Extended Data Figure 2: Kinship analysis.
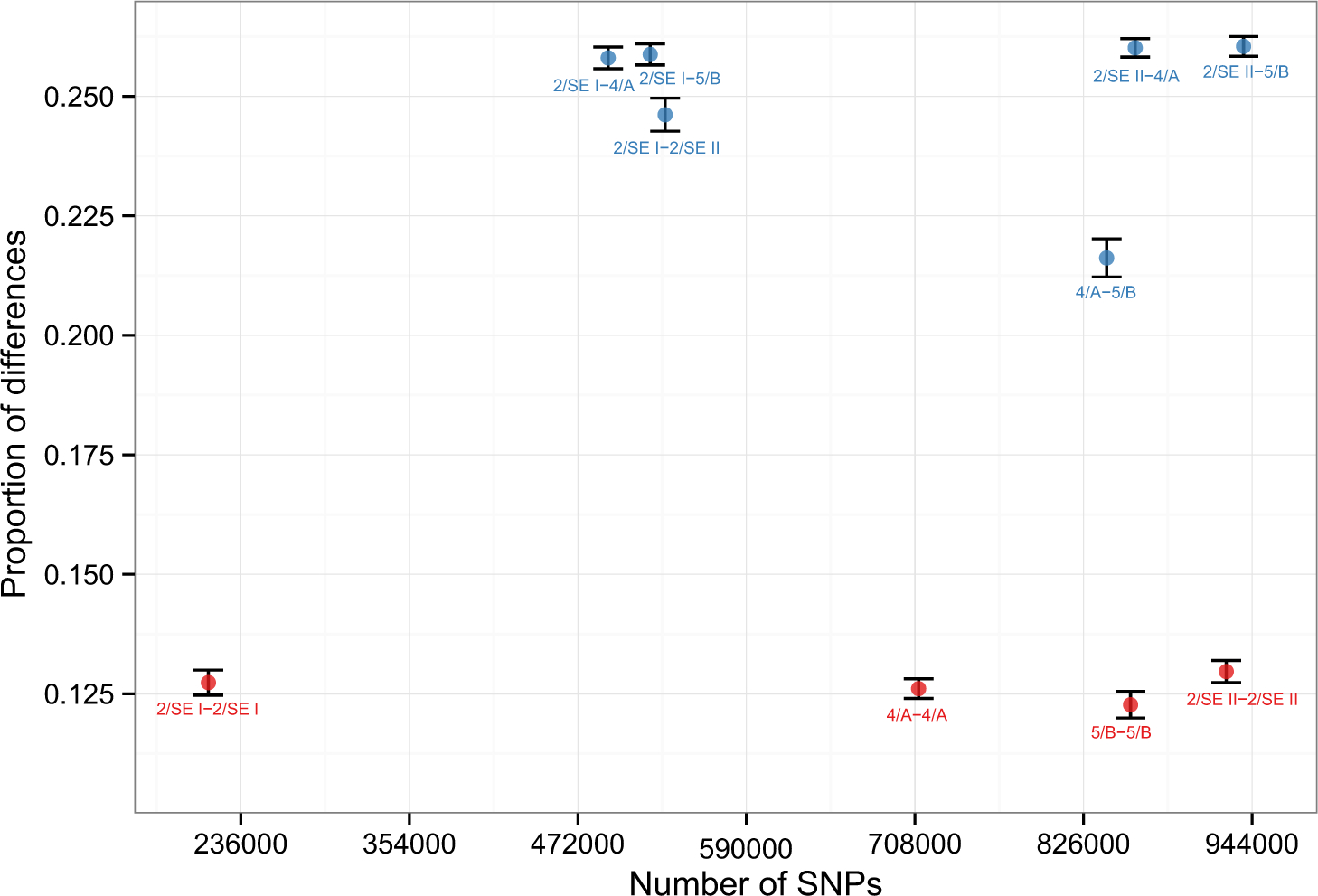
Shown are mean genome-wide allelic mismatch rates for each pair of individuals (blue), as well as intra-individual comparisons (red). We selected one read per individual at random at each targeted SNP (using all 1,233,013 targeted sites). Monozygotic twins (or intra-individual comparisons) are expected to have a value one-half as large as unrelated individuals; first-degree relatives, halfway between monozygotic twins and unrelated individuals; second-degree relatives, halfway between first-degree relatives and unrelated individuals; and so on. The presence of inbreeding also serves to reduce the rates of mismatches. For 4/A and 5/B, because both died as children, we can eliminate a grandparent-grandchild relationship, and the lack of long segments with both homologous chromosomes shared IBD implies that they are not double cousins (the few ostensible double-IBD stretches are likely a result of inbreeding; see Supplementary Information section 2). Thus, we can conclude that they were either uncle and niece (or aunt and nephew) or half-siblings. Bars show 99% confidence intervals (computed by block jackknife).
