keep telomeres within a set range of lengths. We propose that Rap1p molecules are counted by the formation of a transient, highly folded three-dimensional structure stabilized by many weak interactions between Rap1p molecules and negative regulators of telomere length (drawn here as a hairpin for simplicity). When the newly replicated TG1-3 repeats are 325 ± 75 bp, a highly folded structure that links the telomere-nontelomere junction to the chromosome terminus forms. This structure blocks telomerase access to the chromosome terminus, and thus telomere elongation, either by sequestering the end or by recruiting an inhibitory complex (e.g., a complex formed between the Rap1p C terminus, Cdc13p, and Stn1p [9, 34]). When telomeres are short, the structure is not formed, no inhibition occurs, and telomeres are elongated. Lengthening of short telomeres would be enhanced by the Rap1p molecules which are no longer sequestered by the folded structure (38). Subsequent to these events, telomeric heterochromatin is formed and maintained until the next S phase. A simpler model that linked the telomere-nontelomere junction to the chromosome terminus without counting the intervening sequences had been previously proposed for K. lactis telomeres (31).
A number of observations regarding yeast telomere function support our model in limiting telomere length measurement and possible telomere elongation to a period late in S phase that is temporally separated from the establishment of telomere position effect or silencing. Telomere position effect can switch between transcriptionally repressed and active states, and the repressed state can be overcome by transcriptional activators only in S phase (1), indicating that telomeric gene silencing is not stably established at this point in the cell cycle. Late in S phase is when telomeric DNA is replicated (30) and a 50- to 150-bp 3′ extension of the TG1-3 strand is formed (50); therefore, the telomere repeats are undergoing length changes at this time. Slow passage through S phase increases telomeric silencing (22), which is consistent with this hypothesis (Fig. 7) because an increased amount of time after telomere length measurement would allow assembly of telomeric heterochromatin before the chromosome is refolded and the cell cycle proceeds. So the hypothesis that length regulation occurs only in late S phase just after telomeres are replicated, and that afterwards a stable, transcriptionally silenced heterochromatin structure is established and maintained until the next S phase, is consistent with the current data on telomere replication and telomeric silencing.
Our model for telomere length regulation (Fig. 7) can explain all of the results presented here. First, the slight counting differences observed between the different TEF arrays (Table 2) would be due to one set of arrays (e.g., TEF35-6) allowing more efficient structure formation than another (e.g., TEF13-6) because the presentation of Rap1p molecules is slightly different for each array. The more compact Rap1p spacing in the TEF13-6 telomeres may make fewer protein-protein interactions required to form the structure and block telomerase access. As a result, a slightly longer terminal TG1-3 tract is required to introduce an additional Rap1p molecule to stabilize the structure. Second, all TEF telomeres have nearly the same modal telomere length because the positioning of the six Rap1p molecules at the telomere-nontelomere junction is the same in independent transformants. Therefore, the chromatin complex at the terminus (Fig. 7) will see the same chromatin arrangement at the telomere-nontelomere junction in each transformant. The remaining heterogeneity in the TEF telomeres, i.e., the dispersed band formed by the telomeric restriction fragment, results from individual cells having different terminal TG1-3 sequences (48). In contrast, telomeres composed of all TG1-3 sequences have different arrangements of Rap1p molecules at both the telomere-nontelomere junction and the chromosomal terminus in each transformant. Therefore, the chromatin structures of both the telomere-nontelomere junction and the terminus are different in each transformant; thus, structure formation differs in independent transformants, and the modal lengths show large variation (Fig. 6B). Finally, the partial or incomplete counting of the internal 256-bp TG1-3 tract across the 42- and 50-bp spacers (Fig. 3C) would occur because the nontelomeric sequences partially disrupt the protein-protein interactions between the chromosome terminus and the telomere-nontelomere junction and so the structure is not formed. Slight elongation of the terminal TG1-3 tract adds Rap1p-Rap1p interactions that overcome the disruption caused by the 42- or 50-bp spacer. In this model, the 138-bp spacer requires so much elongation of the terminal tract that lengthened TG1-3 repeats can interact more easily with themselves than with the internal tract. This explanation for partial counting can also explain the ∼50 bp of telomere shortening seen in sir3 and sir4 mutants (35). The binding of preexisting Rap1p-Sir3p-Sir4p complexes to random sites in the telomere just after DNA replication would interfere with structure formation in wild-type cells. In sir3 and sir4 mutants, structure formation would be more efficient because more Rap1p molecules can interact with negative regulators of telomere length, and so a shorter tract of TG1-3 and fewer Rap1p molecules would be required to form the structure that blocks telomere lengthening. The result would be slightly shorter telomeres in sir3 and sir4 cells.
While the heterogeneous telomeric sequences in yeast are distinct from the homogeneous repeats found in humans, both of these organisms could share folded, heterogeneous telomeric chromatin structures. Both Rap1p and the human telomeric binding protein TRF1 bend DNA and bind to DNA in a noncooperative fashion (2). Because the human telomeric sequences are homogeneous repeats, the exact positions where telomeric proteins bind within the sequence are not defined. Therefore, the precise arrangement of telomeric binding proteins in a homogeneous sequence can be different at individual telomeres. Thus, the chromatin structures for different human telomeres will be heterogeneous at the protein level, whereas yeast telomeres are also heterogeneous at the DNA sequence level. As both yeast and humans appear to use feedback mechanisms that count telomere binding proteins to measure telomere length (references 29 and 47 and this work), the results discussed here for yeast may apply to humans.
ACKNOWLEDGMENTS
We especially thank Kathleen Berkner and also Bryan Williams, Robert Silverman, Nilanjan Roy, and Rama Kota for comments on the manuscript and Kurt Hotmire for technical support.
K.W.R. was supported by a Junior Faculty Research Award from the American Cancer Society. This work was supported by NIH grant GM50752 to K.W.R.
REFERENCES
- 1.Aparicio O M, Gottschling D E. Overcoming telomeric silencing: a transactivator competes to establish gene expression in a cell cycle-dependent way. Genes Dev. 1994;8:1133–1146. doi: 10.1101/gad.8.10.1133. [DOI] [PubMed] [Google Scholar]
- 2.Bianchi A, Smith S, Chong L, Elias P, de Lange T. TRF1 is a dimer and bends telomeric DNA. EMBO J. 1997;16:1785–1794. doi: 10.1093/emboj/16.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brun C, Marcand S, Gilson E. Proteins that bind to double-stranded regions of telomeric DNA. Trends Cell Biol. 1997;7:317–324. doi: 10.1016/S0962-8924(97)01092-1. [DOI] [PubMed] [Google Scholar]
- 4.Buchman A R, Lue N F, Kornberg R D. Connections between transcriptional activators, silencers, and telomeres as revealed by functional analysis of a yeast DNA-binding protein. Mol Cell Biol. 1988;8:5086–5099. doi: 10.1128/mcb.8.12.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conrad M N, Wright J H, Wolf A J, Zakian V A. RAP1 protein interacts with yeast telomeres in vivo: overproduction alters telomere structure and decreases chromosome stability. Cell. 1990;63:739–750. doi: 10.1016/0092-8674(90)90140-a. [DOI] [PubMed] [Google Scholar]
- 6.Garvik B, Carson M, Hartwell L. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol Cell Biol. 1995;15:6128–6138. doi: 10.1128/mcb.15.11.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gilson E, Roberge M, Giraldo R, Rhodes D, Gasser S M. Distortion of the DNA double helix by RAP1 at silencers and multiple telomeric binding sites. J Mol Biol. 1993;231:293–310. doi: 10.1006/jmbi.1993.1283. [DOI] [PubMed] [Google Scholar]
- 8.Gotta M, Cockell M. Telomeres, not the end of the story. Bioessays. 1997;19:367–370. doi: 10.1002/bies.950190503. [DOI] [PubMed] [Google Scholar]
- 9.Grandin N, Reed S I, Charbonneau M. Stn1, a new Saccharomyces cerevisiae protein, is implicated in telomere size regulation in association with Cdc13. Genes Dev. 1997;11:512–527. doi: 10.1101/gad.11.4.512. [DOI] [PubMed] [Google Scholar]
- 10.Gravel S, Larrivee M, Labrecque P, Wellinger R J. Yeast Ku as a regulator of chromosomal DNA end structure. Science. 1998;280:741–744. doi: 10.1126/science.280.5364.741. [DOI] [PubMed] [Google Scholar]
- 11.Greider C W. Telomere length regulation. Annu Rev Biochem. 1996;65:337–365. doi: 10.1146/annurev.bi.65.070196.002005. [DOI] [PubMed] [Google Scholar]
- 12.Hardy C F J, Balderes D, Shore D. Dissection of a carboxy-terminal region of the yeast regulatory protein RAP1 with effects on both transcriptional activation and silencing. Mol Cell Biol. 1992;12:1209–1217. doi: 10.1128/mcb.12.3.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hardy C F J, Sussel L, Shore D. A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev. 1992;6:801–814. doi: 10.1101/gad.6.5.801. [DOI] [PubMed] [Google Scholar]
- 14.Hecht A, Laroche T, Strahl-Bolsinger S, Gasser S M, Grunstein M. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterchromatin in yeast. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- 15.Hecht A, Strahl-Bolsinger S, Grunstein M. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature. 1996;383:92–95. doi: 10.1038/383092a0. [DOI] [PubMed] [Google Scholar]
- 16.Henry Y A L, Chambers A, Tsang J S H, Kingsman A J, Kingsman S M. Characterisation of the DNA binding domain of the yeast RAP1 protein. Nucleic Acids Res. 1990;18:2617–2623. doi: 10.1093/nar/18.9.2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Konig P, Giraldo R, Chapman L, Rhodes D. The crystal structure of the DNA-binding domain of yeast RAP1 in complex with telomere DNA. Cell. 1996;85:125–136. doi: 10.1016/s0092-8674(00)81088-0. [DOI] [PubMed] [Google Scholar]
- 18.Kramer K M, Haber J E. New telomeres in yeast are initiated with a highly selected subset of TG1-3 repeats. Genes Dev. 1993;7:2345–2356. doi: 10.1101/gad.7.12a.2345. [DOI] [PubMed] [Google Scholar]
- 19.Krauskopf A, Blackburn E H. Control of telomere growth by interactions of RAP1 with the most distal telomere repeats. Nature. 1996;383:354–357. doi: 10.1038/383354a0. [DOI] [PubMed] [Google Scholar]
- 20.Kyrion G, Boakye K A, Lustig A J. C-terminal truncation of RAP1 results in the deregulation of telomere size, stability, and function in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:5159–5173. doi: 10.1128/mcb.12.11.5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kyrion G, Liu K, Liu C, Lustig A J. RAP1 and telomere structure regulate telomere position effects in Saccharomyces cerevisiae. Genes Dev. 1993;7:1146–1159. doi: 10.1101/gad.7.7a.1146. [DOI] [PubMed] [Google Scholar]
- 22.Laman H, Balderes D, Shore D. Disturbance of normal cell cycle progression enhances the establishment of transcriptional silencing in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:3608–3617. doi: 10.1128/mcb.15.7.3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li B, Lustig A J. A novel mechanism for telomere size control in Saccharomyces cerevisiae. Genes Dev. 1996;10:1310–1326. doi: 10.1101/gad.10.11.1310. [DOI] [PubMed] [Google Scholar]
- 24.Lingner J, Hughes T R, Shevchenko A, Mann M, Lundblad V, Cech T R. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 25.Liu C, Mao X, Lustig A J. Mutational analysis defines a C-terminal tail domain of RAP1 essential for telomeric silencing in Saccharomyces cerevisiae. Genetics. 1994;138:1025–1040. doi: 10.1093/genetics/138.4.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lustig A J, Kurtz S, Shore D. Involvement of the silencer and UAS binding protein RAP1 in regulation of telomere length. Science. 1990;250:549–553. doi: 10.1126/science.2237406. [DOI] [PubMed] [Google Scholar]
- 27.Lustig A J, Liu C, Zhang C, Hanish J P. Tethered Sir3p nucleates silencing at telomeres and internal loci in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2483–2495. doi: 10.1128/mcb.16.5.2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Makarov V L, Hirose Y, Langmore J P. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell. 1997;88:657–666. doi: 10.1016/s0092-8674(00)81908-x. [DOI] [PubMed] [Google Scholar]
- 29.Marcand S, Gilson E, Shore D. A protein-counting mechanism for telomere length regulation in yeast. Science. 1997;275:986–990. doi: 10.1126/science.275.5302.986. [DOI] [PubMed] [Google Scholar]
- 30.McCarroll R M, Fangman W L. Time of replication of yeast centromeres and telomeres. Cell. 1988;54:505–513. doi: 10.1016/0092-8674(88)90072-4. [DOI] [PubMed] [Google Scholar]
- 31.McEachern M J, Blackburn E H. Runaway telomere elongation caused by telomerase RNA gene mutations. Nature. 1995;376:403–409. doi: 10.1038/376403a0. [DOI] [PubMed] [Google Scholar]
- 32.McElligott R, Wellinger R J. The terminal DNA structure of mammalian chromosomes. EMBO J. 1997;16:3705–3714. doi: 10.1093/emboj/16.12.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moretti P, Freeman K, Coodly L, Shore D. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev. 1994;8:2257–2269. doi: 10.1101/gad.8.19.2257. [DOI] [PubMed] [Google Scholar]
- 34.Nugent C I, Hughes T R, Lue N F, Lundblad V. Cdc13p: a single-stranded telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science. 1996;274:249–252. doi: 10.1126/science.274.5285.249. [DOI] [PubMed] [Google Scholar]
- 35.Palladino F, Laroche T, Gilson E, Axelrod A, Pillus L, Gasser S M. SIR3 and SIR4 proteins are required for the positioning and integrity of yeast telomeres. Cell. 1993;75:543–555. doi: 10.1016/0092-8674(93)90388-7. [DOI] [PubMed] [Google Scholar]
- 36.Pluta A F, Zakian V A. Recombination occurs during telomere formation in yeast. Nature. 1989;337:429–433. doi: 10.1038/337429a0. [DOI] [PubMed] [Google Scholar]
- 37.Porter S E, Greenwell P W, Ritchie K B, Petes T D. The DNA-binding protein Hdf1p (a putative Ku homologue) is required for maintaining normal telomere length in Saccharomyces cerevisiae. Nucleic Acids Res. 1996;24:582–585. doi: 10.1093/nar/24.4.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ray A, Runge K. The C terminus of the major yeast telomere binding protein Rap1p enhances telomere formation. Mol Cell Biol. 1998;18:1284–1295. doi: 10.1128/mcb.18.3.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Renauld H, Aparicio O M, Zierath P D, Billington B L, Chhablani S K, Gottschling D E. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes Dev. 1993;7:1133–1145. doi: 10.1101/gad.7.7a.1133. [DOI] [PubMed] [Google Scholar]
- 40.Runge K W, Zakian V A. Introduction of extra telomeric DNA sequences into Saccharomyces cerevisiae results in telomere elongation. Mol Cell Biol. 1989;9:1488–1497. doi: 10.1128/mcb.9.4.1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Runge K W, Zakian V A. TEL2, an essential gene required for telomere length regulation and telomere position effect in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:3094–3105. doi: 10.1128/mcb.16.6.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 42a.Schultz, V. Personal communication.
- 43.Shampay J, Blackburn E H. Generation of telomere-length heterogeneity in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1988;85:534–538. doi: 10.1073/pnas.85.2.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- 45.Sussel L, Shore D. Separation of transcriptional activations and silencing functions of the RAP1-encoded repressor/activator protein 1: isolation of viable mutants affecting both silencing and telomere length. Proc Natl Acad Sci USA. 1991;88:7749–7753. doi: 10.1073/pnas.88.17.7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsukamoto Y, Kato J, Ikeda H. Silencing factors participate in DNA repair and recombination in Saccharomyces cerevisiae. Nature. 1997;388:900–903. doi: 10.1038/42288. [DOI] [PubMed] [Google Scholar]
- 47.van Steensel B, de Lange T. Control of telomere length by the human telomeric protein TRF1. Nature. 1997;385:740–743. doi: 10.1038/385740a0. [DOI] [PubMed] [Google Scholar]
- 48.Wang S S, Zakian V A. Sequencing of Saccharomyces telomeres cloned using T4 DNA polymerase reveals two domains. Mol Cell Biol. 1990;10:4415–4419. doi: 10.1128/mcb.10.8.4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wellinger R J, Ethier K, Labrecque P, Zakian V A. Evidence for a new step in telomere maintenance. Cell. 1996;85:423–433. doi: 10.1016/s0092-8674(00)81120-4. [DOI] [PubMed] [Google Scholar]
- 50.Wellinger R W, Wolf A J, Zakian V A. Saccharomyces telomeres acquire single-strand TG1-3 tails late in S phase. Cell. 1993;72:51–60. doi: 10.1016/0092-8674(93)90049-v. [DOI] [PubMed] [Google Scholar]
- 51.Wiley E A, Zakian V A. Extra telomeres, but not internal tracts of telomeric DNA, reduce transcriptional repression at Saccharomyces telomeres. Genetics. 1995;139:67–79. doi: 10.1093/genetics/139.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wotton D, Shore D. A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev. 1997;11:748–760. doi: 10.1101/gad.11.6.748. [DOI] [PubMed] [Google Scholar]
- 53.Wright J, Gottschling D, Zakian V A. Saccharomyces telomeres assume a non-nucleosomal structure. Genes Dev. 1992;6:197–210. doi: 10.1101/gad.6.2.197. [DOI] [PubMed] [Google Scholar]
- 54.Zakian V A. Structure, function and replication of Saccharomyces cerevisiae telomeres. Annu Rev Genet. 1996;30:141–172. doi: 10.1146/annurev.genet.30.1.141. [DOI] [PubMed] [Google Scholar]