Abstract
It has been hypothesized that protein factors may protect CpG islands from methyltransferase during development and that demethylation may involve protein-DNA interactions at demethylated sites. However, direct evidence has been lacking. In this study, demethylation at the EBNA-1 binding sites of the Epstein-Barr virus latent replication origin, oriP, was investigated by using human cells. Several novel findings are discussed. First, there are specific preferential demethylation sites within the oriP region. Second, the DNA sequence of oriP alone is not the target of an active demethylation process. Third, EBNA-1 binding is required for the site-specific demethylation in oriP. Interestingly, CpG sites adjacent to and between the EBNA-1 sites do not become demethylated. Fourth, demethylation of the first DNA strand in oriP at the EBNA-1 binding sites involves a passive (replication-dependent) mechanism. The second-strand demethylation appears to occur through an active mechanism. That is, EBNA-1 protein binding prevents the EBNA-1 binding sites from being remethylated after one round of DNA replication, and it appears that an active demethylase then demethylates these hemimethylated sites. This study provides clear evidence that protein binding specifies sites of DNA demethylation and provides insights into the sequence of steps and the mechanism of demethylation.
CpG methylation plays an important role in mammalian development (31; for a review, see reference 2), and it has been correlated with repression of gene expression (22; for reviews, see reference 4 and 33). Changes in the basic pattern of de novo methylation and demethylation occur throughout development. How DNA regions are targeted for de novo methylation or demethylation and how the de novo methylation and demethylation processes are mediated are not clear.
It has been hypothesized that protein factors may protect CpG islands from methyltransferase (1). However, the data to support this has been indirect. In vivo footprinting and DNA methylation studies on the phosphoglycerate kinase 1 (PGK-1) gene indicated protein-DNA contact in the promoter region of this gene on the active X chromosome but not on the inactive X chromosome (29, 30). It has been proposed by these authors that unidentified protein factors may be involved in keeping specific regions methylation free. In a second system, studies with transgenic mice have indicated that DNA sequences corresponding to Sp1 sites play an important role in protecting the CpG island of the adenine phosphoribosyltransferase (APRT) gene from methyltransferase (3, 24). However, a recent study (26) demonstrated the methylation-free status of the APRT gene in Sp1 knockout mice, and the authors suggested that the methylation-free status may be maintained by the binding of other members, such as Sp3, of the Sp1 family to the Sp1 sites. Hence, the identity of protein factors that might maintain sites of demethylation in this system remains uncertain. Study of the α-actin gene in myoblasts by using transient, nonreplicating plasmids indicated that demethylation of one DNA strand at a specific site occurs within 2 h after DNA enters the cells (28). Although it was not demonstrated directly in their study, Paroush et al. (28) suggested that demethylation may involve protein-DNA interactions based on the specificity of the demethylated sites.
Two possible mechanisms for demethylation have been proposed in the above studies. In the first, a passive mechanism, demethylation is due to the failure of remethylation by maintenance methyltransferase, and hence demethylation of both DNA strands should occur in 50% of the cells after two rounds of replication (32). At least four or five rounds of replication would be required to demethylate about 95% of the DNA on both strands by this mechanism. In the second mechanism, an active mechanism, activity of a demethylase that may require cis- and trans-acting factors (reviewed in reference 38) can lead to demethylation of both strands on all the DNA without any extensive DNA replication. The hemimethylated chicken vitellogenin gene becomes symmetrically demethylated with limited DNA replication at 24 h after hormone stimulation in chicken liver (34). Transient transfection of the α-actin promoter upstream of a reporter gene into a rat cell line indicated that demethylation is a two-step process with a hemimethylated intermediate (28). Studies on δ-crystallin demethylation suggested possible hemimethylated intermediates (10, 36). However, after several rounds of DNA replication, no hemimethylated DNA could be detected by a sensitive PCR assay in an experiment in which in vitro-methylated sequences were injected into mouse zygotes (21). These studies support the hypothesis that demethylation is a multistep process and that a passive mechanism alone without an active mechanism participating at least in some steps cannot achieve complete demethylation.
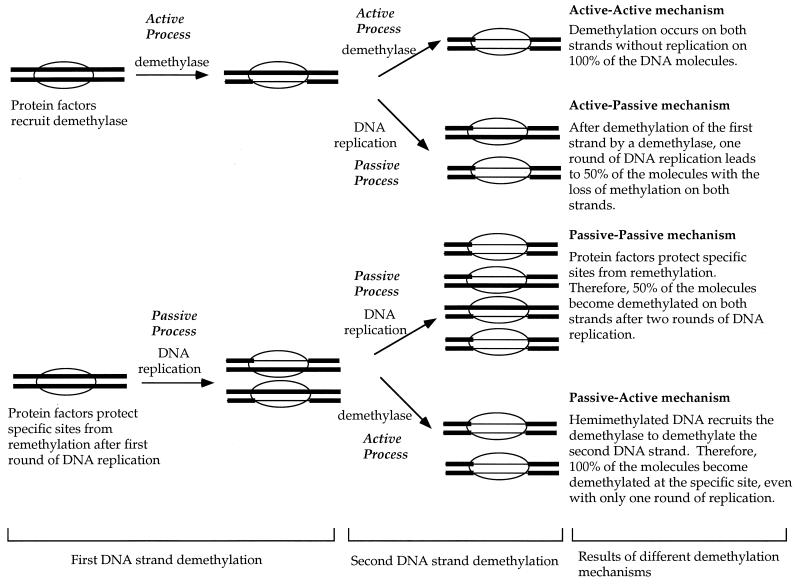
Considering that demethylation of the two DNA strands may involve different (distinguishable consecutive) mechanisms, four possible mechanisms can be postulated for the demethylation of the two DNA strands (Fig. 1): (i) protein binding could recruit a demethylase to demethylate both strands (active-active mechanism); (ii) protein binding could interfere with remethylation after replication, leading to demethylation of both strands on half of the DNA molecules after two rounds of replication (passive-passive mechanism); (iii) protein binding could recruit demethylase to demethylate one DNA strand, and then the first round of replication would lead to demethylation of both strands on half of the molecules (active-passive mechanism); and (iv) protein binding could interfere with remethylation after replication, and then the resulting hemimethylated DNA would recruit a demethylase to demethylate the second DNA strand (passive-active mechanism). While any one of these conceivable pathways may turn out to be the predominant one, it is conceivable that more than one may function during the course of cell differentiation and development at different DNA sites in different cell types.
FIG. 1.
Possible mechanisms of demethylation at specific sites. The term “passive” is used to indicate that the specific protein, such as EBNA-1, binds to its recognition sites and blocks remethylation at these sites by the maintenance methyltransferase after DNA replication. These sites become hemimethylated instead of being restored to the symmetrically methylated configuration after one round of DNA replication. The term “active” is used to indicate that a demethylase removes methyl C independently of any DNA replication. The ovals represent proteins, such as EBNA-1, binding to a specific DNA sequence. Thick lines represent methylated DNA, and thin lines represent unmethylated DNA.
It is well documented that plasmids bearing the latent replication origin of the Epstein-Barr virus (EBV), oriP, can be maintained in human cells expressing the EBV nuclear antigen EBNA-1 (17, 35, 40, 41). We have developed a stable episomal system in which oriP is used to study the dynamics of CpG methylation over time courses of several months in human cells (13). In our system, the CpG methylation patterns on the plasmids generated in vitro by using either FnuDII, HhaI, HpaII, or SssI methylase are maintained for months after transfection into human cells. This indicates that the maintenance methyltransferase can efficiently remethylate the newly synthesized strand at positions opposite the existing sites of CpG methylation. However, we have observed a few specific preferential demethylation sites on the episome. Among these few sites, three are HpaII sites in the oriP region that become demethylated very quickly after transfection into human cells expressing EBNA-1. The demethylation of these HpaII sites appears to be either simultaneous or close to simultaneous.
In the current study, the sites of demethylation in oriP were mapped by using both Southern blot analysis and bisulfite genomic sequencing methods. Furthermore, experiments were designed to explore the mechanism of demethylation in this region. We found that protein binding is required for demethylation of the oriP region. Replication alone does not lead to demethylation of the oriP without EBNA-1 binding. Furthermore, EBNA-1 binding alone also does not lead to demethylation of the oriP before replication. The HpaII sites within the EBNA-1 binding sites remained at least hemimethylated after one round of DNA replication. Moreover, the ability to identify molecules that were not replicated or were replicated once or twice in this system allows the analysis of the steps and mechanism of demethylation in the oriP region. The first-strand demethylation of the oriP region occurs by a passive mechanism, and the second-strand demethylation of these sites is probably processed through an active mechanism. This mechanism offers a logical interpretation of observations on demethylation events of endogenous genes.
MATERIALS AND METHODS
Plasmids.
Plasmids with wild-type oriP sequences used in this study include pCLH22, p291, Δp291, and pHEBo. These plasmids can replicate once per cell cycle in human cells expressing EBNA-1. pCLH22 (13) contains oriP, EBNA-1, the luciferase reporter gene, the hygromycin resistance gene, and the necessary prokaryotic replication sequences. p291 has the simian virus 40 replication origin in addition to the pCLH22 sequences, but it does not have the luciferase reporter gene. pHEBo (35) has only oriP, the hygromycin resistance gene, and the necessary prokaryotic replication sequences. Δp291 was constructed by deleting the HindIII fragment containing the EBNA-1 coding sequence from p291. Several plasmids with defective oriP, dpm1, dpm1+2, and dpm3+4, were used in this study. These plasmids were a generous gift from J. Hearing. They have mutations in the dyad-symmetry (DS) region and were characterized previously (11). In brief, dpm1 has two nucleotides mutated in EBNA-1 binding site 1, dpm1+2 has two nucleotides mutated in each of the EBNA-1 binding sites 1 and 2, and dpm3+4 has two mutations in each of the EBNA-1 binding sites 3 and 4. All mutations, except one in site 3, generate new CpG sites in these binding sites.
In vitro DNA methylation.
DNA was methylated with SssI methylase, which methylates C’s at all CpG sites. The conditions used were those recommended by the manufacturer (New England Biolabs). DNA was extracted with phenol-chloroform and precipitated with ethanol after in vitro methylation. The status of methylation was confirmed by digestion with methylation-sensitive restriction endonucleases.
Cell lines and transfection.
293, a human embryonic kidney carcinoma cell line (American Type Culture Collection), and a derivative of this cell line, 293/EBNA1 (13), were used in this study. A prostate cancer cell line, PC-3, and a derivative, PC-3/EBNA1, with an integrated EBNA-1 gene driven by the cytomegalovirus promoter were also used. The EBNA-1 protein is constitutively expressed in the 293/EBNA1 and the PC-3/EBNA1 cell lines. Throughout this study, the calcium phosphate transfection method (13, 39) was used. All transfections were done in duplicate in each experiment, and all experiments were performed multiple times for confirmation.
Episome recovery and analysis.
In the cases where the plasmids were able to replicate, when the transfected cells reached confluence 2.5% of the cells were replated into a 100-mm plate and the remaining cells were harvested for plasmid DNA extraction. No replating was done in cases where the plasmids were not able to replicate. All the transfection experiments were carried out without any selection for the episomal plasmid.
Plasmid DNA was harvested from the transfected cells by the Hirt method (12). In the time point experiments and in experiments where plasmids do not replicate, plasmid DNA was then recovered by the Hirt method from isolated nuclei. DNA from each harvest was digested with restriction enzymes to determine the methylation status and the status of replication. The digested DNA was fractionated on 0.8 or 1% agarose gels, Southern transferred onto nylon membranes, and probed with a fragment covering the oriP region. The Southern blots were analyzed with a phosphorimager (GS525; Bio-Rad).
Bisulfite genomic sequencing.
Bisulfite genomic sequencing was carried out by the method of Clark et al. (5) with minor modifications. Of the DNA harvested from each transfection, 30% was used for bisulfite genomic sequencing. DNA was digested with MboI and denatured before being treated with a final concentration of 2.3 M sodium bisulfate–0.5 mM hydroquinone at 55°C for 4 h. The bisulfite-treated DNA was purified with the Wizard DNA purification resin (Promega), treated with a final concentration of 0.3 N sodium hydroxide, and precipitated with ethanol. The primers for top-strand amplification were 5′-GTGATAGTTTATGGGGTGGGA (forward) and 5′-CAATCAAAAAAACCTATATAACTAC (reverse). The PCR conditions for the top strand were 3 min at 95°C for initial denaturation followed by 35 cycles of 40 s at 94°C, 40 s at 55°C, and 40 s at 72°C in a Robo 96 cycler (Stratagene). The primers for bottom-strand amplification were 5′-ATAACAACTCATAAAATAAAAAATAT (forward) and 5′-TTAATTAGAGGGGTTTGTGTAG (reverse). The PCR conditions were similar to those for the top-strand amplification but with a reannealing temperature of 52°C. The 245-bp PCR products were gel purified and cloned into T-vector (25) made from pBSK. The clones were sequenced with the ABI PRISM Dye terminator cycle sequencing kit (Perkin-Elmer).
RESULTS
Demethylation occurs at the HpaII sites within the oriP region.
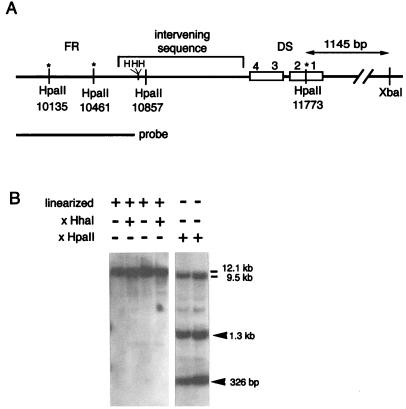
After the SssI-methylated pCLH22 was transfected into 293/EBNA1 cells, the DNA was harvested at various times and digested with HpaII. This 12.1-kb plasmid has the hygromycin resistance gene, the EBNA-1 gene, the luciferase gene, and the oriP segment in addition to the prokaryotic replication sequences and a selectable marker. Stable maintenance of the methylation status at nearly all CpG sites over at least a 2-month interval has been routinely observed in previous experiments (13, 14). However, several specific demethylation sites were observed within a few days after transfection in a large fraction of the minichromosome population (Fig. 2B). By using region-specific probes, these HpaII sites were localized to the oriP region. Three of the four HpaII sites in the oriP region were demethylated on the minichromosome (Fig. 2B). Demethylation at these HpaII sites was also observed when a different plasmid, p291, was used for the same experiment (data not shown). These three HpaII sites are known to remain unmethylated, while the remaining viral DNA is heavily methylated in tumor cells carrying the entire EBV episome (8). Our findings demonstrate that demethylation occurs at specific sites on the minichromosome in human cells and that the demethylation is unique to oriP and is typical of any plasmid that has oriP. Furthermore, the site-specific demethylation appears to be the same for the minichromosome as for the intact EBV viral genome.
FIG. 2.
Demethylation at specific HpaII sites within oriP. (A) HpaII and HhaI sites in the oriP region. The numbers below the HpaII sites are the nucleotide numbers of the HpaII sites on pCLH22. Three HhaI sites in the region are indicated by H. The asterisks indicate the sites of demethylation. (B) Southern blot of DNA harvested 8 days after transfection. The DNA is digested with different enzymes, as indicated above each lane. The probe used is indicated in panel A. The plasmid DNA is linearized by XbaI in the lanes indicated. The 12.1-kb band in the left panel is linearized plasmid. The 9.5-kb band in the right panel is the plasmid backbone hybridized with the portion of the probe upstream from the HpaII site at position 10135. The digestion of oriP by HpaII and the site of linearization resulted in the smaller size of this fragment. The 1.3-kb and 326-bp fragments in the right panel are generated by the digestion at three HpaII sites located at positions 10135, 10461, and 11773. The HpaII site at position 10857 remained methylated and HpaII resistant.
The three demethylated HpaII sites are located within the family of repeats (FR) and the DS regions of oriP, whereas the single HpaII site remaining methylated in the oriP region after several rounds of DNA replication is in the spacer between the FR and the DS region (Fig. 2A). This pattern of demethylation indicates three possible mechanisms. First, the repetitive nature of the DNA at the FR and the DS region may recruit the demethylase and lead to demethylation in these regions. Second, the binding of specific proteins to the EBNA-1 binding sites within these sequences may protect them from remethylation. Third, the involvement of these two regions in the initiation of replication could make them inaccessible to the maintenance methylase.
Demethylation within the oriP region is not due to the DNA sequence alone.
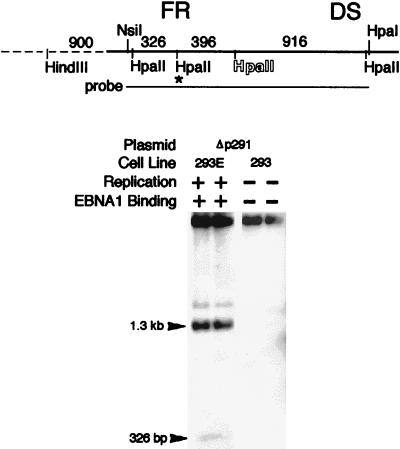
To determine whether the oriP sequence alone without replication or EBNA-1 binding can lead to demethylation of this region, plasmid Δp291 was methylated with SssI and then transfected into either 293 or 293/EBNA1 cells for comparison. Plasmid Δp291 will not replicate in 293 cells because of the absence of EBNA-1, but it will replicate in the 293/EBNA-1 cells. Demethylation of the HpaII sites within the oriP region did not occur when Δp291 DNA was harvested from 293 cells 3 days after transfection (Fig. 3). This strongly suggests that the oriP sequence is not targeted by an active demethylation process without other factors. Demethylation clearly took place in the 293/EBNA1 cells, where replication can occur, during the same time interval (Fig. 3). This indicates that demethylation in the oriP region does not occur without some aspects of the replication process (EBNA-1 binding [6, 15]), bending of the DNA at the origin [6, 15]), and synthesis of a new DNA strand). Therefore, neither the sequence of oriP nor any structural features intrinsic to its repetitive nature leads to demethylation.
FIG. 3.
Demethylation does not occur without EBNA-1 binding and DNA replication. A Southern blot of DNA harvested 3 days after transfection and digested with HindIII (for linearization) and HpaII is shown. The plasmids and cell lines used in the experiments are indicated above the panel. 293E, 293/EBNA1 cell line. The capability of plasmid replication and the presence of EBNA-1 in the cells are also indicated above each lane. The probe used is the NsiI-HpaI fragment, as indicated. For an explanation of the outline letters, see the legend to Fig. 4. The asterisk designates the HpaII site from which the partial digestion product likely derives.
These three HpaII sites appear to all be demethylated on all the molecules that become demethylated within the oriP region. If this were not the case, additional bands at 6.6, 6.3, 2.5, and 1.2 kb should be detected. The absence of these fragments indicates that demethylation of these three sites most probably occurred at the same time.
Replication alone without EBNA-1 binding does not lead to demethylation.
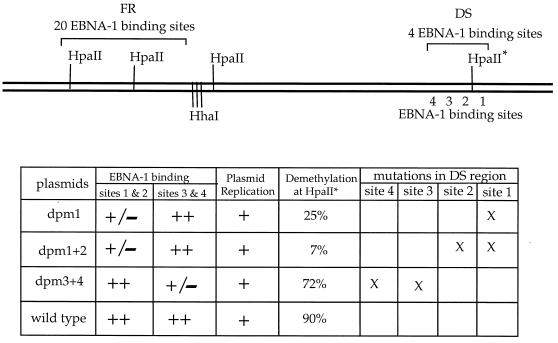
To examine whether EBNA-1 binding plays a role in the demethylation in the oriP region, several mutants with defective EBNA-1 binding sites in the DS region were used. Of the four EBNA-1 binding sites in the DS region, only one pair is required for oriP function (11). The mutant plasmid dpm1 has two point mutations in EBNA-1 binding site 1 in the DS region, and these mutations greatly reduce the binding of EBNA-1 to both sites 1 and 2. Plasmid dpm1+2 has two point mutations in EBNA-1 binding site 2 in addition to dpm1. This plasmid has been shown to replicate in mammalian cells, while the EBNA-1 protein binding to sites 1 and 2 of the DS region is abrogated, as shown in DNase I protection assays (11). Both plasmids dpm1 and dpm1+2 can replicate by using sites 3 and 4 (11). Plasmid dpm3+4 has EBNA-1 binding sites 3 and 4 mutated in the DS region, but it replicates in human cells by using the wild-type EBNA-1 binding sites 1 and 2.
Methylated DNA from these three plasmids and the control plasmid, pHEBo, were transfected into 293/EBNA1 cells. The low-molecular-weight DNA was harvested 10 and 23 days after transfection and analyzed by Southern blotting. The DNA was linearized by EcoRI digestion and then digested with HpaII or MspI. While HpaII and MspI recognize the same DNA sequence, HpaII is CpG methylation sensitive and MspI is CpG methylation insensitive. Subsequently, the Southern blot was probed with an Nsil to HpaI fragment containing the oriP region.
Three completely digested fragments of 326, 396, and 916 bp and a very faint partially digested band containing the 396- and the 326-bp fragments were detected in the MspI digests by using the oriP probe (Fig. 4). In the HpaII digests, a 326-bp fragment of similar intensity to the same fragment in the MspI digest was detected in all the DNA samples (Fig. 4B). Quantitative analysis revealed that the difference in radioactivity in this fragment in the HpaII and MspI digests from the same DNA harvest is less than 10% (range, 2 to 10%). This indicates that the two HpaII sites within the family of repeats (FR) became demethylated on all the molecules regardless of the mutations in the DS region. In contrast, the 396-bp fragment was absent in the HpaII digests of all the DNA samples. This demonstrates that the HpaII site in the spacer region of the oriP remained methylated on all the molecules.
FIG. 4.
Demethylation does not occur without EBNA-1 binding. (A) The diagram indicates the HpaII sites in the oriP region and the HpaII fragment sizes. Outline letters indicate the HpaII site that does not become demethylated and therefore is not digested by HpaII. All the HpaII sites can be digested by a CpG methylation insensitive enzyme, MspI, which is an isoschizomer of HpaII. (B) Southern blot of DNA harvested 10 days after transfection. All of the DNA was linearized by EcoRI digestion before being digested with HpaII (H) or MspI (M). The plasmids used are indicated above the lanes. The 1.6-kb HpaII band and the 727-bp MspI band are most likely to be derived from partial digestion of the HpaII-MspI site indicated by the asterisk in panel A. dpm1, dpm1+2, and dpm3+4 have mutations in the EBNA-1 binding sites in the DS region, as described in Materials and Methods and illustrated in Fig. 6A.
The 1.3-kb HpaII fragment, which contains the 396- and the 916-bp fragments, is a result of demethylation of the HpaII site in the FR that is closer to the DS region and the HpaII site between EBNA-1 binding sites 1 and 2 in the DS region (Fig. 4A). The HpaII site in the FR that is closer to the DS region is clearly digestable on all the plasmids as described above. Therefore, the reduced intensity of the 1.3-kb band is the result of lack of digestion at the HpaII site between EBNA-1 binding sites 1 and 2 in the DS region. The minichromosomes retaining methylation at the HpaII site between EBNA-1 binding sites 1 and 2 in the DS region would have generated a fragment which is indistinguishable from the linearized molecules (7 kb for pHEBo and 6.5 kb for the mutants) instead of the 1.3-kb fragment (Fig. 4B). The intensity of the 1.3-kb band was much stronger than that of the 7-kb band in the harvested dpm3+4 and pHEBo DNA. In contrast, the 7-kb band was much more intense than the 1.3-kb band in the harvested dpm1 and dpm1+2 DNA. This indicates that the majority of the dpm3+4 and pHEBo plasmids were demethylated while most of the dpm1 and dpm1+2 plasmids retained methylation at this HpaII site (Fig. 4B). The difference between these plasmids is the mutations in the EBNA-1 binding sites in the DS region. Mutant plasmids dpm1 and dpm1+2 have mutations in binding site 1 and binding sites 1 and 2, respectively. These plasmids have much reduced EBNA1 binding, while plasmids dpm3+4 (mutations in binding sites 3 and 4) and pHEBo (wild type) have normal EBNA-1 binding at these sites. Therefore, the lack of demethylation at the HpaII site between EBNA-1 binding sites 1 and 2 in the DS region that resulted in the reduced intensity of this 1.3-kb band in plasmid dpm1 is due to the lack of binding by EBNA-1 to sites 1 and 2 in the DS region. These clearly indicate that ENBA-1 binding results in demethylation at HpaII sites within the oriP region.
Further quantitative analysis of the Southern blot was carried out as follows. The 1.3- and 7-kb fragments should hybridize only to the remaining 1.3-kb probe, since the fragments do not include the 326-bp fragment (both HpaII sites in the FR are demethylated as described above). As described above, the lack of demethylation at the HpaII site between EBNA-1 binding sites 1 and 2 in the DS region leads to a decrease of radioactivity in the 1.3-kb band and an increase of radioactivity in the 7-kb band. Therefore, the 1.3-kb band (plus the 1.6-kb partial-digestion band) represents molecules demethylated at the HpaII site between EBNA-1 binding sites 1 and 2 in the DS region and the 7-kb band represents the plasmids retaining methylation at this HpaII site. The radioactivity in the 1.3-kb band (plus the 1.6-kb partial-digestion band) divided by the total radioactivity in the 7- and 1.3-kb bands approximately represents the fraction of plasmids becoming demethylated at the HpaII site between EBNA-1 binding sites 1 and 2 in the DS region. The levels of radioactivity in the 7- band and 1.3-kb bands (including the 1.6-kb partial-digestion band) were quantitated with a phosphorimager (Bio-Rad GS525). Phosphorimager analysis revealed that demethylation at the HpaII site between EBNA-1 binding sites 1 and 2 in the DS region occurs on 25, 7, 72, and 90% of the dpm1, dpm1+2, dpm3+4, and pHEBo plasmids, respectively (Fig. 5). Although there is an alternative way to calculate these fractions, the above calculation is preferred. The 326-bp fragment represents all molecules that have undergone demethylation, since EBNA-1 binding in the FR is not affected by the mutations in the DS region, while the 1.3-kb band represents the fraction of molecules that have undergone demethylation at the HpaII site between EBNA-1 binding sites 1 and 2 in the DS region. Therefore, the fractions can also be calculated by determining the relative radioactivity in the 326-bp band and the 1.3-kb band in each lane. However, the portions of the probe that hybridize to these two fragments are different, and so a greater error can occur.
FIG. 5.
Quantitation of demethylation in oriP. The ratio of radioactivity in the 1.3-kb band (including the 1.6-kb partial-digestion band) to the combined radioactivity in the 7-, 1.3-, and 1.6-kb bands in the HpaII-digested DNA was calculated. Percentages listed in the “demethylation” column represent the fraction of plasmids having undergone demethylation in the HpaII site marked with an asterisk between EBNA-1 binding sites 1 and 2 in the dyad symmetry (see the text for details). The capability of EBNA-1 binding and plasmid replication in eukaryotic cells are indicated: +/−, weak binding; ++, strong binding. The mutated EBNA-1 binding sites in each plasmid are indicated by X. Plasmids dpm1, dpm1+2, and dpm3+4 have mutations in the EBNA-1 binding sites in the DS region, as described in Materials and Methods and illustrated in Fig. 6A.
Similar results were obtained for similar experiments with these plasmids and the PC-3/EBNA1 cell line. These findings indicate that with decreased EBNA-1 binding to sites 1 and 2, the demethylation also dramatically decreased at the HpaII site between these two binding sites. Moreover, this site-specific demethylation of oriP was observed in different cell lines expressing EBNA-1, indicating that it is not a peculiarity of any one cell line or tissue type.
The restriction pattern of EcoRI-HpaII double digests supports the inference that these three HpaII sites are demethylated on all the molecules that become demethylated. If some molecules are demethylated only at one or two of these three HpaII sites, additional bands at 6.1, 5.8, 2.5, and 1.2 kb should be detected in Fig. 4B. The fact that these bands are absent in all the transfections indicates that all the three HpaII sites most probably become demethylated at the same time.
Demethylation of oriP is site specific and is not regionally specific.
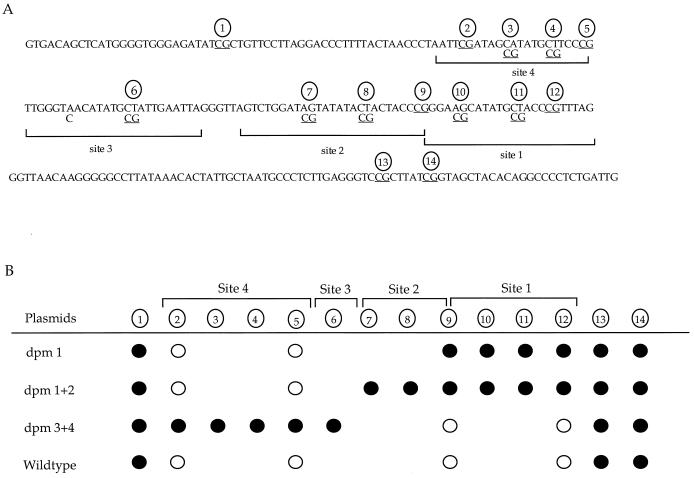
It is intriguing that one of the four HpaII sites within oriP remains methylated. This indicates that demethylation may occur only at CpG sites protected by EBNA-1. Bisulfite genomic sequencing of plasmid DNA harvested from 293/EBNA1 cells transfected with dpm1, dpm1+2, dpm3+4, and pHEBo was carried out to examine the CpG sites within and adjacent to the DS region. The 245-bp sequence examined includes seven CpG sites in the wild-type plasmid, pHEBo (Fig. 6A). Four of these seven sites are located within the EBNA-1 binding sites. There are 9, 11, and 10 CpG sites in dpm1, dpm1+2, and dpm3+4, respectively. The extra CpG sites in these mutants were generated by site-directed mutagenesis in the EBNA-1 binding sites within the DS region (11).
FIG. 6.
Specific CpG site demethylation in the DS region. (A) Nucleotide sequence of the oriP region analyzed by bisulfite genomic sequencing. The wild-type sequence is presented. The mutations in plasmids dpm1, dpm1+2, and dpm3+4 are indicated under the wild-type sequence. The CpG sites are underlined and indicated by a circled number. Brackets indicate sites protected by EBNA-1 in the in vitro DNase I protection assay described by Harrison et al. (11). (B) The CpG sites illustrated in panel A were examined by bisulfite genomic sequencing. The CpG sites in the four EBNA-1 binding sites are indicated by the brackets above the line. •, no conversion by sodium bisulfite treatment (with, the CpG site thus remaining methylated); ○, conversion by sodium bisulfite treatment (with, the CpG site becoming demethylated). Some CpG sites exist only in the mutant plasmids, and circles are absent at these sites in other plasmids. Multiple clones (at least four) from each mutant of each experiment were sequenced, and sequences of the different clones were identical within each mutant. The DNA was harvested 10 days after transfection.
Sodium bisulfite treatment converts unmethylated C to T but does not convert methylated C under these conditions. Sequence analysis of multiple clones from each bisulfite-treated DNA from transfection showed conversion at CpG sites (loss of methylation) protected by EBNA-1 and lack of conversion at CpG sites (methylation) adjacent to the EBNA-1 binding sites (Fig. 6B). Plasmids dpm1 and dpm1+2 carry mutations in EBNA-1 binding site 1 and sites 1 and 2, respectively, and have decreased binding of EBNA-1 to sites 1 and 2. The CpG sites within EBNA-1 binding sites 1 and 2 on these two plasmids were found to remain methylated. In contrast, demethylation (C conversion) was observed at CpG sites within EBNA-1 binding sites 3 and 4 on this two plasmids. Plasmid dpm3+4 has mutations in binding sites 3 and 4 and has decreased binding of EBNA-1 to these two sites. The CpG sites in EBNA-1 binding sites 3 and 4 were found to be methylated (unconverted), while the CpG sites in binding sites 1 and 2 became demethylated (converted) on dpm3+4. On the wild-type plasmid, the CpG sites in all four EBNA-1 binding sites were demethylated (converted) and the three CpG sites adjacent to the binding sites remained methylated (unconverted).
These data clearly illustrate that demethylation in the DS region occurs only at CpG sites protected by EBNA-1 binding, not at CpG sites as close as 32 bases away. Moreover, neither replication nor the DNA bending associated with replication in this region leads to its site-specific demethylation at the subset of sites to which EBNA-1 fails to bind. Therefore, the demethylation process in the oriP region is not due to a functioning replication origin.
Demethylation of the first DNA strand in the EBNA-1 binding sites involves a passive mechanism.
The sequence of oriP alone or DNA replication alone does not lead to demethylation of the EBNA-1 binding sites, as described above. To dissect the possible mechanism underlying the demethylation process, SssI-methylated and unmethylated pCLH22 plasmid was transfected into 293/EBNA1 cells and the DNA was harvested from isolated nuclei at various time points for analysis. In an experiment to determine how quickly the DNA enters the nucleus, DNA was harvested from isolated nuclei at 6 and 12.5 h after transfection. The DNA could be recovered from the nuclei within 6 h after transfection.
In time course experiments, DNA was harvested at 7, 15, 19.5, 24, 40, and 66 h after transfection and analyzed by DpnI and HpaII single digests and Southern blotting to check for replication (DpnI digestion) and CpG methylation (HpaII digestion). DpnI digests plasmid DNA that bears the bacterial dam methylation (methylation of A at GATC) on both strands, but it does not digest DNA that has lost bacterial dam methylation on one or both strands (by virtue of replication in eukaryotic cells). Therefore, plasmid DNA that replicated at least once after entering eukaryotic cells becomes DpnI-resistant. HpaII is sensitive to CpG methylation; therefore, it does not digest hemimethylated or symmetrically methylated DNA. DpnI-resistant (replicated) DNA was first detectable in the 40-h harvest for both unmethylated (Fig. 7A) and methylated (Fig. 7B) pCLH22. This indicates that DNA methylation does not alter the capability or the timing of replication of the episome dramatically.
FIG. 7.
Replication precedes demethylation of the HpaII sites in the oriP region. A Southern blot of DNA harvested 7, 15, 19.5, 24, 40, and 66 h after transfection and digested with DpnI (D) or HpaII (H) is shown. The same probe used in the experiment in Fig. 3 that contains only the oriP region is used here. (A) DNA from cells transfected with unmethylated pCLH22; (B) DNA from cells transfected with SssI-methylated pCLH22 (me-pCLH22). The thin and thick lines above the time designation in panel B represent the possible methylation states within oriP on molecules replicated zero, one, or two times. Thick lines represent methylated DNA strands, and thin lines represent unmethylated DNA strands. The open circle on one end of the line indicates DNA that retains the bacterial dam methylation (at A of GATC). The DpnI-resistant DNA is detected in the DNA harvested at 40 and 66 h after transfection but not at any earlier time points. Plasmid DNA was not linearized in this experiment; therefore, the uncut DNA appears as nicked and supercoiled bands as indicated. The pCLH22 DNA can be digested by HpaII because it is not methylated. A 916-bp band and the smaller 396- and 326-bp, bands which appear as one band, are detected in the HpaII-digested pCLH22 DNA. The methylated pCLH22 DNA is not digestable by HpaII unless demethylation occurs. In the methylated pCLH22 DNA harvested at or before 40 h after transfection, the DNA remains uncut by HpaII. A 1.3-kb band is clearly detected in the methylated pCLH22 DNA harvested at 66 h after transfection. There are two DpnI-MboI sites within the oriP region that generate three complete digest bands at 2.8 kb, 1.0 kb, and 763 bp. Loss of dam methylation at specific sites on some unreplicated molecules, for unknown reasons, generates a strong 3.5-kb band and a weak 1.4-kb band in the transfected DNA (both methylated and unmethylated) in addition to the three bands described above when probed with the oriP fragment.
The restriction enzyme MboI digests only plasmid DNA that has lost the bacterial dam methylation on both strands. Its isoschizomer, DpnI, digests only plasmid DNA with bacterial dam methylation on both strands. Therefore, plasmids which have undergone only one round of DNA replication in eukaryotic cells are resistant to DpnI-MboI double digestion, while all replicated plasmids, regardless of how many times they replicate, are resistant to DpnI single digestion. Quantitation of uncut DNA in the DpnI single digest (Fig. 7B) and DpnI-MboI double digest (data not shown) with a phosphorimager showed that the plasmid DNA harvested at 40 h after transfection had 18% ± 2.6% DpnI-resistant DNA and 15% ± 1.1% DpnI-MboI doubly resistant DNA. Furthermore, all of the plasmid DNA harvested at this time point was MboI resistant (data not shown). These data indicate that most, if not all, of the plasmid that had replicated had gone through only one round of replication at 40 h after transfection. Despite having gone through one round of replication, methylated pCLH22 still remained undigestable by HpaII at 40 h after transfection (Fig. 7B). This indicates that all the plasmids remain methylated at least on one DNA strand at 40 h after transfection. A 1.3-kb band was clearly detectable in the HpaII-digested DNA harvested at 66 h after transfection, and this is the only time point when HpaII-digestable DNA could be detected in the methylated pCLH22 DNA (Fig. 7B). This demonstrates that some plasmids become demethylated on both DNA strands at 66 h after transfection. HpaII digests of transfected pCLH22 showed the expected digestion pattern from unmethylated DNA at all time points (Fig. 7A).
The fact that no replication was detectable at 24 h after transfection indicates that EBNA-1 binding without replication does not lead to demethylation at HpaII sites in the EBNA-1 binding sites after the plasmids have been in the nucleus for at least 18 h (24 h after transfection minus 6 h for DNA to enter the nucleus). Moreover, these HpaII sites are not demethylated on both strands after the first round of replication (40 h after transfection). This demonstrates that EBNA-1 binding plus one round of DNA replication does not lead to demethylation on both strands at HpaII sites within oriP. Therefore, it is highly unlikely that EBNA-1 binding targets oriP for demethylation by an active mechanism which demethylates both DNA strands or by an alternative active process demethylating one DNA strand before DNA replication.
Demethylation of the second DNA strand in the EBNA-1 binding sites most probably involves an active mechanism.
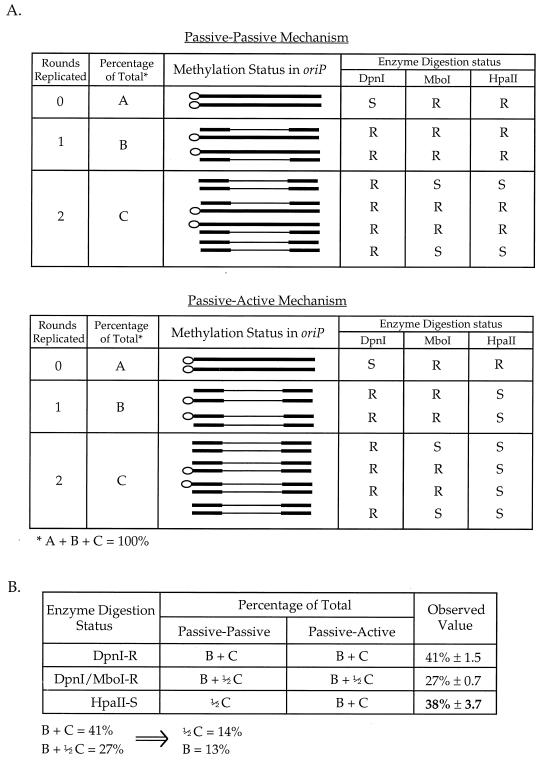
Only plasmids which have lost CpG methylation on both DNA strands in the oriP region will be sensitive to HpaII digestion. If demethylation of the second DNA strand is accomplished by a passive mechanism, only half of the molecules that replicated twice would have lost CpG methylation on both DNA strands in the oriP region and would be digested by HpaII (Fig. 8A). In contrast, if demethylation of the second DNA strand involves an active mechanism, all molecules replicated twice and some molecules replicated once should become demethylated on both DNA strands within oriP; therefore, these molecules would be digestable by HpaII (Fig. 8A).
FIG. 8.
Quantitative analysis of second-strand demethylation. (A) The bacterial and CpG methylation status of molecules in the harvested DNA based on passive-passive and passive-active mechanisms. Thick lines represent CpG-methylated DNA, and thin lines represent DNA demethylated at CpG sites. The open circle on one end of the line indicates DNA that retains the bacterial dam methylation. S, sensitive to enzyme digestion; R, resistant to enzyme digestion. A, plasmids that have undergone no replication; B, plasmids that replicated once; C, plasmids that have replicated twice in plasmids harvested at 66 h after transfection. (B) The fraction of DNA that is DpnI resistant (DpnI-R), DpnI-MboI double resistant (DpnI/MboI-R), and HpaII-sensitive (HpaII-S) in the plasmids harvested at 66 h after transfection. The observed value is derived from quantitation of uncut bands divided by the total hybridization in each of the digests. The percentage of the total for HpaII-sensitive molecules is derived from prediction of methylation status based on each of the two mechanisms as summarized in panel A.
Plasmid DNA harvested at 66 h after transfection consists of molecules that either never replicated, replicated once, or replicated twice. The fractions of these molecules can be quantitated by analyzing the uncut and cut bands in the DpnI digest and DpnI-MboI double digest of transfected DNA by using a phosphorimager. DpnI-resistant DNA accounted for 41% of the total, and this indicates that 41% of the plasmid DNA replicated at least once by 66 h after transfection (Fig. 8B). All plasmids that replicated once and half of the plasmids that replicated twice retained bacterial dam methylation on one DNA strand, and they represent the 27% DpnI-MboI doubly resistant molecules (Fig. 8B). From these results, the percentage of plasmids that have replicated once and the percentage of plasmids that have replicated twice can be determined (B = 13%, and C = 28%).
If the second DNA strand is demethylated by a passive mechanism, the expected molecules that can be digested by HpaII should be 0.5C or 14% of the total. If an active mechanism demethylates the second DNA strand, the HpaII-digestable fraction of the plasmids should be B + C or 41% of the total. The actual fraction of plasmids that become demethylated on both strands may be slightly smaller than predicted, considering the possibility that not all hemimethylated DNA will be demethylated by the active demethylase immediately after replication. Quantitation of the HpaII-digested DNA harvested at 66 h after transfection shows that HpaII-digestable DNA accounts for 38% of the total DNA harvested (Fig. 8B), which is consistent with the passive-active mechanism. Although this finding is indirect, it indicates that demethylation of the second DNA strand is likely to occur by the active process of a demethylase.
DISCUSSION
The major findings in this study are as follows. First, there are specific preferential CpG demethylation sites within the oriP region that are independent of the cell line and of the sequences outside oriP. These sites appear to be demethylated at the same time. Second, the DNA sequence of oriP does not become demethylated without DNA replication and EBNA-1 binding. Third, EBNA-1 binding is required for demethylation at these specific sites. Fourth, demethylation in oriP is not regionally specific but is specified precisely by EBNA-1 binding to its sites. Fifth, EBNA-1 binding with one round of replication does not lead to double-strand demethylation of oriP within 40 h after transfection. Finally, demethylation of the first DNA strand involves a passive mechanism, and demethylation of the second DNA strand most probably involves an active demethylase activity.
It has been reported that the oriP region is unmethylated in the otherwise highly methylated EBV genome in the Burkitt’s lymphoma cell line, RaeI (8). In most cells derived from Burkitt’s lymphomas, EBNA-1 is the only viral protein expressed. It has not been clear whether oriP initially becomes methylated after viral entry and then becomes demethylated some time later or whether it never gets methylated from the start. The HpaII site in the spacer region between the FR and the DS region is unmethylated in the viral genomes from Burkitt’s lymphoma and nasopharyngeal carcinoma cells (7, 8, 16). However, this very same HpaII site did not become demethylated in the present study. This suggests that the oriP region of the EBV genome does not become methylated in the first place in these tumor cells. This is because EBNA-1 binding is inadequate to lead to demethylation at this particular HpaII site if the site were ever to become methylated. It is most likely that EBNA-1 binding can also protect the DNA region from de novo methylation in addition to protecting specific sites from remethylation by the maintenance methylase. It is noteworthy that the region protected from de novo methylation appears to be larger than the region protected from maintenance methylation. This may explain the observation that Sp1 sites can prevent methylation of downstream CpG sites at the APRT gene during development (24).
Although protein-DNA interaction has been suggested to be important for DNA demethylation, this study provides direct evidence that protein binding can specify demethylation sites. It has been demonstrated in this study that demethylation does not take place when EBNA-1 is absent from the cells (therefore, no EBNA-1 binding and no plasmid DNA replication occur). This indicates that oriP demethylation is not specified by the DNA sequence alone. By using oriP mutants, it is clearly shown that the mutated EBNA-1 binding sites remain methylated after many rounds of minichromosome replication. This indicates that replication alone does not lead to demethylation without EBNA-1 binding. Otherwise, demethylation should occur at mutated EBNA-1 binding sites in oriP, regardless of the lack of EBNA-1 binding at those sites. The site of demethylation is specified strictly by EBNA-1 binding based on the observation that only the CpG sites within the EBNA-1 binding sites, not the CpG sites either adjacent to or between them, are demethylated. In summary, EBNA-1 is required for demethylation of the oriP region, and its binding specifies the sites of demethylation. This leaves open the question of mechanism; namely, how does the demethylation occur?
Although the critical role of EBNA-1 in oriP demethylation is clearly defined, the involvement of replication requires more complex analysis. It is difficult to clearly dissect EBNA-1 binding and DNA replication unless plasmid replication does not occur when high EBNA-1 expression and strong binding sites are present. Several experiments were performed to obtain EBNA-1 binding without plasmid replication in the cells. However, a low level of plasmid replication was always observed in the two cell lines, 293/EBNA1 and PC-3/EBNA1, used in this study (results not shown). This is not unexpected, because the use of alternative replication initiation sites has been reported for latent replication of the EBV genome (23). The fact that EBNA-1 binding sites are the components of the functional replication origin limits our ability to directly demonstrate the requirement of replication in demethylation. However, this very feature allows us to analyze the demethylation process in a stepwise manner on each DNA strand in the time course experiments. An entirely different system, containing a protein binding site that is not involved in replication initiation, is currently being developed to address the requirement of replication directly.
The demethylases described by Vairapandi and Duker (37) and Weiss et al. (38) demethylate both DNA strands within a short time (within 6 h) in cell-free enzymatic studies. If confirmed as physiologic demethylating activities, these two enzymes are most likely to be involved in an active-active mechanism (Fig. 1). Moreover, Paroush et al. (28) observed demethylation of the first strand within 2 h after DNA enters the cells. Therefore, demethylation of both strands (making DNA sites HpaII digestable) should be observed within a short period for plasmid entering the nucleus, if the active-active mechanism is operating. If the active-passive mechanism is operating, demethylation of the first DNA strand is most likely to occur much earlier than 40 h after transfection. Therefore, both DNA strands should become demethylated on 50% of the replicated plasmids after the first round of DNA replication while the other 50% of the replicated plasmids become hemimethylated at this time (Fig. 1).
The time course experiments in this study show that replication precedes demethylation of both DNA strands. This is based on the fact that no HpaII-digestable DNA was detected in the DNA harvested at several time points after transfection including the time point (40 h) when replicated DNA was first detected. This clearly demonstrates that although EBNA-1 binding specifies the demethylation sites, binding alone does not lead to demethylation on both DNA strands, at least within 40 h after transfection (34 h in the nucleus). Therefore, demethylation of oriP did not occur through either an active-active mechanism or an active-passive mechanism within 40 h after transfection; if it had, DNA demethylation on both strands should have been observed at this time. Although one can speculate that the active demethylation machinery takes more than 40 h to act on the DNA, this is highly unlikely, based on the findings in the studies referred to above. Moreover, the active demethylase should be able to demethylated both replicated and unreplicated molecules at these HpaII sites. The fact that unreplicated molecules do not become demethylated strongly supports the conclusion that the active mechanism is not involved in the first-strand demethylation in the oriP region and replication is required. This rules out the active-active and the active-passive mechanisms as being the mechanism of the oriP demethylation; hence, the double-strand demethylases described above are most probably not involved in the oriP demethylation.
This study allows us to analyze the order and nature of the steps in oriP demethylation. It provides a compelling indication that DNA replication is required for demethylation of the first DNA strand. The fraction of HpaII-sensitive molecules (38%) and the fraction of DpnI-resistant molecules (41%) are nearly equal in the DNA harvested at 66 h after transfection. This argues that the second DNA strand is demethylated by an active demethylase after the first-strand demethylation. Otherwise, the fraction of HpaII-sensitive molecules should have been much smaller than the observed value.
Observations in this study indicate that demethylation of the oriP is a two-step process. EBNA-1 specifies the sites of demethylation by binding to DNA and interfering with remethylation by the maintenance methylase after replication. This first step generates specific hemimethylated sites, and then the second strand is demethylated by an active demethylase. Interestingly, one of the reported active demethylases, 5-methylcytosine-DNA glycosylase (18, 19), prefers hemimethylated substrates in vitro. This enzyme may target hemimethylated sites specified by protein binding. This model may explain some of the demethylation events in the genome, such as demethylation of CpGs in the regulatory region of the avian vitellogenin gene in chicken liver (34). In vivo experiments that directly address whether protein binding is required for a demethylase to demethylate hemimethylated DNA are under way.
It has been reported that the 5-methylcytosine-DNA glycosylase requires RNA with at least four nucleotides of complementarity to the hemimethylated target (9, 20). One can speculate that transcription factors or other regulatory proteins bind to the regulatory region of genes that are being activated during development and that this leads to hemimethylation at this region after one round of replication and a low level of transcription. This region then becomes demethylated on both strands, with 5-methylcytosine-DNA glycosylase targeting the hemimethylated DNA and being stabilized or activated by the RNA synthesized at the site. Hence, hemimethylated sites without at least a low level of transcription will not be demethylated by this enzyme. Regardless of how the second step of demethylation occurs, the strength of protein binding may play a critical role in targeting specific sites by protecting them from the maintenance methylase in this demethylation process. The requirement of transcription factor binding and DNA replication for demethylation in Xenopus embryos was reported (27) while this paper was in preparation. The findings in the present study are mostly in agreement with the observations reported by Matsuo et al. (27). However, the present study goes considerably further in providing detailed analyses of the sites of demethylation, the role of EBNA-1 binding, the site specificity of demethylation, and the stepwise mechanism of demethylation in mammalian cells. Although the demethylation is clearly a two-step process, this two-step mechanism may be responsible for only a subset of demethylation events. However, nothing is currently known that precludes it from being the predominant and perhaps the only pathway.
ACKNOWLEDGMENTS
I thank U. Grawunder, P. A. Jones, A. Kalb, M. R. Lieber, B. Tracy, R. West, and C. Windham for critical reading of the manuscript. I also thank J. Hearing for her generous gift of the oriP mutant plasmids.
This work was supported by NIH grant GM54781 and CTR grant 4494.
REFERENCES
- 1.Bird A P. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 2.Bird A P. The essentials of DNA methylation. Cell. 1992;70:5–8. doi: 10.1016/0092-8674(92)90526-i. [DOI] [PubMed] [Google Scholar]
- 3.Brandeis M, Frank D, Keshet I, Siegfried Z, Mendelsohn M, Nemes A, Temper V, Razin A, Cedar H. Sp1 elements protect a CpG island from de novo methylation. Nature. 1994;371:435–438. doi: 10.1038/371435a0. [DOI] [PubMed] [Google Scholar]
- 4.Cedar H. DNA methylation and gene activity. Cell. 1988;53:3–4. doi: 10.1016/0092-8674(88)90479-5. [DOI] [PubMed] [Google Scholar]
- 5.Clark S, Harrison J, Paulm C, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dean F B, O’Donnell M. DNA-protein interactions: two steps to binding replication origins? Curr Biol. 1996;6:931–934. doi: 10.1016/s0960-9822(02)00629-2. [DOI] [PubMed] [Google Scholar]
- 7.Ernberg I, Falk K, Minarovits J, Busson P, Tursz T, Masucci M G, Klein G. The role of methylation in the phenotype-dependent modulation of Epstein-Barr nuclear antigen 2 and latent membrane protein genes in cells latently infected with Epstein-Barr virus. J Gen Virol. 1989;70:2989–3002. doi: 10.1099/0022-1317-70-11-2989. [DOI] [PubMed] [Google Scholar]
- 8.Falk K, Ernberg I. An origin of DNA replication (oriP) in highly methylated episomal Epstein-Barr virus DNA localizes to a 4.5-kb unmethylated region. Virology. 1993;195:608–615. doi: 10.1006/viro.1993.1412. [DOI] [PubMed] [Google Scholar]
- 9.Frémont M, Siegmann M, Gaulis S, Matthies R, Hess D, Jost J P. Demethylation of DNA by purified chick embryo 5-methylcytosine-DNA glycosylase requires both protein and RNA. Nucleic Acids Res. 1997;25:2375–2380. doi: 10.1093/nar/25.12.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grainger R M, Hazard-Leonards R M, Samaha F, Hougan L M, Lexk M R, Thomsen G H. Is hypermethylation linked to activation of δ-crystallin genes during lens development? Nature. 1983;306:88–91. doi: 10.1038/306088a0. [DOI] [PubMed] [Google Scholar]
- 11.Harrison S, Fisenne K, Hearing J. Sequence requirements of the Epstein-Barr virus latent origin of DNA replication. J Virol. 1994;68:1913–1925. doi: 10.1128/jvi.68.3.1913-1925.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirt B. Selective extraction of polyoma DNA from infected mouse cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 13.Hsieh C-L. Dependence of transcriptional repression on CpG methylation density. Mol Cell Biol. 1994;14:5487–5494. doi: 10.1128/mcb.14.8.5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsieh C-L. Stability of patch methylation and its impact in regions of transcriptional initiation and elongation. Mol Cell Biol. 1997;17:5897–5904. doi: 10.1128/mcb.17.10.5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsieh D-J, Camiolo S M, Yates J L. Constitutive binding of EBNA1 protein to the Epstein-Barr virus replication origin, oriP, with distortion of DNA structure during latent infection. EMBO J. 1993;12:4933–4944. doi: 10.1002/j.1460-2075.1993.tb06187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu L F, Minarovits J, Cao S L, Contreras-Salazar B, Rymo L, Falk K, Klein G, Ernberg I. Variable expression of latent membrane protein in nasopharyngeal carcinoma can be related to methylation status of the Epstein-Barr virus BNLF-1 5′-flanking region. J Virol. 1991;65:1558–1567. doi: 10.1128/jvi.65.3.1558-1567.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones C H, Hayward S D, Rawlins D R. Interaction of the lymphocyte-derived Epstein-Barr virus nuclear antigen EBNA-1 with its DNA-binding sites. J Virol. 1989;63:101–110. doi: 10.1128/jvi.63.1.101-110.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jost J-P. Nuclear extracts of chicken embryos promote an active demethylation of DNA by excision repair of 5-methyldeoxycytidine. Proc Natl Acad Sci USA. 1993;90:4684–4688. doi: 10.1073/pnas.90.10.4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jost J-P, Siegmann M, Sun L, Leung R. Mechanisms of DNA demethylation in chicken embryos. J Biol Chem. 1995;270:9734–9739. doi: 10.1074/jbc.270.17.9734. [DOI] [PubMed] [Google Scholar]
- 20.Jost J P, Frémont M, Siegmann M, Hofsteenge J. The RNA moiety of chick embryo 5-methylcytosine- DNA glycosylase targets DNA demethylation. Nucleic Acids Res. 1997;25:4545–4550. doi: 10.1093/nar/25.22.4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kafri T, Gao X, Razin A. Mechanistic aspects of genome-wide demethylation in the preimplantation mouse embryo. Proc Natl Acad Sci USA. 1993;90:10558–10562. doi: 10.1073/pnas.90.22.10558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keshet I, Lieman-Hurwitz J, Cedar H. DNA methylation affects the formation of active chromatin. Cell. 1986;44:535–543. doi: 10.1016/0092-8674(86)90263-1. [DOI] [PubMed] [Google Scholar]
- 23.Little R D, Schildkaraut C L. Initiation of latent DNA replication in the Epstein-Barr virus genome can occur at sites other than the genetically defined origin. Mol Cell Biol. 1995;15:2893–2903. doi: 10.1128/mcb.15.5.2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Macleod D, Charlton J, Mullins J, Bird A. Sp1 sites in the mouse aprt gene promoter are required to prevent methylation of the CpG island. Genes Dev. 1994;8:2282–2292. doi: 10.1101/gad.8.19.2282. [DOI] [PubMed] [Google Scholar]
- 25.Marchuk D, Drumm M, Saulino A, Collins F S. Construction of T-vectors, a rapid and general system for direct cloning of unmodified PCr products. Nucleic Acids Res. 1990;19:1154. doi: 10.1093/nar/19.5.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marin M, Karis A, Visser P, Grosveld F, Philipsen S. Transcription factor Sp1 is essential for early embryonic development but dispensable for cell growth and differentiation. Cell. 1997;89:619–628. doi: 10.1016/s0092-8674(00)80243-3. [DOI] [PubMed] [Google Scholar]
- 27.Matsuo K, Silke J, Georgiev O, Marti P, Giovannini N, Rungger D. An embryonic demethylation mechanism involving binding of transcription factors to replicating DNA. EMBO J. 1998;17:1446–1453. doi: 10.1093/emboj/17.5.1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paroush Z, Keshet I, Yisraeli J, Cedar H. Dynamics of demethylation and activation of the a-actin gene in myoblasts. Cell. 1990;63:1229–1237. doi: 10.1016/0092-8674(90)90418-e. [DOI] [PubMed] [Google Scholar]
- 29.Pfeifer G P, Tanguay R L, Steigerwald S D, Riggs A D. In vivo footprint and methylation analysis by PCR-aided genomic sequencing: comparison of active and inactive S chromosomal DNA at the CpG island and promoter of human PGK-1. Genes Dev. 1990;4:1277–1287. doi: 10.1101/gad.4.8.1277. [DOI] [PubMed] [Google Scholar]
- 30.Pfeifer G P, Riggs A D. Chromatin differences between active and inactive X chromosomes revealed by genomic footprinting of permeabilized cells using DNaseI and ligation-mediated PCR. Genes Dev. 1991;5:1102–1113. doi: 10.1101/gad.5.6.1102. [DOI] [PubMed] [Google Scholar]
- 31.Razin A, Cedar H. DNA methylation and gene expression. Microbiol Rev. 1991;55:451–4578. doi: 10.1128/mr.55.3.451-458.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Razin A, Riggs A D. DNA methylation and gene function. Science. 1980;210:604–610. doi: 10.1126/science.6254144. [DOI] [PubMed] [Google Scholar]
- 33.Razin A, Kafri T. DNA methylation from embryo to adult. Prog Nucleic Acid Res Mol Biol. 1994;48:53–81. doi: 10.1016/s0079-6603(08)60853-3. [DOI] [PubMed] [Google Scholar]
- 34.Saluz H P, Jiricny J, Jost J P. Genomic sequencing reveals a positive correlation between the kinetics of strand-specific DNA demethylation of the overlapping estradiol/glucocorticoid-receptor binding sites and the rate of avian vitellogenin mRNA synthesis. Proc Natl Acad Sci USA. 1986;83:7167–7171. doi: 10.1073/pnas.83.19.7167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sugden B, Marsh K, Yates J. A vector that replicates as a plasmid and can be efficiently selected in B-lymphoblasts transformed by Epstein-Barr virus. Mol Cell Biol. 1985;5:410–413. doi: 10.1128/mcb.5.2.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sullivan C H, Grainger R M. δ-Crystallin genes become hypomethylated in postmitotic lens cells during chicken development. Proc Natl Acad Sci USA. 1986;83:329–333. doi: 10.1073/pnas.84.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vairapandi M, Duker N J. Enzymic removal of 5-methylcytosine from DNA by a human DNA-glycosylase. Nucleic Acids Res. 1993;21:5323–5327. doi: 10.1093/nar/21.23.5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weiss A, Keshet I, Razin A, Cedar H. DNA demethylation in vitro: involvement of RNA. Cell. 1996;86:709–718. doi: 10.1016/s0092-8674(00)80146-4. [DOI] [PubMed] [Google Scholar]
- 39.Wigler M, Sweet, R. R, Sim G K, Wold B, Pellicer A, Lacy E, Maniatis T, Silverstein S, Axel R. Transformation of mammalian cells with genes from prokaryotes and eukaryotes. Cell. 1979;16:777–785. doi: 10.1016/0092-8674(79)90093-x. [DOI] [PubMed] [Google Scholar]
- 40.Yates J L, Warren N, Reisman D, Sugden B. A cis-acting element from the Epstein-Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc Natl Acad Sci USA. 1984;81:3804–3810. doi: 10.1073/pnas.81.12.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yates J L, Warren N, Sugden B. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature. 1985;313:812–815. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]