Abstract
Two consecutive cases of Haemophilus influenzae type a sequence type 23 invasive infection in 2 children attending the same daycare in 2019 triggered epidemiologic surveillance of H. influenzae infections in northern Spain. Despite the invasiveness potential of this virus strain, we detected no additional cases for 2013–2020.
Keywords: Haemophilus influenzae, serotype a, ST23, surveillance, respiratory infections, bacteria, Spain
Since the introduction of the Haemophilus influenzae type b (Hib) conjugate vaccine in the infant immunization schedule in 1998, the incidence of invasive H. influenzae (Hi) infections in Spain decreased to 0.7 episodes/100,000 population (1). Higher incidence rates are observed in children ≤2 years of age (1.88/100,000 population) and adults ≥65 years of age (1.89 cases/100,000 population) (2). Invasive disease caused by Hib has nearly disappeared, and most cases are caused by nontypeable strains (3).
Invasive infections caused by H. influenzae type a (Hia) are uncommon in Europe, particularly in Spain. However, Hia incidence is as high in other regions as among indigenous communities in North America (4) and as has emerged in Brazil during the 2000s (5). We describe 2 cases of Hia invasive disease in Gipuzkoa, northern Spain.
Both cases of Hia invasive disease occurred in children in a village with ≈15,000 inhabitants during November 2–3, 2019. The first patient, a 2-year-old boy, was admitted to the pediatric emergency department with good general aspect and persistent low-grade fever without a clear source. The child was not vaccinated according to the routine immunization schedule. Results for pulmonary auscultation and respiratory and cardiac rates were unremarkable, and a chest radiograph showed no abnormalities.
The second patient, a 19-month-old girl, was admitted to the pediatric emergency department with a nonproductive cough and a 39°C fever that was nonresponsive to antipyretics. The infant was vaccinated according to the routine immunization schedule, including Hib vaccination. No dyspnea was observed, and the chest radiograph showed pulmonary infiltrates suggesting pneumonia.
Both children showed increased C-reactive protein, procalcitonin, and white cell counts and had H. influenzae grown in the blood culture taken at admission. The boy was treated with ceftriaxone (50 mg/kg/12 h) for 5 days and the girl with ceftriaxone (50 mg/kg/12 h) for 4 days. Both children were discharged without symptoms or sequelae. Neither patient required additional antibiotic treatment after admission.
Both children attended the same daycare center, where no other children showed symptoms of infection. In Gipuzkoa, no additional cases of Hia invasive infection have been observed since 2013 (Table). However, 1 Hia was isolated 1 week later in the blood-culture of a 51-year-old patient in the adjacent province of Bizkaia.
Table. Epidemiologic and microbiological characteristics of Haemophilus influenzae invasive isolates, Gipuzkoa, Northern Spain, January 2013–December 2020*.
| Year | No. isolates†* | Biotype |
Capsulated serotypes/ST |
Nonencapsulated serotypes (no. isolates) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I | II | III | IV | V | a | b | e | NT | ST‡ | ||||
| 2013 | 7 | 4 | 2 | 1 | 1/ST6 | 6 | 41, 368 (2), 388, 996, 2381§ | ||||||
| 2014 | 5 | 1 | 4 | 1/ST190 | 4 | 105, 249, 1034, 1608 | |||||||
| 2015 | 4 | 3 | 1 | 4 | 40, 155 (2), 2382§ | ||||||||
| 2016 | 7 | 1 | 4 | 1 | 6 | 3, 85, 103, 266, 937, 2383§ | |||||||
| 2017 | 5 | 2 | 2 | 1 | 1/ST6 | 4 | 134, 567, 653, 986 | ||||||
| 2018 | 9 | 2 | 1 | 4 | 1¶ | 6 | 14, 145, 165, 838, 1472, 2384§ | ||||||
| 2019 | 11 | 1 | 6 | 1 | 2 | 2/ST23 | 1/ST995 | 1/ST760 | 6 | 6, 14, 103, 393, 603, 2110 | |||
| 2020 | 5 | 2 | 3 | 5 | 143, 183, 280, 334, 349 | ||||||||
| Total | 53 | 13 | 24 | 5 | 2 | 5 | 2 | 5 | 1 | 41 | |||
*NT, nontypeable; ST, sequence type. †Four isolates were not available for microbiological characterization: 1 in 2016, 2 in 2018, 1 in 2019. ‡If the number of isolates of a specific ST was not 1, the number of isolates corresponding to that ST is indicated in brackets. §New ST found in this study. ¶Serotype was determined but the isolate was not available for multilocus sequence typing.
We identified isolates with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (Beckman Coulter, https://www.beckmancoulter.com). Both case-patients scored >2.000. We biotyped isolates using the API 20E system of bacterial identification and serotyped using multiplex-PCR (6) and confirmed serotypes using BD Difco Haemophilus influenzae Antisera (Fisher Scientific, https://www.fishersci.com). We performed genotyping by multilocus sequenced typing and pulsed-field gel electrophoresis (PFGE) (Figure) after SmaI digestion with the following running conditions: switch angle 120°, 6 V/cm, ramped switch time from 1–30 s over 23 h. The presence of a deletion in the IS1016-bexA genes of the capsular operon was studied by PCR as described (7). We determined antimicrobial susceptibility by broth microdilution method according to EUCAST version 9.0 guidelines and criteria (EUCAST, https://www.eucast.org).
Figure.
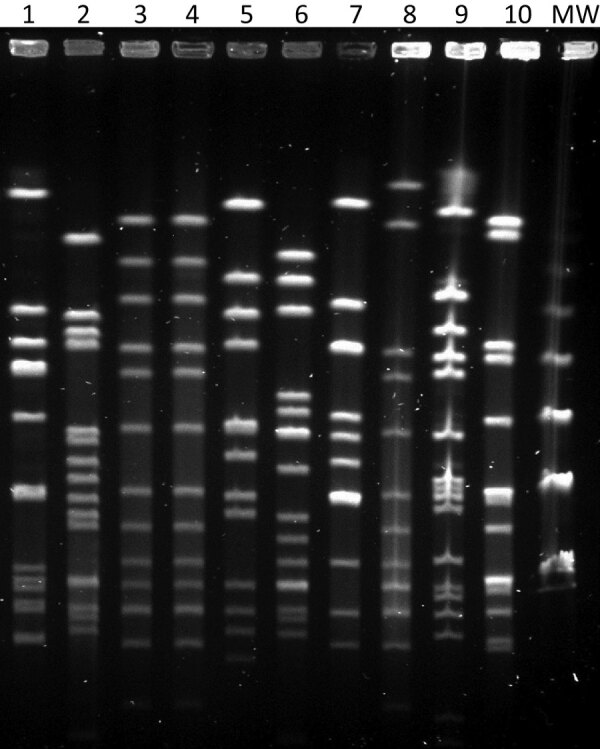
Pulsed-field gel electrophoresis patterns of invasive Haemophilus influenzae isolates collected during 2019–2020, Gipuzkoa, northern Spain. Lane 1, ST103; lane 2, ST760; lanes 3–4, ST23 isolates; lane 5, ST393; lane 6, ST6; lane 7, ST995; lane 8, ST2053; lane 9, ST46; lane 10, control isolate ATCC49766; lane MW, 50 kb DNA ladder. ST, sequence type; MW, molecular weight.
The isolates of both children were biotype II, serotype a, sequence type (ST) 23; showed an indistinguishable PFGE pattern; did not show the IS1016-bexA partial deletion; and were susceptible to ampicillin, azithromycin, and trimethoprim/sulfamethoxazole. The Hia that was isolated 1 week later in Bizkaia was similar to the 2 previous isolates of Gipuzkoa (biotype II, serotype a, not partial IS1016-bexA deletion) but was ST2053 (SLV of ST23) and had a closely related, but not identical, PFGE pattern.
We also characterized all invasive H. influenzae isolates reported since 2013 in Gipuzkoa (Table). Of the 48 isolates, 41 (85.4%) were nontypeable and 5 (10.4%) were serotype b; only the 2 cases described in this article were serotype a. All serotype b isolates were biotype I; showed the IS1016-bexA partial deletion; and belonged to ST6 (n = 2), ST190, or ST995.
Hia ST23 isolates have been described in different parts of the world, especially in Canada (4) and Brazil (5). In Europe, Hia ST23 has been found infrequently in Portugal (8) and recently in Italy (9). The H. influenzae multilocus sequence typing database (https://pubmlst.org/organisms/haemophilus-influenzae) lists only 29 ST23 isolates from the United States, Canada, Malaysia, France, and Spain (the 2 isolates in this article), most of them serotype a from invasive diseases.
Hia ST23 isolates from our region and from Canada did not show the virulence-enhancing IS1016-bexA partial deletion that has been more commonly associated with increased Hia virulence (10). However, isolates from our region only caused a mild and self-limiting infection, as compared with the severe disease observed among native North American Arctic populations that required intensive care unit admission and had notable sequelae (4).
As was the case in Italy, transmission of the highly virulent ST23 clone was substantially limited in Gipuzkoa. Although ST23 is a virulent Hia clone, its sustained spread appears to be limited, primarily among indigenous populations of North America. The origin of the isolates described in this article is unknown because Hia ST23 had not been previously described in Spain.
Biography
Ms. López-Olaizola is a clinical microbiologist in the bacteriology section, University Hospital Donostia. Her research focus on antimicrobial resistance and the epidemiology of infectious agents.
Footnotes
Suggested citation for this article: López-Olaizola M, Aguirre-Quiñonero A, Canut A, Barrios JL, Cilla G, Vicente D, et al. Emergence of Haemophilus influenzae type a ST23, Northern Spain. Emerg Infect Dis. 2021 Sep [date cited]. https://doi.org/10.3201/eid2709.204247
References
- 1.European Centre for Disease Prevention and Control. Haemophilus influenzae—annual epidemiological report for 2017. [cited 2019 Apr 23]. https://www.ecdc.europa.eu/en/publications-data/haemophilus-influenzae-annual-epidemiological-report-2017
- 2.Ciruela P, Martínez A, Izquierdo C, Hernández S, Broner S, Muñoz-Almagro C, et al. ; Microbiological Reporting System of Catalonia Study Group. Epidemiology of vaccine-preventable invasive diseases in Catalonia in the era of conjugate vaccines. Hum Vaccin Immunother. 2013;9:681–91. 10.4161/hv.23266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campos J, Hernando M, Román F, Pérez-Vázquez M, Aracil B, Oteo J, et al. ; Group of Invasive Haemophilus Infections of the Autonomous Community of Madrid, Spain. Analysis of invasive Haemophilus influenzae infections after extensive vaccination against H. influenzae type b. J Clin Microbiol. 2004;42:524–9. 10.1128/JCM.42.2.524-529.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsang RSW, Proulx J-F, Hayden K, Shuel M, Lefebvre B, Boisvert A-A, et al. Characteristics of invasive Haemophilus influenzae serotype a (Hia) from Nunavik, Canada and comparison with Hia strains in other North American Arctic regions. Int J Infect Dis. 2017;57:104–7. 10.1016/j.ijid.2017.02.003 [DOI] [PubMed] [Google Scholar]
- 5.Tuyama M, Corrêa-Antônio J, Schlackman J, Marsh JW, Rebelo MC, Cerqueira EO, et al. Invasive Haemophilus influenzae disease in the vaccine era in Rio de Janeiro, Brazil. Mem Inst Oswaldo Cruz. 2017;112:196–202. 10.1590/0074-02760160391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Falla TJ, Crook DW, Brophy LN, Maskell D, Kroll JS, Moxon ER. PCR for capsular typing of Haemophilus influenzae. J Clin Microbiol. 1994;32:2382–6. 10.1128/jcm.32.10.2382-2386.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leaves NI, Falla TJ, Crook DW. The elucidation of novel capsular genotypes of Haemophilus influenzae type b with the polymerase chain reaction. J Med Microbiol. 1995;43:120–4. 10.1099/00222615-43-2-120 [DOI] [PubMed] [Google Scholar]
- 8.Heliodoro CIM, Bettencourt CR, Bajanca-Lavado MP; Portuguese Group for the Study of Haemophilus influenzae invasive infection. Molecular epidemiology of invasive Haemophilus influenzae disease in Portugal: an update of the post-vaccine period, 2011-2018. Eur J Clin Microbiol Infect Dis. 2020;39:1471–80. 10.1007/s10096-020-03865-0 [DOI] [PubMed] [Google Scholar]
- 9.Giufrè M, Cardines R, Brigante G, Orecchioni F, Cerquetti M. Emergence of invasive Haemophilus influenzae type a disease in Italy. Clin Infect Dis. 2017;64:1626–8. 10.1093/cid/cix234 [DOI] [PubMed] [Google Scholar]
- 10.Kapogiannis BG, Satola S, Keyserling HL, Farley MM. Invasive infections with Haemophilus influenzae serotype a containing an IS1016-bexA partial deletion: possible association with virulence. Clin Infect Dis. 2005;41:e97–103. 10.1086/498028 [DOI] [PubMed] [Google Scholar]


