Abstract
Background and Objective
HuangZhi YiShen Capsule (HZYS) is a Chinese patent herbal drug that protects kidney function in diabetic kidney disease (DKD) patients. However, the pharmacologic mechanisms of HZYS remain unclear. This study would use network pharmacology to explore the pharmacologic mechanisms of HZYS.
Methods
Chemical constituents of HZYS were obtained through the Traditional Chinese Medicine Systems Pharmacology Database (TCMSP) and literature search. Potential targets of HZYS were identified by using the TCMSP and the SwissTarget Prediction databases. DKD-related target genes were collected by using the Online Mendelian Inheritance in Man, Therapeutic Target Database, GeneCards, DisGeNET, and Drugbank databases. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were carried out to further explore the mechanisms of HZYS in treating DKD. Molecular docking was conducted to verify the potential interactions between the prime compounds and the hub genes.
Results
179 active compounds and 620 target genes were obtained, and 571 common targets were considered potential therapeutic targets. The top 10 main active compounds of HZYS were heparin, quercetin, kaempferol, luteolin, methyl14-methylpentadecanoate, methyl (Z)-11-hexadecenoate, 17-hydroxycorticosterone, 4-pregnene-17α, 20β, 21-triol-3, 11-dione, wogonin, and hydroxyecdysone. Hub signaling pathways by which HZYS treating DKD were PI3K-Akt, MAPK, AGE-RAGE in diabetic complications, TNF, and apoptosis. The top 10 target genes associated with these pathways were IL6, MAPK1, AKT1, RELA, BCL2, JUN, MAPK3, MAP2K1, CASP3, and TNF. Quercetin and Luteolin were verified to have good binding capability with the hub potential targets IL6, MAPK1, AKT1 through molecular docking.
Conclusion
HZYS appeared to treat DKD by regulating the inflammatory, oxidative stress, apoptotic, and fibrosis signaling pathways. This study provided a novel perspective for further research of HZYS.
Keywords: HuangZhi YiShen Capsule, diabetic kidney disease, network pharmacology, molecular docking
INTRODUCTION
Diabetic kidney disease (DKD) is a severe microvascular complication of diabetes mellitus (DM).[1] The number of people with diabetes globally was estimated to be 9.3% (463 million people) in 2019, with this figure anticipated to rise to 10.2% (578 million) in 2030 and 10.9% (700 million) in 2045.[2] Approximately 40% of DM patients deteriorate into DKD, the main cause of end-stage renal disease (ESRD).[3] Therefore, DKD is indeed a global health concern. However, the mechanism of DKD is not fully understood. Research has shown that hyperglycemia-induced metabolic impairment and hemodynamic disruptions are involved in the progression of DKD.[4] These changes lead to oxidative stress, inflammation, and generation of advanced glycation end products (AGEs). Currently, DKD treatment methods are mainly tight glucose and blood pressure control, especially to inhibit the renin-angiotensin system by using medications.[5] However, in the United States, for example, the percentage of DKD patients who progress to ESRD has not dropped significantly.[6]
Traditional Chinese medicine (TCM) is guided by its long upheld theories and has the unique characteristics of multiple components, multiple targets, and multiple pathways in treating both simple and complex diseases. In China, herbal medicines, including patent medicines, are used to treat DKD[7,8] and the application scope of many Chinese patent medicines that are effective in treating chronic kidney disease (CKD) has been expanding. One such medicine is HZYS, which was formerly known as Shen Yan Ning, has been found to be effective for CKD. In TCM theory, HZYS nourishes qi and yin, promotes blood circulation and diuresis and can reduce proteinuria and hematuria in chronic nephritis patients who have deficiency of both qi and yin.[9] Clinical trials have found that therapeutic outcomes of HZYS combined with conventional drugs are superior to conventional drugs alone.[10,11] It was reported that HZYS combined with Candesartan Cilexetil tablets had better effect on DKD by reducing proteinuria, improving kidney function than Candesartan Cilexetil tablets.[12] Moreover, clinicians found that HZYS can improve kidney function in patients with DKD.[13] Unfortunately, its clinical effective evidences for treating DKD are relatively rare. The pharmacologic mechanisms and the specific ingredients of HZYS as related to DKD remained unclear due to the limited published clinical and experimental research. Therefore, the aim of this study was to explore the potential key pharmacologic mechanism of HZYS on DKD.
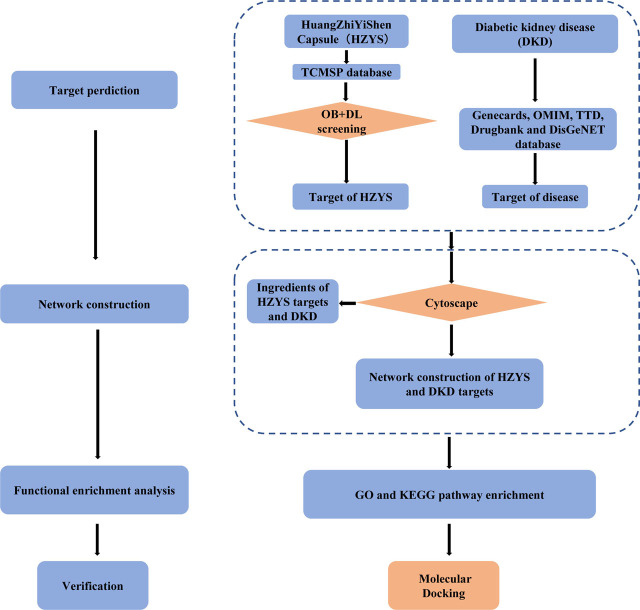
Network pharmacology was firstly proposed by Andrew Hopkins based on chemoinformatics, bioinformatics, network biology, and pharmacology.[14] It can integrate virtual computing technology, public databases, and high-throughput data for revealing the complex network relationship between biologically active ingredients and potential targets and mechanisms of Chinese herbal medicines.[15] HZYS, as a Chinese patent herbal drug, have multiple compounds as traditional Chinese medicine, however, it has not been explored about the specific ingredients of the drug and the potential mechanism in the treatment of DKD. Therefore, the network pharmacology based on a compound-target-disease interaction database and biologic analysis was utilized to explore the relationship between the multiple compounds, targets, and disease in this study.[16] And the key active compounds, important targets and core pathways for HZYS treating DKD were explored by network pharmacology methodology in this study. The study flowchart is shown in Figure 1.
Figure 1.
Schema of network pharmacology analysis of HZYS.
METHODS
Screening Active Compounds and Corresponding Targets
HZYS consists of the following herbs: astragalus root (Astragalus membranaceus (Fisch.) Bunge), leech (Hirudo nipponica Whitman), scrophularia root (Scrophularia ning poensis Hemsl.), glehnia root (Glehnia littoralis F. Schmidtex Miq.), notoginseng root (Panax notoginseng (Burkill) F.H.Chen), Chinese motherwort (Leonurus japonicus Houtt.), achyranthes root (Achyranthes bidentata Blume), goji berry (Lycium barbarum L.), Chinese yam (Dioscorea oppositifolia L.), coix seed (Coix lacryma-jobi L.), eclipta (Eclipta prostrata (L.) L.), human placenta, eucommia bark (Eucommia ulmoides Oliv.), plantain seed (Plantaginis Seme), and cicada molting (Cryptotympana atrata Fabr.) The chemical constituents of HZYS were accessed through the Traditional Chinese Medicine System Pharmacology Database (TCMSP; http://lsp.nwu.edu.cn/tcmsp.php). This database holds information on more than 13,000 pure compounds isolated from 505 Chinese herbs, corresponding targets and associated drug-target-disease networks. In addition, it provides data on absorption, distribution, metabolism, and excretion (ADME) screening for bioactive compounds.[17,18] In this study, the bioactive compounds were retrieved under the retrieval filters of oral bioavailability (OB) ≥ 30% and drug-likeness (DL) ≥ 0.18.[15,19,20] The active compounds of undetected drugs (such as leech, Chinese motherwort, and cicada molting) were obtained from the published literature. The chemical structures of related compounds were obtained from the PubChem database (https://pubchem.ncbi.nlm.nih.gov). Chemical structures that were not available were rendered using ChemDraw software (PerkinElmer; San Jose, CA, USA). All the chemical structure files were in SDF format and uploaded to the Swisstarget prediction server (Swiss Institute of Bioinformatics, http://www.swisstargetprediction.ch) for further target prediction and analysis.[21,22]
Screening Potential Targets for DKD
To find the relevant disease targets more comprehensively, information on DKD-associated target genes were gathered from the following 5 databases: Genecards (https://www.genecards.org),[23] DisGeNET(https://www.disgenet.org/home/),[24] Online Mendelian Inheritance in Man (OMIM; https://omim.org),[25] Therapeutic Target Database (TTD; http://db.idrblab.net/ttd/),[26] and Drugbank (https://www.drugbank.ca).[27] Genecards database furnishes extensive information on human genes including genomic, transcriptomic, proteomic, genetic, clinical, and functional statistics.[23] DisGeNET is a knowledge platform of genes and variants associated with human diseases compiled and integrated from the curated repositories, genome-wide association study catalogs, animal models, and the scientific literature.[24] OMIM is a comprehensive inventory of human genes and genetic phenotypes that has information on all the known mendelian disorders and over 15,000 genes, especially the relationship between phenotype and genotype.[25] TTD has information on therapeutic protein and nucleic acid targets, the targeted disease its matching therapeutic drugs, as well as pathway information.[26] DrugBank is a knowledge base that combines highly specific drug information with extensive drug target data and the related disease target information.
In this study, the keyword “diabetic kidney disease” was used in the aforementioned databases to screen targets related to DKD. The targets were collected and the duplicates were deleted. And the obtained targets were standardized through the Unitprot Knowledgebase (UniprotKB; https://www.uniprot.org), which collects information on functional information on proteins, their sequences, identification, and gene name at each of the targets.[28]
Screening Compound-Disease Common Targets
The screened HZYS chemical targets and DKD targets were imported into R software for statistical computing (version 4.0.2; https://www.r-project.org) and the common targets of compound-disease were obtained as the potential targets for further analysis.[29]
Constructing the Herb-Compound-Disease-Target Network
To find the core compounds of HZYS for treating DKD, a herb-compound-disease-target network was established by using Cytoscape (version 3.7.2; https://cytoscape.org).[30] A text named “Type,” which consists of the herb, compounds, disease, their common targets, and their logic relationship was imported into the Cytoscape software. Besides, a text named “network” was also imported into the Cytoscape software. The resulting node size was used to reflect the number of combined targets (degree). The nodes represented the herb, compounds, disease, and targets, and the edges indicated the interactions among them.
Mapping the Protein-Protein Interaction Network
To find the core targets of HZYS treating DKD, the protein-protein interaction (PPI) network for compound-disease common targets was established by using STRING (version 11.0; https://string-db.org), which collects nearly all the publicly-available sources of PPI information.[31] The species was defined as “Homo sapiens” to obtain the target interaction information. The results were imported into Cytoscape (version 3.7.2; https://cytoscape.org) and R for statistical computing, and the core targets of compound-disease were obtained as the potential targets for further analysis. PPI networks and a bar plot was then drawn.
Analysis of Gene Ontology and KEGG Pathway
To obtain key pathways of HZYS treating DKD, the Database for Annotation, Visualization and Integrated Discovery (DAVID; version 6.8; https://david.ncifcrf.gov) was used to conduct the gene ontology (GO) and KEGG Pathway Enrichment Analysis.[32,33,34,35] Biologic process (BP), molecular function (MF), and cell component (CC) are the general modules that are included in GO term enrichment. The Kyoto Encyclopedia of Genes and Genomes (KEGG) database (https://www.kegg.jp) was used to conduct pathway enrichment analyses. R software was also used for GO functional annotation and KEGG pathway enrichment analyses. Results with P < 0.05 were destined for further analysis.
Molecular Docking
The software AutoDock 4.0 was used to perform molecular docking between the top 4 compounds of HZYS (heparin, quercetin, kaempferol, and luteolin) and the top 3 hub target gene whose crystal structures were screened from the RSCB Protein Data Bank (https://www.rcsb.org/) (IL6, MAPK1, and AKT1, [entry:1ALU,6SLG,4ZZN and 1UNQ, respectively]). In all the dockings, the grid boxes of AKT1, IL6, and MAPK1 were set at 50 × 50 × 50 points, 60 × 50 × 60 points, 60 × 50 × 70 points, and 40 × 40 × 50 points, respectively. The ligand binding sites were the centers of the grid maps, and the grid space was 0.375 Å. Sixteen runs were generated with the Lamarckian genetic algorithm search. Each compound's docked conformation was ranked into clusters according to the binding energy, and the top-ranked conformations whose position with the lowest binding energy (as the most suitable conformation) was selected to be visually analyzed by using Discovery Studio 4.5.
RESULTS
Active Compounds and Corresponding Targets of HZYS
Using the TCMSP database, the active compound targets and corresponding targets of HZYS were searched for each chemical component. A total of 150 compounds were collected with the values of OB ≥ 30% and DL ≥ 0.18 properties, applied to screen the active compounds from HZYS. A total of 53 compounds were retrieved from literature reports. Finally, 179 candidate ingredients were selected from HZYS compounds (Supplementary Table S1) along with obtaining the common ingredients of herbs in HZYS (Table 1). In total, 620 targets were identified after removing the duplicate data (Supplementary Table S2).
Table 1.
Common compounds of herbs in HZYS
| Term | Mol Id | Mol name (compound) | Herb compound is found in common |
|---|---|---|---|
| A1 | MOL000098 | Quercetin | Astragalus root, glehnia root, notoginseng root, Chinese motherwort, achyranthes root, goji berry, eclipta, eucommia bark, plantain seed |
| A2 | MOL000354 | Isorhamnetin | Astragalus root, Chinese motherwort |
| A3 | MOL000422 | Kaempferol | Astragalus mongholicus, motherwort, Radix Achyranthis Bidentatae, Eucommia Astragalus root, Chinese motherwort, achyranthes root, eucommia bark |
| A4 | MOL000211 | Mairin | Astragalus root, Eucommia bark |
| B1 | MOL000358 | Beta-sitosterol | Glehnia root, notoginseng, achyranthes root, goji berry, eucommia bark, scrophularia root |
| B2 | MOL000449 | Stigmasterol | Glehnia root, notoginseng, achyranthes root, goji berry, Chinese yam, coix seed |
| C | MOL001494 | Mandenol | Notoginseng, goji berry, coix seed |
| D1 | MOL000953 | CLR | Goji berry, Chinese yam, coix seed |
| D2 | MOL005438 | Campesterol | Goji berry, Chinese yam, coix seed |
| D3 | MOL001323 | Sitosterol alpha1 | Goji berry, coix seed |
| E | MOL000359 | Sitosterol | Coix seed, plantain seed, scrophularia root |
Retrieval of Potential Compound-Disease Common Targets
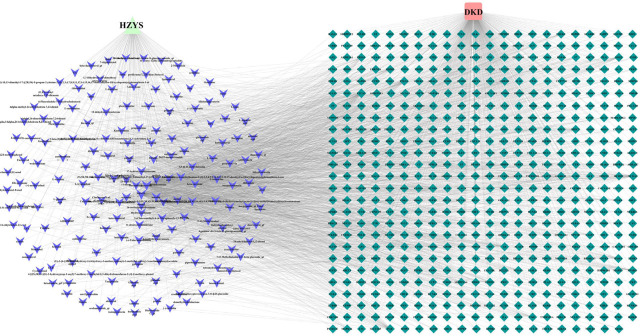
Through the GeneCards, DisGeNET, OMIM, TTD, and Drugbank databases, the retrieved results were integrated to obtain the DKD-related disease protein targets. The potential target genes in HZYS were mapped to the disease target genes using R, and a Venn diagram was drawn (Figure 2). A total of 571 potential drug-disease co-targets were obtained. These targets were sorted according to the degree value in the compound-target network (Supplementary Figure 1) and the target information was identified (Supplementary Table S3).
Figure 2.
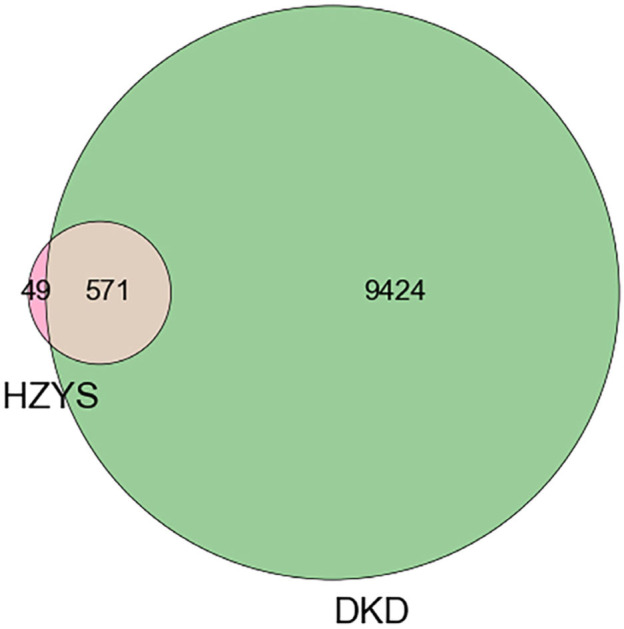
Matching of target genes between DKD and HZYS.
Visual Analysis of the Herb-Compound-Disease-Target Network
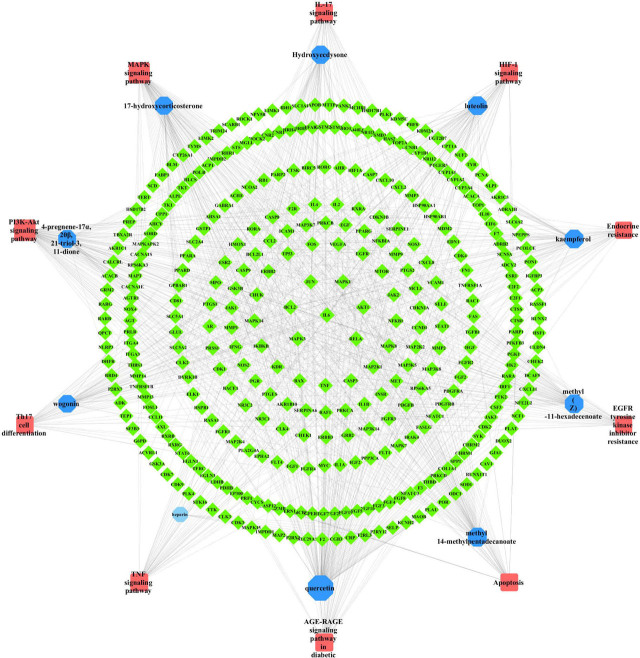
A total of 571 potential targets of DKD-HZYS were imported into Cytoscape for analysis. A herb-compound-disease-target network was constructed to further understand the interactions between the drug, compound, disease, and their targets (Figure 3). The network consisted of 747 nodes and 2787 edges. The sizes and colors of the borders were arranged according to the degree value. The top 10 chemical compounds with high degree values and closeness centralities were heparin, quercetin, kaempferol, luteolin, methyl14-methylpentadecanoate, methyl (Z)-11-hexadecenoate, 17-hydroxycorticosterone, 4-pregnene-17α, 20β, 21-triol-3, 11-dione, wogonin, and hydroxyecdysone. These ten are potentially the main chemical components of HZYS in the treatment of DKD.
Figure 3.
Visual analysis of herb-compound-disease-common target network. Green diamonds represent the targets; purple Vs represent the calculated ingredients; light green triangles represent the calculated drugs. The size and color of the border are arranged according to the degree value. (DKD: diabetic kidney disease; HZYS: Huangzhi Yishen capsule.)
Protein-Protein-Interactions Network
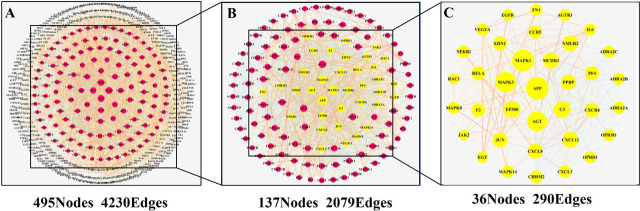
The potential 571 co-target genes associated with DKD were imported into the STRING database for analysis. To establish a high-quality and convincing protein-protein interaction (PPI) network, we selected protein targets with a high confidence score of 0.900 to diagram the network using STRING.[31] A total of 570 nodes and 4230 edges were acquired, and the average node degree was 14.8 (Supplementary Figure 2). To further analyze the results and explore the core targets that HZYS interacts with DKD, the relevant data is uploaded to Cytoscape, a PPI network was mapped using the common potential targets of HZYS capsule and DKD, consisting of 495 nodes and 4230 interaction edges (Figure 4A). 2X median analysis was used to analyze the PPI network to obtain core clusters, consisting of 137 nodes and 2079 interaction edges (Figure 4B). And then 36 hub-targets were screened after being analyzed by Betweenness (BC), Closeness (CC), and Degree (DC) filtering (Figure 4C). The results were then uploaded into R to diagram the bar plot. This led to the acquisition of 10 core targets that have the most connection nodes (Figure 5). Thus, the top 10 targets with the highest number of nodes are the genes APP, MAPK1, MAPK3, AGT, EP300, C3, AKT1, CXCL12, MCHR1, and NMUR2, which correspond to 98, 95, 88, 84, 70, 69, 65, 65, 65, 64 connection nodes respectively, indicating that the targets in the PPI network are significant.
Figure 4.
Common target PPI network between DKD and HZYS.
Figure 5.
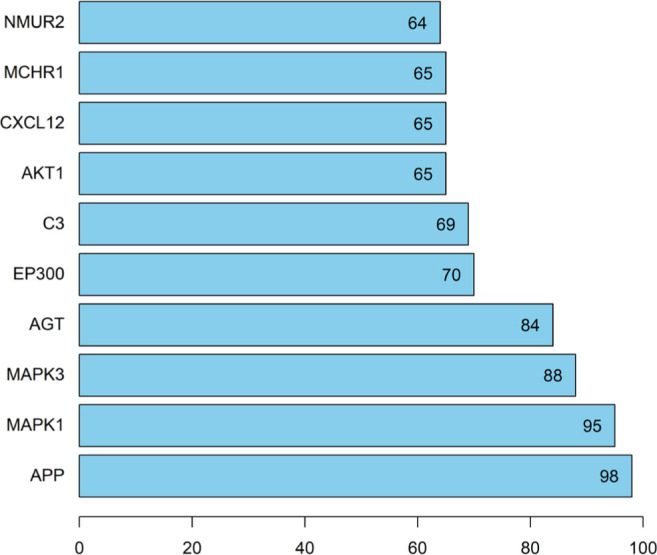
Bar plot of 30 core targets that have more connection nodes based on the PPI network. The number of each bar represents the number of connection nodes associated with the target.
Gene Ontology Enrichment Analysis
The potential 571 co-target genes were uploaded into the DAVID system for gene ontology (GO) analysis, which uncovers the potential co-target's function that are related to biologic processes, molecular functions, and cellular components.
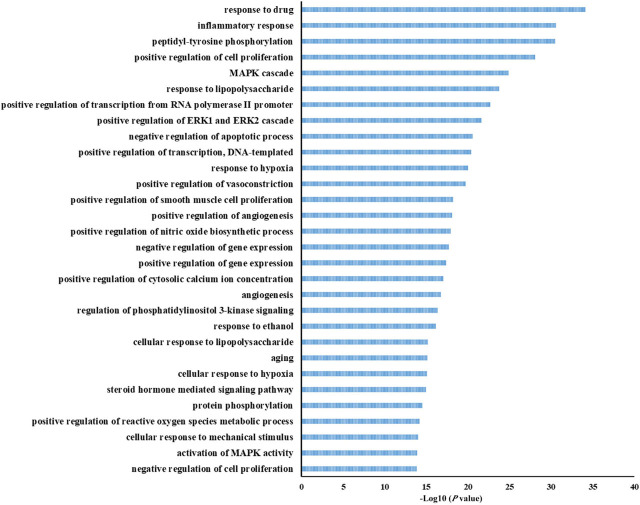
The 1104 biologic processes were then enriched. The top 30 terms that correspond with the criterion of P < 0.05 were selected for further analyses, such as drug response, inflammatory response, positive regulation of cell proliferation, mitogen-activated protein kinase (MAPK) cascade, lipopolysaccharide response, and negative regulation of apoptotic process (Figure 6). Results indicated that HZYS treatment of DKD involves numerous biologic processes, and understanding these processes can help elucidate the mechanisms by which HZYS treats DKD.
Figure 6.
GO enrichment of biologic process (BP) of potential targets of the main active ingredients in HZYS. The top 30 biologic process are shown in this figure. The abscissa is the adjusted P-value, the larger the value, the smaller the P-value.
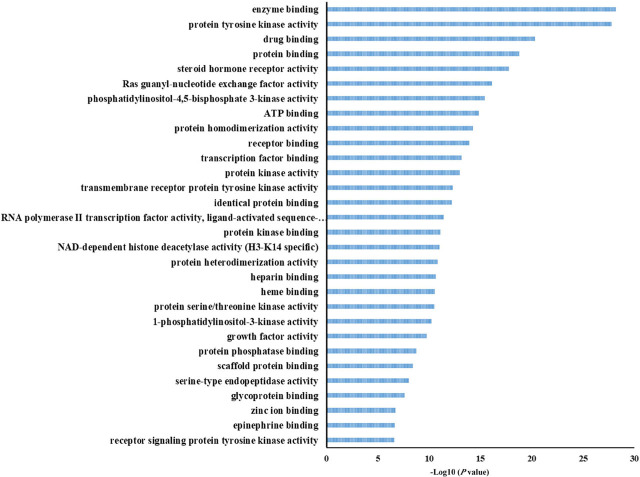
The 145 molecular functions were enriched in using GO and the top 30 terms were selected with the criterion of P < 0.05 for further analysis (Figure 7). The potential 571 co-target genes are involved in molecular functions related to enzyme binding, protein tyrosine kinase activity, drug binding, protein binding, steroid hormone receptor activity, among other functions.
Figure 7.
GO enrichment of molecular function (MF) of potential targets of the main active ingredients in HZYS. The top 30 terms were shown in this figure. The abscissa is the adjusted P-value, the larger the value, the smaller the P-value.
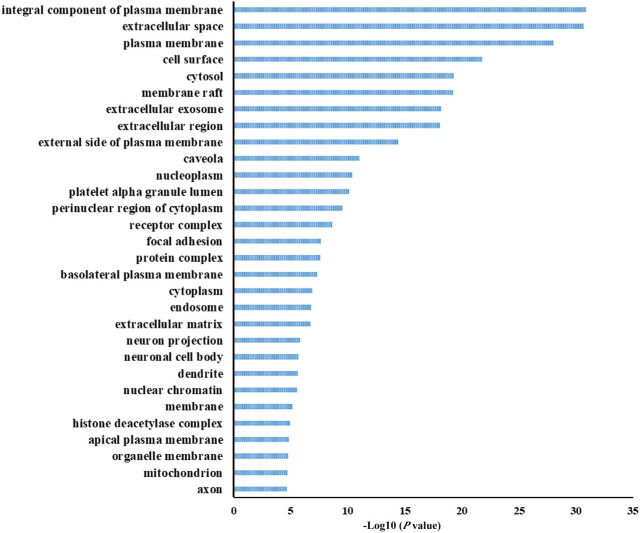
A total of 120 cell components were obtained from GO analysis and the top 30 terms with P < 0.05 underwent further analysis (Figure 8). The resulting potential co-target genes are related to the plasma membrane, extracellular space, cell surface, cytosol, membrane raft, and extracellular exosome. However, the potential mechanisms associated with HZYS treatment of DKD need further exploration.
Figure 8.
GO enrichment of the cell component (CC) of potential targets of the main active ingredients in HZYS. The top 30 terms were shown in this figure. The abscissa is the adjusted P-value, the larger the value, the smaller the P-value.
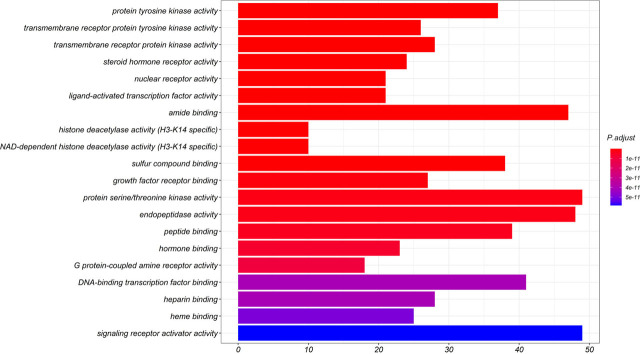
Finally, the data underwent comprehensive analysis using R to obtain the core functions of the co-targets. The top 20 terms with P < 0.05 were selected (Figure 9). The resulting targets are mainly involved with the protein tyrosine kinase activity, transmembrane receptor protein tyrosine kinase activity, transmembrane receptor protein kinase activity, steroid hormone receptor activity, nuclear receptor activity, ligand-activated transcription factor activity, amide binding, sulfur compound binding, and growth factor receptor binding.
Figure 9.
GO enrichment (from R) of potential targets of the main active ingredients in HZYS. Xs represent the number of genes, Ys represent the adjusted P-value. Color of the bar is shown in a gradient from red to blue based on ascending P-value. The top 20 terms are shown in this figure.
KEGG Pathway Enrichment Analysis
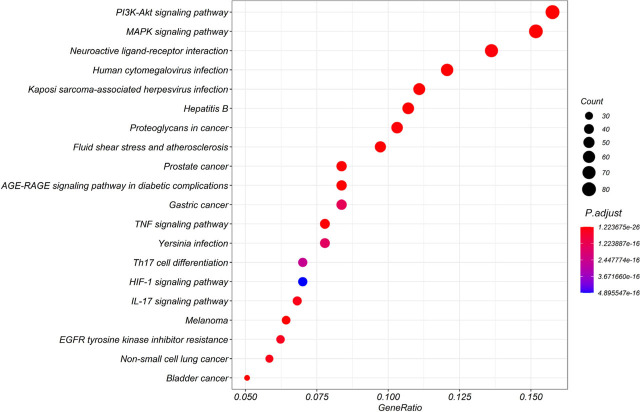
KEGG pathway enrichment analysis was performed using R to further uncover the potential mechanism of HZYS in treating DKD. A total of 174 signaling pathways of the potential co-target genes were enriched (Figure 10), including MAPK, AGE-RAGE in diabetic complications, as well as PI3K-Akt, TNF, IL-17, and HIF-1. These pathways are closely associated with cell proliferation, apoptosis, oxidative stress, and inflammation, and are involved in the development of DKD. Thus, we infer that HZYS may treat DKD by intervening in these pathways. We identified the top 20 pathways that may be associated with the DKD process for further analysis (Table 2). Of interest, signaling pathways correlated with other diseases were revealed, including cancers such as prostate, bladder, melanoma, non-small cell, and gastric as evidenced by the presence of the HIF-1 pathway. In addition, other pathways indicated that HZYS may be a candidate for treatment of other diseases, including hepatitis B, Kaposi sarcoma-associated herpesvirus infection, human cytomegalovirus infection, and yersiniosis.
Figure 10.
KEGG enrichment (from R) of potential targets of the main active ingredients in HZYS. Xs represent the gene ratios, Ys represent the adjusted P-value. The color of the bubble is shown in a gradient from red to blue based on ascending P-value. Size of the bubble is based on the number of genes. The top 20 terms are shown in this figure.
Table 2.
Top 20 pathways associated with DKD
| ID | Pathway description | Gene Ratio | P value | Q value |
|---|---|---|---|---|
| hsa04010 | MAPK signaling pathway | 78/514 | 4.11E-29 | 5.01E-27 |
| hsa04933 | AGE-RAGE signaling pathway in diabetic complications | 43/514 | 1.00E-25 | 3.07E-24 |
| hsa04151 | PI3K-Akt signaling pathway | 81/514 | 1.78E-25 | 3.78E-24 |
| hsa04668 | TNF signaling pathway | 40/514 | 2.03E-20 | 2.26E-19 |
| hsa04657 | IL-17 signaling pathway | 35/514 | 1.11E-18 | 9.45E-18 |
| hsa01521 | EGFR tyrosine kinase inhibitor resistance | 32/514 | 1.81E-18 | 1.38E-17 |
| hsa04659 | Th17 cell differentiation | 36/514 | 1.65E-17 | 1.06E-16 |
| hsa04066 | HIF-1 signaling pathway | 36/514 | 3.29E-17 | 2.01E-16 |
| hsa04210 | Apoptosis | 40/514 | 5.52E-17 | 3.06E-16 |
| hsa01522 | Endocrine resistance | 33/514 | 3.70E-16 | 1.88E-15 |
| hsa04014 | Ras signaling pathway | 50/514 | 8.04E-15 | 3.50E-14 |
| hsa04625 | C-type lectin receptor signaling pathway | 31/514 | 1.33E-13 | 4.79E-13 |
| hsa04660 | T cell receptor signaling pathway | 31/514 | 1.33E-13 | 4.79E-13 |
| hsa04810 | Regulation of actin cytoskeleton | 45/514 | 4.88E-13 | 1.53E-12 |
| hsa04015 | Rap1 signaling pathway | 44/514 | 1.03E-12 | 3.13E-12 |
| hsa04919 | Thyroid hormone signaling pathway | 32/514 | 2.04E-12 | 5.93E-12 |
| hsa04620 | Toll-like receptor signaling pathway | 29/514 | 5.39E-12 | 1.46E-11 |
| hsa04218 | Cellular senescence | 36/514 | 6.71E-12 | 1.74E-11 |
| hsa04062 | Chemokine signaling pathway | 40/514 | 8.30E-12 | 2.07E-11 |
| hsa04020 | Calcium signaling pathway | 41/514 | 1.57149E-11 | 3.76249E-11 |
To further explore the relationships between the predicted pathways, the potential co-target genes, and major compounds, we selected the top 10 pathways that have the highest likelihood to be associated with DKD. A compound-pathway-target network was then established (Figure 11). The top 5 pathways that have the highest degree values are PI3K-Akt, MAPK, AGE-RAGE in diabetic complications, TNF, and apoptosis. These may be the primary pathways by which HZYS treats DKD. Accordingly, the top 10 gene targets were obtained, including IL6, MAPK1, AKT1, RELA, BCL2, JUN, MAPK3, MAP2K1, CASP3, TNF.
Figure 11.
Visual analysis of compound-pathway-target network. Green diamonds represent the targets; blue octagons represent the calculated compounds; red rectangles represent the calculated pathways. The size and color of the border are arranged according to the degree value.
Molecular Docking Verification
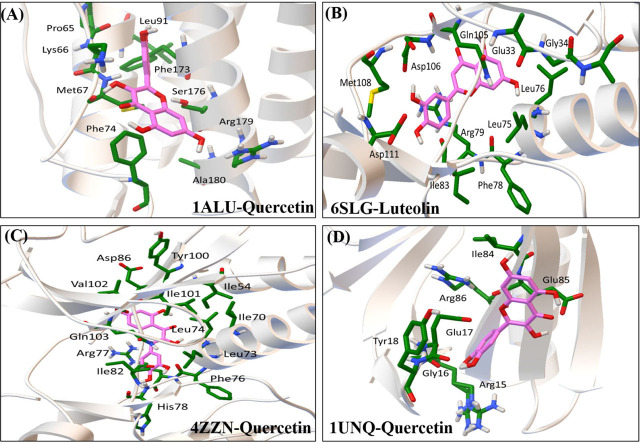
The interactions between the top 4 compounds and IL6, MAPK1, and AKT1 were investigated by molecular docking analysis. We found that all four compounds interacted differently with IL6, MAPK1, and AKT1 (Table 3). Among them, four interactions with lower Ki values and binding energy, which means that they have good bonding ability, were selected out to be further analyzed (Figure 12).
Table 3.
The binding free energy (ΔG) and inhibition constant (Ki) value of the interactions between compounds and target proteins
| IL6 | MAPK1 | AKT1 | ||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| 1ALU | 6SLG | 4ZZN | 1UNQ | |||||
|
| ||||||||
| ΔG[kcal mol−1] | Ki(μmol/L) | ΔG[kcal mol−1] | Ki(μmol/L) | ΔG[kcal mol−1] | Ki(μmol/L) | ΔG[kcal mol−1] | Ki(μmol/L) | |
| kaempferol | −5.89 | 48.41 | −5.21 | 151.77 | −5.39 | 112.34 | −4.21 | 824.62 |
| luteolin | −7.13 | 5.99 | −6.32 | 23..31 | −6.70 | 12.27 | −5.79 | 56.99 |
| methyl | −4.16 | 895.29 | −2.62 | − | −3.80 | − | −3.53 | − |
| quercetin | −7.68 | 2.33 | −6.06 | 36.43 | −6.78 | 10.74 | −5.82 | 54.16 |
Figure 12.
Molecular docking of compound-hub genes. (A) 1ALU to Quercetin (Binding energy = −7.68, Ki = 2.33 μmol/L); (B) 6SLG to Luteolin (Binding energy = −6.32, Ki = 23.31 μmol/L); (C) 4ZZN to Quercetin (Binding energy = −6.78, Ki = 10.74 μmol/L); (D) 1UNQ to Quercetin (Binding energy = −5.82, Ki = 54.16 μmol/L); (1ALU is a crystal structure of IL6; 6SLG is a crystal structure of MAPK1; 4ZZN is a crystal structure of MAPK1; 1UNQ is a crystal structure of AKT1. Interacting amino acids and compound structures are shown in lines, in which amino acids are shown in green, compound structures are shown in pink.)
DISCUSSION
Proper treatment is crucial for patients with DKD. HZYS, a traditional Chinese patent medicine, has long been used with success to treat CKD, including DKD.[2,36,37] Molecular drug targets were developed by the network pharmacology, which aims at biological networks and the links among disease, drug, and targets in those networks.[16,38] Therefore, network pharmacology, as an new approach to study the complex relationship among targets, drugs, diseases, and pathways for drug discovery.[39] In this study, systematic analysis of HZYS was conducted at the molecular level using network pharmacology. Ten potential hub ingredients, 571 HZYS-DKD-common-targets, 10 hub targets, and 10 pathways were acquired, suggesting that they have potential value for HZYS in treatment of DKD.
In this study, some of the active compounds in HZYS appeared to be capable of attenuating proteinuria and oxidative stress, and improving renal function. One such compound was heparin, which was found in leech. Heparin is well known as an anticoagulant; it inactivates the thrombin generating ability of plasma.[40] In DKD model rats, low-molecular weight heparin has been shown to prevent glomerular basement membrane thickening in the kidney, thus reducing proteinuria.[41, 42] In clinical trials, heparin has been found to attenuate proteinuria in both type 1 and type 2 diabetes.[43, 44] Another active compound is quercetin, which is found in several herbs in HZYS, including astragalus root, glehnia root, notoginseng root, Chinese motherwort, achyranthes root, goji berry, eclipta, eucommia bark, and plantain seed. Quercetin appears to decrease insulin resistance by reducing endoplasmic reticulum (ER) stress, oxidative stress, and β-cell death.[45] In streptozotocin-induced DKD rats, quercetin has been shown to improve renal function by downregulating the expression of transforming growth factor beta and connective tissue growth factor.[46] Further studies have discovered that quercetin exhibits several molecular activities. For example, Lu et al. found that quercetin reverses high glucose-induced epithelial-mesenchymal transition of HK-2 and NRK-52E cells by inhibiting mTORC1/p70S6K activation in the diabetic renal cortex.[47] Another research on quercetin has shown that it inhibits the proliferation of high glucose-induced mesangial cells by reactivating the Hippo signaling pathway and suppressing the NF-ϰB signaling pathway.[48,49,50,51] Another active compound in HZYS is kaempferol. It is found in astragalus root, Chinese motherwort, achyranthes root, and eucommia bark which were reported to lower blood sugar level and blood lipid levels.[52,53] In the DKD mouse model, kaempferol has been shown to reduce renal injury and fibrosis by upregulating glucagon-like peptide 1 and insulin.[54] Kaempferol may also reduce the occurrence of complications of diabetes, such as DKD and diabetic retinopathy.[55,56] In addition to the aforementioned active compounds, other compounds in HZYS have also been found in this study to have anti-inflammatory, antioxidant, and antifibrotic activities. Two examples are luteolin and wogonin, which in our study are hub components of HXYS. Zhang et al. found that luteolin suppresses the inflammatory and oxidative responses by suppressing STAT3 activation,[57] Meng et al. discovered that wogonin exerts an antifibrotic effect on renal tubular epithelial cells through smad3-dependent mechanisms.[58] In summary, our network pharmacology results and those of experimental studies indicate that active compounds in HZYS have potential therapeutic value in DKD.
To further evaluate the potential key mechanism of HZYS in the treatment of DKD, 507 compound-disease common targets (chosen from 620 compound targets) underwent GO enrichment analyses. Results indicated that HZYS can potentially treat DKD by regulating the inflammatory response, cell proliferation, apoptotic process, MAPK cascade, and serum lipopolysaccharide activity. In addition, these mechanisms were involved with key targets such as MAPK1, MAPK3, AKT1, AGT, PTGS2.
In this study, KEGG pathway enrichment analysis demonstrated that the potential key mechanism by which HZYS treating DKD was mainly involved with several signaling pathways, including AGE-RAGE in diabetic complications, PI3K-Akt, MAPK, TNF, and apoptosis. The top 10 genes we analyzed from the hub pathways were IL6, MAPK1, AKT1, RELA, BCL2, JUN, MAPK3, MAP2K1, CASP3, and TNF. IL6 is a classic pro-inflammatory cytokine involved in DKD.[59] Human and experimental studies have validated the involvement of the IL-6 signaling pathway in DKD progression. For instance, IL-6 is critically involved in the inflammatory processes of tubular and mesangial cells, and podocytes, which are the resident cells of the kidney.[59] In our study, IL-6 was found to be associated with the AGE-RAGE, PI3K/Akt, HIF-1, and IL17 signaling pathways, which are closely involved with inflammation, apoptosis, and oxidative stress in DKD.[60,61] The PI3K/Akt pathway is also a significant player in autophagy regulation of resident kidney cells.[62] BCL2, CASP3, and TNF are important components of apoptosis and are closely associated with the progression of DKD.[63,64,65] In addition, the AGE-RAGE signaling pathway in diabetic complications is highly involved in the DKD formation process. Previous investigation found that sustained hyperglycemia due to diabetes facilitates increased production of AGEs and stimulates their cell surface receptors, RAGEs,[66] therefore, inducing endoplasmic reticulum (ER) stress and increasing intracellular calcium level and apoptosis rate,[67] thus worsening diabetic kidney disease. In contrast, in RAGE-deficient diabetic mice, development of DKD is delayed, with less inflammatory and fibrosis expressed in renal tissues.[68] Additionally, inflammation and oxidative stress can also induce production of AGEs.[68] Thus, HZYS may treat DKD by alleviating inflammation and oxidative stress to regulate the AGE-RAGE signaling pathway. Moreover, since the PI3K/Akt signaling pathway may be activated by RAGEs,[69] HZYS may suppress RAGEs to regulate the PI3K/Akt pathway to attenuate DKD.[66]
The molecular docking verification presents that the prime compounds of HZYS Quercetin and Luteolin have good binding capability with the hub potential targets IL6, MAPK1, AKT1. Quercetin exhibits a significantly anti-inflammatory effect in kidney disease, among which lessened renal pathologies, and inhibited IL-6 production in mice with LPS-induced AKI.[70] Quercetin alleviated LPS-stimulated HK-2 cell inflammatory injury by regulating miR-124 and reduce IL6 and TNF-α.[71] Quercetin has also been reported to have anti-oxidant effects in kidney through modulating the MAPK and NF-ϰB signaling pathway.[72] And it has also been verified in our study that it has a good binding ability with the crystal structures of MAPK1. Additionally, Quercetin was also reported to regulate the Akt signaling pathway to improve glucose and lipid metabolism of DKD rats.[73] In our study, it was verified to have interaction with AKT1. Luteolin is another prime compound of HZYS. In the past experimental research, luteolin has been proved to have remarkable anti-inflammatory, anti-apoptosis, and anti-oxidant stress effects, which were mainly by regulating MAPK signaling pathway.[74,75,76,77,78,79,80,81,82] Surely, Luteolin did have a good interaction with MAPK in this research.
The results indicated that the screened compounds, targets, and pathways are consistent with current research findings, confirming that HZYS treats DKD by mediating inflammation, oxidative stress, apoptosis, fibrosis, and other signaling pathways. However, the results of our network pharmacology investigation need further verification through in vitro and in vivo experiments to demonstrate the exact mechanism by which HZYS treats DKD.
CONCLUSION
This study provides new insight into the pharmacology of HZYS and the mechanism by which this Chinese herbal formula treating DKD. Our study using network pharmacology revealed that active compounds in HZYS were potentially involved in regulating the inflammatory, oxidative stress, apoptotic, and fibrosis signaling pathways in DKD.
Acknowledgment
The authors thank Nissi S. Wang, MSc, for developmental editing of the manuscript.
Footnotes
Data Availability
Data that support the findings of this study are available from the corresponding author upon request.
Funding
This research was funded by the National Natural Science Foundation of China (Grant No. 81620108031) and was supported by the China-Japan Friendship Hospital project (Grant No. 2020-HX-20).
Conflicts of interest
The authors declare that they have no conflicts of interest.
Authors’ Contributions
Xue-Feng Zhou: conceptualization, design, methodology, software, formal analysis, original draft preparation, visualization, molecular docking; Wei-E Zhou: conceptualization, design, methodology, formal analysis, review and editing, visualization; Wen-Jing Liu: methodology, software, resources; Min-Jing Luo: methodology, resources; Xia-Qing Wu: molecular docking; Ying Wang: methodology, resources; Peng Liu: validation, investigation, review and editing, supervision, project administration, funding acquisition; Yu-Min Wen: validation, funding acquisition; Jia-Lin Li: resources; Ting-Ting Zhao: validation; Hao-Jun Zhang: validation; Hai-Ling Zhao: validation; Ping Li: investigation, review and editing, supervision, project administration, funding acquisition
Supplementary Material.
REFERENCES
- 1.Chowdhury S, Ghosh S, Das AK, Sil PC. Ferulic Acid Protects Hyperglycemia-Induced Kidney Damage by Regulating Oxidative Insult, Inflammation and Autophagy. Front Pharmacol. 2019;10:27. doi: 10.3389/fphar.2019.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saeedi P, Petersohn I, Salpea P, Malanda B, Karuranga S, Unwin N. et al. Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: Results from the International Diabetes Federation Diabetes Atlas, 9 th edition. Diabetes Res Clin Pract. 2019;157 doi: 10.1016/j.diabres.2019.107843. 107843. [DOI] [PubMed] [Google Scholar]
- 3.Wang G, Ouyang J, Li S, Wang H, Lian B, Liu Z. et al. The analysis of risk factors for diabetic nephropathy progression and the construction of a prognostic database for chronic kidney diseases. J Transl Med. 2019;17:264. doi: 10.1186/s12967-019-2016-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Magee C, Grieve DJ, Watson CJ, Brazil DP. Diabetic Nephropathy: a Tangled Web to Unweave. Cardiovasc Drugs Ther. 2017;31:579–92. doi: 10.1007/s10557-017-6755-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haneda M. Diabetic nephropathy. Nihon Rinsho. 2015;73:489–94. [PubMed] [Google Scholar]
- 6.Afkarian M, Zelnick LR, Hall YN, Heagerty PJ, Tuttle K, Weiss NS. et al. Clinical Manifestations of Kidney Disease Among US Adults With Diabetes, 1988–2014. JAMA. 2016;316:602–10. doi: 10.1001/jama.2016.10924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun M, Bu W, Li Y, Zhu J, Zhao J, Zhang P. et al. Danzhi Jiangtang Capsule ameliorates kidney injury via inhibition of the JAK-STAT signaling pathway and increased antioxidant capacity in STZ-induced diabetic nephropathy rats. Biosci Trends. 2019;12:595–604. doi: 10.5582/bst.2018.01255. [DOI] [PubMed] [Google Scholar]
- 8.Chen HY, Pan HC, Chen YC, Chen YC, Lin YH, Yang SH. et al. Traditional Chinese medicine use is associated with lower end-stage renal disease and mortality rates among patients with diabetic nephropathy: a population-based cohort study. BMC Complement Altern Med. 2019;19:81. doi: 10.1186/s12906-019-2491-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li MQ, Ye CH, Shu HQ, Ye XF, Zou LH. Shenyanning Capsule in the Treatment of 62 Cases of Chronic Nephritis Double-blind Double-simulation Randomized Controlled Clinical Trial. Pharmacol Clin Chin Mater Med. 2002;18(6):46–7. [Google Scholar]
- 10.Lin J, Guo M, Guo WJ, Du YL. Clinical study of Huangzhi Yishen Capsules combined with piperazine ferulate in the treatment of chronic glomerulonephritis. Drugs Clin. 2020;35:113–7. [Google Scholar]
- 11.Luo J, Li S, Zhao CY. Analysis of the clinical efficacy of Huangzhi Yishen Capsule in the treatment of nephrotic syndrome type IgA nephropathy. World Latest Med Inf. 2019;(95):17–8. [Google Scholar]
- 12.Liu L, Li LZ. The clinical study of Huangzhi Yishen Capsules combined with Candesartan medoxomil tablets in the treatment of diabetic nephropathy; Annual Meeting of the Renal Disease Professional Committee of the Chinese Society of Integrative Medicine; 2018. [Google Scholar]
- 13.‘Expert consensus on Huangzhi Yishen Capsule in the treatment of chronic kidney disease’ writing group. Expert consensus on Huangzhi Yishen Capsule in the treatment of chronic kidney disease. Chin J Integr Tradit West Nephrol. 2019;20:842–844. [Google Scholar]
- 14.Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol. 2008;4:682–90. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- 15.Mou X, Zhou DY, Zhou D, Liu K, Chen LJ, Liu WH. A bioinformatics and network pharmacology approach to the mechanisms of action of Shenxiao decoction for the treatment of diabetic nephropathy. Phytomedicine. 2020;69 doi: 10.1016/j.phymed.2020.153192. 153192. [DOI] [PubMed] [Google Scholar]
- 16.Yuan H, Ma Q, Cui H, Liu G, Zhao X, Li W. et al. How Can Synergism of Traditional Medicines Benefit from Network Pharmacology? Molecules. 2017;22:1135. doi: 10.3390/molecules22071135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ru J, Li P, Wang J, Zhou W, Li B, Huang C. et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. 2014;6:13. doi: 10.1186/1758-2946-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo MF, Dai YJ, Gao JR, Chen PJ. Uncovering the Mechanism of Astragalus membranaceus in the Treatment of Diabetic Nephropathy Based on Network Pharmacology. J Diabetes Res. 2020;2020 doi: 10.1155/2020/5947304. 5947304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yue SJ, Liu J, Feng WW, Zhang FL, Chen JX, Xin LT. et al. System Pharmacology-Based Dissection of the Synergistic Mechanism of Huangqi and Huanglian for Diabetes Mellitus. Front Pharmacol. 2017;8:694. doi: 10.3389/fphar.2017.00694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu XX, Bi JP, Ping L, Li P, Li F. A network pharmacology approach to determine the synergetic mechanisms of herb couple for treating rheumatic arthritis. Drug Des Devel Ther. 2018;12:967–79. doi: 10.2147/DDDT.S161904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Daina A, Michielin O, Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019;47:W357–64. doi: 10.1093/nar/gkz382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gfeller D, Michielin O, Zoete V. Shaping the interaction landscape of bioactive molecules. Bioinformatics. 2013;29:3073–9. doi: 10.1093/bioinformatics/btt540. [DOI] [PubMed] [Google Scholar]
- 23.Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S. et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr Protoc Bioinformatics. 2016;54:1.30.1–1.30.33. doi: 10.1002/cpbi.5. [DOI] [PubMed] [Google Scholar]
- 24.Yu T, Acharya A, Mattheos N, Li S, Ziebolz D, Schmalz G. et al. Molecular mechanisms linking peri-implantitis and type 2 diabetes mellitus revealed by transcriptomic analysis. Peer J. 2019;7:e7124. doi: 10.7717/peerj.7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amberger JS, Hamosh A. Searching Online Mendelian Inheritance in Man (OMIM): A Knowledgebase of Human Genes and Genetic Phenotypes. Curr Protoc Bioinformatics. 2017;58:1.2.1–1.2.12. doi: 10.1002/cpbi.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, Zhang S, Li F, Zhou Y, Zhang Y, Wang Z. et al. Therapeutic target database 2020: enriched resource for facilitating research and early development of targeted therapeutics. Nucleic Acids Res. 2020;48:D1031–41. doi: 10.1093/nar/gkz981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR. et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46:D1074–82. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47:D506–15. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dessau RB, Pipper CB. [‘’R”--project for statistical computing] Ugeskr Laeger. 2008;170:328–30. [PubMed] [Google Scholar]
- 30.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J. et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–13. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deng JL, Xu YH, Wang G. Identification of Potential Crucial Genes and Key Pathways in Breast Cancer Using Bioinformatic Analysis. Front Gene. 2019;10:695. doi: 10.3389/fgene.2019.00695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang W, Wang T, Wang Y, Piao H, Li B, Zhu Z. et al. Integration of Gene Expression Profile Data to Verify Hub Genes of Patients with Stanford A Aortic Dissection. BioMed Res Int. 2019;2019 doi: 10.1155/2019/3629751. 3629751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 36.Du J, Bao JB. Analysis of clinical efficacy of Shenyanning in the treatment of non-nephrotic syndrome IgA nephropathy. Chin Health Ind. 2014;11:183–4+6. [Google Scholar]
- 37.Zhang CG, Hu XD, Huang XH. Observation of the curative effect of Shenyanning on non-nephrotic syndrome IgA nephropathy. Zhongguo Zhong Xi Yi Jie He Za Zhi. 2013;3:63–4. [Google Scholar]
- 38.Zhou Z, Chen B, Chen S, Lin M, Chen Y, Jin S. et al. Applications of Network Pharmacology in Traditional Chinese Medicine Research. Evid Based Complement Alternat Med. 2020;2020:1–7. doi: 10.1155/2020/1646905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elgoyhen AB, Langguth B, Vanneste S, De Ridder D. Tinnitus: network pathophysiology-network pharmacology. Front Syst Neurosci. 2012;6:1. doi: 10.3389/fnsys.2012.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hemker HC. A century of heparin: past, present and future. J Thromb Haemost. 2016;14:2329–38. doi: 10.1111/jth.13555. [DOI] [PubMed] [Google Scholar]
- 41.Gambaro G, Cavazzana AO, Luzi P, Piccoli A, Borsatti A, Crepaldi G. et al. Glycosaminoglycans prevent morphological renal alterations and albuminuria in diabetic rats. Kidney Int. 1992;42:285–91. doi: 10.1038/ki.1992.288. [DOI] [PubMed] [Google Scholar]
- 42.Gambaro G, Venturini A P, Noonan D M. et al. Treatment with a glycosaminoglycan formulation ameliorates experimental diabetic nephropathy. Kidney Int. 1994;46:797–806. doi: 10.1038/ki.1994.335. [DOI] [PubMed] [Google Scholar]
- 43.Veglio M, Gruden G, Mormile A, Girotto M, Rossetto P, D’Este P. et al. Anticoagulant protein C activity in non-insulin-dependent diabetic patients with normoalbuminuria and microalbuminuria. Acta Diabetol. 1995;32:106–9. doi: 10.1007/BF00569567. [DOI] [PubMed] [Google Scholar]
- 44.Tamsma JT, van der Woude FJ, Lemkes HH. Effect of sulphated glycosaminoglycans on albuminuria in patients with overt diabetic (type 1) nephropathy. Nephrol Dial Transplant. 1996;11:182–5. [PubMed] [Google Scholar]
- 45.Gaballah HH, Zakaria SS, Mwafy SE, Tahoon NM, Ebeid AM. Mechanistic insights into the effects of quercetin and/or GLP-1 analogue liraglutide on high-fat diet/streptozotocin-induced type 2 diabetes in rats. Biomed Pharmacoth. 2017;92:331–9. doi: 10.1016/j.biopha.2017.05.086. [DOI] [PubMed] [Google Scholar]
- 46.Yang LY, Lai PB, Zhang L. Quercetin ameliorates diabetic nephropathy by reducing the expressions of transforming growth factor-β1 and connective tissue growth factor in streptozotocin-induced diabetic rats. Ren Fail. 2012;34:83–7. doi: 10.3109/0886022X.2011.623564. [DOI] [PubMed] [Google Scholar]
- 47.Lu Q, Ji XJ, Zhou YX, Yao XQ, Liu YQ, Zhang F. et al. Quercetin inhibits the mTORC1/p70S6K signaling-mediated renal tubular epithelialmesenchymal transition and renal fibrosis in diabetic nephropathy. Pharmacol Res. 2015;99:237–47. doi: 10.1016/j.phrs.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 48.Dehdashtian E, Pourhanifeh MH, Hemati K, Mehrzadi S, Hosseinzadeh A. Therapeutic application of nutraceuticals in diabetic nephropathy: Current evidence and future implications. Diabetes Metab Res Rev. 2020;36:e3336. doi: 10.1002/dmrr.3336. [DOI] [PubMed] [Google Scholar]
- 49.Lei D, Chengcheng L, Xuan Q, Yibing C, Lei W, Hao Y. et al. Quercetin inhibited mesangial cell proliferation of early diabetic nephropathy through the Hippo pathway. Pharmacol Res. 2019;146 doi: 10.1016/j.phrs.2019.104320. 104320. [DOI] [PubMed] [Google Scholar]
- 50.Chen P, Shi Q, Xu X, Wang Y, Chen W, Wang H. Quercetin suppresses NF-κB and MCP-1 expression in a high glucose-induced human mesangial cell proliferation model. Int J Mol Med. 2012;30:119–25. doi: 10.3892/ijmm.2012.955. [DOI] [PubMed] [Google Scholar]
- 51.Chen P, Chen J, Zheng Q, Chen W, Wang Y, Xu X. Pioglitazone, extract of compound Danshen dripping pill, and quercetin ameliorate diabetic nephropathy in diabetic rats. J Endocrinol Invest. 2013;36:422–7. doi: 10.3275/8763. [DOI] [PubMed] [Google Scholar]
- 52.Bai L, Li X, He L, Zheng Y, Lu H, Li J. et al. Antidiabetic Potential of Flavonoids from Traditional Chinese Medicine: A Review. Am J Chin Med. 2019;47:933–57. doi: 10.1142/S0192415X19500496. [DOI] [PubMed] [Google Scholar]
- 53.Zang Y, Zhang L, Igarashi K, Yu C. The anti-obesity and anti-diabetic effects of kaempferol glycosides from unripe soybean leaves in high-fat-diet mice. Food Funct. 2015;6:834–41. doi: 10.1039/c4fo00844h. [DOI] [PubMed] [Google Scholar]
- 54.Sharma D, Kumar Tekade R, Kalia K. Kaempferol in ameliorating diabetes-induced fibrosis and renal damage: An in vitro and in vivo study in diabetic nephropathy mice model. Phytomedicine. 2020;76 doi: 10.1016/j.phymed.2020.153235. 153235. [DOI] [PubMed] [Google Scholar]
- 55.Sharma D, Gondaliya P, Tiwari V, Kalia K. Kaempferol attenuates diabetic nephropathy by inhibiting RhoA/Rho-kinase mediated inflammatory signalling. Biomed Pharmacoth. 2019;109:1610–9. doi: 10.1016/j.biopha.2018.10.195. [DOI] [PubMed] [Google Scholar]
- 56.Du W, An Y, He X, Zhang D, He W. Protection of Kaempferol on Oxidative Stress-Induced Retinal Pigment Epithelial Cell Damage. Oxid Med Cell Longev. 2018;2018:1–14. doi: 10.1155/2018/1610751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang M, He L, Liu J, Zhou L. Luteolin Attenuates Diabetic Nephropathy through Suppressing Inflammatory Response and Oxidative Stress by Inhibiting STAT3 Pathway. Exp Clin Endocrinol Diabetes. 2020 doi: 10.1055/a-0998-7985. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 58.Meng XM, Ren GL, Gao L, Li HD, Wu WF, Li XF. et al. Anti-fibrotic effect of wogonin in renal tubular epithelial cells via Smad3-dependent mechanisms. Euro J Pharmacol. 2016;789:134–43. doi: 10.1016/j.ejphar.2016.07.014. [DOI] [PubMed] [Google Scholar]
- 59.Feigerlová E, Battaglia-Hsu SF. IL-6 signaling in diabetic nephropathy: From pathophysiology to therapeutic perspectives. Cytokine Growth Factor Rev. 2017;37:57–65. doi: 10.1016/j.cytogfr.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 60.Donate-Correa J, Martín-Núñez E, Muros-de-Fuentes M, Mora-Fernández C, Navarro-González JF. Inflammatory cytokines in diabetic nephropathy. J Diabetes Res. 2015;2015 doi: 10.1155/2015/948417. 948417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Duran-Salgado, Montserrat B. Diabetic nephropathy and inflammation. World J Diabetes. 2014;5:393. doi: 10.4239/wjd.v5.i3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tu Q, Li Y, Jin J, Jiang X, Ren Y, He Q. Curcumin alleviates diabetic nephropathy via inhibiting podocyte mesenchymal transdifferentiation and inducing autophagy in rats and MPC5 cells. Pharm Biol. 2019;57:778–86. doi: 10.1080/13880209.2019.1688843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fu Y, Wang C, Zhang D, Chu X, Zhang Y, Li J. miR-15b-5p ameliorated high glucose-induced podocyte injury through repressing apoptosis, oxidative stress, and inflammatory responses by targeting Sema3A. J Cell Physiol. 2019;234:20869–78. doi: 10.1002/jcp.28691. [DOI] [PubMed] [Google Scholar]
- 64.Chen Y, Liu Q, Shan Z, Zhao Y, Li M, Wang B. et al. The protective effect and mechanism of catalpol on high glucose-induced podocyte injury. BMC Complement Altern Med. 2019;C19:244. doi: 10.1186/s12906-019-2656-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Malik S, Suchal K, Khan SI, Bhatia J, Kishore K, Dinda AK. et al. Apigenin ameliorates streptozotocin-induced diabetic nephropathy in rats via MAPK-NF-κB-TNF-α and TGF-β1-MAPK-fibronectin pathways. Am J Physiol Renal Physiol. 2017;313:F414–22. doi: 10.1152/ajprenal.00393.2016. [DOI] [PubMed] [Google Scholar]
- 66.Sanajou D, Ghorbani Haghjo A, Argani H, Aslani S. AGE-RAGE axis blockade in diabetic nephropathy: Current status and future directions. Euro J Pharmacol. 2018;833:158–64. doi: 10.1016/j.ejphar.2018.06.001. [DOI] [PubMed] [Google Scholar]
- 67.Chen Y, Liu CP, Xu KF, Mao XD, Lu YB, Fang L. et al. Effect of taurine-conjugated ursodeoxycholic acid on endoplasmic reticulum stress and apoptosis induced by advanced glycation end products in cultured mouse podocytes. Am J Nephrol. 2008;28:1014–22. doi: 10.1159/000148209. [DOI] [PubMed] [Google Scholar]
- 68.Hagiwara S, Sourris K, Ziemann M, Tieqiao W, Kantharidis P. RAGE Deletion Confers Renoprotection by Reducing Responsiveness to Transforming Growth Factor-β and Increasing Resistance to Apoptosis. Diabetes. 2018;32:233. doi: 10.2337/db17-0538. [DOI] [PubMed] [Google Scholar]
- 69.Hong JN, Li WW, Wang LL, Guo H, Jiang Y, Gao YJ. Jiangtang decoction ameliorate diabetic nephropathy through the regulation of PI3K/Akt-mediated NF-κB pathways in KK-Ay mice. Chin Med. 2017;12:13. doi: 10.1186/s13020-017-0134-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tan J, He J, Qin W, Zhao L. Quercetin alleviates lipopolysaccharide-induced acute kidney injury in mice by suppressing TLR4/NF-κB pathway. Nan Fang Yi Ke Da Xue Xue Bao. 2019;39:598–602. doi: 10.12122/j.issn.1673-4254.2019.05.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guo S, Sun J, Zhuang Y. Quercetin alleviates lipopolysaccharide-induced inflammatory responses by up-regulation miR-124 in human renal tubular epithelial cell line HK-2. Biofactors. 2020;46:402–10. doi: 10.1002/biof.1596. [DOI] [PubMed] [Google Scholar]
- 72.Liu CM, Sun YZ, Sun JM, Ma JQ, Cheng C. Protective role of quercetin against lead-induced inflammatory response in rat kidney through the ROS-mediated MAPKs and NF-κB pathway. Biochim Biophys Acta. 2012;1820:1693–703. doi: 10.1016/j.bbagen.2012.06.011. [DOI] [PubMed] [Google Scholar]
- 73.Peng J, Li Q, Li K, Zhu L, Lin X, Lin X. et al. Quercetin Improves Glucose and Lipid Metabolism of Diabetic Rats: Involvement of Akt Signaling and SIRT1. J Diabetes Res. 2017;2017 doi: 10.1155/2017/3417306. 3417306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huang WC, Liou CJ, Shen SC, Hu S, Hsiao CY, Wu SJ. Luteolin Attenuates IL-1β-Induced THP-1 Adhesion to ARPE-19 Cells via Suppression of NF-κB and MAPK Pathways. Mediators Inflamm. 2020;2020 doi: 10.1155/2020/9421340. 9421340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Che DN, Shin JY, Kang HJ, Cho BO, Kim YS, Jang SI. Luteolin suppresses IL-31 production in IL-33-stimulated mast cells through MAPK and NF-κB signaling pathways. Int Immunopharmacol. 2020;83 doi: 10.1016/j.intimp.2020.106403. 106403. [DOI] [PubMed] [Google Scholar]
- 76.You Y, Wang R, Shao N, Zhi F, Yang Y. Luteolin suppresses tumor proliferation through inducing apoptosis and autophagy via MAPK activation in glioma. OncoTargets Ther. 2019;12:2383–96. doi: 10.2147/OTT.S191158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xue J, Ye J, Xia Z, Cheng B. Effect of luteolin on apoptosis, MAPK and JNK signaling pathways in guinea pig chondrocyte with osteoarthritis. Cell Mol Biol (Noisy-le-Grand, France) 2019;65:91–5. [PubMed] [Google Scholar]
- 78.Park S, Lim W, You S, Song G. Ameliorative effects of luteolin against endometriosis progression in vitro and in vivo. J Nutr Biochem. 2019;67:161–72. doi: 10.1016/j.jnutbio.2019.02.006. [DOI] [PubMed] [Google Scholar]
- 79.Aziz N, Kim MY, Cho JY. Anti-inflammatory effects of luteolin: A review of in vitro, in vivo, and in silico studies. J Ethnopharmacol. 2018;225:342–58. doi: 10.1016/j.jep.2018.05.019. [DOI] [PubMed] [Google Scholar]
- 80.Lu X, Li Y, Li X, Aisa HA. Luteolin induces apoptosis in vitro through suppressing the MAPK and PI3K signaling pathways in gastric cancer. Oncol Let. 2017;14:1993–2000. doi: 10.3892/ol.2017.6380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim S, Chin YW, Cho J. Protection of Cultured Cortical Neurons by Luteolin against Oxidative Damage through Inhibition of Apoptosis and Induction of Heme Oxygenase-1. Biol Pharm Bull. 2017;40:256–65. doi: 10.1248/bpb.b16-00579. [DOI] [PubMed] [Google Scholar]
- 82.Yu D, Li M, Tian Y, Liu J, Shang J. Luteolin inhibits ROS-activated MAPK pathway in myocardial ischemia/reperfusion injury. Life Sci. 2015;122:15–25. doi: 10.1016/j.lfs.2014.11.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.