ABSTRACT
An estimated 3.5 billion people are colonized by intestinal parasites worldwide. Intestinal parasitic eukaryotes interact not only with the host but also with the intestinal microbiota. In this work, we studied the relationship between the presence of multiple enteric parasites and the community structures of gut bacteria and eukaryotes in an asymptomatic mother-child cohort from a semirural community in Mexico. Fecal samples were collected from 46 mothers and their respective children, with ages ranging from 2 to 20 months. Mothers and infants were found to be multiparasitized by Blastocystis hominis, Entamoeba dispar, Endolimax nana, Chilomastix mesnili, Iodamoeba butshlii, Entamoeba coli, Hymenolepis nana, and Ascaris lumbricoides. Sequencing of bacterial 16S rRNA and eukaryotic 18S rRNA genes showed a significant effect of parasite exposure on bacterial beta-diversity, which explained between 5.2% and 15.0% of the variation of the bacterial community structure in the cohort. Additionally, exposure to parasites was associated with significant changes in the relative abundances of multiple bacterial taxa, characterized by an increase in Clostridiales and decreases in Actinobacteria and Bacteroidales. Parasite exposure was not associated with changes in intestinal eukaryote relative abundances. However, we found several significant positive correlations between intestinal bacteria and eukaryotes, including Oscillospira with Entamoeba coli and Prevotella stercorea with Entamoeba hartmanni, as well as the co-occurrence of the fungus Candida with Bacteroides and Actinomyces, Bifidobacterium, and Prevotella copri and the fungus Pichia with Oscillospira. The parasitic exposure-associated changes in the bacterial community structure suggest effects on microbial metabolic routes, host nutrient uptake abilities, and intestinal immunity regulation in host-parasite interactions.
IMPORTANCE The impact of intestinal eukaryotes on the prokaryotic microbiome composition of asymptomatic carriers has not been extensively explored, especially in infants and mothers with multiple parasitic infections. In this work, we studied the relationship between protist and helminth parasite colonization and the intestinal microbiota structure in an asymptomatic population of mother-child binomials from a semirural community in Mexico. We found that the presence of parasitic eukaryotes correlated with changes in the bacterial gut community structure in the intestinal microbiota in an age-dependent way. Parasitic infection was associated with an increase in the relative abundance of the class Clostridia and decreases of Actinobacteria and Bacteroidia. Parasitic infection was not associated with changes in the eukaryote community structure. However, we observed strong positive correlations between bacterial and other eukaryote taxa, identifying novel relationships between prokaryotes and fungi reflecting interkingdom interactions within the human intestine.
KEYWORDS: parasites, eukaryotes, protists, helminths, microbiota, bacteria, 16S sequencing, 18S sequencing, children, Mexico
INTRODUCTION
Bacteria, viruses, archaea, fungi, and protists inhabiting the mucosal surfaces of the human body have coevolved with the human intestine for millions of years and have broad effects on the physiology of the host. The diverse intestinal bacterial community shares its habitat with a dynamic community of eukaryotes, many of which are well-known parasites. Their interaction can affect the success of parasite colonization, influencing its outcome along the entire parasitism-mutualism spectrum (1). Furthermore, these microbial communities can affect host processes, including metabolism and the normal development and function of the mucosal and systemic immune systems (2–10).
Even though parasites colonize millions of people around the world, the potential damage caused by parasitic colonization is often controlled either by the host or by both the parasite and host, leading to asymptomatic colonization. The influence of intestinal parasites on resident bacterial and eukaryotic community structures has not been fully addressed; some of the previous works have been performed in experimental models of disease (10), but studies in the human host are scarce. Thus, the study of how parasites influence the intestinal microbiota in asymptomatic individuals is an important and relevant topic, especially given the large effect that the microbiome has on the host.
Little is known about the eukaryotic microbiome, or “eukaryome,” and virtually nothing is known about its community structure, particularly in the case of protist colonizations. Morton et al. (5) found a strong association of the presence of Entamoeba with a higher frequency of Firmicutes and a lower frequency of Bacteroidetes in Entamoeba histolytica-positive samples. Nieves-Ramírez et al. (11), working in the same semirural Mexican population as the one of this study, found that asymptomatic colonization in adults with the protist Blastocystis was strongly associated with increases in alpha- and beta-diversities of bacteria and with more discrete changes in the microbial eukaryome.
It has been widely documented that the initial development of the intestinal microbiota in infants has a profound effect on adult intestinal health and disease (12–16). The first 1,000 days of life play an important role in determining the phylogenetic structure of the adult human gut microbiota (16) and factors such as birth mode, breastfeeding, geographical location, household siblings, and pets are important in intestinal microbiome development, especially during the first year of life (17, 18). Early postnatal exposures to parasitic infections could also affect the development of the gut microbiota structure. Given the lack of knowledge on the role of eukaryotes in the establishment of the early-life microbiota, we aimed to study this in a population of 46 mother-child (M-C) asymptomatic binomials from a semirural Mexican population with high levels of intestinal parasite exposure. A comprehensive microbiome assessment was performed using 16S rRNA and 18S rRNA Illumina sequencing analyses for the characterization of bacteria and eukaryotes in the fecal microbiome. The results revealed interesting associations between parasite colonization and distinct microbiome patterns in the intestines of children under 2 years of age.
RESULTS
Characteristics of the individuals studied.
A cohort of 46 mother-child (M-C) binomials was studied. For the study, we selected 11 parasite-positive weaned infants and their parasite-positive mothers, 13 parasite-negative weaned infants and their parasite-negative mothers, 12 parasite-exposed unweaned infants (parasite-negative children) and their parasite-positive mothers, and 10 parasite-unexposed unweaned infants (negative children) and their parasite-negative mothers (Table 1).
TABLE 1.
Demographic data for the children in this study
| Parameter | Value for group |
|||||
|---|---|---|---|---|---|---|
| Unweaned |
Weaned |
|||||
| Parasite exposed (n = 12) | Parasite unexposed (n = 10) | P valuea | Parasite positive (n = 11) | Parasite negative (n = 13) | P valuea | |
| Mean age (mo) | 3.0 | 3.1 | 0.6951 | 19.9 | 17.1 | 0.0189 |
| Mean age at weaning (mo) | 11.5 | 10.5 | 0.4010 | |||
| No. of females/no. of males | 3/9 | 5/5 | 0.2248 | 9/2 | 3/10 | 0.0041 |
| No. of children delivered vaginally/no. delivered by C-section | 12/12 | 10/10 | 5/6 | 7/6 | 0.6820 | |
| Mean age of mother (yrs) | 25.5 | 29.5 | 0.0839 | 25.1 | 29.5 | 0.1225 |
Statistical analysis was performed using chi-square and Fisher’s exact tests for categorical data and a t test for numerical data.
The age of the children ranged from 2 to 20 months (average of 11.04 months). The parasite-positive weaned children were significantly older (P = 0.0189) and were composed of more females (P = 0.0041) than the parasite-negative weaned children. Children were breastfed for an average of 10.9 months (6 to 12 months). Twelve children were delivered by C-section, and 34 were delivered vaginally. All individuals were within a healthy weight range and did not show any stunting or wasting. The mothers’ average age was 27.4 years, with a range of 18 to 47 years. None of the participants reported antibiotic or other drug use, gastrointestinal symptoms (according to the Rome III questionnaire), or inflammatory signs and symptoms (according to medical examination) in the 6 months prior to sampling.
Parasites found in mother-child binomials.
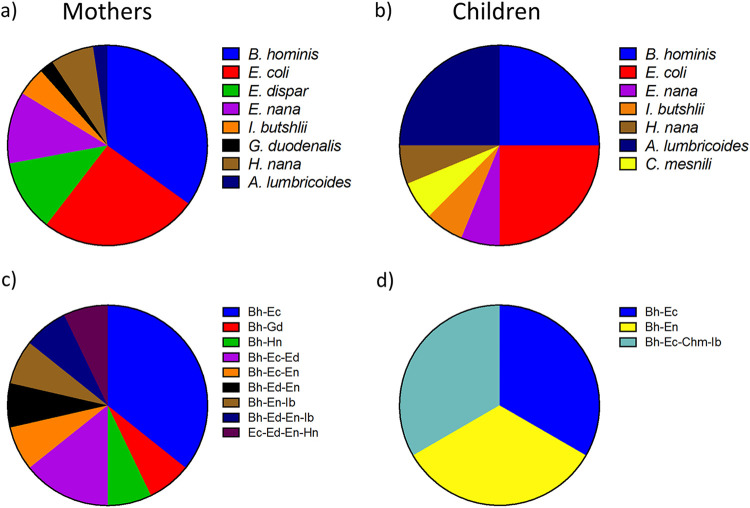
Intestinal parasites found by either microscopy or quantitative PCR (qPCR) in feces of mothers and children included Blastocystis hominis, Entamoeba coli, Entamoeba dispar, Endolimax nana, Iodamoeba butshlii, Giardia duodenalis, Chilomastix mesnili, Hymenolepis nana, and Ascaris lumbricoides (Fig. 1). E. dispar was distinguished from E. histolytica in the taxonomy assignment from 18S sequencing. The parasite most frequently found was B. hominis, present in 32.6% of positive mothers and 8.7% of positive children, followed by E. coli (23.9% and 8.7%, respectively). In the mothers, it was also common to find E. dispar (10.8%), E. nana (10.8%), and H. nana (6.5%). In contrast to mothers, A. lumbricoides was among the most frequent parasites in infants (8.7%). We also found several cases of cocolonizations by two or more intestinal parasites, particularly in the mothers, with B. hominis-E. coli being the most frequently found (10.8% in mothers and 2.2% in infants). Of note, none of the parasite-positive individuals (either mothers or infants) presented gastrointestinal symptoms.
FIG 1.
Proportions of parasites found in fecal samples from mother (a and c)-weaned child (b and d) binomials. (a) The intestinal parasites found in the mothers comprised Blastocystis hominis (Bh) (15 [32.6%]), Entamoeba coli (Ec) (11 [23.9%]), Entamoeba dispar (Ed) (5 [10.8%]), Endolimax nana (En) (5 [10.8%]), Iodamoeba butshlii (Ib) (2 [4.3%]), Giardia duodenalis (Gd) (1 [2.2%]), Chilomastix mesnili (Chm) (0 [0.0%]), Hymenolepis nana (Hn) (3 [6.5%]), and Ascaris lumbricoides (Al) (1 [2.2%]). (b) In children, the parasite frequencies were determined for Blastocystis hominis (4 [8.7%]), Entamoeba coli (4 [8.7%]), Entamoeba dispar (0 [0.0%]), Endolimax nana (1 [2.2%]), Iodamoeba butshlii (1 [2.2%]), Giardia duodenalis (0 [0.0%]), Chilomastix mesnili (1 [2.2%]), Hymenolepis nana (1 [2.2%]), and Ascaris lumbricoides (4 [8.7%]). (c) Cocolonizations by two or more parasites found in the mothers included Blastocystis hominis-Entamoeba coli (5 [10.8%]), Blastocystis hominis-Giardia duodenalis (1 [2.2%]), Blastocystis hominis-Hymenolepis nana (1 [2.2%]), Blastocystis hominis-Entamoeba coli-Entamoeba dispar (2 [4.3%]), Blastocystis hominis-Entamoeba coli-Endolimax nana (1 [2.2%]), Blastocystis hominis-Entamoeba dispar-Endolimax nana (1 [2.2%]), Blastocystis hominis-Endolimax nana-Iodamoeba butshlii (1 [2.2%]), Blastocystis hominis-Entamoeba dispar-Endolimax nana-Iodamoeba butshlii (1 [2.2%]), and Entamoeba coli-Entamoeba dispar-Endolimax nana-Hymenolepis nana (1 [2.2%]). (d) Cocolonizations found in children were Blastocystis hominis-Entamoeba coli (1 [2.2%]), Blastocystis hominis-Endolimax nana (1 [2.2%]), and Blastocystis hominis-Entamoeba coli-Chilomastix mesnili-Iodamoeba butshlii (1 [2.2%]).
Bacterial and eukaryotic diversity in individuals colonized by parasites.
(i) Binomial identity. We determined the fecal bacterial and eukaryotic compositions of the 46 binomials. The parasites found by microscopy or qPCR that were also detected by 18S rRNA gene sequencing were B. hominis, E. coli, E. dispar, E. nana, Iodamoeba, Ascaris, and H. nana.
In order to evaluate if the mother’s intestinal microbiota composition could be a determinant of the intestinal microbiota in children, we tested for differences in Bray-Curtis distances between binomial pairs (intrabinomial) versus all other individuals (interbinomial) with a nonparametric Kruskal-Wallis test. Intrabinomial dissimilarities were not significantly different from interbinomial dissimilarities for both bacterial and eukaryote gut communities (see Fig. S1 in the supplemental material).
Principal-component analysis (PCoA) ordination of the variation in the beta-diversity of human gut bacterial (left) and eukaryote (right) communities in weaned infant (1 year old)-mother binomials (a and b) and unweaned infant (3 months old)-mother binomials (c and d) on Bray-Curtis dissimilarities. Colors represent binomial identity, while shapes represent class (squares for mothers and circles for infants). PERMANOVAs indicate no significant effect of binomial identity. Dark colors indicate no colonization, while light colors indicate colonization. Download FIG S1, TIF file, 0.6 MB (678.7KB, tif) .
Copyright © 2021 Partida-Rodriguez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(ii) Effects on infants less than 5 months old. Because of the known differences in microbiome structure and diversity across the first year of life, as well as due to breastfeeding as a major factor influencing microbiome development over this period (17, 18), we evaluated the effect of parasites in unweaned infants under 1 year of age separately from the rest of the individuals over 1 year of age, including weaned infants. The infants under 1 year of age were all under 5 months of age, and none of them were infected by parasites, but we considered them parasite exposed when their mothers tested positive for parasites.
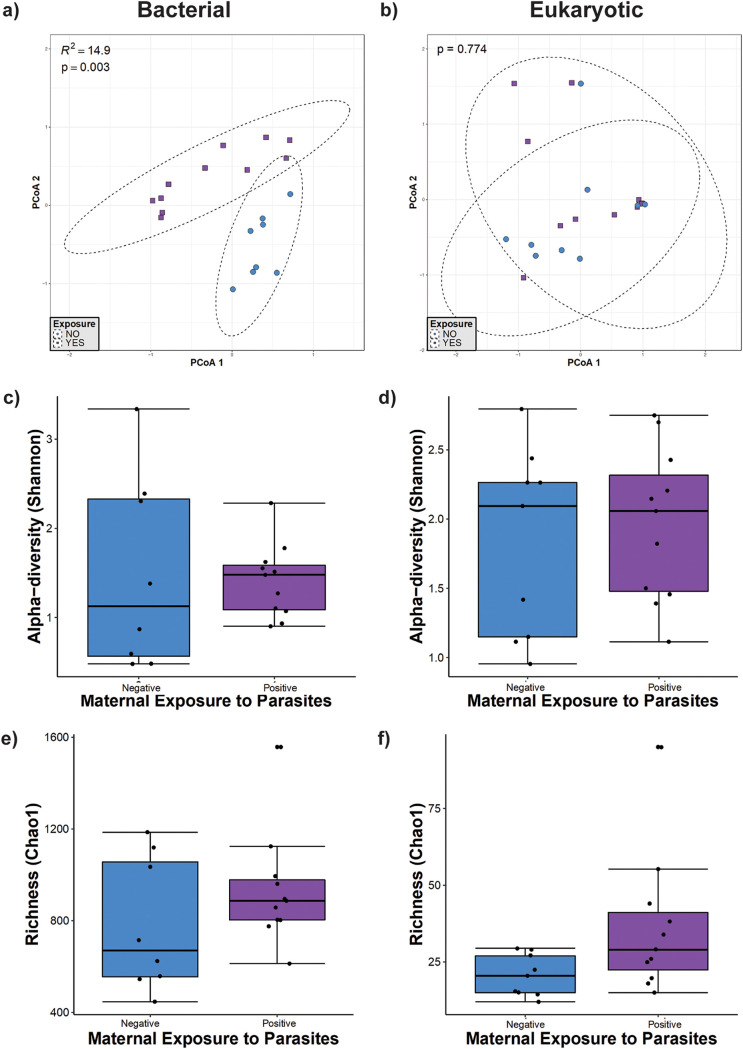
We identified the relationships between bacterial community structure and parasite exposure by conducting permutational multivariate analysis of variance (PERMANOVA) on the community matrix. Based on Bray-Curtis dissimilarities (Fig. 2), we found that the bacterial community structure was significantly related to parasite exposure in infants under 1 year of age (P = 0.003 by PERMANOVA). According to PERMANOVA, parasite exposure explained 15% of the variation in bacterial beta-diversity (Fig. 2a); however, we did not find an effect of parasite exposure on the eukaryote community structure (Fig. 2b). We also calculated the Shannon diversity index and Chao1 richness of bacterial and eukaryotic communities in this group of infants, and we did not find any statistically significant differences between the exposed and nonexposed groups (Fig. 2c to f).
FIG 2.
Microbial diversity in unweaned infants (younger than 5 months of age). (a and b) Principal-component analysis (PCoA) ordination of variation in beta-diversities of human gut bacterial (a) and eukaryote (b) communities based on Bray-Curtis dissimilarities. Color and shape represent maternal exposure to parasites (blue circles represent negative exposure, and purple squares represent positive exposure). PERMANOVAs indicate that maternal exposure to parasites explains 15% (P = 0.003) of the variation in the infant bacterial community structure but is not a significant (P = 0.774) driver of the eukaryote community structure. (c and d) Shannon diversity of gut bacterial (c) and eukaryote (d) community structures. (e and f) Estimated richness of gut bacterial (e) and eukaryote (f) community structures. No significant differences were detected by Mann-Whitney tests for alpha-diversity comparisons between the parasite-positive and -negative groups.
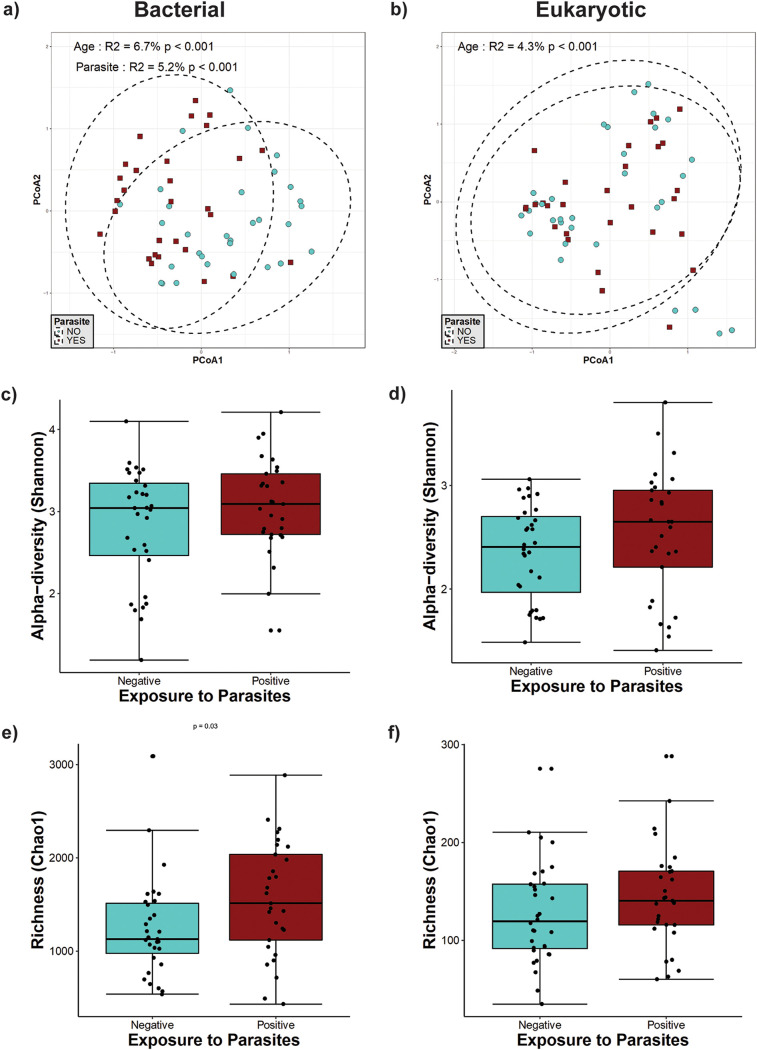
(iii) Individuals older than 1 year of age. In individuals older than 1 year of age (including mothers and weaned children), we found a significant effect of parasite presence on bacterial beta-diversity (Fig. 3a), which explained 5.2% of the variation present in this group. The presence of intestinal parasites was also significantly associated with changes in richness, with an increase in the Chao1 richness for bacterial diversity (P = 0.04) but not eukaryotic diversity (Fig. 3e and f). However, there were no statistically significant differences in bacterial and eukaryote Shannon diversities between parasite-positive and -negative individuals in this age group (Fig. 3c and d), suggesting that changes in beta-diversity are mainly due to a modification of the bacterial taxa present in the gut microbiota. We also observed a significant effect of age on both bacterial (P < 0.001) (Fig. 3a) and eukaryote (P < 0.001) (Fig. 3b) beta-diversities, explaining the variation found in 6.7% and 4.3%, respectively.
FIG 3.
Microbial diversity in individuals older than 1 year of age. (a and b) Principal-component analysis (PCoA) ordination of the variation in the beta-diversity of human gut bacterial (a) and eukaryote (b) communities based on Bray-Curtis dissimilarities. Color and shape represent maternal exposure to parasites (turquoise circles for negative and dark-red squares for positive exposure). PERMANOVAs indicate that maternal exposure to parasites and age explain 5.2% and 6.7% (P < 0.001) of the variation in the bacterial community structure, respectively, while age explains 4.3% (P < 0.001) of the variation in the eukaryote community structure. Ellipses represent the confidence intervals at 95%. (c and d) Shannon diversity of gut bacterial (c) and eukaryote (d) community structures. No significant differences were detected by Mann-Whitney tests for Shannon diversity between the parasite-positive and -negative groups. (e and f) Estimated richness of gut bacterial (e) and eukaryote (f) community structures. A significant difference was detected for bacterial community richness by Mann-Whitney tests for comparisons between two groups.
(iv) Infants from 1 to 2 years of age. In older weaned children between 1 and 2 years of age, we detected an effect of parasite presence on the bacterial community structure (Fig. 4). Parasite exposure explained 8.7% of the bacterial variation, whereas age explained 7.7% (Fig. 4a). Chao1 richness and Shannon diversity indices showed no changes in richness or evenness (Fig. 4c and e). Although there was no significant effect detected on eukaryotic beta-diversity (Fig. 4b) and eukaryotic richness (Fig. 4f), eukaryote alpha-diversity (Shannon index) showed a statistically significant increase in the group colonized by parasites (Fig. 4d).
FIG 4.
Microbial diversity in weaned infants between 1 year and 2 years of age. (a and b) Principal-component analysis (PCoA) ordination of the variation in the beta-diversity of human gut bacterial (a) and eukaryote (b) communities based on Bray-Curtis dissimilarities. Color and shape represent maternal exposure to parasites (turquoise circles for negative and dark-red squares for positive exposure). PERMANOVAs indicate that exposure to parasites and age explain 8.7% (P < 0.01) and 7.7% (P = 0.026) of the variation in the infant gut bacterial community structure, respectively. Ellipses represent the confidence intervals at 95%. No significant effects of age or exposure to parasites were detected for the eukaryote community structure. (c and d) Shannon diversity of gut bacterial (c) and eukaryote (d) community structures. A significant difference was detected only for eukaryote community alpha-diversity by Mann-Whitney tests for comparisons between the two groups. (e and f) Estimated richness of gut bacterial (e) and eukaryote (f) community structures. No significant differences were detected by Mann-Whitney tests for Chao1 estimated richness between the parasite-positive and -negative groups.
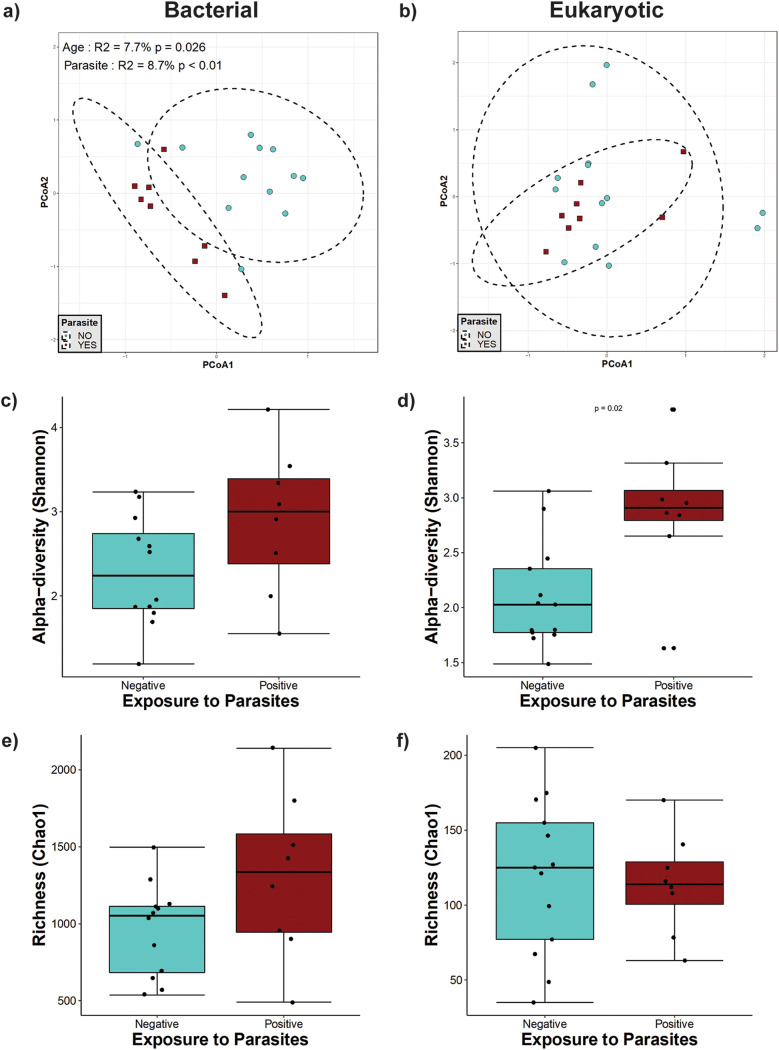
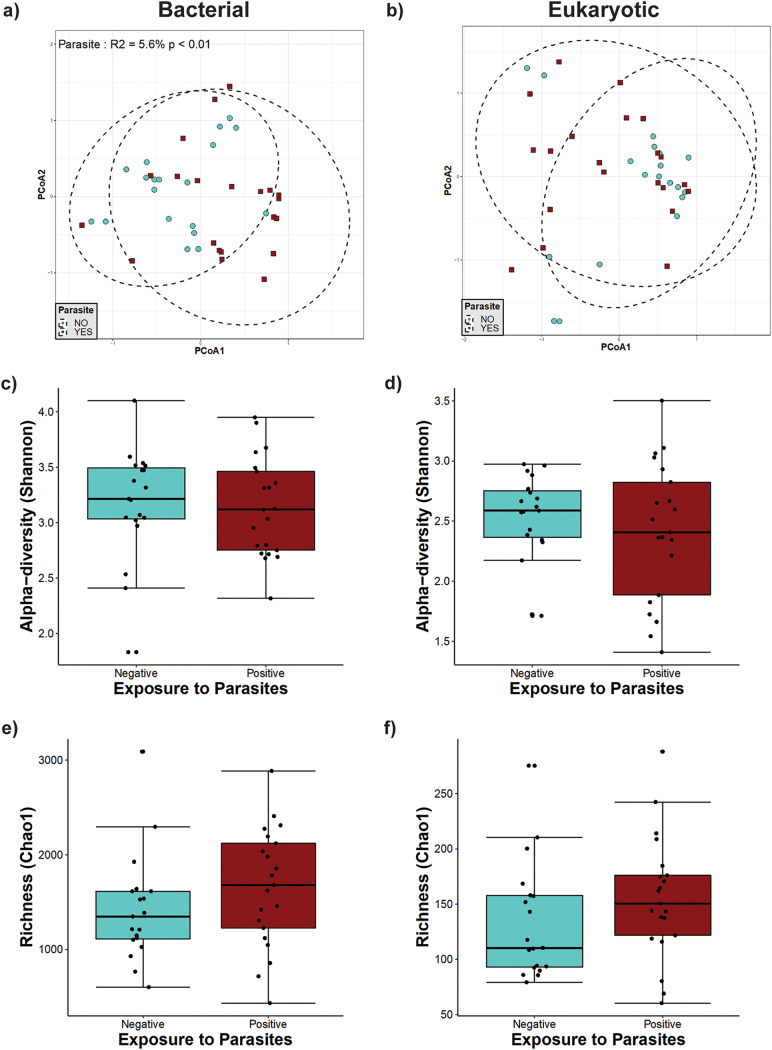
In mothers, we also found a significant effect of parasite exposure on bacterial beta-diversity (Fig. 5a), explaining 5.6% of the variation of the community structure, and no effect on eukaryotic diversity (Fig. 5b). No statistically significant effects were observed in mothers exposed to parasites in bacterial and eukaryote community richness (Chao1) and alpha-diversity (Shannon) (Fig. 5c to f).
FIG 5.
Microbial diversity in mothers. (a and b) Principal-component analysis (PCoA) ordination of the variation in beta-diversity of human gut bacterial (a) and eukaryote (b) communities based on Bray-Curtis dissimilarities. Color and shape represent maternal exposure to parasites (turquoise circles for negative and dark-red squares for positive exposure). PERMANOVAs indicate that exposure to parasites explains 5.6% (P < 0.01) of the variation in mother gut bacterial community structure. Ellipses represent the confidence intervals at 95%. No significant effects of age or exposure to parasites were detected for the eukaryote community structure. (c and d) Shannon diversity of gut bacterial (c) and eukaryote (d) community structures. (e and f) Estimated richness of gut bacterial (e) and eukaryote (f) community structures. No significant differences were detected by Mann-Whitney tests for richness and alpha-diversity between the parasite-positive and -negative groups.
Relative abundances of gut bacteria and eukaryotes.
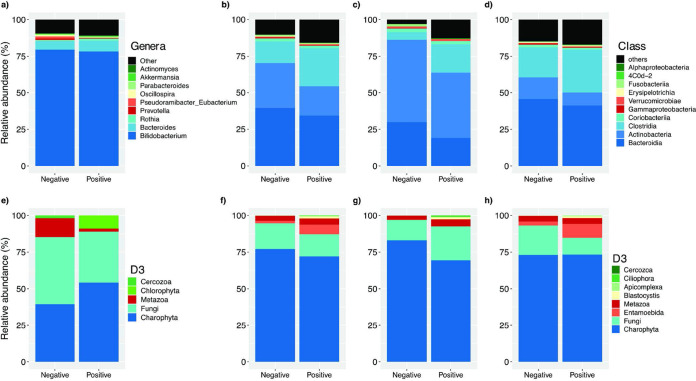
Differential abundance analysis revealed changes in bacterial composition associated with parasite colonization and exposure (Fig. 6). In unweaned infants less than 5 months old exposed to parasites, the most abundant genera were Bifidobacterium and Bacteroides (Fig. 6a), whereas the genera Pseudoramibacter, Eubacterium, Prevotella, and Oscillospira were less abundant than in nonexposed infants. We also found decreases in the taxa Metazoa and Fungi in the unweaned infants exposed to parasites (Fig. 6e).
FIG 6.
Relative abundances of gut bacterial (a to d) and eukaryote (e to h) compositions of parasite-exposed individuals. (a and e) Bacterial and eukaryote compositions in unweaned infants younger than 5 months of age. (b to d and f to h) Relative abundances depending on parasite colonization in individuals over 1 year of age (b and f), only in weaned infants between 1 and 2 years of age (c and g), and only in mothers (d and h).
In individuals older than 1 year of age, only weaned children from 1 to 2 years of age, and only the mothers, the most abundant classes were Bacteroidia, Actinobacteria, Clostridia, and Coriobacteriia, and the presence of parasites was consistently associated with an increase in the relative abundance of Clostridia and decreases in the relative abundances of Actinobacteria and Bacteroidia (Fig. 6b to d). There were also decreases of Fusobacteria and Gammaproteobacteria proportions associated with colonization by parasites in the children from 1 to 2 years of age and the mothers, respectively.
Concerning eukaryotes, in individuals older than 1 year of age, there was a decrease of the taxon Fungi (Fig. 6f), whereas in only children from 1 to 2 years of age, there was an increase in Fungi (Fig. 6g), and in only the mothers, Fungi had a lower abundance in parasite-positive individuals (Fig. 6h).
Eukaryote-bacterium correlations.
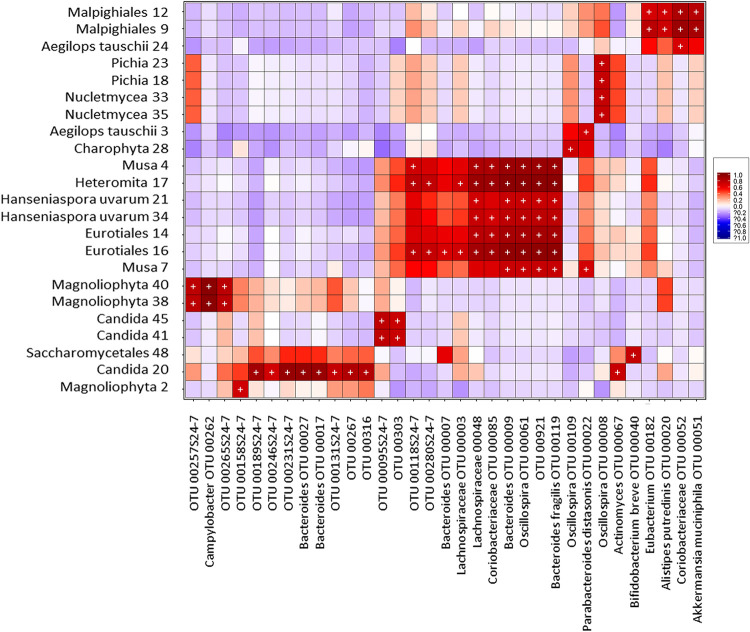
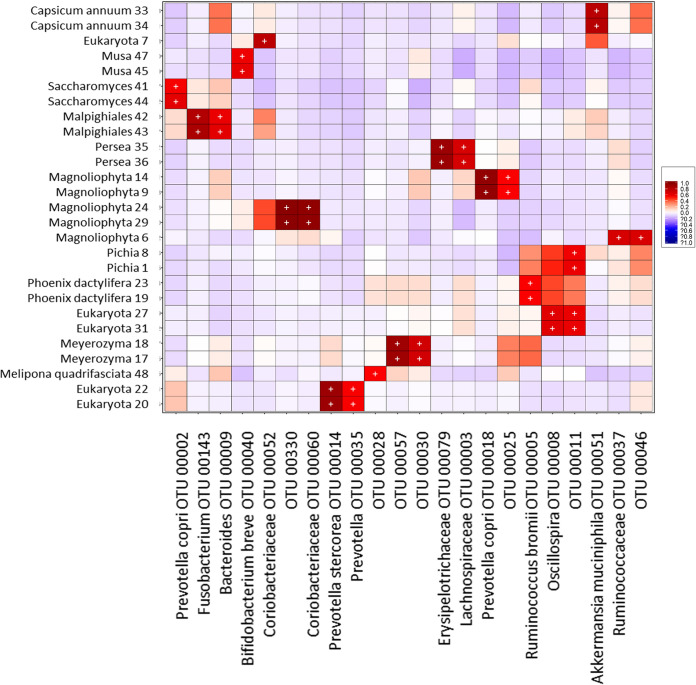
Given the observed role of parasites in the bacterial community structure, we wanted to know whether additional relationships of other eukaryotes with bacteria were present in the intestine. To look for correlations, we created heat maps of biweight correlations between the top 50 bacterial taxon operational taxonomic units (OTUs) and the top 50 eukaryote OTUs in fecal samples, independently of parasite exposure. In the group of unweaned infants less than 5 months of age, we observed numerous statistically significant correlations between bacteria and eukaryotes (Fig. 7), several of which were from food and environmental origins, whereas in both the group of weaned children older than 1 year of age and the group of mothers, there were fewer correlations (Fig. 8).
FIG 7.
Heat map of biweight correlations (Pearson) between the top 50 bacterial (x axis) and the top 50 eukaryote (y axis) taxon OTUs in fecal samples of unweaned infants (∼3 to 4 months old). Colors denote positive (red) and negative (blue) correlation values. Significant correlations are denoted with a plus sign (P < 0.05 [false discovery rate {FDR}]).
FIG 8.
Heat map of biweight correlations (Pearson) between the top 50 bacterial (x axis) and the top 50 eukaryote (y axis) taxon OTUs in fecal samples of weaned children (>1 year old) and mothers. Colors denote positive (red) and negative (blue) correlation values. Significant correlations are denoted with a plus sign (P < 0.05 [FDR]).
We found positive correlations of Bacteroides, Bacteroides fragilis, Lachnospiraceae, Coriobacteriaceae, and Oscillospira with the protist Heteromita. Remarkably, we found positive correlations of Oscillospira with Entamoeba coli and Prevotella stercorea with Entamoeba hartmanni. Additionally, we found various positive correlations of bacteria with fungi, such as Bacteroides and Bifidobacterium with Candida, Aspergillus, and Hanseniaspora; Lachnospiraceae and Coriobacteriaceae with Aspergillus and Hanseniaspora; Oscillospira with Pichia, Aspergillus, and Hanseniaspora; Actinomyces with Candida; and Prevotella copri with Saccharomyces.
DISCUSSION
The most recent reports available estimate that 3.5 billion people are colonized by parasites globally (19, 20). Even though eukaryotes are present in much lower abundances than bacteria, it has been demonstrated that monocolonization by parasites is associated with intestinal microbiota composition changes (5, 11, 21–24). Colonization by parasitic eukaryotes usually does not follow a one-host–one-parasite model (25–29), and very few studies have assessed the intestinal microbiota composition when multiple parasites are present (30). In this project, we studied whether exposure to intestinal parasites in an asymptomatic cohort of mother-child binomials from a semirural community in Mexico is related to changes in the bacterial and eukaryotic intestinal microbiota. Our study revealed important changes in bacterial intestinal microbiota relative abundances associated with exposure to parasites, characterized by an increase in the abundance of the taxon Clostridia and decreases in the abundances of Actinobacteria and Bacteroidia, with no important changes in alpha-diversity indices.
The mother’s microbiota has been determined to be an important microbial source during early colonization of the infant gut (13, 31–41); however, we found no binomial identity effect on either bacterial or eukaryote communities. This may be because most of the above-mentioned studies compared infant samples collected shortly after birth, and the samples analyzed in this study were from older infants. However, it is also possible that the increased dissimilarity between mother-infant binomials in this semirural setting may reflect a larger influence of other individuals or other components of the environment rather than vertical transmission, as has been previously reported for the development of microbiome structure and diversity (32, 41, 42).
We found nine different parasites in the binomials studied, predominated by the protists B. hominis (20.6%), E. coli (16.3%), E. nana (6.5%), and E. dispar (5.4%) and the two helminths A. lumbricoides (5.4%) and H. nana (4.3%). More than half of the exposed individuals (54.8%) were colonized by two or more different parasites. Yet all parasite-positive individuals in our cohort remained asymptomatic. Interestingly, when parasite-positive individuals were compared with parasite-negative ones, we found that the parasite-positive weaned children were significantly older and had a higher proportion of females.
Even though we did not find parasite colonization in the unweaned children, we hypothesize that these infants are exposed to the parasites of their parasite-positive mothers. These exposures originate from birth, breastfeeding, and spending the majority of the time together in the same environment (39). Due to the vertical transmission of bacteria from the mother, the intestinal microbiota of the newborn closely matches the maternal microbiota, and depending on the delivery mode, it resembles the stool, vaginal, or skin microbiota (34–36). It remains unclear why infants in this age group did not become colonized by parasites, but our data clearly indicate that even in the absence of colonization, there are distinct microbiome patterns associated with maternal colonization. These infants were unweaned, and with breastfeeding, the infant bacterial diversity remains limited because the microbiota is dominated by species involved in human milk metabolism in breastfed infants, with greater Bacteroides and Bifidobacterium abundances (16, 38).
In the first 4 months of gut microbe colonization, the infant microbiota gradually differs from the mother’s microbiota, reflecting the progressively increasing gut microbial diversity and complexity of the growing infant over microbes acquired maternally (31, 35). Although we did not find evidence of parasite colonization in the unweaned children, parasites from their positive mothers might still find their entrance to the infant intestine. The parasites are incapable of colonizing, probably also because of the level of maturation of the intestinal microbiota (36). However, these parasites might be temporarily passing through the infant intestine, which may be long enough to interact with the bacteria of the children’s gut and influence its composition. It is also possible that the bacteria that come into contact with the infant from the mother might have already been selected by the parasites in the mother’s gut. These are just hypotheses at this point and require further research.
Our study detected increases in bacterial and eukaryotic richness and alpha-diversity due to parasite colonization only in unweaned infants over 1 year old. Previous reports have associated colonization by parasites with higher intestinal bacterial diversity. A previous study by Morton et al. (5) in rural populations in Cameroon found that the presence of the protist Entamoeba was associated with a significant increase in alpha (intrahost)-diversity. Furthermore, a recent study by our group (11) found dramatic increases in bacterial richness and alpha-diversity in people colonized by Blastocystis from the same community as the one of the present study. The discrepancies between this study and the previous ones, which did not include infants, strongly suggest that the effect of parasite exposure on alpha-diversity could be age dependent and may be attributable to the lower alpha-diversity normally found in infant samples (18, 37). This is supported by a recent study in children from Colombia (30), which also found no differences in bacterial alpha-diversity in children positive for parasites. However, we did not observe an increase in alpha-diversity in the parasite-positive mothers. This finding suggests that in addition to age, other factors may play a role. We think that the common multiparasitic colonization found in our settings may lead to more complex interactions with the resident microbiota, which could affect the bacterial alpha-diversity in the intestine. This may be detected only with further studies with a larger population size and by comparing the effects between monoparasitic and multiparasitic colonization.
Despite the discrepancies with other studies in relation to alpha-diversity, we found that parasite exposure was significantly associated with bacterial intestinal microbiota beta-diversity in both infants and mothers in this study but not with the eukaryotic microbiota. In unweaned children younger than 5 months old, parasite exposure explained 14.9% (P = 0.003) of the variation in bacterial community structure, whereas in the groups of older weaned infants and mothers, parasite exposure was a weaker driver of beta-diversity. This suggests that while parasite exposure is an important factor shaping intestinal bacterial beta-diversity, its effects are more evident during the earlier stages of gut bacterial community establishment.
Exposure to parasites was also associated with changes in specific taxa, including an increase in the class Clostridia and a decrease in the class Bacteroidia. This result agrees with the results of a study by Morton et al. (5), in which there was a strong correlation of Entamoeba with a higher frequency of Firmicutes (particularly the Clostridia class) and a lower frequency of Bacteroidetes (mostly Prevotella) in E. histolytica-positive samples. In the study focused on Blastocystis (11), colonization was also associated with significant increases in the genera Ruminococcus and Oscillospira (members of the Clostridia class) and a reduction in Prevotella (Bacteroidia class).
These changes in relative abundance may be the result of direct parasite-bacterium interactions driven by competition for resources, predation, or the production of molecules that may affect the fitness or survival of the microorganisms involved (1). Protists are well-known bacterivores. Entamoeba and Blastocystis can graze on bacteria (43, 44), and this is an important mechanism for the top-down control of bacterial communities due to their high feeding rates (45). It was also recently reported that Trichuris muris, a nematode from the mouse gut, acquires its intestinal microbiota from the mouse intestine, very likely through ingestion (46). Furthermore, the mechanisms by which bacteria avoid protist predation change their ability to survive and can even promote the emergence of virulence and invasion (45, 47).
Parasites may also influence the bacterial community structure in the intestine through indirect interactions with bacteria. Some protists like Entamoeba and Giardia, which have mucolytic enzymes (48, 49), and helminths like Trichuris trichiura, which stimulate mucin expression or express mucin-like molecules themselves (50–52), can alter the outer mucus layer, changing the bacterial microenvironment and sources of nutrition for certain taxa. Additionally, the parasites may produce metabolites that could influence the regulation of the immune system, a mechanism commonly used in helminths. Trichuris muris and Heligmosomoides polygyrus bakeri can induce the generation of regulatory T cells (Tregs) (53, 54), changing the physical microenvironment by modifying mucus and antimicrobial peptide production, thus potentially promoting the outgrowth of specific taxa among the members of the microbiota (55). The bacterial and eukaryotic taxa identified in this study and other research should be studied in appropriate animal models to further determine the mechanisms involved in these multikingdom interactions.
The effect of exposure to parasites on intestinal eukaryote relative abundance was not as strong as it was with bacteria. However, there were some changes in the abundances of the taxa Chlorophyta, Charophyta, and Fungi, which proved to be age dependent. The correlation analysis identified several significant positive associations between other intestinal eukaryotes and bacteria, most of which were with eukaryotes from food or environmental sources, such as Cucurbita pepo, Capsicum annuum, Musa, Characium sp., and Chlorophyceae. However, among the eukaryotes identified in the sequencing analysis that are known to be gut residents (Entamoeba coli, E hartmanni, Iodamoeba, E. dispar, Ascaris, Trichuris trichiura, Hymenolepis nana, and Blastocystis), we observed positive correlations of two species with bacteria, Entamoeba coli with Oscillospira and E. hartmanni with Prevotella stercorea.
Remarkably, the strongest positive correlations found in the present study were between bacteria and several common fungi of the intestinal microbiota, even though fungal and bacterial abundances in the gut have been negatively correlated, and disruption of the bacterial microbiota is a condition required for fungal overgrowth (56). Agonistic and antagonistic relationships have been described between intestinal fungi and bacteria (56–61). It has been shown that the common yeast Candida albicans suppressed the regrowth of Lactobacillus and promoted the recovery of Bacteroidetes populations during antibiotic recovery (62). Our study also detected the co-occurrence of Candida species with Bacteroides. Other previously unreported co-occurrences of bacteria and fungi were found, such as Oscillospira with Aspergillus, Pichia, and Hanseniaspora; Actinomyces and Bifidobacterium with Candida; and Prevotella copri with Saccharomyces. The mechanisms and functions of these interactions in the gut deserve further studies.
Our work supports previous reports that the presence of intestinal parasites is linked to strong bacterial microbiota community changes. By including mother-child binomials, our work further revealed that these effects occur even in the absence of direct colonization in infants, strongly suggesting that the effect of parasite colonization on the microbiome may also lead to changes in the vertical transmission of bacterial taxa. This implies that colonization by parasites may be an important indirect factor in the inheritable features of the human gut microbiome. How these intestinal microbiota changes associated with parasites may modify the immune system and other aspects of metabolism remains to be elucidated.
MATERIALS AND METHODS
Study population, study design, and ethical considerations.
Xoxocotla is a semirural community of the State of Morelos, Mexico, located 120 km south of Mexico City (longitude, 99°19W; latitude, 18°3N) in an area spanning 29,917 km2 with a tropical climate (warm subhumid). The total population is 5,163 people, whose main sources of income are agriculture and commerce. Sample collection was carried out between April 2011 and January 2013.
In this cross-sectional study of cohorts, every volunteer mother was informed about the characteristics of the project, the objectives, and the advantages of participation, along with the biological samples needed, the sampling procedures, and possible complications that could arise. All the participant mothers signed a written informed consent letter for their children and themselves prior to sample collection. Afterwards, questionnaires to collect sociodemographic, socioeconomic, and health datum antecedents (Rome III questionnaire for gastrointestinal symptoms [63]); nutrition data; and way of delivery and to ensure that there was no use of antibiotics and other drugs for at least 6 months prior to sampling were applied by nurses at the community of Xoxocotla, Morelos, Mexico. All variables were recorded in a database.
All procedures in this study fulfilled the Reglamento de la Ley General de Salud en Materia de Investigación para la Salud of Mexico, in particular the chapters about the ethical aspects of research in human beings, research in communities, research in minors, and research in women of fertile age and pregnant women (Diario Oficial de la Federación, Febrero 1984). All methods were approved by the Ethics Committee of the Faculty of Medicine of the National Autonomous University of Mexico, and research was carried out in accordance with the Declaration of Helsinki.
Sample collection, parasite detection, and DNA extraction from fecal samples.
Fecal samples from mother-child binomials that fulfilled the criteria were collected in sterile plastic containers, immediately placed at 4°C for transport to the laboratory, and stored at −20°C until analysis. The fecal samples were submitted to stool microscopic analysis for the detection of intestinal parasites for the construction of the parasitized and nonparasitized cohorts. The presence of the main intestinal parasites historically found in Xoxocotla (E. histolytica/E. dispar, E. coli, B. hominis, I. butshlii, E. nana, C. mesnili, Giardia intestinalis, Cryptosporidium parvum, A. lumbricoides, and H. nana) was tested in fecal samples by microscopy.
DNA from fecal samples was extracted as previously described (11). Briefly, samples with 50 mg of stool were mechanically lysed using Mo Bio dry bead tubes (Mo Bio Laboratories, Inc.) in a FastPrep homogenizer (MP Biochemicals). DNA isolation was performed using the QIAamp Fast DNA stool minikit (Qiagen) according to the manufacturer’s instructions.
The presence of E. histolytica/E. dispar, B. hominis, G. intestinalis, and C. parvum was confirmed by quantitative PCR (qPCR) as previously reported (11). Briefly, qPCR was performed on an Applied Biosystems 7500 machine using QuantiTect SYBR green master mix (Qiagen) in 10-μl reaction mixture volumes with 6.25 pmol each of primers Ehd-239F–Ehd-88R, BhRDr-RD5, Giardia-80F–Giardia-127R, and CrF-CrR (see Table S1 in the supplemental material). The amplification conditions consisted of 35 cycles of 1 min each at 94°C, 59°C, and 72°C, with an additional step of 95°C for 15 s, 60°C for 1 min, 95°C for 30 s, and 60°C for 15 s (64). Samples previously known to be positive for each parasite as well as standard curves using DNA from each parasite from the ATCC’s enteric protist DNA panel were included as positive controls in the qPCR plates. The difference between the average cycle threshold (CT) value of each parasite qPCR and the average CT value of the 18S rRNA gene reaction was calculated to determine the parasitic loads in each sample.
Primer sequences used for protist determination by qPCR. Download Table S1, DOCX file, 0.03 MB (32.1KB, docx) .
Copyright © 2021 Partida-Rodriguez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Determination of fecal bacterial composition.
The DNA from 46 mother-child binomial fecal samples, isolated as described above, was used for the sequencing of microbial communities. For bacterial determination, samples were amplified by PCR in triplicate using barcoded primer pairs flanking the V4 region of the 16S rRNA gene as previously described (65, 66). Each 50-μl PCR mixture contained 22 μl of water, 25 μl of TopTaq master mix (Qiagen), 0.5 μl of each forward and reverse barcoded primer (66), and 2 μl of template DNA. To ensure that no contamination occurred, controls without template DNA were included. Amplification was performed with an initial DNA denaturation step at 95°C (5 min), 25 cycles of DNA denaturation at 95°C (1 min), an annealing step at 50°C (1 min), an elongation step at 72°C (1 min), and a final elongation step at 72°C (7 min). Amplicons displaying bands at ∼250 bp on a 2% agarose gel were purified using the QIAquick PCR purification kit (Qiagen). Purified samples were quantified with PicoGreen (Invitrogen) in a Tecan M200 plate reader (excitation at 480 nm and emission at 520 nm).
For 16S rRNA gene sequencing, each PCR pool was analyzed on the Agilent Bioanalyzer using the high-sensitivity double-stranded DNA (dsDNA) assay to determine the approximate library fragment size and verify library integrity. Pooled-library concentrations were determined using the TruSeq DNA sample preparation kit, version 2 (Illumina). Library pools were diluted to 4 nM and denatured into single strands using fresh 0.2 N NaOH. The final library loading concentration was 8 pM, with an additional PhiX spike-in of 20%. Sequencing was carried out using a HiSeq 2000 bidirectional Illumina sequencing and cluster kit, version 4 (Macrogen, Inc.).
Determination of fecal eukaryotic composition.
The composition of eukaryotic microorganisms was determined by 18S rRNA gene sequencing. DNA samples were sent to the Integrated Microbiome Resource at Dalhousie University for amplification and sequencing. The 18S rRNA gene was amplified with the primers E572F (5′-YGCGGTAATTCCAGCTC-3′) and E1009R (5′-AYGGTATCTRATCRTCTTYG-3′), and the reaction mixture included a peptide-nucleic acid blocking primer (5′-TCTTAATCATGGCCTCAGTT-3′) to reduce the amplification of mammalian sequences. Amplification was carried out in duplicate, with one reaction mixture using undiluted DNA and the other using DNA diluted 1:10 in PCR water. Amplification was conducted according to previously described protocols (67). PCR products were visualized on E-gels, quantified using Invitrogen Qubit with PicoGreen, and pooled at equal concentrations, according to a previous report (68). PhiX was spiked in at 5%, and the resulting library was sequenced at Dalhousie University on the Illumina MiSeq instrument using the MiSeq 500-cycle reagent kit, version 2 (250 by 2).
Bioinformatics analysis.
Sequences were preprocessed, demultiplexed, denoised, quality filtered, and trimmed and chimeras were removed using Mothur for the 16S rRNA gene (68) or QIIME for the 18S rRNA gene (69).
(i) 16S.
All sequences were processed using Mothur according to standard protocols as previously described (68). Quality sequences were obtained by removing sequences with ambiguous bases, a quality read length, and/or chimeras identified using chimera.uchime. Quality sequences were aligned to the SILVA bacterial reference alignment, and OTUs were generated using a dissimilarity cutoff of 0.03. Sequences were classified using the classify.seqs command.
(ii) 18S.
Demultiplexed reads were trimmed to a uniform length of 250 bp using the FastX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/) and clustered into OTUs using the minimum entropy decomposition (MED) method (70) as implemented in the oligotyping microbial analysis software package (71). MED performs de novo taxonomic clustering using Shannon entropy to separate biologically meaningful patterns of nucleotide diversity from sequencing noise; the processed data are partitioned into phylogenetically homogeneous units (MED nodes) for downstream microbial eukaryotic diversity analyses. This analysis was carried out with the minimum substantive abundance parameter (-M) set at 250 reads. All other parameters were run with default settings; the maximum variation allowed per node (-V) was automatically set at 3 nucleotides.
Representative sequences were classified by clustering against the Greengenes database at 97% similarity (16S rRNA gene [72]) or SILVA release 123 at 99% similarity (18S rRNA gene [73]). The 16S rRNA gene data set was filtered to remove mitochondrion and chloroplast sequences and OTUs present in fewer than three samples. The 18S rRNA gene data set was filtered to remove mammalian and plant sequences and all OTUs present in fewer than three samples. The final data sets contained 5,458,311 and 2,443,357 quality sequences for 16S and 18S, respectively. The 16S data set showed a mean of 64,215 (range, 2,119 to 228,403) sequences per sample identified as 3,046 bacterial OTUs. Samples contained a mean of 508 (range, 180 to 1,073) bacterial OTUs per sample. The 18S data set showed a mean of 28,085 (range, 16 to 223,045) sequences per sample identified as 694 eukaryote OTUs. Samples contained a mean of 86 (range, 6 to 244) eukaryote OTUs per sample.
Statistical analysis.
Differences in frequencies for categorical and continuous variables between cases and controls were evaluated using chi-squared and Student’s t tests, respectively.
Microbial alpha- and beta-diversities as well as the relative abundances of bacterial and eukaryotic taxa were computed using phyloseq (74), along with additional R-based computational tools (75–81). Principal-component analyses (PCoAs) were employed to visualize variation in the microbial community structure (previously transformed by variance-stabilizing transformation to ensure that our statistical results were not an artifact of heteroscedastic dispersion between groups [74]). We quantified the relative influence of various drivers of intestinal microbial beta-diversity by conducting permutational multivariate analysis of variance (PERMANOVA) on Bray-Curtis dissimilarities. The Shannon alpha-diversity and Chao1 richness indices were calculated using phyloseq without excluding singletons and doubletons, and differences between groups were statistically tested by a Mann-Whitney test.
The R packages DESeq2 (81) and MaAsLin (82) were used to calculate differentially abundant OTUs. Correlation analysis was performed using the bicor method in the R package microbiome to correlate the 100 most abundant OTUs from the 16S and 18S rRNA gene data sets. Features in the analysis were included as OTUs and as OTUs combined into taxonomic families.
ACKNOWLEDGMENTS
This work was funded by grant numbers IN226511 and IN218214 from the PAPIIT program at UNAM, grant FIS/IMSS/PROT/1368 from the IMSS, and grant numbers 140990, 272601, 283522, and 257091 from the National Council of Sciences and Technology in Mexico (CONACyT) to C.X. and J.T.; Canadian Institutes for Health Research grants to B.B.F.; and a Human Frontier Science Program grant to L.P. (grant number RGY0078/2015). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. O.P.-R. (proposal number 208253) and M.N.-R. (proposal number 235618) each received a 1-year scholarship from the Estancias Posdoctorales al Extranjero para la Consolidacion de Grupos de Investigación program of CONACyT.
We thank all study participants from Xoxocotla as well as the personnel involved in this cohort.
O.P.-R., M.N.-R., M.C.A., E.M.B., C.X., and B.B.F. designed the study. A.V.-S., P.M., J.T., O.P.-R., M.N.-R., E.G., E.R., U.M., E.H., and C.X. coordinated and facilitated the cohort study in Xoxocotla. P.M., L.R.-V., and C.X. conducted medical examinations and Rome III questionnaires. A.V.-S. curated the database and metadata. M.C.A. and L.P. optimized sequencing strategies. O.P.-R. and M.N.-R. prepared samples for sequencing analysis. M.C.A. and L.P. performed the bioinformatics analysis of sequencing data. M.C.A. and I.L.-L. designed and performed statistical analyses and created figures for the paper. O.P.-R., I.L.-L., and M.C.A. wrote the paper. M.N.-R., I.L.-L., L.P., M.C.A., C.X., and B.B.F. edited.
Contributor Information
Cecilia Ximenez, Email: cximenez@unam.mx.
Hideyuki Tamaki, National Institute of Advanced Industrial Science and Technology.
REFERENCES
- 1.Leung JM, Graham AL, Knowles SCL. 2018. Parasite-microbiota interactions with the vertebrate gut: synthesis through an ecological lens. Front Microbiol 9:843. doi: 10.3389/fmicb.2018.00843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berrilli F, Di Cave D, Cavallero S, D’Amelio S. 2012. Interactions between parasites and microbial communities in the human gut. Front Cell Infect Microbiol 2:141. doi: 10.3389/fcimb.2012.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parfrey LW, Walters WA, Lauber CL, Clemente JC, Berg-Lyons D, Teiling C, Kodira C, Mohiuddin M, Brunelle J, Driscoll M, Fierer N, Gilbert JA, Knight R. 2014. Communities of microbial eukaryotes in the mammalian gut within the context of environmental eukaryotic diversity. Front Microbiol 5:298. doi: 10.3389/fmicb.2014.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lukeš J, Stensvold CR, Jirků-Pomajbíková K, Wegener Parfrey L. 2015. Are human intestinal eukaryotes beneficial or commensals? PLoS Pathog 11:e1005039. doi: 10.1371/journal.ppat.1005039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morton ER, Lynch J, Froment A, Lafosse S, Heyer E, Przeworski M, Blekhman R, Ségurel L. 2015. Variation in rural African gut microbiota is strongly correlated with colonization by Entamoeba and subsistence. PLoS Genet 11:e1005658. doi: 10.1371/journal.pgen.1005658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bär AK, Phukan N, Pinheiro J, Simoes-Barbosa A. 2015. The interplay of host microbiota and parasitic protozoans at mucosal interfaces: implications for the outcomes of infections and diseases. PLoS Negl Trop Dis 9:e0004176. doi: 10.1371/journal.pntd.0004176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iebba V, Santangelo F, Totino V, Pantanella F, Monsia A, Di Cristanziano V, Di Cave D, Schippa S, Berrilli F, D’Alfonso R. 2016. Gut microbiota related to Giardia duodenalis, Entamoeba spp. and Blastocystis hominis infections in humans from Côte d’Ivoire. J Infect Dev Ctries 10:1035–1041. doi: 10.3855/jidc.8179. [DOI] [PubMed] [Google Scholar]
- 8.Burgess SL, Gilchrist CA, Lynn TC, PetriWA, Jr.. 2017. Parasitic protozoa and interactions with the host intestinal microbiota. Infect Immun 85:e00101-17. doi: 10.1128/IAI.00101-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Partida-Rodríguez O, Serrano-Vázquez A, Nieves-Ramírez ME, Moran P, Rojas L, Portillo T, González E, Hernández E, Finlay BB, Ximenez C. 2017. Human intestinal microbiota: interaction between parasites and the host immune response. Arch Med Res 48:690–700. doi: 10.1016/j.arcmed.2017.11.015. [DOI] [PubMed] [Google Scholar]
- 10.Krogsgaard LR, O’Brien Andersen L, Johannesen TB, Engsbro AL, Stensvold CR, Nielsen HV, Bytzer P. 2018. Characteristics of the bacterial microbiome in association with common intestinal parasites in irritable bowel syndrome. Clin Transl Gastroenterol 9:161. doi: 10.1038/s41424-018-0027-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nieves-Ramírez ME, Partida-Rodríguez O, Laforest-Lapointe I, Reynolds LA, Brown EM, Valdez-Salazar A, Morán-Silva P, Rojas-Velázquez L, Morien E, Parfrey LW, Jin M, Walter J, Torres J, Arrieta MC, Ximénez-García C, Finlay BB. 2018. Asymptomatic intestinal colonization with protist Blastocystis is strongly associated with distinct microbiome ecological patterns. mSystems 3:e00007-18. doi: 10.1128/mSystems.00007-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arboleya S, Binetti A, Salazar N, Fernández N, Solís G, Hernández-Barranco A, Margolles A, de Los Reyes-Gavilán CG, Gueimonde M. 2012. Establishment and development of intestinal microbiota in preterm neonates. FEMS Microbiol Ecol 79:763–772. doi: 10.1111/j.1574-6941.2011.01261.x. [DOI] [PubMed] [Google Scholar]
- 13.Matamoros S, Gras-Leguen C, Le Vacon F, Potel G, de La Cochetiere MF. 2013. Development of intestinal microbiota in infants and its impact on health. Trends Microbiol 21:167–173. doi: 10.1016/j.tim.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 14.Tanaka M, Nakayama J. 2017. Development of the gut microbiota in infancy and its impact on health in later life. Allergol Int 66:515–522. doi: 10.1016/j.alit.2017.07.010. [DOI] [PubMed] [Google Scholar]
- 15.Ihekweazu FD, Versalovic J. 2018. Development of the pediatric gut microbiome: impact on health and disease. Am J Med Sci 356:413–423. doi: 10.1016/j.amjms.2018.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robertson RC, Manges AR, Finlay BB, Prendergast AJ. 2019. The human microbiome and child growth—first 1000 days and beyond. Trends Microbiol 27:131–147. doi: 10.1016/j.tim.2018.09.008. [DOI] [PubMed] [Google Scholar]
- 17.Matsuki T, Yahagi K, Mori H, Matsumoto H, Hara T, Tajima S, Ogawa E, Kodama H, Yamamoto K, Yamada T, Matsumoto S, Kurokawa K. 2016. A key genetic factor for fucosyllactose utilization affects infant gut microbiota development. Nat Commun 7:11939. doi: 10.1038/ncomms11939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stewart CJ, Ajami NJ, O’Brien JL, Hutchinson DS, Smith DP, Wong MC, Ross MC, Lloyd RE, Doddapaneni H, Metcalf GA, Muzny D, Gibbs RA, Vatanen T, Huttenhower C, Xavier RJ, Rewers M, Hagopian W, Toppari J, Ziegler AG, She JX, Akolkar B, Lernmark A, Hyoty H, Vehik K, Krischer JP, Petrosino JF. 2018. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 562:583–588. doi: 10.1038/s41586-018-0617-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.WHO. 1987. Prevention and control of intestinal parasitic infections. Report of a WHO expert committee. World Health Organ Tech Rep Ser 749:1–86. https://www.who.int/neglected_diseases/resources/who_trs_749/en/. [PubMed] [Google Scholar]
- 20.WHO Expert Committee. 1987. Public health significance of intestinal parasitic infections. WHO Expert Committee. Bull World Health Organ 65:575–588. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2491073/. [PMC free article] [PubMed] [Google Scholar]
- 21.Walk ST, Blum AM, Ewing SA, Weinstock JV, Young VB. 2010. Alteration of the murine gut microbiota during infection with the parasitic helminth Heligmosomoides polygyrus. Inflamm Bowel Dis 16:1841–1849. doi: 10.1002/ibd.21299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Verma AK, Verma R, Ahuja V, Paul J. 2012. Real-time analysis of gut flora in Entamoeba histolytica infected patients of northern India. BMC Microbiol 12:183. doi: 10.1186/1471-2180-12-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cooper P, Walker AW, Reyes J, Chico M, Salter SJ, Vaca M, Parkhill J. 2013. Patent human infections with the whipworm, Trichuris trichiura, are not associated with alterations in the faecal microbiota. PLoS One 8:e76573. doi: 10.1371/journal.pone.0076573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holm JB, Sorobetea D, Kiilerich P, Ramayo-Caldas Y, Estellé J, Ma T, Madsen L, Kristiansen K, Svensson-Frej M. 2015. Chronic Trichuris muris infection decreases diversity of the intestinal microbiota and concomitantly increases the abundance of lactobacilli. PLoS One 10:e0125495. doi: 10.1371/journal.pone.0125495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steinmann P, Utzinger J, Du Z-W, Zhou X-N. 2010. Multiparasitism a neglected reality on global, regional and local scale. Adv Parasitol 73:21–50. doi: 10.1016/S0065-308X(10)73002-5. [DOI] [PubMed] [Google Scholar]
- 26.Viney ME, Graham AL. 2013. Patterns and processes in parasite co-infection. Adv Parasitol 82:321–369. doi: 10.1016/B978-0-12-407706-5.00005-8. [DOI] [PubMed] [Google Scholar]
- 27.Hellard E, Fouchet D, Vavre F, Pontier D. 2015. Parasite-parasite interactions in the wild: how to detect them? Trends Parasitol 31:640–652. doi: 10.1016/j.pt.2015.07.005. [DOI] [PubMed] [Google Scholar]
- 28.Seppälä O, Jokela J. 2016. Do coinfections maintain genetic variation in parasites? Trends Parasitol 32:930–938. doi: 10.1016/j.pt.2016.08.010. [DOI] [PubMed] [Google Scholar]
- 29.Morrill A, Dargent F, Forbes MR. 2017. Explaining parasite aggregation: more than one parasite species at a time. Int J Parasitol 47:185–188. doi: 10.1016/j.ijpara.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 30.Toro-Londono MA, Bedoya-Urrego K, Garcia-Montoya GM, Galvan-Diaz AL, Alzate JF. 2019. Intestinal parasitic infection alters bacterial gut microbiota in children. PeerJ 7:e6200. doi: 10.7717/peerj.6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. 2010. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A 107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vaishampayan PA, Kuehl JV, Froula JL, Morgan JL, Ochman H, Francino MP. 2010. Comparative metagenomics and population dynamics of the gut microbiota in mother and infant. Genome Biol Evol 2:53–66. doi: 10.1093/gbe/evp057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grönlund MM, Grześkowiak Ł, Isolauri E, Salminen S. 2011. Influence of mother’s intestinal microbiota on gut colonization in the infant. Gut Microbes 2:227–233. doi: 10.4161/gmic.2.4.16799. [DOI] [PubMed] [Google Scholar]
- 34.Pantoja-Feliciano IG, Clemente JC, Costello EK, Perez ME, Blaser MJ, Knight R, Dominguez-Bello MG. 2013. Biphasic assembly of the murine intestinal microbiota during early development. ISME J 7:1112–1115. doi: 10.1038/ismej.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, Li Y, Xia Y, Xie H, Zhong H, Khan MT, Zhang J, Li J, Xiao L, Al-Aama J, Zhang D, Lee YS, Kotowska D, Colding C, Tremaroli V, Yin Y, Bergman S, Xu X, Madsen L, Kristiansen K, Dahlgren J, Wang J. 2015. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 17:852. doi: 10.1016/j.chom.2015.05.012. [DOI] [PubMed] [Google Scholar]
- 36.Dogra S, Sakwinska O, Soh SE, Ngom-Bru C, Brück WM, Berger B, Brüssow H, Karnani N, Lee YS, Yap F, Chong YS, Godfrey KM, Holbrook JD. 2015. Rate of establishing the gut microbiota in infancy has consequences for future health. Gut Microbes 6:321–325. doi: 10.1080/19490976.2015.1078051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Odamaki T, Kato K, Sugahara H, Hashikura N, Takahashi S, Xiao JZ, Abe F, Osawa R. 2016. Age-related changes in gut microbiota composition from newborn to centenarian: a cross-sectional study. BMC Microbiol 16:90. doi: 10.1186/s12866-016-0708-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pannaraj PS, Li F, Cerini C, Bender JM, Yang S, Rollie A, Adisetiyo H, Zabih S, Lincez PJ, Bittinger K, Bailey A, Bushman FD, Sleasman JW, Aldrovandi GM. 2017. Association between breast milk bacterial communities and establishment and development of the infant gut microbiome. JAMA Pediatr 171:647–654. doi: 10.1001/jamapediatrics.2017.0378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferretti P, Pasolli E, Tett A, Asnicar F, Gorfer V, Fedi S, Armanini F, Truong DT, Manara S, Zolfo M, Beghini F, Bertorelli R, De Sanctis V, Bariletti I, Canto R, Clementi R, Cologna M, Crifò T, Cusumano G, Gottardi S, Innamorati C, Masè C, Postai D, Savoi D, Duranti S, Lugli GA, Mancabelli L, Turroni F, Ferrario C, Milani C, Mangifesta M, Anzalone R, Viappiani A, Yassour M, Vlamakis H, Xavier R, Collado CM, Koren O, Tateo S, Soffiati M, Pedrotti A, Ventura M, Huttenhower C, Bork P, Segata N. 2018. Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe 24:133–145.e5. doi: 10.1016/j.chom.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shao Y, Forster SC, Tsaliki E, Vervier K, Strang A, Simpson N, Kumar N, Stares MD, Rodger A, Brocklehurst P, Field N, Lawley TD. 2019. Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth. Nature 574:117–121. doi: 10.1038/s41586-019-1560-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Selma-Royo M, Tarrazó M, García-Mantrana I, Gómez-Gallego C, Salminen S, Collado MC. 2019. Shaping microbiota during the first 1000 days of life. Adv Exp Med Biol 1125:3–24. doi: 10.1007/5584_2018_312. [DOI] [PubMed] [Google Scholar]
- 42.Zhernakova A, Kurilshikov A, Bonder MJ, Tigchelaar EF, Schirmer M, Vatanen T, Mujagic Z, Vila AV, Falony G, Vieira-Silva S, Wang J, Imhann F, Brandsma E, Jankipersadsing SA, Joossens M, Cenit MC, Deelen P, Swertz MA, LifeLines Cohort Study, Weersma RK, Feskens EJ, Netea MG, Gevers D, Jonkers D, Franke L, Aulchenko YS, Huttenhower C, Raes J, Hofker MH, Xavier RJ, Wijmenga C, Fu J. 2016. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 352:565–569. doi: 10.1126/science.aad3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dunn LA, Boreham PF, Stenzel DJ. 1989. Ultrastructural variation of Blastocystis hominis stocks in culture. Int J Parasitol 19:43–56. doi: 10.1016/0020-7519(89)90020-9. [DOI] [PubMed] [Google Scholar]
- 44.Brewer MT, Agbedanu PN, Zamanian M, Day TA, Carlson SA. 2013. Evidence for a bacterial lipopolysaccharide-recognizing G-protein-coupled receptor in the bacterial engulfment by Entamoeba histolytica. Eukaryot Cell 12:1433–1438. doi: 10.1128/EC.00150-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun S, Noorian P, McDougald D. 2018. Dual role of mechanisms involved in resistance to predation by protozoa and virulence to humans. Front Microbiol 9:1017. doi: 10.3389/fmicb.2018.01017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.White EC, Houlden A, Bancroft AJ, Hayes KS, Goldrick M, Grencis RK, Roberts IS. 2018. Manipulation of host and parasite microbiotas: survival strategies during chronic nematode infection. Sci Adv 4:eaap7399. doi: 10.1126/sciadv.aap7399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seiler C, van Velzen E, Neu TR, Gaedke U, Berendonk TU, Weitere M. 2017. Grazing resistance of bacterial biofilms: a matter of predators’ feeding trait. FEMS Microbiol Ecol 93:fix112. doi: 10.1093/femsec/fix112. [DOI] [PubMed] [Google Scholar]
- 48.Hicks SJ, Theodoropoulos G, Carrington SD, Corfield AP. 2000. The role of mucins in host-parasite interactions. Part I—protozoan parasites. Parasitol Today 16:476–481. doi: 10.1016/S0169-4758(00)01773-7. [DOI] [PubMed] [Google Scholar]
- 49.Beatty JK, Akierman SV, Motta JP, Muise S, Workentine ML, Harrison JJ, Bhargava A, Beck PL, Rioux KP, McKnight GW, Wallace JL, Buret AG. 2017. Giardia duodenalis induces pathogenic dysbiosis of human intestinal microbiota biofilms. Int J Parasitol 47:311–326. doi: 10.1016/j.ijpara.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 50.Theodoropoulos G, Hicks SJ, Corfield AP, Miller BG, Carrington SD. 2001. The role of mucins in host-parasite interactions: part II—helminth parasites. Trends Parasitol 17:130–135. doi: 10.1016/S1471-4922(00)01775-X. [DOI] [PubMed] [Google Scholar]
- 51.Sharpe C, Thornton DJ, Grencis RK. 2018. A sticky end for gastrointestinal helminths; the role of the mucus barrier. Parasite Immunol 40:e12517. doi: 10.1111/pim.12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Broadhurst MJ, Ardeshir A, Kanwar B, Mirpuri J, Gundra UM, Leung JM, Wiens KE, Vujkovic-Cvijin I, Kim CC, Yarovinsky F, Lerche NW, McCune JM, Loke P. 2012. Therapeutic helminth infection of macaques with idiopathic chronic diarrhea alters the inflammatory signature and mucosal microbiota of the colon. PLoS Pathog 8:e1003000. doi: 10.1371/journal.ppat.1003000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Finney CA, Taylor MD, Wilson MS, Maizels RM. 2007. Expansion and activation of CD4(+) CD25(+) regulatory T cells in Heligmosomoides polygyrus infection. Eur J Immunol 37:1874–1886. doi: 10.1002/eji.200636751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.D’Elia R, Behnke JM, Bradley JE, Else KJ. 2009. Regulatory T cells: a role in the control of helminth-driven intestinal pathology and worm survival. J Immunol 182:2340–2348. doi: 10.4049/jimmunol.0802767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reynolds LA, Finlay BB, Maizels RM. 2015. Cohabitation in the intestine: interactions among helminth parasites, bacterial microbiota, and host immunity. J Immunol 195:4059–4066. doi: 10.4049/jimmunol.1501432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kapitan M, Niemiec MJ, Steimle A, Frick JS, Jacobsen ID. 2019. Fungi as part of the microbiota and interactions with intestinal bacteria. Curr Top Microbiol Immunol 422:265–301. doi: 10.1007/82_2018_117. [DOI] [PubMed] [Google Scholar]
- 57.Hoffmann C, Dollive S, Grunberg S, Chen J, Li H, Wu GD, Lewis JD, Bushman FD. 2013. Archaea and fungi of the human gut microbiome: correlations with diet and bacterial residents. PLoS One 8:e66019. doi: 10.1371/journal.pone.0066019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sam QH, Chang MW, Chai LYA. 2017. The fungal mycobiome and its interaction with gut bacteria in the host. Int J Mol Sci 18:330. doi: 10.3390/ijms18020330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mukherjee PK, Sendid B, Hoarau G, Colombel JF, Poulain D, Ghannoum MA. 2015. Mycobiota in gastrointestinal diseases. Nat Rev Gastroenterol Hepatol 12:77–87. doi: 10.1038/nrgastro.2014.188. [DOI] [PubMed] [Google Scholar]
- 60.Huseyin CE, O’Toole PW, Cotter PD, Scanlan PD. 2017. Forgotten fungi—the gut mycobiome in human health and disease. FEMS Microbiol Rev 41:479–511. doi: 10.1093/femsre/fuw047. [DOI] [PubMed] [Google Scholar]
- 61.Richard ML, Sokol H. 2019. The gut mycobiota: insights into analysis, environmental interactions and role in gastrointestinal diseases. Nat Rev Gastroenterol Hepatol 16:331–345. doi: 10.1038/s41575-019-0121-2. [DOI] [PubMed] [Google Scholar]
- 62.Mason KL, Erb Downward JR, Mason KD, Falkowski NR, Eaton KA, Kao JY, Young VB, Huffnagle GB. 2012. Candida albicans and bacterial microbiota interactions in the cecum during recolonization following broad-spectrum antibiotic therapy. Infect Immun 80:3371–3380. doi: 10.1128/IAI.00449-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Drossman DA. 2006. The functional gastrointestinal disorders and the Rome III process. Gastroenterology 130:1377–1390. doi: 10.1053/j.gastro.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 64.Nazeer JT, El Sayed Khalifa K, von Thien H, El-Sibaei MM, Abdel-Hamid MY, Tawfik RA, Tannich E. 2013. Use of multiplex real-time PCR for detection of common diarrhea causing protozoan parasites in Egypt. Parasitol Res 112:595–601. doi: 10.1007/s00436-012-3171-8. [DOI] [PubMed] [Google Scholar]
- 65.Bartram AK, Lynch MD, Stearns JC, Moreno-Hagelsieb G, Neufeld JD. 2011. Generation of multimillion-sequence 16S rRNA gene libraries from complex microbial communities by assembling paired-end Illumina reads. Appl Environ Microbiol 77:3846–3852. doi: 10.1128/AEM.02772-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. 2013. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Comeau AM, Douglas GM, Langille MG. 2017. Microbiome helper: a custom and streamlined workflow for microbiome research. mSystems 2:e00127-16. doi: 10.1128/mSystems.00127-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing Mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kuczynski J, Stombaugh J, Walters WA, González A, Caporaso JG, Knight R. 2011. Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr Protoc Bioinformatics Chapter 10:Unit 10.7. doi: 10.1002/0471250953.bi1007s36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Eren AM, Maignien L, Sul WJ, Murphy LG, Grim SL, Morrison HG, Sogin ML. 2013. Oligotyping: differentiating between closely related microbial taxa using 16S rRNA gene data. Methods Ecol Evol 4:1111–1119. doi: 10.1111/2041-210X.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Eren AM, Morrison HG, Lescault PJ, Reveillaud J, Vineis JH, Sogin ML. 2015. Minimum entropy decomposition: unsupervised oligotyping for sensitive partitioning of high-throughput marker gene sequences. ISME J 9:968–979. doi: 10.1038/ismej.2014.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yilmaz P, Parfrey LW, Yarza P, Gerken J, Pruesse E, Quast C, Schweer T, Peplies J, Ludwig W, Glöckner FO. 2014. The SILVA and “All-Species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res 42:D643–D648. doi: 10.1093/nar/gkt1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McMurdie PJ, Holmes S. 2013. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Neuwirth E. 2011. RColorBrewer: ColorBrewer palettes. https://cran.r-project.org/web/packages/RColorBrewer/.
- 76.Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Henry M, Stevens H, Wagner H. 2013. Vegan: community ecology package. https://cran.r-project.org/web/packages/vegan/index.html.
- 77.Ploner A. 2012. Heatplus: heatmaps with row and/or column covariates and colored clusters. https://bioconductor.org/packages/release/bioc/html/Heatplus.html.
- 78.Pollard K, Gilbert HN, Ge Y, Taylor S, Dudoit S. 2005. Multiple testing procedures: R multtst package and applications to genomics, p 249–271. In Gentleman R, Carey V, Huber W, Irizarry R, Dudoit S (ed), Bioinformatics and computational biology solutions using R and Bioconductor. Springer-Verlag, New York, NY. https://www.bioconductor.org/help/publications/books/bioinformatics-and-computational-biology-solutions/. [Google Scholar]
- 79.Wickham H. 2009. ggplot2: elegant graphics for data analysis. Springer-Verlag, New York, NY. https://www.springer.com/gp/book/9780387981413. [Google Scholar]
- 80.Wickham H. 2011. The split-apply-combine strategy for data analysis. J Stat Soft 40:1–29. doi: 10.18637/jss.v040.i01. [DOI] [Google Scholar]
- 81.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, LeLeiko N, Snapper SB, Bousvaros A, Korzenik J, Sands BE, Xavier RJ, Huttenhower C. 2012. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol 13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Principal-component analysis (PCoA) ordination of the variation in the beta-diversity of human gut bacterial (left) and eukaryote (right) communities in weaned infant (1 year old)-mother binomials (a and b) and unweaned infant (3 months old)-mother binomials (c and d) on Bray-Curtis dissimilarities. Colors represent binomial identity, while shapes represent class (squares for mothers and circles for infants). PERMANOVAs indicate no significant effect of binomial identity. Dark colors indicate no colonization, while light colors indicate colonization. Download FIG S1, TIF file, 0.6 MB (678.7KB, tif) .
Copyright © 2021 Partida-Rodriguez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primer sequences used for protist determination by qPCR. Download Table S1, DOCX file, 0.03 MB (32.1KB, docx) .
Copyright © 2021 Partida-Rodriguez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.