ABSTRACT
Pancreatic ductal adenocarcinoma (PDAC) is one of the deadliest malignancies, with poor prognosis resulting mostly from late diagnosis. Surgery remains the most effective treatment and early detection significantly increases the overall survival. Biomarkers used for diagnosis and to monitor the response to treatment, such as carbohydrate antigen 19–9 (CA19-9) and carcinoembryonic antigen (CEA), are not adequate as early detection markers of PDAC, partly due to low sensitivity/specificity. Therefore, new biomarkers for PDAC are critically needed. This review aims at recent advancements in the identification and characterization of new biomarkers, microRNAs, which might prove useful in the early detection of PDAC.
KEYWORDS: Pancreatic ductal adenocarcinoma, early detection, biomarkers, miRNA
1. Introduction
Despite the progress in the design of new therapies in advanced pancreatic cancer, it remains a big challenge to precisely identify the group of patients that will be able to benefit from their use. For some patients, new treatment options may extend their life by several months, for others even by up to two years.1 On the other hand, medical diagnosis without the use of invasive methods makes differentiation between pancreatic ductal adenocarcinoma (PDAC), benign tumors or chronic pancreatitis (CP) difficult.
Biomarkers for PDAC can be classified as diagnostic, prognostic (provide information about the patient’s overall cancer outcome, regardless of therapy), and predictive (give information about the effect of a therapeutic intervention).2 Unfortunately, routine cancer markers (such as carbohydrate antigen 19–9; CA 19–9) do not seem to be reliable in prognosis and detection of early stage of PDAC.1–5 However, new strategies are emerging. Collected data indicate that specificity and sensitivity of diagnosis is optimal when several biomarkers are used in combination. Moreover, the use of newly identified biomarkers together with “classical” CA 19–9 may significantly increase specificity and sensitivity in early PDAC detection.3
An ideal combination of biomarkers should: (1) be capable of distinguishing healthy subjects from patients, (2) be expressed early in the disease progression, (3) be easy to assay and relatively inexpensive, and (4) give reproducible results. There are several factors that may impact the sensitivity and specificity of a biomarker such as: sample type (biofluid vs. tissue), stability of the sample and processing time to assay the biomarker, proper use of negative controls as well as background profiling.
Scientists’ attention has recently been drawn to specific molecules of ribonucleic acid with unique stability. These molecules, called microRNAs (miRNAs), are a group of small non-coding RNAs involved in regulating a range of developmental and physiological processes; their dysregulation has been associated with many diseases, including cancer. Small size as well as the fact that miRNAs bind with proteins may protect them from ribonucleases and thus improve their stability in the blood, which may suggest their utility in biomarker studies. Concurrently, extracellular vesicles (exosomes), which are lipid bilayer-delimited particles that are naturally released from a cell and – unlike the cell – cannot replicate, remain the major source of circulating miRNAs. Exosomes are a type of extracellular vesicles that contain constituents (protein, DNA, and RNA) of the cells that secrete them. Exosomes play an important role in information exchange between cells and once released, they travel throughout the body before releasing their contents into a recipient cell.
Some researchers have shown that miRNAs circulating in the blood can transmit information and regulate the health of cells other than those from which they originate.3,4 Others believe that circulating miRNAs are merely by-products released from cells that are completely unnecessary and have no function.4 Regardless of which of these theories is true, it has become clear that expression levels of certain miRNAs are altered in cancer patients compared with healthy controls.5 Exosomal miRNA profiling could be thus a relatively low-invasive method to detect PDAC, or to monitor its progression or treatment efficacy. However, it also seems obvious that such a mix of miRNAs released from different cells cannot be easily interpreted.
In this review, we focus on miRNAs potential as biomarkers for early detection of PDAC. We summarize the clinical evidence of application of miRNAs in diagnostics and analytical challenges related to their use.
2. Characterization of PDAC – currently available diagnostic tools and treatment options
Pancreatic cancer is a lethal disease with an aggressive biology leading to early metastases in the liver and the gallbladder and low overall survival. The age of peak incidence of pancreatic cancer is between 60 and 80 years of age with male-to-female ratio of 1:1.13.6 The global incidence was 458,918 new cases in 2018, and the trend is increasing (there were 337,872 new cases in 2012) with an estimation of incidence for Europe in 2040 of 171,414 patients and the number of deaths reaching 168,489.7 Late diagnosis is resulting from nonspecific nature of the presenting symptoms, the lack of good biomarkers to identify patients at risk, and limitations of the imaging techniques as well as tissue sampling.8 Continuous efforts to improve diagnostic tools allowed for earlier diagnosis and treatment, which led to an increase in the overall 5-years survival rate to nearly 9%; however, it remains unsatisfactory.6,9
Symptoms of pancreatic cancer are often uncharacteristic and include abdominal pain, loss of appetite, early satiety and nausea. The most common form (90% of cases) of pancreatic cancer is pancreatic ductal adenocarcinoma (PDAC), stromal-rich tumor that develops within the exocrine compartment of the pancreatic gland.10 Because of lack of specific symptoms, except for jaundice, combined with its biological aggressiveness, patients are usually diagnosed late with PDAC, which is in 80–85% present as locally advanced (laPDAC) or distant metastatic disease (mPDAC) with limited therapeutic options, and only a minority of patients (15–20%) are eligible for potentially curative surgical resection.11,12
The systemic therapy has limited effectiveness, partly caused by molecular differences between various PDAC types – defined for example by Collisson E. et al.13 as: classical, quasi-mesenchymal, and exocrine-like, with various therapeutic responses and clinical outcomes. Furthermore, PDAC has extensive, both tumor-promoting and tumor-inhibiting, stromal involvement and Moffitt RA et al.14 investigated tumor- and stroma-specific PDAC gene expression. They have identified four prognostic subtypes of PDAC, being a combination of the two tumor-specific subtypes (classical or basal-like) and two stromal subtypes (normal or activated). The classical tumors with normal stroma subtypes had best prognosis and the basal-like with activated stroma subtypes had worst, although the classical subtype tumors were less responsive to adjuvant therapy compared to basal-like tumors.14 Moreover, many genomic alterations, contributing to PDAC progression were found in PDAC patients, the most frequently altered genes are KRAS (identified in 88% of 3594 PDAC samples), TP53 (74%), CDKN2A (44%), SMAD4 (22%), and CDKN2B (21%).15 The clinical use of this information remained limited until combined with transcriptomic studies, which allowed identification of PDAC subtypes with prognostic and biological relevance.16–18 For example, genetic and epigenetic sequencing revealed the existence of four molecular PDAC subtypes: squamous (31% incidence), pancreatic progenitor (19% incidence), immunogenic (29% incidence) and aberrantly differentiated endocrine exocrine (ADEX) (21% incidence), all having heterogeneous genetic characteristics and different biological behavior.12,19 However, future consensus subtyping is needed to establish the subtypes relevant for clinicians.
PDAC originates from the exocrine glands of the pancreas and the majority of cases are sporadic with lifestyle factors such as smoking, alcohol abuse and obesity, as well as type II diabetes being the known risk factors.20,21 CP is also a risk for PDAC as confirmed in a Danish epidemiological study with a hazard ratio of 6.9 to develop PDAC in patients with CP compared to controls.20,22,23 The key challenge is the differential diagnosis between mass-forming chronic pancreatitis (MFCP) and PDAC as they significantly differ in the management and prognosis, with dramatically worse outcome of the latter. Despite recent progress, the diagnostic imaging significantly overlaps and histopathologic assessment of tissue samples remains the ultimate differentiator between the two.20
The diagnostic as well as decision on the management of PDAC should be performed by a multi-specialist team, consisting of a gastroenterologist, surgeon, radiologist, pathologist and oncologist in high-level reference centers. Standard ultrasound of the abdominal cavity has limited value and a contrast-enhanced computed tomography (CECT) scan of the abdominal cavity and pelvis should be performed, in unclear cases the magnetic resonance imaging (MRI) might be helpful due to its sensitivity and specificity. Positron emission tomography (PET) does not allow to differentiate between PDAC and CP but might be helpful to detect distant metastases or cancer recurrence. The recommended diagnostic method remains the endoscopic ultrasound (EUS) with fine-needle aspiration biopsy. Although the CA19-9 antigen is a recognized prognostic and predictive PDAC biomarker, it has limited value in early diagnostics or screening; however, annual EUS scans and serum CA19-9 are recommended for high-risk patients with hereditary pancreatitis or family history of pancreatic cancer.21
The most effective treatment in early-diagnosed PDAC is pancreatectomy; however, the 5-year survival rate of early stage resected PDAC remains limited to 20–25%.24 The current treatment options for metastatic PDAC include FOLFIRINOX (5-fluorouracil, leucovorin, irinotecan, and oxaliplatin), modified (m)FOLFIRINOX (without bolus fluorouracil) or nab-paclitaxel and gemcitabine in patients with good ECOG performance status (PS 0–1) or gemcitabine mono or combo therapy for patients with ECOG PS 2–3.25 Other therapeutic strategies, esp. immunotherapy, are being investigated and multiple clinical trials are ongoing.
Patients with advanced PDAC should undergo both germline and somatic profiling first and targeted therapy might prove beneficial for some.26 Biomarker-driven strategy, such as platinum-based therapy combined with PARP inhibitor in patients with BRCA-mutated PDAC (4.6–8% incidence) and immune checkpoint inhibitors in patients with microsatellite instability (MSI) may prove beneficial. The pathologic and transcriptomic profiling allows to distinguish between a classical and basal type of PDAC, with increased treatment resistance of the latter.26
Early detection of PDAC would improve survival rate; however, the prevalence of PDAC is relatively low what makes screening within the general population impractical. The crucial task is thus to identify in the general population individuals at risk who would later benefit from surveillance programs, optimally using noninvasive biomarkers complementing diagnostic imaging (CECT, MRI and EUS, of which the latter is considered the most efficient method in early detection).27 The groups at risk include families with germline mutations (BRCA2, BRCA1, PALB2, CDKN2A, ATM, TP53 and mismatch repair genes MLH1, MSH2, MSH6), patients with a history of pancreatitis, with mucinous pancreatic cysts, and elderly patients with new-onset diabetes. The currently investigated biomarker assays for PDAC (proteins, autoantibodies, circulating DNA, miRNAs, methylated DNA, exosomes) undergo testing in a diagnostic setting, but have not yet proved useful for continuous surveillance in asymptomatic individuals.28 Nevertheless, constant efforts are being made worldwide to establish a biomarker-based diagnostic procedure.
3. Recent scientific reports regarding miRNA as biomarkers in PDAC
Micro RNAs (miRNAs) belong to non-coding RNAs (ncRNAs), which, although not translated into proteins, are crucial for DNA replication, translation, RNA splicing and epigenetic regulation. They can be classified based on their localization as cytoplasmic and nuclear or as small (<200 base pairs (bp)) and long (>200 bp) ncRNAs based on nucleotide length. The family of ncRNAs includes miRNAs, PIWI-interacting RNAs (piRNAs), small interfering RNAs (siRNAs), small nucleolar RNAs (snoRNAs), tRNA-derived stress-induced RNAs (tiRNAs), enhancer non-coding RNAs (eRNAs), circular RNAs (circRNAs), and long non-coding RNAs (lncRNAs). ncRNAs were found to regulate tumor progression by affecting cell proliferation, invasion, metastasis, angiogenesis and chemoresistance and ncRNAs with tumor-promoting functions are: ncRNAs (HOTAIR, HOTTIP, MALAT1), lncRNA (H19, PVT1), circ-RNA (ciRS-7, circ-0030235, circ-RNA_100782, circ-LDLRAD3, circ-0007534, circRHOT1, circZMYM2, circ-IARS, PDE8A), miR-21, miR-155, miR-221/222, miR-196b, miR-10a. On the contrary, some ncRNAs, such as GAS5, MEG3, and lncRNA ENST00000480739, has_circ_0001649, miR-34a, miR-100, miR-217, and miR-143 inhibit cell proliferation and invasion of PDAC (Table 1).29
Table 1.
Non-coding RNAs and PDAC tumorigenesis
| PDAC promoting | Aliases for gene (genecards.org/) | Gene location (genenames.org/) | Increased expression promoting other cancers (malacards.org/) |
|---|---|---|---|
| MicroRNA | |||
| miR-21 | MicroRNA 21 | 17q23.1 | Multiple cancers |
| miR-155 | MicroRNA 155 | 21q21.3 | Multiple cancers |
| miR-221/222 | MicroRNA 221/222 | Xp11.3/ Xp11.3 | Multiple cancers |
| miR-196b | MicroRNA 196b | 7p15.2 | Multiple cancers |
| miR-10a | MicroRNA 10a | 17q21.32 | Multiple cancers |
| Circular RNA | |||
| ciRS-7 | CDR1 Antisense RNA | Xq27.1 | Bladder cancer |
| circ-LDLRAD3 | Low Density Lipoprotein Receptor Class A Domain Containing 3 | 11p13 | No data |
| RHOT1 | Ras Homolog Family Member T1 | 17q11.2 | No data |
| ZMYM2 | Zinc Finger MYM-Type Containing 2 | 13q12.11 | Lymphoma, leukemia, myelofibrosis |
| IARS | Isoleucine TRNA Ligase 1, Cytoplasmic | 9q22.31 | Breast ductal adenocarcinoma |
| PDE8A | Phosphodiesterase 8A | 15q25.3 | Melanoma |
| Long non-coding RNA | |||
| H19 | H19 Imprinted Maternally Expressed Transcript | 11p15.5 | Wilms tumor 2, embryonal carcinoma, choriocarcinoma |
| HOTAIR | HOX Transcript antisense RNA | 12q13.13 | Gastrointestinal cancers, triple negative breast cancer |
| HOTTIP | HOXA Transcript At The Distal Tip | 7p15.2 | Glioma, small cell lung cancer |
| MALAT1 | Metastasis Associated In Lung Adenocarcinoma Transcript 1 | 11q13.1 | Acute monocytic leukemia, ovarian endometrial cancer |
| PVT1 | Plasmacytoma Variant Translocation 1 | 8q24.21 | Plasmacytoma, Hodgkin lymphoma, B-cell lymphoma |
| PDAC inhibiting | Aliases for Gene(genecards.org) | Gene location (genenames.org/) | Decreased expression in tumorigenesis of other cancers (malacards.org/) |
| MicroRNA | |||
| miR-34a | MicroRNA 34a | 1p36.22 | Multiple cancers |
| miR-100 | MicroRNA 100 | 11q24.1 | Multiple cancers |
| miR-217 | MicroRNA 217 | 2p16.1 | Chronic lymphocytic leukemia, nasopharyngeal carcinoma |
| miR-143 | MicroRNA 143 | 5q32 | Multiple cancers, incl. PDAC |
| Long non-coding RNA | |||
| GAS5 | Growth Arrest Specific 5 | 1q25.1 | Multiple cancers |
| MEG3 | Maternally Expressed 3 | 14q32.2 | Multiple cancers |
| ENST00000480739 | ribosomal protein L13a pseudogene 23 | 12q13.3 | Osteosarcoma |
miRNAs, discovered in 1993,30 are single-stranded RNAs with 19–25 nucleotides, transcribed and processed into mature miRNAs with 21–23 nucleotides. miRNAs do not encode proteins but regulate approximately 50% of protein-coding genes.31 miRNA molecules are involved in crucial processes for the development and functioning of the body, such as cell division, differentiation and programmed cell death, and blood vessel formation. Due to the ability to regulate gene expression at the post-transcriptional level, miRNAs ultimately affect the number of individual proteins in the body and may be characterized as “guardians” which control the proper course of processes within the cell. Genome damage (translocations, deletions, amplifications, integration of foreign DNA) during oncogenesis or tumorigenesis affects not only the expression of protein genes that are tumor suppressor genes, but also expression of miRNAs. In consequence, a change in the expression level of miRNAs is observed in various types of cancer, e.g., ovarian, breast, liver as well as pancreas. Importantly, normal and cancerous tissues can be differentiated based on the level of miRNAs.
Multiple studies demonstrated that miRNAs are not only highly correlated with tumorigenesis and progression, but are also related to drug resistance, tumor metastasis, angiogenesis, cancer relapse, and poor clinical outcomes.32,33 In PDAC, miRNAs are considered to be responsible for apoptosis escape, proliferation, epithelial mesenchymal transition (EMT; a process by which epithelial cells lose their cell polarity and cell–cell adhesion, and gain migratory and invasive properties to become mesenchymal cells; EMT), metastasis, invasion, and drug resistance, and consequently they might be used as potential biomarkers for PDAC diagnosis.34
EMT is crucial in formation of metastasis in PDAC and miRNAs, particularly miR-10, miR-21 and members of the miR-200 family are closely involved in EMT. Nakamura et al. found relative levels of exosomal miRNAs, ex-miR-21 and ex-miR-155 significantly higher in pancreatic juice of PDAC patients compared to CP patients, with no significant difference in relative levels of miR-21 and miR-155, respectively.35 Hence, the presence of exosomal miRNAs in disease opens up the possibility of using these miRNAs as biomarkers. Exosomal miRNA profiling could be a noninvasive method to detect disease, or a way to monitor progression or treatment efficacy.
Schultz et al.36 found a difference in the miRNA expression in whole blood between patients with pancreatic cancer, patients with CP, and healthy participants and proposed relevant diagnostic panels of miRNAs in pancreatic cancer. Based on microRNAs that were significant in the training cohort, they suggested two diagnostic indices. Index I consisted of four selected microRNAs and was designed to be robust to technical variation as well as contained no model parameters. On the contrary, index II included all significant microRNAs from a multivariable model and corresponded to the upper limit in terms of training and was thus potentially overfitted. These diagnostic panels included four miRNAs in index I (miR-145, miR-150, miR-223, miR-636) and 10 in index II (miR-26b, miR-34a, miR-122, miR-126*, miR-145, miR-150, miR-223, miR-505, miR-636, miR-885.5p) with the potential to distinguish patients with PDAC from healthy controls (Table 2). Here, further research is needed, especially in relation to the levels of serum CA 19–9.36
Table 2.
Index I and II (whole blood levels) – two diagnostic microRNA panels to distinguish patients with PDAC from healthy controls.36.
| Index I | Index II |
|---|---|
| miR-145 | miR-26b |
| miR-150 | miR-34a |
| miR-223 | miR-122 |
| miR-636 | miR-126* |
| miR-145 | |
| miR-150 | |
| miR-223 | |
| miR-505 | |
| miR-636 | |
| miR-885.5p |
Zhu et al.37 found 165 deregulated mature miRNAs in next-generation sequencing (NGS)-analyzed plasma samples from Han Chinese population of PDAC patients, with 75 miRNAs up- and 90 miRNAs down-regulated compared with healthy individuals. The two significantly up-regulated miRNAs were miR-182-5p, known to promote tumor cell proliferation and miR-4732-5p, which interferes with tumor suppressive protein p53. The two significantly down-regulated miRNAs were miR-139-5p and miR-23b-3p. Further prospective investigations in larger cohorts of all four above-mentioned miRNAs circulating in plasma are critically needed.
PDAC is often characterized with elevated levels of pancreatic stellate cells (PSCs), which release exosomes containing high levels of circulating miRNA‑21 (miR‑21). Ma Q. et al.38 evaluated the effects of exosomal miR‑21 on the migratory ability of PDAC cells in order to characterize the underlying molecular mechanisms. Weighted gene correlation network and The Cancer Genome Atlas database analysis revealed that high miR‑21 levels are strongly connected with a poor prognosis in patients with PDAC. Moreover, it has been stated that Ras/ERK signaling pathway may be a potential target of miR‑21. In vitro, PDAC cells were demonstrated to internalize the PSC-derived exosome, resulting in high miR‑21 levels, which subsequently promoted cell migration, induced EMT and increased matrix metalloproteinase‑2/9 activity. In addition, exosomal miR‑21 increased the levels of ERK1/2 and Akt phosphorylation in PDAC cells. Collectively, these results suggested that PSC‑derived exosomal miR‑21 may promote PDAC cell migration and EMT and enhance Ras/ERK signaling activity. Thus, miR‑21 may contribute to the poor prognosis in patients with pancreatic cancer and represent a new diagnosis and treatment target.
The miRNAs, which could be considered as the potential biomarkers for early PDAC detection due to positive initial validation in patients’ material have been summarized in Table 3.37
Table 3.
miRNAs initially tested in human samples as potential biomarkers for PDAC
| Samples tested | Significantly up-regulated | No significant difference | Significantly down-regulated | Potential biomarker for PDAC |
|---|---|---|---|---|
| PDAC pancreatic juice vs. chronic pancreatitis35 | Ex-miR-21 | miR-21 | Ex-miR-21 | |
| Ex-miR-155 | miR-155 | Ex-miR-155 | ||
| Plasma samples from Chinese PDAC patients vs healthy subjects37 | miR-182-5p | miR-139-5p | miR-182-5p | |
| miR-4732-5p | miR-23b-3p | miR-4732-5p (late-stage) miR-23b3p |
Pre-clinical evidence showed that miR-486-5p, which expression is a frequent molecular event in human malignancies, could promote the proliferation of pancreatic cancer cells.39 miR-210, which regulates mitochondrial metabolism and oxidative stress, has been strongly linked with the hypoxia pathway and is upregulated in response to hypoxia-inducible factors which are important transcription factors that regulate oxygen consumption and morphological changes in response to varying oxygen concentrations.
miR-210 is overexpressed in tumors; it is also stated to promote invasion and EMT.40 Yang Z. et al.41 investigated whether exosomes derived from gemcitabine-resistant pancreatic cancer stem cells mediate cell–cell communication between cells that are sensitive or resistant to gemcitabine and, by doing so, regulate drug resistance. Relevant miRNAs associated with gemcitabine resistance were identified and the role of miR-210 in conferring drug resistance was examined in vitro and in vivo. They concluded that exosomes derived from cancer stem cells of gemcitabine-resistant pancreatic cancer cells enhance drug resistance by delivering miR-210.
miR-1246 is considered an oncomiR, i.e., miRNA that is associated with cancer, in various cancer types. However, the origin and biogenesis of miR-1246 remained controversial, with misunderstandings concerning its detection and biological functioning. Consequently, using next-generation small RNA sequencing, clustered regularly interspaced short palindromic repeats Cas-9 (CRISPR-Cas9 knockout, siRNA knockdown and the poly-A tailing SYBR qRT-PCR) Y.-F. Xu et al.42 examined the biogenesis of exosomal miR-1246 in human cancer cell model systems. They found that miR-1246 is highly enriched in exosomes derived from PANC-1 cells – majority of the miR-1246 sequences detected in both PANC-1 cells and PANC-1 exosomes contained the GAGA, and none of them contained AGTG, indicating that the miR-1246 sequence detected is derived from U2 small nuclear RNA (RNU2-1) and not from the precursor miR-1246. Knockdown of RNaseIII enzymes DROSHA and DICER which mediate miRNA maturation, did not reduce exosomal miR-1246 levels, indicating that exosomal miR-1246 is generated in a Drosha- and Dicer-independent manner. Direct digestion of cellular lysate by RNase A and knockdown of the RNU2-1 binding protein SmB/B’ demonstrated that exosomal miR-1246 is a RNU2-1 degradation product. Furthermore, the GCAG motif present in the RUN2-1 transcript was shown to mediate miR-1246 enrichment in human PDAC cells and cancer exosomes – the same was true in other human cells, including MIA-PaCa-2, and hTERT-HPNE lines. Y.-F. Xu et al.42 concluded that exosome miR-1246 is derived from RNU2-1 degradation through a non-canonical miRNA biogenesis process. These findings revealed the origin of an oncomiR in human PDAC cells, providing guidance in understanding miR-1246 detection and biological function as well as potential use as a biomarker. However, clinical evidence is still needed.
A recent study by Khan I.A. et al.43 assessed a panel of miRNAs for their ability to differentiate PDAC from CP, a benign inflammatory condition of the pancreas. Next-generation sequencing was performed to identify miRNAs present in 60 formalin-fixed, paraffin-embedded tissue samples (27 PDAC, 23 CP and 10 normal pancreatic tissues). Four up-regulated (miR-215-5p, miR-122-5p, miR-192-5p, and miR-181a-2-3p) and four down-regulated miRNAs (miR-30b-5p, miR-216b-5p, miR-320b, and miR-214-5p) in PDAC compared to CP were selected based on next-generation sequencing results. Next, the levels of these eight differentially expressed miRNAs were measured by qRT-PCR in 125 serum samples (50 PDAC, 50 CP, and 25 healthy controls (HC)). The results showed significant upregulation of miR-215-5p, miR-122-5p, and miR-192-5p in PDAC serum samples. In contrast, levels of miR-30b-5p and miR-320b were significantly lower in PDAC as compared to CP and HC. Receiver Operator Characteristic analysis showed that these five miRNAs can distinguish PDAC from both CP and HC. Hence, it was concluded that this panel could serve as a noninvasive biomarker for the early detection of PDAC.
A study by Wang C. et al.44 investigated the potential role of serum exosomal miRNA in detection of PDAC and analyzed the correlation between the levels of exosome miRNA and the tumor biology.
It was concluded that the expression of 11 miRNAs showed same trend between PDAC and benign pancreatic lesions, and between PDAC and HC. Six of them were upregulated (miR-203b-5p, miR-342-5p, miR-337-5p, miR-149-5p, miR-877-5p, miR-203a-3p), and five were downregulated (miR-1226-3p, miR-3182, miR-625-3p, miR-624-5p, miR-664a-5p). miR-1226-3p was selected as the candidate exosomal biomarker for the PDAC detection. The expression of serum exosomal miRNA-1226-3p was downregulated in PDACs compared to the benign pancreatic lesions (p = .025). Moreover, miR-1226-3p had acceptable performance in predicting PDAC. Exosomal miRNA-1226-3p level in PDAC with invasion or metastases was lower than that without invasion or metastases (p = .028). Transfection of miRNA-1226-3p significantly inhibited the proliferation of PANC-1 and BXP-3 cells, stimulated cell apoptosis and inhibited cell migration. To conclude, serum exosomal miRNA-1226-3p may be regarded as a potential biomarker in diagnosing and predicting the tumor invasion or metastases of PDAC.
Miao H. et al.45 identified a novel host factor, namely the lncRNA TP73-AS1, as overexpressed in PDAC tissues compared to adjacent healthy tissue samples. The overexpression of TP-73-AS1 was found to correlate with both PDAC stage and lymph node metastasis. To reveal its role in PDAC, they targeted TP73-AS1 using lnRNA inhibitors in a range of PC cell lines. They found that the inhibition of TP73-AS1 led to a loss of MMP14 expression in PC cells and significantly inhibited their migratory and invasive capacity. No effects of TP73-AS1 on cell survival or proliferation were observed. Mechanistically, they found that TP73-AS1 suppressed the expression of the known oncogenic miR-200a. Taken together, these data highlighted the prognostic potential of TP73-AS1 for PC patients and emphasized it as a potential anti-PDAC therapeutic target.
Finally, it has already been stated that four potential miRNAs may affect the progression of PDAC by targeting MET via the PI3K/AKT signaling pathway. Yao LC et al.46 aimed to identify the potential biomarkers of PDAC carcinogenesis and progression using three microarray datasets, GSE15471, GSE16515 and GSE28735, which were downloaded from the Gene Expression Omnibus database. The datasets were analyzed to screen out differentially expressed genes (DEGs) in PDAC tissues and adjacent normal tissues. A total of 143 DEGs were identified, including 132 upregulated genes and 11 downregulated genes. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes functional and signaling pathway enrichment analyses were performed on the DEGs, and the Search Tool for the Retrieval of Interacting Genes/Proteins database was used to construct a protein–protein interaction network. The main functions of DEGs include extracellular matrix degradation, and regulation of matrix metalloproteinase activity and the PI3K-Akt signaling pathway. The five hub genes were subsequently screened using Cytoscape software, and survival analysis demonstrated that abnormal expression levels of the hub genes were associated with poor disease-free survival and overall survival. Biological experiments were performed to confirm whether EMT factors promote the proliferation, migration and invasion of PDAC cells via the PI3K/AKT signaling pathway. In addition, six EMT-targeted miRNAs were identified, four of which had conserved binding sites with EMT. Based on the signaling pathway enrichment analysis of these miRNAs, it is suggested that they can affect the progression of PDAC by targeting EMT via the PI3K/AKT signaling pathway. In conclusion, it was stated that the hub genes and miRNAs that were identified in the study contribute to the molecular mechanisms of PDAC carcinogenesis and progression and may be regarded as candidate biomarkers for early diagnosis and treatment of patients with PDAC.
4. MiRNA as diagnostic tools – limitations
Growing interest in miRNAs as both biomarkers of disease and therapeutic targets drives the need for fast and effective methods for miRNA profiling. Unfortunately, diverse studies have demonstrated the validation of miRNA biomarkers to be unsuccessful, which could be explained by discrepancies in methodology, the lack of standard methods for normalization, miRNA processing and the inability to differentiate among closely related miRNAs. The crucial steps while profiling miRNA that might be challenging are given in Figure 1.
Figure 1.

Steps to miRNA profiling
4.1. Sample preparation
High-quality miRNA can be extracted from both cells and tissue. However, studying miRNAs can be a challenge because of their small size, which requires specialized and dedicated tools for analysis. The cell heterogeneity of the tissue is also an important consideration because many miRNAs are tissue-specific. For this reason, using microdissection methods or other cell-targeting approaches are advisable. Furthermore, miRNAs are often released from blood cells (plasma, red blood cells, white blood cells, and platelets) at the time of storage period, which may result in false outcomes. Finally, the white blood cells and red blood cell hemolysis can also adversely affect the quality and quantity of miRNA extracted.
4.2. miRNA extraction
For some sample types, miRNA extraction methods may require optimization. For example, in their native state, endogenous blood plasma miRNAs are protected from RNase; however, extracted miRNAs that are spiked into plasma degrade quickly. For this reason, it is important that any technique for miRNA extraction from blood plasma should be quick and completely inactivate the RNase activity.
4.3. miRNA quality and quantity control
Evaluating the quality of extracted RNA is important for both the quality and reproducibility of miRNA profiling results. Because most profiling methods use total RNA, it is not usually necessary to quantitate the amount of miRNA present in extracted RNA. Accordingly, the protocols should be standardized for sample collection, storage and processing, e.g., often used anticoagulants while collecting plasma are ethylenediaminetetraacetic acid, citrate or heparin. However, some anticoagulants (e.g. heparin) decrease or even stop the action of the reverse transcriptase as well as DNA polymerase enzymes which are crucial for quantitative studies.47
Importantly, the amount of circulating miRNAs may be affected by many factors such as: gender, age, lifestyle habits (smoking, drugs, diet).48 Knowing how each factor influences miRNAs may improve the characterization and measurement of diagnostic or prognostic values of miRNAs in the specific environment.49
4.4. miRNA qualitive and quantitative measurements
Now that many miRNA sequences are known (cataloged in the miRBase Sequence Database), one of the most common next steps is the analysis of miRNA expression levels between different tissues, developmental stages, or disease states. miRNA expression levels can be studied by several methods: quantitative reverse transcription polymerase chain reaction (qRT-PCR), droplet digital PCR (ddPCR), microarrays and miRNA sequencing (miRNA-seq). miRNA expression by qRT-PCR enables reliable detection and measurement of products generated during each cycle of PCR process and can be performed in an absolute or relative method. However, as there is no invariant validator, relative quantification of miRNA in blood serum or plasma has been faced with severe difficulties.50 Conventionally, relative expression of miRNAs was mainly based on small nuclear (e.g., U6 snRNA) or small nucleolar RNAs (e.g., snoRNA U44, also known as SNORD44) for normalization. However, in a study by Masè et al.50 analysis of the five frequently used reference genes for miRNA research (5S rRNA, U6 snRNA, hsa-miR-16-5p, SNORD48), exposed insufficient and limited performance of widely used normalizers such as U6 snRNAs. Approved and certified miRNA reference genes miR-16, miRs-10b, miR-30a, miR-30d, miR-103, miR-148b, miR-191 and miR-192 have been rumored to be regulated in a different manner in serum compared to plasma of breast cancer patients in several studies.51 Accordingly, it is critical to verify and characterize the expression stability of presumed invariant in each pilot test.50 Of note, when there is no consensus on endogenous controls, it is advisable to perform the absolute quantification of miRNAs.47 miRNAs must be reverse transcribed to cDNA before PCR can be performed, and this step is associated with inherent inefficiencies. ddPCR has been reported to have meaningful potential as a technique to calculate miRNA copy numbers at limiting dilutions without bias.51 However, profiling of hundreds of differentially expressed miRNAs from blood serum or plasma when RNA quantity is limited may cause some technical trouble. It seems that the pre-amplification of cDNA, which is essentially a highly multiplexed PCR reaction performed for a limited number of cycles using the same primer sets that will be used in the downstream qPCR reaction, may address this problem. Still, it has some drawbacks. Microarrays have decreased sensitivity and specificity compared with qRT-PCR and are unreliable at low input. Presently, miRNA-seq is the most appropriate platform for miRNA discovery, which is immensely sensitive and contributes to relative expression data for small RNAs in a sample with a greater dynamic range compared with miRNA microarrays.47
4.5. Data analysis
Analysis of miRNA profiling data is divided into several steps: processing, quality assessment, normalization and differential expression calculations. The best approach to these steps should be dependent on the miRNA profiling platform and the goal of the experiment.
4.6. Other limitations
One of the crucial determinants, which may have an influence on using miRNAs as a diagnostic tool in clinical settings, is correlated with detection of miRNAs in patients with different tumor types. One such example is miR-21, which elevated serum levels have already been found not only in PDAC but also in patients diagnosed with colorectal, lung, breast, prostate, liver, esophageal and endometrial cancers.52 Another problem is that these results are not coherent even among related studies of the same condition.51 Of note, down-regulation of miRNA in tumors may be associated with genomic and epigenomic alterations, but in circulation, this could happen only if the tumor affected the expression of the miRNAs in other cells in an adverse manner or reduced the stability of miRNAs in circulation. This implies that decreased serum levels of circulating miRNAs may be identified with nonspecific responses to the presence of cancer. Additionally, the total lump or mass of tumor tissue carried by an individual with cancer cannot be related with the up-regulated levels of circulating miRNAs. Of note, only in a small number of tumors (mainly late-stage) patients may be characterized with upregulated expression of miRNA in the blood due to dilution effects of blood. Therefore, it is more probable that upregulation of circulating miRNAs contributes to the response(s) to the presence of cancer rather than its absence.51
5. Conclusion
Identification of biomarkers that enable the prediction of treatment efficacy and responses to therapeutics, and which will allow the selection of an appropriate, “tailored” therapy, is not an easy task. An ideal biomarker should be readily available by noninvasive procedures, inexpensive to measure, and very specific and sensitive in the diagnosis of the disease. In addition, an ideal biomarker must not be too susceptible to technical variables and other pathological conditions unrelated to the studied disease but occurring in patients. The most important issue, however, is to reliably identify the disease, preferably before the appearance of its clinical symptoms. The development of PDAC is associated with the accumulation of a number of somatic mutations in the neoplastic genome. These mutations condition the uncontrolled multiplication of tumor cells, which results in clonal expansion leading to tumor progression. Some of the functional somatic mutations in the neoplastic genome cause loss of function of suppressor genes, others are activating, leading to the acquisition of new functions and activation of oncogenes. Hence, we believe that an analysis of somatic mutations in miRNA genes and miRNA biogenesis genes should preferably be the focus before embarking on the quest for the effective biomarkers. Molecular markers open up new diagnostic possibilities and seem easy to interpret, and with circulating miRNAs as relatively noninvasive yet valid biomarkers, there is an opportunity for a screening program in people at higher risk of PDAC. Thus, these people will finally have a chance to receive appropriate medical care at an early-stage disease.
The use of miRNAs as biomarkers has not been validated yet. Most of the studies presented above are preliminary and experimental, in small groups of patients, which do not meet the criteria of validation analyses.
However, certain miRNAs are master regulators of tumorigenesis in some cancers (breast cancer, colorectal cancer, kidney cancer, testicular cancer, etc.), and thus represent powerful therapeutic targets. Moreover, many miRNAs are aberrantly expressed in PDAC and may influence tumor progression. Accumulating studies suggest that multiple miRNAs are actively involved in the PDAC metastasis process. Thus, we aimed to introduce the role of miRNAs in multi-steps of PDAC metastasis, including cancer cell invasion, intravasation, circulation, extravasation, colonization, angiogenesis, and EMT. Of note, several clinical studies are currently ongoing, including a large (5000 patients), prospective, observational, longitudinal study BIOmarkers in Patients With Pancreatic Cancer (BIOPAC), NCT03311776, which has already published initial results with Biomarkers Consensus Signature, although miRNAs have not yet been included.53
What is more, miRNAs also represent a promising target for therapeutic intervention. Potentially, in future, the analysis of miRNAs will be an important stage in diagnostics and an introduction to treatment consistent with the endotype of a given disease entity in personalized medicine.
In order to find validated cancer biomarkers, several important considerations need to be taken in profiling and analyzing circulating miRNAs (Figure 2). Further extensive research in this field is yet warranted to identify an optimal miRNA, or a set of miRNAs, to enable an easy and early diagnosis of PDAC, a difficult to treat disease with low survival rates.
Figure 2.
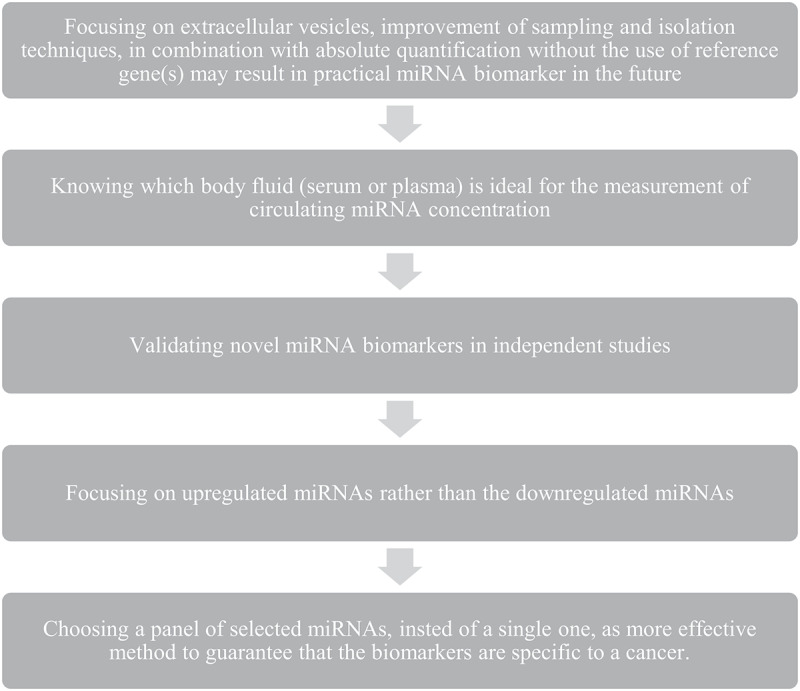
Important considerations necessary in profiling and analyzing circulating miRNAs to find validated cancer biomarkers
Acknowledgments
TM participates in the program “Doktorat wdrożeniowy” from the Polish Ministry of Science and Higher Education.
Funding Statement
This research was funded by the Medical University of Lodz (#503/1-156-04/503-11-001 to JF).
Disclosure statement
No potential conflicts of interest were disclosed.
Author contributions
AT, TM and JF provided the overall concept and framework of the manuscript; AT and TM researched and identified appropriate articles, and wrote the manuscript; AT, TM, EMP and JF revised the manuscript. All authors contributed to the writing of the manuscript and approved the final version.
References
- 1.Gall TMH, Gerrard G, Frampton AE, Castellano L, Ahmad R, Habib N, Spalding D, Pai M, Foroni L, Jiao LR.. Can we predict long-term survival in resectable pancreatic ductal adenocarcinoma? Oncotarget. 2019Jan22;10(7):696–706. doi: 10.18632/oncotarget.26511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kunovsky L, Tesarikova P, Kala Z, Kroupa R, Kysela P, Dolina J, Trna J. The use of biomarkers in early diagnostics of pancreatic cancer. Can J Gastroenterol Hepatol 2018Aug14;2018:5389820. doi: 10.1155/2018/5389820.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azizian A, Rühlmann F, Krause T, Bernhardt M, Jo P, König A, Kleiß M, Leha A, Ghadimi M, Gaedcke J. CA19-9 for detecting recurrence of pancreatic cancer. Sci Rep. 2020Jan28;10(1):1332. doi: 10.1038/s41598-020-57930-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vidigal JA, Ventura A. The biological functions of miRNAs: lessons from in vivo studies. Trends Cell Biol. 2015Mar;25(3):137–147. doi: 10.1016/j.tcb.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang J, Chen J, Sen S. MicroRNA as biomarkers and diagnostics. J Cell Physiol. 2016Jan;231(1):25–30. doi: 10.1002/jcp.25056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hawksworth G, Hales J, Martinez F, Hynes A, Hamilton A, Fernandez V. 2019. Pancreatic cancer trends in Europe: epidemiology and risk factors. Med Stud/Studia Medyczne. 35(2):164–171. doi: 10.5114/ms.2019.86336. [DOI] [Google Scholar]
- 7.Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin DM, Piñeros M, Znaor A, Bray F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019Apr15;144(8):1941–1953. doi: 10.1002/ijc.31937. [DOI] [PubMed] [Google Scholar]
- 8.Lee B, Gibbs P. Inflammation, biomarkers and immuno-oncology pathways in pancreatic cancer. J Pers Med. 2019Apr26;9(2):20. doi: 10.3390/jpm9020020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018Nov;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 10.Haeberle L, Esposito I. Pathology of pancreatic cancer. Transl Gastroenterol Hepatol. 2019Jun27;4:50. doi: 10.21037/tgh.2019.06.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kleeff J, Korc M, Apte M, La Vecchia C, Johnson CD, Biankin AV, Neale RE, Tempero M, Tuveson, Hruban RH, et al. Pancreatic cancer. Nat Rev Dis Primers. 2016Apr21;2:16022. doi: 10.1038/nrdp.2016.22. [DOI] [PubMed] [Google Scholar]
- 12.Lai E, Puzzoni M, Ziranu P, Pretta A, Impera V, Mariani S, Liscia N, Soro P, Musio F, Persano M, et al. New therapeutic targets in pancreatic cancer. Cancer Treat Rev. 2019Dec;81:101926. doi: 10.1016/j.ctrv.2019.101926. [DOI] [PubMed] [Google Scholar]
- 13.Collisson EA, Sadanandam A, Olson P, Gibb WJ, Truitt M, Gu S, Cooc J, Weinkle J, Kim GE, Jakkula L, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med. 2011Apr;17(4):500–503. doi: 10.1038/nm.2344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moffitt RA, Marayati R, Flate EL, Volmar KE, Loeza SG, Hoadley KA, Rashid NU, Williams LA, Eaton SC, Chung AH, et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet. 2015Oct;47(10):1168–1178. doi: 10.1038/ng.3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singhi AD, George B, Greenbowe JR, Chung J, Suh J, Maitra A, Klempner SJ, Hendifar A, Milind JM, Golan T, et al. Real-time targeted genome profile analysis of pancreatic ductal adenocarcinomas identifies genetic alterations that might be targeted with existing drugs or used as biomarkers. Gastroenterology. 2019Jun;156(8):2242–2253.e4. doi: 10.1053/j.gastro.2019.02.037 [DOI] [PubMed] [Google Scholar]
- 16.Connor AA, Denroche RE, Jang GH, Timms L, Kalimuthu SN, Selander I, McPherson T, Wilson GW, Chan-Seng-Yue MA, Borozan I, et al. Association of distinct mutational signatures with correlates of increased immune activity in pancreatic ductal adenocarcinoma. JAMA Oncol. 2017Jun13(6):774–783. doi: 10.1001/jamaoncol.2016.3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P, Johns AL, Miller D, Nones K, Quek K, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015Feb26518(7540):495–501. doi: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Large TYS, Bijlsma MF, Kazemier G, van Laarhoven HWM, Giovannetti E, Jimenez CR. Key biological processes driving metastatic spread of pancreatic cancer as identified by multi-omics studies. Semin Cancer Biol. 2017Jun;44:153–169. doi: 10.1016/j.semcancer.2017.03.008. [DOI] [PubMed] [Google Scholar]
- 19.Torres C, Grippo PJ. Pancreatic cancer subtypes: a roadmap for precision medicine. Ann Med. 2018Jun;50(4):277–287. doi: 10.1080/07853890.2018.1453168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elsherif SB, Virarkar M, Javadi S, Ibarra-Rovira JJ, Tamm EP, Bhosale PR. Pancreatitis and PDAC: association and differentiation. Abdom Radiol (NY). 2020May;45(5):1324–1337. doi: 10.1007/s00261-019-02292-w. [DOI] [PubMed] [Google Scholar]
- 21.Winter K, Talar-Wojnarowska R, Dąbrowski A, Degowska M, Durlik M, Gąsiorowska A, Głuszek S, Jurkowska G, Kaczka A, Lampe P, et al. 2019. Diagnostic and therapeutic recommendations in pancreatic ductal adenocarcinoma. Recommendations of the Working Group of the Polish Pancreatic Club. Prz Gastroenterol. 14(1):1–18. doi: 10.5114/pg.2019.83422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klöppel G, Adsay NV. Chronic pancreatitis and the differential diagnosis versus pancreatic cancer. Arch Pathol Lab Med. 2009Mar;133(3):382–387. doi: 10.1043/1543-2165-133.3.382. [DOI] [PubMed] [Google Scholar]
- 23.Lowenfels AB, Maisonneuve P, Cavallini G, Ammann RW, Lankisch PG, Andersen JR, Dimagno EP, Andrén-Sandberg A, Domellöf L. Pancreatitis and the risk of pancreatic cancer. International Pancreatitis Study Group. N Engl J Med. 1993May20;328(20):1433–1437. doi: 10.1056/NEJM199305203282001. [DOI] [PubMed] [Google Scholar]
- 24.Neoptolemos JP, Stocken DD, Bassi C, Ghaneh P, Cunningham D, Goldstein D, Padbury R, Moore MJ, Gallinger S, Mariette C, et al. European Study Group for Pancreatic Cancer. Adjuvant chemotherapy with fluorouracil plus folinic acid vs gemcitabine following pancreatic cancer resection: a randomized controlled trial. JAMA. 2010Sep8304(10):1073–1081. doi: 10.1001/jama.2010.1275. [DOI] [PubMed] [Google Scholar]
- 25.Singh RR, O’Reilly EM. New treatment strategies for metastatic pancreatic ductal adenocarcinoma. Drugs. 2020May;80(7):647–669. doi: 10.1007/s40265-020-01304-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh RR, Goldberg J, Varghese AM, Yu KH, Park W. Genomic profiling in pancreatic ductal adenocarcinoma and a pathway towards therapy individualization: a scoping review. Cancer Treat Rev. 2019;27–38. doi: 10.1016/j.ctrv.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Canto MI, Hruban RH, Fishman EK, Kamel IR, Schulick R, Zhang Z, Topazian M, Takahashi N, Fletcher J, Petersen G, et al. American Cancer of the Pancreas Screening (CAPS) Consortium. Frequent detection of pancreatic lesions in asymptomatic high-risk individuals. Gastroenterology. 2012Apr;142(4):796–804;quiz e14-5. doi: 10.1053/j.gastro.2012.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singhi AD, Koay EJ, Chari ST, Maitra A. Early detection of pancreatic cancer: opportunities and challenges. Gastroenterology. 2019May;156(7):2024–2040. doi: 10.1053/j.gastro.2019.01.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gong R, Jiang Y. Non-coding RNAs in pancreatic ductal adenocarcinoma. Front Oncol. 2020Mar17;10:309. doi: 10.3389/fonc.2020.00309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993Dec3;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 31.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010Sep;11(9):597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 32.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006Apr;6(4):259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 33.An X, Sarmiento C, Tan T, Zhu H. Regulation of multidrug resistance by microRNAs in anti-cancer therapy. Acta Pharm Sin B. 2017Jan;7(1):38–51. doi: 10.1016/j.apsb.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu R, Chen X, Du Y, Yao W, Shen L, Wang C, Hu Z, Zhuang R, Ning G, Zhang C, et al. Serum microRNA expression profile as a biomarker in the diagnosis and prognosis of pancreatic cancer. Clin Chem. 2012Mar;58(3):610–618. doi: 10.1373/clinchem.2011.172767 [DOI] [PubMed] [Google Scholar]
- 35.Nakamura S, Sadakari Y, Ohtsuka T, Okayama T, Nakashima Y, Gotoh Y, Saeki K, Mori Y, Nakata K, Miyasaka Y, et al. Pancreatic juice exosomal MicroRNAs as biomarkers for detection of pancreatic ductal adenocarcinoma. Ann Surg Oncol. 2019Jul;26(7):2104–2111. doi: 10.1245/s10434-019-07269-z [DOI] [PubMed] [Google Scholar]
- 36.Schultz NA, Dehlendorff C, Jensen BV, Bjerregaard JK, Nielsen KR, Bojesen SE, Calatayud D, Nielsen SE, Yilmaz M, Holländer NH, et al. MicroRNA biomarkers in whole blood for detection of pancreatic cancer. JAMA. 2014Jan22-29311(4):392–404. doi: 10.1001/jama.2013.284664. [DOI] [PubMed] [Google Scholar]
- 37.Zhu Y, Wang J, Wang F, Yan Z, Liu G, Ma Y, Zhu W, Li Y, Xie L, Bazhin AV, et al. Differential MicroRNA expression profiles as potential biomarkers for pancreatic ductal adenocarcinoma. Biochemistry (Mosc). 2019May;84(5):575–582. doi: 10.1134/S0006297919050122 [DOI] [PubMed] [Google Scholar]
- 38.Ma Q, Wu H, Xiao Y, Liang Z, Liu T. Upregulation of exosomal microRNA-21 in pancreatic stellate cells promotes pancreatic cancer cell migration and enhances Ras/ERK pathway activity. Int J Oncol. 2020Apr;56(4):1025–1033. doi: 10.3892/ijo.2020.4986. [DOI] [PubMed] [Google Scholar]
- 39.Wang W, Liu B, Sun S, Lan L, Chen Y, Han S, Li X, Li Z. Downregulation of miR-486-5p enhances the anti-tumor effect of 5-Fluorouracil on pancreatic cancer cells. Onco Targets Ther. 2020Feb24;13:1649–1659. doi: 10.2147/OTT.S231153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takikawa T, Masamune A, Hamada S, Nakano E, Yoshida N, Shimosegawa T. miR-210 regulates the interaction between pancreatic cancer cells and stellate cells. Biochem Biophys Res Commun. 2013Aug2;437(3):433–439. doi: 10.1016/j.bbrc.2013.06.097. [DOI] [PubMed] [Google Scholar]
- 41.Yang Z, Zhao N, Cui J, Wu H, Xiong J, Peng T. Exosomes derived from cancer stem cells of gemcitabine-resistant pancreatic cancer cells enhance drug resistance by delivering miR-210. Cell Oncol (Dordr). 2020Feb;43(1):123–136. doi: 10.1007/s13402-019-00476-6. [DOI] [PubMed] [Google Scholar]
- 42.Xu YF, Hannafon BN, Khatri U, Gin A, Ding WQ. The origin of exosomal miR-1246 in human cancer cells. RNA Biol. 2019Jun;16(6):770–784. doi: 10.1080/15476286.2019.1585738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khan IA, Rashid S, Singh N, Rashid S, Singh V, Gunjan D, Das P, Dash NR, Pandey RM, Chauhan SS, et al. Panel of serum miRNAs as potential non-invasive biomarkers for pancreatic ductal adenocarcinoma. Sci Rep. 2021Feb211(1):2824. doi: 10.1038/s41598-021-82266-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang C, Wang J, Cui W, Liu Y, Zhou H, Wang Y, Chen X, Chen X, Wang Z. Serum exosomal miRNA-1226 as potential biomarker of pancreatic ductal adenocarcinoma. Onco Targets Ther. 2021Feb26;14:1441–1451. doi: 10.2147/OTT.S296816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miao H, Lu J, Guo Y, Qiu H, Zhang Y, Yao X, Li X, Lu Y. LncRNA TP73-AS1 enhances the malignant properties of pancreatic ductal adenocarcinoma by increasing MMP14 expression through miRNA −200a sponging. J Cell Mol Med. 2021Mar8;25(7):3654–3664. doi: 10.1111/jcmm.16425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yao LC, Jiang XH, Yan SS, Wang W, Wu L, Zhai LL, Xiang F, Ji T, Ye L, Tang ZG. Four potential microRNAs affect the progression of pancreatic ductal adenocarcinoma by targeting MET via the PI3K/AKT signaling pathway. Oncol Lett. 2021Apr;21(4):326. doi: 10.3892/ol.2021.12588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moldovan L, Batte KE, Trgovcich J, Wisler J, Marsh CB, Piper M. Methodological challenges in utilizing miRNAs as circulating biomarkers. J Cell Mol Med. 2014Mar;18(3):371–390. doi: 10.1111/jcmm.12236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singh R, Ramasubramanian B, Kanji S, Chakraborty AR, Haque SJ, Chakravarti A. Circulating microRNAs in cancer: hope or hype? Cancer Lett. 2016Oct10;381(1):113–121. doi: 10.1016/j.canlet.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 49.Keller A, Meese E. Can circulating miRNAs live up to the promise of being minimal invasive biomarkers in clinical settings? Wiley Interdiscip Rev RNA. 2016Mar-Apr;7(2):148–156. doi: 10.1002/wrna.1320. [DOI] [PubMed] [Google Scholar]
- 50.Masè M, Grasso M, Avogaro L, D’Amato E, Tessarolo F, Graffigna A, Denti MA, Ravelli F. Selection of reference genes is critical for miRNA expression analysis in human cardiac tissue. A focus on atrial fibrillation. Sci Rep. 2017Jan24;7:41127. doi: 10.1038/srep41127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Witwer KW. Circulating microRNA biomarker studies: pitfalls and potential solutions. Clin Chem. 2015Jan;61(1):56–63. doi: 10.1373/clinchem.2014.221341. [DOI] [PubMed] [Google Scholar]
- 52.Fasching PA, Brucker SY, Fehm TN, Overkamp F, Janni W, Wallwiener M, Hadji P, Belleville E, Häberle L, Taran FA, et al. Biomarkers in patients with metastatic breast cancer and the PRAEGNANT study network. Geburtshilfe Frauenheilkd. 2015Jan;75(1):41–50. doi: 10.1055/s-0034-1396215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mellby LD, Nyberg AP, Johansen JS, Wingren C, Nordestgaard BG, Bojesen SE, Mitchell BL, Sheppard BC, Sears RC, Borrebaeck CAK. Serum biomarker signature-based liquid biopsy for diagnosis of early-stage pancreatic cancer. J Clin Oncol. 2018Oct1;36(28):2887–2894. doi: 10.1200/JCO.2017.77.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]


