Abstract
Polycomb-group (PcG) proteins form large multimeric protein complexes that are involved in maintaining the transcriptionally repressive state of genes. Previously, we reported that RING1 interacts with vertebrate Polycomb (Pc) homologs and is associated with or is part of a human PcG complex. However, very little is known about the role of RING1 as a component of the PcG complex. Here we undertake a detailed characterization of RING1 protein-protein interactions. By using directed two-hybrid and in vitro protein-protein analyses, we demonstrate that RING1, besides interacting with the human Pc homolog HPC2, can also interact with itself and with the vertebrate PcG protein BMI1. Distinct domains in the RING1 protein are involved in the self-association and in the interaction with BMI1. Further, we find that the BMI1 protein can also interact with itself. To better understand the role of RING1 in regulating gene expression, we overexpressed the protein in mammalian cells and analyzed differences in gene expression levels. This analysis shows that overexpression of RING1 strongly represses En-2, a mammalian homolog of the well-characterized Drosophila PcG target gene engrailed. Furthermore, RING1 overexpression results in enhanced expression of the proto-oncogenes c-jun and c-fos. The changes in expression levels of these proto-oncogenes are accompanied by cellular transformation, as judged by anchorage-independent growth and the induction of tumors in athymic mice. Our data demonstrate that RING1 interacts with multiple human PcG proteins, indicating an important role for RING1 in the PcG complex. Further, deregulation of RING1 expression leads to oncogenic transformation by deregulation of the expression levels of certain oncogenes.
During embryogenesis, many different cell types develop from one fertilized egg. Cell type specificity emerges as a result of differential expression of regulatory genes. Notably, cell-specific sets of active and inactive genes determine the cell’s identity. To preserve the identity of the cell, it is important that these specific expression patterns be maintained and stably inherited by daughter cells in a cell-type-specific manner. Therefore, the maintenance of cell type specificity needs to be regulated by a cellular memory system. In Drosophila, for instance, the products of the Polycomb-group (PcG) genes are required for stable repression of gene activity. PcG proteins are evolutionarily conserved, being involved in the inheritably stable repression of homeotic gene expression both in Drosophila and in vertebrates (8, 14, 16, 24, 27).
It has been observed that in Drosophila, different PcG proteins, including Polycomb (Pc), Polyhomeotic (Ph), and Posterior sex combs (Psc), bind in overlapping patterns on polytene chromosomes (18, 36). Based on this observation, it has been proposed that PcG proteins repress gene activity via the formation of multimeric protein complexes. With the genetic yeast two-hybrid system, it is possible to search for direct protein-protein interactions in order to determine the identities of PcG complex components. In this way, several vertebrate PcG homologs have been found to interact. The human homologs of Ph, HPH1 and HPH2, have been found to interact with each other and with BMI1, the vertebrate homolog of the Drosophila PcG protein Psc (9). A human Pc homolog, HPC2, interacts with a RING finger protein, RING1 (21). It has further been found that Pc and Ph coimmunoprecipitate in Drosophila (6). The human HPH1, HPH2, BMI1, HPC2, and RING1 proteins also coimmunoprecipitate, and they colocalize in distinct nuclear domains of mammalian cell lines, termed PcG domains (9, 21). Similar biochemical interactions between homologs of Pc, Ph, Psc, and RING1 have been identified in mice and in Xenopus embryos (1, 10, 19, 21).
Expression analyses of several vertebrate PcG proteins reveal that they are differentially distributed in tissues and cell lines and that the expression of certain PcG proteins in these tissues is dependent on the time of development (4, 9, 15, 19, 21). This finding suggests that different, specific PcG complexes exist with different protein compositions. Direct evidence for the existence of two different vertebrate PcG complexes is gained from the characterization of the vertebrate PcG protein EED. EED coimmunoprecipitates and colocalizes with the mammalian PcG protein Enx1/EZH2 but not with other vertebrate PcG proteins such as HPC2 or BMI1. These findings indicate the existence of different, specific vertebrate PcG complexes that may contribute to specificity for target genes and possibly for different tissues (26, 34).
Recently, we have shown that interference with the function of HPC2 deregulates the expression of the proto-oncogene c-myc. Overexpression of HPC2 results in repression of c-myc. Overexpression of a dominant-negative HPC2 deletion mutant, ΔHPC2, which lacks a conserved C-terminal domain that is crucial for HPC2-mediated gene repression, led to enhanced expression of the c-myc gene in several mammalian cell lines. Concomitantly, overexpression of ΔHPC2 results in cellular transformation and anchorage-independent growth in mammalian cells (22). Although it cannot be concluded whether the effect of HPC2 on c-myc is direct or indirect, these data suggest that one function of the mammalian PcG proteins is to repress the transcription of certain proto-oncogenes. Importantly, HPC2 is not the only PcG member found to be linked with oncogenesis. Two other mammalian PcG proteins, Bmi-1 and mel-18, have also been shown to be involved in tumorigenesis. The mouse PcG gene bmi-1 collaborates with the proto-oncogene c-myc to cause lymphomas (11, 33). Interference with the expression of the mammalian PcG protein mel-18 induces tumors in nude mice (13). These findings indicate that mammalian PcG proteins have oncogenic properties.
Previously we found that the human RING1 protein interacts with HPC2 and is associated with the human PcG protein complex (21). However, little is known about the function of RING1. Here, we analyzed the functions of RING1 in more detail. Using directed two-hybrid and in vitro protein-protein analyses, we found that RING1 is able to interact with multiple human PcG proteins. We also overexpressed RING1 in mammalian cells and analyzed the differences in gene expression patterns. We found that overexpression of RING1 repressed the gene activity of En-2, a mammalian homolog of engrailed, a well-characterized Drosophila PcG target gene. Overexpression of RING1 further deregulated the expression of the proto-oncogenes c-jun and c-fos. Concomitant with the changes in the expression levels of these oncogenes, cellular transformations and the formation of tumors in athymic mice were induced. Our data suggest that RING1 interacts with multiple human PcG proteins and that overexpression of RING1 leads to oncogenic transformations by the deregulation of specific oncogenes.
MATERIALS AND METHODS
Construction of the pAS3 two-hybrid vector.
Using the pAS2 two-hybrid vector (Clontech), we obtained a GAL4 DNA binding domain (DBD) fusion protein in which the GAL4 DBD is positioned at the N terminus of the protein. To generate C-terminally positioned GAL4 DBD fusion proteins, we constructed a new two-hybrid vector in which the GAL4 DBD is placed downstream of the polylinker to create pAS3, a C-terminal fusion protein that is very similar to pAS2. To construct the pAS3 vector, the ADH promoter and the GAL4 DBD domain from pAS2 were recloned. By PCR, we derived the GAL4 DBD fragment, amino acids (aa) 1 to 147, and the ADH promoter, using pAS2 as a template. The GAL4 DBD and the ADH promoter fragments were cloned in pBluescript in a two-step ligation, creating an ADH promoter-polylinker-GAL4 DBD cassette, which has been entirely sequenced. The pAS2 vector was digested with SacI/SalI, releasing the original ADH promoter, GAL4 DBD, polylinker, and CYH2 selection gene and replacing them by the new ADH promoter-polylinker-GAL4 DBD cassette. The 7.5-kb pAS3 vector has the same properties as the pAS2 vector but lacks the CYH2 selection gene.
Analysis of interacting proteins with the two-hybrid system.
Indicated fragments of the cDNAs encoding RING1, BMI1, HPC2, HPH1, HPH2, Enx1, and EED were derived via PCR (Expand; Boehringer). The fragments were subcloned into the pAS2, pAS3, and pGAD10 (GAL4 transactivation domain [TAD]) vectors. The fragments were sequenced over their entire lengths. The resulting plasmids were cotransformed into Saccharomyces cerevisiae Y190. The transformants were plated on medium lacking leucine, tryptophan, and histidine, with or without 30 mM 3-amino-1,2,4-triazole (3-AT). Interactions were scored negative if they failed to grow in the presence of 30 mM 3-AT. Under these nonselective conditions, negative interactions were β-galactosidase negative. Positive interactions meet the two criteria of growing in the presence of 30 mM 3-AT and testing β-galactosidase positive. To exclude the possibility that the negative interactors did not produce either one of the fusion proteins, we Western blotted equal amounts of protein and incubated the blots with monoclonal antibodies that specifically recognize the GAL4 DBD or TAD protein (Clontech, Palo Alto, Calif.). All positive and negative interactors expressed both GAL4 DBD fusions and the GAL4 TAD fusions at approximately the same levels (data not shown).
Construction of GST fusion proteins, protein preparation, and in vitro binding assay.
A 1,131-bp fragment of the RING1 cDNA which encompasses the entire coding region and corresponds to aa 1 to 377 was cloned into pGEX-2TK, thus creating glutathione S-transferase (GST)–RING1. A 990-bp fragment of the bmi-1 cDNA (a gift from M. van Lohuizen) which covers the entire coding sequence and corresponds to aa 1 to 324 was cloned into pGEX-2TK, thus creating GST–Bmi-1. Expression of the GST fusion proteins was induced for 3 h at 30°C with 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG) as instructed by the manufacturer (Pharmacia) (29). The cells were pelleted, resuspended in binding buffer (phosphate-buffered saline containing 1 mM EDTA, 1 mM dithiothreitol, 2 mM phenylmethylsulfonyl fluoride, leupeptin [10 μg/ml], benzamidine [10 μg/ml], trypsin inhibitor [10 μg/ml], and aprotinin [10 μg/ml]) and sonicated. Triton X-100 was added to a final concentration of 1% (vol/vol), and the lysate was incubated for 30 min on ice. Cell debris was removed by centrifugation for 10 min at 14,000 × g, the supernatant was added to glutathione-Sepharose 4B, and the mixture was incubated for 30 min at 4°C. The beads were collected by centrifugation and washed extensively with binding buffer. Capped synthetic HPC2, RING1, and bmi-1 mRNAs were made by in vitro transcription and translated at 20 μg/ml in a rabbit reticulocyte lysate in the presence of [35S]methionine (19). A 10-μl slurry of GST fusion protein (immobilized to glutathione-Sepharose) was preincubated for 30 min on ice in a final volume of 200 μl of binding buffer containing 0.5% Nonidet P-40 and 1 mg of bovine serum albumin per ml. Subsequently, 3 μl of the reticulocyte lysate was added to the mixture and incubated for 30 min at 4°C with rotation. The beads were washed five times with 1 ml of ice-cold binding buffer. The complexes were separated on sodium dodecyl sulfate-polyacrylamide gels, which were subjected to fluorography.
Western blot analysis of RING1.
Expression of the RING1 protein was analyzed in cell lysates of RING1 stably transfected Rat1a (Rat1a/RING1) and control cell lines. For RING1 detection, the blots were incubated with a 1:1,000 dilution of affinity-purified rabbit anti-RING1 antibodies (21). Equal amounts of proteins were loaded, as measured by the bicinchoninic acid method (30) and as visualized by Coomassie staining of a gel.
Atlas cDNA expression array.
Rat1a cells overexpressing wild-type RING1 or pcDNA3 vector, which were used in the soft agar growth assay, were grown, and stably transfected lines were selected by culturing the cells in Dulbecco’s minimal essential medium supplemented with 10% newborn calf serum containing 500 μg of Geneticin (G418; Gibco) per ml for 2 weeks. Surviving cells were clonally expanded in medium containing 250 μg of G418 per ml for 2 to 4 weeks. Individual cell clones were selected and cultured in individual dishes. After five passages, poly(A)+ RNA was isolated and subjected to differential display using the commercial mouse Atlas expression arrays (Clontech). We also blotted poly(A)+ RNA of the selected Rat1a/RING1 clones and control cells and hybridized the blots with probes for GAPDH, c-jun, c-fos, c-myc, and En-2. Isolation of RNA and Northern analysis were performed according to standard procedures. The blots were hybridized with [α-32P]dATP-labeled DNA probes, and the blots were autoradiographed with intensifying screens at −70°C, using preflashed X-ray films.
Soft agar growth assay.
Cell lines were analyzed for anchorage-independent growth as described previously (20, 28, 31). Rat1a cells were transfected by the calcium phosphate transfection procedure with full-length RING1, an N-terminal part of RING1 (RING1 aa 1 to 203), and a C-terminal part of RING1 (RING1 aa 154 to 377), all cloned in the pcDNA3 vector. As a positive control, c-myc cDNA cloned in the pRcCMV vector and the C-terminal deletion mutant of HPC2 (ΔHPC2) (22) were transfected. As a negative control, the pcDNA3 vector alone was transfected. The cells were subjected to Geneticin (G418; 500 μg/ml) selection. Cells were cultured for 14 days. The clones were trypsinized, and cells were counted. Then 5 × 104 cells in 5 ml of 10% Dulbecco’s modified Eagle’s medium containing 0.4% (wt/vol) agarose were seeded in 5-cm-diameter petri dishes which contain 1% (wt/vol) agarose. Plates were inspected 21 to 28 days after seeding of the cells, and colonies were counted. The entire procedure, including transfection of cDNAs, was performed in triplicate.
Metastasis in athymic mice.
For this study, we used athymic nude (nmri/nude) mice that at the time of injection were 4 to 6 weeks of age. All mice were maintained in microisolator cages under HEPA-filtered laminar air. NIH 3T3 cells were transfected with pcDNA3-RING1 and pcDNA3 via calcium phosphate transfection and allowed to grow for 1 week in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum and 250 μg of geneticin (G-418) per ml. Cells were prepared for injection only from cultures in logarithmic growth at the time of harvest. The cells were briefly treated with 0.025% trypsin and 0.1% EDTA in salt solution. The cells were quickly removed from trypsin by centrifugation, resuspended in saline, and injected within 1 h in 0.2 ml in the body cavity with a 26-gauge needle. The mice were maintained under aseptic barrier conditions until the end of the experiment. After 6 weeks, the animals were analyzed for tumors at the surface and in sections of tissues.
RESULTS
Multiple interactions between RING1 and PcG proteins in the two-hybrid system.
Previously, we used the yeast two-hybrid system to identify proteins that interact with components of the multimeric PcG complex. We found that RING1, a previously identified protein with unknown function, interacts with the vertebrate Pc homologs XPc and HPC2 (21). It has been determined that the evolutionarily conserved C-terminal domain of the Pc homologs is the domain of RING1 interaction. The region within the RING1 protein which is responsible for the interaction with HPC2 has not been mapped in detail. However, RING1 contains a well-characterized zinc binding domain, the RING finger, which is not involved in the interaction with HPC2 (21). It has been argued that the RING finger is a domain involved in mediating protein-protein interactions (7). Therefore, it is feasible that RING1 interacts with other proteins besides HPC2. The fact that RING1 is part of a multimeric protein complex suggests that RING1 indeed may interact with more than one protein.
Having already characterized several human PcG proteins (9, 21, 22, 26), we used a directed two-hybrid assay to analyze the interactions of RING1. For this purpose, a so-called two-hybrid grid, containing different constructs of characterized PcG proteins, was designed (Table 1). Previously, differences in two-hybrid interactions were detected and attributed to possible steric hindrance due to the conformation of the two-hybrid fusion proteins (10). Further, the GAL4 DBD can be positioned at the N- or C-terminal end of the protein, and each case a different fusion protein is obtained. The two proteins may differ in three-dimensional conformation, with one hindering a potential protein-protein interaction. In order not to miss a two-hybrid interaction by possible steric hindrance of the GAL4 DBD at the N-terminal end of the protein, we constructed a novel two-hybrid GAL4 DBD fusion vector. In this vector, named pAS3, the DBD is placed at the C-terminal end of the fusion protein. To identify potential RING1 protein interactions, we screened the two-hybrid grid by using both the GAL4 DBD-RING1 (pAS2) and the RING1-GAL4 DBD (pAS3) fusion proteins. Using the RING1-GAL4 DBD construct, an interaction with RING1 itself was detected (Table 1). No RING1-RING1 interaction was detected when RING1 was cloned into the conventional N-terminal GAL4 DBD fusion vector (pAS2). Further, we found that BMI1, as well as HPC2, interacts with GAL4 DBD-RING1. The results are summarized in Table 1. Finally, we found that BMI1 is able to interact with itself. No interactions between RING1 and HPH1, HPH2, EED, and Enx1 could be detected.
TABLE 1.
β-Galactosidase activities of RING1 interactions in the two-hybrid system
| DBD fusion | TAD fusion (aa) | Interactiona |
|---|---|---|
| GAL4 DBD-RING1 | HPC2 (1–558) | + |
| BMI1 (1–326) | + | |
| RING1 (1–377) | − | |
| HPH1 (1–676) | − | |
| HPH1 (713–1013) | − | |
| HPH2 (137–432) | − | |
| Enx1 (1–746) | − | |
| EED (1–535) | − | |
| RING1-GAL4 DBD | HPC2 (1–558) | − |
| BMI1 (1–326) | − | |
| RING1 (1–377) | + | |
| HPH1 (1–676) | − | |
| HPH1 (713–1013) | − | |
| HPH2 (137–432) | − | |
| Enx1 (1–746) | − | |
| EED (1–535) | − |
White colonies were obtained on medium lacking both histidine and 3-AT. Blue colonies were obtained on medium lacking histidine but containing 3-AT.
RING1 interacts with multiple PcG proteins in vitro.
Using the two-hybrid system, we investigated the protein-protein interactions of RING1 and found that RING1 is able to interact with multiple PcG proteins (Table 1). However, different RING1 fusion proteins were used, and it was found that the RING1 protein interactions depend on whether the GAL4 DBD is fused to the N-terminal or C-terminal part of the RING1 protein (Table 1). To rule out the possibility of artifactual positive two-hybrid interactions and confirm the RING1 protein interactions, we performed an independent in vitro protein-protein interaction analysis, the GST pull-down assay.
Fusions of full-length RING1 (aa 1 to 377) and Bmi-1 (aa 1 to 324) proteins to GST were expressed in bacteria. The chimeric GST-RING1 and GST–Bmi-1 proteins were purified and immobilized to GST-Sepharose. Sepharose-bound GST-RING1 was incubated with full-length, in vitro-translated, [35S]methionine-labeled RING1, HPC2, and Bmi-1 proteins, and protein-protein interactions were analyzed. Similarly, interactions between GST–Bmi-1 and RING1 and Bmi-1 were examined.
Full-length HPC2 protein has a molecular mass of approximately 80 kDa (Fig. 1, lane 1), and the in vitro-translated, full-length HPC2 protein was able to bind to the immobilized GST-RING1 (Fig. 1, lane 3) but not to GST-Sepharose alone (Fig. 1, lane 2). Also, in vitro-translated, full-length RING1 (approximately 55 kDa; lane 4) and Bmi-1 (approximately 45 kDa; lane 7) both bound to GST-RING1 (Fig. 1, lanes 6 and 9, respectively). No binding of in vitro-translated RING1 and Bmi-1 with GST-Sepharose was observed (Fig. 1, lanes 5 and 8, respectively). Finally, we found that Bmi-1 is able to interact with itself since in vitro-translated, full-length Bmi-1 binds to immobilized GST–Bmi-1 (Fig. 1, lane 10).
FIG. 1.
Association of RING1 with itself, Bmi-1, and HPC2 and of Bmi-1 with itself in vitro. GST-RING1 fusion protein, immobilized on glutathione-Sepharose, interacted with in vitro-translated, [35S]methionine-labeled HPC2 or RING1. [35S]methionine-labeled HPC2 (lane 1) was incubated with GST-Sepharose alone (lane 2) and with GST-RING1 (lane 3). [35S]methionine-labeled RING1 (lane 4) was incubated with GST-Sepharose alone (lane 5) and with GST-RING1 (lane 6). GST-RING1 and GST–Bmi-1 fusion proteins, immobilized on glutathione-Sepharose, interacted with in vitro-translated, [35S]methionine-labeled Bmi-1. [35S]methionine-labeled Bmi-1 (lane 7) was incubated with GST-Sepharose alone (lane 8), with GST-RING1 (lane 9), and with GST–Bmi-1 (lane 10). All proteins used in the assay are full length. Molecular masses are indicated in kilodaltons. The input (lanes 1, 4, and 7) was 10% of the amount incubated with the GST fusion proteins.
These results confirm our two-hybrid data and also show that in vitro, the RING1 protein is able to interact with itself, Bmi-1, and HPC2 and that Bmi-1 interacts with itself.
RING1 contains two different domains involved in RING1-RING1 interaction.
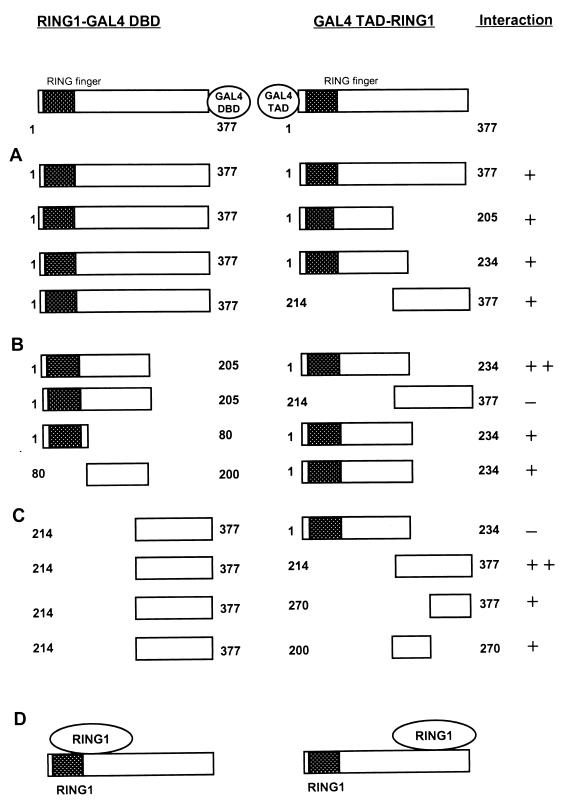
Using the yeast two-hybrid system and the in vitro GST pull-down assay, we found that RING1 is able to interact with itself. We performed a deletion analysis, using the yeast two-hybrid system, to determine the domains within the RING1 protein that are required for RING1-RING1 protein interaction. We found that RING1 contains two regions that are able to associate (Fig. 2). Both the N-terminal region (aa 1 to 205) and the C-terminal region (aa 214 to 377) of the RING1 protein interact with full-length RING1 (aa 1 to 377) (Fig. 2A). The N-terminal region contains the RING finger (aa 1 to 65). Mapping the two interaction domains further, we find that the N-terminal region of RING1 (aa 1 to 205) interacts strongly with the same N-terminal region (aa 1 to 205) but not with the C-terminal half of the RING1 protein (aa 214 to 377) (Fig. 2B). In determining whether the RING finger is involved in mediating this interaction, we made two deletion mutants. One RING1 deletion mutant (aa 1 to 80) still contains the N-terminally located RING finger domain (aa 1 to 65), and the other mutant contains the remaining N-terminal region (aa 80 to 200). We found that both regions are able to interact with RING1 (aa 1 to 205), but not as strongly as the intact N-terminal regions interact with each other (Fig. 2B).
FIG. 2.
Mapping of homodimerization domains of RING1. (A) Full-length RING1 (aa 1 to 377) was fused to the GAL4 DBD, which in all constructs shown is located at the C-terminal end of RING1. The plasmids were cotransformed with different portions of RING1, which were fused to the GAL4 TAD, which is located at the N terminus of RING1. Interactions were positive (+) when the transformants were able to grow on selective medium lacking histidine and when they were also β-galactosidase positive. Relative strength of the interactions is a qualitative indication based on the time needed for blue coloring (++, within 30 min; +, between 30 and 120 min) and the size of the colonies. (B) N-terminal portions of RING1 fused to the GAL4 DBD were tested for interactions with N- and C-terminal portions of RING1. These constructs are fused to the GAL4 TAD. (C) The C-terminal portion of RING1 fused to the GAL4 DBD was tested for interaction with C-terminal portions of RING1 fused to the GAL4 TAD. (D) Schematic representation of the two RING1-RING1 protein interaction domains. The RING finger domain of the RING1 protein is indicated as a hatched black box.
Next, we analyzed the interaction between RING1 and the C-terminal region of RING1 (Fig. 2C). We found that the C-terminal region of RING1 (aa 214 to 377) interacts with the C-terminal region of RING1 (aa 214 to 377) but not with the N-terminal region of the protein (aa 1 to 234) (Fig. 2C). It appeared that the C-terminal region of association is fairly large, as both of the two deletion mutants, RING1 (aa 200 to 270) and RING1 (aa 270 to 377), interact with the C-terminal region of RING1 (aa 214 to 377) (Fig. 2C). However, the interaction of these two deletion proteins is weaker than the interaction of the entire C-terminal region of RING1.
These results show that RING1 contains two different domains, which are both involved in self-binding. The C-terminal region interacts with the C-terminal region, and the N-terminal region interacts with the N-terminal region. Importantly, the C-terminal region does not interact with the N-terminal region. Further, it seems that in both interactions (Fig. 2B and C), several contact sites are involved, as different, separate domains are still able to interact. The RING finger region (aa 1 to 80), for example, is able to interact with the N-terminal half of RING1 (aa 1 to 205) but is not absolutely required, since a different region of the N-terminal-interacting region (aa 80 to 200), outside the RING finger domain, interacts with RING1 (aa 1 to 205). However, the interaction is stronger if the entire region, rather than the different domains, is involved. This finding indicates that several contact sites may be involved in the oligomerization of RING1.
Mapping of the RING1 interaction domain for HPC2.
Previously, RING1 was found to interact in the two-hybrid system with the evolutionarily conserved C-terminal box of vertebrate Pc homologs such as HPC2 (21). The domain within the RING1 protein that interacts with HPC2 has not been mapped precisely. We were interested in analyzing whether the RING1-RING1 and RING1-HPC2 protein interaction domains are identical, since we found that one of the interaction regions of RING1 (C-terminal region) is similar to the region of interaction with HPC2 (Fig. 2). The RING1 two-hybrid clone that we identified as interacting with HPC2 encompasses aa 214 to 377 (Fig. 3B). We made further deletions from this fragment and found that we could narrow down the interaction region only to aa 230 to 377 of RING1 (Fig. 3B). Smaller fragments of RING1 (aa 200 to 270 and aa 270 to 370) which were found to be involved in the self-binding of RING1 did not interact with HPC2. Yet a different fragment of RING1, ranging from aa 230 to 320, also appeared to be insufficient for the association with HPC2.
FIG. 3.
Mapping of interaction domains between RING1 and HPC2. (A) Indicated portions of HPC2 were fused to the GAL4 DBD, which in all constructs shown is located at the N-terminal end of HPC2. The plasmids were cotransformed with full-length RING1 (aa 1 to 377), which is fused to the GAL4 TAD. The GAL4 TAD is located at the N terminus of RING1. (B) Full-length HPC2 (aa 1 to 558) fused to the GAL4 DBD was tested for interaction with various C-terminal region of RING1 fused to the GAL4 TAD. (C) Schematic representation of the interaction domains of HPC2 and RING1. The HPC2 protein contains a chromodomain and a C box, which are indicated as grey and black dotted boxes, respectively. The RING finger domain of the RING1 protein is indicated as a hatched black box.
These data suggest that the C-terminal interaction domain of RING1 is different from the region involved in binding HPC2. A longer region of RING1 (aa 230 to 377) is needed for the association with HPC2 than for the RING1-RING1 interaction. It seems that the RING1-HPC2 interaction involves at least several contact sites which are all requisite for a bona fide interaction.
The RING finger proteins RING1 and BMI1 interact with each other.
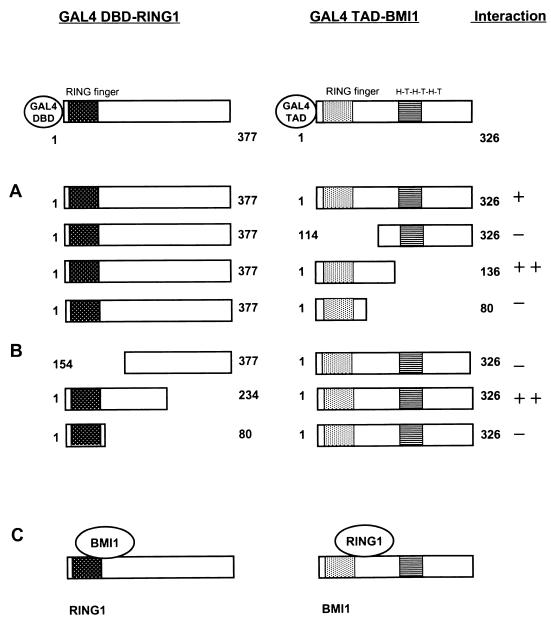
In analyzing the two-hybrid grid and performing the in vitro GST pull-down assay, we detected that RING1 and BMI1 are able to interact physically (Table 1 and Fig. 1). Further two-hybrid deletion analyses (Fig. 4) of different BMI1 regions show that the N-terminal region of BMI1 (aa 1 to 136) containing the RING finger motif is the region of interaction with RING1. The central and C-terminal regions of BMI1 (aa 114 to 326 [Fig. 4A]) containing the putative helix-turn-helix-turn-helix-turn motif is not required for the interaction. This finding suggests that the RING finger of BMI1 is the domain of interaction with RING1. However, the RING finger domain of BMI1 (aa 1 to 80) itself is not sufficient for the interaction (Fig. 4A). It seems, therefore, that both the RING finger and a region of the adjacent C-terminal region of BMI1 are needed for association with RING1.
FIG. 4.
Mapping of interaction domains between RING1 and BMI1. (A) Full-length RING1 (aa 1 to 377) was fused to the GAL4 DBD, which in all constructs shown is located at the N-terminal end of RING1. The plasmid was cotransformed with the indicated portions of BMI1, which is fused to the GAL4 TAD. In all constructs, the GAL4 TAD is located at the N-terminus of BMI1. (B) The indicated portions of RING1 were fused to the GAL4 DBD and tested for interaction with full-length BMI1 fused to the GAL4 TAD. (C) Schematic representation of the interaction domains of RING1 and BMI1. The RING finger domain of the RING1 protein is indicated as a hatched black box. The BMI1 protein has a RING finger domain and a helix-turn-helix-turn-helix-turn (H-T-H-T-H-T) domain, indicated as grey and striped boxes, respectively.
Further, we used a deletion analysis of RING1 to determine the region of RING1 that interacts with BMI1. For RING1, a region similar to that in BMI1 seems to be involved in the RING1-BMI1 association. The N-terminal region of RING1 (aa 1 to 234) showed strong interaction with BMI1. The RING finger domain of RING1 (aa 1 to 80) itself and the central region together with the C-terminal region of RING1 (aa 154 to 377) are not able to interact with BMI1 (Fig. 4B). The RING finger domain of RING1 itself is not sufficient for the interaction with BMI1.
In conclusion, RING1 and BMI1 are found to interact with each other. The domain within RING1 that is responsible for the RING1-BMI1 association seems different from that needed for the RING1-RING1 association. For the latter association, the RING finger itself shows binding activity but is not required for the interaction. In the RING1-BMI1 interaction, both RING fingers are unable to interact on their own, but they do seem to be involved in the interaction together with a region adjacent to the RING finger.
BMI1 is able to interact with itself.
Studying the protein-protein interactions of RING1, we found that RING1 is able to interact with itself. The ability to oligomerize has been detected for several other PcG proteins (9, 10, 19). Therefore, we studied whether BMI1 is also able to interact with itself and found that indeed BMI1 interacts with itself in vitro (Fig. 1). Next, two-hybrid deletion analyses were performed to map the domains of interaction.
Two-hybrid analyses show that different regions of BMI1 are able to interact with full-length BMI1 (Fig. 5A). Both the N-terminal region (aa 1 to 136 and aa 1 to 80), including the RING finger, and the C-terminal region (aa 114 to 326) fused to the GAL4 TAD are able to interact with full-length BMI1 fused to the GAL4 DBD (Fig. 5A). However, the N-terminal region of BMI1 (aa 1 to 136) does not interact with full-length BMI1 when the GAL4 DBD and GAL4 TAD are switched (Fig. 5B). Deletion analysis of the C-terminal part of BMI1 (aa 136 to 326) shows that it interacts with the C-terminal region of BMI1 (aa 114 to 326) but not with the N-terminal region of BMI1 (aa 1 to 136) (Fig. 5C). These results suggest that different regions are involved in the oligomerization of BMI1. The C-terminal region of BMI1 interacts with the C-terminal region of BMI1 but not with the N-terminal region of BMI1. Further, the RING finger domain of BMI1 is also able to associate with the BMI1 protein.
FIG. 5.
Mapping of homodimerization domains of BMI1. (A) Full-length BMI1 (aa 1 to 326) is fused to the GAL4 DBD, which in all constructs shown is located at the C-terminal end of BMI1. The plasmid was cotransformed with different portions of BMI1, which are fused to the GAL4 TAD. In these constructs, the GAL4 TAD is located at the N terminus of BMI1. (B) An N-terminal portion of BMI1 (aa 1 to 136) fused to the GAL4 DBD was tested for interaction with full-length BMI1 (aa 1 to 326) fused to the GAL4 TAD. (C) The C-terminal portion of BMI1 (aa 1 to 136) fused to the GAL4 DBD was tested for interaction with different portions of BMI1 fused to the GAL4 TAD. (D) Schematic representation of the homodimerization domains of BMI1. The BMI1 protein has a RING finger domain and a helix-turn-helix-turn-helix-turn (H-T-H-T-H-T) domain, indicated as grey and striped boxes, respectively.
RING1 overexpression results in repression of engrailed and enhanced expression of c-jun and c-fos.
The PcG protein complex is involved in repression of gene activity. Since RING1 appears to be an integral part of the PcG complex, displaying multiple interactions with PcG proteins, we studied the function of RING1 in regulating gene expression. We overexpressed the RING1 protein in Rat1a fibroblast cells and analyzed differences in gene expression levels.
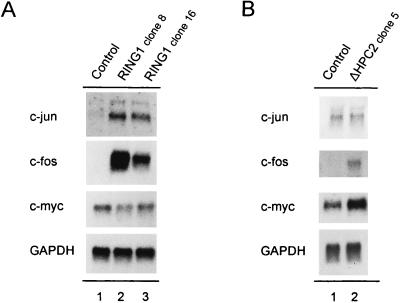
To establish stable cell lines that overexpress the RING1 protein, we transfected Rat1a fibroblast cells. We tested individual clones for proper overexpression of RING1 by Western analysis and found higher levels of RING1 in different clones of Rat1a/RING1 cells than in untransfected cells (Fig. 6A). We selected for further analysis two clones expressing higher levels of RING1 protein (Fig. 6A, lane 2 and 3).
FIG. 6.
Western analysis of stably transfected RING1 and ΔHPC2 proteins from cell extracts of Rat1a cells. Equal amounts of proteins were Western blotted. The blots were incubated with a rabbit anti-RING1 or rabbit anti-HPC2 antibody. (A) Endogenous rat RING1 levels were detected in the untransfected cells (lane 1); elevated levels of RING1 were detected in clone 8 (lane 2) and clone 16 (lane 3). (B) Endogenous rat HPC2 levels were detected in the untransfected cells (lane 1); elevated levels of ΔHPC2 were detected in clone 5 (lane 2). Molecular masses are indicated in kilodaltons.
To analyze differential gene expression levels, we used a mouse Atlas cDNA expression array (Clontech) consisting of two identical filters containing approximately 600 cDNAs of characterized genes. We isolated poly(A)+ mRNA from control cells Rat1a cells and from Rat1a cells overexpressing RING1 (Fig. 6A, lane 1 and 2). The poly(A)+ mRNA isolated from the two cell lines were used to make cDNA, which was labeled and subsequently used for probing the Atlas filters. The filters were autoradiographed, and the films were developed after several days. The strength of the hybridization signal is a measure for the expression level of a gene. Individual gene expression levels can be analyzed by comparing the hybridization signals of a gene from the control filter and the RING1 filter. Hybridization levels were analyzed with a phosphorimager. We found that approximately 20 genes of the 600 on the filter were either upregulated or downregulated due to the overexpression of RING1. Most of these genes are involved in the cell cycle, oncogenesis, or development (data not shown). RING1 overexpression therefore does not affect global gene expression levels, but the targeted genes represent a rather specific selection. We analyzed three of these genes (see below).
To verify the differential expression of genes that we detected in the Atlas cDNA expression arrays, we performed Northern blot analysis with poly(A)+ mRNA of the same cell lines used for the Atlas expression arrays and for the Western analysis of RING1 expression (Fig. 6A). In Rat1a/RING1 cells, we found strong repression of the expression of the mouse engrailed gene, En-2 (Fig. 7). The expression levels of En-2 in the cells overexpressing RING1 are strongly reduced on Northern blots in two independently established clones (Fig. 7, lanes 2 and 3). In Drosophila, the engrailed gene is a direct target gene of PcG proteins (32).
FIG. 7.
Repression of En-2 gene expression activity in RING1-transfected Rat1a cells. Poly(A)+ mRNA isolated from Rat1a control cells (lane 1) and from RING1-transfected clone 8 (lane 2) and clone 16 (lane 3) Rat1a cells was Northern blotted and probed with the En-2 gene. To verify equal RNA loading, the filter was hybridized with a GAPDH probe.
We also found that the expression of two proto-oncogenes, c-fos and c-jun, is strongly enhanced (Fig. 8A). In control cells, the expression of c-fos and c-jun is hardly detectable (Fig. 8A, lane 1). Phosphorimager analysis showed at least a 10-fold increase in the expression level of both proto-oncogenes in cells overexpressing the RING1 protein in two distinct clones (Fig. 8A, lane 2 and 3). In contrast, the c-myc expression level was not changed by the overexpression of RING1 (Fig. 8A).
FIG. 8.
Expression of c-myc, c-jun, and c-fos in RING1- and ΔHPC2-transfected Rat1a cells. (A) Poly(A)+ mRNA isolated from Rat1a control cells (lane 1) and from RING1-transfected Rat1a clone 8 (lane 2) and clone 16 (lane 3) cells was Northern blotted and probed with fragments of c-jun, c-fos, and c-myc. (B) Poly(A)+ mRNA isolated from Rat1a control cells (lane 1) and from ΔHPC2-transfected Rat1a clone 5 (lane 2) cells was Northern blotted and probed with fragments of c-jun, c-fos, and c-myc. To verify equal RNA loading, the filter was hybridized with a GAPDH probe.
Effects of overexpression of ΔHPC2 on proto-oncogene expression.
In a previous study (22), we had shown that overexpression of a C-terminal deletion mutant of HPC2, ΔHPC2 (aa 1 to 530), which is not able to repress gene activity, resulted in elevated expression of c-myc in the mammalian cell lines C57MG and U-2 OS. Surprisingly, overexpression of RING1, which is likely a molecular partner of HPC2 in vivo, does not result in a changed c-myc expression level in Rat1a cells (Fig. 8A). The difference in c-myc gene activity by the overexpression of either RING1 or ΔHPC2 may depend on the difference in cell lines that are used in the two studies. Another plausible explanation is that RING1 and ΔHPC2 affect expression of the c-myc proto-oncogene differently. To address this question, we also analyzed the expression levels of the proto-oncogenes c-myc, c-jun, and c-fos in Rat1a cells stably transfected with ΔHPC2.
Individual clones of Rat1a/ΔHPC2 cells were tested for proper expression of ΔHPC2 by Western analysis, and a representative clone was taken for RNA analysis (Fig. 6B, clone 5). Poly(A)+ mRNA from control Rat1a and from Rat1a/ΔHPC2 clone 5 were blotted, and the mRNA expression levels of c-myc, c-jun, and c-fos were determined. We found that overexpression of ΔHPC2 in Rat1a cells results in a deregulated, enhanced gene expression of both c-myc and c-fos proto-oncogenes (Fig. 8B). However, the expression level of c-jun was not changed by the overexpression of ΔHPC2 in these cell lines (Fig. 8B).
RING1 induces anchorage-independent growth.
Rat1a fibroblast cells are frequently used to determine the neoplastic transformation potential of genes (5, 20, 22, 28, 31). Overexpression of the proto-oncogene c-myc alone in Rat1a cells is sufficient to induce anchorage-independent growth in soft agarose (28, 31). Overexpression of the dominant-negative C-terminal deletion mutant ΔHPC2 enhances the expression of c-myc and induces anchorage-independent growth upon overexpression in Rat1a cells (22). We found that overexpression of RING1 in Rat1a fibroblast cells results in an enhanced expression of the proto-oncogenes c-fos and c-jun but not c-myc (Fig. 8A). We therefore analyzed the potential of the RING1 protein to induce anchorage-independent growth in Rat1a cells.
We found that RING1 induces anchorage-independent growth of Rat1a cells (Table 2 and Fig. 9). Surprisingly, the effect of RING1 on the neoplastic transformation of Rat1a cells seems much stronger than the effect of the positive controls. As positive controls for the induction of anchorage-independent growth, Rat1a cells were transfected with c-myc or the C-terminal deletion mutant of HPC2, which enhances c-myc expression (Fig. 8B). Both the number (approximately 500 colonies/5 × 104 transfected cells) and the size of the colonies are comparable between these cell lines (Table 2 and Fig. 9). However, for Rat1a/RING1 cells, colonies were not only more numerous (approximately 750 versus 500/5 × 104 transfected cells) but also on average approximately twofold larger in diameter (Table 2 and Fig. 9). Further, RING1 and two deletion mutants of RING1, RING1/aa 1 to 205 and RING1/aa 154 to 377, were transfected in Rat1a cells. RING1/aa 1 to 203 contains the RING finger domain but lacks the C-terminal half of the protein, whereas RING1/aa 154 to 377 lacks the RING finger domain. Overexpression of either deletion mutant did not induce colonies of Rat1a cells (Table 2).
TABLE 2.
Colony formation by RING1-transfected Rat1a cells in soft agarose
| Construct (aa) | No. of colonies/5 × 104 transfected cellsa (mean ± SEM) |
|---|---|
| None | 0 |
| pcDNA3 | 0 |
| pRcCMV | 432 ± 67 |
| pcDNA3-ΔHPC2 | 529 ± 39 |
| pcDNA3-RING1 (1–203) | 0 |
| pcDNA3-RING1 (154–377) | 0 |
| pcDNA3-RING1 | 754 ± 112 |
A total of 5 × 104 of each pool of transfected and Geneticin-selected cells was seeded into 0.4% top agarose, and colonies with diameters of >0.1 mm were counted 21 to 28 days after seeding. The entire procedure, including transfection of the cDNAs, was performed in triplicate.
FIG. 9.
Overexpression of RING1 induces anchorage-independent growth in control Rat1a cells (A) and in Rat1a cells transformed with the c-myc oncogene (B), with the C-terminal deletion mutant of HPC2 (C), and with full-length RING1 (D). Bar = 400 μm.
RING1 demonstrates metastatic activity in athymic mice.
Invasion and metastasis have been considered the hallmarks of malignant tumors. NIH 3T3 cells overexpressing oncogenes are found to be metastatic in nude mice (3, 35). This metastatic assay is often used to assess the oncogenic potential of genes. For the PcG proteins Bmi-1 and mel-18, involvement in the formation of tumors in mice has been established (2, 13). Transgenic mice develop lymphomas when overexpressing Bmi-1, which is considered an onco-protein. Nude mice injected with NIH 3T3 cells overexpressing antisense mel-18 also develop tumors (13). We found RING1 to be a potent inducer of anchorage-independent growth in cells and therefore determined the metastatic potential of RING1 by injecting athymic nude mice with NIH 3T3 cells that overexpress RING1.
NIH 3T3 cells were transfected with RING1 (pcDNA3-RING1) and with the empty expression vector (pcDNA3). Nude mice were injected in the body cavity with control cells (NIH 3T3 and NIH 3T3/pcDNA3) and with NIH 3T3 cells transfected with RING1 (NIH 3T3/RING1). Five of the eight mice injected with NIH 3T3/RING1 cells developed tumors; tumors were found throughout the body cavity, predominantly in the liver and epithelial tissues but also on the intestine and kidneys. No tumors were detected in the other three mice injected with NIH 3T3/RING1 cells. Importantly, no tumors were detected in the control groups (four mice per group). These results indicate that RING1 induces the formation of tumors significantly; however, the control mice did not develop any tumors, which suggests that the formation of tumors in NIH 3T3/RING1 cells is a result of RING1 overexpression.
DISCUSSION
RING1 interacts with multiple PcG proteins.
PcG proteins serve as components of multimeric PcG protein complexes, which are involved in the heritable repression of gene activity. RING1 is a newly identified PcG complex-associated protein in mammals. However, the role of RING1 as a component of the PcG complex is unclear. RING1 is found to interact with vertebrate Pc homologs, but a Drosophila RING1 homolog has not been identified. To better understand the role of RING1 as a component of the PcG complex, we investigated the protein-protein interactions of RING1.
Using a combination of directed two-hybrid and in vitro binding analyses, we found that RING1 interacts with multiple PcG proteins: itself, HPC2, and BMI1. (i) RING1 is able to interact with itself via two independent domains. The C-terminal region interacts with the C-terminal region, and the N-terminal region interacts with the N-terminal region; the C-terminal region does not interact with the N-terminal region. (ii) We also mapped the interaction of RING1 with HPC2. A large C-terminal part of RING1 is required for the interaction with HPC2. The interaction domain of RING1 that is responsible for the association with HPC2 is distinct from the domain that interacts with RING1. (iii) For the RING1-BMI1 protein-protein interaction, we found that both the RING1 fingers and the regions adjacent to that motif of RING1 and of BMI1 are needed. These results suggest that HPC2 and BMI1 are able to interact with RING1 at the same time and that RING1 is an integral part of the PcG complex.
Model for distinct PcG complexes.
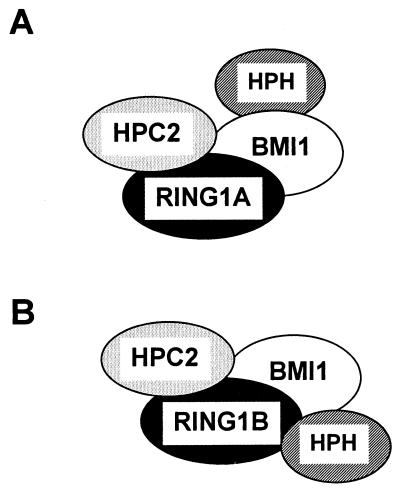
RING1 interacts with multiple PcG proteins and therefore may serve as a central protein for the establishment of a multimeric protein complex. Recently, a human RING1 homolog, dinG, has been identified (12). Two mouse RING homologs have also been identified; mouse Ring1A and Ring1B are homologous to RING1 and dinG, respectively. The dinG protein is 31 aa shorter than and 53% identical with RING1. It contains three regions that are very homologous with RING1. The first 150 aa, including the RING finger, are almost 100% identical; the other two regions of homology are located in the C-terminal part of the proteins and are about 70% identical. Especially the central region, between the RING finger and the C-terminal homology domains, is a region with little homology between the two RING proteins. We found that the N-terminal region of RING1 (aa 1 to 200), which is well conserved between RING1 and dinG, is involved in the self-association of the protein. Therefore, it is likely that dinG, like RING1, can interact with itself via its N-terminal part and that RING1 and dinG are able to interact with each other. The possible interaction of RING1 and dinG would also involve the N-terminal parts of the proteins.
Recently, several mouse proteins have been found to interact with Ring1B/dinG. It has been found that mouse Ring1B/dinG is able to interact with Bmi-1 and with the mouse homolog of HPH2, MPh2 (12). The interaction of Ring1B/dinG with Bmi-1 involves the N-terminal region, including the RING finger. This region is well conserved between RING1 and Ring1B/dinG, and the finding that dinG is involved in the interaction with Bmi-1 is in agreement with our results that RING1 is able to associate with BMI1. Further, it has been found that Ring1B/dinG interacts with MPh2 through its central and C-terminal regions. We have shown that RING1 is not able to interact with human homologs of Ph, HPH1 and HPH2 (Table 1). The region of Ring1B/dinG that is involved in the interaction with MPh2 is not homologous with the corresponding region of RING1. This would imply that Ring1B/dinG contains a specific region that is absent in RING1 and is responsible for the interaction with MPh2. These results indicate that RING1 and dinG are able to interact with different proteins, which could provide specificity for the formation of different PcG protein complexes.
The abilities of RING1/Ring1A and Ring1B/dinG to associate with multiple and different vertebrate PcG proteins suggest that depending on the presence of either RING1/Ring1A or Ring1B/dinG, different vertebrate PcG complexes can be formed. If the RING1/Ring1A protein is present, it can interact with BMI1 but not with HPH2. Next, in the protein-protein association of RING1/Ring1A and BMI1, HPH2 is able to interact with BMI1 (Fig. 10A). If Ring1B/dinG is present, both BMI1 and HPH2 can interact with Ring1B/dinG directly (Fig. 10B). Further, both RING proteins are able to interact with HPC2. It has also been found that the vertebrate PcG proteins BMI1 and HPC2 interact with each other. Mouse homologs of BMI1 and HPC2, Bmi-1 and M33, respectively, have been found to interact in the two-hybrid system (10). Also in Xenopus, direct interactions between homologs of the vertebrate PcG proteins BMI1 and HPC2 have been detected (19). These data suggest that BMI1 is able to interaction with either RING1 or HPC2 in the formation of a multimeric PcG complex. Previously, coimmunoprecipitation experiments presented additional evidence that RING1, BMI1, HPC2, and HPH1 are, indeed, in vivo associated (21). Importantly, here we present models for human PcG complexes in which at least four proteins, RING1, HPC2, BMI1, and HPH2, interact with each other.
FIG. 10.
Model of human PcG multimeric protein complexes. (A) Model of a PcG protein complex which contains RING1/Ring1A; (B) dinG/Ring1B as the central protein for the establishment of a multimeric PcG protein complex.
Deregulated gene activity in RING1-transformed cells.
In Drosophila, PcG proteins have been identified as repressors of homeotic genes (18, 36). PcG proteins have also been found to regulate the expression of gap genes (17) and to self-regulate the expression of certain PcG proteins (18, 36). Previously we have shown that interference with the function of HPC2 deregulates the expression of c-myc. PcG proteins bind to more than 100 loci on the polytene chromosome, of which the majority of the genes have not been determined. It is likely that besides the regulation of genes involved in development, different classes of genes are regulated by PcG proteins. We analyzed the differential expression of genes after overexpression of RING1 in rodent Rat1a fibroblast cells.
Upon the overexpression of RING1, a mouse homolog of engrailed, En-2, is downregulated, resulting in a decreased expression. From these data it cannot be concluded whether the effect of RING1 on En-2 is direct or indirect. However, overexpression of a protein, RING1, that represses gene activity (32) may result in a decreased gene expression. It is therefore feasible that the effect of RING1 on En-2 expression is direct and that En-2 is a direct target gene of RING1. In vivo cross-linking experiments and polytene chromosome binding analyses have shown that the Drosophila engrailed gene is a direct target gene for PcG proteins (32). The functions of different PcG proteins are evolutionarily conserved. For instance, both in Drosophila and in vertebrates, PcG proteins are involved in the regulation of homeotic genes. This suggests that engrailed is also a target gene for mammalian PcG proteins and that En-2 is a direct target gene of RING1.
We have further found that overexpression of RING1 results in a deregulated, strongly enhanced expression of the proto-oncogenes c-jun and c-fos. It can be expected that overexpression of the RING1 gene repressor in general results in a decreased gene expression, as we saw for En-2. The enhanced expression of the proto-oncogenes c-jun and c-fos therefore is not likely to be a direct effect of RING1. Instead, it is more likely that upstream regulator genes are direct target genes of RING1.
Previously, we have observed that overexpression of HPC2 represses the expression of the proto-oncogene c-myc in the mammalian cell lines U-2 OS and C57MG (22). Surprisingly, we did not detect effects on c-myc expression upon the overexpression of RING1. If RING1 has a function in gene expression regulation similar to that of HPC2, a comparable effect on c-myc gene activity would be expected as the result of overexpression of either protein. An explanation for the different effects of RING1 and HPC2 overexpression on c-myc expression may be the difference in cell lines used. In that case, the action of the PcG complexes could be cell type specific. However, comparative analysis of c-myc, c-fos, and c-jun expression upon the overexpression of either RING1 or ΔHPC2 in Rat1a cells (Fig. 8) clearly shows that RING1 and HPC2 have different effects on the expression of at least c-myc and c-jun. These results argue against the idea of a cell-type-specific difference of c-myc expression by PcG proteins and suggest instead that RING1 and HPC2 have distinct regulatory effects on different genes.
Involvement of RING1 in tumorigenesis.
In this study, we investigated the role of RING1 as a component of the vertebrate PcG complex. We found that RING1 interacts with multiple PcG proteins, which indicates that RING1 may function as a central protein in the formation of vertebrate PcG protein complexes. Since several PcG proteins have been implicated in tumorigenesis, we analyzed the potential role of RING1 in this process. We found that overexpression of RING1 results in enhanced expression of the proto-oncogenes c-jun and c-fos but not c-myc. Concomitantly, RING1 is able to induce anchorage-independent growth of Rat1a cells. Moreover, RING1 is a more potent inducer of anchorage-independent growth than are c-myc and ΔHPC2, as measured by both number and size of foci. This observation coincides with the finding that overexpression of ΔHPC2 results in enhanced expression of both c-myc and c-fos but not of c-jun. It suggests (i) that the induction of anchorage-independent growth by RING1 is mediated by elevated c-fos and c-jun expression, whereas the induction of anchorage-independent growth by ΔHPC2 is mediated by elevated c-fos and c-myc expression, and (ii) that the cooperative action of elevated c-fos and c-jun levels is better able than that of enhanced c-fos and c-myc levels to induce neoplastic transformation in Rat1a cells. In this respect, it is of considerable interest that overexpression of human c-jun alone is able to transform Rat1a cells and that this effect is enhanced by the expression of c-fos (25). Finally, the differential enhancement of c-fos, c-jun, and c-myc expression due to overexpression of either RING1 or ΔHPC2 suggests that these proteins cause cellular transformation through different molecular pathways.
As a second assay to determine whether RING1 is involved in tumorigenesis, we studied if RING1 is able to induce metastasis. We found that NIH 3T3 cells overexpressing RING1 form tumors when injected into nude mice. In this respect, RING1 is similar to Bmi-1, another vertebrate PcG protein which is involved in tumorigenesis. Overexpression of Bmi-1 induces the formation of tumors, and therefore bmi-1 is considered to be a proto-oncogene (2). The vertebrate PcG protein mel-18, on the other hand, is considered to be encoded by a tumor suppressor gene since overexpression of antisense DNA but not of sense DNA induces the formation of tumors (13). Taken together, our results support the increasing evidence that chromatin-associated PcG proteins are linked to human diseases like cancer.
ACKNOWLEDGMENTS
We thank Daniel Olson for help with experiments on the induction of tumors in athymic mice. We thank Roel van Driel and Marco Gunster for critically reading the manuscript, and we thank Karien Hamer and Jan den Blaauwen for technical assistance.
REFERENCES
- 1.Alkema M J, Bronk M, Verhoeven E, Otte A P, van’t Veer L J, Berns A, van Lohuizen M. Identification of Bmi1-interacting proteins as constituents of a multimeric mammalian Polycomb complex. Genes Dev. 1997;11:226–240. doi: 10.1101/gad.11.2.226. [DOI] [PubMed] [Google Scholar]
- 2.Alkema M J, Jacobs H, Van Lohuizen M, Berns A. Perturbation of B and T cell development and predisposition to lymphomagenesis in EμBmi1 transgenic mice require the Bmi1 RING finger. Oncogene. 1997;15:899–910. doi: 10.1038/sj.onc.1201262. [DOI] [PubMed] [Google Scholar]
- 3.Bradley M O, Kraynak A R, Storer R D, Gibbs J B. Experimental metastasis in nude mice of NIH 3T3 cells containing various ras genes. Proc Natl Acad Sci USA. 1986;83:5277–5281. doi: 10.1073/pnas.83.14.5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buchenau P, Hodgson J, Strutt H, Arndt-Jovin D J. The distribution of polycomb-group proteins during cell division and development in Drosophila embryos; impact on models for silencing. J Cell Biol. 1998;141:469–481. doi: 10.1083/jcb.141.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen K J, Hanna J S, Prescott J E, Dang C V. Transformation by the Bmi-1 oncoprotein correlates with its subnuclear localization but not its transcriptional suppression activity. Mol Cell Biol. 1996;16:5527–5535. doi: 10.1128/mcb.16.10.5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franke A, DeCamillis M, Zink D, Cheng N, Brock H W, Paro R. Polycomb and polyhomeotic are constituents of a multimeric protein complex in chromatin of Drosophila melanogaster. EMBO J. 1992;11:2941–2950. doi: 10.1002/j.1460-2075.1992.tb05364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freemont P S, Hanson I M, Townsdale J. A novel cysteine-rich sequence element. Cell. 1991;64:483–484. doi: 10.1016/0092-8674(91)90229-r. [DOI] [PubMed] [Google Scholar]
- 8.Gould A. Functions of mammalian Polycomb group and trithorax group related genes. Curr Opin Genet Dev. 1997;7:488–494. doi: 10.1016/s0959-437x(97)80075-5. [DOI] [PubMed] [Google Scholar]
- 9.Gunster M J, Satijn D P E, Hamer K M, den Blaauwen J L, de Bruijn D, Alkema M J, van Lohuizen M, van Driel R, Otte A P. Identification and characterization of interactions between the vertebrate Polycomb-group protein BMI1 and human homologs of Polyhomeotic. Mol Cell Biol. 1997;17:2326–2335. doi: 10.1128/mcb.17.4.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hashimoto N, Brock H W, Nomura M, Kyba M, Hodgson J, Fujita Y, Takihara Y, Shimada K, Higashinakagawa T. RAE28, BMI1, and M33 are members of heterogeneous multimeric mammalian Polycomb group complexes. Biochem Biophys Res Commun. 1998;245:356–365. doi: 10.1006/bbrc.1998.8438. [DOI] [PubMed] [Google Scholar]
- 11.Haupt Y, Alexander W S, Barri G, Klinken S P, Adams J M. Novel zinc finger gene implicated as myc collaborator by retrovirally accelerated lymphogenesis in E(mu)-myc transgenic mice. Cell. 1991;65:753–763. doi: 10.1016/0092-8674(91)90383-a. [DOI] [PubMed] [Google Scholar]
- 12.Hemenway C S, Halligan B W, Levy L S. The Bmi-1 oncoprotein interacts with dinG and Mph2: the role of RING1 finger domains. Oncogene. 1998;16:2541–2547. doi: 10.1038/sj.onc.1202042. [DOI] [PubMed] [Google Scholar]
- 13.Kanno M, Hasegawa M, Ishida A, Isono K, Taniguchi M. mel-18, a Polycomb group-related mammalian gene, encodes a transcriptional negative regulator with tumor suppressive activity. EMBO J. 1995;14:5672–5678. doi: 10.1002/j.1460-2075.1995.tb00254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kingston R E, Bunker C A, Imbalzano A N. Repression and activation by multiprotein complexes that alter chromatin structure. Genes Dev. 1996;10:905–920. doi: 10.1101/gad.10.8.905. [DOI] [PubMed] [Google Scholar]
- 15.Lessard J, Baban S, Sauvageau G. Stage-specific expression of polycomb group genes in human bone marrow cells. Blood. 1998;91:1216–1224. [PubMed] [Google Scholar]
- 16.Lewis E B. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- 17.Pelegri F, Lehmann R. A role of Polycomb group genes in the regulation of Gap gene expression in Drosophila. Genetics. 1994;136:1341–1353. doi: 10.1093/genetics/136.4.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rastelli L, Chan C S, Pirotta V. Related chromosome binding sites for zeste, suppressors of zeste and Polycomb group proteins in Drosophila and their dependence on Enhancer of zeste function. EMBO J. 1993;12:1513–1522. doi: 10.1002/j.1460-2075.1993.tb05795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reijnen M J, Hamer K M, den Blaauwen J L, Lambrechts C, Schoneveld I, van Driel R, Otte A P. Polycomb and bmi-1 homologs are expressed in overlapping patterns in Xenopus embryos and are able to interact with each other. Mech Dev. 1995;53:35–46. doi: 10.1016/0925-4773(95)00422-x. [DOI] [PubMed] [Google Scholar]
- 20.Samuels M L, Weber M J, Bishop J M, McMahon M. Conditional transformation of cells and rapid activation of the mitogen-activated protein kinase cascade by an estradiol-dependent human Raf-1 protein kinase. Mol Cell Biol. 1993;13:6241–6252. doi: 10.1128/mcb.13.10.6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Satijn D P E, Gunster M J, van de Vlag J, Hamer K M, Schul W, Alkema M J, Saurin A J, Freemont P S, van Driel R, Otte A P. RING1 is associated with the Polycomb-group protein complex and acts as a transcriptional repressor. Mol Cell Biol. 1997;17:4105–4113. doi: 10.1128/mcb.17.7.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Satijn D P E, Olson D J, Van der Vlag J, Hamer K M, Lambrechts C, Masselink H, Gunster M J, Sewalt R G A B, Van Driel R, Otte A P. Interference with the expression of a novel human Polycomb protein, HPC2, results in cellular transformation and apoptosis. Mol Cell Biol. 1997;17:6076–6086. doi: 10.1128/mcb.17.10.6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schoorlemmer J, Marcos C, Were F, Martinez R, Garcia E, Satijn D P E, Otte A P, Vidal M. Ring1A is a transcriptional repressor that interacts with the Polycomb-M33 protein and is expressed at rhombomere boudaries in the mouse hindbrain. EMBO J. 1997;16:5930–5942. doi: 10.1093/emboj/16.19.5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schumacher A, Magnuson T. Murine Polycomb- and trithorax-group genes regulate homeotic pathways and beyond. Trends Genet. 1997;13:167–170. [PubMed] [Google Scholar]
- 25.Schutte J, Viallet J, Nau M, Segal S, Fedorko J, Minna J. Jun-B inhibits and c-fos stimulates the transforming and transactivating activities of c-jun. Cell. 1989;59:987–997. doi: 10.1016/0092-8674(89)90755-1. [DOI] [PubMed] [Google Scholar]
- 26.Sewalt R G A B, van der Vlag J, Gunster M J, Hamer K M, den Blaauwen J L, Satijn D P E, Hendrix T, van Driel R, Otte A P. Characterization of interactions between the mammalian Polycomb-group proteins Enx1/EZH2 and EED suggests the existence of different mammalian Polycomb-group protein complexes. Mol Cell Biol. 1998;18:3586–3595. doi: 10.1128/mcb.18.6.3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simon J. Locking in stable states of gene expression: transcriptional control during Drosophila development. Curr Opin Cell Biol. 1995;7:376–385. doi: 10.1016/0955-0674(95)80093-x. [DOI] [PubMed] [Google Scholar]
- 28.Small M B, Hay N, Schwab M, Bishop J M. Neoplastic transformation by the human gene N-myc. Mol Cell Biol. 1987;7:1638–1645. doi: 10.1128/mcb.7.5.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith D B, Johnson K S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 30.Smith P K, Krohn R I, Hermanson G T, Mallia A K, Gartner F H, Provenzano M D, Fujimoto E K, Goeke N M, Olson B J, Klenk D C. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 31.Stone J, de Lange T, Ramsay G, Jokobovits E, Bishop J M, Varmus H E, Lee W M. Definition of regions in human c-myc that are involved in transformation and nuclear localization. Mol Cell Biol. 1987;7:1697–1709. doi: 10.1128/mcb.7.5.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Strutt H, Paro R. The Polycomb group protein complex of Drosophila has different compositions at different target genes. Mol Cell Biol. 1997;17:6773–6783. doi: 10.1128/mcb.17.12.6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Lohuizen M, Verbeek S, Scheijen B, Wientjes E, van der Gulden H, Berns A. Identification of cooperating oncogenes in E(mu)-myc transgenic mice by provirus tagging. Cell. 1991;65:737–752. doi: 10.1016/0092-8674(91)90382-9. [DOI] [PubMed] [Google Scholar]
- 34.Van Lohuizen M, Thijms M, Voncken J W, Schumacher A, Magnuson T, Wientjes E. Interaction of mouse Polycomb-group (Pc-G) proteins Enx1 and Enx2 with Eed: indication for separate Pc-G complexes. Mol Cell Biol. 1998;18:3572–3579. doi: 10.1128/mcb.18.6.3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wallace J S, Hayle A J, Syms A J, Cairney M, Tutty B, Gazzard A, Evans M F, Fleming K A, Tarin D. The ras oncogene and tumour metastasis: observations on murine cells transfected with activated human c-Ha-ras. Differentiation. 1989;41:208–215. doi: 10.1111/j.1432-0436.1989.tb00749.x. [DOI] [PubMed] [Google Scholar]
- 36.Zink B, Paro R. In vivo binding pattern of a transregulator of homeotic genes in Drosophila melanogaster. Nature. 1989;337:468–471. doi: 10.1038/337468a0. [DOI] [PubMed] [Google Scholar]