ABSTRACT
Vibrio cholerae is an aquatic Gram-negative bacterium that causes severe diarrheal cholera disease when ingested by humans. To eliminate competitor cells in both the external environment and inside hosts, V. cholerae uses the type VI secretion system (T6SS). The T6SS is a macromolecular contact-dependent weapon employed by many Gram-negative bacteria to deliver cytotoxic proteins into adjacent cells. In addition to canonical T6SS gene clusters encoded by all sequenced V. cholerae isolates, strain BGT49 encodes another locus, which we named auxiliary (Aux) cluster 4. The Aux 4 cluster is located on a mobile genetic element and can be used by killer cells to eliminate both V. cholerae and Escherichia coli cells in a T6SS-dependent manner. A putative toxin encoded in the cluster, which we name TpeV (type VI permeabilizing effector Vibrio), shares no homology to known proteins and does not contain motifs or domains indicative of function. Ectopic expression of TpeV in the periplasm of E. coli permeabilizes cells and disrupts the membrane potential. Using confocal microscopy, we confirm that susceptible target cells become permeabilized when competed with killer cells harboring the Aux 4 cluster. We also determine that tpiV, the gene located immediately downstream of tpeV, encodes an immunity protein that neutralizes the toxicity of TpeV. Finally, we show that TpeV homologs are broadly distributed across important human, animal, and plant pathogens and are localized in proximity to other T6SS genes. Our results suggest that TpeV is a toxin that belongs to a large family of T6SS proteins.
IMPORTANCE Bacteria live in polymicrobial communities where competition for resources and space is essential for survival. Proteobacteria use the T6SS to eliminate neighboring cells and cause disease. However, the mechanisms by which many T6SS toxins kill or inhibit susceptible target cells are poorly understood. The sequence of the TpeV toxin that we describe here is unlike any previously described protein. We demonstrate that it has antimicrobial activity by permeabilizing cells, eliminating membrane potentials, and causing severe cytotoxicity. TpeV homologs are found near known T6SS genes in human, animal, and plant bacterial pathogens, indicating that the toxin is a representative member of a broadly distributed protein family. We propose that TpeV-like toxins contribute to the fitness of many bacteria. Finally, since antibiotic resistance is a critical global health threat, the discovery of new antimicrobial mechanisms could lead to the development of new treatments against resistant strains.
KEYWORDS: Vibrio cholerae, antimicrobial agents, secretion systems, toxins
INTRODUCTION
The type VI secretion system (T6SS) is a common contact-dependent antibacterial weapon employed by many Gram-negative species (1–3). Cells with an active T6SS (“killer cells” here) translocate toxic protein effectors into adjacent target cells. However, the outcome of T6SS-mediated aggression is influenced by the presence of immunity proteins in target cells, the external environment, and target cell stress responses (4–7). The harpoon-like proteinaceous apparatus is anchored to the membrane of killer cells by the membrane complex, which spans the inner membrane and periplasm (8–10). VasK, a component of the membrane complex, is essential for the assembly of the T6SS (8, 11). Hcp (hemolysin-coregulated protein) hexamers stack to form an inner tube that is capped at the distal end by a trimer of VgrG (valine-glycine repeat protein G) tip-forming proteins (2, 12, 13). PAAR (proline-alanine-alanine-arginine) proteins also interact with VgrGs and expand the toxin repertoire (14, 15). Furthermore, T6SS adaptor proteins (Taps) with a DUF4123 domain function as chaperones that deliver effectors to the apparatus (16–18). The T6SS uses a contraction mechanism that propels the inner tube and exports the toxic payload (19–21).
Vibrio cholerae is a wide-spread gastrointestinal pathogen that has caused seven cholera pandemics (22). The bacterium is found in polymicrobial marine ecosystems in association with copepods, fish, and insects (23–25). V. cholerae employs T6SS effectors that disrupt cell envelopes and contribute to pathogenicity in hosts (26–33). T6SS genes are distributed across a large cluster and two auxiliary clusters in all sequenced V. cholerae isolates (34, 35). In clinical strains like V52 and C6706, the large gene cluster encodes a VgrG tip-forming protein with a C-terminal peptidoglycan-degrading domain (32). Auxiliary clusters 1 and 2 encode the TseL lipase and VasX colicin-like effectors, respectively (28–31). An auxiliary cluster 3 is found in a subset of V. cholerae isolates and contains a peptidoglycan-degrading toxin (6, 36, 37).
Most clinical V. cholerae strains encode T6SS effectors with conserved activities (28, 34, 35). By contrast, many isolates obtained from sources other than patients harbor toxins with diverse predicted biochemical functions and may carry additional T6SS toxins compared to clinical isolates (34, 35, 38–40). We previously identified auxiliary 5 (Aux 5) T6SS clusters, which encode predicted phospholipase effectors (34, 40). Recently, several V. cholerae strains have been shown to possess an Aux 6 T6SS cluster with antibacterial activity (39). We and others have also reported that many isolates (but not C6706) contain an additional gene cluster with putative T6SS components, which we named Aux 4 (34, 40, 41). However, the ability of V. cholerae cells to use the Aux 4 cluster in T6SS-mediated bacterial competition has not been validated, and the role played by the putative effector in intoxicating target cells has not been examined.
Here, we demonstrate that the Aux 4 cluster can be used by V. cholerae to kill bacterial cells in a T6SS-dependent manner. We report that the toxin found within the cluster permeabilizes cells and disrupts the membrane potentials when expressed in the periplasm of Escherichia coli cells. A protein encoded by a gene immediately downstream of the effector neutralizes its toxicity and acts as a protective immunity factor. Finally, we show that homologs of the Aux 4 effector are found in diverse bacterial species, including human, animal, and plant pathogens. The potent antimicrobial activity of the protein and broad distribution of identified homologs suggest the toxins confer significant competition advantages to bacteria that harbor them.
RESULTS
The Aux 4 tpeV-tpiV are an active effector-immunity pair in strain BGT49.
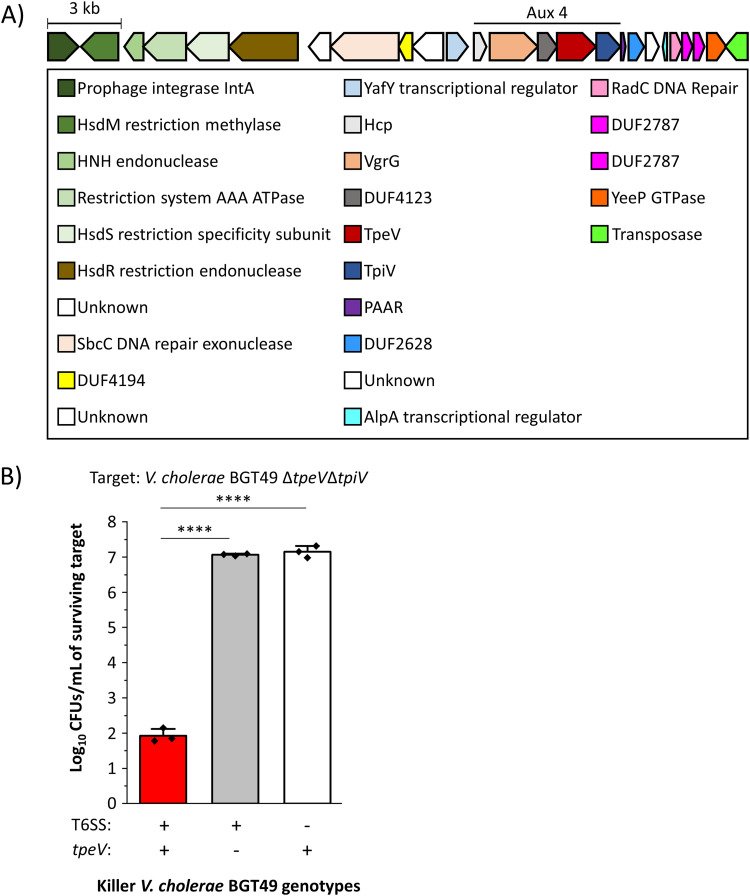
V. cholerae strain BGT49 encodes the Aux 4 cluster in addition to the canonical T6SS large operon and auxiliary clusters 1 and 2 (Fig. 1A). The Aux 4 cluster contains the following predicted T6SS genes: an hcp, a vgrG, a DUF4123 chaperone, and a paar (11) (Fig. 1A). Genes coding for a putative effector toxin (which we name type VI permeabilizing effector Vibrio [tpeV]; see below) and a putative immunity protein (which we name type VI permeabilizing immunity Vibrio [tpiV]; see below) are also found within the cluster (Fig. 1A) (16, 41, 42). The Aux 4 VgrG does not contain a toxic C-terminal domain as described for the V. cholerae VgrG-1 or VgrG-3 (32, 43, 44).
FIG 1.
Vibrio cholerae strain BGT49 encodes the Aux 4 T6SS cluster and efficiently eliminates target bacteria in a TpeV- and T6SS-dependent manner. (A) The Aux 4 cluster encodes predicted hcp, vgrG, DUF4123-containing chaperone, effector, immunity, and paar genes. The cluster is found on a predicted mobile genetic element, flanked by integrase and transposase genes. (B) Target V. cholerae BGT49 ΔtpeV ΔtpiV (CC170) was cocultured with either WT, ΔtpeV (CC167), or ΔvasK T6SS− (CC168) BGT49. A one-way analysis of variance (ANOVA) with a post hoc Tukey honestly significant difference (HSD) test was used to determine significance. ****, P < 0.0001.
Genes for a restriction modification system are found upstream of the Aux 4 cluster (Fig. 1A). Both the Aux 4 T6SS cluster and restriction modification system genes are flanked upstream by a predicted integrase and downstream by a predicted transposase (Fig. 1A). Attachment (att) sites similar to those found in the Vibrio pathogenicity island 1 (VPI-1) also flank the region (41, 45).
To experimentally determine that the tpeV gene encodes a T6SS toxin, we engineered a ΔtpeV ΔtpiV target BGT49 strain (CC170). The ΔtpeV ΔtpiV target strain was then cocultured separately with the wild-type (WT) BGT49 killer, an isogenic ΔtpeV mutant (lacking the TpeV effector, CC167) and an isogenic ΔvasK nonkiller (T6SS−, CC168). The recovery of the ΔtpeV ΔtpiV target strain was significantly reduced (by approximately 5 orders of magnitude) when cocultured with wild-type BGT49 killer cells compared to the ΔtpeV or ΔvasK strains (Fig. 1B). This result suggests that TpeV is a T6SS effector that is actively used by V. cholerae strain BGT49 to eliminate susceptible cells that lack the TpiV immunity protein.
The Aux 4 cluster can be expressed in trans in a clinical V. cholerae strain where it confers competitive advantages.
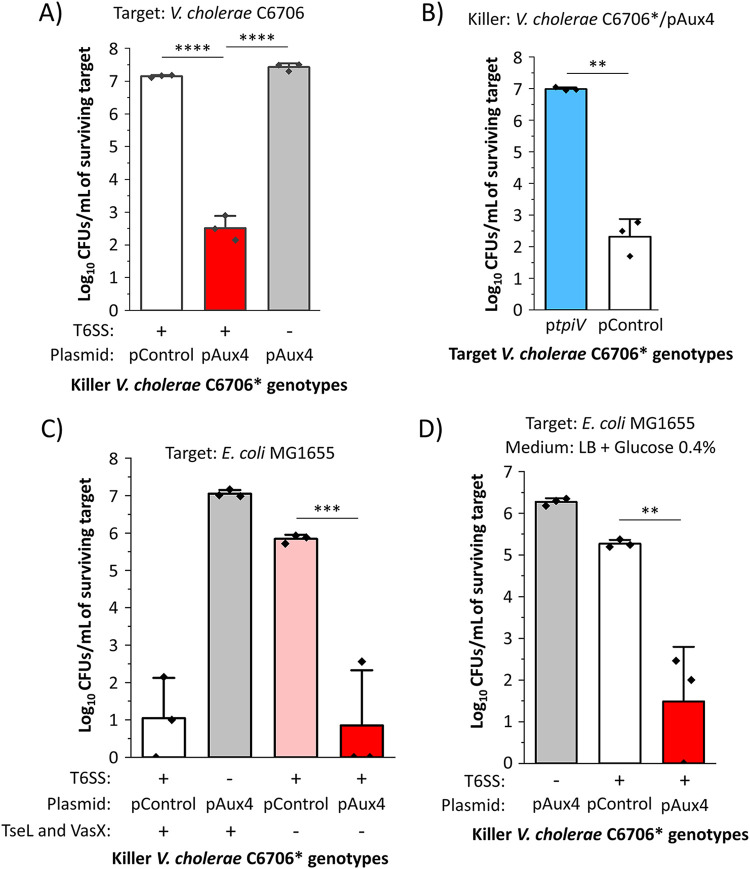
Since we observed that the Aux 4 cluster is located on a predicted mobile genetic element, we hypothesized that it can be used by other V. cholerae strains to eliminate competitor cells in a T6SS-dependent manner. In V. cholerae C6706, the QstR protein is a gene regulator that is required and sufficient to induce expression of T6SS genes (46–48). We cloned the Aux4 vgrG, tap, tpeV, tpiV, and paar genes on a plasmid (pAux4) under the control of the Ptac promoter. We then introduced the pAux4 plasmid into V. cholerae strain C6706*, which constitutively expresses the QstR protein but does not possess Aux 4 cluster genes on its chromosomes (33, 46–48). The V. cholerae C6706* killer with the Aux 4 cluster on a plasmid (C6706*/pAux4) efficiently eliminates the parental target strain, unlike a killer C6706* strain carrying a plasmid control (Fig. 2A). By contrast, a C6706*/pAux4 T6SS− strain cannot eliminate the parental target strain (Fig. 2A). To provide further evidence that TpiV can confer immunity, we introduced the tpiV gene into target V. cholerae C6706 and cocultured the strain with killer C6706*/pAux4 cells. V. cholerae C6706*/pAux4 kills V. cholerae target cells with a plasmid control but not when they encode the tpiV gene (Fig. 2B).
FIG 2.
V. cholerae C6706* can use the Aux 4 cluster to eliminate target cells in a T6SS-dependent manner. (A) V. cholerae C6706* (T6SS+ or T6SS−) with a plasmid control or a plasmid encoding the Aux 4 cluster was cocultured with target parental V. cholerae C6706. A one-way ANOVA with a post hoc Tukey HSD test was used to determine significance. (B) Killer V. cholerae C6706* with the Aux 4 cluster was cocultured with target C6706 cells with a plasmid control or a plasmid encoding tpiV. Welch’s t test was used to determine significance. (C) V. cholerae C6706* with deletions in the known tseL and vasX T6SS effectors containing either a plasmid control or a plasmid with Aux 4 was cocultured with E. coli MG1655 cells. A one-way ANOVA with a post hoc Tukey HSD test was used to determine significance. (D) V. cholerae C6706* with a plasmid control or a plasmid encoding the Aux 4 cluster was cocultured with E. coli MG1655 on LB medium with 0.4% glucose. A one-way ANOVA with a post hoc Tukey HSD test was used to determine significance. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01.
We next inquired whether the Aux 4 cluster can be used by V. cholerae to kill other target bacterial species. A C6706* strain that lacks the native TseL and VasX effectors poorly eliminates E. coli cells compared to a C6706* strain that harbors both toxins (Fig. 2C) (49, 50). However, the introduction of the Aux 4 cluster into the C6706* strain lacking TseL and VasX effectors restores its ability to efficiently eliminate E. coli cells (Fig. 2C). We recently reported that target E. coli cells are protected against T6SS attacks from strain C6706* when cocultured on LB medium supplemented with 0.4% glucose (4). By contrast, we observed that killer C6706*/pAux4 cells bypass the glucose-mediated resistance and efficiently eliminate E. coli cells when the coculture is performed on LB medium with glucose (Fig. 2D). These results confirm that the Aux 4 cluster can be used by V. cholerae to intoxicate competitor cells.
TpeV permeabilizes cells and disrupts the membrane potential.
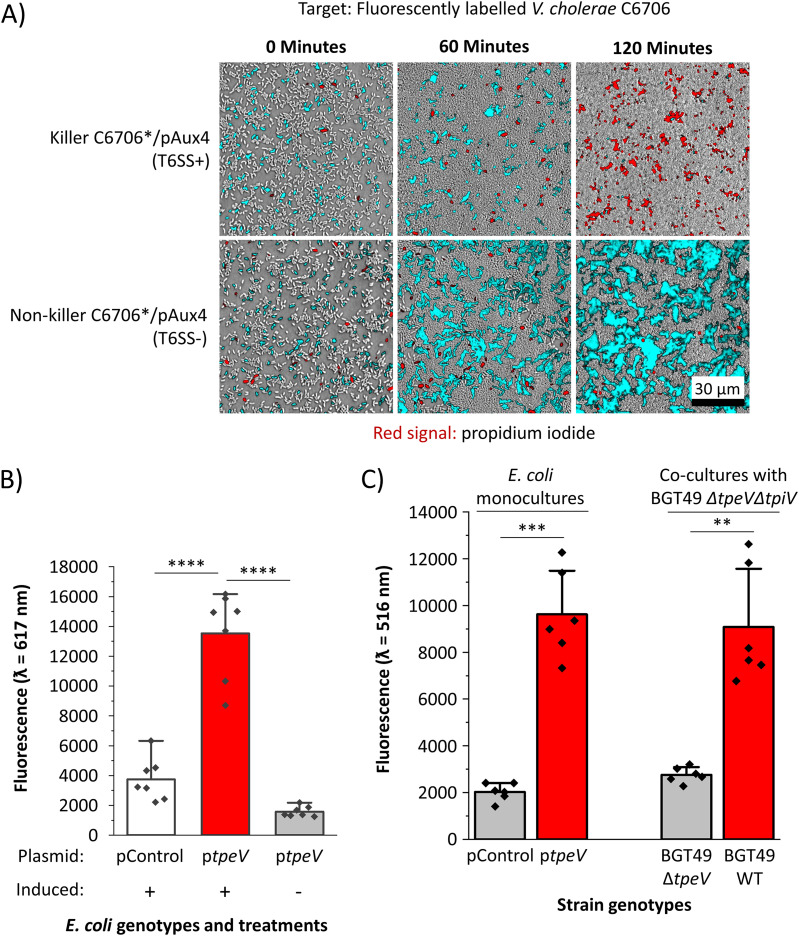
We used confocal microscopy to examine cocultures between fluorescently labeled target V. cholerae C6706 cells (shown as cyan) and unlabeled killer C6706*/pAux4 cells (Fig. 3A). To each coculture, we added propidium iodine (PI), a molecule that cannot penetrate cells with intact membranes but exhibits high fluorescence when bound to the DNA of cells with compromised membranes. Fluorescently labeled V. cholerae target cells are successfully eliminated when cocultured with killer C6706*/pAux4 cells but remain viable when competing cells cannot assemble the T6SS apparatus (T6SS−) (Fig. 3A). Furthermore, a robust PI signal (depicted with red) is detectable when target cells are cocultured with C6706*/pAux4 cells (Fig. 3A). These results provide evidence that V. cholerae harboring the Aux 4 cluster can permeabilize target cells in a T6SS-dependent manner.
FIG 3.
TpeV permeabilizes target cells and disrupts the membrane potential, leading to cytotoxicity. (A) Confocal microscopy was used to visualize a coculture between C6706* cells with Aux 4 (T6SS− or T6SS+) and fluorescently labeled target C6706 cells in the presence of propidium iodide. Scale bar = 30 μm. (B) E. coli cells carrying a periplasmic tpeV construct or plasmid control were incubated with propidium iodide. Fluorescence readings were taken at an excitation ƛ of 535 nm and emission ƛ of 617 nm. A one-way ANOVA with a post hoc Tukey HSD test was used to determine significance. (C) E. coli cells carrying a periplasmic tpeV construct or plasmid control, or V. cholerae BGT49 cocultures between target ΔtpeV ΔtpiV and wild-type or ΔtpeV killer cells were incubated with the membrane potential-sensitive DiBAC4(3) dye. Fluorescence readings were taken at an excitation ƛ of 490 nm and emission ƛ of 516 nm. Welch’s t tests were used to determine significance. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01.
We next sought to further characterize the activity of the TpeV effector. The protein does not share primary sequence homology to known toxins and does not contain motifs or domains indicative of its toxicity. Tertiary structural prediction algorithms also fail to detect significant homologs with known functions. TpeV has 11 cysteine residues, suggesting that multiple disulfide bonds could play roles in stabilizing the protein. Transmembrane prediction algorithms TMHMM and Phobius do not detect extensive transmembrane regions, and SignalP 5.0 does not predict a signal sequence (see Fig. S1 and S2 in the supplemental material) (51–53). We also attempted to identify TpeV homologs using the secondary structure predictor JPred (54). While most homologs are hypothetical proteins with unknown functions, some contain domains similar to the peptidoglycan-binding C-terminal regions of the OmpA protein (55, 56). OmpA proteins are involved in pathogenesis and have diverse functions that include formation of porins and channels (57, 58). Because target V. cholerae cells have a substantial PI signal when cocultured with killer cells harboring the Aux 4 cluster, we hypothesized that TpeV might permeabilize target cells when delivered to the periplasm.
TpeV Phobius transmembrane helix prediction. The amino acid sequence of TpeV was analyzed using the Phobius transmembrane predictor. Download FIG S1, TIF file, 0.9 MB (908.2KB, tif) .
Copyright © 2021 Crisan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
TpeV SignalP prediction. The amino acid sequence of TpeV was analyzed using the SignalP 5.0 transmembrane predictor. Download FIG S2, TIF file, 1.6 MB (1.6MB, tif) .
Copyright © 2021 Crisan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To test this prediction, we introduced plasmid-borne tpeV with a periplasmically directing pelB sequence under the control of an inducible promoter into E. coli cells. A significantly higher PI signal is detected when E. coli cells are induced to express periplasmic TpeV compared to that of cells that harbor a plasmid control (Fig. 3B). We also hypothesized that TpeV disrupts the bacterial cell membrane potential (28, 59, 60). To test this hypothesis, we used the bis-(1,3-dibutylbarbituric acid) trimethine oxonol [DiBAC4(3)] potential-sensitive dye, which is excluded from cells with a normal membrane potential but exhibits fluorescence in depolarized cells (60–62). E. coli cells that express periplasmically delivered TpeV have significantly higher DiBAC4(3) uptake than E. coli cells that express a plasmid control (Fig. 3C). As a positive membrane depolarization control, we exposed E. coli harboring plasmid control or TpeV to the carbonyl cyanide m-chlorophenyl hydrazone (CCCP) ionophore (see Fig. S3 in the supplemental material). E. coli cells expressing periplasmically delivered TpeV have comparable DiBAC4(3) fluorescence to E. coli cells treated with CCCP (Fig. 3C; see also Fig. S3). To probe whether TpeV can disrupt the membrane potential in a T6SS-dependent manner, we cocultured V. cholerae BGT49 ΔtpeV ΔtpiV (CC170) target cells with either the BGT49 wild-type or BGT49 ΔtpeV (CC167) strain. Following cocultures with the wild-type but not the ΔtpeV strain, bacterial membrane potentials are disrupted as cells display an elevated DiBAC4(3) signal (Fig. 3C). Taken together, these findings demonstrate that TpeV permeabilizes cells and disrupts the membrane potential of target bacteria.
CCCP disrupts the membrane potential of E. coli cells. E. coli cells harboring either plasmid control or periplasmically delivered TpeV were first incubated with CCCP and then with the membrane potential-sensitive DiBAC4(3) dye. Fluorescence readings were taken at an excitation ƛ = 490 nm and emission ƛ = 516 nm. A one-way ANOVA was used to determine significance. NS, not significant. Download FIG S3, TIF file, 0.9 MB (876.8KB, tif) .
Copyright © 2021 Crisan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
TpeV belongs to a large family of T6SS proteins and is spread widely across V. cholerae isolates.
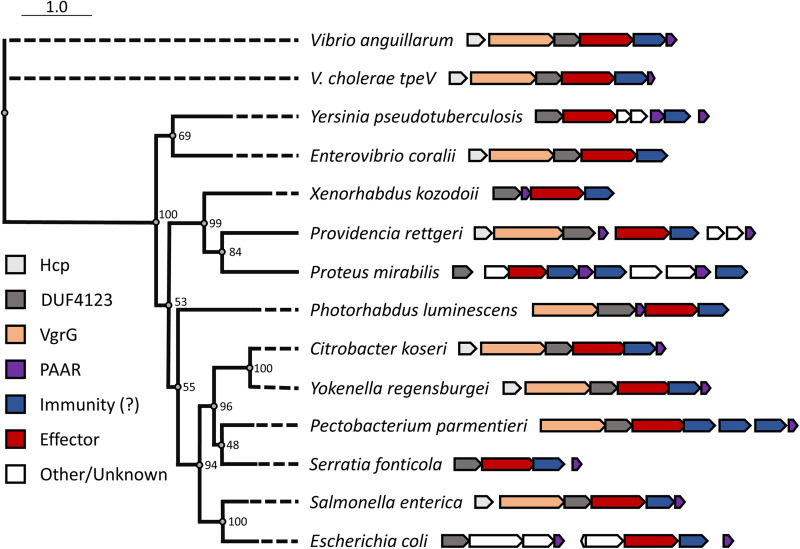
Since the sequence or predicted structure of TpeV shares no homology to known toxins (including known permeabilizing toxins), we used PHMMER to search for homologs in other bacterial species (63). We identified tpeV-like genes across diverse Gammaproteobacteria (Fig. 4; see also Table S1 in the supplemental material). In all selected species, genes coding for TpeV homologs are found near known T6SS genes like hcp, vgrG, DUF4123-containing chaperones, or other structural components (Fig. 4). Furthermore, we observed that tpeV-tpiV sequences are present in both patient and environmental V. cholerae genomes and are broadly distributed in countries across Africa, Asia, Europe, North America, and South America (see Table S2 in the supplemental material). Our results indicate that TpeV is a representative member of a family of T6SS toxins with antimicrobial activity that allows cells to eliminate competitor bacteria.
FIG 4.
TpeV homologs are found in many bacterial species near other T6SS genes. TpeV homologs were identified using PHMMER, and selected sequences were aligned using MUSCLE. A phylogenetic tree was constructed with 100 bootstraps.
Complete list of all identified TpeV protein homologs using the PHMMER algorithm. Download Table S1, XLS file, 0.05 MB (50.5KB, xls) .
Copyright © 2021 Crisan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
V. cholerae strains from diverse geographical locations harbor tpeV-tpiV modules. Download Table S2, DOCX file, 0.02 MB (16.1KB, docx) .
Copyright © 2021 Crisan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
Here, we show that many bacterial species encode homologs of a previously undescribed T6SS protein that intoxicates, permeabilizes, and disrupts the membrane potential of target cells. While studies have examined antibacterial effectors from clinical V. cholerae isolates, such as C6706 and V52, we and others found that strains isolated from sources other than patients encode a more diverse set of putative T6SS toxins (8, 28, 31, 34, 35, 41). Many T6SS toxins degrade components of the cell envelope; lipases and peptidoglycan-degrading enzymes target lipid membranes and cell walls, respectively (6, 30, 32, 49, 64).
Pore-forming proteins represent another functional class of T6SS toxins that act in the bacterial cell envelope (28, 59, 60, 65). The V. cholerae VasX effector encoded in the Aux 2 cluster is a large protein that contains a C-terminal colicin domain effective at eliminating both bacterial and eukaryotic cells (28, 29). Since it is predicted to form large pores, VasX permeabilizes cells and allows passage of molecules like PI into the cell (28). The Pseudomonas aeruginosa Tse4 and the Serratia marcescens Ssp6 effectors are both relatively small proteins that form ion-selective pores but do not allow larger molecules like PI to enter cells (59, 60). Recently, Vibrio parahaemolyticus has also been shown to harbor T6SS effectors that disrupt cellular membranes (65).
Importantly, the V. cholerae TpeV T6SS effector that we describe in this study does not contain known features, and its sequence does not share homology to any previously characterized proteins. We provide evidence that TpeV is a T6SS toxin that can be used by V. cholerae cells to permeabilize target cells and disrupt the cell membrane potential (Fig. 2 and 3). The cell membrane potential is essential for ATP synthesis, cell division, and membrane transport (66–68). Therefore, TpeV-mediated toxicity is likely to inflict substantial damage to target cells by perturbing multiple essential processes.
We hypothesize that TpeV could permeabilize cells by forming pores. Pore-forming toxins (PFTs) are widespread among all kingdoms of life (69–73). Based on the secondary structure of the membrane-spanning domain, two major classes of PFTs have been described: α-PFTs and β-PFTs (69, 71, 74). α-PTFs include the E. coli colicin and cytolysin A families, while β-PFTs are found in many Gram-positive bacterial species and contribute to the virulence of pathogens like Staphylococcus aureus and Clostridium perfringens (71, 73–75). Our homology predictions suggest that TpeV might harbor a peptidoglycan-binding OmpA-like domain (55). RmpM is a Neisseria meningitidis periplasmic protein that also possesses an OmpA-like domain (56). Experimental evidence suggests that RmpM stabilizes oligomeric porins in the outer membrane (56, 76). Rather than form new pores, it is also possible that TpeV might interact with and disrupt the normal functions of existing porins or channels in the membranes of target bacteria. Future experiments will determine whether TpeV forms pores or employs other mechanisms that damage membranes and permeabilize cells.
In strain BGT49, the Aux 4 cluster and a restriction modification system are found near a prophage integrase and a transposase (Fig. 1A). This suggests that the genes are located on a mobile genetic element that can be transferred between bacterial cells to confer advantages against phages and other bacteria (41). The V. cholerae T6SS Aux 3 cluster was also experimentally validated to be located on a mobile genetic element (37). Our results show that V. cholerae strain C6706* can use the Aux 4 cluster to kill parental cells and support the hypothesis that the Aux 4 cluster can be transferred to confer competitive advantages. This hypothesis is further supported by our observation that TpeV homologs are found close to other T6SS genes in many bacterial species, including human pathogens (Providencia rettgeri, Proteus mirabilis, Citrobacter koseri, Yokenella regensburgei, Serratia fonticola, Salmonella enterica, and E. coli), animal pathogens (Vibrio anguillarum and Photorhabdus luminescens), and plant pathogens (Pectobacterium parmentieri) (77–84) (Fig. 4). TpeV homologs found in other bacteria are also located near transposase-like genes (data not shown).
We also hypothesized that the paar gene downstream of tpiV might be required for tpeV-mediated toxicity (Fig. 1). However, we observed that a BGT49 strain with an Aux 4 paar deletion (Δpaar, CC179) eliminated susceptible ΔtpeV ΔtpiV cells in a similar manner to wild-type BGT49, suggesting that the PAAR protein is not required for TpeV-mediated killing (see Fig. S4 in the supplemental material). All known T6SS toxic effectors are neutralized by cognate immunity proteins, which are generally encoded by genes adjacent to effectors (28, 85). We found that tpiV, the gene immediately downstream of tpeV, confers immunity to target cells against tpeV-mediated toxicity (Fig. 1B and 2B). SignalP predicts that TpiV encodes a periplasmic Sec-tag, which is expected since TpeV exhibits its toxicity when delivered to the periplasm of target cells (Fig. 3B and C; see also Fig. S5 in the supplemental material). In other species, we observed that multiple putative immunity proteins can be found near TpeV homologs (Fig. 4). Additional studies are required to confirm which predicted TpiV-like proteins are the cognate immunity factors for the TpeV homologs.
The Aux 4 paar gene is not required for TpeV-mediated killing. Target V. cholerae BGT49 ΔtpeVΔtpiV (CC170) was cocultured with either Δpaar (CC179), WT, ΔvasK (T6SS−, CC168), or ΔtpeV (CC167) BGT49. Download FIG S4, TIF file, 1.8 MB (1.8MB, tif) .
Copyright © 2021 Crisan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
TpiV SignalP prediction. The amino acid sequence of TpiV was analyzed using the SignalP 5.0 transmembrane predictor. Download FIG S5, TIF file, 1.6 MB (1.6MB, tif) .
Copyright © 2021 Crisan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
In conclusion, we demonstrate that the T6SS Aux 4 cluster found in many V. cholerae isolates encodes a toxin that can be used to eliminate competitor bacteria. TpeV is a T6SS effector that permeabilizes target bacteria and disrupts the membrane potential, leading to severe cellular intoxication. However, target cells expressing TpiV are protected and resist TpeV-mediated toxicity. Finally, we find that TpeV homologs are widespread among Gram-negative bacteria, suggesting that the protein represents a novel and potent antimicrobial agent of interest for further studies. Understanding the molecular mechanisms of antimicrobial toxins that drive competition and antagonism could lead to the development of novel biotechnology and medical applications.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Plasmids were constructed using standard molecular biology techniques. Gibson mix reagents, restriction enzymes, and polymerases were used as recommended by manufacturers (Promega and New England BioLabs). Plasmids were verified by PCR and Sanger sequencing (Eurofins and Eton Bioscience). V. cholerae C6706 mutant strains were made using pKAS allelic exchange methods as described previously (86). V. cholerae BGT49 mutant strains were made using natural transformation as described previously with modifications (87, 88). Briefly, overnight cultures were back-diluted in fresh LB medium for approximately 1 h and then statically incubated overnight at 30°C in liquid LB medium with a sterile crab shell fragment. Crab shells were transferred to fresh LB medium containing 30 to 50 μg of a plasmid engineered to encode ∼1,000-bp flanking regions to replace the desired genes with an antibiotic cassette. Cells were incubated statically overnight at 30°C and then spread on antibiotic plates to select for transformants. BGT49 mutants were confirmed by PCR and antibiotic resistance. Bacterial strains and plasmids used are listed in Table S3 in the supplemental material.
Strains and plasmids used in this study. Download Table S3, DOCX file, 0.02 MB (18.8KB, docx) .
Copyright © 2021 Crisan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Bacterial competition assays.
Bacterial cultures were grown overnight in liquid LB medium at 37°C with shaking. Overnight cultures were back-diluted and incubated in liquid LB medium at 37°C with shaking for 3 h. Bacterial cultures were then normalized to an optical density at 600 nm (OD600) of 1. If strains harbored plasmids, cultures were grown overnight with antibiotics to maintain plasmids and 100 μM isopropyl-β-d-thiogalactopyranoside (IPTG) if plasmids contained an inducible promoter. If strains were grown in medium containing antibiotics, liquid cultures were then washed three times with fresh LB medium before they were cocultured. For bacterial competitions performed on LB agar medium with 0.4% glucose, overnight and back-diluted cultures were also grown in LB medium supplemented with 0.4% glucose. A 50-μl mixture aliquot of a 10:1 killer/target cell ratio was spotted on a 0.22-μm-pore-size filter paper, which was placed on LB agar medium and incubated at 37°C. After 3 h, filters were vortexed in 5 ml of sterile LB medium for 30 s. One hundred microliters of serial dilutions were then spread (or 3 μl or serial dilutions were spotted) on plates containing the required antibiotic to select for target cells. Data from three cocultures were used to determine significance. Results are representative of at least two independent experiments.
Confocal microscopy.
Overnight cultures were back-diluted 1:100 for 3 h in liquid LB medium. Samples were then normalized to an OD600 of 1. A 2-μl aliquot of 10:1 killer/target cell mixture was spotted on top of a dry 8-μl aliquot of propidium iodide (100 μg/ml) on an LB agar pad. A Nikon A1R confocal microscope using a Perfect Focus System with a 40× objective (Plan Fluor ELWD 40× DIC M N1) was used to stabilize the focus in the plane of the colony growth. Cells were imaged at 90 to 100% humidity and 37°C. Images were processed using ImageJ. Results are representative of at least three independent experiments.
Membrane permeabilization assays.
Bacterial cultures of E. coli Shuffle T7 Express (New England BioLabs) cells carrying either a control plasmid or a periplasmic tpeV construct were grown overnight in liquid LB medium supplemented with 0.2% glucose and ampicillin at 37°C with shaking. Cells were washed three times with LB and 100× back-dilutions were made in fresh liquid LB medium with 500 μM IPTG (or 0.2% glucose for uninduced controls) and ampicillin. Strains were incubated at 37°C for 2 h, washed three times with phosphate-buffered saline (PBS), and normalized to an OD600 of 1. One hundred microliters of each culture was incubated with 1 μl propidium iodide (1 mg/ml) for 15 to 30 min. Fluorescence values were taken on a Synergy BioTek plate reader using an excitation ƛ of 535 nm and emission ƛ of 617 nm and normalized by subtracting the average values from samples with propidium iodide but no cells. Data represent the averages obtained from seven biological replicates from two independent experiments.
Membrane potential assays.
Bacterial cultures of E. coli Shuffle T7 Express (New England BioLabs) cells carrying either a control plasmid or a periplasmic tpeV construct were grown overnight with shaking at 37°C in liquid LB medium supplemented with 0.2% glucose and ampicillin. Cells were washed three times with LB, and 100× back-dilutions were incubated at 37°C for 2 h in fresh liquid LB medium with 500 μM IPTG and ampicillin. Cells were again washed three times with PBS and normalized to an OD600 of 1 in PBS. Cells were incubated for 30 min in the dark with DiBAC4(3) at a final concentration of 10 μM and washed three times with PBS. For positive depolarization controls, E. coli cells were incubated with 10 μM carbonyl cyanide m-chlorophenyl hydrazone for 10 min in the dark prior to staining with DiBAC4(3). Fluorescence values were taken on a Synergy BioTek plate reader using an excitation ƛ of 490 nm and emission ƛ of 516 nm. Data represent the averages obtained from six biological replicates from two independent experiments.
For coculture measurements of membrane potentials, overnight cultures of the indicated V. cholerae BGT49 strains were back-diluted and incubated in liquid LB medium at 37°C with shaking for 3 h. Bacterial cultures were then normalized to an OD600 of 10. A 50-μl mixture aliquot with a ratio of 1:1 killer/target cells was spotted on a 0.22-μm-pore-size filter paper, which was placed on LB agar medium and incubated at 37°C. After 2 h, filters were vortexed in sterile LB medium for 30 s. Cells were washed three times with PBS, normalized to an OD600 of 1, incubated for 30 min in the dark with DiBAC4(3) at a final concentration of 10 μM and washed three times with PBS. Fluorescence values were taken on a Synergy BioTek plate reader using an excitation ƛ of 490 nm and emission ƛ of 516 nm. Data represent the averages obtained from six biological replicates from two independent experiments.
Bioinformatic analyses.
The PHMMER server was used to search for homologs of TpeV in the UniProtKB database (63, 89). Obsolete or duplicate hits were removed from Table S1. Selected homologs were aligned using MUSCLE and were used to retrieve genomes from the NCBI database (90). A phylogenetic tree was constructed using PhyML with 100 bootstrap values and visualized using PRESTO (91–93). Putative immunity proteins were predicted based on homology to TpiV and genomic location. Truncated vgrG-like genes encoding stop codons were observed in some species but were excluded from Fig. 4.
The tpeV-tpiV genes were used to retrieve homologous sequences from the NCBI RefSeq Genome Database (94). Selected strains harboring homologs to the tpeV-tpiV gene module are displayed in Table S2 in the supplemental material.
ACKNOWLEDGMENTS
We thank Jacob Thomas, Mackenzie Martin, Athéna Patterson-Orazem, Dustin Huard, and Tong Yu for advice, experimental help, and useful discussions.
B.K.H. would also like to thank funding from the Georgia Institute of Technology School of Biological Sciences, NSF (MCB 1149925 and BMAT-2003721) and BSF (2015103). G.S. would like to thank the German National Academy of Natural Sciences Leopoldina (LDPS 2017-03).
We declare no competing interests.
Contributor Information
Brian K. Hammer, Email: brian.hammer@biology.gatech.edu.
Craig D. Ellermeier, University of Iowa
REFERENCES
- 1.Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC, Heidelberg JF, Mekalanos JJ. 2006. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci U S A 103:1528–1533. doi: 10.1073/pnas.0510322103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, Goodman AL, Joachimiak G, Ordoñez CL, Lory S, Walz T, Joachimiak A, Mekalanos JJ. 2006. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312:1526–1530. doi: 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyer F, Fichant G, Berthod J, Vandenbrouck Y, Attree I. 2009. Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: what can be learned from available microbial genomic resources? BMC Genomics 10:104. doi: 10.1186/1471-2164-10-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crisan CV, Nichols HL, Wiesenfeld S, Steinbach G, Yunker PJ, Hammer BK. 2021. Glucose confers protection to Escherichia coli against contact killing by Vibrio cholerae. Sci Rep 11:2935. doi: 10.1038/s41598-021-81813-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steinbach G, Crisan C, Ng SL, Hammer BK, Yunker PJ. 2020. Accumulation of dead cells from contact killing facilitates coexistence in bacterial biofilms. J R Soc Interface 17:20200486. doi: 10.1098/rsif.2020.0486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hersch SJ, Watanabe N, Stietz MS, Manera K, Kamal F, Burkinshaw B, Lam L, Pun A, Li M, Savchenko A, Dong TG. 2020. Envelope stress responses defend against type six secretion system attacks independently of immunity proteins. Nat Microbiol 5:706–714. doi: 10.1038/s41564-020-0672-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamal F, Liang X, Manera K, Pei T-T, Kim H, Lam LG, Pun A, Hersch SJ, Dong TG. 2020. Differential cellular response to translocated toxic effectors and physical penetration by the type VI secretion system. Cell Rep 31:107766. doi: 10.1016/j.celrep.2020.107766. [DOI] [PubMed] [Google Scholar]
- 8.Zheng J, Ho B, Mekalanos JJ. 2011. Genetic analysis of anti-amoebae and anti-bacterial activities of the type VI secretion system in Vibrio cholerae. PLoS One 6:e23876. doi: 10.1371/journal.pone.0023876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aschtgen MS, Thomas MS, Cascales E. 2010. Anchoring the type VI secretion system to the peptidoglycan: TssL, TagL, TagP… what else? Virulence 1:535–540. doi: 10.4161/viru.1.6.13732. [DOI] [PubMed] [Google Scholar]
- 10.Rapisarda C, Cherrak Y, Kooger R, Schmidt V, Pellarin R, Logger L, Cascales E, Pilhofer M, Durand E, Fronzes R. 2019. In situ and high‐resolution cryo‐EM structure of a bacterial type VI secretion system membrane complex. EMBO J 38:e100886. doi: 10.15252/embj.2018100886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cascales E, Cambillau C. 2012. Structural biology of type VI secretion systems. Philos Trans R Soc Lond B Biol Sci 367:1102–1111. doi: 10.1098/rstb.2011.0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ballister ER, Lai AH, Zuckermann RN, Cheng Y, Mougous JD. 2008. In vitro self-assembly of tailorable nanotubes from a simple protein building block. Proc Natl Acad Sci U S A 105:3733–3738. doi: 10.1073/pnas.0712247105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Silverman JM, Agnello DM, Zheng H, Andrews BT, Li M, Catalano CE, Gonen T, Mougous JD. 2013. Haemolysin coregulated protein is an exported receptor and chaperone of type VI secretion substrates. Mol Cell 51:584–593. doi: 10.1016/j.molcel.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cianfanelli FR, Alcoforado Diniz J, Guo M, De Cesare V, Trost M, Coulthurst SJ. 2016. VgrG and PAAR proteins define distinct versions of a functional type VI secretion system. PLoS Pathog 12:e1005735. doi: 10.1371/journal.ppat.1005735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wood TE, Howard SA, Wettstadt S, Filloux A. 2019. PAAR proteins act as the ‘sorting hat’ of the type VI secretion system. Microbiology (Reading) 165:1203–1218. doi: 10.1099/mic.0.000842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Unterweger D, Kostiuk B, Ötjengerdes R, Wilton A, Diaz‐Satizabal L, Pukatzki S. 2015. Chimeric adaptor proteins translocate diverse type VI secretion system effectors in Vibrio cholerae. EMBO J 34:2198–2210. doi: 10.15252/embj.201591163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saak CC, Gibbs KA. 2016. The self-identity protein IdsD is communicated between cells in swarming Proteus mirabilis colonies. J Bacteriol 198:3278–3286. doi: 10.1128/JB.00402-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zepeda-Rivera MA, Saak CC, Gibbs KA. 2018. A proposed chaperone of the bacterial type VI secretion system functions to constrain a self-identity protein. J Bacteriol 200:e00688-17. doi: 10.1128/JB.00688-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bröms JE, Ishikawa T, Wai SN, Sjöstedt A. 2013. A functional VipA-VipB interaction is required for the type VI secretion system activity of Vibrio cholerae O1 strain A1552. BMC Microbiol 13:96. doi: 10.1186/1471-2180-13-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bönemann G, Pietrosiuk A, Diemand A, Zentgraf H, Mogk A. 2009. Remodelling of VipA/VipB tubules by ClpV-mediated threading is crucial for type VI protein secretion. EMBO J 28:315–325. doi: 10.1038/emboj.2008.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kube S, Kapitein N, Zimniak T, Herzog F, Mogk A, Wendler P. 2014. Structure of the VipA/B type VI secretion complex suggests a contraction-state-specific recycling mechanism. Cell Rep 8:20–30. doi: 10.1016/j.celrep.2014.05.034. [DOI] [PubMed] [Google Scholar]
- 22.Sack DA, Sack RB, Nair GB, Siddique A. 2004. Cholera. Lancet 363:223–233. doi: 10.1016/S0140-6736(03)15328-7. [DOI] [PubMed] [Google Scholar]
- 23.de Magny GC, Mozumder PK, Grim CJ, Hasan NA, Naser MN, Alam M, Sack RB, Huq A, Colwell RR. 2011. Role of zooplankton diversity in Vibrio cholerae population dynamics and in the incidence of cholera in the Bangladesh Sundarbans. Appl Environ Microbiol 77:6125–6132. doi: 10.1128/AEM.01472-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Senderovich Y, Izhaki I, Halpern M. 2010. Fish as reservoirs and vectors of Vibrio cholerae. PLoS One 5:e8607. doi: 10.1371/journal.pone.0008607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sela R, Hammer BK, Halpern M. 2021. Quorum‐sensing signaling by chironomid egg masses’ microbiota, affect haemagglutinin/protease (HAP) production by Vibrio cholerae. Mol Ecol 30:1736–1746. doi: 10.1111/mec.15662. [DOI] [PubMed] [Google Scholar]
- 26.Logan SL, Thomas J, Yan J, Baker RP, Shields DS, Xavier JB, Hammer BK, Parthasarathy R. 2018. The Vibrio cholerae type VI secretion system can modulate host intestinal mechanics to displace gut bacterial symbionts. Proc Natl Acad Sci U S A 115:E3779–E3787. doi: 10.1073/pnas.1720133115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao W, Caro F, Robins W, Mekalanos JJ. 2018. Antagonism toward the intestinal microbiota and its effect on Vibrio cholerae virulence. Science 359:210–213. doi: 10.1126/science.aap8775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyata ST, Unterweger D, Rudko SP, Pukatzki S. 2013. Dual expression profile of type VI secretion system immunity genes protects pandemic Vibrio cholerae. PLoS Pathog 9:e1003752. doi: 10.1371/journal.ppat.1003752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyata ST, Kitaoka M, Brooks TM, McAuley SB, Pukatzki S. 2011. Vibrio cholerae requires the type VI secretion system virulence factor VasX to kill Dictyostelium discoideum. Infect Immun 79:2941–2949. doi: 10.1128/IAI.01266-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Russell AB, LeRoux M, Hathazi K, Agnello DM, Ishikawa T, Wiggins PA, Wai SN, Mougous JD. 2013. Diverse type VI secretion phospholipases are functionally plastic antibacterial effectors. Nature 496:508–512. doi: 10.1038/nature12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ho BT, Fu Y, Dong TG, Mekalanos JJ. 2017. Vibrio cholerae type 6 secretion system effector trafficking in target bacterial cells. Proc Natl Acad Sci U S A 114:9427–9432. doi: 10.1073/pnas.1711219114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brooks TM, Unterweger D, Bachmann V, Kostiuk B, Pukatzki S. 2013. Lytic activity of the Vibrio cholerae type VI secretion toxin VgrG-3 is inhibited by the antitoxin TsaB. J Biol Chem 288:7618–7625. doi: 10.1074/jbc.M112.436725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crisan CV, Hammer BK. 2020. The Vibrio cholerae type VI secretion system: toxins, regulators and consequences. Environ Microbiol 22:4112–4122. doi: 10.1111/1462-2920.14976. [DOI] [PubMed] [Google Scholar]
- 34.Crisan CV, Chande AT, Williams K, Raghuram V, Rishishwar L, Steinbach G, Watve SS, Yunker P, Jordan IK, Hammer BK. 2019. Analysis of Vibrio cholerae genomes identifies new type VI secretion system gene clusters. Genome Biol 20:163. doi: 10.1186/s13059-019-1765-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Unterweger D, Miyata ST, Bachmann V, Brooks TM, Mullins T, Kostiuk B, Provenzano D, Pukatzki S. 2014. The Vibrio cholerae type VI secretion system employs diverse effector modules for intraspecific competition. Nat Commun 5:3549. doi: 10.1038/ncomms4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Altindis E, Dong T, Catalano C, Mekalanos J. 2015. Secretome analysis of Vibrio cholerae type VI secretion system reveals a new effector-immunity pair. mBio 6:e00075-15. doi: 10.1128/mBio.00075-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Santoriello FJ, Michel L, Unterweger D, Pukatzki S. 2020. Pandemic Vibrio cholerae shuts down site-specific recombination to retain an interbacterial defence mechanism. Nat Commun 11:6246. doi: 10.1038/s41467-020-20012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kirchberger PC, Unterweger D, Provenzano D, Pukatzki S, Boucher Y. 2017. Sequential displacement of type VI secretion system effector genes leads to evolution of diverse immunity gene arrays in Vibrio cholerae. Sci Rep 7:45133. doi: 10.1038/srep45133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Drebes Dörr NC, Blokesch M. 2020. Interbacterial competition and anti-predatory behaviour of environmental Vibrio cholerae strains. Environ Microbiol 22:4485–4504. doi: 10.1111/1462-2920.15224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hussain NA, Kirchberger PC, Case RJ, Boucher YF. 2021. Modular molecular weaponry plays a key role in competition within an environmental Vibrio cholerae population. Front Microbiol 12:671092. doi: 10.3389/fmicb.2021.671092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Labbate M, Orata FD, Petty NK, Jayatilleke ND, King WL, Kirchberger PC, Allen C, Mann G, Mutreja A, Thomson NR, Boucher Y, Charles IG. 2016. A genomic island in Vibrio cholerae with VPI-1 site-specific recombination characteristics contains CRISPR-Cas and type VI secretion modules. Sci Rep 6:36891. doi: 10.1038/srep36891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shneider MM, Buth SA, Ho BT, Basler M, Mekalanos JJ, Leiman PG. 2013. PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature 500:350–353. doi: 10.1038/nature12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Durand E, Derrez E, Audoly G, Spinelli S, Ortiz-Lombardia M, Raoult D, Cascales E, Cambillau C. 2012. Crystal structure of the VgrG1 actin cross-linking domain of the Vibrio cholerae type VI secretion system. J Biol Chem 287:38190–38199. doi: 10.1074/jbc.M112.390153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma AT, Mekalanos JJ. 2010. In vivo actin cross-linking induced by Vibrio cholerae type VI secretion system is associated with intestinal inflammation. Proc Natl Acad Sci U S A 107:4365–4370. doi: 10.1073/pnas.0915156107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karaolis DKR, Johnson JA, Bailey CC, Boedeker EC, Kaper JB, Reeves PR. 1998. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc Natl Acad Sci U S A 95:3134–3139. doi: 10.1073/pnas.95.6.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watve SS, Thomas J, Hammer BK. 2015. CytR is a global positive regulator of competence, type VI secretion, and chitinases in Vibrio cholerae. PLoS One 10:e0138834. doi: 10.1371/journal.pone.0138834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lo Scrudato M, Blokesch M. 2013. A transcriptional regulator linking quorum sensing and chitin induction to render Vibrio cholerae naturally transformable. Nucleic Acids Res 41:3644–3658. doi: 10.1093/nar/gkt041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jaskolska M, Stutzmann S, Stoudmann C, Blokesch M. 2018. QstR-dependent regulation of natural competence and type VI secretion in Vibrio cholerae. Nucleic Acids Res 46:10619–10634. doi: 10.1093/nar/gky717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dong TG, Ho BT, Yoder-Himes DR, Mekalanos JJ. 2013. Identification of T6SS-dependent effector and immunity proteins by Tn-seq in Vibrio cholerae. Proc Natl Acad Sci U S A 110:2623–2628. doi: 10.1073/pnas.1222783110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liang X, Kamal F, Pei T, Xu P, Mekalanos JJ, Dong TG. 2019. An onboard checking mechanism ensures effector delivery of the type VI secretion system in Vibrio cholerae. Proc Natl Acad Sci U S A 116:23292–23298. doi: 10.1073/pnas.1914202116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Almagro Armenteros JJ, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, Brunak S, von Heijne G, Nielsen H. 2019. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol 37:420–423. doi: 10.1038/s41587-019-0036-z. [DOI] [PubMed] [Google Scholar]
- 52.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. J Mol Biol 305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 53.Käll L, Krogh A, Sonnhammer ELL. 2004. A combined transmembrane topology and signal peptide prediction method. J Mol Biol 338:1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 54.Drozdetskiy A, Cole C, Procter J, Barton GJ. 2015. JPred4: a protein secondary structure prediction server. Nucleic Acids Res 43:W389–W394. doi: 10.1093/nar/gkv332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Samsudin F, Ortiz-Suarez ML, Piggot TJ, Bond PJ, Khalid S. 2016. OmpA: a flexible clamp for bacterial cell wall attachment. Structure 24:2227–2235. doi: 10.1016/j.str.2016.10.009. [DOI] [PubMed] [Google Scholar]
- 56.Grizot S, Buchanan SK. 2004. Structure of the OmpA-like domain of RmpM from Neisseria meningitidis. Mol Microbiol 51:1027–1037. doi: 10.1111/j.1365-2958.2003.03903.x. [DOI] [PubMed] [Google Scholar]
- 57.De Mot R, Proost P, Van Damme J, Vanderleyden J. 1992. Homology of the root adhesin of Pseudomonas fluorescens OE 28.3 with porin F of P. aeruginosa and P. syringae. Mol Gen Genet 231:489–493. doi: 10.1007/BF00292721. [DOI] [PubMed] [Google Scholar]
- 58.Smith SGJ, Mahon V, Lambert MA, Fagan RP. 2007. A molecular Swiss army knife: OmpA structure, function and expression. FEMS Microbiol Lett 273:1–11. doi: 10.1111/j.1574-6968.2007.00778.x. [DOI] [PubMed] [Google Scholar]
- 59.LaCourse KD, Peterson SB, Kulasekara HD, Radey MC, Kim J, Mougous JD. 2018. Conditional toxicity and synergy drive diversity among antibacterial effectors. Nat Microbiol 3:440–446. doi: 10.1038/s41564-018-0113-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mariano G, Trunk K, Williams DJ, Monlezun L, Strahl H, Pitt SJ, Coulthurst SJ. 2019. A family of type VI secretion system effector proteins that form ion-selective pores. Nat Commun 10:5484. doi: 10.1038/s41467-019-13439-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sánchez E, García S, Heredia N. 2010. Extracts of edible and medicinal plants damage membranes of Vibrio cholerae. Appl Environ Microbiol 76:6888–6894. doi: 10.1128/AEM.03052-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ji X, Zou J, Peng H, Stolle A-S, Xie R, Zhang H, Peng B, Mekalanos JJ, Zheng J. 2019. Alarmone Ap4A is elevated by aminoglycoside antibiotics and enhances their bactericidal activity. Proc Natl Acad Sci U S A 116:9578–9585. doi: 10.1073/pnas.1822026116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Finn RD, Clements J, Eddy SR. 2011. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 39:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Flaugnatti N, Le TTH, Canaan S, Aschtgen M-S, Nguyen VS, Blangy S, Kellenberger C, Roussel A, Cambillau C, Cascales E, Journet L. 2016. A phospholipase A1 antibacterial type VI secretion effector interacts directly with the C-terminal domain of the VgrG spike protein for delivery. Mol Microbiol 99:1099–1118. doi: 10.1111/mmi.13292. [DOI] [PubMed] [Google Scholar]
- 65.Fridman CM, Keppel K, Gerlic M, Bosis E, Salomon D. 2020. A comparative genomics methodology reveals a widespread family of membrane-disrupting T6SS effectors. Nat Commun 11:1085. doi: 10.1038/s41467-020-14951-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Benarroch JM, Asally M. 2020. The microbiologist’s guide to membrane potential dynamics. Trends Microbiol 28:304–314. doi: 10.1016/j.tim.2019.12.008. [DOI] [PubMed] [Google Scholar]
- 67.Maloney PC, Kashket ER, Wilson TH. 1974. A protonmotive force drives ATP synthesis in bacteria. Proc Natl Acad Sci U S A 71:3896–3900. doi: 10.1073/pnas.71.10.3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Strahl H, Hamoen LW. 2010. Membrane potential is important for bacterial cell division. Proc Natl Acad Sci U S A 107:12281–12286. doi: 10.1073/pnas.1005485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Peraro MD, Van Der Goot FG. 2016. Pore-forming toxins: ancient, but never really out of fashion. Nat Rev Microbiol 14:77–92. doi: 10.1038/nrmicro.2015.3. [DOI] [PubMed] [Google Scholar]
- 70.Hinds MG, Zhang W, Anderluh G, Hansen PE, Norton RS. 2002. Solution structure of the eukaryotic pore-forming cytolysin equinatoxin II: implications for pore formation. J Mol Biol 315:1219–1229. doi: 10.1006/jmbi.2001.5321. [DOI] [PubMed] [Google Scholar]
- 71.Cascales E, Buchanan SK, Duché D, Kleanthous C, Lloubès R, Postle K, Riley M, Slatin S, Cavard D. 2007. Colicin biology. Microbiol Mol Biol Rev 71:158–229. doi: 10.1128/MMBR.00036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yamashita K, Kawai Y, Tanaka Y, Hirano N, Kaneko J, Tomita N, Ohta M, Kamio Y, Yao M, Tanaka I. 2011. Crystal structure of the octameric pore of staphylococcal γ-hemolysin reveals the β-barrel pore formation mechanism by two components. Proc Natl Acad Sci U S A 108:17314–17319. doi: 10.1073/pnas.1110402108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Savva CG, Da Costa SPF, Bokori-Brown M, Naylor CE, Cole AR, Moss DS, Titball RW, Basak AK. 2013. Molecular architecture and functional analysis of NetB, a pore-forming toxin from Clostridium perfringens. J Biol Chem 288:3512–3522. doi: 10.1074/jbc.M112.430223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mueller M, Grauschopf U, Maier T, Glockshuber R, Ban N. 2009. The structure of a cytolytic α-helical toxin pore reveals its assembly mechanism. Nature 459:726–730. doi: 10.1038/nature08026. [DOI] [PubMed] [Google Scholar]
- 75.Song L, Hobaugh MR, Shustak C, Cheley S, Bayley H, Gouaux JE. 1996. Structure of staphylococcal α-hemolysin, a heptameric transmembrane pore. Science 274:1859–1865. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 76.Maharjan S, Saleem M, Feavers IM, Wheeler JX, Care R, Derrick JP. 2016. Dissection of the function of the RmpM periplasmic protein from Neisseria meningitidis. Microbiology (Reading) 162:364–375. doi: 10.1099/mic.0.000227. [DOI] [PubMed] [Google Scholar]
- 77.Yoh M, Matsuyama J, Ohnishi M, Takagi K, Miyagi H, Mori K, Park KS, Ono T, Honda T. 2005. Importance of Providencia species as a major cause of travellers’ diarrhoea. J Med Microbiol 54:1077–1082. doi: 10.1099/jmm.0.45846-0. [DOI] [PubMed] [Google Scholar]
- 78.Endimiani A, Luzzaro F, Brigante G, Perilli M, Lombardi G, Amicosante G, Rossolini GM, Toniolo A. 2005. Proteus mirabilis bloodstream infections: risk factors and treatment outcome related to the expression of extended-spectrum β-lactamases. Antimicrob Agents Chemother 49:2598–2605. doi: 10.1128/AAC.49.7.2598-2605.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dzeing-Ella A, Szwebel TA, Loubinoux J, Coignard S, Bouvet A, Le Jeunne C, Aslangul E. 2009. Infective endocarditis due to Citrobacter koseri in an immunocompetent adult. J Clin Microbiol 47:4185–4186. doi: 10.1128/JCM.00957-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Abbott SL, Janda JM. 1994. Isolation of Yokenella regensburgei ('Koserella trabulsii’) from a patient with transient bacteremia and from a patient with a septic knee. J Clin Microbiol 32:2854–2855. doi: 10.1128/jcm.32.11.2854-2855.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Di Martino ML, Ek V, Hardt WD, Eriksson J, Sellin ME. 2019. Barcoded consortium infections resolve cell type-dependent Salmonella enterica serovar Typhimurium entry mechanisms. mBio 10:e00603-19. doi: 10.1128/mBio.00603-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Frans I, Michiels CW, Bossier P, Willems KA, Lievens B, Rediers H. 2011. Vibrio anguillarum as a fish pathogen: virulence factors, diagnosis and prevention. J Fish Dis 34:643–661. doi: 10.1111/j.1365-2761.2011.01279.x. [DOI] [PubMed] [Google Scholar]
- 83.Ciche TA, Ensign JC. 2003. For the insect pathogen Photorhabdus luminescens, which end of a nematode is out? Appl Environ Microbiol 69:1890–1897. doi: 10.1128/AEM.69.4.1890-1897.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bellieny-Rabelo D, Tanui CK, Miguel N, Kwenda S, Shyntum DY, Moleleki LN. 2019. Transcriptome and comparative genomics analyses reveal new functional insights on key determinants of pathogenesis and interbacterial competition in Pectobacterium and Dickeya spp. Appl Environ Microbiol 85:e02050-18. doi: 10.1128/AEM.02050-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Russell AB, Hood RD, Bui NK, LeRoux M, Vollmer W, Mougous JD. 2011. Type VI secretion delivers bacteriolytic effectors to target cells. Nature 475:343–347. doi: 10.1038/nature10244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Skorupski K, Taylor RK. 1996. Positive selection vectors for allelic exchange. Gene 169:47–52. doi: 10.1016/0378-1119(95)00793-8. [DOI] [PubMed] [Google Scholar]
- 87.Marvig RL, Blokesch M. 2010. Natural transformation of Vibrio cholerae as a tool - optimizing the procedure. BMC Microbiol 10:155. doi: 10.1186/1471-2180-10-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Watve SS, Bernardy EE, Hammer BK. 2014. Vibrio cholerae: measuring natural transformation frequency. Curr Protoc Microbiol 2014:6A.4.1–6A.4.12. doi: 10.1002/9780471729259.mc06a04s35. [DOI] [PubMed] [Google Scholar]
- 89.The UniProt Consortium. 2021. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res 49:D480–D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed]
- 90.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 92.Lefort V, Longueville JE, Gascuel O. 2017. SMS: smart model selection in PhyML. Mol Biol Evol 34:2422–2424. doi: 10.1093/molbev/msx149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lemoine F, Correia D, Lefort V, Doppelt-Azeroual O, Mareuil F, Cohen-Boulakia S, Gascuel O. 2019. NGPhylogeny.fr: new generation phylogenetic services for non-specialists. Nucleic Acids Res 47:W260–W265. doi: 10.1093/nar/gkz303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.O'Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, Astashyn A, Badretdin A, Bao Y, Blinkova O, Brover V, Chetvernin V, Choi J, Cox E, Ermolaeva O, Farrell CM, Goldfarb T, Gupta T, Haft D, Hatcher E, Hlavina W, Joardar VS, Kodali VK, Li W, Maglott D, Masterson P, McGarvey KM, Murphy MR, O'Neill K, Pujar S, Rangwala SH, Rausch D, Riddick LD, Schoch C, Shkeda A, Storz SS, Sun H, Thibaud-Nissen F, Tolstoy I, Tully RE, Vatsan AR, Wallin C, Webb D, Wu W, Landrum MJ, Kimchi A, et al. 2016. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 44:D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
TpeV Phobius transmembrane helix prediction. The amino acid sequence of TpeV was analyzed using the Phobius transmembrane predictor. Download FIG S1, TIF file, 0.9 MB (908.2KB, tif) .
Copyright © 2021 Crisan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
TpeV SignalP prediction. The amino acid sequence of TpeV was analyzed using the SignalP 5.0 transmembrane predictor. Download FIG S2, TIF file, 1.6 MB (1.6MB, tif) .
Copyright © 2021 Crisan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
CCCP disrupts the membrane potential of E. coli cells. E. coli cells harboring either plasmid control or periplasmically delivered TpeV were first incubated with CCCP and then with the membrane potential-sensitive DiBAC4(3) dye. Fluorescence readings were taken at an excitation ƛ = 490 nm and emission ƛ = 516 nm. A one-way ANOVA was used to determine significance. NS, not significant. Download FIG S3, TIF file, 0.9 MB (876.8KB, tif) .
Copyright © 2021 Crisan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Complete list of all identified TpeV protein homologs using the PHMMER algorithm. Download Table S1, XLS file, 0.05 MB (50.5KB, xls) .
Copyright © 2021 Crisan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
V. cholerae strains from diverse geographical locations harbor tpeV-tpiV modules. Download Table S2, DOCX file, 0.02 MB (16.1KB, docx) .
Copyright © 2021 Crisan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The Aux 4 paar gene is not required for TpeV-mediated killing. Target V. cholerae BGT49 ΔtpeVΔtpiV (CC170) was cocultured with either Δpaar (CC179), WT, ΔvasK (T6SS−, CC168), or ΔtpeV (CC167) BGT49. Download FIG S4, TIF file, 1.8 MB (1.8MB, tif) .
Copyright © 2021 Crisan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
TpiV SignalP prediction. The amino acid sequence of TpiV was analyzed using the SignalP 5.0 transmembrane predictor. Download FIG S5, TIF file, 1.6 MB (1.6MB, tif) .
Copyright © 2021 Crisan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Strains and plasmids used in this study. Download Table S3, DOCX file, 0.02 MB (18.8KB, docx) .
Copyright © 2021 Crisan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.