Abstract
In vivo proton magnetic resonance spectroscopy and spectroscopic imaging (MRS/MRSI) are valuable tools to study normal and abnormal human brain physiology. However, they are sensitive to motion, due to strong crusher gradients, long acquisition times, reliance on high magnetic field homogeneity, and particular acquisition methods such as spectral editing. The effects of motion include incorrect spatial localization, phase fluctuations, incoherent averaging, line broadening, and ultimately quantitation errors. Several retrospective methods have been proposed to correct motion-related artifacts. Recent advances in hardware also allow prospective (real-time) correction of the effects of motion, including adjusting voxel location, center frequency and magnetic field homogeneity. This article reviews prospective and retrospective methods available in the literature and their implications for clinical MRS/MRSI. In combination, these methods can attenuate or eliminate most motion-related artifacts and facilitate the acquisition of high-quality data in the clinical research setting.
Keywords: MRS, MRSI, motion, prospective correction, retrospective correction, navigated spectroscopy sequence
Introduction
Proton (1H) magnetic resonance spectroscopy (MRS) is a widely used non-invasive method to measure levels of endogenous metabolites and neurochemicals in the human brain [1]. 1H-MRS provides clinicians and researchers with a powerful tool to study disease and perform longitudinal studies of disease progression. Prior to the emergence of MRS, neurometabolic or neurochemical measurements were generally limited to analysis of the cerebrospinal fluid (CSF), which provides an indirect measure of the chemical milieu of the brain. Although CSF measurements are precise and provide an abundant amount of information [2,3], they cannot be attributed to a particular region of the brain or used to evaluate localized metabolism or regional pathology. In vivo 1H-MRS in the human brain circumvents these shortcomings and allows the quantitative localized evaluation of metabolites associated with the brain tissue of interest or particular cell types (e.g., neurons or glia). Metabolite levels may reflect normal and abnormal brain development [4–7], brain diseases and disorders [8–12], and drug-mediated effects [13–15]. A number of chemical compounds can be measured from a 1H spectrum from the brain. These neurochemicals include glucose, glycerol, scyllo-inositol, ethanol, 2-hydroxyglutarate, phenylalanine, N-acetylaspartate (NAA), choline-containing compounds (Cho) such as glycerophosphocholine (GPC) and phosphocholine (PCh), creatine-containing compounds or total creatine (tCr), including phosphocreatine (PCr) and creatine (Cr), Myo-Inositol (MI), N-acetylaspartylglutamate (NAAG), lactate (Lac), γ-aminobutyric acid (GABA), ascorbate (Asc), aspartate (Asp), glutathione (GSH), glutamate (Glu), and glutamine (Gln) [1,16–19].
1H-MRS can be performed using single-voxel (SV) or spectroscopic imaging (SI) sequences. Commonly used SV localization sequences are stimulated echo acquisition mode (STEAM) [20], point resolved spectroscopy (PRESS) [21], spin-echo full-intensity acquired localized spectroscopy (SPECIAL) [22], and (semi-) localization by adiabatic selective refocusing (semi-LASER and LASER) [23–25]. Almost all SV spectroscopy (SVS) sequences use orthogonal slice-selective radio-frequency (RF) pulses to localize a region of interest (voxel), which typically ranges between 5 and 30 cc. Magnetic resonance spectroscopic imaging (MRSI) sequences allow measurements of metabolites from multiple voxels simultaneously, commonly as small as 1 cc. MRSI involves signal excitation of a large brain region in combination with spatial-encoding gradients applied in two or three directions.
Signals from some molecules of interest, such as GABA and GSH, are overlapped by signals from other metabolites with a much higher concentration (e.g., tCr and Cho). Spectral editing methods, such as Mescher-Garwood (MEGA-) PRESS/SPECIAL/semi-LASER/LASER [26–29], exploit known J-coupling relationships within molecules of interest to separate their signals from the overlying signals. Metabolites that benefit from spectral editing include GABA, GSH, Asc, Lac, NAAG, and 2-hydroxyglutarate (2HG). The current status and applications of 1H-MRS, using edited and non-edited SVS and MRSI sequences, have been reviewed extensively [30–36]. For a basic understanding of MRS, challenges related to clinical MRS, post-processing of MRS data, and issues about spectral quality and quality assessment, readers may refer to the following published review articles [37–39].
In order to attain spectra with sufficient signal-to-noise ratio (SNR) and acceptable quality for quantification, relatively long MRS/MRSI scan durations are commonly needed, ranging from a few minutes for unedited SVS, approximately 10 minutes for edited SVS, and 20–30 minutes for MRSI (at 3T). Therefore, the spectral peaks commonly show degraded (increased) wide line widths, due to some degree of subject motion, especially in incompliant subjects, children [40], the elderly, and patients with movement disorders [41]. The presence of strong crusher gradients (which support signal localization) sensitizes pulse sequences to motion. Subject motion during MRS scans can have adverse effects on MR spectra beyond broader spectral lines, such as insufficient water suppression, reduced SNR [42], lipid contamination and baseline fluctuation [43], and incorrect quantification [39,44]. Even when MR spectra contain no visible artifacts, subject motion can lead to measurements at a brain region different from the region of interest, and hence reduce the specificity of the data acquired. Therefore, it is desirable to have methods to correct for subject motion during MRS/MRSI studies.
This paper reviews the various mechanisms by which motion can interfere with the integrity of MRS measurements and summarizes a variety of prospective (real-time) and retrospective (offline) techniques to correct errors due to subject motion. These are largely presented from the viewpoint of PRESS-localized SVS, but the general principles apply to other MRS or MRSI sequences. This review will not cover the all available methods that can minimize subject motion, of which full advantage should be taken.
Overview
Table 1 provides an overview of the various effects of subject motion during MRS scans. Subject movements can alter the sampled brain region, and change the transmitter frequency (F0), main magnetic field (B0) homogeneity, and RF field (B1) strength, as the brain moves relative to the stationary scanner (and RF) reference frame. In turn, these effects can result in incorrect spatial localization, line broadening or signal losses and interfere with frequency-selective water suppression and/or editing pulses. Most effects are dependent upon the degree of displacement of the brain. Some motion-related artifacts are identifiable visually. For instance, larger changes in the head position may lead to improper voxel localization, such that readily identifiable large lipid signals from the skull or scalp dominate a spectrum. However, smaller motions may cause subtle alterations in the final spectrum that are difficult or impossible to identify with typical quality assurance measures. For instance, continuous small head movements may cause signal attenuations and apparent changes in metabolite concentrations if uncorrected. In general terms, motion impacts MRS data in three distinct ways: 1) localization errors; 2) frequency and phase shifts; and 3) decreased field homogeneity. These are discussed separately in the next few paragraphs.
Table 1:
Overview of the various effects of head movements on PRESS-localized MRS acquisitions.
| Cause | Specifics | Effect | Correction Method |
|---|---|---|---|
| Position changes | Pose | Incorrect spatial localization | Adaptive motion correction |
| Intra-scan translations | Translational velocity | Phase shifts | Phase-navigator |
| Intra-scan rotations | Rotational velocity | Unbalanced gradient moments and signal attenuation | Amplitude navigator |
| Change in frequency | Pose | Line (frequency) shift | Frequency navigator |
| Change in shim due to pose change | Pose | Line broadening | Dynamic shimming |
A “pose” refers to the six parameters for a rigid body transformation (three translations and three rotations). B1: RF field strength
Motion-induced localization errors
MRS data acquired from a moving subject may be attained from a region other than the region of interest and thereby provide inaccurate metabolite measurements. These errors depend upon the degree of spatial variability in metabolites of interest, and are generally more pronounced for small anatomical targets. Importantly, motion during MRS scans may not cause distinct artifacts and may go unnoticed during quality control (QC). Motion-induced localization errors can be eliminated with prospective motion correction (PMC) methods, which provide real-time tracking of head movements and dynamic updates of voxel positions. PMC approaches have been demonstrated for both MRI [45,46] and MRS (and MRSI), using optical cameras [43,47] or image-based navigators [42,48]. The type of motion in the human body can either be rigid (translation and rotation) or non-rigid (involving scaling or shearing). Since this review focuses on the brain, it is assumed that only rigid-body motion occurs, which is characterized by 6 parameters: three translations and three rotation angles. Here, we will refer to the six parameters of a rigid body transformation as “Pose”. Changes in pose during a scan are described as velocities, and it is helpful to separately consider translational and rotational velocities.
Motion-induced frequency and phase shifts
Motion during a scan commonly changes the F0 and zero-order phase of individual FIDs. For all MRS sequences, these shot-to-shot fluctuations can result in degraded spectral quality if signal averaging is performed on uncorrected transients. Many post-processing methods have been proposed for frequency and phase correction of single-voxel data (see below). However, for phase-encoded MRSI data, such phase fluctuations are more challenging to address. Motion-induced changes in F0 can also cause problems for methods that rely on frequency-selective RF pulses, such as water suppression modules or spectral editing approaches. These methods may benefit from a prospective field-frequency lock [49,50], which is most commonly applied for spectral editing methods.
Motion-induced decreases in B0 field homogeneity
In addition to frequency shifts, head motions may degrade B0 homogeneity across the acquisition volume. Poor shimming (i.e. poor B0 homogeneity) is associated with broadened metabolite signals, increased signal overlap, and ultimately biased quantification of metabolites. Even with prospective motion-corrected MRS or MRSI sequences, moderate head movements can degrade spectral quality [51,52] and necessitate prospective shimming to produce high-quality spectra [48,53].
Theory of intra-scan motions
Interaction between rigid-body motion and gradients
Many of the motion effects summarized above are caused by changes in the pose of the head across a given MRS study (e.g. over multiple TR periods). However, faster movements can also change the evolution of the spin phase within a single TR period, i.e. between excitation and signal readout. The current section describes the effects of such “intra-scan motions”. These faster movements result in a dynamic situation where spins are changing their position while simultaneously magnetic field gradients are being switched. We assume that the object to be scanned (brain) undergoes a continuous rigid body motion. The issue of motion during MR acquisitions has been studied before [54]. However, since many MR spectroscopists may be unfamiliar with the effects of intra-scan motions on the spin system, we briefly summarize these effects.
For rigid body motion, the positional state of the object at a time t is determined by 6 parameters; 3 for translations and 3 for rotations. More specifically, we represent the translational state of the object at time t by the 3-dimensional vector . We further assume that the spin system is excited at t = 0, and that (0) = (0 0 0). Likewise, we represent the rotational state of the object at time t by the 3 × 3 rotation matrix R(t), and assume that R(0) = .
Next, we describe the motion of a single spin that is at position at time t = 0. The spin will continuously change its position along with the entire rigid object. We chose a particular mathematical representation where we first apply a rotation matrix and then a translation; therefore the position (t, ) of this spin (a 3-D vector) at time t can be calculated as follows:
| [1] |
Of note, one may choose other mathematical representations of the trajectory, for instance, first apply the translation and then the rotation, but parameterization [1] is the most convenient and most common one.
Simultaneously with the objects’ rigid body movement, the magnetic field gradients of the MR scanner change continuously. We represent the time course of the gradients by the time-dependent 3-dimensional vector (t).
The changing position of each spin interacts with the time-dependent gradient field (t), which induces phase errors during intra-scan motions. Therefore, we calculate the phase evolution of the spin originally at position . In the rotating frame (at Larmor frequency), the frequency experienced by this spin at time t is determined by the scalar product of its position with the gradient field vector:
γ is the gyromagnetic ratio Substituting Eq. [1] yields:
| [2] |
The incremental phase change dφ(t, ) at time t associated with the frequency f(t, ) during the interval dt is:
| [3] |
We can then calculate the total phase φ(T, ) accumulated at time T by the spin originally at position , by integrating Eq. [3] over time:
| [4] |
Substitution of Eqs. [2] and [3] into [4] yields
| [5a] |
| [5b] |
Therefore, the effect of rigid body motion on the phase of a single spin can be split into that of a pure translation [5a; phase is dependent on translational trajectory (t)] and that of a pure rotation [5b; phase is dependent on rotational trajectory R(t)]. Of note, these two effects do not interact, due to the linearity of rigid body motion and due to the particular parameterization of the motion chosen in Eq. [1]. In the next two sections, we will assess the effects of translational and rotational motions on an ensemble of spins that are spatially distributed across a rigid object of interest.
Effect of intra-scan translational motion on MR spectra
The effects of the intra-scan translational component (t) of motions on the phase is described by the first term of Eq. [5a]. We note that this term is independent of . Consequently, translations induce the same phase shift for all spins independently of their original position at time t = 0, i.e. intra-scan translational movements cause a global phase shift. Therefore, for intra-scan translational movements, if S(ω) denotes an ideal noiseless spectrum (with ω as frequency), then the spectrum of average k (1 < k < Naverages) is:
| [6] |
with nk(ω) being the spectral noise of shot k and φk denoting a translation-dependent global phase for shot k as per Eq. [5a].
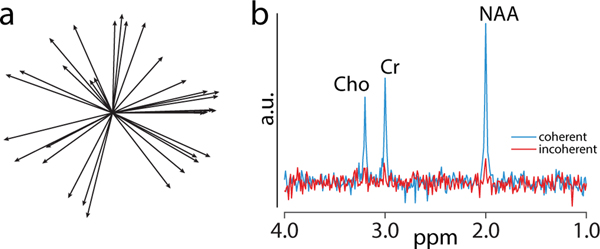
For instance, Figure 1a shows a plot of 32 water signal vectors in the complex plane during simulated head tremor motion. A volunteer rapidly shook his head up-and-down (frequency ~1 Hz and amplitude approximately a few degrees), while a non-water-suppressed acquisition was performed in the mid-frontal gray matter, resulting in rather high velocities but minimal net head offsets. The phases of the vectors are random, leading to almost complete signal loss if the signals are averaged without phase correction (simulated incoherent averaging; red spectrum in Figure 1b). Eq. [6] suggests that the solution to the problem is to determine the global phase φk for each shot from the raw data and reverse the phase from each shot prior to averaging (simulated coherent averaging; blue spectrum in Figure 1b). The phase can be determined using signals from major metabolites, such as Cr [55], or under-suppressed and/or over-suppressed water [56]. The coherently averaged spectrum in Figure 1b was simulated using the Lorentzian function in Gannet [57] with the following parameters: NAA/Cr/Cho areas and positions 1.3/0.8/0.7 a.u. and 2.0/3.0/3.2 ppm; phase 0 radians; linewidth at full-width half-maximum ~2 Hz; and random (background) noise generated using the randn function with the mean 0 a.u. and standard deviation 4 a.u. For the incoherently averaged spectrum, the same simulation parameters were used except amplitude and phases, which were estimated from the vectors (length and phase) in Figure 1a and incorporated in the Lorentzian function.
Figure 1:
a) Plot of water signal vectors during head motion. Each solid line represents the water signal of a single FID in the complex plane (total 32 FIDs). The length of some of the vectors is decreased (1a), probably due to motion-induced attenuation during fast movements; b) MR spectrum from coherent (blue) and incoherent averaging (red). When using the individual signals represented in 1a (incoherent averaging), the phase of the individual FIDs is random, leading to incoherent averaging.
Effect of intra-scan rotational motion on spin phase
The effects of the intra-scan rotational component (t) of motions on the spin phase is described by the second term of Eq. [5b]:
| [5b] |
Due to the orthogonality of the rotation matrix, this can be rewritten as:
| [7] |
Therefore, the effect of the rotational component R(t) on the phase is linearly dependent upon the original spin position .
The term under the integral has the unit of a gradient moment (mT/m · ms). If no intra-scan rotations occur, i.e. R(t) = 1 for all t, then the integral yields the zero-order gradient moment for stationary spins. This zero-order gradient moment is zero for typical MRS sequences, since gradients are balanced. However, as per Eq. [7], rotational intra-scan movements modify the gradient moment in the form
| [8] |
where Aeff defines an “effective gradient moment” due to rotational motion. Therefore, intra-scan rotations cause unbalanced zero-order gradient moments. Non-zero gradient moments alter the phase of individual spins within a voxel as a function of their original position (as per Eq. [7]), leading to incoherent averaging across the volume of interest. Ultimately, this can lead to signal attenuation for individual FIDs, especially due to the relatively large dimension of MRS voxels. This effect is visible in Figure 1a, as a decrease in length of some of the magnetization vectors.
Changes in voxel position: primary effects and prospective motion correction
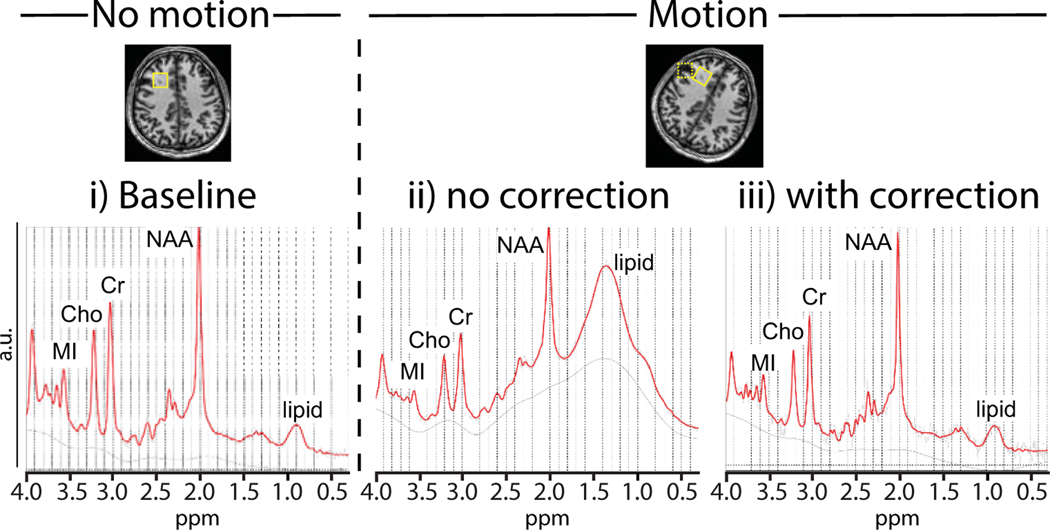
During an MRS scan, the localization volume is defined by slice-selective gradients and RF frequency offsets in the scanner coordinate frame. Motion of the head relative to the fixed location of the prescribed volume results in sampling the incorrect region of interest. This can lead to lipid artifacts (as in Figure 2), as well as a mismatch between the data acquired and the intended sampling volume. Such localization errors can be eliminated by using prospective motion correction (PMC) techniques, which were recently reviewed by Maclaren et al. [46]. PMC methods involve a two-step approach: 1) continuously measure the head position and rotation; and 2) use the tracking information to dynamically adjust scan planes such that they are locked relative to the mobile head reference frame [58]. Motion tracking can be achieved using navigator scans [59–61], MR images [58], or external tracking devices, including optical techniques [62,63]. 1H-MRS is well suited to imaging navigators because of its relatively long repetition times (TRs 1.5–3 s), a substantial portion of which are needed for recovery of longitudinal magnetization and is available for navigator acquisition. SV 1H-MRS also particularly benefits from PMC due to its lack of anatomical information.
Figure 2:
In vivo PRESS-localized data with i) baseline acquisition with no intentional motion performed in frontal white matter (solid box); ii) motion without correction shifted the voxel and intersected with the scalp (dashed box); and iii) motion with correction using motion navigators.
MRI-based motion tracking was initially implemented for 3D MRI [64]. For instance, three orthogonal, low-flip angle, low-resolution spiral navigator scans allow estimation of translations and rotations in real-time. For use with localized MRS, the PMC module is played out once per TR period prior to the water suppression block, and pose updates are applied to the three subsequent slice-selective RF pulses defining the MRS voxel [52]. Figure 2 shows a frontal white matter spectrum acquired at rest (2i; baseline), with head motion but no PMC (2ii), and with head motion plus motion correction (2iii). PMC (2ii, solid box) eliminated the strong lipid signal that occurred in the uncorrected scan when the stationary MRS voxel intersected with the skull or scalp (dashed box) during subject movements (2iii versus 2ii). A similar approach using echo-planar imaging based (EPI) volume navigators (vNavs) [42] improved data quality and line widths in MRS scans of 59 school-aged children [65].
Several groups have used external tracking for the PMC of MRS scans. Zaitsev et al. [66] used a stereovision system to track head motion via a marker mounted on a bite-bar, and then continuously adjusted MRS voxel poses. The implementation was based on a PRESS sequence and included additional interleaved reference scans (IRS) of the water signal. In three volunteers, moderate head movements (approximately 5° rotations) resulted in lipid-artifacts that were eliminated when PMC was enabled. Use of IRS water signals to perform shot-to-shot phase and frequency corrections improved spectral line widths and SNR. However, PMC plus IRS did not fully recover these two measures of spectral quality compared to scans without motion. The incomplete recovery was most likely due to the rather large movements, which changed the B0 homogeneity (shim) within MRS voxels.
Another study used an in-bore camera and optical marker for tracking head motion [51]. Five subjects performed two types of motion during short-TE PRESS acquisitions, with and without PMC. In a first set, the voxel was placed near the top of the brain, and subjects were instructed to slide along the bore 10–15 mm during the acquisition. Without PMC, this large z-translation resulted in substantial lipid artifacts, since some of the individual FIDs were acquired from the skull or scalp. PMC nearly eliminated these artifacts. In a second set, the voxel was placed in the medial frontal gray matter, and volunteers were trained to tilt their heads towards the left. Without PMC, this z-rotation effectively moved the sampling volume from gray matter to white matter. In turn, this alteration in voxel composition increased the choline-to-creatine (Cho/Cr) ratio by 15%. Conversely, when PMC was enabled, these z-rotations did not cause significant changes in Cho/Cr across the five subjects (mean change 1%). Overall, these findings suggest that PMC not only reduced the presence of significant lipid artifacts but also ensured proper voxel composition (e.g., gray versus white matter) during head movements.
Finally, using a similar optical tracking system and marker, Deelchand et al [67] performed prospective motion and shim correction in the human brain using the semi-LASER sequence at 7T; more details are described below (in the section on Dynamic updates of frequency and first-order shims) [67].
Secondary effects of changes in voxel position
As noted above, prospective motion correction eliminates errors in voxel location by maintaining the pose of the MRS voxel relative to the mobile head reference frame. However, changes in head pose relative to the stationary scanner reference frame may lead to secondary alterations in F0, shim (B0 homogeneity), and B1-field amplitude. These changes are dependent upon the magnitude of displacement induced by subject motion. Various techniques have been reported and implemented to correct for these errors (summarized in Table 1).
Alterations in resonance frequency (F0)
When PMC is enabled, changes in the voxel position relative to the scanner reference frame alter the B0 field at the voxel, and consequently F0. However, for small head movements, the resulting frequency changes may be negligible. Specifically, as long as the frequency changes (Δf) are much smaller than the bandwidth of chemical-shift selective (fcs) water suppression RF pulses (), frequency alterations do not interfere with the acquisition of non-edited spectra, and can be corrected shot-by-shot during spectral reconstruction, using water navigators. However, editing pulses require higher F0 stability in order to ensure efficient and consistent editing of the metabolites during acquisition. Therefore, in the presence of motion and/or scanner-induced B0 instabilities, the scanner F0 needs to be updated dynamically to ensure proper functioning of chemical-shift selective and editing pulses.
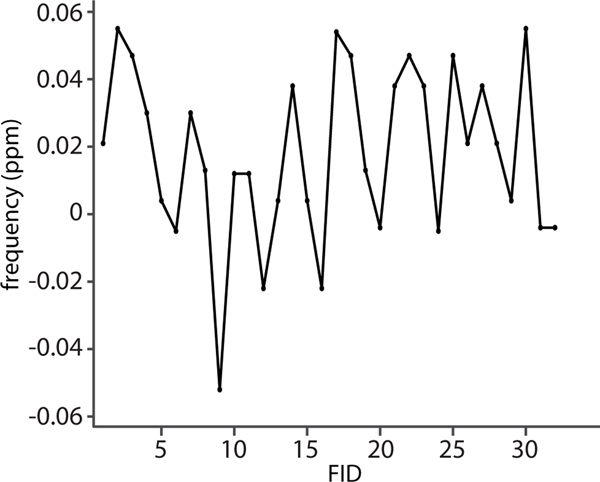
For instance, Figure 3 shows changes in the water frequency for a frontal gray matter voxel (anterior cingulate) in a subject who moved his head up and down by about 10 mm (“nodding”). This moderate movement induced frequency shifts of about ± 0.06 ppm (~15 Hz at 3T) peak-to-peak, consistent with previously reported changes [68]. Since larger displacements are likely to cause greater changes in F0, it is necessary to adjust F0 continuously during head movements. Additionally, dynamic updates in the linear shim settings, as described below, will change the F0 for off-center voxels; however, these changes can be predicted and updated accordingly.
Figure 3:
Changes in water frequency of a mid-frontal voxel during continuous up-down head movements (“nodding”) of about ±10 mm translation and ±10° rotation for 32 individual FIDs. The nodding caused changes in the water frequency of approximately ± 0.06 ppm
Finally, there is a subtle secondary effect of motion-induced frequency changes, in that frequency changes alter the exact position of an MRS voxel. However, as noted, head movements typically change the frequency by 10s of Hz, which is much lower than the bandwidth of slice-selective RF pulses used to define MRS voxels (typically kHz). Consequently, frequency-induced changes in voxel position may amount to a few percent of the voxel size only, which may be negligible. Furthermore, dynamic adjustment of F0 will completely eliminate these residual localization errors.
Alterations in B0 homogeneity
Due to the narrow range of the chemical shift in 1H spectra, the B0 homogeneity is optimized, or “shimmed”, prior to each MRS/MRSI acquisition to minimize the line width of the spectral peaks. These adjustments always include the linear shim terms, as well as higher-order shim terms on some systems. Shimming can be achieved by acquiring a B0 field map, and then fitting the shim terms to minimize the residual field variation across the voxel [69,70]. Another approach involves the acquisition of a series of spatial projections through a given voxel, from which the correct shim settings can be calculated [71,72]. However, due to the relatively small voxel size of typical SVS acquisitions (~20-mm voxel linear dimensions) relative to the whole brain, most of the improvement in field homogeneity and line widths is typically achieved by optimizing the linear terms. It is worth noting that the use of non-linear shim terms to maximize within-voxel homogeneity can result in more rapid field changes outside the voxel and make F0 more sensitive to motion.
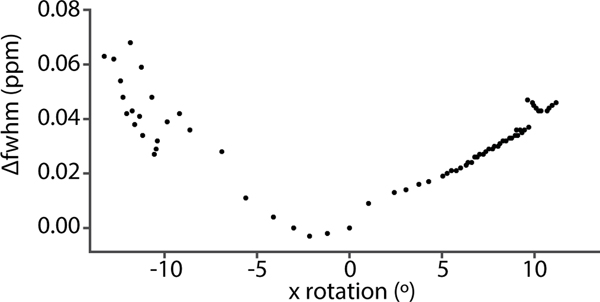
During prospective motion correction, as the MRS voxel tracks a subject’s head movement, its position changes relative to the scanner reference frame. This change causes an alteration in the magnetic field distribution within the voxel, due to changes in the B0 field of the scanner, as well as changes in the induced field distribution in the brain as a result of head rotations [73]. One study demonstrated that clinically realistic head rotations (~5°) induced spatially approximately linear field alterations, with a slope of about 0.01 ppm/cm in the center of the brain [68]. Likewise, Figure 4 demonstrates that head rotations of 10° increase metabolite line widths by ~0.04 ppm in a medial frontal gray matter voxel, which is undesirable since it would approximately double the typical line widths.
Figure 4:
Changes in water line width as a function of x-rotation (“nodding”) during an MRS scan of a mid-frontal voxel. Prospective motion correction was enabled to ensure the voxel was locked relative to the moving brain. Δfwhm: change in full-width half-maximum.
However, more severe B0 inhomogeneities occur between the brain tissue and air from the nasal and auditory passages, causing localized B0 distortions in the frontal and temporal lobes [74]. Subject motion can alter these distortions, leading to frequency and non-linear shim changes. Figure 1 of Hess et al. [75] demonstrates these changes as a result of left-right or up-down head motions. These movements caused varying frequency and shim changes in the brain, especially in frontal, temporal, and parietal regions. These motion-induced changes can affect MRSI studies, and especially edited MRSI studies, more severely than SVS acquisitions. More specifically, each MRSI voxel experiences unique alterations in B0 homogeneity and F0, and consequently in water suppression, spectral quality, and apparent metabolite concentrations. In the case of spectral editing, the editing efficiency is worsened and varies among voxels in MRSI data, resulting in sub-optimal editing of metabolites, subtraction artifacts, baseline distortion, and ultimately erroneous metabolite levels [29].
To counteract these motion-induced effects, many real-time motion correction approaches continuously measure and update the first-order (linear) shim settings (see below). Conversely, the higher-order shim terms are usually not updated dynamically for the following reasons: 1) the absence of eddy-current compensation associated with the terms and relatively slow updates; 2) B0 changes during head movements are predominantly linear (within single voxels) [68]; and 3) the higher-order shim settings require the acquisition of multiple projections [71,72], which would be difficult to accomplish within one TR.
Alterations in B1 strength
Changes in the voxel position can also cause alterations in the B1-field strength, both during transmission and signal detection. The changes in the field strength occur because the position of the voxel relative to the RF coils changes, leading to alterations in the signal amplitude. However, to our knowledge, there are no published MRS studies to correct motion-induced B1 changes, possibly since these might become relevant only for large movements.
Dynamic updates of frequency and first-order shims
As noted, while PMC techniques provide efficient motion tracking and correction and eliminate localization errors, head movements can alter B0 homogeneity (shim) across MRS voxels as well as induce frequency offsets. To address these issues, Keating et al. [76] proposed a method for rapidly mapping linear variations in B0 across a small volume using two-dimensional excitations. The technique was integrated into a PMC-enabled PRESS sequence and successfully performed real-time correction of both frequency and first-order shim terms. Hess et al. [77] added a pair of echo-planar imaging (EPI) volume navigators (vNavs) into a PRESS sequence to correct in real-time the volume of interest, frequency, and first-order shim terms. The navigator minimally affected the signals of metabolites with high concentration and maintained spectral quality when a subject moved during the scan. EPI vNavs have been implemented with 2D- [75] and 3D-LASER-MRSI [78] sequences. By applying real-time motion and shim corrections, the navigated MRSI sequences yielded data with good spectral quality in the presence of scanner and subject instabilities compared with the non-navigated sequences. Lastly, Deelchand et al. [67] used optical tracking and FASTMAP-like navigators to perform prospective motion and shim correction in the human brain using the semi-LASER sequence at 7T. Again, in the presence of large movements, motion correction alone was not sufficient to prevent degradation of data quality, but data quality remained excellent with simultaneous motion and shim correction [67].
Prospective frequency- and B0-correction techniques are important for spectral editing sequences because they ensure that fcs pulses are applied correctly at the editing target (e.g., 4.56 ppm for GSH editing) and minimize spectral interference with co-edited signals (e.g., GSH-edited signal at 2.95 ppm and co-edited NAA signal at 2.6 ppm) [79]. Bogner et al. [48] used EPI vNavs in an MRSI-MEGA-LASER sequence for GABA+ (GABA + macromolecules MM) editing, and performed real-time corrections along with selective reacquisition of data [80]. If the head translation or rotation between edit ON and OFF scans exceeded a threshold, the affected pair was discarded and immediately reacquired. As a result, the navigated sequence attained 3D GABA images with higher SNR and lower subtraction errors than the unnavigated sequence. Some editing sequences apply fcs pulses spectrally close to each other to reduce overlapping signals. For instance, fcs pulses are applied symmetrically at about 1.7 ppm (i.e., 1.9 ppm in one TR and at 1.5 ppm in another TR) to reduce the MM contamination in the GABA-edited signal at 3 ppm [81]. Similarly, to separate NAAG from NAA at 3T, fcs pulses are applied symmetrically at about 4.49 ppm (i.e., 4.61 ppm in one TR and at 4.38 ppm in another TR) [82]. These editing methods require high F0 and B0 stability to ensure the reliable measurement of edited metabolites. Accordingly, Saleh et al. [83] incorporated EPI vNavs into the MEGA-SPECIAL sequence to correct for motion, F0 and B0 instabilities in real-time during the MM-suppressed GABA editing [83]. The navigated sequence yielded spectra with superior spectral quality and reduced subtraction artifacts relative to scans with no motion correction, even in the presence of large movements. This sequence also yielded reproducible measurements of MM-suppressed GABA using three different analytical software approaches [84]. Another method to remove MM from GABA+ is through subtracting MM attained from a separate metabolite nulled scan using an inversion pulse prior to MEGA-edited sequences [85,86]. However, this method results in greater sensitivity to subject and scanner instabilities because of the long acquisition times and subtraction of two non-interleaved measurements [32]. To address these instabilities, Moser et al. [87] prospectively corrected for subject motion and scanner instabilities using a volumetric navigated MRSI sequence at 7T, MEGA-semi-LASER with 2D phase-encoding gradients, with the selective reacquisition of corrupted data [80], and with and without an inversion pulse. The navigated sequence performed efficient MEGA-editing and yielded high-quality maps of GABA+ (without the inversion pulse), MM (with the inversion pulse), and GABA in the human brain. Recently developed multiplexed editing methods apply multiple fcs pulses to simultaneously edit multiple metabolites (with negligible crosstalk) [88–90], along with the separation of NAA and NAAG and the reduction of MM [91] in a single MR examination. Integration of prospective motion, F0, and B0 correction techniques with multiplexed editing methods would likely increase the reliability and reproducibility of metabolite measurements.
Retrospective correction
Although prospective motion, F0, and B0 correction modules are beneficial to attain data with higher SNR and narrower line widths, they may not be able to adequately correct all shot-to-shot frequency and phase errors in the MRS data, either due to the presence of non-rigid body physiological motions (e.g., cardiac and respiratory motion), head movements at higher velocities, or errors in real-time adjustments. Also, the resolution of MR scanner software may not be able to handle frequency updates in a non-integer format, which may result in cumulative frequency errors. Felbliger et al. [44] proposed ECG-gated sequences to reduce phase dispersions related to cardiac motion; however, group differences in heart rate caused TR to vary, and as a result, introduce systematic differences in signal saturation (e.g., T1 effects). Fortunately, artifacts from physiological motion or errors in correction modules can be reduced by correcting frequency and phase errors in every transient before signal averaging using retrospective (post-processing) methods. These methods become more essential for clinical MRS studies acquired without prospective-correction modules.
As described above, an important effect of head motion is motion-induced variation in the phase of MRS signals from shot to shot, which may lead to reductions in signal-to-noise ratio due to incoherent averaging [92]. One strategy is to use the phase of the water signal to restore phase coherence during signal averaging [93]. The water signal may also be used to correct frequency variations in the metabolite signals, resulting in improved (reduced) line widths in the final spectrum [93]. Since optimal water suppression may result in a residual water signal that is too small or too unreliable to provide a valid reference signal, the use of weak water suppression is advantageous [94]. Of note, since the residual water signal in partially-suppressed acquisitions mirrors the evolution of metabolite spins during the pulse sequence, starting with longitudinal magnetization prior to excitation, the partially suppressed water truly reflects the effect of subject motion as per Eq. 5a and 5b. Alternatively, the unsuppressed water signal [95,96] might be used as a reference; however, the resulting spectra may show poor baseline definition due to the extremely large water signal amplitude.
Thiel et al. [97] proposed the use of a second PRESS acquisition with a low excitation flip angle and no water suppression immediately after each MRS acquisition; the resulting water signal can be used for shot-to-shot phase and frequency correction. Alternately, Ernst et al. [56] proposed a water-cycling scheme where the residual water is alternated between under-suppressed and over-suppressed from shot-to-shot. The residual water provides a robust phase and frequency reference for each acquisition, while simultaneously eliminating residual water in the final (summed) spectrum, resulting in a well-defined baseline.
Another method that does not rely on water referencing is based on fitting the Cr signal at 3 ppm to a Lorentzian function, from which the frequency and phase information can be estimated [55]. This method and its variants [55] have been implemented successfully, but they rely strongly on the SNR of the single-shot Cr signal and are therefore not applicable to every MRS application. Near et al. [98] proposed a “Spectral Registration” method, which involves fitting each spectral transient to a reference scan in the time domain by adjusting the frequency and phase terms. The reference scan could either be the average of transients or the first transient. One significant benefit of this method is that it does not rely on the collection of any navigator echoes or the presence of any specific resonances such as the residual water or Cr. Wilson [99] developed a new method “Robust Alignment to a Target Spectrum” (RATS), which aligns individual transients to a reference spectrum in the frequency domain using variable projection (VARPRO) [100,101] with baseline modeling. RATS offers improved accuracy and stability for large frequency shifts and unstable baseline.
Edited spectra again present unique challenges. They require the subtraction of large metabolite signals to reveal the underlying small metabolite signals, e.g., subtraction of Cr to reveal edited-GABA and edited-GSH signals. Post-processing edited MRS data is essential to reduce subtraction artifacts in difference spectra. Although several methods have been developed to address the alignment of sub-spectra [55,94,102,103], the optimal strategy depends on the metabolite edited. For example, GSH-edited sub-spectra cannot be aligned using the residual water signal because it is suppressed during the GSHON step (at 4.56 ppm). Similarly, GABA-edited data may not be well aligned using Cr in the final GABA-edited signal [102] or NAA, which is suppressed during the GABAON step (at 1.9 ppm). In the case of multiplexed editing methods, such as HERMES [90], there are now four sub-spectra that do not match in spectral appearances, in terms of the water, NAA, or Cr signals. To address this challenge, Mikkelsen et al. [104] proposed a multi-step correction method based on “Spectral Registration” of subspectra, which improves the correction of frequency and phase errors in the multiplexed editing of GABA and GSH. However, this method has some limitations. For example, the alignment among sub-spectra relies on Cho and Cr signals, which may not be available in the other multiplexed editing methods. Also, this method may not be applicable to edited MRSI data. Overall, as the multiplexed editing methods become more complex [89,105], the multi-step correction may require modification, or new methods may need to be developed [106].
Challenges and available solutions in MRSI
MRSI provides metabolic information in the multiple regions of the brain within one acquisition. Although the information gathered is of high quality and spatial resolution [107], MRSI typically requires long measurement times (≥ 20 min), rendering standard MRSI sequences susceptible to subject motion and scanner instabilities as a result of heating of gradient coil and passive shims that cause F0 drifts [35]. Subject motion may result in the measurement of metabolites in the unintended region, phase artifacts, and lipid contaminations. In some cases, motion-related artifacts cannot be recognized, and thus may bias the measurement outcome [39]. Motion can also cause each voxel to experience a different level of B0 homogeneity and F0, and thus further exacerbate MRSI acquisition problems [75]. Advancements in the field of MRSI has led to accelerated sequences – such as proton/echo-planar spectroscopic imaging (PEPSI or EPSI) and spiral encoded MRSI [108–110] – attaining 3D coverage with high spatial resolution in clinically feasible times, rendering these sequences somewhat less susceptible to motion. However, the use of heavy-duty cycle in sequences such as EPSI or spirals may result in B0 inhomogeneity and F0 drifts [111]. As discussed earlier, incorporation of prospective motion, F0, and shim correction in MRSI sequences can address these drawbacks and generate high-quality metabolite maps on both high-field and ultra-high-field scanners [29,75,78,87].
Further improvements might be achieved using retrospective correction methods. However, such methods are challenging to utilize on MRSI data due to the use of phase-encoding gradients during acquisition, which has generally precluded the application of such methods in clinical MRSI. To our knowledge, only a few published papers applied retrospective correction on MRSI data. Haupt et al. [112] used external spatial reference markers for motion detection during in vivo MRSI acquisitions. The spatial positions of the markers at each encoding step provided information on in-plane rotation and translation of the subject that were used during spatial reconstruction of MRSI data to reduce image artifacts. Ebel et al. [111] presented a method that performed interleaved measurement of the instantaneous resonance frequency of a reference water signal during a volumetric MRSI acquisition. These measurements were used during post-processing to correct the frequency and phase of the MRSI data. The corrections largely removed line shape distortions, recovered metabolite signal loss, and improved spectral quality. Edited MRSI poses a yet greater challenge, since motion-related distortion of even a few points in k-space can lead to subtraction artifacts in the final edited spectra. Chan et al. [113] proposed a method that either corrects or removes transients that do not share the same properties as others at the same point in k-space. This method reduced subtraction artifacts in GABA-edited spectra and showed promising results for GSH-edited spectra. Further improvements of this method may substantially benefit multiplexed edited MRSI [114].
Implications for clinical investigations
1H-MRS and MRSI allow quantitative measurements of brain metabolites that provide useful markers for disease investigation and longitudinal evaluations. MRS has been applied to study numerous brain conditions and disorders, including brain development [7,115], drug discovery and evaluation [14], and several diseases and disorders, such as HIV/AIDS [10,116–119], Alzheimer’s disease and mild cognitive impairment (MCI) [120], multiple sclerosis [121,122], schizophrenia [123], Parkinson’s disease (PD) [124,125], amyotrophic lateral sclerosis [126], fetal alcohol spectrum disorders [127], Tourette syndrome [128], and autism [129,130].
As discussed earlier, MRS/MRSI acquisitions are necessarily long to attain data with sufficient SNR, rendering these acquisitions susceptible to subject and scanner instabilities. Motion can be a major problem when scanning children, the elderly, and patients with cognitive impairments, movement disorders or reduced impulse control [131,132]; hence, motion represents an unaddressed confound of many MRS studies. Heckova et al. [133] compared involuntary head motion, F0 and B0 shim changes, and the subsequent effects on data quality during prospective navigated 3D GABA-edited MRSI [29] in patients with MCI and PD, as well as in young and elderly healthy volunteers. The extent of unprompted head movements during the acquisition was 1.5 to 2.5-fold higher for the elderly healthy volunteers and patients compared to young healthy volunteers. These motions resulted in increased frequency offsets and B0-shim changes in the data from the patients. The SNR and spectral line width were better for young healthy controls compared with those in the elderly healthy volunteers and patients.
To demonstrate the benefits of tracking and subsequent correction of head motion and accompanying F0 and B0 changes, Hess et al. [134] compared the quality and variance in 1H-MRS brain data from a longitudinal study involving 59 non-sedated school-aged children with fetal alcohol spectrum disorders, using initially a standard PRESS sequence and a year later a motion, F0, and shim-navigated PRESS sequence [42]. Both datasets were retrospectively frequency-and-phase corrected before averaging. Compared to the standard sequence, the navigated sequence provided ~50% more data with suitable quality for spectral analysis. Significant improvements (p < 0.05) in SNR and line widths, and reduced variance in metabolite measurements were observed using the navigated sequence. In a separate study, Hess et al. [40] assessed the repeatability of metabolite concentrations (average 231 days apart) using the navigated PRESS sequence in three brain regions of 12 children enrolled in a longitudinal HIV study. The navigated sequence corrected for a substantial number of head movements along with F0 and B0-shim changes during acquisition. Data were also retrospectively frequency-and-phase corrected before averaging, which resulted in narrow line widths and low variation in metabolite concentration differences.
In another study, Hnilicová et al. [135] determined the reproducibility and reliability of GABA measurements using a volumetric navigated 3D GABA-edited MRSI sequence [29] in 14 healthy adults on a 3T scanner. These subjects were scanned in two sessions approximately 30 minutes apart. The navigated sequence successfully corrected for subject motions, and F0 and B0-shim changes. It yielded a high-quality 3D map of GABA+ (GABA+MM) in the human brain with a 3 cm3 nominal resolution in about 20 min. Both intra-subject and inter-subject variability was low for GABA+ ratios across all voxels, and excellent reliability was achieved between repeated measurements. To our knowledge, this work is the first study to demonstrate that a navigated edited-MRSI sequence can attain a 3D map of GABA+ with robust reproducibility and reliability, demonstrating the utility of the navigated sequence in clinical studies.
In the clinical setting, motion-related artifacts, such as broader spectral lines, insufficient water suppression, baseline distortion, and reduced SNR, affect the quality and interpretation of data. As noted, some artifacts in SVS studies can be reduced relatively easily through retrospective-correction methods, but these might not be an option for MRSI data. Prospective motion correction methods are primarily available in the research domain and not readily used in the clinical environment, although they might be of substantial benefit not only for improved clinical care but also for financial reasons since the need to repeat corrupted scans due to motion will be minimized. To estimate the financial implications of motion during acquisition, Andre et al. [136] assessed the prevalence and degree of motion-related artifacts in clinical MRI and found that ~59% of the data showed various levels of motion-related artifacts. Of these motion-corrupted data, ~28% were moderately to severely affected by artifacts, which rendered them marginally diagnostic to non-diagnostic and should (or must) be repeated. Repeated examinations require additional scan and patient time, and a low estimate based on these factors could cause lost revenues of over $100,000 per scanner per year [136]. Such high revenue loss demonstrates that motion during MRS studies may cause a significant financial burden to hospitals and imaging centers even when accounting for the lower percentage of clinical MRS/MRSI studies.
As demonstrated above, the amalgamation of prospective and retrospective correction can substantially increase the efficacy of MRS/MRSI sequences, data quality, reproducibility, and, more importantly, the accuracy of metabolite quantitation. The motion-, F0-, and shim-corrected edited and non-edited MRS/MRSI sequences allow robust investigation of multiple neurochemical and neuroanatomical systems in any subject population, adding scope and impact to the method beyond the above-reported clinical MRS applications. Given the important roles of many of these brain metabolites and neurochemicals in normal and disordered brain function, accurate measurements of these compounds with optimized correction schemes will provide improved clinical diagnoses, clinical evaluations and disease monitoring in future clinical trials for a vast number of brain conditions.
Conclusions
Recent technical progress towards eliminating the deleterious effects of head motion on in vivo 1H-MRS and MRSI acquisitions show significant benefits. Several studies, including clinical research investigations, demonstrate that prospective motion correction along with real-time F0 and B0 shim corrections are needed to diminish artifacts arising from subject and scanner instabilities. Additional efforts are required to increase the dissemination and implementation of these methods into the clinical and clinical research settings.
Acknowledgements
The work presented here was in part supported by NIH grants 1R01 DA021146, U54 56883 (SNRP), K02-DA16991, and G12 RR003061-21 (RCMI). RE/MS: P41 EB015909 R01 EB016089 R01 EB023963.
Abbreviations:
- 1H
proton
- SVS
single-voxel spectroscopy
- MRS
magnetic resonance spectroscopy
- MRSI
magnetic resonance spectroscopy imaging
- RF
radiofrequency
- F0
center/transmitter frequency
- fcs
chemical-shift selective
- B0
main magnetic field
- B1
radiofrequency field
- ppm
parts per million
- CV
coefficient of variation
- ICC
intra-class correlation coefficient
- cc
cubic centimeter
References
- 1.De Graaf RA. In vivo NMR spectroscopy: principles and techniques: John Wiley & Sons; 2019. [Google Scholar]
- 2.Jasperse B, Jakobs C, Eikelenboom MJ, Dijkstra CD, Uitdehaag BM, Barkhof F, Polman CH, Teunissen CE. N-acetylaspartic acid in cerebrospinal fluid of multiple sclerosis patients determined by gas-chromatography-mass spectrometry. Journal of neurology. 2007;254:631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Teunissen C, Iacobaeus E, Khademi M, Brundin L, Norgren N, Koel-Simmelink M, Schepens M, Bouwman F, Twaalfhoven H, Blom H. Combination of CSF N-acetylaspartate and neurofilaments in multiple sclerosis. Neurology. 2009;72:1322–1329. [DOI] [PubMed] [Google Scholar]
- 4.Schmitz B, Wang X, Barker PB, Pilatus U, Bronzlik P, Dadak M, Kahl KG, Lanfermann H, Ding XQ. Effects of aging on the human brain: a proton and phosphorus MR spectroscopy study at 3T. Journal of Neuroimaging. 2018;28:416–421. [DOI] [PubMed] [Google Scholar]
- 5.Gao F, Edden RA, Li M, Puts NA, Wang G, Liu C, Zhao B, Wang H, Bai X, Zhao C, Wang X, Barker PB. Edited magnetic resonance spectroscopy detects an age-related decline in brain GABA levels. Neuroimage. 2013;78:75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Porges EC, Woods AJ, Edden RA, Puts NA, Harris AD, Chen H, Garcia AM, Seider TR, Lamb DG, Williamson JB. Frontal gamma-aminobutyric acid concentrations are associated with cognitive performance in older adults. Biological Psychiatry: Cognitive Neuroscience and Neuroimaging. 2017;2:38–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vigneron DB. Magnetic resonance spectroscopic imaging of human brain development. Neuroimaging Clinics. 2006;16:75–85. [DOI] [PubMed] [Google Scholar]
- 8.Burlina AP, Aureli T, Bracco F, Conti F, Battistin L. MR spectroscopy: a powerful tool for investigating brain function and neurological diseases. Neurochem Res. 2000;25:1365–1372. [DOI] [PubMed] [Google Scholar]
- 9.Bonavita S, Di Salle F, Tedeschi G. Proton MRS in neurological disorders. Eur J Radiol. 1999;30:125–131. [DOI] [PubMed] [Google Scholar]
- 10.Barker PB, Lee RR, McArthur JC. AIDS dementia complex: evaluation with proton MR spectroscopic imaging. Radiology. 1995;195:58–64. [DOI] [PubMed] [Google Scholar]
- 11.Dager SR, Oskin N, Richards TL, Posse S. Research applications of magnetic resonance spectroscopy (MRS) to investigate psychiatric disorders. Topics in magnetic resonance imaging: TMRI. 2008;19:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Öz G, Alger JR, Barker PB, Bartha R, Bizzi A, Boesch C, Bolan PJ, Brindle KM, Cudalbu C, Dinçer A. Clinical proton MR spectroscopy in central nervous system disorders. Radiology. 2014;270:658–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Licata SC, Renshaw PF. Neurochemistry of drug action: insights from proton magnetic resonance spectroscopic imaging and their relevance to addiction. Annals of the New York Academy of Sciences. 2010;1187:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mason GF, Krystal JH. MR spectroscopy: its potential role for drug development for the treatment of psychiatric diseases. NMR in Biomedicine: An International Journal Devoted to the Development and Application of Magnetic Resonance In vivo. 2006;19:690–701. [DOI] [PubMed] [Google Scholar]
- 15.Hellem T, Shi X, Latendresse G, Renshaw PF. The utility of magnetic resonance spectroscopy for understanding substance use disorders: a systematic review of the literature. Journal of the American Psychiatric Nurses Association. 2015;21:244–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Govind V, Young K, Maudsley AA. Corrigendum: Proton NMR chemical shifts and coupling constants for brain metabolites. Govindaraju V, Young K, Maudsley AA, NMR Biomed. 2000; 13: 129–153.NMR Biomed. 2015;28:923–924. [DOI] [PubMed] [Google Scholar]
- 17.Govindaraju V, Young K, Maudsley AA. Proton NMR chemical shifts and coupling constants for brain metabolites. NMR Biomed. 2000;13:129–153. [DOI] [PubMed] [Google Scholar]
- 18.Terpstra M, Cheong I, Lyu T, Deelchand DK, Emir UE, Bednařík P, Eberly LE, Öz G. Test retest reproducibility of neurochemical profiles with short echo, single voxel MR spectroscopy at 3T and 7T. Magn Reson Med. 2016;76:1083–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saleh MG, Wang M, Mikkelsen M, Hui SC, Oeltzschner G, Boissoneault J, Stennett B, Edden RA, Porges EC. Simultaneous edited MRS of GABA, glutathione, and ethanol. NMR Biomed. 2020:e4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frahm J, Merboldt K-D, Hänicke W. Localized proton spectroscopy using stimulated echoes. Journal of Magnetic Resonance (1969). 1987;72:502–508. [Google Scholar]
- 21.Bottomley PA. Spatial localization in NMR spectroscopy in vivo. Proc Natl Acad Sci U S A. 1987;508:333–348. [DOI] [PubMed] [Google Scholar]
- 22.Mlynarik V, Gambarota G, Frenkel H, Gruetter R. Localized short-echo-time proton MR spectroscopy with full signal-intensity acquisition. Magn Reson Med. 2006;56:965–970. [DOI] [PubMed] [Google Scholar]
- 23.Scheenen TW, Klomp DW, Wijnen JP, Heerschap A. Short echo time 1H-MRSI of the human brain at 3T with minimal chemical shift displacement errors using adiabatic refocusing pulses. Magn Reson Med. 2008;59:1–6. [DOI] [PubMed] [Google Scholar]
- 24.Slotboom J, Bovee W. Adiabatic slice selective rf pulses and a single shot adiabatic localization pulse sequence. Concepts in Magnetic Resonance. 1995;7:193–217. [Google Scholar]
- 25.Garwood M, DelaBarre L. The return of the frequency sweep: designing adiabatic pulses for contemporary NMR. J Magn Reson. 2001;153:155–177. [DOI] [PubMed] [Google Scholar]
- 26.Mescher M, Merkle H, Kirsch J, Garwood M, Gruetter R. Simultaneous in vivo spectral editing and water suppression. NMR Biomed. 1998;11:266–272. [DOI] [PubMed] [Google Scholar]
- 27.Near J, Simpson R, Cowen P, Jezzard P. Efficient gamma-aminobutyric acid editing at 3T without macromolecule contamination: MEGA-SPECIAL. NMR Biomed. 2011;24:1277–1285. [DOI] [PubMed] [Google Scholar]
- 28.Andreychenko A, Boer VO, Arteaga de Castro CS, Luijten PR, Klomp DW. Efficient spectral editing at 7 T: GABA detection with MEGA sLASER. Magn Reson Med. 2012;68:1018–1025. [DOI] [PubMed] [Google Scholar]
- 29.Bogner W, Gagoski B, Hess AT, Bhat H, Tisdall MD, van der Kouwe AJ, Strasser B, Marjańska M, Trattnig S, Grant E. 3D GABA imaging with real-time motion correction, shim update and reacquisition of adiabatic spiral MRSI. Neuroimage. 2014;103:290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu H, Barker PB. MR spectroscopy and spectroscopic imaging of the brain. Magnetic resonance neuroimaging: Springer; 2011. p 203–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buonocore MH, Maddock RJ. Magnetic resonance spectroscopy of the brain: a review of physical principles and technical methods. Reviews in the Neurosciences. 2015;26:609–632. [DOI] [PubMed] [Google Scholar]
- 32.Mullins PG, McGonigle DJ, O’Gorman RL, Puts NA, Vidyasagar R, Evans CJ, Cardiff Symposium on MRSoG, Edden RA. Current practice in the use of MEGA-PRESS spectroscopy for the detection of GABA. Neuroimage. 2014;86:43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Puts NA, Edden RA. In vivo magnetic resonance spectroscopy of GABA: a methodological review. Prog Nucl Magn Reson Spectrosc. 2012;60:29–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harris AD, Saleh MG, Edden RA. Edited 1H magnetic resonance spectroscopy in vivo: Methods and metabolites. Magn Reson Med. 2017;77:1377–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bogner W, Hangel G, Esmaeili M, Andronesi OC. 1D-spectral editing and 2D multispectral in vivo1H-MRS and 1H-MRSI-Methods and applications. Analytical biochemistry. 2017;529:48–64. [DOI] [PubMed] [Google Scholar]
- 36.Landheer K, Schulte RF, Treacy MS, Swanberg KM, Juchem C. Theoretical description of modern 1H in Vivo magnetic resonance spectroscopic pulse sequences. J Magn Reson Imaging. 2019. [DOI] [PubMed] [Google Scholar]
- 37.Drost DJ, Riddle WR, Clarke GD. Proton magnetic resonance spectroscopy in the brain: report of AAPM MR Task Group# 9. Medical physics. 2002;29:2177–2197. [DOI] [PubMed] [Google Scholar]
- 38.Jiru F. Introduction to post-processing techniques. Eur J Radiol. 2008;67:202–217. [DOI] [PubMed] [Google Scholar]
- 39.Kreis R. Issues of spectral quality in clinical 1H magnetic resonance spectroscopy and a gallery of artifacts. NMR Biomed. 2004;17:361–381. [DOI] [PubMed] [Google Scholar]
- 40.Hess AT, van der Kouwe AJ, Mbugua KK, Laughton B, Meintjes EM. Quality of 186 child brain spectra using motion and B0 shim navigated single voxel spectroscopy. J Magn Reson Imaging. 2014;40:958–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adler CH, Ahlskog JE. Parkinson’s disease and movement disorders: diagnosis and treatment guidelines for the practicing physician: Springer Science & Business Media; 2000. [Google Scholar]
- 42.Hess AT, Dylan Tisdall M, Andronesi OC, Meintjes EM, van der Kouwe AJ. Real time motion and B0 corrected single voxel spectroscopy using volumetric navigators. Magn Reson Med. 2011;66:314–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zaitsev M, Speck O, Hennig J, Büchert M. Single voxel MRS with prospective motion correction and retrospective frequency correction. NMR Biomed. 2010;23:325–332. [DOI] [PubMed] [Google Scholar]
- 44.Felblinger J, Kreis R, Boesch C. Effects of physiologic motion of the human brain upon quantitative 1H MRS: analysis and correction by retro gating. NMR in Biomedicine: An International Journal Devoted to the Development and Application of Magnetic Resonance In Vivo. 1998;11:107–114. [DOI] [PubMed] [Google Scholar]
- 45.Godenschweger F, Kägebein U, Stucht D, Yarach U, Sciarra A, Yakupov R, Lüsebrink F, Schulze P, Speck O. Motion correction in MRI of the brain. Physics in Medicine & Biology. 2016;61:R32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maclaren J, Herbst M, Speck O, Zaitsev M. Prospective motion correction in brain imaging: a review. Magn Reson Med. 2013;69:621–636. [DOI] [PubMed] [Google Scholar]
- 47.Zaitsev M, Dold C, Sakas G, Hennig J, Speck O. Magnetic resonance imaging of freely moving objects: prospective real-time motion correction using an external optical motion tracking system. Neuroimage. 2006;31:1038–1050. [DOI] [PubMed] [Google Scholar]
- 48.Bogner W, Gagoski B, Hess AT, Bhat H, Tisdall MD, van der Kouwe AJ, Strasser B, Marjanska M, Trattnig S, Grant E, Rosen B, Andronesi OC. 3D GABA imaging with real-time motion correction, shim update and reacquisition of adiabatic spiral MRSI. Neuroimage. 2014;103:290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Henry PG, van de Moortele PF, Giacomini E, Nauerth A, Bloch G. Field frequency locked in vivo proton MRS on a whole body spectrometer. Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine. 1999;42:636–642. [DOI] [PubMed] [Google Scholar]
- 50.Edden RA, Oeltzschner G, Harris AD, Puts NA, Chan KL, Boer VO, Schär M, Barker PB. Prospective frequency correction for macromolecule suppressed GABA editing at 3T. J Magn Reson Imaging. 2016;44:1474–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Andrews Shigaki BC, Armstrong BS, Zaitsev M, Ernst T. Prospective motion correction for magnetic resonance spectroscopy using single camera retro grate reflector optical tracking. J Magn Reson Imaging. 2011;33:498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Keating B, Deng W, Roddey JC, White N, Dale A, Stenger VA, Ernst T. Prospective motion correction for single voxel 1H MR spectroscopy. Magn Reson Med. 2010;64:672–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Keating B, Ernst T. Real-time dynamic frequency and shim correction for single voxel magnetic resonance spectroscopy. Magn Reson Med. 2012;68:1339–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nehrke K, Börnert P. Prospective correction of affine motion for arbitrary MR sequences on a clinical scanner. Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine. 2005;54:1130–1138. [DOI] [PubMed] [Google Scholar]
- 55.Waddell KW, Avison MJ, Joers JM, Gore JC. A practical guide to robust detection of GABA in human brain by J-difference spectroscopy at 3 T using a standard volume coil. Magnetic resonance imaging. 2007;25:1032–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ernst T, Li J. A novel phase and frequency navigator for proton magnetic resonance spectroscopy using water suppression cycling. Magn Reson Med. 2011;65:13–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Edden RA, Puts NA, Harris AD, Barker PB, Evans CJ. Gannet: A batch processing tool for the quantitative analysis of gamma aminobutyric acid–edited MR spectroscopy spectra. J Magn Reson Imaging. 2014;40:1445–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thesen S, Heid O, Mueller E, Schad L. Prospective acquisition correction for head motion with image-based tracking for real-time fMRI. Magnetic Resonance in Medicine. 2000;44:457–465. [DOI] [PubMed] [Google Scholar]
- 59.Felmlee J, Ehman R, Riederer S, Korin H. Adaptive motion compensation in MRI: accuracy of motion measurement. Magnetic Resonance in Medicine. 1991;18:207–213. [DOI] [PubMed] [Google Scholar]
- 60.Ward HA, Riederer SJ, Grimm RC, Ehman RL, Felmlee JP, Jack CRj. Prospective multiaxial motion correction for fMRI. Magnetic Resonance in Medicine. 2000;43:459–469. [DOI] [PubMed] [Google Scholar]
- 61.Moriguchi H, Lewin J, Duerk J. Novel interleaved spiral imaging motion correction technique using orbital navigators. Magnetic Resonance in Medicine. 2003;50:423–428. [DOI] [PubMed] [Google Scholar]
- 62.Zaitsev M, Dold C, Sakas G, Hennig J, Speck O. Magnetic resonance imaging of freely moving objects: prospective real-time motion correction using an external optical motion tracking system. Neuroimage. 2006;31:1038–1050. [DOI] [PubMed] [Google Scholar]
- 63.Armstrong B, Andrews-Shigaki B, Barrows R, Kusik T, Ernst T, Speck O. Performance of Stereo-Vision and Retro-Grate Reflector Motion Tracking Systems in the Space Constraints of an MR Scanner. 2009; Honolulu. p 4641. [Google Scholar]
- 64.White N, Roddey C, Shankaranarayanan A, Han E, Rettmann D, Santos J, Kuperman J, Dale A. PROMO: Real-time prospective motion correction in MRI using image-based tracking. Magn Reson Med. 2010;63:91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hess AT, Jacobson SW, Jacobson JL, Molteno CD, van der Kouwe AJ, Meintjes EM. A comparison of spectral quality in magnetic resonance spectroscopy data acquired with and without a novel EPI-navigated PRESS sequence in school-aged children with fetal alcohol spectrum disorders. Metabolic brain disease. 2014;29:323–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zaitsev M, Speck O, Hennig J, Buchert M. Single-voxel MRS with prospective motion correction and retrospective frequency correction. NMR Biomed. 2010;23:325–332. [DOI] [PubMed] [Google Scholar]
- 67.Deelchand DK, Joers JM, Auerbach EJ, Henry PG. Prospective motion and B0 shim correction for MR spectroscopy in human brain at 7T. Magn Reson Med. 2019;82:1984–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ward HA, Riederer S, Jack CR. Real-time autoshimming for echo planar timecourse imaging. Journal of Magnetic Resonance Imaging. 2002;48:771–780. [DOI] [PubMed] [Google Scholar]
- 69.Kim DH, Adalsteinsson E, Glover GH, Spielman DM. Regularized higher-order in vivo shimming. Magn Reson Med. 2002;48:715–722. [DOI] [PubMed] [Google Scholar]
- 70.Mackenzie IS, Robinson EM, Wells AN, Wood B. A simple field map for shimming. Magn Reson Med. 1987;5:262–268. [DOI] [PubMed] [Google Scholar]
- 71.Automatic Gruetter R., localized in vivo adjustment of all first- and second-order shim coils. Magn Reson Med. 1993;29:804–811. [DOI] [PubMed] [Google Scholar]
- 72.Shen J, Rycyna RE, Rothman DL. Improvements on an in vivo automatic shimming method [FASTERMAP]. Magn Reson Med. 1997;38:834–839. [DOI] [PubMed] [Google Scholar]
- 73.Li S, Williams GD, Frisk TA, Arnold BW, Smith MB. A computer simulation of the static magnetic field distribution in the human head. Magnetic Resonance in Medicine. 1995;34:268–275. [DOI] [PubMed] [Google Scholar]
- 74.Juchem C, de Graaf RA. B0 magnetic field homogeneity and shimming for in vivo magnetic resonance spectroscopy. Analytical biochemistry. 2017;529:17–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hess AT, Andronesi OC, Dylan Tisdall M, Gregory Sorensen A, van der Kouwe AJ, Meintjes EM. Real time motion and B0 correction for localized adiabatic selective refocusing (LASER) MRSI using echo planar imaging volumetric navigators. NMR Biomed. 2012;25:347–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Keating B, Ernst T. Real-time dynamic frequency and shim correction for single-voxel magnetic resonance spectroscopy. Magn Reson Med. 2012;68:1339–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hess AT, Tisdall MD, Andronesi OC, Meintjes EM, van der Kouwe AJ. Real-time motion and B0 corrected single voxel spectroscopy using volumetric navigators. Magn Reson Med. 2011;66:314–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bogner W, Hess AT, Gagoski B, Tisdall MD, van der Kouwe AJ, Trattnig S, Rosen B, Andronesi OC. Real-time motion-and B0-correction for LASER-localized spiral-accelerated 3D-MRSI of the brain at 3 T. Neuroimage. 2014;88:22–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.An L, Zhang Y, Thomasson DM, Latour LL, Baker EH, Shen J, Warach S. Measurement of glutathione in normal volunteers and stroke patients at 3T using J-difference spectroscopy with minimized subtraction errors. Journal of magnetic resonance imaging : JMRI. 2009;30:263–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tisdall MD, Hess AT, Reuter M, Meintjes EM, Fischl B, van der Kouwe AJ. Volumetric navigators for prospective motion correction and selective reacquisition in neuroanatomical MRI. Magn Reson Med. 2012;68:389–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Henry PG, Dautry C, Hantraye P, Bloch G. Brain GABA Editing Without Macromolecule Contamination. Magn Reson Med. 2001;45:517–520. [DOI] [PubMed] [Google Scholar]
- 82.Edden RA, Pomper MG, Barker PB. In vivo differentiation of N-acetyl aspartyl glutamate from N-acetyl aspartate at 3 Tesla. Magn Reson Med. 2007;57:977–982. [DOI] [PubMed] [Google Scholar]
- 83.Saleh MG, Alhamud A, Near J, Kouwe AJ, Meintjes EM. Volumetric navigated MEGA SPECIAL for real time motion and shim corrected GABA editing. NMR Biomed. 2016;29:248–255. [DOI] [PubMed] [Google Scholar]
- 84.Saleh MG, Near J, Alhamud A, Robertson F, van der Kouwe AJ, Meintjes EM. Reproducibility of macromolecule suppressed GABA measurement using motion and shim navigated MEGA-SPECIAL with LCModel, jMRUI and GANNET. Magn Reson Mater Phy. 2016;29:863–874. [DOI] [PubMed] [Google Scholar]
- 85.Terpstra M, Ugurbil K, Gruetter R. Direct in vivo measurement of human cerebral GABA concentration using MEGA editing at 7 Tesla. Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine. 2002;47:1009–1012. [DOI] [PubMed] [Google Scholar]
- 86.Bhattacharyya P, Phillips M, Stone L, Lowe M. In-vivo MRS measurement of gray-matter and white-matter GABA concentration in sensorimotor cortex using a motion-controlled MEGA-PRESS Sequence. Magnetic resonance imaging. 2011;29:374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Moser P, Hingerl L, Strasser B, Považan M, Hangel G, Andronesi OC, van der Kouwe A, Gruber S, Trattnig S, Bogner W. Whole-slice mapping of GABA and GABA+ at 7T via adiabatic MEGA-editing, real-time instability correction, and concentric circle readout. Neuroimage. 2019;184:475–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chan KL, Puts NA, Schar M, Barker PB, Edden RA. HERMES: Hadamard encoding and reconstruction of MEGA-edited spectroscopy. Magn Reson Med. 2016;76:11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Oeltzschner G, Saleh MG, Rimbault D, Mikkelsen M, Chan KL, Puts NA, Edden RA. Advanced Hadamard-encoded editing of seven low-concentration brain metabolites: Principles of HERCULES. Neuroimage. 2019;185:181–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Saleh MG, Oeltzschner G, Chan KL, Puts NA, Mikkelsen M, Schär M, Harris AD, Edden RA. Simultaneous edited MRS of GABA and glutathione. Neuroimage. 2016;15:576–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Oeltzschner G, Chan KL, Saleh MG, Mikkelsen M, Puts NA, Edden RA. Hadamard editing of glutathione and macromolecule suppressed GABA. NMR Biomed. 2018;31:e3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Felblinger J, Kreis R, Boesch C. Effects of physiologic motion of the human brain upon quantitative 1H-MRS: analysis and correction by retro-gating. NMR in Biomedicine. 1998;11:107–114. [DOI] [PubMed] [Google Scholar]
- 93.Helms G, Piringer A. Restoration of motion-related signal loss and line-shape deterioration of proton MR spectra using the residual water as intrinsic reference. Magnetic Resonance in Medicine. 2001;46:395–400. [DOI] [PubMed] [Google Scholar]
- 94.Helms G, Piringer A. Restoration of motion related signal loss and line shape deterioration of proton MR spectra using the residual water as intrinsic reference. Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine. 2001;46:395–400. [DOI] [PubMed] [Google Scholar]
- 95.Hurd RE, Gurr D, Sailasuta N. Proton spectroscopy without water suppression: the oversampled J-resolved experiment. Magn Reson Med. 1998;40:343–347. [DOI] [PubMed] [Google Scholar]
- 96.Clayton DB, Elliott MA, Leigh JS, Lenkinski RE. 1H spectroscopy without solvent suppression: characterization of signal modulations at short echo times. J Magn Reson. 2001;153:203–209. [DOI] [PubMed] [Google Scholar]
- 97.Thiel T, Czisch M, Elbel G, Hennig J. Phase coherent averaging in magnetic resonance spectroscopy using interleaved navigator scans: compensation of motion artifacts and magnetic field instabilities. Magnetic Resonance in Medicine. 2002;47:1077–1082. [DOI] [PubMed] [Google Scholar]
- 98.Near J, Edden R, Evans CJ, Paquin R, Harris A, Jezzard P. Frequency and phase drift correction of magnetic resonance spectroscopy data by spectral registration in the time domain. Magn Reson Med. 2015;73:44–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wilson M. Robust retrospective frequency and phase correction for single voxel MR spectroscopy. Magn Reson Med. 2019;81:2878–2886. [DOI] [PubMed] [Google Scholar]
- 100.Golub GH, Pereyra V. The differentiation of pseudo-inverses and nonlinear least squares problems whose variables separate. SIAM Journal on numerical analysis. 1973;10:413–432. [Google Scholar]
- 101.Van der Veen J, De Beer R, Luyten P, Van Ormondt D. Accurate quantification of in vivo 31P NMR signals using the variable projection method and prior knowledge. Magn Reson Med. 1988;6:92–98. [DOI] [PubMed] [Google Scholar]
- 102.Evans CJ, Puts NA, Robson SE, Boy F, McGonigle DJ, Sumner P, Singh KD, Edden RA. Subtraction artifacts and frequency (Mis ) alignment in J difference GABA editing. J Magn Reson Imaging. 2013;38:970–975. [DOI] [PubMed] [Google Scholar]
- 103.Wiegers EC, Philips BW, Heerschap A, van der Graaf M. Automatic frequency and phase alignment of in vivo J-difference-edited MR spectra by frequency domain correlation. Magn Reson Mater Phy. 2017;30:537–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mikkelsen M, Saleh MG, Near J, Chan KL, Gong T, Harris AD, Oeltzschner G, Puts NA, Cecil KM, Wilkinson ID. Frequency and phase correction for multiplexed edited MRS of GABA and glutathione. Magn Reson Med. 2018;80:21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Saleh MG, Wang M, Mikkelsen M, Hui SCN, Oeltzschner G, Boissoneault J, Stennett B, Edden RAE, Porges EC. Simultaneous edited MRS of GABA, glutathione and ethanol. NMR in Biomedicine. DOI: 10.1002/nbm.4227.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mikkelsen M, Near J, Saleh MG, Mostofsky SH, Puts NAJ, RAE E. Robust Correction of Frequency and Phase Errors in Edited MRS Data. Proceedings of the 27th annual meeting of ISMRM, Montreal, Canada. 2018. [Google Scholar]
- 107.Bogner W, Gruber S, Trattnig S, Chmelik M. High resolution mapping of human brain metabolites by free induction decay 1H MRSI at 7 T. NMR Biomed. 2012;25:873–882. [DOI] [PubMed] [Google Scholar]
- 108.Adalsteinsson E, Irarrazabal P, Topp S, Meyer C, Macovski A, Spielman DM. Volumetric spectroscopic imaging with spiral based k space trajectories. Magn Reson Med. 1998;39:889–898. [DOI] [PubMed] [Google Scholar]
- 109.Maudsley AA, Domenig C, Govind V, Darkazanli A, Studholme C, Arheart K, Bloomer C. Mapping of brain metabolite distributions by volumetric proton MR spectroscopic imaging (MRSI). Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine. 2009;61:548–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Posse S, Tedeschi G, Risinger R, Ogg R, Bihan DL. High speed 1H spectroscopic imaging in human brain by echo planar spatial spectral encoding. Magn Reson Med. 1995;33:34–40. [DOI] [PubMed] [Google Scholar]
- 111.Ebel A, Maudsley AA. Detection and correction of frequency instabilities for volumetric 1H echo-planar spectroscopic imaging. Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine. 2005;53:465–469. [DOI] [PubMed] [Google Scholar]
- 112.Haupt CI, Kiefer AP, Maudsley AA. In plane motion correction for MR spectroscopic imaging. Magn Reson Med. 1998;39:749–753. [DOI] [PubMed] [Google Scholar]
- 113.Chan KL, Barker PB. Retrospective motion compensation for edited MR spectroscopic imaging. Neuroimage. 2019;202:116141. [DOI] [PubMed] [Google Scholar]
- 114.Chan KL, Oeltzschner G, Saleh MG, Edden RA, Barker PB. Simultaneous editing of GABA and GSH with Hadamard encoded MR spectroscopic imaging. Magn Reson Med. 2019;82:21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kreis R, Ernst T, Ross BD. Development of the human brain: in vivo quantification of metabolite and water content with proton magnetic resonance spectroscopy. Magn Reson Med. 1993;30:424–437. [DOI] [PubMed] [Google Scholar]
- 116.Chang L, Lee P, Yiannoutsos C, Ernst T, Marra C, Richards T, Kolson D, Schifitto G, Jarvik J, Miller E. A multicenter in vivo proton-MRS study of HIV-associated dementia and its relationship to age. Neuroimage. 2004;23:1336–1347. [DOI] [PubMed] [Google Scholar]
- 117.Chang L, Ernst T, Leonido-Yee M, Walot I, Singer E. Cerebral metabolite abnormalities correlate with clinical severity of HIV-1 cognitive motor complex. Neurology. 1999;52:100–100. [DOI] [PubMed] [Google Scholar]
- 118.Mohamed MA, Lentz MR, Lee V, Halpern EF, Sacktor N, Selnes O, Barker PB, Pomper MG. Factor analysis of proton MR spectroscopic imaging data in HIV infection: metabolite-derived factors help identify infection and dementia. Radiology. 2010;254:577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mbugua KK, Holmes MJ, Cotton MF, Ratai E-M, Little F, Hess AT, Dobbels E, Van der Kouwe AJ, Laughton B, Meintjes EM. HIV-associated CD4/8 depletion in infancy is associated with neurometabolic reductions in the basal ganglia at age 5 years despite early antiretroviral therapy. AIDS (London, England). 2016;30:1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gao F, Barker PB. Various MRS application tools for Alzheimer disease and mild cognitive impairment. AJNR Am J Neuroradiol. 2014;35:S4–S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Llufriu S, Kornak J, Ratiney H, Oh J, Brenneman D, Cree BA, Sampat M, Hauser SL, Nelson SJ, Pelletier D. Magnetic resonance spectroscopy markers of disease progression in multiple sclerosis. JAMA neurology. 2014;71:840–847. [DOI] [PubMed] [Google Scholar]
- 122.Marshall I, Thrippleton MJ, Bastin ME, Mollison D, Dickie DA, Chappell FM, Semple SI, Cooper A, Pavitt S, Giovannoni G. Characterisation of tissue-type metabolic content in secondary progressive multiple sclerosis: a magnetic resonance spectroscopic imaging study. Journal of neurology. 2018;265:1795–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Arnold DL, Matthews PM, Francis G, Antel J. Proton magnetic resonance spectroscopy of human brain in vivo in the evaluation of multiple sclerosis: assessment of the load of disease. Magn Reson Med. 1990;14:154–159. [DOI] [PubMed] [Google Scholar]
- 124.Tuura RLG, Baumann CR, Baumann-Vogel H. Neurotransmitter activity is linked to outcome following subthalamic deep brain stimulation in Parkinson’s disease. Parkinsonism & related disorders. 2018;50:54–60. [DOI] [PubMed] [Google Scholar]
- 125.Gong T, Xiang Y, Saleh MG, Gao F, Chen W, Edden RA, Wang G. Inhibitory motor dysfunction in parkinson’s disease subtypes. J Magn Reson Imaging. 2018;47:1610–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Weerasekera A, Peeters R, Sima D, Dresselaers T, Sunaert S, De Vocht J, Claeys K, Van Huffel S, Van Damme P, Himmelreich U. Motor cortex metabolite alterations in amyotrophic lateral sclerosis assessed in vivo using edited and non-edited magnetic resonance spectroscopy. Brain Res. 2019;1718:22–31. [DOI] [PubMed] [Google Scholar]
- 127.du Plessis L, Jacobson JL, Jacobson SW, Hess AT, van der Kouwe A, Avison MJ, Molteno CD, Stanton ME, Stanley JA, Peterson BS, Meintjes E. An in vivo 1H magnetic resonance spectroscopy study of the deep cerebellar nuclei in children with fetal alcohol spectrum disorders. Alcohol Clin Exp Res. 2014;38:1330–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Puts NA, Harris AD, Crocetti D, Nettles C, Singer HS, Tommerdahl M, Edden RA, Mostofsky SH. Reduced GABAergic inhibition and abnormal sensory symptoms in children with Tourette syndrome. J Neurophysiol. 2015;114:808–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Drenthen GS, Barendse EM, Aldenkamp AP, van Veenendaal TM, Puts NA, Edden RA, Zinger S, Thoonen G, Hendriks MP, Kessels RP. Altered neurotransmitter metabolism in adolescents with high-functioning autism. Psychiatry Research: Neuroimaging. 2016;256:44–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Puts NA, Wodka EL, Harris AD, Crocetti D, Tommerdahl M, Mostofsky SH, Edden RA. Reduced GABA and altered somatosensory function in children with autism spectrum disorder. Autism Research. 2017;10:608–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Flory JD, Manuck SB. Impulsiveness and cigarette smoking. Psychosomatic Medicine. 2009;71:431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Paydary K, Mahin Torabi S, SeyedAlinaghi S, Noori M, Noroozi A, Ameri S, Ekhtiari H. Impulsivity, sensation seeking, and risk-taking behaviors among HIV-positive and HIV-negative heroin dependent persons. AIDS research and treatment. 2016;2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Heckova E, Považan M, Strasser B, Krumpolec P, Hnilicová P, Hangel GJ, Moser PA, Andronesi OC, van der Kouwe AJ, Valkovic P, Ukropcova B, Trattnig S, Bogner W. Real-time correction of motion and imager instability artifacts during 3D γ-aminobutyric acid–edited MR spectroscopic imaging. Radiology. 2017;286:666–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hess AT, Jacobson SW, Jacobson JL, Molteno CD, van der Kouwe AJ, Meintjes EM. A comparison of spectral quality in magnetic resonance spectroscopy data acquired with and without a novel EPI-navigated PRESS sequence in school-aged children with fetal alcohol spectrum disorders. Metabolic brain disease. 2014;29:323–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hnilicová P, Považan M, Strasser B, Andronesi OC, Gajdošík M, Dydak U, Ukropec J, Dobrota D, Trattnig S, Bogner W. Spatial variability and reproducibility of GABA edited MEGA LASER 3D MRSI in the brain at 3 T. NMR Biomed. 2016;29:1656–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Andre JB, Bresnahan BW, Mossa-Basha M, Hoff MN, Smith CP, Anzai Y, Cohen WA. Toward quantifying the prevalence, severity, and cost associated with patient motion during clinical MR examinations. Journal of the American College of Radiology. 2015;12:689–695. [DOI] [PubMed] [Google Scholar]