ABSTRACT
Bacillus amyloliquefaciens is considered the most successful biological control agent due to its ability to colonize the plant rhizosphere and phyllosphere where it outgrows plant pathogens by competition, antibiosis, and inducing plant defense. Its antimicrobial function is thought to depend on a diverse spectrum of secondary metabolites, including peptides, cyclic lipopeptides, and polyketides, which have been shown to target mostly fungal pathogens. In this study, we isolated and characterized the catecholate siderophore bacillibactin by B. amyloliquefaciens MBI600 under iron-limiting conditions and we further identified its potential antibiotic activity against plant pathogens. Our data show that bacillibactin production restrained in vitro and in planta growth of the nonsusceptible (to MBI600) pathogen Pseudomonas syringae pv. tomato. Notably, it was also related to increased antifungal activity of MBI600. In addition to bacillibactin biosynthesis, iron starvation led to upregulation of specific genes involved in microbial fitness and competition.
IMPORTANCE Siderophores have mostly been studied concerning their contribution to the fitness and virulence of bacterial pathogens. In the present work, we isolated and characterized for the first time the siderophore bacillibactin from a commercial bacterial biocontrol agent. We proved that its presence in the culture broth has significant biocontrol activity against nonsusceptible bacterial and fungal phytopathogens. In addition, we suggest that its activity is due to a new mechanism of action, that of direct antibiosis, rather than by competition through iron scavenging. Furthermore, we showed that bacillibactin biosynthesis is coregulated with the transcription of antimicrobial metabolite synthases and fitness regulatory genes that maximize competition capability. Finally, this work highlights that the efficiency and range of existing bacterial biocontrol agents can be improved and broadened via the rational modification of the growth conditions of biocontrol organisms.
KEYWORDS: Bacillus amyloliquefaciens, siderophores, antibiotics, iron starvation, plant pathogens, bactericides, plant-pathogenic bacteria
INTRODUCTION
Iron is a vital element required by every living organism that functions as a cofactor in numerous cellular processes, such as the tricarboxylic acid cycle, electron transport chain, and oxidative phosphorylation, or the biosynthesis of vitamins, antibiotics, toxins, and other Fe-containing aromatic compounds (1). It also plays an important role in microbial biofilm formation by regulating the surface motility of microorganisms (2, 3). Under certain environmental conditions, the level of physiologically available concentrations of iron can drop far below 1 mM (4). As a survival strategy, many bacteria, fungi, and monocotyledonous plants synthesize and secrete high-affinity extracellular siderophores to acquire iron (5, 6). Additional functions of bacterial siderophores include the alteration of the soil microbial community (7), the promotion of plant growth (8), the potential use as biocontrol agents (9, 10), and the enhancement of bioremediation of heavy metals (11, 12). Siderophores have also been exploited as biosensors (13, 14) and as selective mediators of antibiotic clinical bacteria (Trojan horse strategy) (15).
Siderophores are low-molecular-weight molecules in the range of approximately 400 to 1,500 Da. They usually form hexadentate or octahedral complexes with ferric iron and typically employ hydroxamates, α-hydroxycarboxylates and catechols as extremely effective Fe3+ ligands; thus, they can be classified as hydroxamate, hydroxycarboxylate, or catecholate type siderophores, respectively (16). The hydroxamate-type siderophores are produced by Pseudomonas spp. (17), while the catecholate types are produced by Gram-negative Escherichia coli, Salmonella enterica serovar Typhimurium, and Klebsiella pneumonia, which produce enterochelin (18), and Gram-positive Bacillus spp. that produce bacillibactin (19).
Bacillus species dominate the biopesticide market (20). Among them, B. amyloliquefaciens is an exceptionally pervasive species due to its ability to colonize different ecological niches such as soil, water, a variety of surfaces, and the rhizospheres of many plants (21). This adaptation in a wide range of environments derives from its impressive ability to produce a diverse spectrum of antagonistic secondary metabolites, including peptides, cyclic lipopeptides, and polyketides. Such peptides can facilitate root colonization and interaction with the host plant and prime plant defense responses (22, 23).
B. amyloliquefaciens has been considered the most competitive strain among biological control agents (BCAs) based on its antifungal activity against agronomically important fungal phytopathogens (24–26). However, little is known about its bactericidal activity and mode of action. B. amyloliquefaciens has been reported to control a few economically important bacterial diseases, such as the bacterial blight and bacterial leaf streak of rice caused by Xanthomonas oryzae (27), fire blight disease (Erwinia amylovora) (28), and tomato wilting (Ralstonia solanacearum) (29). More recent studies have shown biocontrol activity against some Pseudomonas syringae pathovars on sugar beet (30) and tomato plants (31, 32). Serifel is a commercial formulation of Bacillus amyloliquefaciens subsp. plantarum MBI600 endospores. Our group has previously shown that MBI600 can interact with the plant host by triggering a signaling network that induces systemic plant defense responses (33), including resistance to viral pathogens of tomato (34).
In this study, the ability of B. amyloliquefaciens MBI600 to produce and secrete the catecholate siderophore bacillibactin is demonstrated. In parallel, bactericidal efficacy evaluation experiments showed that iron starvation induced production of bacillibactin, which significantly enhanced, in vitro and in planta, the bactericidal activity of MBI600 against the phytopathogenic species Pseudomonas syringae pv. tomato. It also broadened or enhanced the antibiotic activity against phytopathogenic fungi. Finally, it is shown that under iron starvation conditions, the expression of genes involved in bacillibactin biosynthesis is coordinated with that of genes involved in the synthesis of secondary metabolites as well as with antibiotic activity, cell mobility, and chemotaxis.
RESULTS
Characterization of siderophores that are secreted by B. amyloliquefaciens MBI600 under iron starvation.
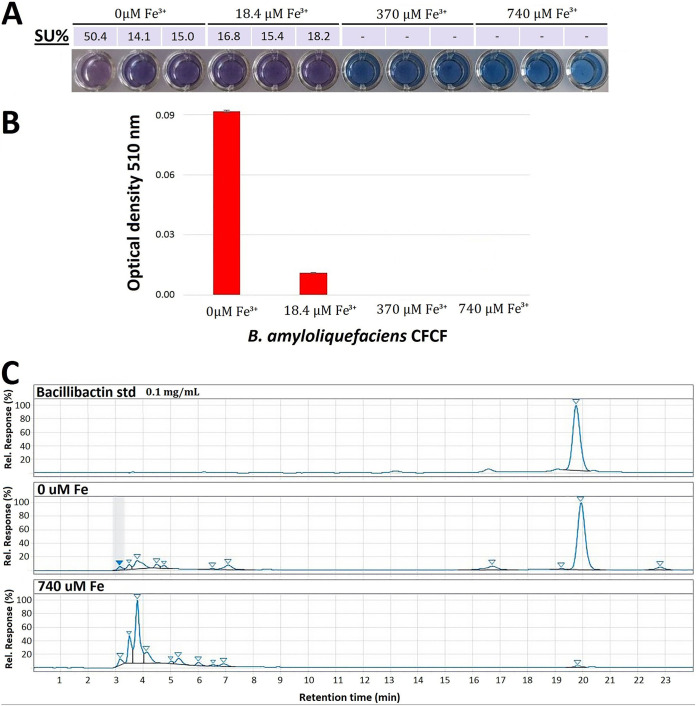
We screened for siderophore production in the supernatants of MBI600 cultures grown in chemically defined low-iron medium (CDLIM) (35) at various Fe3+ stress levels (0 and 18.4 μM added Fe3+ [iron starvation], 370 μM added Fe3+ [low iron stress], and 740 μM added Fe3+ [no iron stress]) using the chrome azurol S (CAS) assay (32). Under such iron stress conditions, MBI600 was capable of successfully growing (see Fig. S7 in the supplemental material). Siderophore production in the culture supernatants was clearly detected only under the iron starvation conditions of 0 and 18.4 μM Fe3+ (Fig. 1A). Catecholate groups, and the possible presence of bacillibactin, were identified using Arnow’s assay (increased absorbance at λmax of 510 nm in the supernatants with 0 μM Fe3+), as shown in Fig. 1B.
FIG 1.
Production and identification of bacillibactin by B. amyloliquefaciens MBI600. (A) Chrome azurol S (CAS) liquid assay of MBI600 culture supernatants in increasing Fe3+ concentrations and respective siderophore unit (SU%) calculations. (B) Determination of the catecholate siderophore type by Arnow’s assay. (C) RP-HPLC chromatograms of bacillibactin commercial standard and siderophore extracts from MBI600 culture supernatants, in the Discovery BIO wide-pore C18-10 preparative column.
B. amyloliquefaciens MBI600’s growth curve under iron starvation conditions (0, 18.5, and 370 μM Fe3+). Optical densities at 600 nm were measured for 48 h at 37°C using a multidetection microplate reader and automatically recorded for each well every 29 min. Each ribbon depicts growth under a specific Fe3+ concentration; full line represents mean values and bars represent SEs from three independent biological experiments. Download FIG S7, EPS file, 0.5 MB (534.1KB, eps) .
Copyright © 2021 Dimopoulou et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Bacillibactin production in the culture supernatant under Fe3+ stress conditions (0 μM Fe3+) was verified through a two-stage reverse-phase high-performance liquid chromatography (RP-HPLC) analysis coupled with nuclear magnetic resonance (NMR) analysis. A clear peak at 315 nm and 19.72 min, corresponding to bacillibactin standard, was detected in the chromatograms of the first preparative column and only in the 0 μΜ Fe3+ supernatants (Fig. 1C). The peak was collected, concentrated, and applied in a second analytical column for further purification. A very clear peak was detected (see Fig. S1), which was collected for NMR analysis.
HPLC chromatograms of the purified peak of bacillibactin from 0 μM Fe3+ supernatant and the commercial standard at 315 nm in the Poroshell 120 EC-C18 analytical column. (Inset) The corresponding NMR spectra. Download FIG S1, EPS file, 0.3 MB (287.8KB, eps) .
Copyright © 2021 Dimopoulou et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
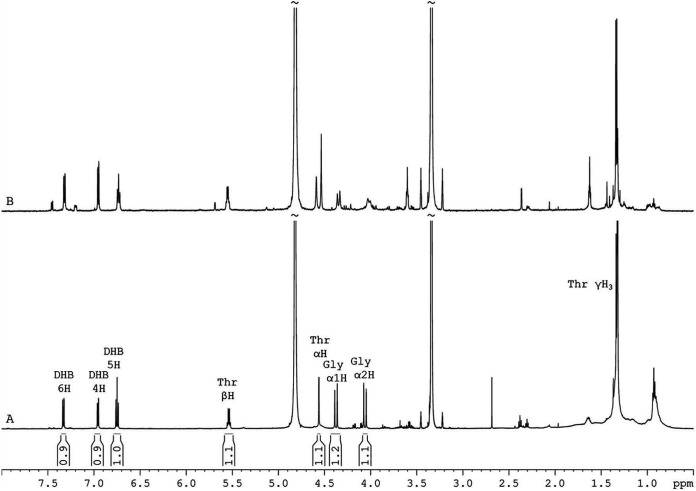
Following vacuum evaporation, the dried final peak was reconstituted in deuterated methanol for structure elucidation by 1H and 13C NMR spectroscopy. In the 1H one-dimensional (1D) NMR spectrum of the purified peak of 0 μM Fe3+ supernatant extract (Fig. 2A), all the characteristic resonances of bacillibactin structure were detected. The structure was further confirmed by a two-dimensional (2D) correlation spectroscopy (COSY) spectrum, where all the expected 1H-1H correlations were observed (Fig. S2A), and by a heteronuclear single quantum coherence-distortionless enhancement by polarization transfer (HSQC-DEPT) 1H-13C (Fig. S2B) spectrum, where protons are correlated with the corresponding carbon. All spectroscopic data, 1H and 13C chemical shifts, and 1H-1H and 1H-13C connectivities are in accordance with the literature (36, 37). For comparison reasons, the 1H 1D (Fig. 2B) and 2D NMR spectra were recorded also for the bacillibactin standard. 1H and 13C chemical shifts and coupling constants are provided in detail in Table 1.
FIG 2.
NMR analysis of the purified peak of 0 μM Fe3+ extract. Region (8.00 to 0.50 ppm) of 1H 1D NMR spectra of bacillibactin (BB) extracted from 0 μM Fe3+ supernatants of MBI600 (A) and commercially purchased bacillibactin (B) in deuterated methanol at 305 K. Peak integral values are noted between the x (ppm) axis and BB spectrum.
TABLE 1.
1H and 13C chemical shifts of the bacillibactin extracted from 0 μM Fe3+ supernatants of MBI600a
| Proton | 1H shiftb (ppm) | Multiplicityc and coupling constant (Hz)c | 13C shiftd (ppm) |
|---|---|---|---|
| DHBe H-4 | 6.96 | dd, 7.8/1.3 | 119.8 |
| DHB H-5 | 6.75 t | t, 8.0 | 119.9 |
| DHB H-6 | 7.33 | dd, 8.1/1.4 | 119.6 |
| Thr αH | 4.56 | d, 1.7 | 59.2 |
| Thr βH | 5.54 | qd, 6.7/1.7 | 72.5 |
| Thr γH | 1.33 | d, 6.6 | 17.3 |
| Gly αH | 4.38/4.06 | d, 16.2 | 44.3 |
The spectra were recorded in deuterated methanol (MeOD) (1H, 3.34 ppm; 13C, 49.15 ppm) at 305 K.
1H shift, 600.11 MHz.
d, doublet; dd, doublet of doublets; t, triplet; qd, quartet of doublets.
13C shift, 150.9 MHz.
DHB, 2,3-dihydroxybenzoic acid.
1H-1H COSY (8.00 to 0.50 ppm) (A) and 1H-13C HSQC-DEPT (8.00 to 0.50 ppm; 130 to 10 ppm) (B) NMR spectra of bacillibactin extracted from 0 μM Fe3+ supernatants of MBI600, in deuterated methanol, at 305 K. In HSQC-DEPT spectrum, methylene protons are colored red, while methine and methylene protons are black. Download FIG S2, EPS file, 0.1 MB (127.8KB, eps) .
Copyright © 2021 Dimopoulou et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Iron starvation increases the susceptibility of P. syringae pv. tomato to MBI600.
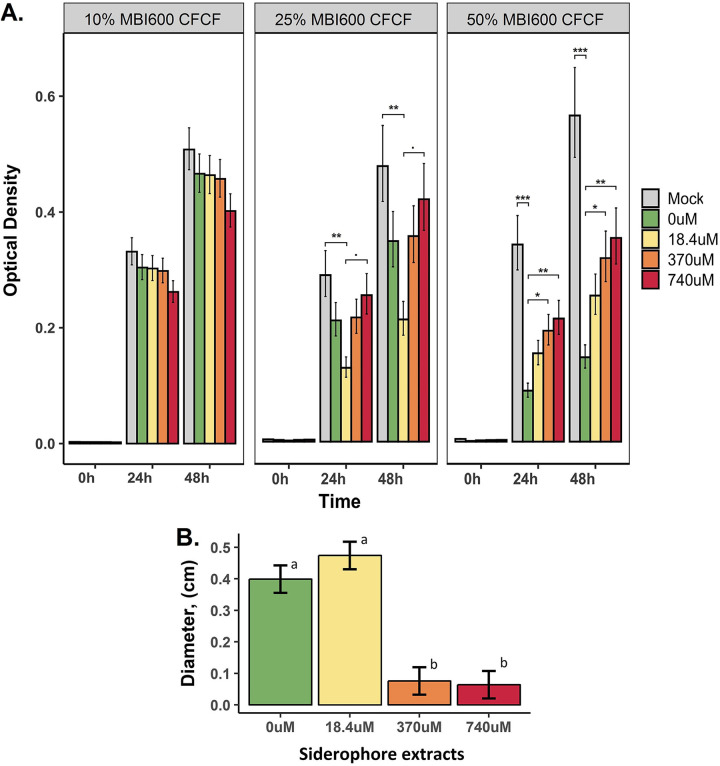
(i) In vitro bactericidal efficacy of cell-free culture filtrate.P. syringae pv. tomato was found to be marginally nonsusceptible to MBI600 in broth and plate susceptibility testing (see Fig. S3). Furthermore, it has been reported to assimilate xenosiderophores (38). It was, therefore, selected to assess the antibacterial activity of bacillibactin. P. syringae pv. tomato susceptibility was evaluated in dose-response treatments of MBI600 cell-free culture filtrate (CFCF) extracted from cultures grown under a range of iron starvation conditions (0, 18.4, 370, or 740 μM Fe3+) (Fig. 3A). P. syringae pv. tomato growth was not affected by the presence of 10% MBI600 CFCF in the case of all Fe3+ conditions and at both time points (24 and 48 h). It was only negatively affected by the 25% treatment with MBI600 CFCF extracted from iron-limiting conditions (0 μM and 18.4 μM Fe3+). However, under all iron conditions, except 18.4 μΜ Fe3+ (P = 0.0865 and 0.0102 for control and mock, respectively), results were not statistically significant compared to those for control (740 mM) and mock (LB) conditions, at both time points tested (Fig. 3A). The 50% CFCF treatment significantly decreased P. syringae pv. tomato growth in comparisons between MBI600 iron starvation conditions and control (0 μΜ Fe3+, P = 0.002) or mock (0 μΜ Fe3+, P < 0.0001; 18.4 μΜ Fe3+, P = 0.012) conditions. The effect of iron starvation was also confirmed in experiments using a different carbon source (succinic acid) (see Fig. S4) to exclude medium interference. In this case, there was a significant delay in P. syringae pv. tomato growth in the presence of 50% CFCF from MBI600 grown in iron-depleted broth compared to that grown in the iron-containing broth. Finally, iron starvation increased MBI600 bactericidal efficacy in agar plates containing a P. syringae pv. tomato lawn (see Fig. S5). The inhibition zone formed around MBI600 colonies was negatively correlated with the iron concentration of the agar medium.
FIG 3.
P. syringae pv. tomato becomes susceptible to MBI600 metabolites produced under iron starvation conditions. (A) In vitro bactericidal efficacy of CFCF. Susceptibility was tested against increasing MBI600 CFCF concentrations (0%, 2%, 10%, 25%, and 50% [vol/vol]). CFCF was extracted from MBI600 cells grown in a siderophore-producing medium (CDLIM) supplemented with increasing Fe3+ concentrations (0, 18.4, 370, and 740 μM) for 48 h. Observed effects were evaluated based on optical density at 600 nm (OD600) readings at 0, 24, and 48 hpi in microtiter plates. Estimated marginal means and their standard errors are plotted. ·, P < 0.1; *, P < 0.05; **, P < 0.01; ***, P < 0.001. (B) Inhibitory activity of bacillibactin. Estimated marginal means and standard errors of inhibition zone measurements in P. syringae pv. tomato agar cultures are plotted. Different lowercase letters represent statistically different estimates at a P value of ≤0.05 according to Tukey’s post hoc comparisons.
In vitro bactericidal efficacy of B. amyloliquefaciens MBI600. (Left) P. syringae pv. tomato broth susceptibility was tested against increasing MBI600 cell free culture filtrate (CFCF) concentrations (0, 0.5, 2, 10, 25, and 50% vol/vol). CFCF was extracted from MBI600 cells grown in LB medium. Observed effects were evaluated based on OD600 readings at 0, 24, and 48 hpi in microtiter plates. Estimated marginal means and their standard errors are plotted. (Right) No inhibition zone was formed around MBI600 culture grown on 9-mm Whatman discs that were placed on agar medium inoculated with P. syringae pv. tomato. Download FIG S3, EPS file, 1.4 MB (1.5MB, eps) .
Copyright © 2021 Dimopoulou et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Effect on growth of Pseudomonas syringae pv. tomato (Pto) in LB medium supplemented with 10% (a), 25% (b), or 50% (c) B. amyloliquefaciens (Bam) CFCF (cell-free culture filtrate) supplemented with different iron concentrations (0, 18.4, 148, and 370 μM Fe3+). Optical densities at 600 nm were measured for 48 h at 28°C using a multidetection microplate reader. Estimated marginal means and their standard errors are plotted. Download FIG S4, EPS file, 2.4 MB (2.4MB, eps) .
Copyright © 2021 Dimopoulou et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Inhibition zones formed in Pseudomonas syringae pv. tomato (Pto) agar cultures in the presence of B. amyloliquefaciens (Bam). Pto inoculum suspension was adjusted to 109 CFU/ml and allowed to grow in agar medium containing increasing iron concentrations (0, 18.4, and 148 μM Fe3+). Values represent the averages of 24 inhibition zone measurements for each iron concentration. Different letters represent statistically different data points at a P value of ≤0.05 according to Tukey’s post hoc comparisons. Download FIG S5, EPS file, 0.7 MB (730.9KB, eps) .
Copyright © 2021 Dimopoulou et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(ii) Bactericidal efficacy of siderophore extracts.
We tested the bactericidal efficacy of ethyl acetate siderophore extracts in agar cultures of P. syringae pv. tomato. Inhibition clear zones were observed and measured at 0.39 and 0.47 cm in the case of 0 and 18.4 μM Fe3+ conditions, respectively (Fig. 3B). When the iron supply was 370 or 740 μM Fe3+, that is, when bacillibactin was not detected, there was not a significant difference compared to the mock treatment (water, zero diameter). Both treatments leading to bacillibactin production (i.e., 0 and 18.4 μM Fe3+) led to statistically significant bactericidal efficacy compared to that with both 370 and 740 μM Fe3+ and controls.
(iii) In planta bactericidal efficacy of CFCF.
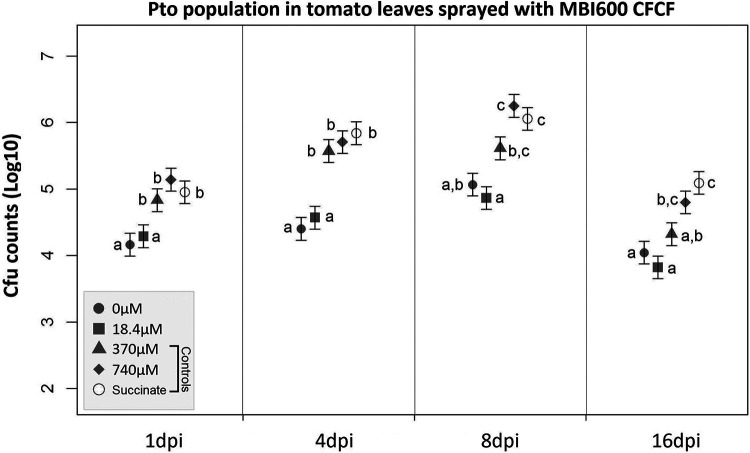
In planta bactericidal activity of CFCF was based on P. syringae pv. tomato total population in leaves of tomato plants sprayed with MBI600 CFCF over a period of 16 days postinoculation (dpi). At 1 dpi, the P. syringae pv. tomato population was significantly reduced after treatment with CFCF from iron-depleted media (0 and 18.4 μM Fe3+) compared to that in the controls (370 and 740 μM Fe3+ and the succinate control). Such differences were amplified at 4 dpi and persisted for several days before disease development (Fig. 4).
FIG 4.
P. syringae pv. tomato population growth in tomato leaves sprayed with succinic acid medium (SM) (control) or CFCF of MBI600 grown in increasing Fe3+ concentrations (0, 18.4, 340, or 740 μM). Estimated means and corresponding standard errors of the logarithm of the bacterial population from 4 samples are shown. Different lowercase letters represent statistically different data points at a P value of ≤0.05 according to Tukey’s post hoc comparisons.
Iron starvation broadens the antifungal activity of MBI600.
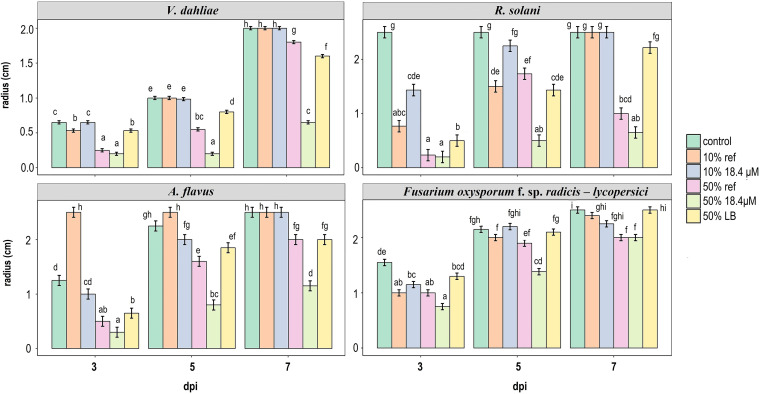
Based on the known antifungal range of MBI600 (Serifel label claims), we selected 7 strains of nonsusceptible fungal species (see Table S1) as well as Verticillium dahliae and oomycete Phytophthora cactorum strains, which are of intermediate susceptibility. Low susceptibility was confirmed using a treatment of MBI600 CFCF grown in LB (instead of the iron-limiting CDLIM) at the 50% (vol/vol) dose (nominated LB treatment). Susceptibility of all species was then tested against three doses/treatments (0, 10, and 50% [vol/vol]) of CFCF extracted from MBI600 cultures grown under 18.4 or 740 μΜ Fe3+ conditions as well as the LB treatment. Effects were attributed to bacillibactin only if comparisons between the 18.4 μM Fe3+ condition and each of the three controls (740 μΜ Fe3+ condition, no treatment, and LB treatment) were statistically significant. We found that iron starvation resulted in an inhibition of fungal growth, which was positively correlated with treatment dose and time (Fig. 5). Fungal growth was not affected by low CFCF doses (10%). On the other hand, all pathogenic species were found to be susceptible to the 50% treatment under the 18.4 μM Fe3+ condition. Growth inhibition was statistically significant in the cases of Fusarium oxysporum f. sp. radicis-lycopersici and Rhizoctonia solani at 5 dpi as well as in the cases of Aspergillus flavus and V. dahliae at 5 and 7 dpi (Fig. 5). In the cases of all other tested species, the 18.4 μM Fe3+ condition showed statistically significant differences compared with control (0%) or LB treatments but not with 50% 740 μM Fe3+ (see Fig. S6).
FIG 5.
Iron starvation expands the range of antifungal activity of MBI600. Radial growth of 4 fungal species in minimal medium (MM) agar plates and in response to the following treatments: no treatment (control), 10% and 50% CFCF of MBI600 grown in 18.4 or 740 (ref) μM Fe+3 conditions, and 50% CFCF of MBI600 grown in LB. Values represent the estimated marginal means and corresponding standard errors of inhibition zone measurements. Different lowercase letters within each panel represent statistically different data points at a P value of ≤0.05 according to Tukey’s post hoc comparisons. Comparisons are among all treatments and among time points for each fungal species.
Iron starvation expands the range of antifungal activity of MBI600. Radial growth of 4 fungal species in minimal medium (MM) agar plates and in response to the following treatments: no treatment (control), 10% and 50% CFCF of MBI600 grown under either 740 μM Fe+3 (ref) or 18.4 μM Fe+3 conditions, and 50% CFCF of MBI600 grown in LB. Values represent the estimated marginal means and corresponding standard errors of inhibition zone measurements. ***, P < 0.001; **, P < 0.01; *, P < 0.05; ·, P < 0.1 according to Tukey’s post hoc comparisons. Download FIG S6, EPS file, 0.9 MB (943.9KB, eps) .
Copyright © 2021 Dimopoulou et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
List of bacterial, fungal, and oomycete strains used in this study. Download Table S1, DOCX file, 0.1 MB (14.7KB, docx) .
Copyright © 2021 Dimopoulou et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Determination of MBI600 gene expression in different iron concentrations.
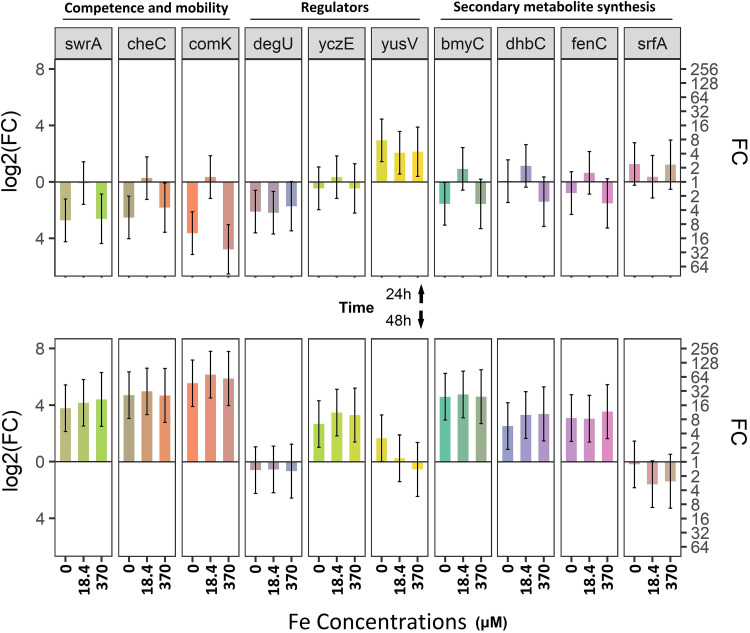
Using the aforementioned iron-depleted media, we showed that bacillibactin synthase (dhbC) is activated in iron-depleted cultures found in late static phase (48 h) but not under the 740 μM Fe3+ condition (Fig. 6), thus confirming siderophore production assays and the notion that 740 μM Fe3+ exceeds the threshold for iron deficiency (Fig. 1). Genes involved in B. amyloliquefaciens competence (comK), chemotaxis (cheC), and swarming (swrA) were significantly downregulated under all conditions compared to expression in the control at 24 h postinoculation (hpi). At 48 hpi, all 3 genes showed a significant upregulation under all iron conditions compared to expression in the control (baseline log2 fold change [FC]) (Fig. 6). A 2-fold upregulation of srfAC occurred at 24 hpi and in the cases of 0 and 370 μM Fe3+ conditions. Concerning the regulatory genes of these two groups of genes, those for competence and mobility, yusV was upregulated at 24 hpi under all iron conditions compared to expression in the control (740 μΜ Fe3+) and remained elevated at 48 hpi only in the case of the 0 μM Fe3+ condition. The upregulation of yczE occurred later (48 hpi) under all conditions compared to expression in the control, while the regulatory gene degU was not activated upon iron depletion.
FIG 6.
Iron starvation activates genes involved in MBI600 fitness. MBI600 cells were collected at 24 h (top) and 48 h (bottom) postinoculation of SM broth supplemented with increasing Fe3+ concentrations (0, 18.4, 340, or 740 μM). RT-qPCR analysis was performed using specific primers for the following genes: dhbC, siderophore synthase; yusV, siderophore transport system. Fold change (FC) represents the relative difference in expression between each treatment and control (740 μM Fe3+). gyrA was used as a reference gene. Bars indicate the 95% confidence interval (2 × standard error [SE]). Estimates whose confidence interval includes the baseline value 0 (1 in fold change scale) are not significantly different from control at an α of 0.05. All other differences are significant.
DISCUSSION
Bacillus amyloliquefaciens is known to produce a broad range of antimicrobial metabolites (22). Analysis of the B. amyloliquefaciens genome revealed 11 gene clusters, which occupy 9% of its genome, that are involved in the synthesis of antimicrobial metabolites (39). Three gene clusters are involved in nonribosomal synthesis of polyketides such as difficidin, macrolactin, and bacillaene, which are considered to have antibacterial activities (40, 41). Another five clusters are involved in nonribosomal synthesis of cyclic lipopeptides such as surfactin, bacillomycin, fengycin and bacillibactin (42). Surfactins are antibiotic compounds that show hemolytic, antimicrobial, and antiviral activities by altering membrane integrity (4, 43), whereas fengycin is specifically active against filamentous fungi and inhibits phospholipase A2 (44, 45). Bacillomycin D is a member of the iturin family, with a wide range of antibacterial and antifungal properties and hemolytic activities (26). Finally, bacillibactin is thought to contribute to the virulence of animal pathogens (46, 47), but its role in microbial competition remains to be elucidated.
Even if the role of siderophores in the biological control mechanism was first suggested 40 years ago (9), their antimicrobial activity has been reported only sporadically (48, 49). Siderophores have been studied for their roles in fitness and pathogenicity rather than biocontrol, with pyoverdine-producing pseudomonads being the model system (50). Lately, numerous studies correlate biological control and siderophore production by plant growth-promoting rhizobacteria (PGPRs). For example, Ghazy and El-Nahrawy suggested that two siderophore-producing strains, Bacillus subtilis MF497446 and Pseudomonas koreensis MG209738, reduced infection by Cephalosporium maydis in maize plant (51). Furthermore, Prema and Selvarani suggested microbial siderophores as potential biocontrol agents against plant pathogens (52). In this work, siderophore extract and bacillibactin were isolated from MBI600 cultures and tested for their bactericidal activity. This is the first report of bacillibactin isolation from commercial B. amyloliquefaciens species and characterization by NMR. We developed an HPLC protocol to isolate and purify bacillibactin from MBI600 culture supernatant, and its structure was confirmed by NMR analysis. All spectroscopic data were in accordance not only with the literature (33, 34) but also with our purchased bacillibactin standard (EMC Microcollections, Germany). Interestingly, NMR spectra revealed that isolated bacillibactin from this study showed higher purity than the purchased standard, highlighting the effectiveness of the developed isolation and purification protocol.
The combination of agar and broth tests suggested that P. syringae pv. tomato is susceptible to MBI600 cells and CFCF grown under iron-limited conditions. P. syringae pv. tomato inhibition was negatively correlated with the iron concentration of the MBI600 diet. Since MBI600 metabolites secreted in iron-rich media failed to inhibit Pseudomonas growth, it is concluded that iron starvation-elicited production of bacillibactin is responsible for this gain of function. This hypothesis was confirmed by the formation of inhibition zones around a bacillibactin extract in Pseudomonas-containing agar media. Production of siderophores was correlated not just with an extension of the biocontrol activity range of MBI600 against a nonsusceptible pseudomonad but also with biocontrol activity against nonsusceptible fungal pathogens. The strongest inhibition was observed in the cases of A. flavus, R. solani, and V. dahliae, while Botrytis cinerea, F. oxysporum f. sp. radicis-lycopersici, and P. cactorum growth was delayed in the presence of MBI600 siderophores. Results suggest that nutrition plays a crucial role in MBI600 antifungal activity, though in planta experiments are needed to confirm this notion. The fact that bacillibactin inhibited the growth of pathogens in iron-containing media suggests that its antibiotic function does not rely on iron scavenging to starve competing species. In this work, we identify, for the first time, the antibiotic potential of bacillibactin. Moreover, siderophores have been associated with iron scavenging (10). We suggest that their role in microbial competition should be reconsidered. Previous studies reported that several bacteria, including Pseudomonas, are able to utilize siderophores of other species (xenosiderophores) (38). It is, therefore, possible that bacillibactin is absorbed by Pseudomonas syringae pv. tomato and that its antibiotic function is intercellular.
Combined evidence supports the notion that to determine the range of biocontrol activity of BCAs in vitro, with the maximum correlation to in planta outcomes, it is necessary to test them in different minimal media in which both BCA and pathogens can grow. Such media would best mimic conditions of the nutrient-poor phyllosphere but also allow antagonists to fully exploit their antimicrobial arsenal. This hypothesis was confirmed in planta. Growth of MBI600 cultures under iron starvation conditions induced bacillibactin synthesis and secretion to a CFCF that was capable of reducing P. syringae pv. tomato population and delay the infection of tomato plants. Siderophores have already been hypothesized to contribute to the antifungal activity of B. subtilis in planta (10), but this is the first report that provides direct evidence of the antibacterial and antifungal activities of bacillibactin in planta.
The enhanced susceptibility of Penicillium sp. and Aspergillus niger to CFCF extracted from MBI600 cultures grown in both iron-depleted and iron-rich minimal media suggested that the nutrient composition activated the production of fungicidal metabolites in addition to bacillibactin. Expression analyses of (i) genes involved in secondary metabolite synthesis (dhbC, siderophore synthase; bmyC, bacillomycin [iturin] synthase; yczE, positive regulator of bacillomycin [iturin] synthesis; fenC, fengycin synthase; and sfpAC, surfactin) and (ii) comΚ (competition), cheC (chemotaxis), swrA (swarming), deqU (positive regulator of bacillomycin [iturin]), and yusV (probable siderophore transport system) confirmed the activation of the metabolic network responsible for B. amyloliquefaciens antibiosis, competence, and mobility.
It was previously shown that Bacillus subtilis has a strong antifungal activity due to the synergistic actions of surfactin, bacillomycin D, and fengycin (53, 54). Production of either of the latter two is articulately delayed (late stationary phase) compared with that of surfactin (transition between exponential and stationary growth) (55, 56). Accordingly, it was found that iron enhanced the expression of the surfactin biosynthetic gene srfAC at 24 hpi, while expression of bacillomycin synthase C (bmyC) and fengycin synthase C (fenC) was enhanced at 48 hpi. In support, yczE, a positive regulator of bacillomycin, was also activated at 48 hpi under low/depleted-iron conditions. These data suggest that MBI600 coordinately induces its offense to prevail against other organisms in iron-depleted habitats. Such a hypothesis is supported by the activation of comK during stationary phase, as previously reported (57, 58). In parallel, iron depletion was detected by chemotaxis and stimulated swarming motility toward more favorable conditions (59–61), as indicated by the overexpression of cheC and swrA, respectively. The fact that degU, which regulates motility and biofilm formation (62, 63), is not overexpressed under iron stress is expected, as its function is thought to depend on its protein phosphorylation (64). Expression of genes involved in biosynthetic pathways is an indication for the production of endpoint metabolites, but their actual levels can only be confirmed by quantification experiments. Production of bacillibactin was confirmed by activation of dhbC, an isochorismate synthase of the siderophore synthase operon (65), under iron depletion conditions at 48 hpi. In support, yusV, a gene involved in siderophore uptake (66), was overexpressed at 24 hpi in the cases of all treatments compared to expression with the reference treatment.
B. amyloliquefaciens MBI600 has been mostly used commercially against fungal diseases. This study confirmed that its bactericidal activity is broadened by iron starvation. The production of bacillibactin enhanced the biocontrol activity of MBI600. This is the first study showing that a member of Bacillus spp. can restrain P. syringae pv. tomato growth in vitro and in planta. Other studies showed that B. amyloliquefaciens restrained pathogen growth in vitro against other pseudomonads and in planta growth of Pseudomonas syringae pv tomato. In the case of the latter, biocontrol could rely in phylloplane competition or induction of host defenses (33, 34) rather than direct actions.
Evidence that elucidates the understanding of the elaborated arsenal of BCA antimicrobial metabolites could facilitate the improvement of biopesticides and their application methods. The former could be achieved by breeding BCA strains under specific conditions and utilizing novel active compounds produced. Enhancement of application methods can be achieved by combinational use of biopesticides with agents that might simultaneously enhance BCA aggression and pathogen susceptibility. Clinicians treat patients with transferrin-mediated iron sequestration therapy to maximize antibiotic impact and avoid the emergence of resistance (67). A similar approach could be adopted in agriculture with combinational use of chelating agents of fertilizers (e.g., EDTA) and biopesticides.
MATERIALS AND METHODS
Growth conditions of microorganisms.
B. amyloliquefaciens was routinely grown on tryptic soy agar (TSA; Oxoid) agar and tryptic soy broth (TSB; Oxoid) media at 37°C. Pseudomonas syringae pv. tomato was routinely grown on LB and nutrient agar (NA) media at 28°C for 48 h. Fungal and oomycete strains were grown on solid (12 g · liter−1 agar) minimal medium (MM) that consisted of salt solution supplemented with glucose (10 g · liter−1) and ammonium tartrate (1 g · liter−1) as carbon and nitrogen sources, respectively (68). The pH was adjusted to 6.8, and plates were incubated at 25°C. Bacterial species/pathovars and fungal and oomycete strains are listed in Table S1 in the supplemental material
Siderophore production and purification.
All glassware (Borosil/Agarwal) used for growth medium and reagent preparation were cleaned, prior to use, with 6 N HCl to remove residual iron, rinsed with deionized water, and oven dried.
(i) Growth conditions.
Chemically defined low-iron medium (CDLIM) consisted of (g · liter−1): KHSO4, 1.5; K2HPO4, 3; NaCl, 1; and NH4Cl, 5; pH 6.5. The medium was supplemented with 2 mg · liter−1 thiamine and 2% (wt/vol) glucose as an energy source (35). Finally, the following trace elements were added (mg · liter−1 in the final medium): CaCl2·2H2O, 100; MgSO4·7H2O, 80; ZnSO4·7H2O, 2; MnSO4, 0.0035; CuSO4, 0.005. CDLIM was further supplemented with various final Fe3+ concentrations (0, 18.4, 370, and 740 μM) using FeCl3. The MBI600 inoculum for siderophore production (2% [vol/vol] in the final culture) was prepared from 10 ml of an overnight culture in LB that was pelleted, washed once, and resuspended in 10 ml CDLIM. Flask cultures were incubated at 28°C and 200 rpm for 48 h. Cell mass was separated by centrifugation at 4,000 rpm for 15 min, and the supernatant was aspired for further treatment.
(ii) Siderophore detection and type determination.
Siderophore production in the supernatant was detected by the chrome azurol S (CAS) method as described by Schwyn and Neilands (69) and quantitated in terms of percent siderophore units (70). Catecholate presence in the supernatants was performed through the Arnow’s test (71).
(iii) Siderophore extraction.
A modified version of the siderophore extraction protocol developed by Patel et al. (35) was used. In brief, 200 ml of culture supernatant was acidified to pH 2.0 with 12 M HCl and extracted 2 times with equal volumes of ethyl acetate. Ethyl acetate fractions were pooled, concentrated to 2 ml using a rotary vacuum evaporator (Buchi), and stored at −20°C until further analysis.
HPLC analysis of siderophore extracts.
Bacillibactin was quantified and purified from the various culture supernatants through reverse-phase HPLC on an Agilent 1220 Infinity LC system using a two-step purification protocol and an HPLC-grade bacillibactin standard (EMC Microcollections GmbH; Tubingen, Germany). The concentrated aqueous product from the ethyl acetate extraction was initially fractionated in a preparative column (Discovery BIO wide-pore C18-10, 25 cm by 10 mm, 10 μm; SUPELCO Analytical) in 250-μl batches and a 4.5-ml/min flowrate. The elution gradient was applied follows: 0 to 10 min, 100% solution A (sol.A) (acetonitrile [ACN]-H2O, 20:80, plus 0.1% trifluoroacetic acid [TFA]); 10- to 30-min linear gradient from 0% to 40% solution B (sol.B) (ACN-H2O, 80:20 plus 0.1% TFA). The principal sample peaks from the various runs that corresponded to bacillibactin standard (detection at 315 nm) were collected and concentrated to dryness in a rotary evaporator and resuspended in HPLC-grade methanol. The concentrate from the preparative column was further purified in 20-μl batches on an analytical column (Poroshell 120 EC-C18, 4 μm, 4.6 by 150 mm) at a flowrate of 0.8 ml/min. The elution gradient was applied as follows: 0 to 6.5 min with sol.A followed by a linear gradient (6.5 to 26.5 min) from 0% to 25% sol.B. Bacillibactin, eluted as a highly symmetrical peak, was collected, concentrated, and stored in HPLC-grade methanol at −20°C until NMR analysis.
NMR analysis.
NMR spectra were recorded on a Bruker AVANCE III 600 spectrometer (Bruker, Karlsruhe, Germany) operating at 600.11 MHz for 1H and 150.9 MHz for 13C nuclei. Spectra were recorded at 305 K in deuterated methanol. The 1H 1D spectrum was recorded with analysis of 64,000 data points, for a spectral width of 10 ppm with 64 scans. The 2D COSY spectrum (pulse sequence cosygpqf, Bruker library) was recorded for a spectral width of 10 ppm with 2,000 data points in F2 dimension and 150 increments of 32 repetitions each. 1H-13C single bond correlations were detected in a HSQC-DEPT spectrum (hsqcedetgpsisp2.3, Bruker library) recorded with the standard parameters (1JH-X = 145) for spectral widths of 12 ppm for 1H and 185 ppm for 13C nuclei, 2,000 data points in F2 dimension, and 150 increments of 32 repetitions each. The solvent signal was used for axis calibration. Data manipulations were performed with the software TopSpin v3.1 (Bruker, Germany).
Cell-free culture filtrate preparation.
B. amyloliquefaciens MBI600 cells were allowed to grow in LB or siderophore-producing medium, CDLIM or succinic acid (72), for 24 or 48 h, respectively. CFCF was prepared as previously described (33). Siderophore production was higher in CDLIM than in the succinic acid medium that was initially used.
Broth microdilution method.
The Pseudomonas syringae pv. tomato susceptibility test against B. amyloliquefaciens MBI600 was performed in 96-well microplates (Greiner CELLSTAR 96-well microplates, F-bottom) as previously described (73, 74). In brief, P. syringae pv. tomato was grown exponentially in overnight cultures and was streaked on LB plates to check its purity. Culture aliquots were then adjusted to reach a final concentration of 5 × 105 CFU/ml, while MBI600 CFCF and LB medium concentrations were adjusted to reach final concentrations of 0, 0.5, 2, 10, 25, and 50% CFCF and 1× LB in a total volume of 100 μl in each well. Blank wells (treatments and medium only) containing each test concentration were also included in duplicates. Plates were incubated at 30°C for 48 h. Three independent biological experiments (inocula, CFCF extracts, and tested plates) were performed. Absorbance measurements were recorded at 0, 24, and 48 hpi at 30°C using triplicate readings from a multidetection microplate reader (Fluostar Galaxy; BMG Labtech) at 600 nm.
Disk diffusion susceptibility tests.
To prepare dual cultures, 20 ml of overnight-grown naive stationary-phase cultures of each bacterial pathogen were adjusted to a concentration of 108 CFU · ml−1 and added to 180 ml LB agar before the mixture was poured into petri dishes and incubated overnight at 30°C. A 40-μl MBI600 suspension, adjusted to 5 × 106 cells, was then pipetted on 9-mm antibiotic assay discs (Whatman) that were placed on the agar cultures of each pathogen. As a control, sterile LB was used in place of the MBI600 suspension. Each plate was quadruplicated in 3 independent experiments and incubated at 30°C for 2 days before the diameter of the clear halo surrounding the disk was measured on three different axes. A similar protocol was developed to evaluate the effect of siderophore extracts, only this time, a 20-μl aliquot of concentrated siderophore extract was pipetted directly onto LB agar plates containing P. syringae pv. tomato pathogen suspension. As a control, a noninoculated liquid CDLIM for each Fe3+ concentration was used.
Fungal susceptibility evaluation.
MBI600 CFCFs extracted from succinic acid broth or LB medium supplemented with increasing FeCl3 concentrations (18.4 or 740 μM Fe3+) were mixed with MM agar in ratios of 1:2 or 1:10 (vol/vol) and poured into petri dishes. MM agar was used as control. MM agar containing 10% or 50% MBI600 CFCF grown in minimal medium supplemented with 740 μM Fe3+ was used as the reference (ref). The technique of point inoculation was used for both treatment plates and controls (68). Each treatment was repeated three times. Plates were incubated at 25°C, and the radius of fungal growth was recorded daily on three different axes.
In planta experiments.
Tomato plants were grown and inoculated with P. syringae pv. tomato as previously described (75). Plants were inoculated at the developmental stage of fully developed 4th true leaves. Pathogen inoculum was prepared from 5 ml overnight (18 h) cultures in 10 mM MgCl2 at a final concentration of 108 CFU · ml−1. Two independent experiments were performed, each in 5 plots of 12 plants/plot. CFCF was harvested as aforementioned from MBI600 succinate broth cultures in which the Fe3+ concentration was adjusted to 0, 18.4, 370, or 740 μM Fe3+. Plain medium was used as a control. Pathogen inoculum and CFCF were simultaneously sprayed to run off using hand-pump sprayers and allowed to dry before sampling.
The pathogen population was estimated as previously described (75), based on cell counts in 4 pooled leaf disc samples, each containing 2 discs from 3 different plants per treatment. Samples were collected at 1, 4, 8, and 16 dpi from 4 upper leaves of plants at the 6 true leaf stage. Leaves were homogenized for 60 s with a tissue homogenizer (HCT Shaping System SA) in Bioreba extraction bags containing 10 ml of sterile distilled water. Aliquots of 0.1 ml from a 10-fold dilution series were plated on nutrient agar (NA) medium supplemented with 5 μg/ml rifampicin.
Gene expression assays.
B. amyloliquefaciens MBI600 cultures were incubated in succinic acid medium with increasing FeCl3 concentrations (0, 18.4, 370, and 740 μM Fe3+) at 37°C and 200 rpm for 48 h. Total RNA was extracted from each sample using TRI reagent (Ambion) according to the manufacturer’s instructions. The concentration and purity of RNA were estimated using a P330 nanophotometer (IMPLEN). DNase-treated RNA (RNase-free DNase; New England BioLabs) was reverse transcribed into cDNA using Superscript II reverse transcriptase and oligo(dT) as primers (Invitrogen). Reverse transcription-quantitative PCR (RT-qPCR) was performed as previously described (33). Transcript levels of target genes were normalized relative to the level of gyrA. Primer sequences are listed in Table S2 in the supplemental material.
List of primers used in this study. Download Table S2, DOCX file, 0.1 MB (13.4KB, docx) .
Copyright © 2021 Dimopoulou et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data analysis.
(i) Broth microdilution method. Broth microdilution method analysis was performed using the method described in previous works (73, 74).
(ii) Disk diffusion and mycelial growth susceptibility tests.
For the analysis of disk diffusion (bacterial) or mycelial growth (fungal) susceptibility, radial growth was modeled using linear mixed models (LMMs) in R language (76). In the case of bacterial diffusion susceptibility tests, before the estimation of marginal means, diameter estimates of the control assays (noninoculated liquid CDLIM) were subtracted from the experimental values. The corrected diameter was the dependent variable, and CFCF concentration was the independent fixed variable. For the fungal susceptibility tests, the radius of growth inhibition was the dependent variable, while CFCF concentration, time, and their interaction were the independent fixed variables. Repeated axial measurements nested within three replicated experimental plates were the random factors in both cases. Estimated marginal means and their standard errors were plotted, and Tukey’s post hoc comparisons were applied.
(iii) Bacterial population assessment.
Pathogen growth in planta was assessed as previously described (75, 77). CFU counts were modeled using LMMs to log10-transformed data (number of bacterial CFU for each time point). The times after inoculation and treatment as well as their interaction were modeled as fixed factors, while plant individuals nested within a block comprised the random factor. Tukey’s tests were applied for post hoc comparisons of estimated marginal means.
(iv) Gene expression data analysis.
RT-qPCR analysis was performed as previously described (33, 34). Threshold cycle (CT) values were modeled in the context of a LMM and comprised the independent variables, while time, gene, Fe concentrations, and their interactions were included as the dependent variables. Samples nested within experiments were the random factors of the model. Contrasts were formulated according to equation 5 reported by Steibel et al. (78), so that the reference gene (gyrA) and reference treatment (control, 740 μM Fe3+) represented the baselines of all relevant comparisons.
ACKNOWLEDGMENTS
We thank BASF SE (Germany) for providing the MBI600 strain and its genome sequence.
This study was partially supported by BASF SE funds (BIPoD, grant no. PAR00415-1). Participation by D.B. and D.G.H. in this project was partially supported by the projects Synthetic biology: from omics technologies to genomic engineering (OMIC-ENGINE) (MIS 5002636: NSRF 2014-2020), Reinforcement of the research and innovation infrastructure, and the operational program Competitiveness, entrepreneurship and innovation, cofinanced by Greece and the European Union (European Regional Development Fund).
Contributor Information
Nicholas Skandalis, Email: skandali@usc.edu.
Maria L. Marco, University of California, Davis
REFERENCES
- 1.Abbaspour N, Hurrell R, Kelishadi R. 2014. Review on iron and its importance for human health. J Res Med Sci 19:164–174. [PMC free article] [PubMed] [Google Scholar]
- 2.Glick BR. 2010. Using soil bacteria to facilitate phytoremediation. Biotechnol Adv 28:367–374. doi: 10.1016/j.biotechadv.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Rizzi A, Roy S, Bellenger JP, Beauregard PB. 2019. Iron homeostasis in Bacillus subtilis requires siderophore production and biofilm formation. Appl Environ Microbiol 85:e02439-18. doi: 10.1128/AEM.02439-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heerklotz H, Seelig J. 2001. Detergent-like action of the antibiotic peptide surfactin on lipid membranes. Biophys J 81:1547–1554. doi: 10.1016/S0006-3495(01)75808-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quadri LEN. 2000. Assembly of aryl-capped siderophores by modular peptide synthetases and polyketide synthases. Mol Microbiol 37:1–12. doi: 10.1046/j.1365-2958.2000.01941.x. [DOI] [PubMed] [Google Scholar]
- 6.Su C, Zhang M, Lin L, Yu G, Zhong H, Chong Y. 2020. Reduction of iron oxides and microbial community composition in iron-rich soils with different organic carbon as electron donors. Int Biodeterior Biodegradation 148:104881. doi: 10.1016/j.ibiod.2019.104881. [DOI] [Google Scholar]
- 7.Sullivan TS, Ramkissoon S, Garrison VH, Ramsubhag A, Thies JE. 2012. Siderophore production of African dust microorganisms over Trinidad and Tobago. Aerobiologia (Bologna) 28:391–401. doi: 10.1007/s10453-011-9243-x. [DOI] [Google Scholar]
- 8.Masalha J, Kosegarten H, Elmaci O, Mengel K. 2000. The central role of microbial activity for iron acquisition in maize and sunflower. Biol Fertil Soils 30:433–439. doi: 10.1007/s003740050021. [DOI] [Google Scholar]
- 9.Kloepper JW, Leong J, Teintze M, Schroth MN. 1980. Enhanced plant growth by siderophores produced by plant growth-promoting rhizobacteria. Nature 286:885–886. doi: 10.1038/286885a0. [DOI] [Google Scholar]
- 10.Yu X, Ai C, Xin L, Zhou G. 2011. The siderophore-producing bacterium, Bacillus subtilis CAS15, has a biocontrol effect on Fusarium wilt and promotes the growth of pepper. Eur J Soil Biol 47:138–145. doi: 10.1016/j.ejsobi.2010.11.001. [DOI] [Google Scholar]
- 11.Rajkumar M, Ae N, Narasimha M, Prasad V, Freitas H. 2010. Potential of siderophore-producing bacteria for improving heavy metal phytoextraction. Trends Biotechnol 28:142–149. doi: 10.1016/j.tibtech.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 12.O’Brien S, Hodgson DJ, Buckling A. 2014. Social evolution of toxic metal bioremediation in Pseudomonas aeruginosa. Proc Biol Sci 281:28120140858. doi: 10.1098/rspb.2014.0858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orcutt KM, Jones WS, McDonald A, Schrock D, Wallace KJ. 2010. A lanthanide-based chemosensor for bioavailable Fe3+ using a fluorescent siderophore: an assay displacement approach. Sensors (Basel) 10:1326–1337. doi: 10.3390/s100201326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta M, Yoshioka H, Ohnishi K, Mizumoto H, Hikichi Y, Kiba A. 2013. A translationally controlled tumor protein negatively regulates the hypersensitive response in Nicotiana benthamiana. Plant Cell Physiol 54:1403–1414. doi: 10.1093/pcp/pct090. [DOI] [PubMed] [Google Scholar]
- 15.Huang Y, Jiang Y, Wang H, Wang J, Cheol M, Byun Y, He H, Liang Y, Yang VC. 2013. Curb challenges of the “Trojan Horse” approach: smart strategies in achieving effective yet safe cell-penetrating peptide-based drug delivery. Adv Drug Deliv Rev 65:1299–1315. doi: 10.1016/j.addr.2012.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neilands JB. 1995. Siderophores: structure and function of microbial iron transport compounds. J Biol Chem 270:26723–26726. doi: 10.1074/jbc.270.45.26723. [DOI] [PubMed] [Google Scholar]
- 17.Dave BP, Anshuman K, Hajela P. 2006. Siderophores of halophilic archaea and their chemical characterization. Indian J Exp Biol 44:340–344. [PubMed] [Google Scholar]
- 18.Dertz EA, Xu J, Stintzi A, Raymond KN. 2006. Bacillibactin-mediated iron transport in Bacillus subtilis. J Biol Inorg Chem 11:1087–1097. doi: 10.1007/s00775-006-0151-4. [DOI] [PubMed] [Google Scholar]
- 19.Miethke M, Schmidt S, Marahiel MA. 2008. The major facilitator superfamily-type transporter YmfE and the multidrug-efflux activator Mta mediate bacillibactin secretion in Bacillus subtilis. J Bacteriol 190:5143–5152. doi: 10.1128/JB.00464-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fravel DR. 2005. Commercialization and implementation of biocontrol. Annu Rev Phytopathol 43:337–359. doi: 10.1146/annurev.phyto.43.032904.092924. [DOI] [PubMed] [Google Scholar]
- 21.Connor N, Sikorski J, Rooney AP, Kopac S, Koeppel AF, Burger A, Cole SG, Perry EB, Krizanc D, Field NC, Slaton M, Cohan FM. 2010. Ecology of Speciation in the Genus Bacillus. Appl Environ Microbiol 76:1349–1358. doi: 10.1128/AEM.01988-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen XH, Koumoutsi A, Scholz R, Schneider K, Vater J, Süssmuth R, Piel J, Borriss R. 2009. Genome analysis of Bacillus amyloliquefaciens FZB42 reveals its potential for biocontrol of plant pathogens. J Biotechnol 140:27–37. doi: 10.1016/j.jbiotec.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 23.Chowdhury SP, Uhl J, Grosch R, Alqu S, Pittroff S, Dietel K, Schmitt-Kopplin P, Borriss R, Hartmann A. 2015. Cyclic lipopeptides of Bacillus amyloliquefaciens subsp. plantarum colonizing the lettuce rhizosphere enhance plant defense responses toward the bottom rot pathogen Rhizoctonia solani. Mol Plant Microbe Interact 28:984–995. doi: 10.1094/MPMI-03-15-0066-R. [DOI] [PubMed] [Google Scholar]
- 24.Chowdhury SP, Hartmann A, Gao XW, Borriss R. 2015. Biocontrol mechanism by root-associated Bacillus amyloliquefaciens FZB42 - a review. Front Microbiol 6:780. doi: 10.3389/fmicb.2015.00780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao H, Shao D, Jiang C, Shi J, Li Q, Huang Q, Rajoka MSR, Yang H, Jin M. 2017. Biological activity of lipopeptides from Bacillus. Appl Microbiol Biotechnol 101:5951–5960. doi: 10.1007/s00253-017-8396-0. [DOI] [PubMed] [Google Scholar]
- 26.Gu Q, Yang Y, Yuan Q, Shi G, Wu L, Lou Z, Huo R, Wu H, Borriss R, Gao X. 2017. Bacillomycin D produced by Bacillus amyloliquefaciens is involved in the antagonistic interaction with the plant pathogenic fungus Fusarium graminearum. Appl Environ Microbiol 83:e01075-17. doi: 10.1128/AEM.01075-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu L, Wu H-J, Qiao J, Gao X, Borriss R. 2015. Novel routes for improving biocontrol activity of Bacillus based bioinoculants. Front Microbiol 6:1395. doi: 10.3389/fmicb.2015.01395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen XH, Scholz R, Borriss M, Junge H, Mogel G, Kunz S, Borriss R. 2009. Difficidin and bacilysin produced by plant-associated Bacillus amyloliquefaciens are efficient in controlling fire blight disease. J Biotechnol 140:38–44. doi: 10.1016/j.jbiotec.2008.10.015. [DOI] [PubMed] [Google Scholar]
- 29.Cao Y, Pi H, Chandrangsu P, Li Y, Wang Y, Zhou H, Xiong H, Helmann JD, Cai Y. 2018. Antagonism of two plant-growth promoting Bacillus velezensis isolates against Ralstonia solanacearum and Fusarium oxysporum. Sci Rep 8:4360. doi: 10.1038/s41598-018-22782-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nikolić I, Berić T, Dimkić I, Popović T, Lozo J, Fira D, Stanković S. 2019. Biological control of Pseudomonas syringae pv. aptata on sugar beet with Bacillus pumilus SS-10.7 and Bacillus amyloliquefaciens (SS-12.6 and SS-38.4) strains. J Appl Microbiol 126:165–176. doi: 10.1111/jam.14070. [DOI] [PubMed] [Google Scholar]
- 31.Lanna Filho R, De Souza RM, Ferreira A, Quecine MC, Alves E, De Azevedo JL. 2013. Biocontrol activity of Bacillus against a GFP-marked Pseudomonas syringae pv. tomato on tomato phylloplane. Australas Plant Pathol 42:643–651. doi: 10.1007/s13313-013-0233-z. [DOI] [Google Scholar]
- 32.Fousia S, Paplomatas EJ, Tjamos SE. 2016. Bacillus subtilis QST 713 confers protection to tomato plants against Pseudomonas syringae pv tomato and induces plant defence-related genes. J Phytopathol 164:264–270. doi: 10.1111/jph.12455. [DOI] [Google Scholar]
- 33.Dimopoulou A, Theologidis I, Liebmann B, Kalantidis K, Vassilakos N, Skandalis N. 2019. Bacillus amyloliquefaciens MBI600 differentially induces tomato defense signaling pathways depending on plant part and dose of application. Sci Rep 9:19120. doi: 10.1038/s41598-019-55645-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beris D, Theologidis I, Skandalis N, Vassilakos N. 2018. Bacillus amyloliquefaciens strain MBI600 induces salicylic acid dependent resistance in tomato plants against Tomato spotted wilt virus and Potato virus Y. Sci Rep 8:10320. doi: 10.1038/s41598-018-28677-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patel AK, Deshattiwar MK, Chaudhari BL, Chincholkar SB. 2009. Production, purification and chemical characterization of the catecholate siderophore from potent probiotic strains of Bacillus spp. Bioresour Technol 100:368–373. doi: 10.1016/j.biortech.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 36.Zhou M, Liu F, Yang X, Jin J, Dong X, Zeng KW, Liu D, Zhang Y, Ma M, Yang D. 2018. Bacillibactin and bacillomycin analogues with cytotoxicities against human cancer cell lines from marine Bacillus sp. PKU-MA00093 and PKU-MA00092. Mar Drugs 16:22. doi: 10.3390/md16010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Budzikiewicz H, Bössenkamp A, Taraz K, Pandey A, Meyer J-M. 1997. Corynebactin, a cyclic catecholate siderophore from Corynebacterium glutamicum ATCC 14067 (Brevibacterium sp. DSM 20411). Zeitschrift Für Naturforsch C 52:551–554. doi: 10.1515/znc-1997-7-820. [DOI] [Google Scholar]
- 38.Crosa JH, Mey AR, Payne SM. 2004. Iron transport in bacteria. ASM Press, Washington, DC. doi: 10.1128/9781555816544. [DOI] [Google Scholar]
- 39.Chen XH, Vater J, Piel J, Franke P, Scholz R, Schneider K, Koumoutsi A, Hitzeroth G, Grammel N, Strittmatter AW, Gottschalk G, Süssmuth RD, Borriss R. 2006. Structural and functional characterization of three polyketide synthase gene clusters in Bacillus amyloliquefaciens FZB 42. J Bacteriol 188:4024–4036. doi: 10.1128/JB.00052-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schneider K, Chen XH, Vater J, Franke P, Nicholson G, Borriss R, Süssmuth RD. 2007. Macrolactin is the polyketide biosynthesis product of the pks2 cluster of Bacillus amyloliquefaciens FZB42. J Nat Prod 70:1417–1423. doi: 10.1021/np070070k. [DOI] [PubMed] [Google Scholar]
- 41.Chen XH, Koumoutsi A, Scholz R, Borriss R. 2009. More than anticipated - production of antibiotics and other secondary metabolites by Bacillus amyloliquefaciens FZB42. J Mol Microbiol Biotechnol 16:14–24. doi: 10.1159/000142891. [DOI] [PubMed] [Google Scholar]
- 42.Koumoutsi A, Chen X, Henne A, Hitzeroth G, Franke P, Vater J, Borriss R, Liesegang H. 2004. Structural and functional characterization of gene clusters directing nonribosomal synthesis of bioactive cyclic lipopeptides in Bacillus amyloliquefaciens strain FZB42. J Bacteriol 186:1084–1096. doi: 10.1128/JB.186.4.1084-1096.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peypoux F, Bonmatin JM, Wallach J. 1999. Recent trends in the biochemistry of surfactin. Appl Microbiol Biotechnol 51:553–563. doi: 10.1007/s002530051432. [DOI] [PubMed] [Google Scholar]
- 44.Falardeau J, Wise C, Novitsky L, Avis TJ. 2013. Ecological and mechanistic insights into the direct and indirect antimicrobial properties of Bacillus subtilis lipopeptides on plant pathogens. J Chem Ecol 39:869–878. doi: 10.1007/s10886-013-0319-7. [DOI] [PubMed] [Google Scholar]
- 45.Tang Q, Bie X, Lu Z, Lv F, Tao Y, Qu X. 2014. Effects of fengycin from Bacillus subtilis fmbJ on apoptosis and necrosis in Rhizopus stolonifer. J Microbiol 52:675–680. doi: 10.1007/s12275-014-3605-3. [DOI] [PubMed] [Google Scholar]
- 46.Weinberg ED. 2009. Iron availability and infection. Biochim Biophys Acta 1790:600–605. doi: 10.1016/j.bbagen.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 47.Eijkelkamp BA, Hassan KA, Paulsen IT, Brown MH. 2011. Investigation of the human pathogen Acinetobacter baumannii under iron limiting conditions. BMC Genomics 12:126. doi: 10.1186/1471-2164-12-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schenk PM, Carvalhais LC, Kazan K. 2012. Unraveling plant-microbe interactions: can multi-species transcriptomics help? Trends Biotechnol 30:177–184. doi: 10.1016/j.tibtech.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 49.Beneduzi A, Ambrosini A, Passaglia LMP. 2012. Plant growth-promoting rhizobacteria (PGPR): their potential as antagonists and biocontrol agents. Genet Mol Biol 35:1044–1051. doi: 10.1590/s1415-47572012000600020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Franza T, Expert D. 2013. Role of iron homeostasis in the virulence of phytopathogenic bacteria: an “à la carte” menu. Mol Plant Pathol 14:429–438. doi: 10.1111/mpp.12007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ghazy N, El-Nahrawy S. 2021. Siderophore production by Bacillus subtilis MF497446 and Pseudomonas koreensis MG209738 and their efficacy in controlling Cephalosporium maydis in maize plant. Arch Microbiol 203:1195–1209. doi: 10.1007/s00203-020-02113-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Prema PP, Selvarani MS. 2012. Microbial siderophore as a potent biocontrol agent for plant pathogens. Int J Sci Res 2:521–523. [Google Scholar]
- 53.Zhao X, Wang R, Shang Q, Hao H, Li Y, Zhang Y, Guo Z, Wang Y, Xie Z. 2016. The new flagella-associated collagen-like proteins ClpB and ClpC of Bacillus amyloliquefaciens FZB42 are involved in bacterial motility. Microbiol Res 184:25–31. doi: 10.1016/j.micres.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 54.Khan N, Maymon M, Hirsch A. 2017. Combating Fusarium infection using Bacillus-based antimicrobials. Microorganisms 5:75. doi: 10.3390/microorganisms5040075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hofemeister J, Conrad B, Adler B, Hofemeister B, Feesche J, Kucheryava N, Steinborn G, Franke P, Grammel N, Zwintscher A, Leenders F, Hitzeroth G, Vater J. 2004. Genetic analysis of the biosynthesis of non-ribosomal peptide- and polyketide-like antibiotics, iron uptake and biofilm formation by Bacillus subtilis A1/3. Mol Genet Genomics 272:363–378. doi: 10.1007/s00438-004-1056-y. [DOI] [PubMed] [Google Scholar]
- 56.Stein T. 2005. Bacillus subtilis antibiotics: structures, syntheses and specific functions. Mol Microbiol 56:845–857. doi: 10.1111/j.1365-2958.2005.04587.x. [DOI] [PubMed] [Google Scholar]
- 57.Yüksel M, Power JJ, Ribbe J, Volkmann T, Maier B. 2016. Fitness trade-offs in competence differentiation of Bacillus subtilis. Front Microbiol 7:888. doi: 10.3389/fmicb.2016.00888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miras M, Dubnau D. 2016. A DegU-P and DegQ-dependent regulatory pathway for the K-state in Bacillus subtilis. Front Microbiol 7:1868. doi: 10.3389/fmicb.2016.01868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kirby JR, Kristich CJ, Saulmon MM, Zimmer MA, Garrity LF, Zhulin IB, Ordal GW. 2001. CheC is related to the family of flagellar switch proteins and acts independently from CheD to control chemotaxis in Bacillus subtilis. Mol Microbiol 42:573–585. doi: 10.1046/j.1365-2958.2001.02581.x. [DOI] [PubMed] [Google Scholar]
- 60.Kearns DB, Chu F, Rudner R, Losick R. 2004. Genes governing swarming in Bacillus subtilis and evidence for a phase variation mechanism controlling surface motility. Mol Microbiol 52:357–369. doi: 10.1111/j.1365-2958.2004.03996.x. [DOI] [PubMed] [Google Scholar]
- 61.Calvio C, Celandroni F, Ghelardi E, Amati G, Salvetti S, Ceciliani F, Galizzi A, Senesi S. 2005. Swarming differentiation and swimming motility in Bacillus subtilis are controlled by swrA, a newly identified dicistronic operon. J Bacteriol 187:5356–5366. doi: 10.1128/JB.187.15.5356-5366.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kobayashi K. 2007. Gradual activation of the response regulator DegU controls serial expression of genes for flagellum formation and biofilm formation in Bacillus subtilis. Mol Microbiol 66:395–409. doi: 10.1111/j.1365-2958.2007.05923.x. [DOI] [PubMed] [Google Scholar]
- 63.Verhamme DT, Kiley TB, Stanley-Wall NR. 2007. DegU co-ordinates multicellular behaviour exhibited by Bacillus subtilis. Mol Microbiol 65:554–568. doi: 10.1111/j.1365-2958.2007.05810.x. [DOI] [PubMed] [Google Scholar]
- 64.Murray EJ, Kiley TB, Stanley-Wall NR. 2009. A pivotal role for the response regulator DegU in controlling multicellular behaviour. Microbiology (Reading) 155:1–8. doi: 10.1099/mic.0.023903-0. [DOI] [PubMed] [Google Scholar]
- 65.Rowland BM, Taber HW. 1996. Duplicate isochorismate synthase genes of Bacillus subtilis: regulation and involvement in the biosyntheses of menaquinone and 2,3-dihydroxybenzoate. J Bacteriol 178:854–861. doi: 10.1128/jb.178.3.854-861.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baichoo N, Wang T, Ye R, Helmann JD. 2002. Global analysis of the Bacillus subtilis fur regulon and the iron starvation stimulon. Mol Microbiol 45:1613–1629. doi: 10.1046/j.1365-2958.2002.03113.x. [DOI] [PubMed] [Google Scholar]
- 67.Luna BM, Ershova K, Yan J, Ulhaq A, Nielsen TB, Hsieh S, Pantapalangkoor P, Vanscoy B, Ambrose P, Rudin S, Hujer K, Bonomo RA, Actis L, Skaar EP, Spellberg B. 2019. Adjunctive transferrin to reduce the emergence of antibiotic resistance in Gram-negative bacteria. J Antimicrob Chemother 74:2631–2639. doi: 10.1093/jac/dkz225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pontecorvo G, Roper JA, Hemmons LM, Macdonald KD, Bufton AWJ. 1953. The genetics of Aspergillus nidulans. Adv Genet 5:141–238. doi: 10.1016/s0065-2660(08)60408-3. [DOI] [PubMed] [Google Scholar]
- 69.Schwyn B, Neilands JB. 1987. Universal assay for the detection and determination of siderophores. Anal Biochem 160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 70.Payne SM. 1994. Detection, isolation, and characterization of siderophores. Methods Enzymol 235:329–344. doi: 10.1016/0076-6879(94)35151-1. [DOI] [PubMed] [Google Scholar]
- 71.Arnow LE. 1937. Colometric determination of the components of 3,4-dihydroxyphenylalanine-tyrosine mixtures. J Biol Chem 118:531–538. doi: 10.1016/S0021-9258(18)74509-2. [DOI] [Google Scholar]
- 72.Bharucha U, Patel K, Trivedi U. 2013. Antifungal activity of catecholate type siderophore produced by Bacillus sp. Int J Res Sci 4:528–531. [Google Scholar]
- 73.Varympopi A, Dimopoulou A, Theologidis I, Karamanidou T, Kerou AK, Vlachou A, Karfaridis D, Papafotis D, Hatzinikolaou DG, Tsouknidas A, Skandalis N. 2020. Bactericides based on copper nanoparticles restrain growth of important plant pathogens. Pathogens 9:1024. doi: 10.3390/pathogens9121024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Skandalis N, Dimopoulou A, Georgopoulou A, Gallios N, Papadopoulos D, Tsipas D, Theologidis I, Michailidis N, Chatzinikolaidou M. 2017. The effect of silver nanoparticles size, produced using plant extract from Arbutus unedo, on their antibacterial efficacy. Nanomaterials 7:178. doi: 10.3390/nano7070178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Skandalis N, Dimopoulou A, Beri D, Tzima A, Malandraki I, Theologidis I, Bitivanos S, Varveri C, Klitsinaris T, Vassilakos N. 2016. Effect of pyraclostrobin application on viral and bacterial diseases of tomato. Plant Dis 100:1321–1330. doi: 10.1094/PDIS-10-15-1216-RE. [DOI] [PubMed] [Google Scholar]
- 76.R Core Development Team. 2020. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.r-project.org. [Google Scholar]
- 77.Papadakis G, Skandalis N, Dimopoulou A, Glynos P, Gizeli E. 2015. Bacteria murmur: application of an acoustic biosensor for plant pathogen detection. PLoS One 10:e0132773. doi: 10.1371/journal.pone.0132773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Steibel JP, Poletto R, Coussens PM, Rosa GJM. 2009. A powerful and flexible linear mixed model framework for the analysis of relative quantification RT-PCR data. Genomics 94:146–152. doi: 10.1016/j.ygeno.2009.04.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
B. amyloliquefaciens MBI600’s growth curve under iron starvation conditions (0, 18.5, and 370 μM Fe3+). Optical densities at 600 nm were measured for 48 h at 37°C using a multidetection microplate reader and automatically recorded for each well every 29 min. Each ribbon depicts growth under a specific Fe3+ concentration; full line represents mean values and bars represent SEs from three independent biological experiments. Download FIG S7, EPS file, 0.5 MB (534.1KB, eps) .
Copyright © 2021 Dimopoulou et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
HPLC chromatograms of the purified peak of bacillibactin from 0 μM Fe3+ supernatant and the commercial standard at 315 nm in the Poroshell 120 EC-C18 analytical column. (Inset) The corresponding NMR spectra. Download FIG S1, EPS file, 0.3 MB (287.8KB, eps) .
Copyright © 2021 Dimopoulou et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
1H-1H COSY (8.00 to 0.50 ppm) (A) and 1H-13C HSQC-DEPT (8.00 to 0.50 ppm; 130 to 10 ppm) (B) NMR spectra of bacillibactin extracted from 0 μM Fe3+ supernatants of MBI600, in deuterated methanol, at 305 K. In HSQC-DEPT spectrum, methylene protons are colored red, while methine and methylene protons are black. Download FIG S2, EPS file, 0.1 MB (127.8KB, eps) .
Copyright © 2021 Dimopoulou et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
In vitro bactericidal efficacy of B. amyloliquefaciens MBI600. (Left) P. syringae pv. tomato broth susceptibility was tested against increasing MBI600 cell free culture filtrate (CFCF) concentrations (0, 0.5, 2, 10, 25, and 50% vol/vol). CFCF was extracted from MBI600 cells grown in LB medium. Observed effects were evaluated based on OD600 readings at 0, 24, and 48 hpi in microtiter plates. Estimated marginal means and their standard errors are plotted. (Right) No inhibition zone was formed around MBI600 culture grown on 9-mm Whatman discs that were placed on agar medium inoculated with P. syringae pv. tomato. Download FIG S3, EPS file, 1.4 MB (1.5MB, eps) .
Copyright © 2021 Dimopoulou et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Effect on growth of Pseudomonas syringae pv. tomato (Pto) in LB medium supplemented with 10% (a), 25% (b), or 50% (c) B. amyloliquefaciens (Bam) CFCF (cell-free culture filtrate) supplemented with different iron concentrations (0, 18.4, 148, and 370 μM Fe3+). Optical densities at 600 nm were measured for 48 h at 28°C using a multidetection microplate reader. Estimated marginal means and their standard errors are plotted. Download FIG S4, EPS file, 2.4 MB (2.4MB, eps) .
Copyright © 2021 Dimopoulou et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Inhibition zones formed in Pseudomonas syringae pv. tomato (Pto) agar cultures in the presence of B. amyloliquefaciens (Bam). Pto inoculum suspension was adjusted to 109 CFU/ml and allowed to grow in agar medium containing increasing iron concentrations (0, 18.4, and 148 μM Fe3+). Values represent the averages of 24 inhibition zone measurements for each iron concentration. Different letters represent statistically different data points at a P value of ≤0.05 according to Tukey’s post hoc comparisons. Download FIG S5, EPS file, 0.7 MB (730.9KB, eps) .
Copyright © 2021 Dimopoulou et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Iron starvation expands the range of antifungal activity of MBI600. Radial growth of 4 fungal species in minimal medium (MM) agar plates and in response to the following treatments: no treatment (control), 10% and 50% CFCF of MBI600 grown under either 740 μM Fe+3 (ref) or 18.4 μM Fe+3 conditions, and 50% CFCF of MBI600 grown in LB. Values represent the estimated marginal means and corresponding standard errors of inhibition zone measurements. ***, P < 0.001; **, P < 0.01; *, P < 0.05; ·, P < 0.1 according to Tukey’s post hoc comparisons. Download FIG S6, EPS file, 0.9 MB (943.9KB, eps) .
Copyright © 2021 Dimopoulou et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
List of bacterial, fungal, and oomycete strains used in this study. Download Table S1, DOCX file, 0.1 MB (14.7KB, docx) .
Copyright © 2021 Dimopoulou et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
List of primers used in this study. Download Table S2, DOCX file, 0.1 MB (13.4KB, docx) .
Copyright © 2021 Dimopoulou et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.