Abstract
Bacteriophages are an invaluable source of novel genetic diversity. Sequencing of phage genomes can reveal new proteins with potential uses as biotechnological and medical tools, and help unravel the diversity of biological mechanisms employed by phages to take over the host during viral infection. Aiming to expand the available collection of phage genomes, we have isolated, sequenced, and assembled the genome sequences of four phages that infect the clinical pathogen Klebsiella pneumoniae: vB_KpnP_FBKp16, vB_KpnP_FBKp27, vB_KpnM_FBKp34, and Jumbo phage vB_KpnM_FBKp24. The four phages show very low (0–13%) identity to genomic phage sequences deposited in the GenBank database. Three of the four phages encode tRNAs and have a GC content very dissimilar to that of the host. Importantly, the genome sequences of the phages reveal potentially novel DNA packaging mechanisms as well as distinct clades of tubulin spindle and nucleus shell proteins that some phages use to compartmentalize viral replication. Overall, this study contributes to uncovering previously unknown virus diversity, and provides novel candidates for phage therapy applications against antibiotic-resistant K. pneumoniae infections.
Keywords: bacteriophage, Jumbo phage, comparative genomics, phage therapy
1. Introduction
Bacteriophages or phages are ubiquitous viruses of prokaryotes that exert an enormous influence over the microbial biosphere, playing a critical role in the nutrient and energy cycles,1–3 in the evolution of bacterial pathogens,4 and in shaping gut microbial communities.5 Phages have also contributed immensely to the field of molecular biology, having been at the core of the discovery of central features such as DNA as the genetic material,6 the triplet genetic code,7 messenger RNA,8 restriction enzymes,9 and recombinant DNA.10 Phages and their interactions with prokaryotic hosts led also to the evolution of CRISPR-Cas and development of programmable genome editing tools, one of the most revolutionary tools in biology that enables tailored engineering of genomic sequences in a range of species including humans.11 There is also a rekindled interest in the therapeutic use of phages—phage therapy—to control bacterial pathogens, as a consequence of the alarming rise of antibiotic-resistant infections observed in recent years.12–14 The study of phages and their genomes is therefore inherently valuable to advance our understanding in a diversity of fields including molecular biology, ecology, evolution, bacterial pathogenesis, biotechnology, and health. Understanding phage genomes will certainly create opportunities to translate novel phage proteins and phages themselves into potent biotechnological15 and medical tools.16 Here we isolated and sequenced the genomes of four novel phages infecting Klebsiella pneumoniae, an increasingly relevant pathogen identified by the World Health Organization as priority for the development of new antibiotics.17 These phages have little to no sequence similarity to known phages, but a series of genomics and phylogenetic analysis revealed interesting features that could aid the expansion of our understanding of the hidden genetic treasures in phage biology.
2. Materials and methods
2.1. Bacteriophage isolation
Four clinical isolates of K. pneumoniae isolated at the University Medical Centre Utrecht (UMCU) were used for phage isolation: K6310 (blood culture from a 77-year-old patient with obstructive cholangitis due to disseminated pancreatic carcinoma), K6592 (infected total hip prosthesis from a 74-year-old patient), L923 (blood culture from a 67-year-old kidney transplant patient with an urinary tract infection and sepsis), and K6453 (cerebrospinal fluid taken post-mortem from a healthy 57-year-old woman with unexplained sudden out of hospital cardiac arrest). As phage source, approximately 5 l of sewage water were sequentially filtered with coffee filters, membrane filters (0.45 and 0.2 µm PES), and 10× concentrated using a tangential flow cassette (100 kDa PES Vivaflow 200, Sartorius, Germany). Approximately 5 ml of the concentrated virome were added to 20 ml of Lysogeny Broth (LB), inoculated with 100 µl of each of the overnight grown K. pneumoniae strains, and incubated overnight at 37°C with rocking. Samples were centrifuged at 16,000 × g for 5 min and filter-sterilized (0.2 µm PES). The phage-containing supernatant was serially diluted in SM buffer (100 mM NaCl, 8 mM MgSO4, 50 mM Tris–HCl pH 7.5) and spotted on double layer agar (DLA) plates of the isolation strains for the detection of phages. Single plaques with distinct morphologies were picked with sterile toothpicks and spread with sterile paper strips into fresh bacterial lawns. The procedure was repeated as needed to obtain a consistent plaque morphology. Average plaque size was determined by measurement of the diameter of 10 individual plaques using ImageJ software.18 Phages from purified plaques were then produced in liquid media with their respective host, centrifuged, filter-sterilized, and stored as phage lysates (>108 pfu/ml) at 4°C, and at −80°C with 50% (v/v) glycerol.
2.2. Transmission electron microscopy
One ml of each phage lysate at >109 pfu/ml was sedimented at 21,000 × g for 1 h, washed, and re-suspended in 1 ml of MilliQ water. Phages (3.5 µl) were deposited and incubated for 1 min on transmission electron microscopy (TEM) grids (Carbon Type-B 400 mesh, TED PELLA). Grids were washed thrice with 40 µl of MilliQ water and stained with 3.5 µl of 2% (w/v) uranyl acetate (pH 4.0) for 30 s. Grids were examined using a JEM-1400 plus (JEOL) TEM. The capsid diameter and the tail length and width of 10 phage particles were measured using EMMENU v4.0.9.8.7 (Tietz Video & Image Processing Systems GmbH, Gauting, Germany) and used to calculate the average dimensions of each phage.
2.3. Antibiotic susceptibility
The antibiotic susceptibility of the K. pneumoniae clinical isolates was measured using the Phoenix M50 system (BD Biosciences). Minimal inhibitory concentrations were interpreted as sensitive, resistant, or intermediate using EUCAST susceptibility breakpoints v8.1 (https://eucast.org), as shown in Supplementary Table S1.
2.4. Bacteriophage host range
Ten-fold serial dilutions of the phages were spotted onto DLA plates of K. pneumoniae strains K6310, K6592, L923, K6453 (isolation strains), K6500 (blood culture, UMCU), K5962 (tissue, UMCU), ATCC 11296, Staphylococcus aureus ATCC 19685, Pseudomonas aeruginosa PAO1, Acinetobacter baumannii DSM 30007, and Escherichia coli BL21-AI and K12 MG1655. The plates were incubated overnight at 37°C, and the phage plaques were observed to distinguish productive infection (lysis with individual phage plaques formed) and lysis from without19 (lysis without individual phage plaques). Efficiency of platting was determined as the ratio between phage titre in test strain and phage titre in host strain.
2.5. Temperature and pH stability
For temperature stability, 1 ml of each phage at 107 pfu/ml in LB was incubated at −80°C, −20°C, 4°C, 16°C, 22°C, 37°C, 50°C, and 80°C for 24 h. For pH stability, 1 ml of each phage at 107 pfu/ml in LB adjusted to a pH of 2, 4, 6, 7, 8, 10, and 12 with either hydrochloric acid or sodium hydroxide, was incubated for 24 h at room temperature. Phages were 10-fold serially diluted and spotted on DLA plates of the isolation strains for quantification.
2.6. Killing assays
Overnight cultures of the four isolation strains were diluted in LB to an optical density (600 nm) of 0.1–0.2. The bacterial suspension was distributed into wells of 96-well microtitre plates, and 10-fold dilutions of the phages were added to the wells in triplicates. A control was used where LB was added instead of phages. The plates were incubated in a plate reader (BioTek Epoch 2 microplate reader) at 37°C with rocking, and optical density measurements at 600 nm were taken every 10 min for 24 h.
2.7. One step growth curve
Isolation strains were grown in LB at 37°C with rocking up to an optical density (600 nm) of 0.4–0.5. The cultures were centrifuged at 8,000 × g for 10 min, and the cell pellet was re-suspended to half of the initial volume in LB. Phages were added at a multiplicity of infection (MOI) of 0.001 and let to adsorb for 10 min at 37°C with rocking. Cultures were centrifuged and the cell pellet containing the adsorbed phages was re-suspended to the initial volume in LB and incubated at 37°C with rocking for a total of 120 min. Samples were taken at time 0 and at 5-min intervals during the first 30 min, and 10-min intervals thereafter. The samples were immediately 10-fold serially diluted and spotted on DLA plates of the isolation strains for quantification.
2.8. Bacteriophage and bacteria genome sequencing
Phage DNA was extracted using phenol–chloroform as previously described.20 Phage genomic DNA was fragmented by Covaris 55 µl series Ultrasonicator, and used to construct DNA nanoball-based libraries by rolling circle replication. DNA was sequenced using the BGI MGISEQ-2000 platform (BGI Shenzhen, China) with paired-end 100 nt strategy, generating 4.6–19.2 Gb sequencing data for each sample with sequencing depth >10,000×. Bacterial genomic DNA was extracted using the GeneJET Genomic DNA Purification kit (Thermo Fisher), fragmented by Covaris 55 µl series Ultrasonicator, and used to construct paired-end libraries with an insert size of 200–400 bp. Bacterial genomes were sequenced on the BGISEQ-500 (MGI, BGI-Senzhen) platform, generating 1.4–2.0 Gb sequencing data for each sample with sequencing depth >100×. Quality control of the raw data was performed using FastP21 and Soapnuke22 with default parameters to remove low quality reads and duplications, and the reads were trimmed and processed using Seqtk.23 The filtered reads were assembled into the final genomes with SPAdes v3.13.0.24
2.9. Bacteriophage genome annotation and comparative genomics
Open reading frames (ORFs) of phage genomes were predicted and automatically annotated using the RAST server v2.0.25 with taxonomy ID 573, genetic code 11, and default options for the annotation pipeline. Long (>100 bp) genomic regions without predicted ORFs were manually confirmed using NCBI ORFfinder (https://www.ncbi.nlm.nih.gov/orffinder/) with a minimal ORF length of 75 nt and genetic code 11, using ATG and alternative initiation codons. Additional putative functions were assigned to ORFs by BlastP v.2.10.026 and Hmmer v3.3.127 when predicted with high similarity and in multiple hits. Domains identified by Hmmer were included as ‘Notes’ in the annotation files. Proteins annotated as host binding were also analysed by HHpred28 to further identify possible depolymerase activity. tRNAs were predicted with tRNAscan-SE v2.029 using sequence source ‘Bacterial’ and default options. Genomic comparisons were performed using BlastN v.2.10.0.26 Schematics of phage genomes were built with the Linear Genome Plot tool available at CTP Galaxy (https://cpt.tamu.edu/galaxy-pub).
2.10. Bacteria genome annotation and analysis
Bacterial genomes were annotated using Prokka30 with enabled search for ncRNAs (summary data in Supplementary Table S2). The capsular type of the K. pneumoniae strains was determined using Kaptive v0.7.3,31,32 and the sequence type was determined using MLST 2.0.33
2.11. Evolutionary analysis of phage proteins
The genome packaging strategy of the phages was predicted by phylogenetic analysis of the large terminase subunit as previously described,34 and PhageTerm was used to determine the phage genome termini.35 Evolutionary relationships of phage tubulin spindle and nucleus shell proteins were investigated by building unrooted phylogenetic trees with proteins found by psi-Blast (with five iterations) and Hmmer to be homologous to the tubulin spindle (Supplementary Table S3) and nucleus shell (Supplementary Table S4) proteins of Pseudomonas phage 201phi2-1, with an e-value equal or less than 1e−5. For all trees, proteins were aligned using MAFFT v7.308 with default settings, and the trees built using RAxML 7.2.8 with bootstrapping set to 100, without outgroup. A consensus tree was obtained using Consensus Tree Builder in Geneious v9.1.8.
2.12. Codon usage analysis
Codon usage of the bacteriophages and the K. pneumoniae HS11286 reference genome (GenBank RefSeq: NC_016845.1) was analysed with Cusp from EMBOSS (last accessed January 2021).36
3. Results and discussion
3.1. General morphological and phenotypic features
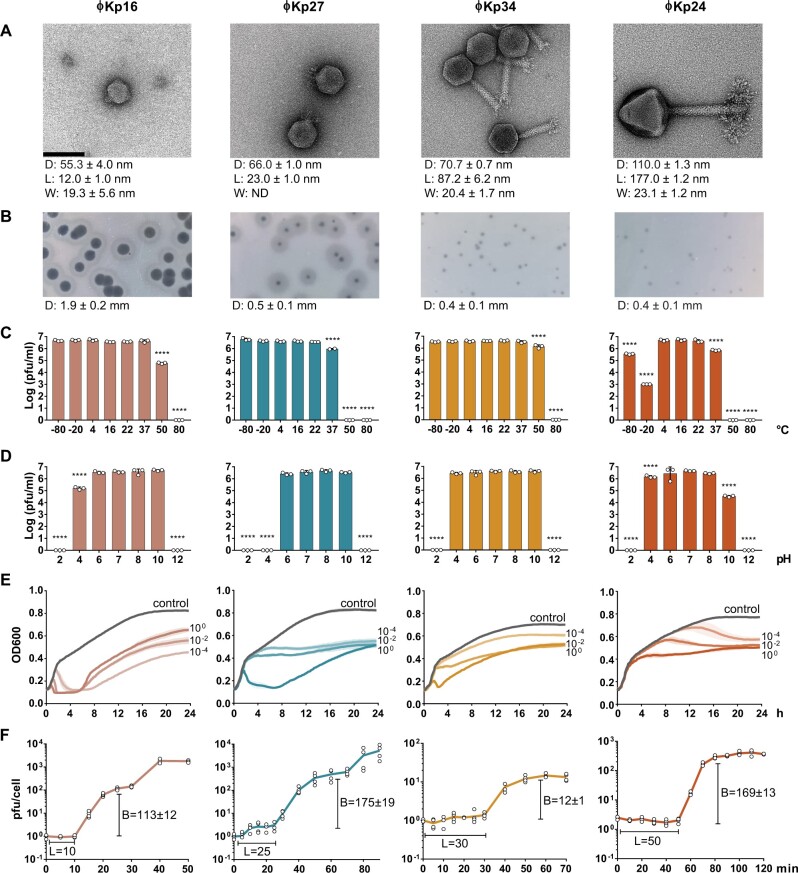
We have isolated four phages infecting K. pneumoniae from sewage samples: vB_KpnP_FBKp16 (ϕKp16), vB_KpnP_FBKp27 (ϕKp27), vB_KpnM_FBKp34 (ϕKp34), and vB_KpnM_FBKp24 (ϕKp24). The four phages have a tail and therefore belong to the Caudovirales order of phages with double stranded DNA (Fig. 1a). Phages ϕKp16 and ϕKp27 (Fig. 1a) have short tails and encode an RNA polymerase (Supplementary Tables S5 and S6), features that classify these phages in the Autographiviridae family.37 Phage ϕKp34 has a long contractile tail and a small baseplate with tail spikes or small tail fibres, and belongs to the Myoviridae family. Phage ϕKp24 also has a long contractile tail, but with a complex tail fibre structure at the baseplate, and a capsid that is 1.5 times larger than that of ϕKp34. These morphological features and the large ≈307 kb genome (Table 1) indicate that ϕKp24 is a Jumbo Myoviridae.38
Figure 1.
Morphological and phenotypic features of four newly isolated Klebsiella pneumoniae bacteriophages. (a) Transmission electron microscopy images of Autographiviridae ϕKp16, Autographiviridae ϕKp27, Myoviridae ϕKp34, and Myoviridae ϕKp24. Bacteriophages were negatively stained with 2% uracyl acetate. The diameter (D) of the capsid, and the length (L), and width (W) of the tail are given in nanometres below each phage as the average dimensions of 10 phage particles. Bar: 100 nm. All micrographs were taken at ×200,000 magnification. (b) Morphology of phage plaques. The diameter (D) of the plaque is given in millimetres below each phage as the average dimension of 10 phage plaques. (c) Viability of the phages at different temperatures. (d) Viability of the phages at different pH values. (e) Effect of the phages on cell growth of their isolation strain, measured as optical density at 600 nm. Control represents the growth of the strain without phage, while the remaining curves represent growth of the strain when infected with phage at a multiplicity of infection of 100, 10−2, or 10−4. (f) One step growth curve of the phages. The burst size (B) and latency period (L) are given in pfu and min, respectively.
Table 1.
Morphological and genomic features of the bacteriophages isolated in this work.
| vB_KpnP_FBKp16 | vB_KpnP_FBKp27 | vB_KpnM_FBKp34 | vB_KpnM_FBKp24 | |
|---|---|---|---|---|
| Short name | ϕKp16 | ϕKp27 | ϕKp34 | ϕKp24 |
| K. pneumoniae host | K6310 | L923 | K6453 | K6592 |
| Family | Autographiviridae | Autographiviridae | Myoviridae | Myoviridae |
| Genome size (bp) | 44,010 | 76,339 | 141,376 | 307,210 |
| Best Blast hit (query coverage, identity) | Salmonella phage BP12B (13%, 77.2%)a | Pectobacterium phage Nepra (2%, 75.4%)a | Proteus phage Mydo (3%, 87.5%) | No hit |
| GC content (%) | 51.9 | 44.2 | 36.0 | 45.1 |
| Number of CDS | 51 | 93 | 248 | 372 |
| Number of hypothetical proteins | 35 (69%) | 68 (73%) | 194 (78%) | 292 (79%) |
| Possible host receptor binding proteins | 1 | 3 | 1 | 9 |
| gp045 | gp091, gp093 | gp164 | gp196, gp300, gp303, gp304, gp306, gp308, gp309, gp310, gp357 | |
| Possible depolymerases | 0 | 2 | 0 | 8 |
| Pectate lyase | — | gp093 | — | gp300, gp308 |
| Glycosidase | — | gp091 | — | gp304, gp309, gp310 |
| Hydrolase | — | — | — | gp196, gp303, gp306 |
| DNA packaging | ||||
| Phylogenetic analysis | T7-like short direct terminal repeats | Undetermined | Undetermined | phiKZ-like headful |
| PhageTerm | T7-like short direct terminal repeats (length 238 bp; position 4,031–4,268 bp | T7-like short direct terminal repeats (length 349 bp; position 40,914–41,262 bp) | T7-like short direct terminal repeats (367 bp, 12,982–13,347 bp) | Undetermined |
| Structure of genome | Linear, redundant ends | Linear, redundant ends | Linear, redundant ends | Circularly permuted, terminally redundant |
| tRNA genes | 0 | 6 | 18 | 9 |
During the revision of this report, the sequence of Proteus phage PmP19 has been deposited on Genbank, which has 88% query cover and 91.68% identity to phage ϕKp16; and the sequence of Klebsiella phage vB_KpnP_P184 has been deposited, with 89% query cover and 95.38% identity to phage ϕKp27.
Autographiviridae ϕKp16 and ϕKp27 both form plaques with double halos, commonly associated with the activity of depolymerases,39 while the Myoviridae ϕKp34 and ϕKp24 form small plaques with no visible double halo (Fig. 1b). The four phages show variable temperature stability (Fig. 1c). Phages ϕKp16 and ϕKp34 are stable at temperatures ranging from −80°C to 50°C, although with viability reduction at 50°C. Viability of phage ϕKp27 is affected at 37°C and completely lost at higher temperatures. Similarly to phage ϕKp27, ϕKp24 is unstable at high temperatures, and it also shows loss of viability at −80°C and especially at −20°C. In terms of pH stability (Fig. 1d), phage ϕKp34 is the most stable, showing no viability changes at pH 4–10. Phages ϕKp16 and ϕKp24 are also relatively stable at pH 4–10, with some viability loss at pH 4 and 10, respectively. Phage ϕKp27 is stable at pH 6–10. Killing assays (Fig. 1e) show that Autographiviridae ϕKp16 and ϕKp24 are the fastest at killing the host bacteria, especially at higher MOI, but no phage results in complete bacterial clearance. One step growth curves of the phages (Fig. 1f) show different infection patterns, with latency periods ranging from 10 (ϕKp16) to 50 (ϕKp24) min. All phages have a burst size of more than 100 phages per infected cell, with the exception of phage ϕKp34 with a burst size of 12. Interestingly, phage ϕKp24 has a small plaque size (Fig. 1b) despite its large burst size, suggesting a poor diffusion in the agar plate as a consequence of its large size (Fig. 1a).
We tested the host range of the phages against a set of K. pneumoniae strains of distinct capsular types, as well as against other bacterial species (Table 2). As expected, the four phages were highly specific to K. pneumoniae, with ϕKp16, ϕKp27 and ϕKp34 infecting one capsular type (KL110, KL30, and KL38, respectively), and ϕKp24 infecting three capsular types (KL61, KL64, and KL46).
Table 2.
Host range of the bacteriophages isolated in this work, shown as efficiency of platting plating.
| Strain | Capsular type | Sequence type | ϕKp16 | ϕKp27 | ϕKp34 | ϕKp24 |
|---|---|---|---|---|---|---|
| Klebsiella pneumoniae | ||||||
| K6310 | KL110 | ST1958 | 1 | — | — | — |
| L923 | KL30 | Unknown (nearest ST1329, ST4863) | — | 1 | — | — |
| K6453 | KL38 | ST308 | — | — | 1 | — |
| K6592 | KL64 | ST147 | — | — | — | 1 |
| K6500 | KL61 | ST1411 | — | — | — | 0.01 |
| K5962 | KL46 | Unknown (nearest ST2118) | — | — | — | 0.21 |
| ATCC 11296 | KL4 | ST91 | — | — | — | — |
| Other species | ||||||
| Staphylococcus aureus ATCC 19685 | — | — | — | — | — | — |
| Pseudomonas aeruginosa PAO1 | — | ST549 | — | — | — | — |
| Acinetobacter baumannii DSM 30007 | KL3 | ST52 | — | — | — | — |
| Escherichia coli BL21-AI | — | ST93 | — | — | — | — |
| Escherichia coli K12 MG1655 | — | ST10 | — | — | — | — |
3.2. General genomic features
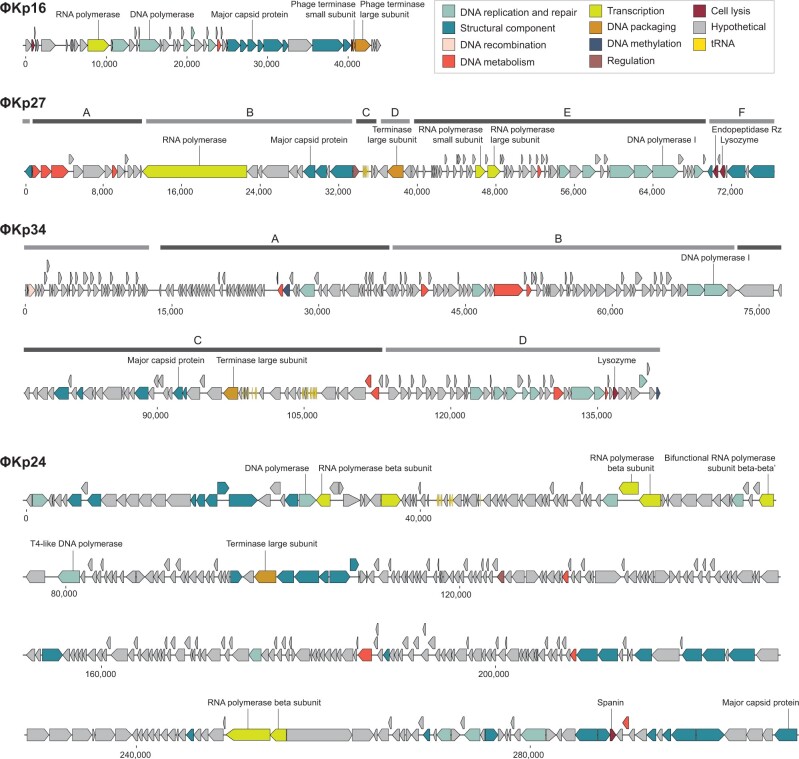
The four phages share a very low sequence similarity to each other and to phage genome sequences deposited in Genbank (Table 1). The genome of phage ϕKp24 is of particular highlight since no similarity was found to any genome sequence in Genbank, underlining its novelty. The genomes of phages are often organized in clusters of functionally related genes. While this analysis is made difficult due to the high number of hypothetical proteins with unassigned function (69–79%, Table 1), it is still possible to observe functional gene clustering.
Phage ϕKp16: The linear genome (Table 1) has all genes but one oriented in the same direction and organized in functional groups, especially evidenced by DNA replication and repair, and structural component genes (Fig. 2, Supplementary Table S5).
Figure 2.
Linear genome maps of the four newly isolated Klebsiella pneumoniae bacteriophages, ϕKp16, ϕKp27, ϕKp34, and ϕKp24. ORFs are coloured according to predicted function as shown in the key. Clusters depict clear gene operons located in the same strand. Clusters are not shown for ϕKp16 and ϕKp24 since most genes are located in the same strand.
Phage ϕKp27: The linear genome (Table 1) has genes organized in clusters of different orientation that group genes of related functions (Fig. 2, Supplementary Table S6). Cluster A groups genes involved in DNA metabolism, while Cluster B groups genes for transcription (an RNA polymerase) and a first set of genes for structural components related to capsid and tail tape measure proteins. Cluster C contains genes involved in regulation and six tRNAs, while Cluster D has only one gene with function identified for DNA packaging. Cluster E groups a second set of genes for transcription (a second RNA polymerase) as well as genes involved in DNA metabolism and DNA replication and repair. Finally, cluster F groups genes for a second set of structural components related to tail and host binding proteins, as well as genes for cell lysis.
Phage ϕKp34: The linear genome (Table 1) is organized in clusters of opposing orientation grouping genes of related functions, although genes for similar functions appear in more than one cluster (Fig. 2, Supplementary Table S7). Cluster A groups genes related to DNA methylation, DNA metabolism, and DNA replication and repair. Cluster B also groups genes for DNA metabolism and DNA replication and repair, as well as multiple genes seemingly related to bacterial metabolism (Supplementary Table S7). Cluster C groups all genes identified as structural components, as well as genes involved in DNA packaging, DNA metabolism and tRNAs, and Cluster D groups most genes related to DNA replication and repair, as well as genes for DNA metabolism, DNA methylation, DNA recombination, and cell lysis.
Phage ϕKp24: The circularly permuted genome (Table 1) has genes mostly oriented in the same direction and in some cases the predominant gene orientation is reversed by individual or small groups of genes in the opposite orientation (Fig. 2, Supplementary Table S8). The large genome size and the high percentage (79%) of proteins with unassigned functions in phage ϕKp24 make it difficult to define functional gene groups. Still, it is possible to identify three major groups of genes coding for structural components, as well as small groups of genes involved in DNA metabolism, transcription and DNA replication and repair, evidencing the functional clustering of genes commonly observed in phages.
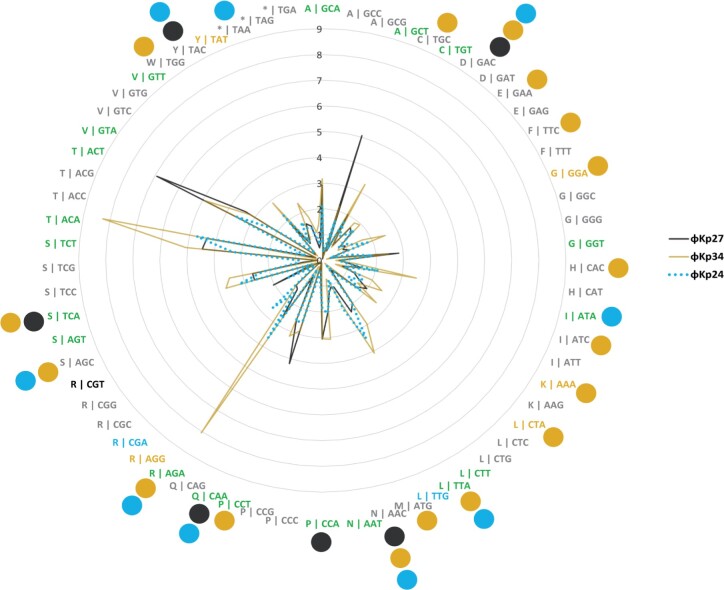
Phages ϕKp27, ϕKp34, and ϕKp24 encode 6, 18, and 9 tRNA genes, respectively (Table 1). As of yet, there is no clear explanation for the presence of tRNA genes in phage genomes.40–42 A number of studies have proposed that tRNA-containing phages have a codon bias that diverges from that of the bacterial host, therefore using the tRNAs to compensate for a metabolic difference.43,44 However, other studies have shown that this is not an universal observation. Here, we observe that less than half of the tRNAs encoded in phages ϕKp27 (3 of 6), ϕKp34 (7 of 18), and ϕKp24 (4 of 9) associate with codons that are more used in the phage than in the bacterial host (Fig. 3, Supplementary Table S9), suggesting that codon bias is not the (only) explanation for the presence of tRNAs in phages. It has also been suggested that tRNAs in phages may be beneficial to overcome the codon bias of different hosts,45 but this is difficult to assess since it is virtually impossible to determine the full range of species and strains that a phage can infect. Interestingly, 67% (18 of 27) of the codons more frequently used by the phages are shared by at least two phages, with 44% (12 of 27) being shared by the three. It will be interesting to explore common features of codon usage among phages of a certain species, rather than the similarity of codon usage between phage and host, as a feature to help predict the host in the future. Of note is also the presence of a suppressor tRNA (tRNA-Sup-TTA, Supplementary Table S8) in ϕKp24. Suppressor tRNAs arise when a mutation changes the tRNA anticodon, allowing it to recognize a stop codon and, instead of terminating, insert an amino acid at that position in the polypeptide chain.46 The tRNA-Sup-TTA of ϕKp24 has an arginine identity and is likely able to suppress opal (TGA) stop codons47 by inserting an arginine, but experimental validation is required to confirm the translational read-through. By inserting an amino acid where translation would otherwise stop, suppressor tRNAs can give rise to abnormally long proteins and produce metabolic changes.48 In phages, suppressor tRNAs have been shown to alleviate nonsense mutations (formation of a non-functional protein due to the premature appearance of a terminator codon in mRNA) that sometimes appear due to the rapid mutation rate of phages.49 Suppressor tRNAs have also been suggested to support the use of alternative genetic codes in megaphages (genomes >500 kb).50 Whether the suppressor tRNA of phage ϕKp24 serves a similar or different (e.g. interfere with host protein expression for host takeover) purpose, requires further investigation.
Figure 3.
Codon usage by Klebsiella pneumoniae phages ϕKp27, ϕKp24, and ϕKp34 as compared to the codon usage of K. pneumoniae HS11286. Codon usage is represented as the fraction between the frequency of codon usage in the phage divided by that of the bacteria. Codons are represented as X | YYY, in which X is the amino acid codified by codon YYY. Codons expressed at least 2-fold higher in phages are coloured black if overexpressed only in ϕKp27, mustard if only in ϕKp34, blue if only in ϕKp24, and green if overexpressed in at least two of the three phages. Coloured circles indicate codons for which the phages encode a tRNA.
The GC content of the phages (36.0–51.9%) is lower than the median GC content of K. pneumoniae (57.2%), a feature that is particularly prominent for phage ϕKp34 (36.0%) (Table 1). These results corroborate previous studies that show the GC content of phage genomes to accurately (>95%) predict the host associated with a phage at the phyla level but not at lower taxonomic levels.51 In fact, the divergence in GC content between phages and their bacterial hosts has been previously observed for phages infecting different species.52–55 Curiously, phage ϕKp34 has the lowest GC content and encodes for the largest number of tRNA genes, while phage ϕKp16 has the GC content closest to its host and encodes no tRNA genes, suggesting a connection worth exploring in future work.
3.3. Bacteriophage ϕKp16 has internal virion proteins
Phage ϕKp16 encodes two proteins annotated as putative internal virion proteins (gp042 and gp044), i.e. proteins that are encapsidated with the phage genome in the phage capsid during phage assembly inside the cell. In particular, gp044 holds similarity to gp37 of Enterobacteria phage SP6 (99% query cover, 75.71% identity). This protein is a homologue of protein gp16 of Enterobacteria phage T7, which forms part of the ejectosome complex that degrades the bacterial cell wall prior to DNA ejection, by forming an inner pore in the inner membrane to allow entry of the phage DNA into the cell.56 Phage ϕKp16 most likely uses a similar mechanism in which proteins encapsidated with the genome are ejected to form a transmembrane channel through which the phage genome can cross to enter the cell cytoplasm. The proteins in ϕKp16 are however distantly related to those of phage T7 and even SP6, suggesting a possible variant mode of channel formation.
3.4. Bacteriophages ϕKp27 and ϕKp34 have potentially novel genome packaging mechanisms
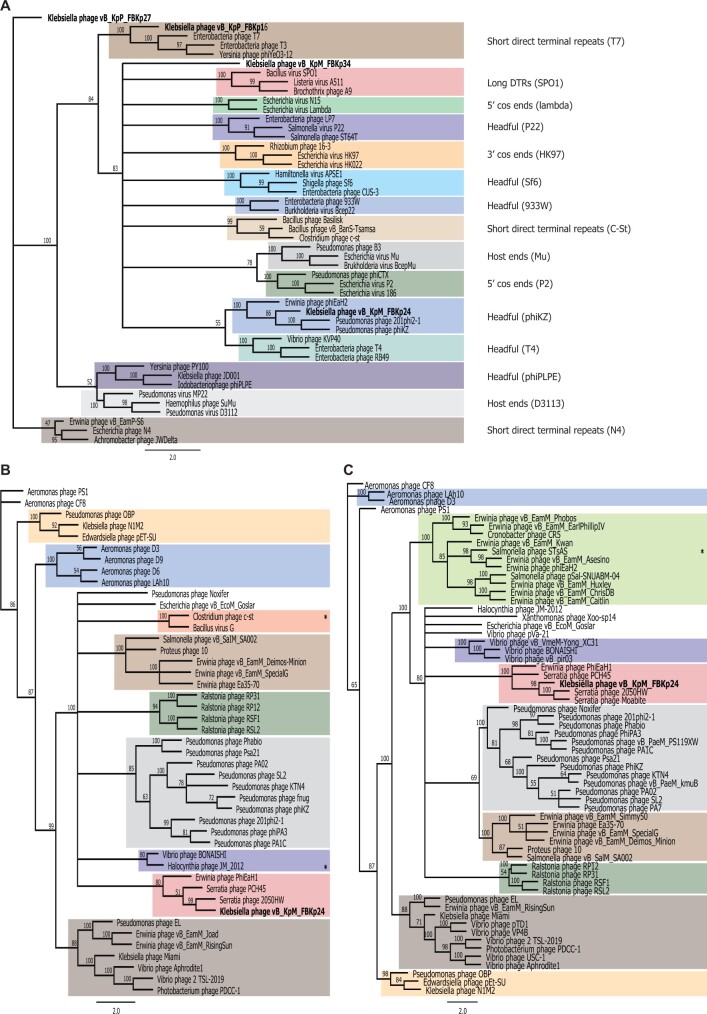
Genome packaging is a critical step in the assembly of Caudovirales and is carried out by a protein known as the large terminase.57 Using PhageTerm35 and a phylogenetics approach34 with the large terminase subunits of our phages and phages with well-characterized packaging mechanisms, we could infer the termini and packaging mechanisms used by our phages (Fig. 4a, Table 1, Supplementary Fig. S1). Both approaches suggest that phage ϕKp16 uses a packaging mechanism based on T7-like short direct terminal repeats. Phage ϕKp24 was shown by PhageTerm to have a circularly permuted and terminally redundant genome, which is associated with headful packaging, as inferred from the phylogenetics approach. The terminases of phages ϕKp27 and ϕKp34 formed their own clades in the phylogenetic tree, suggesting packaging mechanisms distinct from those currently known. The terminase of ϕKp34 has a C-terminal domain (PF17289) and an N-terminal domain (PF03237) commonly found in the terminase large subunit of phages such as T4, suggesting a potential headful packaging mechanism. However, PhageTerm analysis suggests that ϕKp34 has T7-like short direct terminal repeats. These contradicting outputs may indicate the use of a packaging mechanism not yet characterized. The terminase of ϕKp27 appears as the most distanced in the phylogenetic tree (Fig. 4a), but a Blastp analysis revealed high similarity (99% query cover, >70% identity) to the large terminase of N4-like phages,58,59 suggesting a packaging mechanism of short direct terminal repeats, as predicted also by PhageTerm (Table 1).
Figure 4.
Phylogenetic trees of selected phage proteins. (a) Analysis of large terminase subunits using proteins from phages of well-known packaging mechanisms. (b) Analysis of the tubulin spindle protein of phage ϕKp24 and all protein homologues to the tubulin spindle of phage 201phi2-1 found by psi-Blast and Hmmer. (c) Analysis of the nucleus shell protein of phage ϕKp24 and all protein homologues to the nucleus shell protein of phage 201phi2-1 found by psi-Blast and Hmmer. Trees were built from MAFFT alignments using RAxML with bootstrapping of 100. Identical colours were used in panels (b) and (c) to identify similar phage clusters. All phages in panels (b) and (c) have genomes above 200 kb (Jumbo phages) with the exception of those marked with *, which have a genome size of 167–197 kb.
3.5. Bacteriophage ϕKp34 encodes genes with possible anti-viral functions
Phage ϕKp34 encodes an insertion sequence of the IS200/IS605 family (gp188) that is commonly found in bacterial and prophage genomes.24,60 IS sequences contribute majorly to bacterial genome diversification, and have also been suggested to play a role in the inactivation and immobilization of other invading phages.61 Interestingly, phage ϕKp34 also contains a cluster of genes similar to terC (gp124), terF C-terminal vWA domain (gp131), and terD (gp132, gp133) from the terZABCDEF system, and one gene similar to telA (gp135) from the telAB system. The terZABCDEF and telAB operons seem to constitute a membrane-linked chemical stress response and anti-viral defence system in bacteria.62–64 The subset of genes from the original operons present in ϕKp34 seems to constitute a functional hub found in most major bacterial lineages,64 and has also been reported in virulent phages.65
3.6. Bacteriophage ϕKp24 has multiple depolymerases and tubulin and nuclear shell proteins
Phage ϕKp24 has a distinctive complex structure at its baseplate (Fig. 1d) possibly composed of nine host binding proteins (Table 1), in comparison with one host binding protein in phages ϕKp16 and ϕKp34, and two in phage ϕKp27. Eight of the nine possible host binding proteins of ϕKp24 have putative depolymerase domains (Table 1) of pectase lyase, glycosidase, and hydrolase activity, while ϕKp16 and ϕKp34 have no predicted depolymerases. The two host binding proteins of ϕKp27 have a predicted glycosidase (gp091) and pectate lyase (gp093) activity. Depolymerases are used by phages to degrade the capsule of bacteria and to gain access to their secondary receptor (e.g. outer membrane protein, lipopolysaccharide) on the host’s surface. Depolymerases tend to be specific to a capsular type, and the presence of multiple depolymerases with different activities suggests that phage ϕKp24 can interact and degrade different capsular types,54 likely expanding the phage’s host range. This is corroborated by phage ϕKp24 binding to strains of capsular types KL61, KL64, and KL46, whereas the other phages bind to only one of the capsular types tested (Table 2).
Interestingly, and akin to other Jumbo phages, phage ϕKp24 codes for tubulin spindle (gp094) and nucleus shell (gp083) proteins that function to enhance phage reproduction.66,67 The nucleus shell protein forms a proteinaceous barrier that encloses viral DNA, separating phage DNA replication and transcription from other cellular functions, and providing a protective physical barrier against DNA-targeting CRISPR-Cas systems;68,69 while the tubulin spindle positions the phage nucleus structure at the cell centre.70 A phylogenetic analysis of the tubulin spindle and nucleus shell proteins of ϕKp24 and all protein homologues to those of phage 201phi2-1 (where these proteins were first reported) found by psi-Blast and Hmmer search (Fig. 4b and c) reveals that the proteins of ϕKp24 cluster with those of Serratia phage 2050HW, Serratia phage PCH45, and Erwinia phage PhiEaH1. Interestingly, clusters formed by tubulin spindle and nucleus shell proteins are identical, suggesting that these proteins have co-evolved, and seem to group according to the bacterial species infected. It is also curious that only three of the phages encoding tubulin spindle and nucleus shell proteins (Fig. 4b and c, Supplementary Tables S3 and S4) have genomes smaller than 200 kb (167–197 kb), further underpinning the exclusive use of these proteins by Jumbo phages.
4. Conclusion
The genomic analysis of the four K. pneumoniae phages isolated in this study revealed potential novel packaging mechanisms (ϕKp27 and ϕKp34), the presence of possible anti-viral strategies in phage genomes (ϕKp34), and distinctive and novel clades of tubulin spindle and nucleus shell proteins (ϕKp24) that will help shed light into the evolution of compartmentalization in prokaryotes and eukaryotes. Phages ϕKp16, ϕKp27, ϕKp34, and ϕKp24 are strong candidates for phage therapy against antibiotic-resistant K. pneumoniae infections. Further exploration of phage genomes will help elucidate the origins, genetic diversity and evolutionary mechanisms of phages, and contribute to a better understanding of the broader biology of microbial populations and how their genomic characteristics contribute to observable features. This knowledge and the study of individual genes and proteins will certainly also be translated into innovative tools with biotechnological and medical applications.
Supplementary Material
Funding
This work was supported by donations from University Fund from the Delft University of Technology to Fagenbank, as well as generous donations from the public. A.R.C. is supported by the Netherlands Organisation for Scientific Research (NWO) NWA Startimpuls grant 17.366. S.J.J.B. is supported by NWO Vici grant VI.C182.027. Genome sequencing and assembly is supported by China National GeneBank and the Global Phage Hub initiated by BGI-Shenzhen.
Accession numbers
The assembled and annotated phage genome sequences have been deposited in Genbank (https://www.ncbi.nlm.nih.gov/genbank/) under accession numbers MW394389 (ϕKp16), MW394388 (ϕKp27), MW394391 (ϕKp24), and MW394390 (ϕKp34). The assembled bacterial genomes have been deposited in Genbank under BioProject accession number PRJNA745534. The data are also available at the China National GeneBank (CNGB, https://db.cngb.org/) under accession number CNP0000861.
Conflict of interest
None declared.
References
- 1. Weinbauer M.G. 2004, Ecology of prokaryotic viruses, FEMS Microbiol. Rev., 28, 127–81. [DOI] [PubMed] [Google Scholar]
- 2. Danovaro R., Corinaldesi C., Dell'anno A., et al. 2011, Marine viruses and global climate change, FEMS Microbiol. Rev., 35, 993–1034. [DOI] [PubMed] [Google Scholar]
- 3. Proctor L.M., Fuhrman J.A.. 1990, Viral mortality of marine bacteria and cyanobacteria, Nature, 343, 60–2. [Google Scholar]
- 4. Brüssow H., Canchaya C., Hardt W.-D.. 2004, Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion, Microbiol. Mol. Biol. Rev., 68, 560–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sausset R., Petit M.A., Gaboriau-Routhiau V., De Paepe M.. 2020, New insights into intestinal phages, Mucosal Immunol., 13, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hershey A.D., Chase M.. 1952, Independent functions of viral protein and nucleic acid in growth of bacteriophage, J. Gen. Physiol., 36, 39–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Crick F.H.C., Barnett L., Brenner S., Watts-Tobin R.J.. 1961, General nature of the genetic code for proteins, Nature, 192, 1227–32. [DOI] [PubMed] [Google Scholar]
- 8. Brenner S., Jacob F., Meselson M.. 1961, An unstable intermediate carrying information from genes to ribosomes for protein synthesis, Nature., 190, 576–81. [DOI] [PubMed] [Google Scholar]
- 9. Arber W., Linn S.. 1969, DNA modification and restriction, Annu. Rev. Biochem., 38, 467–500. [DOI] [PubMed] [Google Scholar]
- 10. Lobban P.E., Kaiser A.D.. 1973, Enzymatic end-to-end joining of DNA molecules, J. Mol. Biol., 78, 453–71. [DOI] [PubMed] [Google Scholar]
- 11. Pickar-Oliver A., Gersbach C.A.. 2019, The next generation of CRISPR–Cas technologies and applications, Nat. Rev. Mol. Cell Biol., 20, 490–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. O’Neill J. Tackling drug-resistant infections globally: Final report and recommendations. HM Gov. Welcome Trust UK. 2016. [Google Scholar]
- 13. Schroven K., Aertsen A., Lavigne R.. 2021, Bacteriophages as drivers of bacterial virulence and their potential for biotechnological exploitation, FEMS Microbiol. Rev., 45, fuaa041. [DOI] [PubMed] [Google Scholar]
- 14. Pires D.P., Costa A.R., Pinto G., Meneses L., Azeredo J.. 2020, Current challenges and future opportunities of phage therapy, FEMS Microbiol. Rev., 44, 684–700. [DOI] [PubMed] [Google Scholar]
- 15. Zampara A., Sørensen M.C.H., Gencay Y.E., et al. 2021, Developing innolysins against campylobacter jejuni using a novel prophage receptor-binding protein, Front. Microbiol., 12, 619028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dedrick R.M., Guerrero-Bustamante C.A., Garlena R.A., et al. 2019, Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus, Nat. Med., 25, 730–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tacconelli E., Carrara E., Savoldi A., WHO Pathogens Priority List Working Group., et al. 2018, Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis, Lancet Infect. Dis., 18, 318–27. [DOI] [PubMed] [Google Scholar]
- 18. Schneider C.A., Rasband W.S., Eliceiri K.W.. 2012, NIH Image to ImageJ: 25 years of image analysis, Nat. Methods, 9, 671–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Abedon S.T. 2011, Lysis from without, Bacteriophage, 1, 46–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sambrook J., Russell D.W.. 2001, Molecular Cloning : A Laboratory Manual, Cold Spring Harbor Laboratory Press, New York. [Google Scholar]
- 21. Chen S., Zhou Y., Chen Y., Gu J.. 2018, Fastp: an ultra-fast all-in-one FASTQ preprocessor, Bioinformatics, 34, i884–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen Y., Chen Y., Shi C., et al. 2018, SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data, Gigascience, 7, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li H. 2012, seqtk, Toolkit for processing sequences in FASTA/Q formats, GitHub., 767, 69. [Google Scholar]
- 24. Bankevich A., Nurk S., Antipov D., et al. 2012, SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing, J. Comput. Biol., 19, 455–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Aziz R.K., Bartels D., Best A.A., et al. 2008, The RAST Server: rapid annotations using subsystems technology, BMC Genomics, 9, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J.. 1990, Basic local alignment search tool, J. Mol. Biol., 215, 403–10. [DOI] [PubMed] [Google Scholar]
- 27. Mistry J., Finn R.D., Eddy S.R., Bateman A., Punta M.. 2013, Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions, Nucleic Acids Res., 41, e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zimmermann L., Stephens A., Nam S.-Z., et al. 2018, A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core, J. Mol. Biol., 430, 2237–43. [DOI] [PubMed] [Google Scholar]
- 29. Chan P.P., Lowe T.M.. 2019, tRNAscan-SE: searching for tRNA genes in genomic sequences, Methods Mol. Biol., 1962, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Seemann T. 2014, Prokka: rapid prokaryotic genome annotation, Bioinformatics, 30, 2068–9. [DOI] [PubMed] [Google Scholar]
- 31. Wyres K.L., Wick R.R., Gorrie C., et al. 2016, Identification of Klebsiella capsule synthesis loci from whole genome data, Microb. Genom., 2, e000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wick R.R., Heinz E., Holt K.E., Wyres K.L.. 2021, Kaptive Web: user-friendly capsule and lipopolysaccharide serotype prediction for Klebsiella genomes, J. Clin. Microbiol., 56, e00197–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Larsen M.V., Cosentino S., Rasmussen S., et al. 2012, Multilocus sequence typing of total-genome-sequenced bacteria, J. Clin. Microbiol., 50, 1355–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Merrill B.D., Ward A.T., Grose J.H., Hope S.. 2016, Software-based analysis of bacteriophage genomes, physical ends, and packaging strategies, BMC Genomics, 17, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Garneau J.R., Depardieu F., Fortier L.-C., Bikard D., Monot M.. 2017, PhageTerm: a tool for fast and accurate determination of phage termini and packaging mechanism using next-generation sequencing data, Sci. Rep., 7, 8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rice P., Longden I., Bleasby A.. 2000, EMBOSS: the European molecular biology open software suite, Trends Genet., 16, 276–7. [DOI] [PubMed] [Google Scholar]
- 37. Adriaenssens E.M., Sullivan M.B., Knezevic P., et al. 2020, Taxonomy of prokaryotic viruses: 2018–2019 update from the ICTV Bacterial and Archaeal Viruses Subcommittee, Arch. Virol., 165, 1253–60. [DOI] [PubMed] [Google Scholar]
- 38. Hendrix R.W. 2009, Jumbo bacteriophages, Curr. Top. Microbiol. Immunol., 328, 229–40. [DOI] [PubMed] [Google Scholar]
- 39. Knecht L.E., Veljkovic M., Fieseler L.. 2019, Diversity and function of phage encoded depolymerases, Front. Microbiol., 10, 2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Samson J.E., Moineau S.. 2010, Characterization of lactococcus lactis phage 949 and comparison with other lactococcal phages, Appl. Environ. Microbiol., 76, 6843–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dreher T.W., Brown N., Bozarth C.S., et al. 2011, A freshwater cyanophage whose genome indicates close relationships to photosynthetic marine cyanomyophages, Environ. Microbiol., 13, 1858–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gervasi T., Curto R.L., Narbad A., Mayer M.J.. 2013, Complete genome sequence of ΦCP51, a temperate bacteriophage of Clostridium perfringens, Arch. Virol., 158, 2015–7. [DOI] [PubMed] [Google Scholar]
- 43. Bailly-Bechet M., Vergassola M., Rocha E.. 2007, Causes for the intriguing presence of tRNAs in phages, Genome Res., 17, 1486–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kunisawa T. 2000, Functional role of mycobacteriophage transfer RNAs, J. Theor. Biol., 205, 167–70. [DOI] [PubMed] [Google Scholar]
- 45. Delesalle V.A., Tanke N.T., Vill A.C., Krukonis G.P.. 2016, Testing hypotheses for the presence of tRNA genes in mycobacteriophage genomes, Bacteriophage, 6, e1219441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Eggertsson G., Soll D.. 1988, Transfer ribonucleic acid-mediated suppression of termination codons in Escherichia coli, Microbiol. Rev., 52, 354–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Palma M., Lejeune F.. 2021, Deciphering the molecular mechanism of stop codon readthrough, Biol. Rev. Camb. Philos. Soc., 96, 310–29. [DOI] [PubMed] [Google Scholar]
- 48. Herring C.D., Blattner F.R.. 2004, Global transcriptional effects of a suppressor tRNA and the inactivation of the regulator frmR, J. Bacteriol., 186, 6714–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. McClain W.H. 1970, UAG suppressor coded by bacteriophage T4, FEBS Lett., 6, 99–101. [DOI] [PubMed] [Google Scholar]
- 50. Devoto A.E., Santini J.M., Olm M.R., et al. 2019, Megaphages infect Prevotella and variants are widespread in gut microbiomes, Nat. Microbiol., 4, 693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Edwards R.A., McNair K., Faust K., Raes J., Dutilh B.E.. 2016, Computational approaches to predict bacteriophage-host relationships, FEMS Microbiol. Rev., 40, 258–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Marinelli L.J., Fitz-Gibbon S., Hayes C., et al. 2012, Propionibacterium acnes bacteriophages display limited genetic diversity and broad killing activity against bacterial skin isolates, mBio., 3, e00279–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Simoliunas E., Kaliniene L., Truncaite L., et al. 2012, Genome of Klebsiella sp.-infecting bacteriophage vB_KleM_RaK2, J. Virol., 86, 5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pan Y.-J. Lin T.-L., Chen C.-C., et al. 2017, Klebsiella phage ΦK64-1 encodes multiple depolymerases for multiple host capsular types, J. Virol., 91, e02457–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dupuis M.È., Moineau S.. 2010, Genome organization and characterization of the virulent lactococcal phage 1358 and its similarities to Listeria phages, Appl. Environ. Microbiol., 76, 1623–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Leptihn S., Gottschalk J., Kuhn A.. 2016, T7 ejectosome assembly: a story unfolds, Bacteriophage, 6, e1128513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wangchuk J., Prakash P., Bhaumik P., Kondabagil K.. 2018, Bacteriophage N4 large terminase: expression, purification and X-ray crystallographic analysis, Acta Crystallogr. F Struct. Biol. Commun., 74, 198–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Buttimer C., Hendrix H., Lucid A., et al. 2018, Novel N4-like bacteriophages of pectobacterium atrosepticum, Pharmaceuticals., 11, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shi X., Zhao F., Sun H., et al. 2020, Characterization and complete genome analysis of Pseudomonas aeruginosa bacteriophage vB_PaeP_LP14 belonging to genus Litunavirus, Curr. Microbiol., 77, 2465–74. [DOI] [PubMed] [Google Scholar]
- 60. Kuno S., Yoshida T., Kamikawa R., Hosoda N., Sako Y.. 2010, The distribution of a phage-related insertion sequence element in the cyanobacterium, Microcystis aeruginosa, Microbes Environ., 25, 295–301. [DOI] [PubMed] [Google Scholar]
- 61. Ooka T., Ogura Y., Asadulghani M., et al. 2009, Inference of the impact of insertion sequence (IS) elements on bacterial genome diversification through analysis of small-size structural polymorphisms in Escherichia coli O157 genomes, Genome Res., 19, 1809–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Whelan K.F., Colleran E., Taylor D.E.. 1995, Phage inhibition, colicin resistance, and tellurite resistance are encoded by a single cluster of genes on the IncHI2 plasmid R478, J. Bacteriol., 177, 5016–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Walter E.G., Thomas C.M., Ibbotson J.P., Taylor D.E.. 1991, Transcriptional analysis, translational analysis, and sequence of the kilA-tellurite resistance region of plasmid RK2Te(r), J. Bacteriol., 173, 1111–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Anantharaman V., Iyer L.M., Aravind L.. 2012, Ter-dependent stress response systems: novel pathways related to metal sensing, production of a nucleoside-like metabolite, and DNA-processing, Mol. Biosyst., 8, 3142–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Frampton R.A., Taylor C., Holguín Moreno A.V., et al. 2014, Identification of bacteriophages for biocontrol of the kiwifruit canker phytopathogen Pseudomonas syringae pv, Appl. Environ. Microbiol., 80, 2216–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kraemer J.A., Erb M.L., Waddling C.A., et al. 2012, A phage tubulin assembles dynamic filaments by an atypical mechanism to center viral DNA within the host cell, Cell., 149, 1488–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Erb M.L., Kraemer J.A., Coker J.K.C., et al. 2014, A bacteriophage tubulin harnesses dynamic instability to center DNA in infected cells, Elife., 3, e03197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Mendoza S.D., Nieweglowska E.S., Govindarajan S., et al. 2020, A bacteriophage nucleus-like compartment shields DNA from CRISPR nucleases, Nature, 577, 244–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Malone L.M., Warring S.L., Jackson S.A., et al. 2020, A jumbo phage that forms a nucleus-like structure evades CRISPR–Cas DNA targeting but is vulnerable to type III RNA-based immunity, Nat. Microbiol., 5, 48–55. [DOI] [PubMed] [Google Scholar]
- 70. Chaikeeratisak V., Nguyen K., Egan M.E., et al. 2017, The phage nucleus and tubulin spindle are conserved among large Pseudomonas phages, Cell Rep., 20, 1563–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.