Abstract
The regulation of the c-src N1 exon is mediated by an intronic splicing enhancer downstream of the N1 5′ splice site. Previous experiments showed that a set of proteins assembles onto the most conserved core of this enhancer sequence specifically in neuronal WERI-1 cell extracts. The most prominent components of this enhancer complex are the proteins hnRNP F, KSRP, and an unidentified protein of 58 kDa (p58). This p58 protein was purified from the WERI-1 cell nuclear extract by ammonium sulfate precipitation, Mono Q chromatography, and immunoprecipitation with anti-Sm antibody Y12. Peptide sequence analysis of purified p58 protein identified it as hnRNP H. Immunoprecipitation of hnRNP H cross-linked to the N1 enhancer RNA, as well as gel mobility shift analysis of the enhancer complex in the presence of hnRNP H-specific antibodies, confirmed that hnRNP H is a protein component of the splicing enhancer complex. Immunoprecipitation of splicing intermediates from in vitro splicing reactions with anti-hnRNP H antibody indicated that hnRNP H remains bound to the src pre-mRNA after the assembly of spliceosome. Partial immunodepletion of hnRNP H from the nuclear extract partially inactivated the splicing of the N1 exon in vitro. This inhibition of splicing can be restored by the addition of recombinant hnRNP H, indicating that hnRNP H is an important factor for N1 splicing. Finally, in vitro binding assays demonstrate that hnRNP H can interact with the related protein hnRNP F, suggesting that hnRNPs H and F may exist as a heterodimer in a single enhancer complex. These two proteins presumably cooperate with each other and with other enhancer complex proteins to direct splicing to the N1 exon upstream.
Alternative RNA splicing is a process that allows the production of multiple mRNAs from a single gene through the selection of different combinations of splice sites within a precursor mRNA (pre-mRNA). This process is an important mechanism in the developmental and cell-type-specific control of gene expression. Although the control of alternative splicing is poorly understood, specific regulatory proteins have been identified in some systems. These splicing regulatory proteins are thought to bind to sequence elements in a pre-mRNA and positively or negatively affect spliceosome assembly at nearby splice sites (1, 9, 66, 67).
cis-acting RNA sequences can serve to either enhance or repress the use of certain splice sites. Splicing enhancers are classified by their location in either exons or introns. Exonic splicing enhancers interact with trans-acting factors including an important class of splicing regulators called serine-arginine (SR) proteins (28, 66). SR proteins each possess one or two RNA recognition motif-type RNA binding domains and a C-terminal domain containing numerous SR dipeptides (20, 39). The exonic splicing enhancer in the Drosophila doublesex pre-mRNA is bound by two specific regulatory proteins, Transformer (Tra) and Transformer-2 (Tra-2) (38). Tra and Tra-2 bind to multiple elements of the enhancer and recruit specific SR proteins to assemble a large enhancer complex. This exonic enhancer complex is thought to activate splicing by recruiting the essential splicing factor U2AF to the 3′ splice site upstream, although the mechanism of this recruitment is not clear (56, 69, 72).
In addition to exonic enhancers, there are also intronic splicing enhancers. Intronic enhancer sequences are found downstream of many short or tissue-specific exons and are required for the splicing of these exons (2, 4, 11, 16, 31, 33, 50, 57, 61, 68). These elements are diverse in sequence and tissue-specific activity, and how intronic enhancers control splice site selection is largely unknown.
The hnRNPs are a large group of proteins that associate with pre-mRNAs in eukaryotic cells. The most abundant of these proteins have been characterized and designated hnRNP A1 through hnRNP U (18). These proteins each contain one or more RNA binding domains, usually of the type called the RNA recognition motif or RNP consensus sequence, as well as various auxiliary domains (7). The biological functions of these hnRNP proteins are not well understood. However, it is within hnRNP complexes that pre-mRNAs are processed to mature mRNAs before export from the nucleus (18).
hnRNPs A1, A2, B1, B2, C1, and C2 are the most abundant and the best-characterized hnRNPs. These proteins form specific multimeric assemblies that bind to nearly any RNA sequence to form RNP complexes (46). In addition to this general packaging of RNA, some of these proteins have affinity for specific RNA sequences, and several are implicated in more precise nuclear functions (8, 24, 65). For example, antibodies to the C1 protein have been shown to inhibit pre-mRNA splicing in vitro (15). The hnRNP A1 protein has two different activities of note. hnRNP A1 is known to shuttle from nucleus to cytoplasm and back, and it may play a role in the nucleocytoplasmic transport of mRNAs (19, 32, 53, 60). hnRNP A1 is also known to affect pre-mRNA splicing; the concentration of hnRNP A1 relative to the splicing factor SF2/ASF strongly affects the choice of certain alternative 5′ splice sites both in vitro and in vivo (10, 12, 21, 45, 71).
In addition to the major A, B, and C proteins, there are a number of other proteins that have been purified from bulk hnRNP complexes (42, 43, 52, 54). These proteins, including hnRNPs D through U, fall into diverse structural classes (18). They differ in their types and numbers of RNA binding domains and in the additional domains they carry. The different hnRNPs also vary in their affinities for different ribohomopolymers, and certain proteins have high affinity for specific RNA sequences (23, 34, 62, 65). These hnRNPs assemble in different subsets or combinations onto different RNA transcripts, implying that they have transcript-specific functions (3, 41). However, little is known about the natural RNA binding sites for these proteins or about how they cooperatively assemble with nascent RNA transcripts into hnRNP complexes.
We are studying the mouse c-src transcript as a model for understanding neuron-specific splicing regulation. The src primary transcript contains an 18-nucleotide exon (N1) that is inserted between the constitutive exons 3 and 4 in neurons but is skipped in other cells (37, 40). Analysis of src splicing both in vivo and in vitro indicates that the neuronal inclusion of the N1 exon requires an intronic splicing enhancer sequence lying between 17 and 142 nucleotides downstream of the exon (4, 50). The central, most conserved portion of this enhancer sequence (nucleotides 38 to 70) is called the downstream control sequence (DCS). The DCS binds to a complex of regulatory proteins that is important in allowing N1 splicing in vitro (48). Gel mobility shift assays show that the DCS complex assembles onto the RNA in neuronal WERI-1 (hereafter called WERI) cell nuclear extract but not in nonneuronal HeLa extract. This finding indicates the presence in WERI cells of protein factors that are distinct from those in HeLa cells. UV cross-linking experiments indicate that several proteins within the DCS complex bind directly to the RNA. Two of these proteins were identified as hnRNP F and the KH-type splicing regulatory protein (KSRP) (48, 49). These proteins appear to be required for exon N1 splicing and are present in both cell types.
In UV cross-linking assays, a third prominent protein of 58 kDa (p58) was seen binding to the DCS. We have now purified p58. We show here that this component of the DCS complex is hnRNP H and that, like hnRNP F and KSRP, hnRNP H is needed for src N1 splicing in vitro.
MATERIALS AND METHODS
UV cross-linking reactions and gel mobility shift assays.
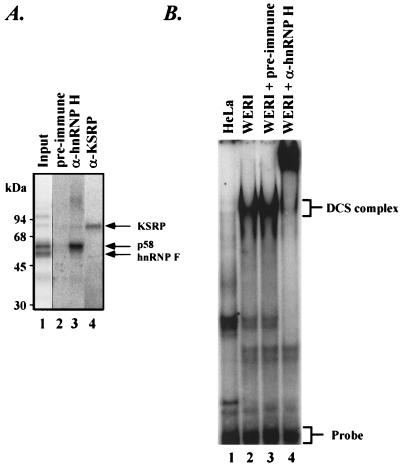
The reaction conditions for the UV cross-linking and gel mobility shift assays were described previously (48). For the antibody supershift of the DCS complex (Fig. 4B), 3 μg of protein A-purified total immunoglobulin G (IgG) from either preimmune or anti-hnRNP H antiserum was added to each binding reaction mixture and incubated for an additional 8 min at 30°C before loading onto a native gel.
FIG. 4.
hnRNP H is a component of the DCS complex. (A) Immunoprecipitation of the p58 protein cross-linked to the DCS RNA by anti-hnRNP H antibody. The ASP40 fraction of the WERI nuclear extract was UV cross-linked to radiolabeled DCS RNA and then subjected to RNase digestion. The mixture (lane 1) was immunoprecipitated with preimmune serum (lane 2), anti-hnRNP H antibody (lane 3), and anti-KSRP antibody (lane 4), followed by SDS-PAGE and autoradiography. (B) Supershift of the DCS complex by anti-hnRNP H antibody. The radiolabeled DCS RNA was incubated with the ASP40 fraction of the HeLa (lane 1) or WERI (lanes 2 to 4) nuclear extract. The WERI fraction was preincubated with nothing (lane 2), preimmune serum (lane 3), or anti-hnRNP H serum (lane 4). After incubation with protein, the DCS RNA was analyzed on a native electrophoretic gel. The free probe and the previously characterized DCS complex are indicated at the right.
Isolation of p58 protein component.
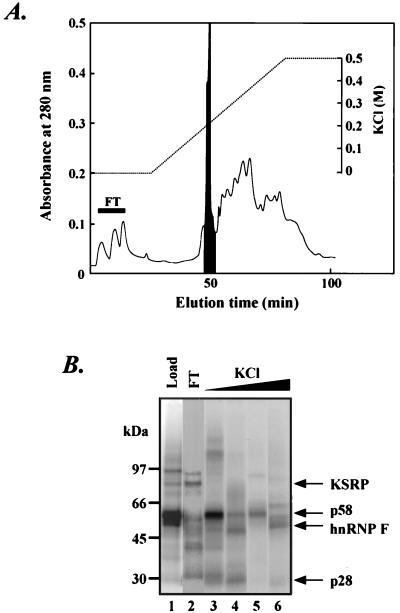
Unless otherwise indicated, all operations were carried out at 4°C. The WERI and HeLa cell nuclear extracts were prepared as described previously (4, 17). The WERI nuclear extract (20 ml from 1010 cells) was subjected to ultracentrifugation at 360,000 × g for 30 min. Ammonium sulfate was added to the supernatant to a final concentration of 40%. After centrifugation at 75,000 × g for 30 min, the pellet was resuspended in 15 ml of DG buffer (4) and dialyzed against the same buffer overnight. After removal of the precipitate by centrifugation, the supernatant was loaded on a Mono Q column (HR 5/5; Pharmacia) equilibrated in DG buffer at a flow rate of 0.25 ml/min. After being washed with the same buffer, the column was eluted with a linear KCl gradient from 0 to 0.5 M in the same buffer (total volume 25 ml). The peak fraction shown in Fig. 1A was collected and incubated overnight with 0.5 ml of GammaBind G Sepharose (Pharmacia) carrying immobilized anti-Sm monoclonal antibody Y12 (provided by J. Steitz) on a rotatory platform. The gel slurry was packed into an empty Bio-spin column (0.64 by 5 cm; Bio-Rad). The column was washed with 5 ml of DG buffer, followed by elution with the same buffer containing 2% sodium dodecyl sulfate (SDS). The eluted protein sample was directly subjected to SDS-polyacrylamide gel electrophoresis (PAGE) analysis. The gel was stained with Coomassie blue. The 58-kDa protein band was excised from the gel and subjected to in-gel tryptic digestion as described elsewhere (59). The eluted tryptic peptides were separated by reverse-phase high-pressure liquid chromatography (HPLC) on a Vydac C18 column, and individual peptides were sequenced on an automated protein sequencer (Perkin-Elmer/ABI model 492). These peptide sequences (YGDGGSTFQSTT, HTGPNSPDTAND, FFSDCK, GLPYR, and FIYTR) all match the published sequence of hnRNP H.
FIG. 1.
Isolation of p58 by Mono Q chromatography. (A) The ASP40 fraction from the WERI nuclear extract was loaded on a Mono Q column. Proteins were eluted by a linear KCl gradient from 0 to 0.5 M, and the UV absorbance at 280 nm was determined. The flowthrough (FT) is indicated with a solid bar, and the peak fraction containing the bulk of the p58 is shaded. (B) SDS-PAGE analysis of the Mono Q fractions. Proteins from the WERI ASP40 fraction (lane 1), the flowthrough fraction (lane 2), the peak fraction eluting at 0.2 M (lane 3), the 0.2 to 0.3 M fraction (lane 4), the 0.3 to 0.4 M fraction (lane 5), and the 0.4 to 0.5 M fraction (lane 6) were UV cross-linked to radiolabeled DCS RNA. These samples were then RNase treated, separated by SDS-PAGE, and detected by autoradiography.
Cloning and expression of recombinant hnRNPs H and F.
The hnRNP H cDNA was cloned by reverse transcription-PCR from WERI poly(A)+ RNA with primers 5′-GGCTCGAGATGATGTTGGGCACGGAAGGTG-3′ and 5′-TTGGATCCCTATGCAATGTTTGATTGAAAATCACTG-3′. The hnRNP F cDNA was cloned by reverse transcription-PCR from WERI poly(A)+ RNA with primers 5′-GGCTCGAGATGGGGATGCTGGGCCCTG-3′ and 5′-TTGGATCCCTAGTCATAGCCACCCATG-3′. Both PCR products were digested with XhoI and BamHI and then cloned into the expression vector pET 15b (Novagen) at the same sites. The cloned inserts were then completely sequenced. The recombinant N-terminal histidine-tagged proteins were expressed in Escherichia coli BL21(DE3). The bacterial cells were grown in LB medium containing ampicillin (100 μg/ml) at 37°C to an optical density of 0.7 at 600 nm before induction with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 3 h. For the purification of hnRNPs H and F, 1 liter each of the IPTG-induced cells was pelleted by centrifugation and washed with 50 mM Tris-HCl (pH 8.0) containing 0.1 M NaCl. The cells were lysed in the same buffer with a French press (Amicon). After centrifugation, the pellets were dissolved in the same buffer containing 6 M urea. The protein samples were loaded on a Ni-nitrilotriacetic acid (NTA) column (0.5 by 10 cm) and refolded by a linear urea gradient from 6 to 1 M. The renatured proteins were then eluted with DG buffer containing 0.25 M imidazole and dialyzed against DG buffer containing 1 mM dithiothreitol.
Anti-hnRNP H antibody production.
An anti-hnRNP H antiserum against the C-terminal peptide of hnRNP H (amino acid residues 435 to 449) coupled to keyhole limpet hemocyanin was raised in rabbits by Anaspec, Inc. Total IgG was purified from serum on a protein A column according to the standard protocol (26).
In vitro splicing and immunodepletion assays.
WERI nuclear extracts were prepared, and in vitro splicing reactions using the BS7 src pre-mRNA were carried out as described previously (14). Immunodepletion and reconstitution experiments were performed as described by Zuo and Maniatis (72). Briefly, 0.5 ml of protein A-Sepharose beads (Pharmacia) was mixed with an equal volume of anti-hnRNP H or preimmune serum containing 5 mg of bovine serum albumin per ml for 1 h at 4°C. The gel slurry was packed into a column, and the column was washed with 5 ml of DG buffer. One milliliter of WERI nuclear extract was passed through the column five times at room temperature. The eluate was then passed through a fresh protein A column (0.25-ml bed volume) twice to remove residual IgG present in the hnRNP H-depleted nuclear extract. To have the amount of residual hnRNP H in the immunodepleted nuclear extract be minimal while keeping the mock-depleted nuclear extract functioning, we set up splicing reactions using 6 μl of either hnRNP H-depleted or mock-depleted nuclear extract and containing either src or adenovirus major late (Ad) pre-mRNA as the substrate.
Immunoprecipitation and immunoblotting.
For immunoprecipitation of UV cross-linked protein components, protein A-Sepharose beads (10 μl; Pharmacia) were bound to antibodies for 1 h at 4°C. The beads were washed three times with immunoprecipitation buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.1% Triton X-100). Two standard UV cross-linking reaction mixtures (50 μl) as described above were added to the antibody-bound beads, allowed to mix for 1 h at 4°C, and washed as described above. The beads were boiled in SDS-PAGE loading buffer for 5 min, and the proteins were resolved by SDS-PAGE. Immunoprecipitation of splicing complexes from in vitro splicing reactions was performed as described elsewhere (6) except for the following changes. Protein A-Sepharose beads (15 μl; Pharmacia) were precoated with 50 μl of anti-hnRNP H or preimmune serum for 1 h at 4°C. Two splicing reactions (50 μl total) were used for each immunoprecipitation assay.
Immunoblotting was performed as follows. After SDS-PAGE, proteins were transferred to a nitrocellulose membrane. The membrane was blocked with 5% nonfat milk in phosphate-buffered saline containing 0.1% Tween 20 and probed with antibodies. The bound antibodies were detected with peroxidase-conjugated goat anti-rabbit IgG antibodies and visualized by the Amersham ECL detection system.
In vitro binding studies.
The hnRNP F cDNA was cloned into the in vitro translation vector pSPUTK (Stratagene) at the NcoI and BamHI sites. The KSRP cDNA was cloned as described previously (49) and subcloned into the pSPUTK vector at the NcoI and SmaI sites. In vitro-translated proteins were prepared by using the Promega nuclease-treated rabbit reticulocyte lysate system in the presence of [35S]methionine. [35S]methionine-labeled proteins were incubated with 1 μg of histidine-tagged hnRNP H immobilized on 15 μl of Ni-NTA agarose beads in 300 μl of binding buffer (20 mM Tris-HCl [pH 7.5], 100 mM NaCl, 0.1% Nonidet P-40) containing 5 mg of bovine serum albumin per ml at 4°C for 1 h. After beads were washed three times in the binding buffer plus 25 mM imidazole, proteins were eluted in SDS, resolved by SDS-PAGE, and visualized with a PhosphorImager (Molecular Dynamics). For RNase A treatment experiments, beads were incubated with RNase A (2 μg/ml) for 30 min at 37°C prior to the wash step (70). For the coimmunoprecipitation of [35S]methionine-labeled proteins with hnRNP H, 15 μl of protein A beads coupled to the anti-hnRNP H antibody was used under the above conditions except that no imidazole was introduced for the wash steps.
RESULTS
Isolation of p58/hnRNP H.
Previous RNA electrophoretic mobility shift and UV cross-linking experiments showed that a set of proteins, including a p75 doublet, p58, p55, and p28, assemble onto the DCS enhancer RNA (48). We identified p75 as KSRP and p55 as hnRNP F (48). To identify other components, we used a Mono Q anion-exchange column to separate the proteins of the 40% ammonium sulfate pellet (ASP40) fraction of WERI nuclear extract. This ASP40 fraction assembles the DCS complex and contains all of its identified components. As shown in Fig. 1A, there is a peak of 280-nm absorbance that is eluted at 0.2 M KCl. UV cross-linking of the eluted fractions to the DCS RNA indicated that the majority of the p58 protein was in this peak fraction. In contrast, the KSRP was in the flowthrough fraction and hnRNP F eluted at high salt concentrations (Fig. 1B).
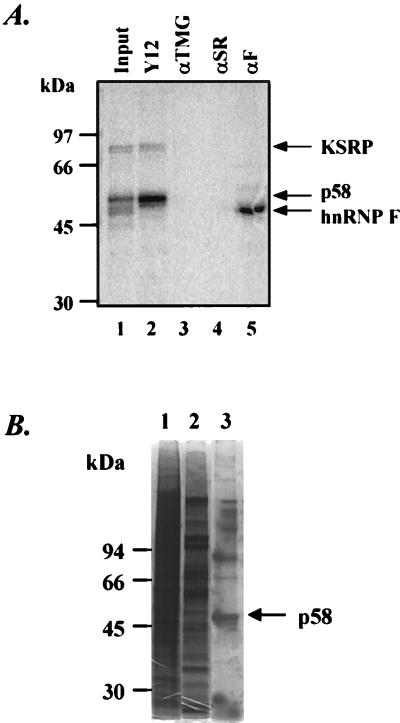
In activating splicing at the N1 exon, the proteins assembled on the enhancer presumably interact with components of the spliceosome and alter their assembly. To look for such interactions, we examined whether any of the DCS complex proteins were associated with the spliceosomal snRNPs. These proteins were tested for coimmunoprecipitation with the abundant U snRNPs. We used monoclonal antibody Y12 specific to the Sm antigens common to these snRNPs (36), and the anti-trimethyl guanosine (αTMG) antibody reactive with the cap structure on the U snRNAs (35). We found that antibody Y12 precipitated the p58 protein and KSRP, whereas αTMG did not (Fig. 2A, lanes 2 and 3). This indicated that p58 and KSRP either were associated with protein carrying the Sm epitope or contained the epitope themselves. This association with Sm antigens does not necessarily include an snRNA because αTMG did not precipitate these proteins. Similarly, an antibody reactive with the SR family of splicing factors (16H3) (51) did not react with any of the DCS proteins (Fig. 2A, lane 4). As shown previously (22, 48), antibodies against hnRNP F efficiently precipitated the F protein cross-linked in the complex (lane 5).
FIG. 2.
The p58 protein is immunoprecipitable with anti-Sm monoclonal antibody Y12. (A) Immunoprecipitation of proteins UV cross-linked to the DCS RNA. The WERI ASP40 fraction was UV cross-linked with radiolabeled DCS RNA and then subjected to RNase digestion. The mixture was immunoprecipitated with anti-Sm monoclonal antibody Y12 (lane 2), αTMG (lane 3), anti-SR protein antibody 16H3 (αSR; lane 4), and anti-hnRNP F antibody (αF; lane 5). The total cross-linked proteins are shown in lane 1. The immunoprecipitated radiolabeled proteins were separated by SDS-PAGE and detected with a PhosphorImager (Molecular Dynamics). (B) SDS-PAGE of the p58 protein isolated by anti-Sm immunoaffinity column. An immunoaffinity column was prepared by using the anti-Sm monoclonal antibody Y12. The Mono Q peak fraction was incubated with the Y12 beads. After the column was washed with buffer DG, the proteins were eluted with 2% SDS. Lane 1, nuclear extract; lane 2, peak fraction after Mono Q chromatography; lane 3, 2% SDS-eluted proteins from the anti-Sm column. Proteins were resolved by SDS-PAGE on a 10% gel in the absence of β-mercaptoethanol and stained with Coomassie blue. The p58 protein band is indicated with an arrow. This band was cut out and subjected to in-gel tryptic digestion, and the resulting peptides were separated by reverse-phase HPLC and sequenced by automated Edman degradation.
Although these experiments did not uncover an interaction with a specific snRNP, the reactivity of p58 with antibody Y12 allowed us to isolate the protein. The Mono Q peak fraction was incubated with antibody Y12 immobilized on protein G beads. After binding, the beads were loaded in a column and eluted with 2% SDS to release the bound proteins, and the eluate was resolved by SDS-PAGE. To prevent the immunoglobulin heavy chain from obscuring other isolated proteins, electrophoresis was carried out in the absence of reducing agent; this causes the immunoglobulin chains to migrate together high in the gel. As shown in Fig. 2B, there is a distinct band in the immunoprecipitate of approximate molecular mass of 58 kDa (lane 3). This protein band was excised from the gel and subjected to tryptic digestion. The peptide mixture was separated by reverse-phase HPLC, and individual peptides were subjected to amino acid sequence analysis. Five tryptic peptide sequences were obtained, and all of them were identical to peptides in hnRNP H.
hnRNP H is a component of the DCS complex.
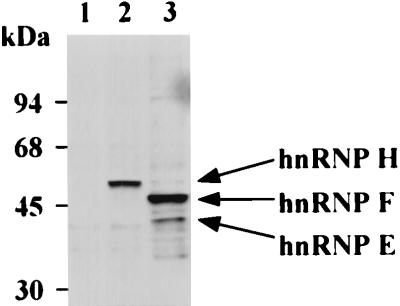
We showed earlier that hnRNP F is in the DCS complex (48). hnRNP F and hnRNP H are 78% identical in protein sequence and are immunologically related (30, 44). To confirm that hnRNP H is also a component of the DCS complex, we generated an antibody that reacts only with hnRNP H. Alignment of the hnRNP F and hnRNP H protein sequences indicates that hnRNP H contains two peptide sequences that are not present in hnRNP F: an extra copy of a 19-residue repeat present in the C-terminal domain and the 15 residues at the extreme C terminus of hnRNP H (30). We raised rabbit polyclonal antibodies against the C-terminal peptide of hnRNP H. As shown in Fig. 3, Western blot analysis of the WERI cell nuclear extract indicates that this antibody specifically recognizes the 58-kDa hnRNP H (lane 2), compared to a polyclonal anti-hnRNP F serum which recognizes hnRNP F (53 kDa) and hnRNP E (40 kDa) and weakly reacts with hnRNP H (58 kDa) (lane 3) (22).
FIG. 3.
Antibody to the C-terminal peptide sequence of hnRNP H reacts with hnRNP H but not hnRNP F. Shown is Western blot analysis of WERI nuclear extract with preimmune serum (lane 1), C-terminal peptide antibodies specific to hnRNP H (lane 2), and anti-hnRNP F antibodies (lane 3).
To confirm that hnRNP H is the p58 DCS binding protein, the WERI nuclear extract was UV cross-linked with radiolabeled DCS RNA and then subjected to RNase A digestion. The radiolabeled DCS cross-linked proteins were immunoprecipitated with anti-hnRNP H antibodies and then subjected to SDS-PAGE analysis. As shown in Fig. 4A, the DCS-labeled p58 was immunoprecipitated with the anti-hnRNP H antibody, indicating that the p58 DCS binding protein is indeed hnRNP H.
We previously identified by gel mobility shift analysis a protein complex that assembles onto the DCS RNA in WERI extract but not HeLa extract (48). This DCS complex contains hnRNP F, p58, and KSRP as well as other proteins. To prove that hnRNP H is a component of the DCS complex, we performed gel mobility shift assays in the presence of the hnRNP H antibody. As shown in Fig. 4B, the DCS complex can be observed by gel shift assay in the ASP40 fraction of the WERI extract (lane 2) but not in the same fraction of the HeLa extract (lane 1). When the anti-hnRNP H antibody was added to the WERI ASP40 fraction, the DCS complex was supershifted in the native gel (lane 4), indicating that hnRNP H is indeed a protein component of the DCS splicing enhancer complex.
hnRNP H is associated with the src pre-mRNA assembled into spliceosomes.
Many experiments indicate that the DCS complex is required for src N1 exon splicing. Short DCS RNAs competitively inhibit N1 exon splicing in vitro (48). Antibodies to the DCS complex proteins hnRNP F and KSRP both strongly inhibit splicing of the N1 exon, and purified KSRP can restore splicing activity to an extract inhibited with the anti-KSRP antibody (49). However, it has not been directly shown that hnRNP H or other DCS complex proteins bind to the full-length pre-mRNA during in vitro splicing. The binding of proteins to the N1 splicing enhancer has always been tested by using short DCS RNA probes. Moreover, it was not clear whether these proteins remained bound to the pre-mRNA after the spliceosome assembled or only functioned early in the assembly process. Indeed, studies have shown that some hnRNPs are displaced from the pre-mRNA during spliceosome assembly (3, 63). To look at these issues, we examined whether hnRNP H remained associated with the pre-mRNA during the splicing reaction.
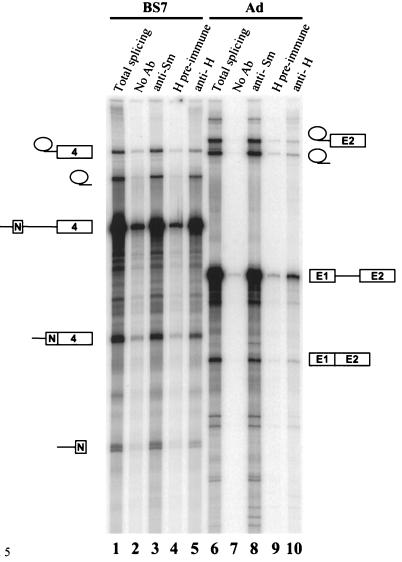
The hnRNP H antibody was used to immunoprecipitate splicing complexes assembled in vitro on either src (BS7) or Ad pre-mRNA substrate (Fig. 5). The positive control, anti-Sm monoclonal antibody Y12, recognizes the common Sm snRNPs. As seen previously, Y12 immunoprecipitates the unspliced pre-mRNA as well as the intermediates and products of the splicing reaction for either the src or Ad pre-mRNA (lanes 3 and 8) (5, 13, 25). The protein A beads alone or the preimmune serum showed trace amounts of nonspecific binding that was 20-fold below that seen with the anti-Sm serum (lanes 2, 4, 7, and 9). Strikingly, the anti-hnRNP H antibody also efficiently immunoprecipitated the RNA species from the src pre-mRNA splicing reaction (lane 5). For the src RNAs, this anti-hnRNP H immunoprecipitation was nearly as efficient as the anti-Sm reaction. In contrast, the Ad RNAs showed much weaker reactivity with anti-hnRNP H than with anti-Sm (lanes 8 and 10). These results indicate that hnRNP H is indeed bound to the full-length src pre-mRNA. Moreover, the immunoprecipitation of the reaction intermediates and products indicates that hnRNP H is present in the src pre-mRNA complexes containing the spliceosome and does not come off during spliceosome assembly and catalysis.
FIG. 5.
Anti-hnRNP H antibody immunoprecipitates src pre-mRNA splicing complexes from the in vitro splicing reaction. WERI nuclear extract was incubated with 32P-labeled src BS7 (lanes 1 to 5) or Ad (lanes 6 to 10) pre-mRNA substrate under splicing conditions. Reaction mixtures were aliquoted to protein A-Sepharose beads carrying no antibody (Ab) (lanes 2 and 7), anti-Sm antibody (lanes 3 and 8), preimmune serum for the anti-hnRNP H antibody (lanes 4 and 9), or anti-hnRNP H antibody (lanes 5 and 10), and the RNAs were recovered after immunoprecipitation. RNAs extracted from the total splicing reaction are in lanes 1 and 6. These lanes contain RNA from one-fourth of the amount of extract used in the immunoprecipitations. RNAs were analyzed by electrophoresis on an 8% polyacrylamide gel containing 8 M urea.
hnRNP H is required for efficient N1 exon splicing in vitro.
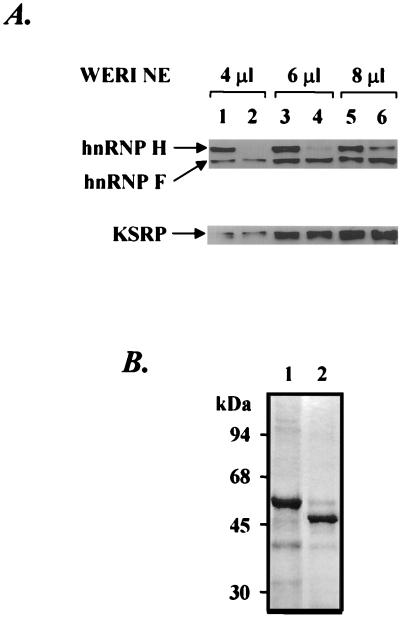
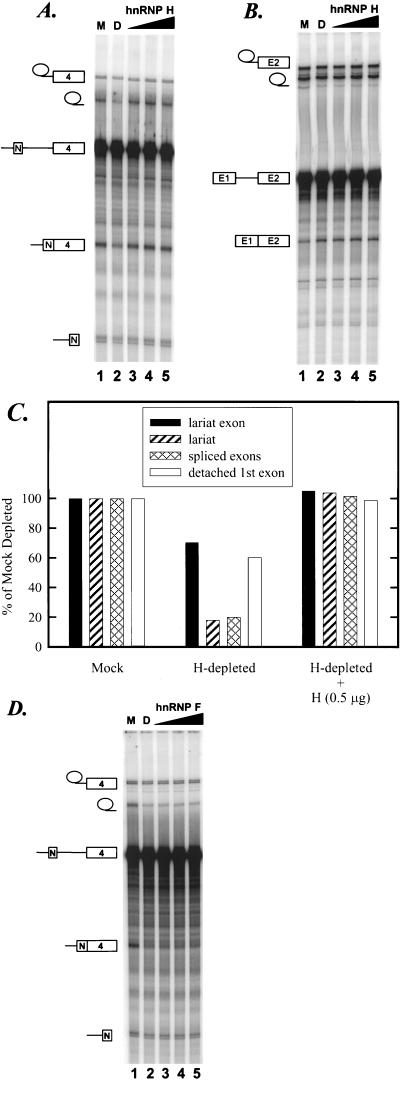
We also tested whether the antibody to the C-terminal peptide of hnRNP H would inhibit splicing when added directly to the splicing extract similar to the anti-hnRNP F antibody examined previously. Unfortunately, although the anti-hnRNP H serum inhibited splicing, so did the preimmune serum from the same rabbit (data not shown). Thus, the effect of the anti-hnRNP H antibody on splicing could not be assessed. As an alternative means to directly examine the involvement of hnRNP H in src N1 splicing, immunodepletion-reconstitution experiments were performed. An affinity column was prepared with the anti-hnRNP H antibody to deplete hnRNP H from the WERI nuclear extract. After repeated passage of the nuclear extract through affinity columns carrying either anti-hnRNP H or preimmune serum, the extent of the hnRNP H depletion was determined by Western blotting. As shown in Fig. 6A, the anti-hnRNP H antibody significantly reduced but did not fully deplete the hnRNP H in the nuclear extract. The hnRNP H in the depleted extract was at 18% of the level in the mock-depleted nuclear extract (compare lanes 5 and 6). hnRNP F and KSRP were not detectably reduced. As shown in Fig. 7A and C, the splicing of src BS7 pre-mRNA was decreased four- to fivefold in the hnRNP H-depleted nuclear extract, corresponding well with the amount of residual H protein (compare lanes 1 and 2). The remaining H protein in the depleted extract is still in excess of the input pre-mRNA for in vitro splicing. Thus, the activity remaining after depletion probably results from this residual protein.
FIG. 6.
(A) Western blot analysis of different amounts of mock-depleted (lanes 1, 3, and 5) or hnRNP H-depleted (lanes 2, 4, and 6) WERI nuclear extract (NE) probed with anti-hnRNP H and anti-hnRNP F antibodies (top) or anti-KSRP antibody (bottom). (B) SDS-PAGE of recombinant hnRNPs H (lane 1) and F (lane 2) stained with Coomassie blue. N-terminal histidine-tagged hnRNPs H and F were expressed in E. coli in an inducible T7 RNA polymerase-based system. After overexpression and cell lysis, protein was dissolved in 6 M urea, loaded on a Ni-NTA column, and refolded in a linear gradient of 6 to 1 M urea. The renaturated protein was then eluted in 0.25 M imidazole.
FIG. 7.
hnRNP H restores src pre-mRNA splicing in hnRNP H-depleted extract. (A) Splicing of the src BS7 pre-mRNA. In a 25-μl volume, reaction mixtures contained 6 μl of mock-depleted extract (M; lane 1), hnRNP H-depleted extract (D; lane 2), or hnRNP H-depleted extract supplemented with increasing amounts (0.1, 0.5, and 1 μg) of recombinant hnRNP H (lanes 3 to 5). (B) Splicing of the Ad pre-mRNA in mock-depleted extract (lane 1), hnRNP H-depleted extract (lane 2), or hnRNP H-depleted extract supplemented with increasing amounts (0.1, 0.5, and 1 μg) of recombinant hnRNP H (lanes 3 to 5). (C) Quantitation of hnRNP H depletion-reconstitution levels of src BS7 RNA species shown in panel A. (D) Splicing of the src BS7 pre-mRNA in mock-depleted extract (lane 1), hnRNP H-depleted extract (lane 2), or hnRNP H-depleted extract supplemented with increasing amounts (0.1, 0.5, and 1 μg) of recombinant hnRNP F (lanes 3 to 5).
Unexpectedly, the second step of the splicing reaction which generates released lariat and the ligated exon product was affected more strongly by hnRNP H depletion than the first step (Fig. 7A). The cause of this difference is not clear. As seen in Fig. 5, the enhancer complex continues to interact with spliceosomal components after the first step, and so the absence of hnRNP H could well affect later rearrangements in the spliceosome. However, from the point of view of controlling splice site choice, one would expect an effect on assembly of the spliceosome and hence the first step. Since the interaction of hnRNP H with pre-mRNA can be seen in the absence of ATP (data not shown), we presume that hnRNP H binds to the splicing substrate prior to spliceosome assembly. It may be that the hnRNP H depletion affects both steps of splicing, but under the conditions tested here the first step is not rate limiting for the reaction. The depletion of hnRNP H had no effect on splicing of the Ad pre-mRNA (Fig. 7B, lane 2) or a β-globin pre-mRNA (data not shown).
To confirm the function of hnRNP H in src splicing, we reconstituted the hnRNP H-depleted nuclear extract with bacterially expressed hnRNP H. Recombinant hnRNP H was purified from E. coli and shown to be homogeneous (Fig. 6B, lane 1). After renaturation and purification, this recombinant hnRNP H was fully active for binding to poly(rG) RNA (data not shown). Addition of this recombinant hnRNP H to the immunodepleted extract restored the splicing activity for the src BS7 pre-mRNA (Fig. 7A, lanes 3 to 5; Fig. 7C). Again, there was no effect of added hnRNP H on the splicing of the Ad pre-mRNA (Fig. 7B).
The restoration of splicing activity was specific to hnRNP H. Recombinant hnRNP F was also produced in E. coli (Fig. 6B, lane 2). The addition of recombinant hnRNP F to the hnRNP H-depleted extract did not restore the splicing activity (Fig. 7D, lanes 3 to 5). Thus, the src pre-mRNA has a specific requirement for hnRNP H for full splicing activity.
hnRNP F and hnRNP H form heterodimers.
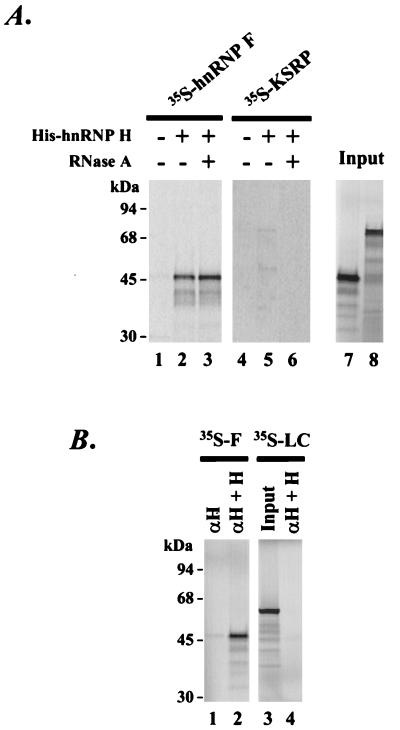
So far we have identified hnRNP F, KSRP, and hnRNP H as components of the DCS splicing enhancer complex. We next examined the interactions of these proteins with each other in the absence of RNA. We tested whether in vitro-translated hnRNP F and KSRP bound to histidine-tagged recombinant hnRNP H coupled to Ni-NTA agarose. As shown in Fig. 8A, the in vitro-translated hnRNP F bound to the resin coupled hnRNP H (lane 2) but not to the resin alone (lane 1). The in vitro-translated KSRP showed a weak interaction with hnRNP H (Fig. 8A, lane 5). To show that these proteins were directly interacting with hnRNP H and were not binding to the resin through an intervening in RNA, the resin bound proteins were treated with RNase A (70). After RNase treatment and washing with buffer, the hnRNP F remained associated with hnRNP H (Fig. 8A, lane 3). However, KSRP lost its interaction with hnRNP H upon RNase treatment (Fig. 8A, lane 6), indicating that the weak coprecipitation of KSRP and hnRNP H is presumably through an intervening RNA molecule. The interaction between hnRNPs H and F can also be observed in immunoprecipitation experiments where the in vitro-translated hnRNP F was coimmunoprecipitated with hnRNP H by the anti-hnRNP H antibody (Fig. 8B, lane 2). hnRNP F alone was not immunoprecipitable with the hnRNP H-specific antibody (lane 1). A control luciferase protein also showed no interaction with hnRNP H (Fig. 8B, lane 4).
FIG. 8.
hnRNP H interacts with hnRNP F. (A) Recombinant hnRNP F and KSRP were translated in a rabbit reticulocyte lysate and labeled with [35S]methionine (lanes 7 and 8). Radiolabeled hnRNP F and KSRP were incubated with histidine-tagged recombinant hnRNP H bound to Ni-NTA agarose beads. After incubation, beads were either treated with RNase A (lanes 3 and 6) or not treated (lanes 2 and 5). As negative controls, 35S-labeled hnRNP F and KSRP were incubated with Ni-NTA beads in the absence of hnRNP H (lanes 1 and 4). (B) Coimmunoprecipitation of hnRNP F with hnRNP H by anti-hnRNP H antibody. Radiolabeled hnRNP F (35S-F) was incubated with anti-hnRNP H-coupled protein A beads in the absence (lane 1) or presence (lane 2) of hnRNP H. Radiolabeled luciferase (35S-LC) was used as negative control (lane 4). After washing, the proteins were recovered from the beads in 2% SDS and resolved by SDS-PAGE.
The above experiments were carried out in roughly physiological salt (100 mM NaCl), and the interaction between hnRNPs H and F was lost when the NaCl concentration was above 200 mM (data not shown). These results indicate that hnRNP F can interact with hnRNP H under standard splicing conditions and that these proteins could be present in the DCS complex as a heterodimer. Further experiments will be needed to determine the exact stoichiometry of the proteins in the DCS complex.
DISCUSSION
Splicing of the c-src N1 exon is controlled by a splicing enhancer sequence in the downstream intron. The conserved core of this sequence (called the DCS) assembles a complex of regulatory proteins. We previously identified two proteins in this complex, hnRNP F and KSRP, and showed that they were needed for N1 splicing in vitro. We have now isolated a third major component of the DCS complex and identified it as hnRNP H. hnRNP H is needed for efficient splicing of the N1 exon in vitro and is likely to play a critical role in the assembly of the DCS complex. Thus, one cellular function for hnRNP H is in regulating alternative splicing patterns. This does not preclude other functions.
We isolated hnRNP H by Mono Q and anti-Sm (Y12) affinity chromatography. Since antibody Y12 had not previously been shown to bind hnRNP H, we thought it likely that hnRNP H coimmunoprecipitated with Sm proteins in the Mono Q peak fraction. However, Western blot analysis of the Mono Q fraction with antibody Y12 detected none of the standard Sm proteins. Instead, a band with the same mass as hnRNP H was observed (data not shown). After cloning the protein, we confirmed that hnRNP H cross-reacts with antibody Y12 by immunoprecipitation of the in vitro-translated hnRNP H. Interestingly, the in vitro-translated hnRNP F was not immunoprecipitable with antibody Y12 (data not shown). The Sm snRNPs share a sequence motif that could serve as the epitope to antibody Y12 (27, 29, 58). However, we have not found this motif in the hnRNP H sequence, and thus the basis for the antibody Y12 reactivity is still not clear.
We previously showed that hnRNP F is in the DCS complex by using a monoclonal antibody to specifically precipitate hnRNP F cross-linked to the DCS RNA (48). This antibody also had reactivity to hnRNP H by Western blot analysis (44). However, this hnRNP H reactivity was much weaker than with hnRNP F, which may explain why we did not immunoprecipitate the H protein and identify it previously.
After comparing the sequences of hnRNPs F and H, we raised an antibody to the unique C-terminal peptide of hnRNP H. This antibody is specific to hnRNP H by both Western blotting and immunoprecipitation assays. Using this hnRNP H antibody, we depleted the hnRNP H and not hnRNP F from the nuclear extract. The immunodepleted nuclear extract was significantly reduced in N1 splicing. The addition of recombinant hnRNP H to the depleted extract restored the splicing activity, indicating a role for hnRNP H in N1 exon splicing. Because both the depletion of hnRNP H and the inhibition of splicing were only partial, there are limits to the interpretation of this experiment. We cannot say that hnRNP H is required for any splicing activity, only that it is needed for full activity. The residual splicing after the hnRNP H depletion does not likely result from the highly related hnRNP F protein substituting for hnRNP H, as addition of hnRNP F to the hnRNP H-depleted extract did not restore activity. Moreover, the residual 18% of the hnRNP H remaining in the depleted extract is still in excess of the pre-mRNA in the in vitro splicing reaction, and this depletion produces a fourfold reduction in splicing. It thus seems likely that hnRNP H is indeed essential to the reaction and that a complete depletion, if it were achieved, would show a complete inhibition of splicing.
Although they are very similar in protein sequence (78% identical), several studies have revealed functional distinctions between hnRNP F and hnRNP H. Phorbol ester treatment of cultured cells strongly down-regulates the expression of hnRNP F but not the expression of hnRNP H (30). Yeast two-hybrid screening identified an interaction between hnRNP F and the nuclear cap-binding protein complex (CBC). Subsequent gel mobility shift assays showed that CBC-RNA complexes bind preferentially to hnRNP F over hnRNP H (22). These authors also showed that partial immunodepletion of hnRNP F led to partial inhibition of splicing in vitro. Far-Western blotting analysis identified hnRNP F, but not hnRNP H, as interacting with transportin 1, a mediator of nucleocytoplasmic transport for certain hnRNPs (55, 60). Studies of the rat β-tropomyosin gene transcript have implicated hnRNP H in the regulation of alternative splicing in that system (26a). These results imply important activity differences between the two proteins H and F.
Given that hnRNP H can bind directly to hnRNP F, hnRNPs H and F may at times function as a heterodimer. Gel mobility shift results indicate that both hnRNP F and hnRNP H are present in the DCS complex simultaneously. The complex is known to contain F and yet can be completely supershifted by the hnRNP H-specific antibody. Determining the stoichiometry and interactions of each protein in the DCS complex will be important in identifying any functional differences between these two highly homologous proteins.
The DCS is 33 nucleotides long and is composed of at least three different functional elements, GGGGGCUG, CUCUCU, and UGCAUG (14, 50, 50a). Additional elements outside the DCS are also required for enhancer function. The G tract in the DCS is likely to be the binding site for hnRNP H, since hnRNP H is known to bind tightly to poly(rG) (44). A similar element GGGGGAUG is also present upstream of the DCS, and G-rich elements have been identified within several other intronic splicing regulatory elements (11, 47). In the chicken β-tropomyosin pre-mRNA, UV cross-linking experiments identified a 55-kDa protein binding to intronic (A/U)GGG enhancer elements that activate alternative splicing (61). It is not clear yet whether this protein is hnRNP H.
Spliceosome assembly is a very dynamic process. The interactions between the different components of the splicing apparatus change at the different steps of assembly (64). Some interactions with the pre-mRNA are transient, occurring only at a specific step in the pathway. It has been shown that many hnRNPs that initially bind to the pre-mRNA in splicing extract are displaced upon assembly of the spliceosome (63). However, immunoprecipitation of splicing products and intermediates with anti-hnRNP H antibody indicates that hnRNP H is present in the spliceosome of the src pre-mRNA. This enhancer protein evidently maintains its interaction with the src pre-mRNA throughout splicing.
So far, we have identified hnRNP F, hnRNP H, and KSRP as components of the DCS complex, responsible for activating N1 exon splicing. All of these proteins are non-cell-type specific factors. This is not unexpected, as the enhancer has some activity in nonneural cells (50). However, the enhancer is stronger in neuronal cells, and in vitro the assembly of the full-sized DCS complex is specific to neuronal extract. It is not clear what causes the neuron-specific assembly of the complex. The availability of recombinant hnRNPs F and H and KSRP will allow us to develop assays for the cooperative assembly of these proteins into a splicing enhancer complex. Ultimately, one would like to examine how these proteins interact with each other to assemble onto a specific RNA sequence and how the assembled enhancer complex interacts with the spliceosome.
ACKNOWLEDGMENTS
We thank Iain Mattaj, Chiara Gamberi, Mei-Di Shu, Joan Steitz, Mark Roth, and Karla Neugebauer for antisera, and we thank members of the Black lab for advice and discussions. We are also grateful to one of the anonymous reviewers for suggesting an important control.
This work was supported by NIH grant R29 GM49662. Douglas Black is an associate investigator of the Howard Hughes Medical Institute and a David and Lucile Packard Foundation Fellow.
REFERENCES
- 1.Adams M D, Tarng R S, Rio D C. The alternative splicing factor PSI regulates P-element third intron splicing in vivo. Genes Dev. 1997;11:129–138. doi: 10.1101/gad.11.1.129. [DOI] [PubMed] [Google Scholar]
- 2.Balvay L, Libri D, Gallego M, Fiszman M Y. Intronic sequence with both negative and positive effects on the regulation of alternative transcripts of the chicken beta tropomyosin transcripts. Nucleic Acids Res. 1992;20:3987–3992. doi: 10.1093/nar/20.15.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett M, Pinol-Roma S, Staknis D, Dreyfuss G, Reed R. Differential binding of heterogeneous nuclear ribonucleoproteins to mRNA precursors prior to spliceosome assembly in vitro. Mol Cell Biol. 1992;12:3165–3175. doi: 10.1128/mcb.12.7.3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Black D L. Activation of c-src neuron-specific splicing by an unusual RNA element in vivo and in vitro. Cell. 1992;69:795–807. doi: 10.1016/0092-8674(92)90291-j. [DOI] [PubMed] [Google Scholar]
- 5.Black D L, Chabot B, Steitz J A. U2 as well as U1 small nuclear ribonucleoproteins are involved in premessenger RNA splicing. Cell. 1985;42:737–750. doi: 10.1016/0092-8674(85)90270-3. [DOI] [PubMed] [Google Scholar]
- 6.Blencowe B J, Nickerson J A, Issner R, Penman S, Sharp P A. Association of nuclear matrix antigens with exon-containing splicing complexes. J Cell Biol. 1994;127:593–607. doi: 10.1083/jcb.127.3.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burd C G, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 8.Burd C G, Dreyfuss G. RNA binding specificity of hnRNP A1: significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J. 1994;13:1197–1204. doi: 10.1002/j.1460-2075.1994.tb06369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caceres J F, Krainer A R. Mammalian pre-mRNA splicing factors. Oxford, England: Oxford University Press; 1997. pp. 174–212. [Google Scholar]
- 10.Caceres J F, Stamm S, Helfman D M, Krainer A R. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science. 1994;265:1706–1709. doi: 10.1126/science.8085156. [DOI] [PubMed] [Google Scholar]
- 11.Carlo T, Sterner D A, Berget S M. An intron splicing enhancer containing a G-rich repeat facilitates inclusion of a vertebrate micro-exon. RNA. 1996;2:342–353. [PMC free article] [PubMed] [Google Scholar]
- 12.Chabot B, Blanchette M, Lapierre I, La Branche H. An intron element modulating 5′ splice site selection in the hnRNP A1 pre-mRNA interacts with hnRNP A1. Mol Cell Biol. 1997;17:1776–1786. doi: 10.1128/mcb.17.4.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chabot B, Steitz J A. Multiple interactions between the splicing substrate and small nuclear ribonucleoproteins in spliceosomes. Mol Cell Biol. 1987;7:281–93. doi: 10.1128/mcb.7.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan R C, Black D L. Conserved intron elements repress splicing of a neuron-specific c-src exon in vitro. Mol Cell Biol. 1995;15:6377–6385. doi: 10.1128/mcb.15.11.6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi Y D, Grabowski P J, Sharp P A, Dreyfuss G. Heterogeneous nuclear ribonucleoproteins: role in RNA splicing. Science. 1986;231:1534–1539. doi: 10.1126/science.3952495. [DOI] [PubMed] [Google Scholar]
- 16.Del Gatto F, Plet A, Gesnel M C, Fort C, Breathnach R. Multiple interdependent sequence elements control splicing of a fibroblast growth factor receptor 2 alternative exon. Mol Cell Biol. 1997;17:5106–5116. doi: 10.1128/mcb.17.9.5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dignam J D. Preparation of extracts from higher eukaryotes. Methods Enzymol. 1990;182:194–203. doi: 10.1016/0076-6879(90)82017-v. [DOI] [PubMed] [Google Scholar]
- 18.Dreyfuss G, Matunis M J, Pinol-Roma S, Burd C G. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- 19.Fridell R A, Truant R, Thorne L, Benson R E, Cullen B R. Nuclear import of hnRNP A1 is mediated by a novel cellular cofactor related to karyopherin-beta. J Cell Sci. 1997;110:1325–1331. doi: 10.1242/jcs.110.11.1325. [DOI] [PubMed] [Google Scholar]
- 20.Fu X D. The superfamily of arginine/serine-rich splicing factors. RNA. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- 21.Fu X D, Mayeda A, Maniatis T, Krainer A R. General splicing factors SF2 and SC35 have equivalent activities in vitro, and both affect alternative 5′ and 3′ splice site selection. Proc Natl Acad Sci USA. 1992;89:11224–11228. doi: 10.1073/pnas.89.23.11224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gamberi C, Izaurralde E, Beisel C, Mattaj I W. Interaction between the human nuclear cap-binding protein complex and hnRNP F. Mol Cell Biol. 1997;17:2587–2597. doi: 10.1128/mcb.17.5.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghetti A, Pinol-Roma S, Michael W M, Morandi C, Dreyfuss G. hnRNP I, the polypyrimidine tract-binding protein: distinct nuclear localization and association with hnRNAs. Nucleic Acids Res. 1992;20:3671–3678. doi: 10.1093/nar/20.14.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorlach M, Burd C G, Dreyfuss G. The determinants of RNA-binding specificity of the heterogeneous nuclear ribonucleoprotein C proteins. J Biol Chem. 1994;269:23074–23078. [PubMed] [Google Scholar]
- 25.Grabowski P J, Seiler S R, Sharp P A. A multicomponent complex is involved in the splicing of messenger RNA precursors. Cell. 1985;42:345–353. doi: 10.1016/s0092-8674(85)80130-6. [DOI] [PubMed] [Google Scholar]
- 26.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 26a.Helfman, D. Personal communication.
- 27.Hermann H, Fabrizio P, Raker V A, Foulaki K, Hornig H, Brahms H, Luhrmann R. snRNP Sm proteins share two evolutionarily conserved sequence motifs which are involved in Sm protein-protein interactions. EMBO J. 1995;14:2076–2088. doi: 10.1002/j.1460-2075.1995.tb07199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hertel K J, Lynch K W, Maniatis T. Common themes in the function of transcription and splicing enhancers. Curr Opin Cell Biol. 1997;9:350–357. doi: 10.1016/s0955-0674(97)80007-5. [DOI] [PubMed] [Google Scholar]
- 29.Hirakata M, Craft J, Hardin J A. Autoantigenic epitopes of the B and D polypeptides of the U1 snRNP. Analysis of domains recognized by the Y12 monoclonal anti-Sm antibody and by patient sera. J Immunol. 1993;150:3592–3601. [PubMed] [Google Scholar]
- 30.Honore B, Rasmussen H H, Vorum H, Dejgaard K, Liu X, Gromov P, Madsen P, Gesser B, Tommerup N, Celis J E. Heterogeneous nuclear ribonucleoproteins H, H′, and F are members of a ubiquitously expressed subfamily of related but distinct proteins encoded by genes mapping to different chromosomes. J Biol Chem. 1995;270:28780–28789. doi: 10.1074/jbc.270.48.28780. [DOI] [PubMed] [Google Scholar]
- 31.Huh G S, Hynes R O. Elements regulating an alternatively spliced exon of the rat fibronectin gene. Mol Cell Biol. 1993;13:5301–5314. doi: 10.1128/mcb.13.9.5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Izaurralde E, Jarmolowski A, Beisel C, Mattaj I W, Dreyfuss G, Fischer U. A role for the M9 transport signal of hnRNP A1 in mRNA nuclear export. J Cell Biol. 1997;137:27–35. doi: 10.1083/jcb.137.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawamoto S. Neuron-specific alternative splicing of nonmuscle myosin II heavy chain-B pre-mRNA requires a cis-acting intron sequence. J Biol Chem. 1996;271:17613–17616. [PubMed] [Google Scholar]
- 34.Kiledjian M, Dreyfuss G. Primary structure and binding activity of the hnRNP U protein: binding RNA through RGG box. EMBO J. 1992;11:2655–2664. doi: 10.1002/j.1460-2075.1992.tb05331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krainer A R. Pre-mRNA splicing by complementation with purified human U1, U2, U4/U6 and U5 snRNPs. Nucleic Acids Res. 1988;16:9415–9429. doi: 10.1093/nar/16.20.9415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lerner E A, Lerner M R, Janeway C A, Jr, Steitz J A. Monoclonal antibodies to nucleic acid-containing cellular constituents: probes for molecular biology and autoimmune disease. Proc Natl Acad Sci USA. 1981;78:2737–2741. doi: 10.1073/pnas.78.5.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levy J B, Dorai T, Wang L H, Brugge J S. The structurally distinct form of pp60c-src detected in neuronal cells is encoded by a unique c-src mRNA. Mol Cell Biol. 1987;7:4142–4145. doi: 10.1128/mcb.7.11.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lynch K W, Maniatis T. Assembly of specific SR protein complexes on distinct regulatory elements of the Drosophila doublesex splicing enhancer. Genes Dev. 1996;10:2089–2101. doi: 10.1101/gad.10.16.2089. [DOI] [PubMed] [Google Scholar]
- 39.Manley J L, Tacke R. SR proteins and splicing control. Genes Dev. 1996;10:1569–1579. doi: 10.1101/gad.10.13.1569. [DOI] [PubMed] [Google Scholar]
- 40.Martinez R, Mathey-Prevot B, Bernards A, Baltimore D. Neuronal pp60c-src contains a six-amino acid insertion relative to its non-neuronal counterpart. Science. 1987;237:411–415. doi: 10.1126/science.2440106. [DOI] [PubMed] [Google Scholar]
- 41.Matunis E L, Matunis M J, Dreyfuss G. Association of individual hnRNP proteins and snRNPs with nascent transcripts. J Cell Biol. 1993;121:219–228. doi: 10.1083/jcb.121.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matunis E L, Matunis M J, Dreyfuss G. Characterization of the major hnRNP proteins from Drosophila melanogaster. J Cell Biol. 1992;116:257–269. doi: 10.1083/jcb.116.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matunis M J, Matunis E L, Dreyfuss G. Isolation of hnRNP complexes from Drosophila melanogaster. J Cell Biol. 1992;116:245–255. doi: 10.1083/jcb.116.2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matunis M J, Xing J, Dreyfuss G. The hnRNP F protein: unique primary structure, nucleic acid-binding properties, and subcellular localization. Nucleic Acids Res. 1994;22:1059–1067. doi: 10.1093/nar/22.6.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mayeda A, Krainer A R. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell. 1992;68:365–375. doi: 10.1016/0092-8674(92)90477-t. [DOI] [PubMed] [Google Scholar]
- 46.McAfee J G, Huang M, Soltaninassab S, Rech J E, Iyengar S, Lestourgeon W M. The packaging of pre-mRNA. Oxford, England: Oxford University Press; 1997. pp. 68–102. [Google Scholar]
- 47.McCullough A J, Berget S M. G triplets located throughout a class of small vertebrate introns enforce intron borders and regulate splice site selection. Mol Cell Biol. 1997;17:4562–4571. doi: 10.1128/mcb.17.8.4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Min H, Chan R C, Black D L. The generally expressed hnRNP F is involved in a neural-specific pre-mRNA splicing event. Genes Dev. 1995;9:2659–2671. doi: 10.1101/gad.9.21.2659. [DOI] [PubMed] [Google Scholar]
- 49.Min H, Turck C W, Nikolic J M, Black D L. A new regulatory protein, KSRP, mediates exon inclusion through an intronic splicing enhancer. Genes Dev. 1997;11:1023–1036. doi: 10.1101/gad.11.8.1023. [DOI] [PubMed] [Google Scholar]
- 50.Modafferi E F, Black D L. A complex intronic splicing enhancer from the c-src pre-mRNA activates inclusion of a heterologous exon. Mol Cell Biol. 1997;17:6537–6545. doi: 10.1128/mcb.17.11.6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50a.Modafferi, E. F., and D. L. Black. Unpublished data.
- 51.Neugebauer K M, Stolk J A, Roth M B. A conserved epitope on a subset of SR proteins defines a larger family of pre-mRNA splicing factors. J Cell Biol. 1995;129:899–908. doi: 10.1083/jcb.129.4.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pinol-Roma S, Choi Y D, Dreyfuss G. Immunological methods for purification and characterization of heterogeneous nuclear ribonucleoprotein particles. Methods Enzymol. 1990;181:317–325. doi: 10.1016/0076-6879(90)81132-e. [DOI] [PubMed] [Google Scholar]
- 53.Pinol-Roma S, Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992;355:730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- 54.Pinol-Roma S, Swanson M S, Matunis M J, Dreyfuss G. Purification and characterization of proteins of heterogeneous nuclear ribonucleoprotein complexes by affinity chromatography. Methods Enzymol. 1990;181:326–331. doi: 10.1016/0076-6879(90)81133-f. [DOI] [PubMed] [Google Scholar]
- 55.Pollard V W, Michael W M, Nakielny S, Siomi M C, Wang F, Dreyfuss G. A novel receptor-mediated nuclear protein import pathway. Cell. 1996;86:985–994. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- 56.Rudner D Z, Breger K S, Rio D C. Molecular genetic analysis of the heterodimeric splicing factor U2AF: the RS domain on either the large or small Drosophila subunit is dispensable in vivo. Genes Dev. 1998;12:1010–1021. doi: 10.1101/gad.12.7.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ryan K J, Cooper T A. Muscle-specific splicing enhancers regulate inclusion of the cardiac troponin T alternative exon in embryonic skeletal muscle. Mol Cell Biol. 1996;16:4014–4023. doi: 10.1128/mcb.16.8.4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Seraphin B. Sm and Sm-like proteins belong to a large family: identification of proteins of the U6 as well as the U1, U2, U4 and U5 snRNPs. EMBO J. 1995;14:2089–2098. doi: 10.1002/j.1460-2075.1995.tb07200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 60.Siomi M C, Eder P S, Kataoka N, Wan L, Liu Q, Dreyfuss G. Transportin-mediated nuclear import of heterogeneous nuclear RNP proteins. J Cell Biol. 1997;138:1181–1192. doi: 10.1083/jcb.138.6.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sirand-Pugnet P, Durosay P, Brody E, Marie J. An intronic (A/U)GGG repeat enhances the splicing of an alternative intron of the chicken beta-tropomyosin pre-mRNA. Nucleic Acids Res. 1995;23:3501–3507. doi: 10.1093/nar/23.17.3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Soulard M, Della Valle V, Siomi M C, Pinol-Roma S, Codogno P, Bauvy C, Bellini M, Lacroix J C, Monod G, Dreyfuss G, et al. hnRNP G: sequence and characterization of a glycosylated RNA-binding protein. Nucleic Acids Res. 1993;21:4210–4217. doi: 10.1093/nar/21.18.4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Staknis D, Reed R. SR proteins promote the first specific recognition of pre-mRNA and are present together with the U1 small nuclear ribonucleoprotein particle in a general splicing enhancer complex. Mol Cell Biol. 1994;14:7670–7682. doi: 10.1128/mcb.14.11.7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Staley J P, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 65.Swanson M S, Dreyfuss G. Classification and purification of proteins of heterogeneous nuclear ribonucleoprotein particles by RNA-binding specificities. Mol Cell Biol. 1988;8:2237–2241. doi: 10.1128/mcb.8.5.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang J, Manley J L. Regulation of pre-mRNA splicing in metazoa. Curr Opin Genet Dev. 1997;7:205–211. doi: 10.1016/s0959-437x(97)80130-x. [DOI] [PubMed] [Google Scholar]
- 67.Wang Y-C, Selvakumar M, Helfman D. Alternative pre-mRNA splicing. Oxford, England: Oxford University Press; 1997. pp. 242–279. [Google Scholar]
- 68.Wei N, Lin C Q, Modafferi E F, Gomes W A, Black D L. A unique intronic splicing enhancer controls the inclusion of the agrin Y exon. RNA. 1997;3:1–14. [PMC free article] [PubMed] [Google Scholar]
- 69.Wu J Y, Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- 70.Xiao S H, Manley J L. Phosphorylation of the ASF/SF2 RS domain affects both protein-protein and protein-RNA interactions and is necessary for splicing. Genes Dev. 1997;11:334–344. doi: 10.1101/gad.11.3.334. [DOI] [PubMed] [Google Scholar]
- 71.Yang X, Bani M R, Lu S J, Rowan S, Ben-David Y, Chabot B. The A1 and A1B proteins of heterogeneous nuclear ribonucleoparticles modulate 5′ splice site selection in vivo. Proc Natl Acad Sci USA. 1994;91:6924–6928. doi: 10.1073/pnas.91.15.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zuo P, Maniatis T. The splicing factor U2AF35 mediates critical protein-protein interactions in constitutive and enhancer-dependent splicing. Genes Dev. 1996;10:1356–1368. doi: 10.1101/gad.10.11.1356. [DOI] [PubMed] [Google Scholar]