ABSTRACT
Autophagy (a process of cellular self-eating) is a conserved cellular degradative process that plays important roles in maintaining homeostasis and preventing nutritional, metabolic, and infection-mediated stresses. Surprisingly, little attention has been paid to the role of this cellular function in species of agronomical interest, and the details of how autophagy functions in the development of phenotypes of agricultural interest remain largely unexplored. Here, we first provide a brief description of the main mechanisms involved in autophagy, then review our current knowledge regarding autophagy in species of agronomical interest, with particular attention to physiological functions supporting livestock animal production, and finally assess the potential of translating the acquired knowledge to improve animal development, growth and health in the context of growing social, economic and environmental challenges for agriculture.
Abbreviations: AKT: AKT serine/threonine kinase; AMPK: AMP-activated protein kinase; ASC: adipose-derived stem cells; ATG: autophagy-related; BECN1: beclin 1; BNIP3: BCL2 interacting protein 3; BVDV: bovine viral diarrhea virus; CALCOCO2/NDP52: calcium binding and coiled-coil domain 2; CMA: chaperone-mediated autophagy; CTSB: cathepsin B; CTSD: cathepsin D; DAP: Death-Associated Protein; ER: endoplasmic reticulum; GFP: green fluorescent protein; Gln: Glutamine; HSPA8/HSC70: heat shock protein family A (Hsp70) member 8; IF: immunofluorescence; IVP: in vitro produced; LAMP2A: lysosomal associated membrane protein 2A; LMS: lysosomal membrane stability; MAP1LC3/LC3: microtubule associated protein 1 light chain 3; MDBK: Madin-Darby bovine kidney; MSC: mesenchymal stem cells; MTOR: mechanistic target of rapamycin kinase; MTORC1: MTOR complex 1; NBR1: NBR1 autophagy cargo receptor; NDV: Newcastle disease virus; NECTIN4: nectin cell adhesion molecule 4; NOD1: nucleotide-binding oligomerization domain 1; OCD: osteochondritis dissecans; OEC: oviduct epithelial cells; OPTN: optineurin; PI3K: phosphoinositide-3-kinase; PPRV: peste des petits ruminants virus; RHDV: rabbit hemorrhagic disease virus; SQSTM1/p62: sequestosome 1; TEM: transmission electron microscopy.
KEYWORDS: Agriculture, autophagy, cellular and tissue homeostasis, farm animals, physiology and animal production
Introduction
Autophagy, a lysosomal degradation pathway by which the cell self-digests its own components, has been recognized over the past decade as an essential process of cellular metabolism and a main contributor to homeostasis [1]. Autophagy rids the cell of excessive or damaged organelles, misfolded proteins, and invading microorganisms, and provides nutrients to maintain crucial cellular functions. Modulation of autophagy is linked to numerous fundamental physiological functions. Its dysfunction is an important common denominator in many diseases affecting a wide range of organs and systems [2]. Consequently, the past two decades have witnessed an explosion of research on the molecular mechanisms of autophagy in the context of diverse human diseases including heart failure, liver inflammation, impaired long-term humoral immunity, Parkinson disease, and cancers [1–3].
Surprisingly, whereas it is now clearly established that autophagy plays a central role in differentiation and development, cellular and tissue homeostasis, protein and organelle quality control, metabolism and immunity, little attention has been paid to the role of this cellular function in species of agronomical interest. The details of how autophagy functions in fertility, development and growth of the livestock animals, tolerance to nutrient deficiency and/or imbalances, abiotic stresses or pathogens, and quality of the animal products remain largely unexplored.
Autophagy is a key process of cellular metabolism and represents an interesting target to design new feeding/breeding strategies to optimize animal growth while maintaining products of high nutritional value and quality and to lower environmental impacts. In this article, after a brief description of the main mechanisms involved in autophagy, we review the current knowledge regarding autophagy in species of agronomical interest, with particular attention to the processes of reproduction and production of animal products, as well as the potential role of this cellular function in the adaptation of animals to farming conditions. Finally, we conclude on the relevance to consider (as new metabolic criteria) and/or manipulate this function in these species to face new social and environmental challenges for agriculture in the near future.
Molecular description of autophagic pathways
The term autophagy encompasses different types of mechanisms (known as macroautophagy, chaperone-mediated autophagy, microautophagy, and endosomal microautophagy) depending on the delivery route of cytoplasmic material to the lysosomal lumen (Figure 1). Here, we provide a brief description of each of these autophagic pathways, but readers interested in a more complete description are encouraged to consult review articles that discuss these pathways in detail [4–8].
Figure 1.
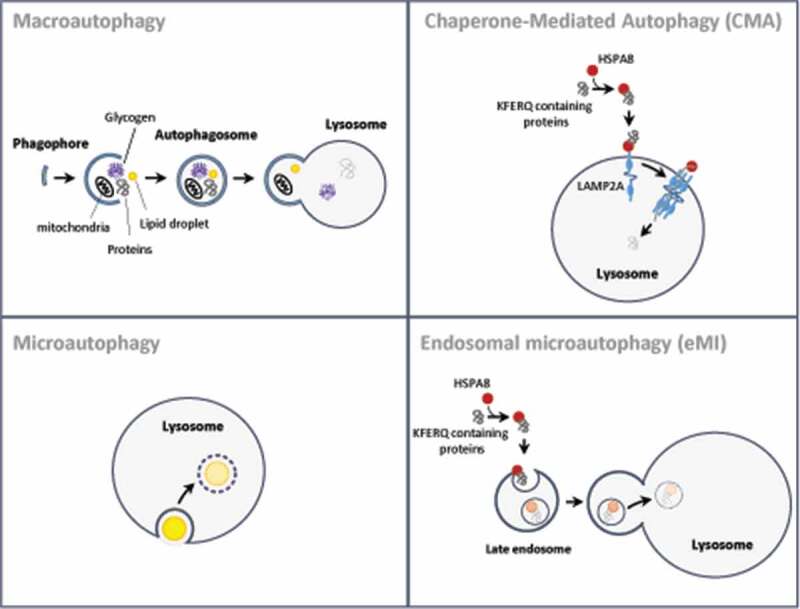
Illustration of the main steps of the different pathways of mammalian autophagy. In macroautophagy, double-membrane organelles called autophagosomes engulf intracellular components and then fuse with lysosomes. In CMA, cytosolic proteins carrying a KFERQ-like motif bind the chaperone HSPA8/HSC70 (heat shock protein family A [Hsp70] member 8/heat-shock cognate protein of 70 kDa) and are subsequently translocated into the lysosomes in a LAMP2A-dependent mechanism. In microautophagy, cytoplasmic substrates intended for degradation are absorbed by the lysosome by direct membrane invagination. In endosomal microautophagy, the complex HSPA8-KFERQ-containing protein is targeted to the endosomal membrane to be internalized into multivesicular bodies, which then fuse with lysosomes to allow degradation of their content
Macroautophagy
Macroautophagy is the variant of autophagy best characterized so far. This system relies on the formation of specialized double-membrane vesicles called autophagosomes, which engulf and transport cell components to the lysosome. The autophagosomes do not bud from a preexisting compartment; they form de novo by the coordinated action of over 20 evolutionary conserved autophagy-related (ATG) proteins [9], originally discovered in yeast genetic screens [10]. Upon autophagy induction, ATGs are hierarchically recruited to lipo-proteic cores called “phagophore,” close to specific endoplasmic reticulum (ER) subdomains [11], where they play essential roles during autophagosome formation (Figure 1). These involve (1) the de novo assembly of the phagophore, which precisely shapes like a cup, (2) the elongation of the phagophore membranes by accommodating incoming phospholipids from various sources including the ER, recycling endosomes, and mitochondria, (3) the sequestration of substrates (referred to as cargos) and (4) the complete closure of the vesicle to form a novel double-membrane compartment with a distinct identity and functions. Upon sealing of the autophagosome membrane, these cargo-containing vesicles move along microtubules to fuse with the lysosome and deliver the sequestered cargo in this hydrolytic organelle [12]. This whole process is fine-tuned in time and space and is fundamental to ensure developmental and environmental plasticity in eukaryotes [13–15].
Macroautophagic responses can result in nonselective degradation of cytoplasmic components. Upon lysosomal degradation, these autophagy substrates fuel bioenergetic metabolism or repair processes [16,17]. However, it is now clear that macroautophagy also contributes to intracellular homeostasis by selectively degrading cargo material such as aggregated proteins, damaged mitochondria, excess peroxisomes or invading pathogens [18]. Several targets of selective macroautophagy have been described, such as aggregated proteins (aggrephagy), mitochondria (mitophagy), peroxisomes (pexophagy), ribosomes (ribophagy), endoplasmic reticulum (reticulophagy), lipid droplets (lipophagy), glycogen (glycophagy) and even invading pathogens (xenophagy) [19–22]. In these cases, the formation of the phagophore occurs in the vicinity of the cargo through specific receptors, such as SQSTM1/p62 (sequestosome 1), NBR1, CALCOCO2/NDP52 and OPTN [18]. Most of these receptors share an evolutionarily conserved domain known as the MAP1LC3/LC3 (microtubule associated protein 1 light chain 3)‐interacting region (LIR), which allows them to bring macroautophagy cargo in the proximity of forming autophagosomes through the simultaneous binding of the cargo and LC3, a key component of the autophagy machinery.
Chaperone-mediated autophagy (CMA)
CMA has been described as a selective mechanism for the degradation of specific soluble proteins within lysosomes [23]. Besides a well-described role in protein quality control (resulting from its ability to selectively target damaged or nonfunctional proteins for degradation), the diversity of the sub-proteome degraded by CMA also associates this function with the regulation of transcriptional programs [24], cell death and cell survival mechanisms [25–27], DNA repair and cell cycle progression [28] as well as a variety of intracellular processes related to the control of cellular energetics [29–33]. Not surprisingly, CMA emerged these last years as a key factor in the control of cellular homeostasis [33].
CMA involves the direct delivery of cytosolic proteins targeted for degradation to the lysosome [5]. During this process, cytosolic proteins containing a CMA-targeting motif (KFERQ-related sequences) are first recognized by the chaperone HSPA8/HSC70 [34,35] (Figure 1). This substrate/chaperone complex then docks at the lysosomal membrane through specific binding to the cytosolic tail of the LAMP2A (lysosomal associated membrane protein 2A), the only one of the three spliced isoforms of the LAMP2 gene that has been shown to be essential and limiting for CMA activity [36]. Multimerization of LAMP2A will then result in the formation of a translocation complex, and promote translocation of substrate proteins [37]. After unfolding, substrates subsequently translocate across the lysosomal membrane and are degraded.
There is still little information on the regulation of this autophagic route. Available data indicate that CMA activity depends on the levels of the LAMP2A protein at the lysosomal membrane [38] as well as on the efficiency of its assembly and disassembly in this compartment [39]. CMA is active in most cells under basal conditions, but it is maximally activated in response to oxidative stress [40], hypoxia [25,41] or nutrient deprivation [42].
Microautophagy and endosomal microautophagy
Microautophagy is defined as a lysosomal (vacuolar) membrane dynamics that directly wraps and transports cytosolic components into the lumen of the lytic organelle [43]. Microautophagy is probably the least studied form of autophagy and the molecular signature of the process remains, for the most part, unknown. Most of what is known about this degradative pathway originate from studies in yeast where it has been involved in the degradation of multiple substrates, including peroxisomes [44], portions of the nucleus [45], damaged mitochondria [46], and lipid droplets [47].
Interestingly, a similar mechanism has been recently identified in late endosomes of cells from Drosophila melanogaster [48] and mammals [49,50]. During this process, commonly known as endosomal microautophagy (or eMI), proteins harboring the KFERQ-like motif bind the chaperone HSPA8/HSC70 and are directed to the endosomal membrane to be subsequently internalized into multivesicular bodies in endosomal sorting complexes required for transport (ESCRT)-dependent mechanism.
Despite the marked differences between these autophagic pathways, they all contribute to the preservation of cellular homeostasis. In physiological conditions, they all proceed at baseline levels, avoiding the accumulation of potentially cytotoxic entities that may cause abnormal cellular functions (e.g., damaged mitochondria) [7,17,51]. In addition, they are also sensitive to perturbations of intracellular or extracellular homeostasis. Thus, disturbances of different nature (nutritional, metabolic, chemical, physical or hormonal) can induce an autophagy response that involves complex interactions between the different autophagy routes that remain largely misunderstood.
Current knowledge on autophagy in species of agronomical interest
As stated above, research on molecular mechanisms of autophagy in the context of various human diseases has increased dramatically over the past 20 years. In contrast, much less attention has been paid to autophagy in species of agronomical interest and to the relevance of considering and/or manipulating this cellular function in these species to face new social and environmental challenges for agriculture. In order to synthesize the data available on autophagy in livestock animals and to single out present and future research subjects, we conducted an exhaustive keyword-based bibliographic search in the Web Of Science, one of the most comprehensive bibliographic databases that provide extensive and well-formatted bibliographic publication records in numerous disciplines including agriculture, biological sciences, engineering, medical and life sciences. The lists of keywords included or excluded from the analysis and the method used are detailed in Figure 2 and Data S1. A total of 945 articles published between 1975 and 2019 have been extracted. However, after the exclusion of irrelevant references that do not relate to issues or functions of agronomic interest or that, although the term “autophagy” is present in the keywords, are not or only marginally related to this topic, only 295 articles were considered as relevant for our analysis. In the next subsections, we will describe the results obtained for each species or group of species and the main insights gained from them. They mainly concern macroautophagy hereafter referred to as autophagy.
Figure 2.
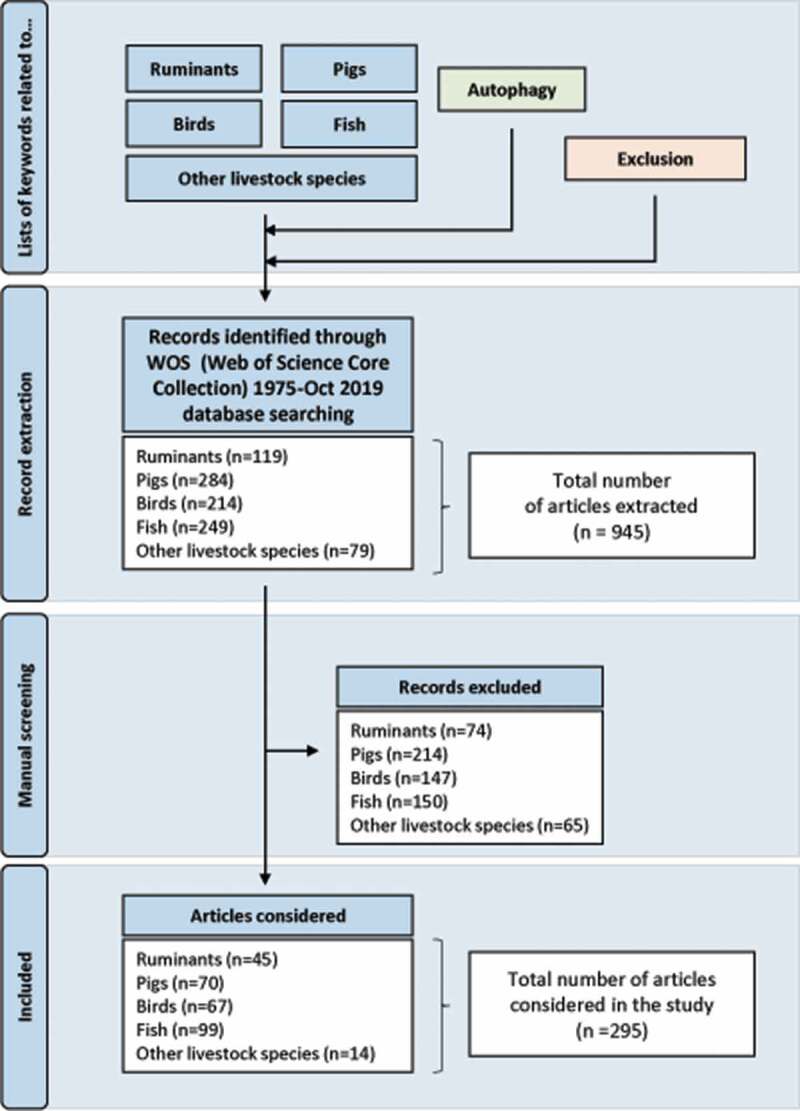
Flow chart of the bibliographic search. The list of keywords related to each species and autophagy as well as that of terms to be excluded are detailed in supplementary data. The manual curation aimed to exclude irrelevant references that do not relate to issues or functions of agronomic interest (specified in supplementary data) or that, although the term “autophagy” is present in the keywords, are not or only marginally related to this topic
Autophagy, meat and dairy ruminants
We extracted 119 publications related to autophagy in ruminants (Figure 2). Of these, however, only 45 publications brought knowledge on the autophagy processes monitored by at least one method proposed by the recent guidelines for studying autophagy [52]. At a glance, these few publications highlight that autophagy is a key cellular process that contributes to the health and many normal developmental processes that sustain ruminant productions.
Role of autophagy in pathogen infections in ruminants
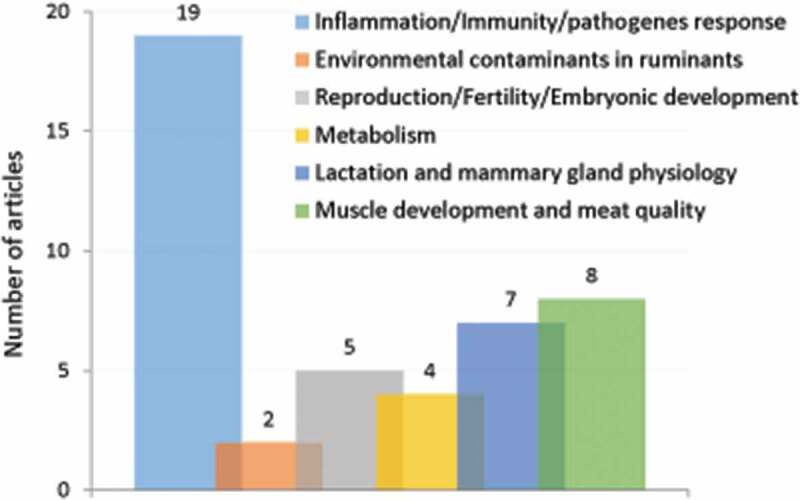
Of the 45 publications, 19 reported how interactions of microorganisms or viruses with the autophagy system of host cells contribute to their pathogenicity (Figure 3). Notably, several ruminant viruses have developed strategies to use autophagy machinery to promote their replication. This was well documented for bovine viral diarrhea virus (BVDV) infection. Immuno-co-localization of BVDV antigens with the autophagy marker LC3 revealed that BVDV replication was associated with autophagosomes in Madin-Darby bovine kidney (MDBK) cells [53]. BVDV infection induced the autophagy flux in these cells [54]. The authors examined the autophagy flux in cells expressing a fusion construct comprising green fluorescent protein (GFP) tagged to LC3 (GFP-LC3) and by using chloroquine as an inhibitor of the autophagic process. The increase in the number of double- or single-membrane vesicles, the rapid redistribution of GFP-LC3 in puncta and the accumulation of autophagosome-associated LC3 protein (known as LC3-II) upon BVDV infection supported autophagy induction. This was associated with the promotion of virus replication (as already inferred by Fu et al. [55]) and inhibition of the interferon-mediated signaling pathway and apoptosis. Interestingly, not only BVDV envelope proteins but also a non-structural protein (NS4B) alone could induce autophagosomes formation [56].
Figure 3.
Number of original articles related to autophagy in ruminants according to agronomic fields
There are several other examples of interplay of viruses with autophagy. Infection by peste des petits ruminants virus (PPRV) [57] and Bluetongue virus [58] also triggered an autophagic response as shown by the appearance of double- and single-membrane autophagy-like vesicles, increased ratio of LC3-II:LC3-I, and SQSTM1 degradation. Yang et al. reported that PPRV induced a first wave of autophagy in infected cells through the cellular pathogen receptor NECTIN4 and the AKT-MTOR-dependent pathway [59]. They also described that a later wave resulted from interaction of viral proteins and the immunity-related protein IRGM and the heat shock protein HSPA1A and was sustained during replication.
Immune positivity for LC3B was also correlated to the distribution and intensity of Brucella spp antigen in several tissues of bovine fetuses aborted due to natural infection with Brucella spp [60]. The immunopositivity of LC3B and cleaved CASP3 (an apoptotic marker) was extremely high in the lung, thymus, spleen, kidney, and liver tissues and was highly similar to the distribution of Brucella spp antigens, suggesting that autophagy may be activated by this infectious agent.
In prion diseases, Lopez-Perez et al. investigated the relationship between gene expression and protein distribution of different autophagy-related markers and prion-associated lesions in several areas of the central nervous system [61]. In sheep infected with classical scrapie, while the expression of autophagy markers ATG5 and ATG9 was downregulated in some areas of the brain, ATG5 protein accumulated in the medulla oblongata (the most inferior part of the brainstem) and positively correlated with prion deposition and scrapie-related lesions. In the late phases of the disease, downregulation of autophagy-related genes (ATG5 and ATG9) and increase in the levels of ATG5 and SQSTM1 proteins were observed in highly affected areas, suggesting an arrest of the autophagy machinery that would facilitate prion replication. Conversely, the autophagy machinery was still intact in less affected areas, particularly in specific neuronal cells such as Purkinje cells, and consequently would constitute a neuroprotective mechanism against prion-induced toxicity. In sheep experimentally infected with atypical scrapie prions, while ATG5 protein showed a similar distribution to that observed in the classical form, expression of the isoforms LC3B and LC3A of the LC3 protein did not change in any brain region. Overexpression of SQSTM1 in the most prion-affected areas, including Purkinje cells [62], confirmed the association between prion-related lesions and autophagy downregulation in a different form of the disease.
Autophagy and environmental contaminants in ruminants
Two publications of the corpus provide pieces of evidence on the role of autophagy in crops pests- or chemicals-induced toxicity of ruminants [63,64]. The hepatotoxicity of Saanen goats induced by the perennial herbaceous plant Eupatorium adenophorum was associated with an excessive autophagy that triggers apoptosis that leads to cell death. Specifically, Eupatorium adenophorum was shown to suppress the PI3 K-AKT-MTOR pathway and activate AMP kinase, promoting the expression of autophagic proteins LC3-II and BECN1 (beclin 1), puncta formation and autophagic vacuole accumulation in hepatocytes [63]. Higher rates of autophagy markers, such as autophagosome formation and increased expression of autophagy-related genes and proteins were also evidenced in bisphenol A-mediated toxicity of goat Sertoli cells [64]. According to the authors, autophagy might act as a transitory survival pathway in goat Sertoli cells under bisphenol A treatment but might be deleterious when it occurs excessively or when it is impaired and might lead to activation of apoptosis mediated cell death.
Role of autophagy in fertility and reproduction in ruminants
In ruminants, the gestation/lactation cycle must be efficient for the renewing of herd and for milk production, which implies an optimal fertility, reproduction and mammary gland development. Of the 45 publications selected, five publications suggest that autophagy is involved in many facets related to fertility or reproduction in ruminants (Figure 3). The expression of autophagy-related genes (LC3A, LC3B, ATG3, and ATG7), the levels of LC3-II as well as the lysosomal activity were thus shown to increase during the regression of bovine Corpus Luteum, a temporary endocrine structure in female ovaries involved in the production of progesterone and required during the estrus cycle [65]. High levels of genes involved in all stages of autophagy – from initial step of vesicle elongation (ATG9, LC3, ATG5) to autophagosome-lysosome fusion (RAB7) – as well as a high number of double membrane-enclosed vesicles (autophagosomes) with engulfed portions of cytoplasm were also observed in the placental tissues after the transfer of in vitro-produced (IVP) sheep embryos [66]. The authors of this study conclude that high autophagic activity in IVP placentae can be a successful temporary counterbalance to the delayed vasculogenesis and the reduction of fetal growth observed in pregnancies after the transfer of IVP embryos. In goats, the variation in the abundance of canonical markers of cellular autophagy was used to propose that autophagy pathway is part of the signaling involved in the endometrial function, the conceptus elongation and thus the pregnancy success [67].
However, increased autophagy has also been related to detrimental reproductive performance due to oviduct inflammation. Notably, in bovine oviduct epithelial cells (OECs) incubated with palmitic acid (a major saturated fatty acid known to exert a negative effect on the physiological functions of bovine OECs), LC3 and SQSTM1 proteins content were increased. This effect was associated with induction of CXCL8/IL8 (C-X-C motif chemokine ligand 8) secretion, suggesting a role for autophagy in the regulation of inflammation induced by palmitic acid [68].
Role of autophagy in lactation and mammary gland physiology, and early weaning in ruminants
Over the 45 publications selected for ruminants, seven reported that autophagy is involved in the lactation-driven mammary gland development or involution (Figure 3). Results from GFP-LC3 fluorescence microscopy, accumulation of autophagosomes measured by transmission electron microscopy (TEM) and variations in the abundances of LC3 and BECN1 in bovine mammary epithelial cells support that autophagy is increased during the remodeling of the mammary gland induced by the drying-off in cows (for review see Klionsky et al. [52]), as well as during the mammary gland development under the control of ovarian hormones [69]. Besides mammary gland, autophagy can be considered as part of the adaptive metabolism that sustains milk production. To face the negative energy balance classically experienced by early-lactating ruminants, dairy cows mobilize fat and muscle tissues. A key adaptive metabolic organ is the liver that interconverts mobilized molecules and nutrients thanks to increased gluconeogenesis and ketogenesis. A decrease in the abundance of the ratio of LC3-II:LC3-I in liver was reported in early lactating cows intravenously treated with hyperketonemia treatments [70]. In muscle, phosphorylation of AKT and MTORC1 substrates was decreased whereas macroautophagy assayed by the ratio of LC3-II:LC3-I was upregulated at 21 d post-partum comparatively to 28 d before calving [71]. Therefore, autophagy may be closely associated with metabolic changes that balance the partition of nutrients between milk and whole-body metabolism.
As an indirect consequence regarding milk production, the early weaning may be used increasingly. Indeed, early weaning is widely used in livestock farming; especially dairy calves are weaned early to reduce milk costs. However, their gastrointestinal tract is not ready for early weaning. Glutamine (Gln) supplementation was proposed as a strategy to protect the intestinal barrier integrity. Gln infusion increased growth rates, villus height, and crypt depth in the duodenum of early-weaned calves [72]. Low-dose infusion of Gln induced autophagy, which is presumed to reduce cellular stress, and decreased the level of apoptosis in the duodenum of early-weaned calves [73]. The Gln-induced autophagy was mainly dependent on the inhibition of MTOR phosphorylation.
Role of autophagy in ruminant muscle development and the conversion of muscle in meat
Muscle development and the transformation of muscle into meat are major components of meat production and qualities, especially tenderness. Meat conversion is a complex mechanism dependent on changes in the architecture and the integrity of the muscle fibers. The process of conversion includes biochemical changes, degradation and oxidation of myofibrillar proteins, and breakdown of high-energy compounds. The effective conversion of the muscle into meat after slaughter includes the pre-rigor mortis phase, the rigor mortis establishment, and the tenderization phase where muscle fiber degradation occurs due to proteolytic enzyme activity. The proteolytic changes occurring in the architecture of the myofibril and its associated proteins are a result of the activity of endogenous proteases (e.g., the calpain proteinase system, cathepsins, and caspases). Meat scientists have considered the role of apoptosis in the hours following animal killing as a crucial early process during meat conversion [74]. More recently, Sierra and Oliván have proposed that autophagy should also be considered in the pre-rigor phase (within the first 24 h postmortem), but this is still debated [75]. Garcia-Macia et al. reported time variations in the autophagic-lysosomal pathway in postmortem longissimus dorsi muscle of beef breeds differing in their production purposes: a meat-specialized breed (Asturiana de los Valles) vs. a small- to medium-sized rustic breed (Asturiana de la Montaña) [76]. They analyzed the pattern of several autophagic markers and reported an increase in abundances of BECN1, LC3, CTSD (cathepsin D) and CTSB along the 2–12 h postmortem as well as in the ratio of the CTSD:CTSB activity (a marker of lysosomal viability) at 2 h postmortem in both beef breeds studied. Based on significantly higher LC3-II and LC3-I abundance at 2 h postmortem, the authors concluded to an earlier autophagic activation in the muscle of the rustic breed. Early autophagic activation is an adaptive response to rescue muscle cells from stress conditions after slaughter (sudden arrest of blood flow, deprivation of oxygen and nutrients, oxidative challenge). According to several reports showing that rustic beef breeds exhibit a slower tenderization pattern than that of meat-specialized breeds, the authors proposed that the early autophagy activation results in postponing cellular degradation and, eventually, meat maturation. Longo et al. did not confirm the contribution of autophagy to the control of meat tenderization by studying the temporal evolution of the proteome, phosphoproteome and metabolome of the Piedmontese longissimus thoracis muscle undergoing muscle-to-meat conversion [77]. However, they found compelling pieces of evidence of the contribution of apoptosis in this process.
Conclusion
Despite the poor availability of specific antibodies for ruminants, most of the knowledge on autophagy in these species was obtained thanks to the quantification of autophagy-related protein abundances. A more in-depth analysis of autophagic processes will be challenging because of the inability to utilize genetic approaches and the prohibitive costs of administering pharmacological inhibitors. There are pieces of evidence for the implication of autophagy as a regulator of tissue remodeling, plasticity, and functioning that sustain ruminant productions and health. The identification of the conditions (including genetics and nutritional feeding strategy) that modulate autophagy-based favorable traits will help benefit from them.
Autophagy in pigs
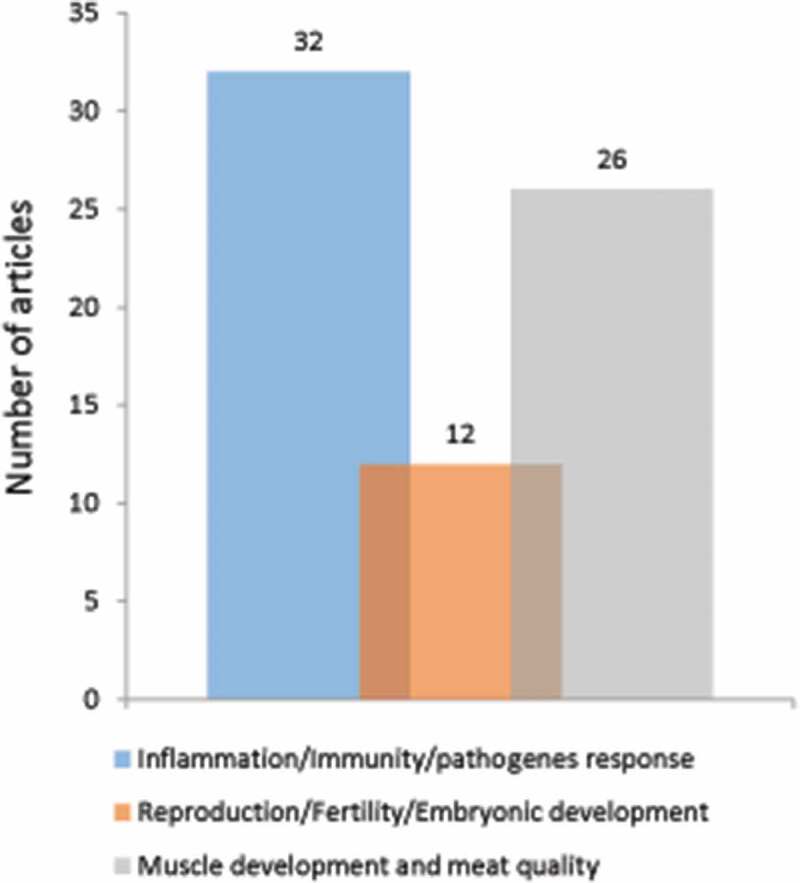
In this section, we provide an overview of the literature on autophagy in pigs (284 articles extracted). We eliminated 214 articles that did not directly address issues of agronomic interest (Figure 2). In the remaining 70 articles, the role of autophagy has mainly been studied in the context of (i) pathogen infections (32 articles), (ii) reproductive disorders (12 articles), and (iii) muscle development and the transformation of muscle in meat (26 articles) (Figure 4). We describe here the main information provided by these articles.
Figure 4.
Number of original articles related to autophagy in pigs according to agronomic fields
Role of autophagy in pathogen infections in pigs
In pigs, numerous studies have highlighted that autophagy is involved in many processes of infection through different pathogens (viruses and bacteria). Indeed, many of them suggest that some viruses are able to exploit autophagic processes for their propagation as already mentioned above. In particular, viruses, such as poliovirus or hepatitis C, use autophagosomes to gather their replication complexes into the cytoplasm of infected cells. Recently, Gou et al. have also shown the involvement of autophagy in the death of T lymphocytes in the spleen of pigs infected with the classical swine virus fever [78]. Compared to healthy animals, the T lymphocytes of infected pigs expressed significantly more BECN1, ATG5 and LC3-II. Other studies have also revealed that autophagy was strongly activated in the context of respiratory pathologies [79].
Role of autophagy in fertility and reproduction in pigs
A significant part of the literature (12 publications) also addressed the role of autophagy in the ovarian follicles during their maturation. Indeed, a successful estrus cycle requires the remodeling of the follicular cells and the disappearance of 99% of them. Kim et al. have shown that several autophagy-related genes (LC3A, ATG5, BECN1) are induced during follicular development [80]. During embryonic development, variations in the microenvironment (calcium, ceramide, ROS) are also able to induce mitochondrial disturbances and autophagy. In 2011, Xu et al. have thus shown that the LC3B and LAMP2 genes were overexpressed in pigs during early embryonic pre-implantation [81]. In addition, a study showed that heat stress (HS), well known to compromise follicular and early embryo development in a variety of agriculturally important species [82–86], induced a decrease in ATG12–ATG5 complexes and an increase in anti-apoptotic signals BCL2 and BCL2L1 in oocyte and early follicles in pig [87]. These data thus contributed to the biological understanding of how HS acts as an environmental stress to affect follicular development and negatively impacts reproduction.
Role of autophagy in early weaning in pigs
Weaning is the most challenging period that has significant bearings on pig welfare and growth performance in swine industry. During weaning period, piglets are immediately imposed on a number of environmental and psychosocial stressors that predispose them to diarrhea and gut damage, which can adversely affect their survival at a very early and most vulnerable stage. Zhang et al. were the first to highlight that autophagic activity was significantly upregulated in the liver, spleen and skeletal muscle tissues of piglets upon early-weaning [88]. More recently, Liao et al. examined the effect of chloroquine as a feed additive agent to modulate autophagy after early weaning [89]. Throughout the experiment, chloroquine treatment was beneficial to alleviate early weaning stress as well as intestinal and immune system dysfunctions.
Role of autophagy in muscle development and the transformation of muscle in meat
In pigs, many studies (26 publications) also focused on the role of autophagy in the development of muscle and adipose tissue in response to nutrition. Rapid muscle growth in the newborn has thus been correlated to the positive balance between protein synthesis and degradation [90]. The authors highlighted the important role of insulin and amino acids (in particular leucine), as signal molecules, to modulate several important factors of muscle protein synthesis and degradation after birth, including the autophagosome-related proteins LC3 and ULK1. In adult pigs, autophagy has also been shown to be downregulated in re-feeding conditions after a period of food restriction, again supporting the role of this cellular function in muscle development [91].
Finally, autophagy acts on postmortem muscle metabolism during meat maturation. Rubio-Gonzalez et al. have revealed that pig rearing strategies and stress related to animal transport impact postmortem muscle autophagy [92]. The authors propose that monitoring the evolution of several biomarkers of autophagy (e.g., BECN1, LC3-II:LC3-I ratio) in the muscle within the first 24 h postmortem may help the detection of animal stress and its potential effect on the postmortem muscle metabolism and meat maturation.
In conclusion, a significant proportion of studies on autophagy in pigs focuses on its role in controlling key physiological functions (e.g., reproduction and muscle development) but also meat quality in the final product. However, there are still many studies to be carried out to understand precisely how this cellular function works in pigs, but also to determine how to apply the knowledge acquired to improve animal development, growth, and health.
Autophagy in animal species used in poultry production
Poultry or domestic fowl classically includes, among other species, chickens, turkeys, ducks and geese. In this section, we will discuss the existing data on autophagy in these species of economic interest, but not in wild birds. Available data on autophagy in the chicken used as a model for neuroscience, embryonic development, medicine will also be not considered here: although they are obtained on same species or strains, the information they provide is outside the scope of the review because it is not related to issues or physiological functions supporting livestock animal production such as reproduction, growth, immune system … (37 articles excluded). In these conditions, 67 articles were selected concerning chickens, ducks, and geese, but also quails and ostrich from the 214 extracted publications initially (Figure 2). The most represented species was the chicken (Gallus gallus) studied in 55 articles (about 80% of the total number of extracted articles) (Figure 5A). Only one other species reached a noticeable representation, ducks (Anas platyrhynchos and Cairina moschata) with 7 (10%) articles. For other species, i.e., ostrich (Struthio camelus), quail (Coturnix coturnix Japonica), and goose (Anser cygnoides), the number of items found was 1 to 3, and, except for geese, autophagy was generally not the central subject. Overall, the selected articles covered research in infectiology-immunity with 25 articles (37% of the total number of extracted articles), environmental stress with 18 (27%) articles, reproduction with 9 (13%) articles, metabolism with 11 (17%) articles and muscle growth-meat quality with 4 (6%) articles (Figure 5A).
Figure 5.
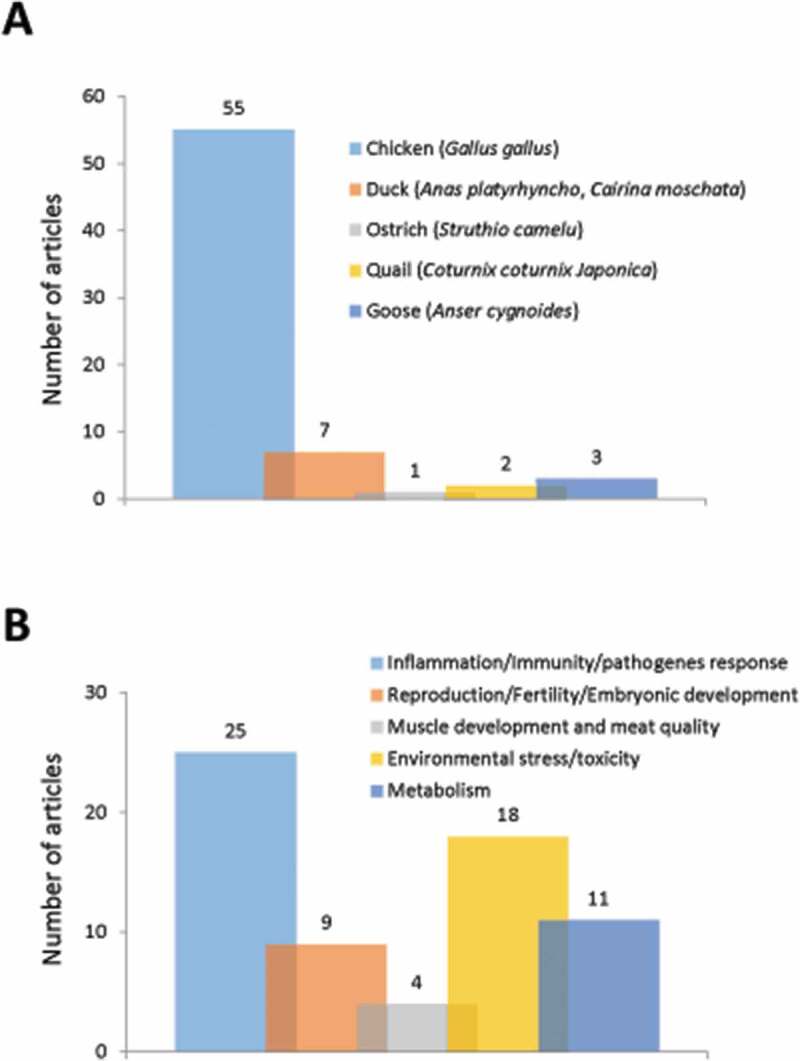
Number of original articles related to autophagy in (A) species used in poultry production and (B) according to agronomic fields
Role of autophagy in pathogen infections and immunity in chickens and ducks
Autophagy has been largely investigated in chickens and ducks due to the major detrimental impacts of pathogen infection on poultry production. The corpus of articles includes studies in which pathogens cause liver disease or egg drop syndrome (avian adenovirus) [93,94], neoplastic disease and immunosuppression (avian leucosis virus) [95–97], respiratory diseases and/or malabsorption syndrome (avian reovirus [98–105], Newcastle disease virus [106,107], avian metapneumovirus [108], avian infectious bronchitis virus [109,110]), and acute disease with high mortality rates (duck enteritis virus [111–114] and duck hepatitis virus type 1 [115]). In domestic fowl, as in non-avian species, there is clear evidence that autophagy is involved in the processes related to infectiology [116–118]. Autophagy participates in the control of infections acting as an anti-viral (via the degradation of intracellular pathogens once internalized in the host cell and the regulation of the immune response) and pro-viral pathway, the latter being the most studied in recent years in poultry. Indeed, the roles of autophagy depend on the virus and species because of host-pathogen coevolution. Some pathogens have developed strategies to counteract autophagy (e.g., decreased autophagy in case of avian leucosis virus infection [95]), or alternately, to exploit autophagy to their own profit and promote their replication, as reported for numerous viruses in chickens (Newcastle disease virus [106,107]; avian metapneumovirus [108]; reovirus [98,99]) and ducks (egg drop syndrome virus [94]; reovirus [105]; duck enteritis virus [111–113]). However, despite the fact that these findings can be illustrated in avian species as in ruminants and pigs (present review), it is important to avoid over-generalization and study the mechanism of autophagy and its involvement in the different economically important avian diseases. For example, classical autophagy may not play a major role in the replication cycle of infectious bronchitis virus as reported by Maier et al. in avian cells [109].
Most of the studies extracted here used in vitro models, such as mainly duck and chick embryo fibroblasts, and chicken embryo fibroblast DF-1 cell line. This in vitro approach facilitates the use of various and specific approaches to investigate autophagy, allowing, for example, the quantification of key proteins (LC3, SQSTM1, BECN1, ATG5, ATG7) and associated signaling pathways (AKT, MTOR, AMPK) by western blot and even autophagic flux measurements [107]. This also allows the visualization of the phagophores and autophagosomes by TEM and particularly by immunofluorescence (IF) with antibodies against LC3, or the GFP-LC3 construct. The GFP-LC3 co-localization with LAMP1 was also investigated by IF [94,107,109]. Only three articles reported quantification of gene expression level by RT-qPCR and/or transcriptomic approach [97,103,106]. Two of them aimed to clarify the interactions between viruses and host, with a particular interest for genes related to autophagy, apoptosis, antiviral defense, innate or adaptive immunity and inflammation [97,103]. The last article, combining the quantification of gene expression and different other approaches in studies performed in vitro and in vivo, aimed to understand the role of autophagy during Newcastle disease virus (NDV) infection [106]. The authors first demonstrated that NDV induced autophagy in chicken cells (higher autophagosome formation shown by TEM, enhanced conversion of LC3-I to LC3-II, and degradation of SQSTM1) and that autophagy inhibition by chloroquine enhanced apoptosis in NDV-infected chickens cells. The potential roles of autophagy during NDV infection were then investigated in vivo by determining the levels of the autophagy-related genes that are involved in autophagosome initiation, elongation and maturation steps. The results indicated that autophagy induction by rapamycin resulted in upregulation of almost all autophagy-related genes in the NDV-infected spleen and lung tissues, whereas the inhibition of autophagy by chloroquine decreased the expression levels of ATGs in the tissues targeted by NDV, suggesting that autophagy plays a crucial role in the host response to NDV infection.
Some relatively few studies included additional in vivo and tissue measurements in order to quantify the impact of disease, for example, liver integrity and function due to adenovirus [93], and pathogenesis, virus infectivity and/or apoptosis in target tissues after Newcastle disease virus infection [106,107]. Interestingly, the knowledge on autophagy in domestic fowl completed by in vivo and tissue measurements can also help to propose protection strategies and test their efficacy by determining the protection against infection by avian leucosis virus using an autophagy-targeted vaccine [96], and the survival of reovirus-infected chicken embryos following treatment with autophagy inhibitors [102].
Environmental stress studied in chickens and ostrich
Environmental stress is considered here as representing a series of factors independent of the animal and generally related to oxidative stress or inflammation. It has been shown to induce autophagy in chickens and ostrich. In poultry, the main factors considered are 1) environmental toxic pollutants (arsenic, copper, cadmium, lead), since the water system and soil can contain elevated concentrations of these toxic elements (11 articles), 2) some trace elements present in excess or deficiency in the diet (iron, selenium, boron; 5 articles) and 3) aflatoxins that contaminate a large variety of tropical and subtropical foodstuffs (2 articles). For example, it has been reported that copper and/or arsenic induced autophagy while inhibiting AKT/MTOR pathway in chicken skeletal muscle [119] and cadmium modulated MIR-33-AMPK axis and led to BNIP3-dependent autophagy in chicken spleen [120]. In this domain, autophagy was mostly studied at the tissue level, more precisely in muscle [119,121], intestine [122], gizzard [123], heart [124], brain [125], liver [126–128], kidney [129], testis [130], and immune organs [120,131–133]. The methods combined the quantification of key proteins involved in macroautophagy (LC3, SQSTM1, BECN1, ATG4B, ATG5, ATG12), sometimes mitophagy (MFN1, MFN2, BNIP3) and associated signaling pathways by western blot and/or their transcript by RT-qPCR and ultrastructure imaging by TEM. Interestingly, the use of the autofluorescent compound monodansylcadaverine was reported in two studies performed in chicken hepatocytes for labeling of autophagic vacuoles (IF imaging) [134,135]. Due to the link between environmental and oxidative stress, numerous studies also explored antioxidant system, oxidative stress cascades, inflammation or immune system. Moreover, some of them tested potential strategies (dietary selenium supplementation, astragalus polysaccharide supply) to limit the deleterious effects of environmental stress by enhancing antioxidant systems and/or promoting immunomodulatory effects as mechanisms of protection against pollutant-induced autophagy [130,134,136].
Reproduction in chickens, geese and quails
In poultry, the role of autophagy on reproduction has mainly been analyzed on egg production-related functions; only two articles focused on other facets related to reproduction, i.e., spermiogenesis and blastoderm dormancy [137,138], but autophagy was not the central subject. Egg production-related functions present peculiarities in domestic fowl. The characteristic feature of avian species includes the presence of an ovarian follicular hierarchy containing follicles at all stages of maturation, and consequently the possibility of having one egg per day in laying hens, the existence of molt induction (period without egg production) to rejuvenate the egg-laying performance of commercial laying hen flocks and the existence of broodiness, a specific maternal behavior of poultry, which can result in reduced egg-lay. Using TEM, it has been shown that autophagy participates in the involution of isthmus (section of the oviduct) in molting hens [139,140] and follicular atresia (i.e., follicular degeneration and resorption) in the ovaries of quails [141] and geese [142]. The transcriptome analysis of follicles also reveals the importance of autophagy in broodiness in goose [143]. The increase in the formation of autophagic vacuoles in broody goose follicles was confirmed by the elevated expression of the autophagy-related genes LC3, BECN1, ATG9, ATG12 as well as the abundance of LC3-II [142], and further supported by the involvement of MTOR as regulator [144]. In laying period, autophagy was shown to be involved in the regression of the ruptured follicle after ovulation, especially in the theca layer (i.e., the envelope of connective tissue surrounding the granulosa cells) [145]: BECN1 was distributed mainly in theca cells and coupled with LC3-II accumulation (IF imaging); moreover, LC3-II and BECN1 increased and SQSTM1 declined with the regression of post-ovulatory follicles (western blot).
Metabolism and growth-meat quality in chickens and quails
There is a relative paucity of information on the role of autophagy in relation to metabolism, growth and meat quality in domestic fowl (15 articles). Scientific research in this area concerns the response to food deprivation or nutrient depletion, the regulation of feed efficiency (i.e. weight gain per amount of feed consumed), body composition and meat quality.
The effect of food deprivation was investigated about 20 years ago by TEM in the intestine of hens [146] and in chickens [147] demonstrating autophagic vacuoles in the duodenal epithelial cells of fasted animals. These findings were linked to changes in the morphology of intestinal villi, and therefore intestinal functions. More recent studies integrating western blot analyses highlighted the effect of fasting on LC3-II:total LC3 (i.e., increased ratio) in skeletal muscle [148], as well as the effect of nutrient depletion on LC3-II and GABARAP in chicken embryo fibroblasts [149].
Regarding autophagy and functions more geared toward production efficiency, articles mainly reported quantification of gene expression level by RT-qPCR and/or omic approaches (global gene and gene and/or protein expression analyses). This includes the characterization of autophagy-related genes (involved in autophagosome initiation and elongation) in chickens and quails [150], and the detection of differentially expressed genes or proteins related to autophagy in the breast muscle of chickens varying in dietary methionine provision (e.g., increased expression of the genes ATG5 and BECN1 with slight methionine deficiency [151]) and in feed efficiency (pedigree broiler model) [152–154]. In the last model, the autophagy pathway was enriched in the breast muscle of high feed efficiency broilers with an increased expression of several genes involved in autophagosome initiation, elongation and maturation steps.
Little data is available concerning mechanisms underlying skeletal muscle growth, even if DAP (death associated protein; a negative regulator of autophagy) was shown to be required for regulating myogenesis and apoptosis of chicken satellite cells, which may affect muscle mass accretion [155]. Two articles concern muscle differentiation [156] and skeletal development [157], but autophagy was not the central subject. Meat quality was the subject of only one article [158], in which the authors investigated the relationship between meat quality and oxidative damage in the breast muscle of broiler chickens. The expression of autophagy-related genes (LC3, BECN1) and the abundance of protein LC3-II increased by oxidative stress induced by H2O2 (in agreement with the findings presented in the section Environmental stress studied in chickens and ostrich), while a decline was found in histomorphology and meat quality traits such as the ultimate pH and tenderness.
Interestingly, two recent papers focused on the regulation of autophagy by the endocrine system by studying the effect of either LEP (leptin) or corticosterone [159,160]. Besides reducing feed intake, intracerebroventricular administration of LEP increased the expression of autophagy-related genes and/or proteins (LC3, BECN1, ATG3, ATG5, ATG7) in hypothalamus, liver and muscle [159]. These findings were corroborated by testing the direct effect of LEP treatment in hypothalamic organotypic culture as well as muscle and hepatocyte cell cultures by using the same techniques (RT-qPCR and western blot) and IF. The potential downstream mediator of the effect of LEP was also investigated. Overall, this study demonstrates that AMPK mediates the effect of LEP on autophagy in a tissue-specific manner. Otherwise, corticosterone induced a decrease of LC3-II and LAMP1 protein levels as well as of autophagic vacuoles in chicken liver [160]. This study also demonstrated that dietary supplementation of betaine to laying hens could prevent the cortiscosterone-induced inhibition of these autophagy- and lysosome-related factors in the offspring. The authors propose that this effect of maternal betaine supplementation on autophagy could partially contribute to alleviate the corticosterone-induced hepatic lipid accumulation by enhancing lipophagy, a specific autophagy-related process through which lipid droplets in the hepatocytes can be degraded.
Conclusion
The research effort on autophagy in poultry appears relatively unbalanced according to the agronomic domains, both in number of studies and tools or approaches developed. A very significant expansion has been observed in recent years in infectiology and environmental stresses, thereby allowing us to imagine some potential protection strategies. On the other hand, too few studies have been devoted to the role of autophagy in relation to metabolism, growth and quality of meat, and the underlying mechanisms must be investigated to control these functions by new nutritional and breeding strategies.
Autophagy in animal species used in aquaculture
In this section, we discuss the existing data on autophagy in the most important fish and shellfish species cultured worldwide (FAO 2011–2018). Available data on autophagy in the model fish species zebrafish (Danio rerio) are therefore not covered here, and readers are referred to review articles focusing on this specie in detail [161–164].
Overall, our analysis allowed extracting 99 articles devoted and/or related to autophagy in fish and shellfish species of economic interest. These articles focused on research in infectiology-immunity (30 articles), environmental stress (33 articles), reproduction (4 articles), metabolism (17 articles) and muscle growth-meat quality (16 articles) (Figure 6A). They concerned 24 species, including 7 shellfish species (5 mollusks and 2 crustaceans) and 17 fish species (Figure 6B). Among these farmed aquatic species, two are over-represented: the blue mussel (Mytilus edulis), which has been studied in 23 articles (23% of the total number of extracted articles) and rainbow trout (Oncorhynchus mykiss) covered in 23 articles (23% of the total number of extracted articles). Two other species reach a noticeable representation, although much less than the former two, namely the common carp (Cyprinus carpio) and the Pacific oyster (Crassostrea gigas) with 8 (8%) and 7 (7%) articles, respectively. For other species, the number of articles found is 1 to 5, and autophagy for most of them is not the central subject. Here, we outline the data available for the 2 most significant species, the blue mussels (Mytilus edulis) and the rainbow trout (Oncorhynchus mykiss).
Figure 6.
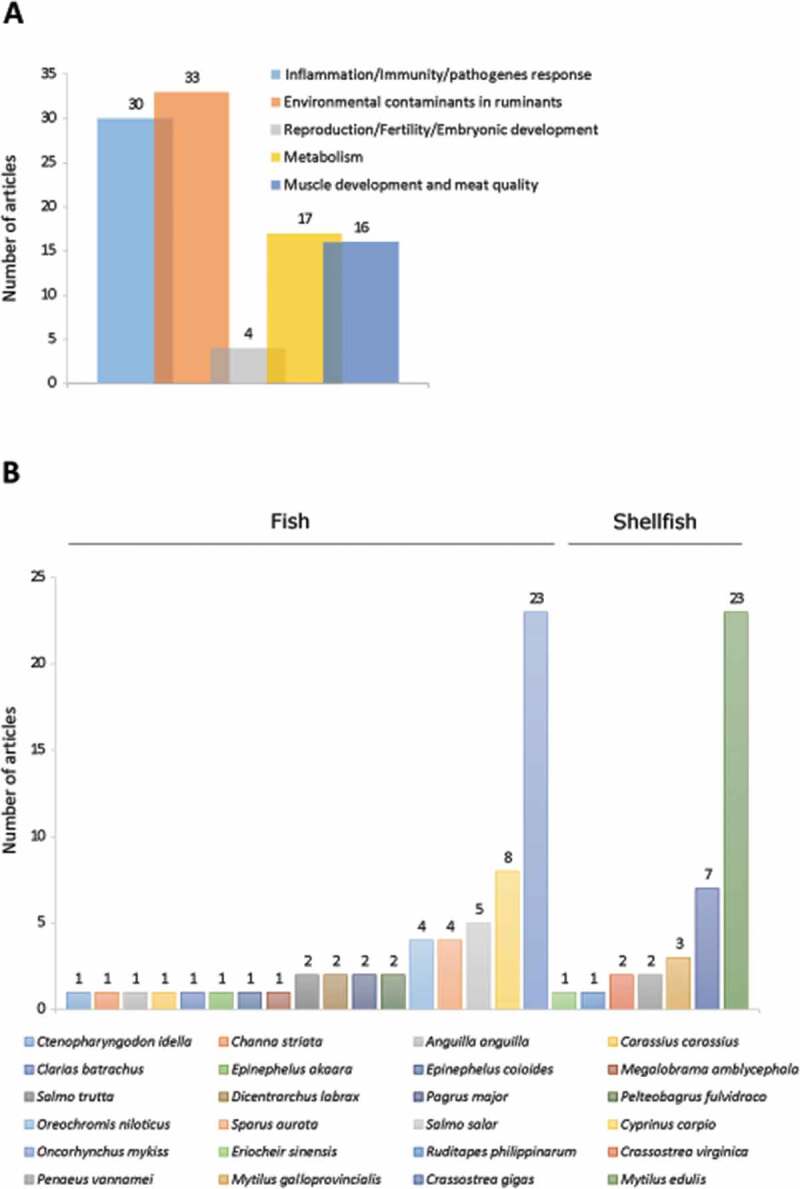
Number of original articles related to autophagy in animal species used in aquaculture classified by (A) agronomic field or (B) species
Blue mussels (Mytilus edulis)
Blue mussels are produced in fluctuating environments such as estuaries where they are subjected to variable salinity, temperature, nutritional condition, and chemical pollutants, as well as repeated air exposure (which results in hypoxia) and reimmersion in seawater [14,165–170]. Consequently, this essentially stressful fluctuating environment has been shown to induce the autophagy-lysosomal system, which, by removing altered proteins and/or damaged organelles, results in more efficient cellular housekeeping and helps to minimize the formation of harmful lipofuscin and other aggregates [13–15].
A well-established tool to assess the autophagy-lysosomal response of mussels’ cells to environmental stress is the lysosomal membrane stability (LMS) assay (based on cytochemically determined lysosomal enzyme latency or, alternatively, lysosomal retention of neutral red dye) [170]. Lysosomes respond to a broad range of pollutants and/or environmental stresses with a significant increase in permeability of their membranes [171–173]. The evaluation of LMS has thus emerged as an easy and relevant method to estimate the degree of stress or disease in mussels [174]. In this regard, LMS but also the size of lysosomal membrane has been correlated to site-specific contamination levels [175–177], as well as to natural stress factors such as temperature, hypoxia, salinity or food availability and quality [178–181]. In addition to these anthropogenic and natural stressors, specific physiological and/or physiopathological conditions such as reproductive status, infection or disease were reported to affect LMS [182,183].
Other biomarkers suitable to assess the autophagy-lysosomal system reactions to environmental stressors in blue mussels have been developed in complement to LMS. They include, among others, lipofuscin levels [184], lysosomal neutral lipid content [184], and more recently LC3 puncta [185], CTSB and CTSD activities [186], and the phosphorylation of the master autophagy inhibitor MTOR [187].
Overall, the development of these markers increased our understanding of the response of the autophagic-lysosomal system to environmental changes and/or during particular physiological or physiopathological situations in mollusks. Recent studies used a network modeling approach to integrate multi-biomarker data and provide a more comprehensive view of the interconnected autophagic and lysosomal processes, together with anti-oxidant protective system, enable animals to respond and adapt to changes in their environment [187–189].
Rainbow trout (Oncorhynchus mykiss)
Rainbow trout is probably one of the most deeply studied fish species with a long history of research carried out in physiology, nutrition, ecology, genetics, pathology, carcinogenesis, and toxicology [190]. In addition, as a carnivorous species, it has unusual metabolic characteristics (i.e., a high requirement for dietary protein combined with an apparent inability to metabolize dietary carbohydrates) that make it a particularly relevant model organism for studying the metabolic role of autophagy.
The first data concerning autophagy in this species date from the ‘90s. An in vitro study conducted on isolated rainbow trout hepatocytes revealed by TEM the existence of autophagic vacuoles containing organelles (such as mitochondria) or lipid droplets [191]. Since then, we and others identified and studied the regulation of the expression of several critical rainbow trout atg genes in muscle and cultured myocytes as well as, more recently, in RTgill-W1 epithelial cells [192–199]. The expression of these genes was shown to be induced in catabolic situations (e.g., fish subjected to prolonged fasting or during sexual maturation or cells subjected to nutrient restriction) and suppressed by refeeding [192,196,199,200]. In this regard, we reported an unexpected role of amino acids in the regulation of the expression of atg genes, thus providing new evidence for understanding the nutritional control of this cellular function [194]. However, different effects of fasting and refeeding on the expression of these genes have been recorded between diploid and triploid rainbow trout [197], suggesting that ploidy affects the mechanisms of atg gene regulation. Further characterizing these mechanisms will contribute to the development of ploidy-specific feeding strategies, diet formulations, or husbandry practices that optimize nutrient utilization and growth performance.
Recently, we also demonstrated that both broodstock nutritional history and an early acute hypoxia applied at embryo stage induced significant changes in both the mRNA levels and the methylation of several CpG sites of bnip3a and bnip3lb1, two genes involved in the autophagy-mediated mitochondrial degradation (known as mitophagy), in whole-body fry [201]. Similarly, a recent study also demonstrated that early feeding of rainbow trout with a methionine-deficient diet over a 2-week period increases the abundance of several canonical markers of mitophagy at later juvenile stages [202]. These findings thus raised the interesting possibility of directing autophagy-related functions in trout and open up new opportunities toward the optimization of nutrition and health of trout by the mean of nutritional programming.
However, as autophagy machinery is primarily regulated at post-translational level, a number of tools have also been developed for accurate monitoring of autophagy activity. Most components of autophagy, autophagy-related functions (e.g., mitophagy) and associated signaling pathways (Akt, Foxo, Ampk, Mtor) are evolutionary conserved and can be studied in rainbow trout by western blot through the use of commercially available antibodies produced against their mammalian orthologs [193,203,204]. The detection of Lc3-II, which is the gold standard method for estimating autophagosome quantity within the cell, can also be performed by western blot or by IF in trout using commercially available antibodies [194,199,204–207]. Overall, these tools allowed studying both in vivo and in vitro the response of autophagy in rainbow trout to different factors including nutrient restriction or imbalance [199,202,204–206] or exposure to viruses [207]. Recently, antibodies specific to rainbow trout Lc3 and Atg12 were also produced with the objective of exploring the main effect of follicle autophagy on gonadal degeneration in triploid female rainbow trout [208]. These antibodies were proved successful for western blot (Lc3 and Atg12) and immunochemistry (Lc3) analyses, further increasing the arsenal of tools available to study this function in trout.
However, despite the increase in the number of tools available to monitor autophagy in trout, most of the available data remains descriptive and few studies addressed the specific role of this cellular function in this species. Recently, the respective role of both the proteasome and autophagy to total protein degradation has been studied in fasted trout myotubes by using specific inhibitors [209]. The results demonstrated that autophagy plays a major role in this process (around 50% of total protein degradation) compared to the proteasome (which only accounts for 17%), at least in this model. By using the autophagic flux inhibitor colchicine, autophagy was also shown to contribute to the maintenance of blood glucose and amino acid levels as well as the lipid stores in the liver of this species [210]. Similarly, the treatment of fasted trout hepatocytes with the autophagy inhibitor bafilomycin Al has been shown to decrease the mRNA levels of many of the gluconeogenesis-related genes and increased those of genes involved in intracellular lipid stores [211]. Together, these two studies corroborated the reported role of autophagy in the regulation of glucose homeostasis and intracellular lipid stores, highlighting the importance of this function in the control of hepatic metabolism in rainbow trout.
Collectively, these studies led to significant advances in the characterization of autophagy in trout and in our understanding of the main mechanisms involved in its control, especially by nutrients. However, studies on the role of this function in fish are still very limited, and details on how autophagy functions in fertility, development and growth of trout (as well as of other aquaculture fish species) but also tolerance to dietary imbalances, abiotic or pathogenic stresses and finally in the quality of the flesh consumed remain widely unexplored.
Autophagy in other livestock animals: horse, rabbit
This section gathered publications dealing with autophagy in two species: horse and rabbit. Over 79 original articles, only 14 were related to autophagy in 5 agronomic fields: reproduction (5 studies), inflammation/immunity/pathogens response (4 studies), metabolism (2 studies), growth (2 studies), environmental stress/toxicology (1 study) and distributed in each animal as shown in Figure 7.
Figure 7.
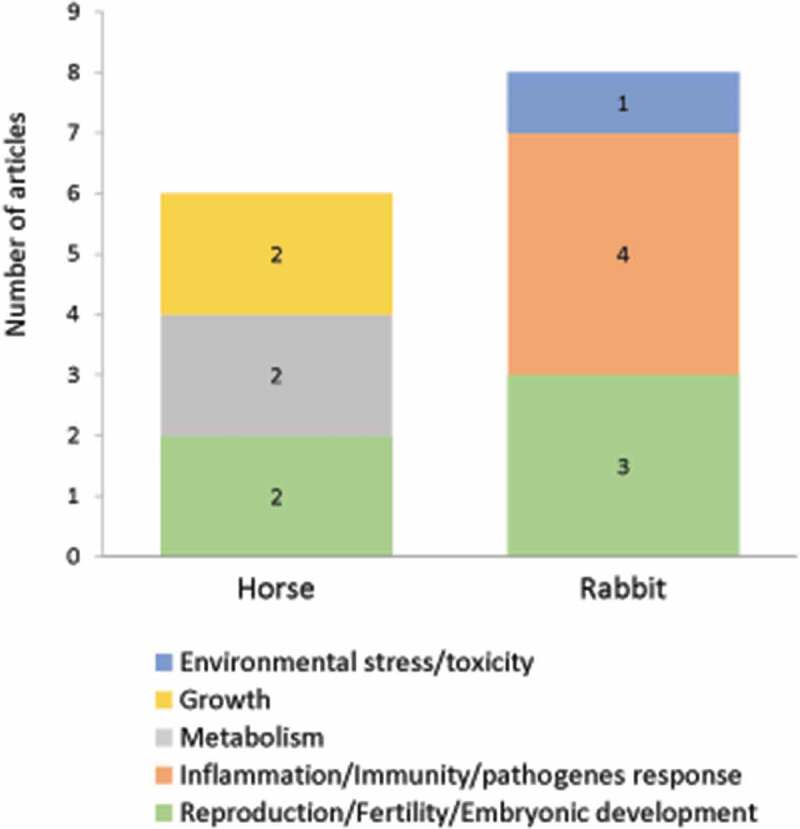
Number of original articles related to autophagy in horse and rabbit according to agronomic fields
Horses
In the context of equine science, we identified six publications related to autophagy in metabolism, growth, and reproduction.
It has thus been observed that the aging of equine triceps brachii muscle is accompanied by a decrease in the level of autophagy-related proteins ATG5 and LC3-II and an accumulation of SQSTM1, suggesting an age-associated decline of autophagosome formation in this tissue [212].
In a different context, Marycz et al. demonstrated that autophagy is one of the mechanisms underlying the biological properties of mesenchymal stem cells (MSCs) and chondrocytes in equine metabolic syndrome (an endocrinopathy affecting horses and ponies) and osteoarticular disease, respectively [213]. For many years, MSCs have gained popularity in equine veterinary medicine for regenerative purposes because of their self-renewal ability and their multilineage differentiation potential, conferring them a high value in tissue engineering. In this respect, the study of Marycz et al. showed that adipose-derived stem cells (ASCs) isolated from horses affected by equine metabolic syndrome were markedly characterized by impaired proliferative and metabolic activity (deterioration of mitochondrial dynamics), which is related to lower mitochondrial metabolism and induced autophagy process. The authors showed that the differentiation potential of these cells relies on the induction of autophagy (evaluated by mRNA expression of LC3, BECN1 and LAMP2). The increase in autophagy was found to alleviate the energy production crisis caused by dysfunctional mitochondria and excessive oxidative stress induced by the metabolic syndrome in those cells [213]. Currently, various pretreatment conditions are evaluated to improve the regenerative potential of equine adipose-derived stromal cells. Indeed, it has been demonstrated that calystegines (polyhydroxylated alkaloids) extracted from Hyoscyamus albus can rescue ASCs affected by metabolic syndrome by promoting their survival by blocking apoptosis and improving the dynamic status of mitochondria [214].
Osteochondrosis (OC) is a condition commonly found in sport and racing horses, usually accompanied by osteochondritis dissecans (OCD), characterized by a defect of ossification in young animals, which negatively affects joint homeostasis. Kornicka et al. investigated the cytophysiological properties of equine OCD chondrocytes at molecular level including expression of chondrogenic genes, apoptosis, mitochondria dynamics and autophagy [215]. OCD chondrocytes were characterized by increased apoptosis and senescence. The expression of LC3 mRNA is diminished in OCD cells while there is no change in BECN1 expression.
Finally, in the field of reproduction, autophagy was investigated in relation to sperm conservation and quality, but the results are still highly debated. Thus, in a first study, the authors suggested that the increase in LC3-II levels could be considered as a marker of sperm survival when evaluating different cooling media [216]. However, in a second study, the increase in LC3-I to -II processing was considered as a marker of stress during spermatozoa freezing, and inhibition of autophagy was shown to improve sperm viability [217].
Rabbit
The rabbit is an essential model in scientific research, particularly in the study of several human diseases, because it is phylogenetically closer to humans than rodents. Rabbits remained nevertheless a model for studies in agronomic field. Among the 63 listed publications related to autophagy in rabbit, only 8 publications were selected, dealing with animal diseases (due to viral or bacterial infection), reproduction and toxicology.
Several examples illustrated the involvement of autophagy in pathogen infections in rabbits. Ferreira et al. provided pieces of evidence for the presence of autophagic vesicles during hepatocellular degeneration induced by the rabbit hemorrhagic disease virus (RHDV) [218]. A more recent study demonstrated that RHDV induced the formation of autophagosomes and autolysosomes, as well as the expression of several autophagy-related proteins or markers [219]. Such autophagic activity induced by the RHDV declined in late periods of infection in parallel to the increase in apoptosis. These results suggest that virally induced autophagy might contribute to limit cell death triggered by RHDV infection. Another example illustrates the involvement of autophagy in the host response to bacterial infection. Guo et al. characterized the rabbit NOD1 (rNOD1; nucleotide-binding oligomerization domain 1) member of the NOD-like receptors, involved in the mammalian innate immune system [220]. In particular, they showed that rNOD1 induced autophagy in cells infected with E. coli. These findings could contribute to therapeutic strategies such as targets of new vaccine adjuvants or drugs. The implications are important due to the increasing use of rabbits for research and food production and the risk of further interspecies pathogen transmission.
In the field of reproduction, the sexual differentiation involves programmed cell death of epithelial cells in Mullerian and Wolffian ducts and the chronology of such event has been found to vary from one species to another in mammals. Two studies further provide important new insights concerning the role of autophagy in the process of progressing and regressing genital ducts in the rabbit [221,222]. In vivo, as well as in organ culture, the ultrastructural modifications observed in the epithelial cells during the sexual differentiation in genital ducts of fetal rabbit were an increase in lysosome numbers followed by subsequent accumulation of autophagic vacuoles. In turn, the duct cell autophagy can be stopped if the formation of lysosomes was prevented. These studies suggest that autophagy is involved in cell degeneration of the genital ducts.
Understanding the mechanism by which cells become sensitive to toxicity of chemicals has important implications in veterinary medicine. The study of Bozzatto et al. demonstrates the occurrence of autophagic cell death processes in the tissues of rabbits infested with Rhipicephalus sanguineus ticks and exposed to the action of different selamectin (an acaricide) concentrations [223]. Based on increased concentrations of hydrolytic enzymes (acid phosphatase) in tissues exposed to selamectin, the authors established that selactamin is a dose-dependent toxic agent.
Conclusion
Although research on autophagy expanded dramatically in the past 15–20 years, it has been mainly focused on rodent, particularly in the context of human diseases. Thus, due to the limited amount of information available for other vertebrate taxa, it remains unknown whether the rodent autophagy genes repertoire as well as the related expression patterns, regulations and functions is even representative of the other vertebrate species. In this regard, recent findings demonstrated that while 90% of the “autophagy core” genes are conserved across all eukaryotic species, those coding for proteins involved in recognition of substrates to be degraded by autophagy (known as autophagy receptors) display rather low conservation in ancient taxa, suggesting a specialization of this function during evolution [224]. Concerning fish in particular, it is possible that the whole genome duplication occurred 300–350 million years ago in the common ancestor of teleosts (ray-finned fishes gathering 99.8% of current fish species) has strongly modulated the genetic structure and therefore the function of autophagy in this group of vertebrates compared to mammals. It is, therefore, possible that some variations (even marginal) in the repertoire of autophagy genes or their expression pattern, regulation and function between species may have important consequences in the development of phenotypes of agricultural interest that remains to be characterized and studied.
In latest years, the number of studies on autophagy in farm animals has increased significantly. The recent availability of the genome of these species allowed to characterize the main factors involved in this function and to establish a solid knowledge base on its regulation and role in these species. Thus, physiological functions supporting livestock animal production (reproduction, growth, immune system, and meat maturation) have been shown to be regulated by autophagy. However, much work is still needed to understand how autophagy works in these species and how to apply the acquired knowledge to improve animal development, growth and health, as the quality of their products, in the context of growing social and environmental challenges for agriculture. For example, while it is now clearly established in pig, cow or chicken that many viruses use the autophagosomal machinery to spread, the lack of precise data on the mechanisms involved prevents the definition of effective antiviral strategies. Similarly, a better knowledge of the mechanisms at play in the response of autophagy to environmental, nutritional and/or oxidative stresses in these species would allow the definition of new strategies adapted to farming constraints and challenges. It will also be essential to decipher the respective role of the different autophagic route (macroautophagy, microautophagy and CMA) in the development of phenotypes of agricultural interest.
Improving our knowledge of the mechanisms involved in autophagy in farm animals will require the development of appropriate tools. Indeed, the lack or the limited tools available (e.g., specific antibodies) to study autophagy in these species represent the main limitation in the field nowadays. The ability to manipulate or study autophagy as a dynamic process is also particularly limited in large animals due to financial, infrastructural and ethic constraints associated with the use of pharmacological agents and/or genome editing tools in these species. In this context, one of the major challenges in the future will be the development of reliable tools or assays to accurately monitor and quantify in these species the different stages of the autophagic process as well as the different subtypes of selective autophagy, including mitophagy or CMA, which have emerged these last few years as a major core component in the control of cellular homeostasis and energetics but remain barely studied in livestock animals.
In the meantime, we strongly recommend authors to multiply the approaches and tools currently available to report the studied autophagic events, in accordance with the Guidelines for the use and interpretation of assays for monitoring autophagy [52]. Below are some basic principles that we believe to be critical when designing studies and/or considering/reviewing the literature on autophagy in farm animals:
Be aware that the ability to study autophagy as a dynamic process in large animals is often limited because the use of inhibitors or transgenic animals is not feasible.
Consider the alternative and/or complementary cell culture approach that allows manipulating or studying autophagy as a dynamic process.
Be aware that most of the measurements per se are not reflective of dynamic autophagy process as they rely on the interpretation of markers of different steps of this process (initiation or degradation of autophagosomes, for example), and therefore consider the use of different approaches/tools available for each species to report the studied autophagic events.
Avoid the over-interpretation of the data obtained. For example, the induction of the expression of autophagy-related genes does not necessarily reflect induction of autophagy.
Supplementary Material
Acknowledgments
This paper was supported by the INRAE “Animal Physiology and Livestock Systems” Division. We apologize to the authors of several high-quality scientific articles that have contributed significantly to the development of the field but could not be cited.
Funding Statement
This work was supported by the INRAE “Animal Physiology and Livestock Systems” Division [ACI_PHASE2018_autoPHAgoSomE].
Disclosure statement
There are no contractual agreements for the presented data, which might cause conflicts of interest.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Mizushima N.A brief history of autophagy from cell biology to physiology and disease. Nat Cell Biol. 2018;20:521–527. [DOI] [PubMed] [Google Scholar]
- [2].Levine B, Kroemer G. Biological functions of autophagy genes: a disease perspective. Cell. 2019;176:11–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Dikic I, Elazar Z. Mechanism and medical implications of mammalian autophagy. Nat Rev Mol Cell Biol. 2018;19:349–364. [DOI] [PubMed] [Google Scholar]
- [4].Galluzzi L, Baehrecke EH, Ballabio A, et al. Molecular definitions of autophagy and related processes. EMBO J. 2017;36:1811–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kaushik S, Cuervo AM. The coming of age of chaperone-mediated autophagy. Nat Rev Mol Cell Biol. 2018;19:365–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Tekirdag K, Cuervo AM. Chaperone-mediated autophagy and endosomal microautophagy: joint by a chaperone. J Biol Chem. 2018;293:5414–5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Li W, Li J, Bao J. Microautophagy: lesser-known self-eating. Cell Mol Life Sci. 2012;69:1125–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Oku M, Sakai Y. Three distinct types of microautophagy based on membrane dynamics and molecular machineries. Bioessays. 2018;40:e1800008. [DOI] [PubMed] [Google Scholar]
- [9].Klionsky DJ, Cregg JM, Dunn WA, et al. A unified nomenclature for yeast autophagy-related genes. Dev Cell. 2003;5:539–545. [DOI] [PubMed] [Google Scholar]
- [10].Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. [DOI] [PubMed] [Google Scholar]
- [11].Molino D, Nascimbeni AC, Giordano F, et al. ER-driven membrane contact sites: evolutionary conserved machineries for stress response and autophagy regulation? Commun Integr Biol. 2017;10:e1401699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Shen H-M MN. At the end of the autophagic road: an emerging understanding of lysosomal functions in autophagy. Trends Biochem Sci. 2014;39:61–71. [DOI] [PubMed] [Google Scholar]
- [13].Di Bartolomeo S, Nazio F, Cecconi F. The role of autophagy during development in higher eukaryotes. Traffic. 2010;11:1280–1289. [DOI] [PubMed] [Google Scholar]
- [14].Moore MN, Allen JI, Somerfield PJ. Autophagy: role in surviving environmental stress. Mar Environ Res. 2006;62:S420–S425. [DOI] [PubMed] [Google Scholar]
- [15].Pellacani C, Costa LG. Role of autophagy in environmental neurotoxicity. Environ Pollut. 2018;235:791–805. [DOI] [PubMed] [Google Scholar]
- [16].Liu EY, Xu N, O’Prey J, et al. Loss of autophagy causes a synthetic lethal deficiency in DNA repair. Proc Natl Acad Sci USA. 2015;112:773–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Sica V, Galluzzi L, Bravo-San Pedro JM, et al. Organelle-specific initiation of autophagy. Mol Cell. 2015;59:522–539. [DOI] [PubMed] [Google Scholar]
- [18].Stolz A, Ernst A, Dikic I. Cargo recognition and trafficking in selective autophagy. Nat Cell Biol. 2014;16:495–501. [DOI] [PubMed] [Google Scholar]
- [19].Anding AL, Baehrecke EH. Cleaning house: selective autophagy of organelles. Dev Cell. 2017;41:10–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Delbridge LMD, Mellor KM, Taylor DJR, et al. Myocardial autophagic energy stress responses–macroautophagy, mitophagy, and glycophagy. Am J Physiol Heart Circ Physiol. 2015;308:H1194–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Liu K, Czaja MJ. Regulation of lipid stores and metabolism by lipophagy. Cell Death Differ. 2013;20:3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Nakatogawa H, Mochida K. Reticulophagy and nucleophagy: new findings and unsolved issues. Autophagy. 2015;11:2377–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kaushik S, Cuervo AM. Chaperones in autophagy. Pharmacol Res. 2012;66:484–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Valdor R, Mocholi E, Botbol Y, et al. Chaperone-mediated autophagy regulates T cell responses through targeted degradation of negative regulators of T cell activation. Nat Immunol. 2014;15:1046–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ferreira JV, Fôfo H, Bejarano E, et al. STUB1/CHIP is required for HIF1A degradation by chaperone-mediated autophagy. Autophagy. 2013;9:1349–1366. [DOI] [PubMed] [Google Scholar]
- [26].Xie W, Zhang L, Jiao H, et al. Chaperone-mediated autophagy prevents apoptosis by degrading BBC3/PUMA. Autophagy. 2015;11:1623–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Yang Q, She H, Gearing M, et al. Regulation of neuronal survival factor MEF2D by chaperone-mediated autophagy. Science. 2009;323:124–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Park C, Suh Y, Cuervo AM. Regulated degradation of Chk1 by chaperone-mediated autophagy in response to DNA damage. Nat Commun. 2015;6:6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kaushik S, Cuervo AM. Degradation of lipid droplet-associated proteins by chaperone-mediated autophagy facilitates lipolysis. Nat Cell Biol. 2015;17:759–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Lv L, Li D, Zhao D, et al. Acetylation targets the M2 isoform of pyruvate kinase for degradation through chaperone-mediated autophagy and promotes tumor growth. Mol Cell. 2011;42:719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Schneider JL, Suh Y, Cuervo AM. Deficient chaperone-mediated autophagy in liver leads to metabolic dysregulation. Cell Metab. 2014;20:417–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Xia H-G, Najafov A, Geng J, et al. Degradation of HK2 by chaperone-mediated autophagy promotes metabolic catastrophe and cell death. J Cell Biol. 2015;210:705–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Tasset I, Cuervo AM. Role of chaperone-mediated autophagy in metabolism. Febs J. 2016;283:2403–2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Chiang HL, Terlecky SR, Plant CP, et al. A role for a 70-kilodalton heat shock protein in lysosomal degradation of intracellular proteins. Science. 1989;246:382–385. [DOI] [PubMed] [Google Scholar]
- [35].Kirchner P, Bourdenx M, Madrigal-Matute J, et al. Proteome-wide analysis of chaperone-mediated autophagy targeting motifs. PLoS Biol. 2019;17:e3000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Cuervo AM, Dice JF. A receptor for the selective uptake and degradation of proteins by lysosomes. Science. 1996;273:501–503. [DOI] [PubMed] [Google Scholar]
- [37].Bandyopadhyay U, Kaushik S, Varticovski L, et al. The chaperone-mediated autophagy receptor organizes in dynamic protein complexes at the lysosomal membrane. Mol Cell Biol. 2008;28:5747–5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Cuervo AM, Dice JF. Unique properties of lamp2a compared to other lamp2 isoforms. J Cell Sci. 2000;113(Pt 24):4441–4450. [DOI] [PubMed] [Google Scholar]
- [39].Bandyopadhyay U, Sridhar S, Kaushik S, et al. Identification of regulators of chaperone-mediated autophagy. Mol Cell. 2010;39:535–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kiffin R, Christian C, Knecht E, et al. Activation of chaperone-mediated autophagy during oxidative stress. Mol Biol Cell. 2004;15:4829–4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Hubbi ME, Hu H, Ahmed I, et al. Chaperone-mediated autophagy targets hypoxia-inducible factor-1α (HIF-1α) for lysosomal degradation. J Biol Chem. 2013;288:10703–10714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Cuervo AM, Knecht E, Terlecky SR, et al. Activation of a selective pathway of lysosomal proteolysis in rat liver by prolonged starvation. Am J Physiol. 1995;269:C1200–1208. [DOI] [PubMed] [Google Scholar]
- [43].Mijaljica D, Prescott M, Devenish RJ. Microautophagy in mammalian cells: revisiting a 40-year-old conundrum. Autophagy. 2011;7:673–682. [DOI] [PubMed] [Google Scholar]
- [44].Farré J-C SS. Peroxisome turnover by micropexophagy: an autophagy-related process. Trends Cell Biol. 2004;14:515–523. [DOI] [PubMed] [Google Scholar]
- [45].Kvam E, Goldfarb DS. Nucleus-vacuole junctions and piecemeal microautophagy of the nucleus in S. cerevisiae. Autophagy. 2007;3:85–92. [DOI] [PubMed] [Google Scholar]
- [46].Kissová I, Salin B, Schaeffer J, et al. Selective and non-selective autophagic degradation of mitochondria in yeast. Autophagy. 2007;3:329–336. [DOI] [PubMed] [Google Scholar]
- [47].Vevea JD, Garcia EJ, Chan RB, et al. Role for lipid droplet biogenesis and microlipophagy in adaptation to lipid imbalance in yeast. Dev Cell. 2015;35:584–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Mukherjee A, Patel B, Koga H, et al. Selective endosomal microautophagy is starvation-inducible in Drosophila. Autophagy. 2016;12:1984–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Sahu R, Kaushik S, Clement CC, et al. Microautophagy of cytosolic proteins by late endosomes. Dev Cell. 2011;20:131–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Uytterhoeven V, Lauwers E, Maes I, et al. Hsc70-4 deforms membranes to promote synaptic protein turnover by endosomal microautophagy. Neuron. 2015;88:735–748. [DOI] [PubMed] [Google Scholar]
- [51].Cuervo AM, Wong E. Chaperone-mediated autophagy: roles in disease and aging. Cell Res. 2014;24:92–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Klionsky DJ, Abdelmohsen K, Abe A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy. 2016;12:1–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Rajput MKS, Abdelsalam K, Darweesh MF, et al. Both cytopathic and non-cytopathic bovine viral diarrhea virus (BVDV) induced autophagy at a similar rate. Vet Immunol Immunopathol. 2017;193:1–9. [DOI] [PubMed] [Google Scholar]
- [54].Zhou YL, Ren YC, Cong YL, et al. Autophagy induced by bovine viral diarrhea virus infection counteracts apoptosis and innate immune activation. Arch Virol. 2017;162:3103–3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Fu Q, Shi HJ, Zhang H, et al. Autophagy during early stages contributes to bovine viral diarrhea virus replication in MDBK cells. J Basic Microbiol. 2014;54:1044–1052. [DOI] [PubMed] [Google Scholar]
- [56].Suda Y, Murakami S, Horimoto T. Bovine viral diarrhea virus non-structural protein NS4B induces autophagosomes in bovine kidney cells. Arch Virol. 2019;164:255–260. [DOI] [PubMed] [Google Scholar]
- [57].Yang B, Xue QH, Qi XF, et al. Autophagy enhances the replication of peste des petits ruminants virus and inhibits caspase-dependent apoptosis in vitro. Virulence. 2018;9:1176–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Lv S, Xu QY, Sun EC, et al. Autophagy activated by bluetongue virus infection plays a positive role in its replication. Viruses-Basel. 2015;7:4657–4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Yang B, Xue QH, Guo JN, et al. Autophagy induction by the pathogen receptor NECTIN4 and sustained autophagy contribute to peste des petits ruminants virus infectivity. Autophagy 2020;16:842–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Ozkaraca M, Ceribasi S, Ceribasi AO, et al. The role of apoptosis and autophagy in bovine abortions associated with Brucella spp. Acta Vet-Beogr. 2016;66:37–50. [Google Scholar]
- [61].Lopez-Perez O, Otero A, Filali H, et al. Dysregulation of autophagy in the central nervous system of sheep naturally infected with classical scrapie. Sci Rep [Internet]. 2019;9:WOS:000458571300019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Lopez-Perez O, Bolea R, Marin B, et al. Autophagy impairment in highly prion-affected brain areas of sheep experimentally infected with atypical scrapie. Vet Microbiol. 2019;233:78–84. [DOI] [PubMed] [Google Scholar]
- [63].He YJ, Mo Q, Luo B, et al. Induction of apoptosis and autophagy via mitochondria- and PI3K/Akt/mTOR-mediated pathways by E. adenophorum in hepatocytes of saanen goat. Oncotarget. 2016;7:54537–54548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Zhang YL, Han L, Yang H, et al. Bisphenol A affects cell viability involved in autophagy and apoptosis in goat testis sertoli cell. Environ Toxicol Pharmacol. 2017;55:137–147. [DOI] [PubMed] [Google Scholar]
- [65].Aboelenain M, Kawahara M, Balboula AZ, et al. Status of autophagy, lysosome activity and apoptosis during corpus luteum regression in cattle. J Reprod Dev. 2015;61:229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Toschi P, Czernik M, Zacchini F, et al. Evidence of placental autophagy during early pregnancy after transfer of in vitro produced (IVP) sheep embryos. PLoS One [Internet]. 2016;11:WOS:000378212400025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Yang DQ, Jiang TT, Liu JG, et al. CREB3 regulatory factor - mTOR-autophagy regulates goat endometrial function during early pregnancy. Biol Reprod. 2018;98:713–721. [DOI] [PubMed] [Google Scholar]
- [68].Ohtsu A, Tanaka H, Seno K, et al. Palmitic acid stimulates interleukin-8 via the TLR4/NF-kappa B/ROS pathway and induces mitochondrial dysfunction in bovine oviduct epithelial cells. Am J Reprod Immunol [Internet]. 2017;77:WOS:000403366400005. [DOI] [PubMed] [Google Scholar]
- [69].Zielniok K, Motyl T, Gajewska M. Functional Interactions between 17 beta-estradiol and progesterone regulate autophagy during acini formation by bovine mammary epithelial cells in 3D cultures. Biomed Res Int [Internet]. 2014;2014:382653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Mann S, Yepes FAL, Wakshlag JJ, et al. The effect of different treatments for early-lactation hyperketonemia on liver triglycerides, glycogen, and expression of key metabolic enzymes in dairy cattle. J Dairy Sci. 2018;101:1626–1637. [DOI] [PubMed] [Google Scholar]
- [71].Mann S, Abuelo A, Nydam DV, et al. Insulin signaling and skeletal muscle atrophy and autophagy in transition dairy cows either overfed energy or fed a controlled energy diet prepartum. J Comp Physiol B-Biochem Syst Environ Physiol. 2016;186:513–525. [DOI] [PubMed] [Google Scholar]
- [72].Hu ZY, Su HW, Li SL, et al. Effect of parenteral administration of glutamine on autophagy of liver cell and immune responses in weaned calves. J Anim Physiol Anim Nutr. 2013;97:1007–1014. [DOI] [PubMed] [Google Scholar]
- [73].Dong XS, Zhai RN, Liu ZL, et al. The effect of intravenous infusions of glutamine on duodenal cell autophagy and apoptosis in early-weaned calves. Animals [Internet]. 2019;9:WOS:000478611800028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Becila S, Hernan Herrera-Mendez C, Coulis G, et al. Postmortem muscle cells die through apoptosis. Eur Food Res Technol. 2010;231:485–493. [Google Scholar]
- [75].Sierra V, Olivan M.. Role of mitochondria on muscle cell death and meat tenderization [Internet]. 2013. [cited 2019 Oct 24]. Available from: https://www.ingentaconnect.com/content/ben/emi/2013/00000007/00000002/art00005 [DOI] [PubMed]
- [76].Garcia-Macia M, Sierra V, Palanca A, et al. Autophagy during beef aging. Autophagy. 2014;10:137–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Longo V, Lana A, Bottero MT, et al. Apoptosis in muscle-to-meat aging process: the omic witness. J Proteomics. 2015;125:29–40. [DOI] [PubMed] [Google Scholar]
- [78].Gou HC, Zhao MQ, Fan SQ, et al. Autophagy induces apoptosis and death of T lymphocytes in the spleen of pigs infected with CSFV. Sci Rep [Internet]. 2017;7:WOS:000413191500016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Sun MX, Huang L, Wang R, et al. Porcine reproductive and respiratory syndrome virus induces autophagy to promote virus replication. Autophagy. 2012;8:1434–1447. [DOI] [PubMed] [Google Scholar]
- [80].Kim SH, Min KS, Kim NH, et al. Differential expression of programmed cell death on the follicular development in normal and miniature pig ovary. PLoS One [Internet]. 2012;7:WOS:000309831500030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Xu YN, Cui XS, Sun SC, et al. Mitochondrial dysfunction influences apoptosis and autophagy in porcine parthenotes developing in vitro. J Reprod Dev. 2011;57:143–150. [DOI] [PubMed] [Google Scholar]
- [82].Dutt RH, Hamm PT. Effect of exposure to high environmental temperature and shearing on semen production of rams in winter. J Anim Sci. 1957;16:328–334. [Google Scholar]
- [83].Tompkins EC, Heidenreich CJ, Stob M. Effect of post-breeding thermal stress on embyronic mortality in swine. J Anim Sci. 1967;26:377–380. [Google Scholar]
- [84].Omtvedt IT, Nelson RE, Edwards RL, et al. Influence of heat stress during early, mid and late pregnancy of gilts. J Anim Sci. 1971;32:312–317. [DOI] [PubMed] [Google Scholar]
- [85].Ealy AD, Drost M, Hansen PJ. Developmental changes in embryonic resistance to adverse effects of maternal heat stress in cows1. J Dairy Sci. 1993;76:2899–2905. [DOI] [PubMed] [Google Scholar]
- [86].Ozawa M, Tabayashi D, Latief TA, et al. Alterations in follicular dynamics and steroidogenic abilities induced by heat stress during follicular recruitment in goats. Reproduction. 2005;129:621–630. [DOI] [PubMed] [Google Scholar]
- [87].Hale BJ, Hager CL, Seibert JT, et al. Heat stress induces autophagy in pig ovaries during follicular development. Biol Reprod. 2017;97:426–437. [DOI] [PubMed] [Google Scholar]
- [88].Zhang SJ, Li X, Li L, et al. Autophagy up-regulation by early weaning in the liver, spleen and skeletal muscle of piglets. Br J Nutr. 2011;106:213–217. [DOI] [PubMed] [Google Scholar]
- [89].Liao S, Tang S, Chang M, et al. Chloroquine downregulation of intestinal autophagy to alleviate biological stress in early-weaned piglets. Animals. 2020;10:290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Suryawan A, Davis TA. Regulation of protein degradation pathways by amino acids and insulin in skeletal muscle of neonatal pigs. J Anim Sci Biotechnol [Internet]. 2014;5:WOS:000333189600001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Ballester M, Amills M, Gonzalez-Rodriguez O, et al. Role of AMPK signalling pathway during compensatory growth in pigs. BMC Genomics [Internet]. 2018;19:WOS:000444902700006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Rubio-Gonzalez A, Potes Y, Illan-Rodriguez D, et al. Effect of animal mixing as a stressor on biomarkers of autophagy and oxidative stress during pig muscle maturation. Animal. 2015;9:1188–1194. [DOI] [PubMed] [Google Scholar]
- [93].Niu YJ, Sun QQ, Zhang GH, et al. Fowl adenovirus serotype 4-induced apoptosis, autophagy, and a severe inflammatory response in liver. Vet Microbiol. 2018;223:34–41. [DOI] [PubMed] [Google Scholar]
- [94].Wang XP, Qi XF, Yang B, et al. Autophagy benefits the replication of egg drop syndrome virus in duck embryo fibroblasts. Front Microbiol [Internet]. 2018;9:WOS:000433323400002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Liu HX, Cao WS, Li YH, et al. Subgroup J avian leukosis virus infection inhibits autophagy in DF-1 cells. Virol J [Internet]. 2013;10:WOS:000322130300001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Dai ZK, Huang JF, Lei XY, et al. Efficacy of an autophagy-targeted DNA vaccine against avian leukosis virus subgroup. Vaccine. 2017;35:808–813. [DOI] [PubMed] [Google Scholar]
- [97].Chen GH, Li ZT, Su S, et al. Identification of key genes fluctuated induced by avian leukemia virus (ALV-J) infection in chicken cells. In Vitro Cell Dev Biol-Anim. 2018;54:41–51. [DOI] [PubMed] [Google Scholar]
- [98].Li CX, Wei HC, Yu LP, et al. Nuclear localization of the p17 protein of avian reovirus is correlated with autophagy induction and an increase in viral replication. Arch Virol. 2015;160:3001–3010. [DOI] [PubMed] [Google Scholar]
- [99].Chi PI, Huang WR, Lai IH, et al. The p17 nonstructural protein of avian reovirus triggers autophagy enhancing virus replication via activation of phosphatase and tensin deleted on chromosome 10 (PTEN) and AMP-activated protein kinase (AMPK), as well as dsRNA-dependent protein kinase (PKR)/eIF2 alpha signaling pathways. J Biol Chem. 2013;288:3571–3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Meng SS, Jiang K, Zhang XR, et al. Avian reovirus triggers autophagy in primary chicken fibroblast cells and Vero cells to promote virus production. Arch Virol. 2012;157:661–668. [DOI] [PubMed] [Google Scholar]
- [101].Lin P-Y, Chang C-D, Chen Y-C, et al. RhoA/ROCK1 regulates avian reovirus S1133-induced switch from autophagy to apoptosis. BMC Vet Res [Internet]. 2015;11(1):WOS:000354349200001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Duan SP, Cheng JH, Li CX, et al. Autophagy inhibitors reduce avian-reovirus-mediated apoptosis in cultured cells and in chicken embryos. Arch Virol. 2015;160:1679–1685. [DOI] [PubMed] [Google Scholar]
- [103].Niu XS, Wang YY, Li M, et al. Transcriptome analysis of avian reovirusmediated changes in gene expression of normal chicken fibroblast DF-1 cells. BMC Genomics [Internet]. 2017;18:WOS:000416131200009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Niu XS, Zhang CC, Wang YY, et al. Autophagy induced by avian reovirus enhances viral replication in chickens at the early stage of infection. BMC Vet Res [Internet]. 2019;15:WOS:000468924200005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Wu YJ, Cui LP, Zhu EP, et al. Muscovy duck reovirus sigma NS protein triggers autophagy enhancing virus replication. Virol J [Internet]. 2017;14:WOS:000396761100002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Kang YF, Yuan RY, Xiang B, et al. Newcastle disease virus-induced autophagy mediates antiapoptotic signaling responses in vitro and in vivo. Oncotarget. 2017;8:73981–73993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].YJ S, Yu SQ, Ding N, et al. Autophagy benefits the replication of Newcastle disease virus in chicken cells and tissues. J Virol. 2014;88:525–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Hou L, Wei L, Zhu SS, et al. Avian metapneumovirus subgroup C induces autophagy through the ATF6 UPR pathway. Autophagy. 2017;13:1709–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Maier HJ, Cottam EM, Stevenson-Leggett P, et al. Visualizing the autophagy pathway in avian cells and its application to studying infectious bronchitis virus. Autophagy. 2013;9:496–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Chen BY, Itakura C. Cytopathology of chick renal epithelial cells experimentally infected with avian infectious bronchitis virus. Avian Pathol. 1996;25:675–690. [DOI] [PubMed] [Google Scholar]
- [111].Yin HC, Zhao LL, Jiang XJ, et al. DEV induce autophagy via the endoplasmic reticulum stress related unfolded protein response. PLoS One [Internet]. 2017;12:WOS:000418651500010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Yin HC, Zhao LL, Li SQ, et al. Autophagy activated by duck enteritis virus infection positively affects its replication. J Gen Virol. 2017;98:486–495. [DOI] [PubMed] [Google Scholar]
- [113].Yin HC, Zhao LL, Li SQ, et al. Impaired cellular energy metabolism contributes to duck-enteritis-virus-induced autophagy via the AMPK-TSC2-MTOR signaling pathway. Front Cell Infect Microbiol [Internet]. 2017;7:WOS:000411721200001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Yin HC, Zhao LL, Wang YP, et al. Duck enteritis virus activates CaMKK beta-AMPK to trigger autophagy in duck embryo fibroblast cells via increased cytosolic calcium. Virol J [Internet]. 2018;15:WOS:000441121300001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Ming K, Yuan WJ, Chen Y, et al. PI3KC3-dependent autophagosomes formation pathway is of crucial importance to anti-DHAV activity of Chrysanthemum indicum polysaccharide. Carbohydr Polym. 2019;208:22–31. [DOI] [PubMed] [Google Scholar]
- [116].Pleet ML, Branscome H, DeMarino C, et al. Autophagy, EVs, and Infections: a perfect question for a perfect time. Front Cell Infect Microbiol. 2018;8:362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Viret C, Rozieres A, Faure M. Autophagy during early virus-host cell interactions. J Mol Biol. 2018;430:1696–1713. [DOI] [PubMed] [Google Scholar]
- [118].Jackson WT. Viruses and the autophagy pathway. Virology. 2015;479:450–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Wang Y, Zhao HJ, Shao YZ, et al. Copper or/and arsenic induces autophagy by oxidative stress-related PI3K/AKT/mTOR pathways and cascaded mitochondrial fission in chicken skeletal muscle. J Inorg Biochem. 2018;188:1–8. [DOI] [PubMed] [Google Scholar]
- [120].Chen MH, Li XJ, Fan RF, et al. Cadmium induces BNIP3-dependent autophagy in chicken spleen by modulating miR-33-AMPK axis. Chemosphere. 2018;194:396–402. [DOI] [PubMed] [Google Scholar]
- [121].Wang Y, Zhao HJ, Shao YZ, et al. Copper (II) and/or arsenite-induced oxidative stress cascades apoptosis and autophagy in the skeletal muscles of chicken. Chemosphere. 2018;206:597–605. [DOI] [PubMed] [Google Scholar]
- [122].Gou ZY, Li L, Fan QL, et al. Effects of oxidative stress induced by high dosage of dietary iron ingested on intestinal damage and caecal microbiota in Chinese yellow broilers. J Anim Physiol Anim Nutr. 2018;102:924–932. [DOI] [PubMed] [Google Scholar]
- [123].Fei DX, Zhao HJ, Wang Y, et al. The disturbance of autophagy and apoptosis in the gizzard caused by copper and/or arsenic are related to mitochondrial kinetics. Chemosphere. 2019;231:1–9. [DOI] [PubMed] [Google Scholar]
- [124].Li SW, Zhao HJ, Wang Y, et al. Regulation of autophagy factors by oxidative stress and cardiac enzymes imbalance during arsenic or/and copper induced cardiotoxicity in Gallus gallus. Ecotox Environ Safe. 2018;148:125–134. [DOI] [PubMed] [Google Scholar]
- [125].Sun X, Li JL, Zhao HJ, et al. Synergistic effect of copper and arsenic upon oxidative stress, inflammation and autophagy alterations in brain tissues of Gallus gallus. J Inorg Biochem. 2018;178:54–62. [DOI] [PubMed] [Google Scholar]
- [126].Wang WZ, Zhang T, Lin HJ, et al. Role of hydrogen sulfide on autophagy in liver injuries induced by selenium deficiency in chickens. Biol Trace Elem Res. 2017;175:194–203. [DOI] [PubMed] [Google Scholar]
- [127].Wang XH, Li W, Han MY, et al. Water-soluble substances of wheat: a potential preventer of aflatoxin B1-induced liver damage in broilers. Poult Sci. 2019;98:136–149. [DOI] [PubMed] [Google Scholar]
- [128].Liu CP, Fu J, Liu C, et al. The role of nitric oxide and autophagy in liver injuries induced by selenium deficiency in chickens. RSC Adv. 2015;5:50549–50556. [Google Scholar]
- [129].Shi QX, Jin X, Fan RF, et al. Cadmium-mediated miR-30a-GRP78 leads to JNK-dependent autophagy in chicken kidney. Chemosphere. 2019;215:710–715. [DOI] [PubMed] [Google Scholar]
- [130].Huang H, Wang Y, An Y, et al. Selenium alleviates oxidative stress and autophagy in lead-treated chicken testes. Theriogenology. 2019;131:146–152. [DOI] [PubMed] [Google Scholar]
- [131].Khoso PA, Pan TR, Wan N, et al. Selenium deficiency induces autophagy in immune organs of chickens. Biol Trace Elem Res. 2017;177:159–168. [DOI] [PubMed] [Google Scholar]
- [132].Huang HB, Xiao K, Lu S, et al. Increased thymic cell turnover under boron stress may bypass TLR3/4 pathway in African ostrich. PLoS One [Internet]. 2015;10:WOS:000355955300150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Liu JJ, Zhao HJ, Wang Y, et al. Arsenic trioxide and/or copper sulfate induced apoptosis and autophagy associated with oxidative stress and perturbation of mitochondrial dynamics in the thymus of Gallus gallus. Chemosphere. 2019;219:227–235. [DOI] [PubMed] [Google Scholar]
- [134].Zhang C, Lin J, Ge J, et al. Selenium triggers Nrf2-mediated protection against cadmium-induced chicken hepatocyte autophagy and apoptosis. Toxicol Vitro. 2017;44:349–356. [DOI] [PubMed] [Google Scholar]
- [135].Chen BL, Li DY, Li M, et al. Induction of mitochondria-mediated apoptosis and PI3K/Akt/mTOR-mediated autophagy by aflatoxin B-2 in hepatocytes of broilers. Oncotarget. 2016;7:84989–84998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Xie WQ, Ge M, Li GX, et al. Astragalus polysaccharide protect against cadmium-induced cytotoxicity through the MDA5/NF-kappa B pathway in chicken peripheral blood lymphocytes. Molecules [Internet]. 2017;22:WOS:000414670600038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Gunawardana VK. Lysosomes in the seminiferous tubules of the domestic-fowl as revealed by acid-phosphatase-activITY. Acta Histochem Cytochem. 1992;25:91–96. [Google Scholar]
- [138].Hwang YS, Ko MH, Kim YM, et al. The avian-specific small heat shock protein HSP25 is a constitutive protector against environmental stresses during blastoderm dormancy. Sci Rep [Internet]. 2016;6:WOS:000387541100001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Mpango MM, Madekurozwa MC. Comparative histomorphological and ultrastructural study of the luminal epithelium of the isthmus in laying and moulting domestic fowls (Gallus domesticus). Anat Histol Embryol. 2018;47:444–455. [DOI] [PubMed] [Google Scholar]
- [140].Madekurozwa MCN, Mpango MM. Ultrastructure of the tubular glands in the isthmus region of the oviduct in laying and natural moulting commercial egg-type chickens. Anat Histol Embryol. 2018;47:493–497. [DOI] [PubMed] [Google Scholar]
- [141].Dherde K, DePrest B, Roels F. Subtypes of active cell death in the granulosa of ovarian atretic follicles in the quail (Coturnix coturnix japonica). Reprod Nutr Dev. 1996;36:175–189. [DOI] [PubMed] [Google Scholar]
- [142].Yu J, Lou YP, He K, et al. Goose broodiness is involved in granulosa cell autophagy and homeostatic imbalance of follicular hormones. Poult Sci. 2016;95:1156–1164. [DOI] [PubMed] [Google Scholar]
- [143].Yu J, Lou YP, Zhao A. Transcriptome analysis of follicles reveals the importance of autophagy and hormones in regulating broodiness of Zhedong white goose. Sci Rep [Internet]. 2016;6:WOS:000387477300001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Lou YP, Yu WS, Han L, et al. ROS activates autophagy in follicular granulosa cells via mTOR pathway to regulate broodiness in goose. Anim Reprod Sci. 2017;185:97–103. [DOI] [PubMed] [Google Scholar]
- [145].Lin X, Liu XT, Ma YF, et al. Coherent apoptotic and autophagic activities involved in regression of chicken postovulatory follicles. Aging-US. 2018;10:819–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Yamauchi K, Kamisoyama H, Isshiki Y. Effects of fasting and refeeding on structures of the intestinal villi and epithelial cells in white leghorn hens. Br Poult Sci. 1996;37:909–921. [DOI] [PubMed] [Google Scholar]
- [147].Yamauchi K, Tarachai P. Changes in intestinal villi, cell area and intracellular autophagic vacuoles related to intestinal function in chickens. Br Poult Sci. 2000;41:416–423. [DOI] [PubMed] [Google Scholar]
- [148].Saneyasu T, Kimura S, Inui M, et al. Differences in the expression of genes involved in skeletal muscle proteolysis between broiler and layer chicks during food deprivation. Comp Biochem Physiol B-Biochem Mol Biol. 2015;186:36–42. [DOI] [PubMed] [Google Scholar]
- [149].Maynard S, Ghosh R, Wu Y, et al. GABARAP is a determinant of apoptosis in growth-arrested chicken embryo fibroblasts. J Cell Physiol. 2015;230:1475–1488. [DOI] [PubMed] [Google Scholar]
- [150].Piekarski A, Khaldi S, Greene E, et al. Tissue distribution, gender-and genotype-dependent expression of autophagy-related genes in avian species. PLoS One [Internet]. 2014;9:WOS:000345250400063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Zeitz JO, Mohrmann S, Kading SC, et al. Effects of methionine on muscle protein synthesis and degradation pathways in broilers. J Anim Physiol Anim Nutr. 2019;103:191–203. [DOI] [PubMed] [Google Scholar]
- [152].Bottje WG, Lassiter K, Piekarski-Welsher A, et al. Proteogenomics reveals enriched ribosome assembly and protein translation in pectoralis major of high feed efficiency pedigree broiler males. Front Physiol [Internet]. 2017;8:WOS:000403252900002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Bottje WG. BOARD INVITED REVIEW: oxidative stress and efficiency: the tightrope act of mitochondria in health and disease. J Anim Sci. 2019;97:3169–3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Piekarski-Welsher A, Greene E, Lassiter K, et al. Enrichment of autophagy and proteosome pathways in breast muscle of feed efficient pedigree male broilers. Front Physiol. 2018;9:1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Shin J, McFarland DC, Strasburg GM, et al. Function of death-associated protein 1 in proliferation, differentiation, and apoptosis of chicken satellite cells. Muscle Nerve. 2013;48:777–790. [DOI] [PubMed] [Google Scholar]
- [156].Possidonio ACB, Miranda M, Gregoracci GB, et al. Cholesterol depletion induces transcriptional changes during skeletal muscle differentiation. BMC Genomics [Internet]. 2014;15:WOS:000338593300001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Zhang JL, Huang SC, Tong XL, et al. Chlorogenic acid alleviates thiram-induced tibial dyschondroplasia by modulating caspases, BECN1 expression and ECM degradation. Int J Mol Sci [Internet]. 2019;20:WOS:000477041100048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Chen XX, Zhang L, Li JL, et al. Hydrogen peroxide-induced change in meat quality of the breast muscle of broilers is mediated by ROS generation, apoptosis, and autophagy in the NF-kappa B signal pathway. J Agric Food Chem. 2017;65:3986–3994. [DOI] [PubMed] [Google Scholar]
- [159].Piekarski A, Nagarajan G, Ishola P, et al. AMP-activated protein kinase mediates the effect of leptin on avian autophagy in a tissue-specific manner. Front Physiol [Internet]. 2018;9:WOS:000432408700002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Hu Y, Sun QW, Hu Y, et al. Corticosterone-induced lipogenesis activation and lipophagy inhibition in chicken liver are alleviated by maternal betaine supplementation. J Nutr. 2018;148:316–325. [DOI] [PubMed] [Google Scholar]
- [161].Fodor E, Sigmond T, Ari E, et al. Methods to study autophagy in zebrafish. Meth Enzymol. 2017;588:467–496. [DOI] [PubMed] [Google Scholar]
- [162].Wager K, Russell C. Mitophagy and neurodegeneration: the zebrafish model system. Autophagy. 2013;9:1693–1709. [DOI] [PubMed] [Google Scholar]
- [163].Fleming A, Rubinsztein DC. Zebrafish as a model to understand autophagy and its role in neurological disease. Biochim Biophys Acta. 2011;1812:520–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Mathai BJ, Meijer AH, Simonsen A. Studying autophagy in zebrafish. Cells. 2017;6:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Moore MN, Viarengo A, Donkin P, et al. Autophagic and lysosomal reactions to stress in the hepatopancreas of blue mussels. Aquat Toxicol. 2007;84:80–91. [DOI] [PubMed] [Google Scholar]
- [166].Kirchin M, Moore M, Dean R, et al. The role of oxyradicals in intracellular proteolysis and toxicity in mussels. Mar Environ Res. 1992;34:315–320. [Google Scholar]
- [167].Moore MN. Diet restriction induced autophagy: a lysosomal. protective system against oxidative- and pollutant-stress and cell injury. Mar Environ Res. 2004;58:603–607. [DOI] [PubMed] [Google Scholar]
- [168].Moore MN, Allen JI, McVeigh A. Environmental prognostics: an integrated model supporting lysosomal stress responses as predictive biomarkers of animal health status. Mar Environ Res. 2006;61:278–304. [DOI] [PubMed] [Google Scholar]
- [169].Moore MN, Allen JI, McVeigh A, et al. Lysosomal and autophagic reactions as predictive indicators of environmental impact in aquatic animals. Autophagy. 2006;2:217–220. [DOI] [PubMed] [Google Scholar]
- [170].Moore MN. Autophagy as a second level protective process in conferring resistance to environmentally-induced oxidative stress. Autophagy. 2008;4:254–256. [DOI] [PubMed] [Google Scholar]
- [171].Moore M, Lowe D, Fieth P. Responses of lysosomes in digestive cells of common mussel, mytilus-edulis, to sex steroids and cortisol. Cell Tissue Res. 1978;188:1–9. [DOI] [PubMed] [Google Scholar]
- [172].Viarengo A, Moore M, Pertica M, et al. A simple procedure for evaluating the protein-degradation rate in mussel (mytilus-Galloprovincialis-Lam) tissues and its application in a study. Comp Biochem Physiol B-Biochem Mol Biol. 1992;103:27–32. [Google Scholar]
- [173].Marigomez I, Baybay-Villacorta L. Pollutant-specific and general lysosomal responses in digestive cells of mussels exposed to model organic chemicals. Aquat Toxicol. 2003;64:235–257. [DOI] [PubMed] [Google Scholar]
- [174].Allen JI, Moore MN. Environmental prognostics: is the current use of biomarkers appropriate for environmental risk evaluation?. Mar Environ Res. 2004;58:227–232. [DOI] [PubMed] [Google Scholar]
- [175].Broeg K, Zander S, Diamant A, et al. The use of fish metabolic, pathological and parasitological indices in pollution monitoring - 1. North Sea Helgoland Mar Res. 1999;53:171–194. [Google Scholar]
- [176].Broeg K, Kohler A, Von Westernhagen H. Disorder and recovery of environmental health monitored by means of lysosomal stability in liver of European flounder (Platichthys flesus L.). Mar Environ Res. 2002;54:569–573. [DOI] [PubMed] [Google Scholar]
- [177].Kagley AN, Snider RG, Krishnakumar PK, et al. Assessment of seasonal variability of cytochemical responses to contaminant exposure in the blue mussel Mytilus edulis (complex). Arch Environ Contam Toxicol. 2003;44:43–52. [DOI] [PubMed] [Google Scholar]
- [178].Marigomez J, Soto M, Angulo E. Responses of winkles digestive cells and their lysosomal system to environmental salinity changes. Cell Mol Biol. 1991;37:29–39. [PubMed] [Google Scholar]
- [179].Tremblay R, PellerinMassicotte J. Effect of the tidal cycle on lysosomal membrane stability in the digestive gland of Mya arenaria and Mytilus edulis L. Comp Biochem Physiol A-Physiol. 1997;117:99–104. [Google Scholar]
- [180].Abele D, Burlando B, Viarengo A, et al. Exposure to elevated temperatures and hydrogen peroxide elicits oxidative stress and antioxidant response in the Antarctic intertidal limpet Nacella concinna. Comp Biochem Physiol B-Biochem Mol Biol. 1998;120:425–435. [Google Scholar]
- [181].McVeigh A, Moore M, Allen JI, et al. Lysosomal responses to nutritional and contaminant stress in mussel hepatopancreatic digestive cells: A modelling study. Mar Environ Res. 2006;62:S433–S438. [DOI] [PubMed] [Google Scholar]
- [182].Bignell JP, Dodge MJ, Feist SW, et al. Mussel histopathology: and effects of season, disease and species. Aquat Biol. 2008;2:1–15. [Google Scholar]
- [183].Brenner M, Broeg K, Frickenhaus S, et al. Multi-biomarker approach using the blue mussel (Mytilus edulis L.) to assess the quality of marine environments: season and habitat-related impacts. Mar Environ Res. 2014;95:13–27. [DOI] [PubMed] [Google Scholar]
- [184].Koukouzika N, Raftopoulou EK, Dimitriadis VK. Seasonal differences of lysosomal, lipid and lipofuscin parameters in the digestive gland of the mussel Mytilus galloprovincialis. J Molluscan Stud. 2009;75:261–267. [Google Scholar]
- [185].Brenner M, Broeg K, Wilhelm C, et al. Effect of air exposure on lysosomal tissues of Mytilus edulis L. from natural intertidal wild beds and submerged culture ropes. Comp Biochem Physiol A-Mol Integr Physiol. 2012;161:327–336. [DOI] [PubMed] [Google Scholar]
- [186].Lysenko L, Sukhovskaya I, Borvinskaya E, et al. Detoxification and protein quality control markers in the mussel Mytilus edulis (Linnaeus) exposed to crude oil: salinity-induced modulation. Estuar Coast Shelf Sci. 2015;167:220–227. [Google Scholar]
- [187].Sforzini S, Moore MN, Oliveri C, et al. Role of mTOR in autophagic and lysosomal reactions to environmental stressors in molluscs. Aquat Toxicol. 2018;195:114–128. [DOI] [PubMed] [Google Scholar]
- [188].Moore MN. Is toxicological pathology characterised by a loss of system complexity? Marine Environ Res. 2010;69:S37–S41. [DOI] [PubMed] [Google Scholar]
- [189].Moore MN, Shaw JP, Adams DRF, et al. Anti-oxidative cellular protection effect of fasting-induced autophagy as a mechanism for hormesis. Mar Environ Res. 2015;107:35–44. [DOI] [PubMed] [Google Scholar]
- [190].Thorgaard GH, Bailey GS, Williams D, et al. Status and opportunities for genomics research with rainbow trout. Comp Biochem Physiol B-Biochem Mol Biol. 2002;133:609–646. [DOI] [PubMed] [Google Scholar]
- [191].Braunbeck T, Storch V. Senescence of hepatocytes isolated from rainbow-trout (Oncorhynchus-mykiss) in primary culture - an ultrastructural-study. Protoplasma. 1992;170:138–159. [Google Scholar]
- [192].Seiliez I, Gutierrez J, Salmeron C, et al. An in vivo and in vitro assessment of autophagy-related gene expression in muscle of rainbow trout (Oncorhynchus mykiss). Comp Biochem Physiol B-Biochem Mol Biol. 2010;157:258–266. [DOI] [PubMed] [Google Scholar]
- [193].Seiliez I, Panserat S, Skiba-Cassy S, et al. Effect of acute and chronic insulin administrations on major factors involved in the control of muscle protein turnover in rainbow trout (Oncorhynchus mykiss). Gen Comp Endocrinol. 2011;172:363–370. [DOI] [PubMed] [Google Scholar]
- [194].Seiliez I, Gabillard JC, Riflade M, et al. Amino acids downregulate the expression of several autophagy-related genes in rainbow trout myoblasts. Autophagy. 2012;8:364–375. [DOI] [PubMed] [Google Scholar]
- [195].Cleveland BM, Weber GM. Effects of sex steroids on indices of protein turnover in rainbow trout (Oncorhynchus mykiss) white muscle. Gen Comp Endocrinol. 2011;174:132–142. [DOI] [PubMed] [Google Scholar]
- [196].Cleveland BM, Kenney PB, Manor ML, et al. Effects of feeding level and sexual maturation on carcass and fillet characteristics and indices of protein degradation in rainbow trout (Oncorhynchus mykiss). Aquaculture. 2012;338:228–236. [Google Scholar]
- [197].Cleveland BM, Weber GM. Effects of triploidy on growth and protein degradation in skeletal muscle during recovery from feed deprivation in juvenile rainbow trout (Oncorhynchus mykiss). Comp Biochem Physiol A-Mol Integr Physiol. 2013;166:128–137. [DOI] [PubMed] [Google Scholar]
- [198].Cleveland BM. In vitro and in vivo effects of phytoestrogens on protein turnover in rainbow trout (Oncorhynchus mykiss) white muscle. Comp Biochem Physiol C-Toxicol Pharmacol. 2014;165:9–16. [DOI] [PubMed] [Google Scholar]
- [199].Balmori-Cedeno J, Liu JT, Misk E, et al. Autophagy-related genes in rainbow trout Oncorhynchus mykiss (Walbaum) gill epithelial cells and their role in nutrient restriction. J Fish Dis. 2019;42:549–558. [DOI] [PubMed] [Google Scholar]
- [200].Paneru B, Ali A, Al-Tobasei R, et al. Crosstalk among lncRNAs, microRNAs and mRNAs in the muscle “degradome” of rainbow trout. Sci Rep [Internet]. 2018;8:WOS:000433539800014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Lescat L, Herpin A, Mourot B, et al. CMA restricted to mammals and birds: myth or reality? Autophagy. 2018;14:1267–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Seite S, Masagounder K, Heraud C, et al. Early feeding of rainbow trout (Oncorhynchus mykiss) with methionine-deficient diet over a 2 week period: consequences for liver mitochondria in juveniles. J Exp Biol [Internet]. 2019;222:WOS:000488952100006. [DOI] [PubMed] [Google Scholar]
- [203].Seiliez I, Gabillard J-C, Skiba-Cassy S, et al. An in vivo and in vitro assessment of TOR signaling cascade in rainbow trout (Oncorhynchus mykiss). Am J Physiol Regul Integr Comp Physiol. 2008;295:R329–R335. [DOI] [PubMed] [Google Scholar]
- [204].Seite S, Mourier A, Camougrand N, et al. Dietary methionine deficiency affects oxidative status, mitochondrial integrity and mitophagy in the liver of rainbow trout (Oncorhynchus mykiss). Sci Rep [Internet]. 2018;8:WOS:000437413200007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Belghit I, Panserat S, Sadoul B, et al. Macronutrient composition of the diet affects the feeding-mediated down regulation of autophagy in muscle of rainbow trout (O. mykiss). PLoS One [Internet]. 2013;8:WOS:000326240100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Belghit I, Skiba-Cassy S, Geurden I, et al. Dietary methionine availability affects the main factors involved in muscle protein turnover in rainbow trout (Oncorhynchus mykiss). Br J Nutr. 2014;112:493–503. [DOI] [PubMed] [Google Scholar]
- [207].Nombela I, Requena-Platek R, Morales-Lange B, et al. Rainbow trout red blood cells exposed to viral hemorrhagic septicemia virus up-regulate antigen-processing mechanisms and MHC I&II, CD86, and CD83 antigen-presenting cell markers. Cells [Internet]. 2019;8:WOS:000470994400006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Huang TQ, Sun HZ, Wang YN, et al. Effect of follicle cell autophagy on gonadal development of triploid female rainbow trout (Oncorhynchus mykiss). Fish Physiol Biochem. 2018;44:185–196. [DOI] [PubMed] [Google Scholar]
- [209].Seiliez I, Dias K, Cleveland BM. Contribution of the autophagy-lysosomal and ubiquitin-proteasomal proteolytic systems to total proteolysis in rainbow trout (Oncorhynchus mykiss) myotubes. Am J Physiol Regul Integr Comp Physiol. 2014;307:R1330–R1337. [DOI] [PubMed] [Google Scholar]
- [210].Seiliez I, Belghit I, Gao YJ, et al. Looking at the metabolic consequences of the colchicine-based in vivo autophagic flux assay. Autophagy. 2016;12:343–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Seite S, Pioche T, Ory N, et al. The autophagic flux inhibitor bafilomycine A1 affects the expression of intermediary metabolism-related genes in trout hepatocytes. Front Physiol [Internet]. 2019;10:WOS:000461731500001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Li C, White SH, Warren LK, et al. Skeletal muscle from aged American quarter horses shows impairments in mitochondrial biogenesis and expression of autophagy markers. Exp Gerontol. 2018;102:19–27. [DOI] [PubMed] [Google Scholar]
- [213].Marycz K, Kornicka K, Marezdziak M, et al. Equine metabolic syndrome impairs adipose stem cells osteogenic differentiation by predominance of autophagy over selective mitophagy. J Cell Mol Med. 2016;20:2384–2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Bourebaba L, Bedjou F, Rocken M, et al. Nortropane alkaloids as pharmacological chaperones in the rescue of equine adipose-derived mesenchymal stromal stem cells affected by metabolic syndrome through mitochondrial potentiation, endoplasmic reticulum stress mitigation and insulin resistance alleviation. Stem Cell Res Ther [Internet]. 2019;10:WOS:000472113100003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Kornicka K, Al Naem M, Rocken M, et al. Osteochondritis dissecans (OCD)-derived chondrocytes display increased senescence, oxidative stress, chaperone-mediated autophagy and, in co-culture with adipose-derived stem cells (ASCs), enhanced expression of MMP-13. J Clin Med [Internet]. 2019;8:WOS:000464442900002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Gallardo Bolanos JM, Miro Moran A, Balao da Silva CM, et al. During cooled storage the extender influences processed autophagy marker light chain 3 (LC3B) of stallion spermatozoa. Anim Reprod Sci. 2014;145:40–46. [DOI] [PubMed] [Google Scholar]
- [217].Aparicio IM, Munoz PM, Salido GM, et al. The autophagy-related protein LC3 is processed in stallion spermatozoa during short-and long-term storage and the related stressful conditions. Animal. 2016;10:1182–1191. [DOI] [PubMed] [Google Scholar]
- [218].Ferreira PG, Costa-e-Silva A, Monteiro E, et al. Liver enzymes and ultrastructure in rabbit haemorrhagic disease (RHD). Vet Res Commun. 2006;30:393–401. [DOI] [PubMed] [Google Scholar]
- [219].San-Miguel B, Crespo I, Vallejo D, et al. Melatonin modulates the autophagic response in acute liver failure induced by the rabbit hemorrhagic disease virus. J Pineal Res. 2014;56:313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [220].Guo MJ, Wu FH, Zhang ZF, et al. Characterization of rabbit nucleotide-binding oligomerization domain 1 (NOD1) and the role of NOD1 signaling pathway during bacterial infection. Front Immunol [Internet]. 2017;8:WOS:000412636300001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [221].Djehiche B, Segalen J, Chambon Y. Ultrastructure of mullerian and wolffian ducts of fetal rabbit in-vivo and in organ-culture. Tissue Cell. 1994;26:323–332. [DOI] [PubMed] [Google Scholar]
- [222].Djehiche B, Segalen J, Chambon Y. Inhibition of autophagy of fetal rabbit gonoducts by puromycin, tunicamycin and chloroquin in organ culture. Tissue Cell. 1996;28:115–121. [DOI] [PubMed] [Google Scholar]
- [223].Bozzatto V, De Oliveira PR, Furquim KCS, et al. The occurrence of autophagic cell death in the tegument of rabbits pre-infested with rhipicephalus sanguineus and exposed to selamectin (Active principle of Acaricide pfizer revolution (R)). Microsc Res Tech. 2013;76:1171–1176. [DOI] [PubMed] [Google Scholar]
- [224].Till A, Saito R, Merkurjev D, et al. Evolutionary trends and functional anatomy of the human expanded autophagy network. Autophagy. 2015;11:1652–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


