ABSTRACT
Cellular stress response mechanisms typically increase organellar quantity and volume. To restore cellular homeostasis and organellar integrity, the surplus organelles are cleared by macroautophagy/autophagy, an intracellular process that shuttles cytoplasmic material to the lysosomes for degradation. The degradation is mediated by autophagy receptors that selectively link the degradable cargo to the autophagy machinery. Studies have identified receptors for the degradation of mitochondria, endoplasmic reticulum, lysosomes, and peroxisomes. The autophagic degradation of the Golgi, named Golgiphagy, however, has remained undefined. The Golgi is essential for the processing, sorting and trafficking of proteins and lipids in the secretory pathway. In a recent study, we identified CALCOCO1 as a Golgiphagy receptor in response to nutrient deprivation. CALCOCO1 interacts with Golgi membranes by binding to cytoplasmic Ankyrin repeat (AR) domains of Golgi resident ZDHHC17 and ZDHHC13 palmitoyltransferases (PATs) via a defined zDHHC-AR-binding motif (zDABM) to recruit autophagy machinery. Lack of CALCOCO1 in cells causes an impaired Golgiphagy and expansion of the Golgi.
KEYWORDS: Autophagy receptor, CALCOCO1, Golgi, Golgiphagy, Golgiphagy receptor, zDABM motif, ZDHHC17
In a previous study, we identified CALCOCO1 as a reticulophagy receptor, an adaptor protein that targets specific endoplasmic reticulum (ER) portions to the lysosome for degradation. CALCOCO1 is composed of an N-terminal SKICH domain, an atypical LIR motif and a C-terminal region harboring a zinc finger domain. We found that it also contains a UDS-interacting region (UIR) that binds to Atg8-family proteins, and a FFAT-like motif. For reticulophagy, CALCOCO1 binds degradable cargo by interacting with the ER-resident VAPA and VAPB proteins via the FFAT-like motif and then recruits autophagy machinery by interacting with Atg8-family proteins via the LIR and UIR motifs. In a recent study [1], we identified a zDABM motif as a new interaction feature in the C-terminal half of CALCOCO1. The zDABM motif was originally identified in ZDHHC17-interacting proteins and is characterized by a (VIAP)(VIT)XXQP core consensus sequence where X is any amino acid. The motif in CALCOCO1 encompasses the 574VVISQP579 core sequence. Surprisingly, we also identified and validated a similar motif in the CALCOCO1 paralog TAX1BP1, encompassing 673VVCSQP678 core sequence. PATs are transmembrane enzymes that catalyze the reversible addition of fatty acids, typically palmitate, to cysteine residues of proteins. There are 24 PATs in mammals but only ZDHHC17 and ZDHHC13 possess an AR domain. The AR domain interacts with proteins bearing a zDABM motif to recruit them either as substrates or for palmitoylation-independent functions. Our studies revealed that CALCOCO1 interacts with ZDHHC17 and ZDHHC13 to mediate Golgiphagy during starvation-induced stress, thereby uncovering a novel function of the AR-zDABM interaction.
Nutrient starvation in cell culture causes an upregulation of Golgi matrix and membrane proteins leading to disassembly of the Golgi into fragments that are then engulfed within autophagosomes, followed by autophagic degradation of the proteins. Both the engulfment and the degradation of the Golgi-resident GOLGA2/GM130, TMEM165 and ZDHHC13 proteins are significantly reduced by depletion of CALCOCO1. The degradation is, however, restored by ectopic expression of CALCOCO1, suggesting a dependency on CALCOCO1. Conversely, the degradation cannot be restored by ectopic expression of CALCOCO1 that either has a mutated zDABM motif or is incapable of interacting with Atg8-family proteins, indicating that concurrent interaction with ZDHHC17 and autophagy machinery is required for CALCOCO1-mediated Golgiphagy. Deletion of the FFAT-like motif does not significantly reduce CALCOCO1-mediated Golgiphagy, nor does mutation of the zDABM motif impair CALCOCO1-mediated reticulophagy, suggesting that the two motifs are functionally independent. Hence, CALCOCO1 can act as a receptor both for reticulophagy and Golgiphagy by distinguishing the different cargos through distinct specific interactions (Figure 1).
Figure 1.
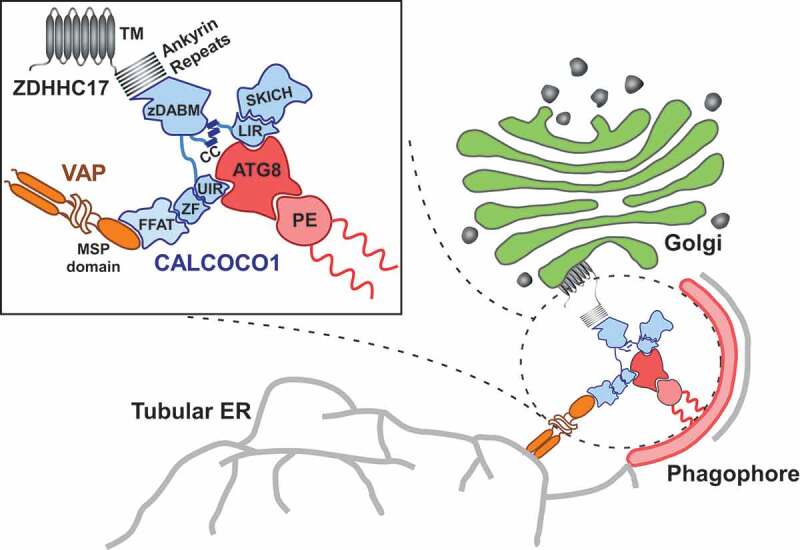
Role of CALCOCO1 during Golgiphagy. Upon nutrient stress, CALCOCO1 mediates the degradation of Golgi fragments through interaction with the Golgi-localized transmembrane palmitoyl transferase ZDHHC17. CALCOCO1 binds to the ankyrin repeats of ZDHHC17 (or ZDHHC13) via its zDABM motif and recruits the autophagy machinery via LIR-LDS- and UIR-UDS-mediated interactions with Atg8-family proteins (ATG8) to initiate phagophore formation. The Golgi fragments are subsequently sequestrated within autophagosomes and degraded when the autophagosomes fuse with lysosomes. The dual ability of CALCOCO1 to also degrade ER via FFAT-motif interactions with ER-localized VAP proteins is also indicated
Because stress signaling increases the amount of the Golgi components, we envisage that Golgiphagy is triggered by the need to degrade damaged or excess Golgi in order to restore pre-stress status. During nutrient deprivation, our data suggest that CALCOCO1, already bound to specific Golgi subdomains via ZDHHC17 and ZDHHC13, recruits the autophagy machinery via interaction with Atg8-family proteins to initiate autophagic capture of the degradable cargo that is then shuttled to the lysosomes for degradation. The recruitment of the autophagy machinery is very likely propagated by the upregulation of ZDHHC17 and ZDHHC13 during stress via increased interaction with CALCOCO1. Because the zDABM motif has been identified in many proteins, it is probable that the interaction of CALCOCO1 with ZDHHC17 and ZDHHC13 is transient under normal conditions and it is the recruitment of the autophagy machinery that likely stabilizes the interactions for cargo capture.
8Nutrient starvation is likely not the only trigger for CALCOCO1-mediated Golgiphagy. Microbial infections, toxic insults and neurodegenerative diseases also cause an upregulation of Golgi-resident proteins and Golgi fragmentation. Because our data suggest that CALCOCO1-mediated Golgiphagy is induced by the need to remove excess Golgi components generated during stress in order to restore the pre-stress status, it is probable that CALCOCO1-mediated Golgiphagy is involved also in the removal of damaged and excess Golgi components during these pathological conditions. An important question for future investigations is whether Golgiphagy has any prognostic role in these pathologies.
In conclusion, our study identified CALCOCO1 as a Golgiphagy receptor during nutrient starvation through interaction with Golgi-resident ZDHHC17 and ZDHHC13 PATs. The interaction occurs by CALCOCO1 binding to the AR domain of ZDHHC17 and ZDHHC13 via a novel zDABM motif on CALCOCO1. The presence of a zDABM motif in TAX1BP1 suggests that it is similarly involved in Golgiphagy as a receptor and therefore it is a prime candidate for further studies on Golgiphagy.
Funding Statement
This work was funded by The Research Council of Norway [grant number 249884] and the Norwegian Cancer Society [grant number 190214] to TJ.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Reference
- [1].Nthiga TM, Shrestha BK, Bruun J-A, et al. Regulation of Golgi turnover by CALCOCO1-mediated selective autophagy. J Cell Biol. 2021;220:e202006128. [DOI] [PMC free article] [PubMed] [Google Scholar]


