Abstract
SAGA, a recently described protein complex in Saccharomyces cerevisiae, is important for transcription in vivo and possesses histone acetylation function. Here we report both biochemical and genetic analyses of members of three classes of transcription regulatory factors contained within the SAGA complex. We demonstrate a correlation between the phenotypic severity of SAGA mutants and SAGA structural integrity. Specifically, null mutations in the Gcn5/Ada2/Ada3 or Spt3/Spt8 classes cause moderate phenotypes and subtle structural alterations, while mutations in a third subgroup, Spt7/Spt20, as well as Ada1, disrupt the complex and cause severe phenotypes. Interestingly, double mutants (gcn5Δ spt3Δ and gcn5Δ spt8Δ) causing loss of a member of each of the moderate classes have severe phenotypes, similar to spt7Δ, spt20Δ, or ada1Δ mutants. In addition, we have investigated biochemical functions suggested by the moderate phenotypic classes and find that first, normal nucleosomal acetylation by SAGA requires a specific domain of Gcn5, termed the bromodomain. Deletion of this domain also causes specific transcriptional defects at the HIS3 promoter in vivo. Second, SAGA interacts with TBP, the TATA-binding protein, and this interaction requires Spt8 in vitro. Overall, our data demonstrate that SAGA harbors multiple, distinct transcription-related functions, including direct TBP interaction and nucleosomal histone acetylation. Loss of either of these causes slight impairment in vivo, but loss of both is highly detrimental to growth and transcription.
The process of transcriptional activation hinges on the ability of various factors to facilitate the function of the transcription complex. In eukaryotes, chromatin structure is a major obstacle to transcription by RNA polymerase II: DNA is wound around histone proteins to form a repeating array of nucleosomes (81), making it inaccessible to the transcriptional machinery (53, 55). Nucleosomes must therefore be perturbed at promoter regions prior to or during activation, and such remodeling has been observed in a number of studies (29, 69, 71).
In recent years, a variety of factors relevant to transcription have been identified as involved in nucleosome remodeling or modification. ATP-dependent remodeling of nucleosomes has been demonstrated by a number of large protein complexes isolated from several eukaryotes (43). The Swi-Snf complex, identified in yeast and human cells, was shown to remodel chromatin in vivo and in vitro and stimulate activator and basal factor binding to nucleosomal DNA (15, 35, 38, 45, 52). The Drosophila Nurf (73), CHRAC (76), and ACF (39) complexes and the yeast RSC complex (10) are believed to carry out similar functions, also through the hydrolysis of ATP.
Histone acetylation is another mechanism by which nucleosomes are modified. The core histones (H2A, H2B, H3, and H4) can be acetylated on the lysine side chains of their amino-terminal tail regions (7), reducing their positive charge and presumably reducing their affinity for negatively charged DNA or other chromatin proteins. Substantial evidence suggests that acetylated nucleosomes are more permissive for transcription. For example, in vivo, histones associated with active chromosomal loci were shown to be hyperacetylated (34), while those at inactive or heterochromatin regions were shown to be hypoacetylated (8, 42, 50, 74). In vitro, histone acetylation results in increased binding of transcriptional activators to their sites in nucleosomal DNA (77).
A more definitive link between histone acetylation and transcriptional activation was realized with the discovery that the yeast Saccharomyces cerevisiae transcriptional adaptor Gcn5 is a histone acetyltransferase (HAT) (9). Gcn5 is one of a group of adaptors (also known as mediators or coactivators) that were hypothesized to provide a physical bridge between upstream DNA-bound activators and the transcriptional machinery at a promoter (28). Since the discovery of the HAT activity of Gcn5, additional transcriptional cofactors in yeast and higher eukaryotes, including the TATA-binding protein (TBP)-associated factor TAFII250 (the human homologue of yeast TafII145/130 [49]), p300/CBP (2, 51), and P/CAF (p300/CBP-associated factor [82]), have been identified as HATs, suggesting that acetylation may be important in activation. In yeast, genes encoding adaptor proteins were originally identified by mutations that suppress the toxicity caused by a high level of the acidic activator, Gal4-VP16 (5). Besides Gcn5 (48), these proteins include Ada1 (37), Ada2 (5), Ada3 (57), and Ada5 (47), and they were subsequently demonstrated to interact physically and functionally in vivo and in vitro (11, 36, 48). The ability of the adaptors to associate with activation domains (3, 14, 68, 75) and TBP (3), a component of TFIID, further indicated their function as part of a complex involved in activated transcription.
The crucial role of the HAT activity for Gcn5 function was recently demonstrated through the creation and analysis of HAT substitution mutants. Specific alanine substitutions (44, 78) in the previously identified HAT domain of Gcn5 (13) lower HAT activity, and loss of activity strongly correlates with defects in growth and transcription in vivo. Moreover, acetylation of histones at the promoter of the HIS3 gene correlates with gene activity, and the acetylation at this promoter is reduced in the presence of substitution mutations in Gcn5 that impair its HAT activity (44). In addition, mutations in the HAT domain of Gcn5 correlate with perturbation of the normal chromatin structure at the PHO5 promoter (27).
Gcn5 alone acetylates only free histones; however, as a component of native yeast complexes, Gcn5 acetylates histones in nucleosomes (24, 63). In one study, Gcn5 was shown to be a component of two of four distinct nucleosome-acetylating activities (24). These two Gcn5-containing complexes, which acetylate histones H3 and H2B in nucleosomes, also contained the adaptors Ada2 and Ada3. One of these Gcn5-dependent HAT complexes had a molecular size of 0.8 MDa and was termed the Ada complex.
The other Gcn5-dependent HAT complex, with a molecular size of about 1.8 MDa, possessed the adaptor components as well as four Spt proteins. This complex was therefore named SAGA (Spt-Ada-Gcn5-acetyltransferase). The Spt proteins known to be present in the SAGA complex include Spt3, Spt7, Spt8, and Spt20. Spt7 (20) and Spt20 (60) are apparently vital to the structure of SAGA, since null mutations in either gene causes complete disruption of the SAGA complex and severe growth defects (24). Interestingly, spt20 null mutants also have an Ada− phenotype: the gene was independently identified as ADA5 in a genetic screen for ada mutants (47).
The SPT genes were originally identified as suppressors of Ty or δ insertions in the promoter regions of the yeast HIS4 and LYS2 genes. The insertion mutations abolish or otherwise alter transcription of the adjacent gene, resulting in a His− or Lys− phenotype, and strains with spt mutations restore functional transcription of the adjacent gene (reviewed in reference 79). A subset of five SPT genes have been grouped together based on common mutant phenotypes. This group includes SPT15, which encodes TBP (19, 30). The other four members of this group encode the Spt proteins contained in SAGA. Spt3 and Spt8 in particular seem to have the most direct relationship to TBP, since specific spt3 and spt15 mutations suppress each other in an allele-specific fashion (17), implying physical interaction. Genetic evidence also has suggested that Spt8 is required for the functional interaction between Spt3 and TBP (18). Furthermore, TBP, as well as other SAGA components, were pulled down via expressed glutathione S-transferase (GST)–Spt20 from yeast extract (59). Interestingly, TBP also coimmunoprecipitated from yeast extract with Ada3, another SAGA subunit (64). A further potential relationship between SAGA and TBP was identified most recently with the discovery that a subset of five TafIIs are also present within SAGA (25). Interestingly, the TafII possessing HAT activity, TafII145, is not associated with SAGA. The precise mechanism of SAGA interaction with TBP and its role in vivo remain to be determined.
Mutations in different ADA and SPT SAGA genes cause distinct classes of genetic and biochemical phenotypes (26). Two classes of SAGA mutants have moderate sets of phenotypes. Gcn5, Ada2, and Ada3 form one distinct class within SAGA because (i) they are also present in the Ada complex, (ii) they are required for HAT activity within both complexes, and (iii) genetic disruptions of them exhibit Ada− but not Spt− phenotypes. A second potential subgroup within SAGA is Spt3/Spt8, mutations of which exhibit a significantly less severe range of phenotypes than disruptions of either Spt20 or Spt7, and in particular exhibit an Spt−, but not Ada−, phenotype. The third subgroup is Spt20/Spt7 mutations, which have severe phenotypes and exhibit both Spt− and Ada− phenotypes (47, 60). Loss of Spt20 or Spt7 completely disrupts the SAGA complex.
Further genetic support for multiple functions within SAGA comes from an analysis of double-mutant phenotypes between mutations in SAGA genes and mutations in genes that encode other transcription regulatory factors (59). Specifically, spt20Δ and spt7Δ are synthetically lethal in combination with mutations in genes that encode members of the Swi-Snf or Srb-mediator complex. In contrast, null mutations in the SAGA genes GCN5, SPT3, and SPT8 do not cause inviability in combination with mutations in these other transcription factor genes.
In this study, we sought to establish the basic functional organization of SAGA, using genetic and biochemical analyses of certain key components. Our results suggest that the multiple discrete biochemical functions relate in a direct way to the distinct genetic phenotypes and that mutant SAGA complexes, lacking certain components, contain subfunctional activity. These data suggest a model for overall SAGA function in the process of transcriptional activation.
MATERIALS AND METHODS
Yeast strains and media.
The yeast strains used in this study are listed in Table 1. All FY strains are congenic and were originally derived from the S288C derivative FY2 (80). To introduce the snf5Δ2 null mutant allele (46) into S288C background, integrating plasmid pLY17 was used for transplacement (1, 66) in haploid strain FY37. All other deletion derivatives except ada mutations have been described previously (reference 59 and references therein). Null mutants were constructed by transforming diploids with a restriction fragment designed to delete the gene being studied (62). In each case, the null mutation contained a selectable nutritional marker. Transformants were then sporulated, and tetrads were dissected to generate haploid mutant progeny (1). In the ada1Δ::HIS3, ada2Δ::HIS3, and ada3Δ::HIS3 alleles, the entire open reading frame of each corresponding gene was replaced with the HIS3 gene, using a fragment generated by PCR (1). The following PCR primer pairs were used to create and verify correct integration of the corresponding ada knockout mutations: ADA1KOLEFT (5′-ATAGGGAAAACAAGCCCAGTAGTTTTGATTTCTTCTATCCTGTGCGGTATTTCACACCG-3′), ADA1KORITE (5′-TATAATTACAACATACCGCATACACACACTTTTTATACAAGATTGTACTGAGAGTGC AC-3′), ADA1LCHECK (5′-TGTGCCTTGAACATCGAACTTAC-3′), ADA1RCHECK (5′-GCTCACTTAAGCTCGGTCACA-3′), ADA2KOLEFT (5′-ATCAGCGTAGTCTGAAAATATATACATTAAGCAAAAAGATGCGGTATT TCACACCG-3′), ADA2KORITE (5′-ACTAGTGACAATTGTAGTTACTTTTCAATTTTTTTTTTGAGATTGTACTGAGAGTGCAC-3′), ADA2LCHECK (5′-CCAGACGTTCCAAACAAATAAGTG-3′), ADA2RCHECK (5′-CAAGGTCCCTTTATGACTTGGCC-3′), ADA3KOLEFT (5′-TAACAAAGACGGAGCGACGAGAAGTATTGGACAGGACATCTGTGCGGTATTTCAC ACCG-3′), ADA3KORITE (5′-GTTATTATGCTACGTATTTTTCCTTAGAGTTCGTATATTAGATTGTACTGAGAGTGCAC-3′), ADA3LCHECK (5′-GAGCTCCGACGTGCAACGCGA-3′), and ADA3RCHECK (5′-CCCGCGATCGGAAGTTCATGC-3′). For each ADA gene, the corresponding KOLEFT and KORITE primers were used in a standard PCR to amplify the HIS3 gene from pRS403 (67). The resulting PCR fragments, which contained the HIS3 gene flanked by 78 bp of 5′ and 3′ noncoding sequence from the respective ADA gene, were used to transform diploid S288C yeast strains. We verified the correct integration event for each mutation by using PCR with three different primer pairs (data not shown).
TABLE 1.
S. cerevisiae strains used
| Strain | Genotype |
|---|---|
| FY2 | MATα ura3-52 |
| FY37 | MATa his3Δ200 lys2-128δ ura3-52 |
| FY293 | MATα spt3Δ202 his4-917δ lys2-173R2 ura3-52 |
| FY294 | MATa spt3Δ202 his4-917δ leu2Δ1 lys2-173R2 trp1Δ63 ura3-52 |
| FY297 | MATα spt3Δ202 his4-917δ lys2-173R2 trp1Δ63 ura3-52 |
| FY462 | MATa spt8Δ302::LEU2 his4-917δ leu2Δ1 lys2-173R2 trp1Δ63 |
| FY463 | MATα spt8Δ302::LEU2 his4-917δ leu2Δ1 lys2-173R2 trp1Δ63 ura3-52 |
| FY602 | MATa his3Δ200 leu2Δ1 lys2-128δ trp1Δ63 ura3-52 |
| FY630 | MATα his4-917δ leu2Δ1 lys2-173R2 trp1Δ63 ura3-52 |
| FY631 | MATa his4-917δ leu2Δ1 lys2-173R2 trp1Δ63 ura3-52 |
| FY1009 | MATa spt7-502 his4-917δ leu2Δ1 ura3-52 |
| FY1098 | MATa spt20Δ100::URA3 his3Δ200 leu2Δ1 ura3-52 |
| FY1106 | MATα spt20Δ100:URA3 his4-917δ leu2Δ1 lys2-173R2 trp1Δ63 ura3-52 |
| FY1253 | MATa sin4Δ::TRP1 his3Δ200 leu2Δ1 lys2-173R2 trp1Δ63 ura3-52 |
| FY1254 | MATα swi1Δ::LEU2 his3Δ200 leu2Δ1 lys2-128δ ura3-52 |
| FY1286 | MATa gcn5Δ::HIS3 arg4-12 his3Δ200 leu2Δ1 lys2-173R2 ura3-52 |
| FY1299 | MATα spt8Δ302::LEU2 his3Δ200 leu2Δ1 lys2-173R2 trp1Δ63 ura3-52 |
| FY1359 | MATα srb2Δ::HIS3 arg4-12 his3Δ200 lys2-173R2 ura3-52 |
| FY1370 | MATα gcn5Δ::HIS3 his3Δ200 leu2Δ1 ura3-52 |
| FY1440 | MATα gcn5Δ::HIS3 spt3Δ202 arg4-12 his3Δ200 leu2Δ1 lys2-173R2 ura3-52 |
| FY1441 | MATa gcn5Δ::HIS3 spt3Δ202 his3Δ200 leu2Δ1 lys2-173R2 ura3-52 |
| FY1542 | MATa ada3Δ::HIS3 his3Δ200 leu2Δ1 lys2-173R2 trp1Δ63 ura3-52 |
| FY1545 | MATα ada3Δ::HIS3 arg4-12 his3Δ200 lys2-173R2 trp1Δ63 ura3-52 |
| FY1547 | MATa ada3Δ::HIS3 his3Δ200 lys2-173R2 trp1Δ63 ura3-52 |
| FY1548 | MATa ada2Δ::HIS3 his3Δ200 leu2Δ1 lys2-128δ ura3-52 |
| FY1553 | MATa ada2Δ::HIS3 his3Δ200 his4-917δ leu2Δ1 lys2-173R2 ura3-52 |
| FY1554 | MATa ada2Δ::HIS3 his3Δ200 lys2-173R2 trp1Δ63 ura3-52 |
| FY1557 | MATα ada1Δ::HIS3 his3Δ300 leu2Δ1lys2-128δ ura3-52 |
| FY1559 | MATa ada1Δ::HIS3 his3Δ200 his4-917δ leu2Δ1 lys2-173R2 trp1Δ63 ura3-52 |
| FY1599 | MATa gcn5Δ::HIS3 arg4-12 his3Δ200 his4-917δ leu2Δ1 lys2-173R2 trp1Δ63 ura3-52 |
| FY1656 | MATα snf2Δ::LEU2 his3Δ200 leu2Δ1 ura3-52 |
| FY1657 | MATα gal11Δ::TRP1 arg4-12 his3Δ200 lys2-173R2 trp1Δ63 ura3-52 |
| FY1658 | MATa snf5Δ2 his3Δ200 lys2-128δ ura3-52 |
| FY1719 | MATa gcn5Δ::HIS3 spt8Δ302::LEU2 arg4-12 his3Δ200 leu2Δ1 lys2-173R2 trp1Δ63 ura3-52 |
| FY1720 | MATa gcn5Δ::HIS3 spt8Δ302::LEU2 his4-917δ leu2Δ1 lys2-173R2 ura3-52 |
| L937 | MATα srb5Δ::URA3hisG arg4-12 his3Δ200 leu2 trp1Δ63 ura3-52 |
| PSY316 | MATα ade2-101 his3Δ200 leu2-3, 2-112 lys2 ura3-52 |
| SB11 | MATα ada1Δ::HIS3 ade2-101 his3Δ200 leu2-3, 2-112 lys2 trp1Δ ura3-52 |
| SB326 | MATa gcn5Δ spt7-502 his4-917δ leu2Δ1 ura3-52 |
ADA1 deletion mutant SB11 was created by PCR amplification of the HIS3 gene with chimeric primers that included sequence upstream from or near the end of the ADA1 gene; these oligonucleotides were 5′-CGTTGCTCTTTTCATTCGTCTGTTCGTTATCATCATTGGATAGGGCTTACTCTTGGCCTCCT CTAG-3′ (upstream) and 5′-CATCCAAAGGCGTGAAAGTCGGCTTTTCATTTAAGAAGTCCGGCAAGTCGTCGCCTCGTTCAGAATGACACG-3′ (downstream). The resulting PCR product was transformed into a trp1Δ derivative of strain PSY316 (12), and ada1Δ clones were selected on His− selective plates.
To investigate the relationship between the Gcn5 and Spt7 bromodomains, GCN5 deletion strain SB326 was produced from spt7 ΔBrD strain FY1009 (20) by transformation with the BglII/XhoI fragment of gcn5Δ::URA3 hisG plasmid pyGCN5.KO (12) followed by selection on 5-fluoroorotic acid (5-FOA) plates to remove the URA3 gene (6). This strain was then used for integration of pRS306-PADH-yGCN5 1-439 and 1-280 by the methods described below.
Rich (YPD), minimal, synthetic complete (SC), 5-FOA, and sporulation media were prepared as described previously (61). Media for testing Gal and Ino phenotypes were as described by Gansheroff et al. (20). For caffeine (12 mM) and hydroxyurea (150 mM) plates, SC medium was used. Standard protocols for transformation were used in strain constructions (61).
Plasmid constructs.
For integration of wild-type and bromodomain truncation mutant GCN5 into yeast, constructs were prepared in which the appropriate open reading frames followed the ADH1 promoter in the vector pRS306 (67). By standard recombinant DNA techniques (65), wild-type GCN5 (coding for residues 1 to 439), and the longer of the two mutant genes (with residues 1 to 350) were obtained as KpnI/PvuII fragments from pPC87-yGCN5 plasmids (13) and subcloned into KpnI/SmaI-cut pRS306. For the other mutant plasmid (with residues 1 to 280), a vector, pRS306-PADH, was first created by subcloning a KpnI/PvuII fragment of pPC87 (containing the ADH1 promoter and terminator) into pRS306 that had been digested with BstXI, blunted by the exonuclease activities of T4 and T7 DNA polymerase, and digested with KpnI. The resulting plasmid was cut with NotI, and truncated GCN5 was inserted as an EagI fragment of the pSP64-yGCN5 clone for the 1-280 mutant (13). The pRS306-PADH-yGCN5 1-439, 1-350, and 1-280 constructs were then linearized with NsiI and transformed into the gcn5Δ mutant FY1370. Clones were selected on Ura− medium; since pRS306 has no origin of replication, these represent plasmid integrations into the genome at the URA3 locus.
Plasmids BD1 (SWI1) and FB565 (GAL11), used to recover otherwise lethal progeny in double-mutant analysis (Table 2), have been described previously (56, 59). Plasmid pGEX-3X, for bacterial expression of GST, was from Pharmacia, and pGST-TBP was provided by R. H. Reeder (Fred Hutchinson Cancer Research Center, Seattle, Wash.).
TABLE 2.
Phenotypes of ada mutants
| Double mutanta | Phenotypee |
|---|---|
| ada1Δ swi1Δb,d | Dead |
| ada1Δ snf5Δ2c | Dead |
| ada1Δ srb5Δc | Dead |
| ada1Δ gal11Δb,d | Dead |
| ada2Δ swi1Δ | Alive, sick |
| ada2Δ snf2Δd | Alive, sick |
| ada2Δ srb5Δ | Alive, slow growing |
| ada2Δ srb2Δ | Alive, slow growing |
| ada2Δ gal11Δ | Alive |
| ada3Δ swi1Δd | Alive, sick |
| ada3Δ snf2Δ | Alive, sick |
| ada3Δ srb5Δ | Alive, slow growing |
| ada3Δ srb2Δ | Alive, slow growing |
| ada3Δ gal11Δd | Alive |
| ada3Δ sin4Δ | Alive |
In each cross, genetic markers allowed us to unambiguously identify all relevant markers in every tetrad. The parents for the crosses in the order listed are as follows: FY602 × FY1254, FY1658 × FY1557, FY1557 × L937, FY602 × FY1657, FY1548 × FY1254, FY602 × FY1656, FY1548 × L937, FY1548 × FY1359, FY1554 × FY1657, FY602 × FY1254, FY1542 × FY1656, FY1542 × L937, FY1542 × FY1359, FY602 × FY1657, and FY1253 × FY1545.
Viability was scored by the ability of the double mutant to lose a URA3-marked wild-type SWI1 (BD1) or GAL11 (FB565) plasmid as determined by growth on 5-FOA medium (alive, growth on 5-FOA; dead, no growth on 5-FOA).
Viability was determined by failure to recover any double mutants after dissecting 20 tetrads.
For all crosses in which FY602 is listed as the MATa parent, the double mutant was constructed by first creating a diploid between FY602 and the corresponding MATα parent. This diploid was then transformed with a PCR fragment containing the relevant ADA ORF completely disrupted by the HIS3 gene. Those single purified His+ diploid transformants that displayed correct ADA knockout integration (as described in Materials and Methods) were used in tetrad analysis.
Sick, extremely small colonies after 2 to 3 days of growth at 30°C. Slow growing, colonies were moderately smaller than those of wild-type strains after 2 to 3 days of growth at 30°C.
HAT complex purification and assays.
Nucleosomal HAT complexes were prepared from ada1Δ, gcn5Δ, gcn5Δ spt3Δ, gcn5Δ spt8Δ, and PSY316 wild-type cells and from wild-type and bromodomain-truncated GCN5 integrants by the previously described purification scheme (24): growth in 4 liters of YPD to an optical density of 2 to 2.5 at 600 nm, cell breakage with glass beads, incubation of extract with Ni2+-agarose and recovery of a 300 mM imidazole eluate, and purification on a Mono Q column with a 100 to 500 mM NaCl gradient. For spt3Δ, spt8Δ, and accompanying wild-type complex preparation, starting material was a 6-liter culture, and an additional Superose 6 column step was employed. Peak Mono Q fractions of SAGA were pooled and concentrated to 0.6 ml in a Centriprep-30 concentrator (Amicon). Samples were loaded on a Superose 6 HR 10/30 column (Pharmacia) equilibrated in 250 mM NaCl. The calibration of this column was as follows: thyroglobulin (669 kDa), fraction 24; ferritin (440 kDa), fraction 27; adolase (158 kDa), fraction 30; and ovalbumin (45 kDa), fraction 33/34.
HAT assays were performed by previously described methods (24): 30-μl reaction mixtures contained 2 μl of enzyme sample, 2 μg of oligonucleosomes or 10 μg of free histones, and 0.25 μCi of 3H-labeled acetyl coenzyme A in HAT buffer (50 mM Tris-HCl [pH 8.0], 50 mM KCl, 5% glycerol, 0.1 mM EDTA, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 10 mM sodium butyrate) and were incubated 30 min at 30°C. Oligonucleosomes were prepared as described previously (16), and free histones (purchased from Sigma) were from calf thymus. Half of each reaction was spotted on P81 filter paper (Whatman) for histone binding, washed, and used for liquid scintillation counting; the other half was heated in sodium dodecyl sulfate (SDS) sample buffer with β-mercaptoethanol and run on an SDS–18% polyacrylamide gel, which was fluorographed with Enhance (Dupont NEN) and dried. Fluorography was performed with Kodak X-Omat film at −70°C, with exposure times of 2.5 days for nucleosome assays and 1 to 1.5 days for free histone assays. Quantitation of histone H3 and H2B acetylated by SAGA was achieved by cutting out these bands from the dried gels, pulverizing them, and using them for liquid scintillation counting; five sets of counts were averaged for each, and error was 8% or less.
Antibodies and Western blotting.
For Western blot experiments, samples were boiled in SDS–β-mercaptoethanol sample buffer, electrophoresed on SDS–8 or 10% polyacrylamide gels, electroblotted to nitrocellulose, and visualized immunochemically by standard methods (32). Anti-Ada2, anti-Gcn5, and anti-Spt3 antisera and anti-Spt20 affinity-purified antibodies were as described previously (24); primary antibody dilutions used were typically 1:4,000, 1:4,000, 1:500, and 1:2,000, respectively. Anti-TafII90, anti-TafII68, and anti-TafII60 antisera (25) were used at dilutions of 1:3,000, 1:6,000, and 1:6,000, respectively. Rabbit anti-Spt8 antibodies were raised against a synthetic peptide spanning the amino-terminal 20 amino acids (aa) of Spt8 (MDEVDDILINNQVVDDEEDD) by Cocalico Biologicals; this antiserum was used at a dilution of 1:2,000. Immunodetection was performed with a secondary antibody (goat anti-rabbit immunoglobulin G-horseradish peroxidase conjugate; Bio-Rad) and an enhanced chemiluminescence kit (Amersham). Some blots were stripped of antibody before reprobing; this was accomplished with 62.5 mM Tris (pH 6.8)–2% SDS–100 mM β-mercaptoethanol at 50°C for 30 min.
In vivo RNA analysis.
Yeast strains used for analyzing transcriptional effects of the bromodomain were FY1370 (gcn5Δ) and GCN5 wild-type and bromodomain deletion integrants as described above; for HAT domain experiments, strains were FY1370 plasmid integrants containing wild-type GCN5, empty vector (gcn5Δ), or GCN5 with HAT domain substitution mutations (78). Total RNA was isolated from cultures grown in SC medium to an optical density at 600 nm of 0.8 to 1.2 by the hot phenol method as described previously (40). To derepress HIS3 transcription, cells were grown in SC medium lacking histidine, and 3-aminotriazole was added to 40 mM 2 h (for bromodomain study) or 6 h (for HAT domain study) prior to RNA isolation. RNA concentration was quantitated spectrophotometrically at 260-nm wavelength; 50 μg of each RNA sample was hybridized to a completion with an excess of 32P-end-labeled HIS3 and tRNAw oligonucleotides and treated with S1 nuclease as described elsewhere (40, 54). HIS3 RNA levels were quantitated on a PhosphorImager (Molecular Dynamics). S1 assays were performed in duplicate several times (exception for mutants LKN, IFQ, and YIA, performed once), and PhosphorImager quantitation showed less than 15% error. tRNAw probe (bromodomain study) and PMA1 RNA (HAT domain study) were used as internal controls for equal loading.
GST-TBP binding study.
Escherichia coli DH5α, grown in LB medium with 100 μg of ampicillin per ml, was used for induction of GST and GST-TBP. Expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside for 4 h at 37°C, and cell lysates were prepared by sonication in phosphate-buffered saline (65) containing protease inhibitors and rotated for 30 min at 4°C with glutathione-Sepharose beads (Pharmacia). Samples of these beads (about 15 μl containing several μg of GST or GST-TBP) were then rotated alone or with Superose 6-purified wild-type (fraction 20), spt3Δ (fraction 21), or spt8Δ (fraction 21) SAGA complexes for 1 h at 4°C. Amounts of SAGA samples used (30 μl for wild-type and spt8Δ strains and 20 μl for spt3Δ strains) were adjusted to have similar amounts of protein, based on prior Western blotting. Each reaction (150 μl, final volume) was adjusted to a final NaCl concentration of approximately 60 mM and contained 100 μl of binding buffer (20 mM HEPES [pH 7.3], 5 mM MgCl2, 0.5 mM EDTA, 15% glycerol, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride). The resulting beads were washed three times with 500 μl of binding buffer with 50 mM NaCl, boiled in SDS sample buffer, and loaded onto an SDS–10% polyacrylamide gel, which was used for Western blot analysis with anti-Ada2, anti-Spt20, and anti-Spt3 antibodies.
RESULTS
Loss of Spt3 or Spt8 has moderate effects on SAGA structure.
Our previous results have indicated that severe phenotypes correspond to disruption in SAGA structure; i.e., the major growth defects seen in spt20Δ and spt7Δ mutants correlate with a complete loss of the SAGA complex. In contrast, the mild gcn5Δ phenotype corresponds to a subtly altered SAGA complex. To test further the hypothesis that severity in mutant phenotype relates directly to biochemical integrity of the SAGA complex, we examined SAGA structure in the moderately defective spt3Δ and spt8Δ and the severe ada1Δ mutants and then compared their mutant phenotypes.
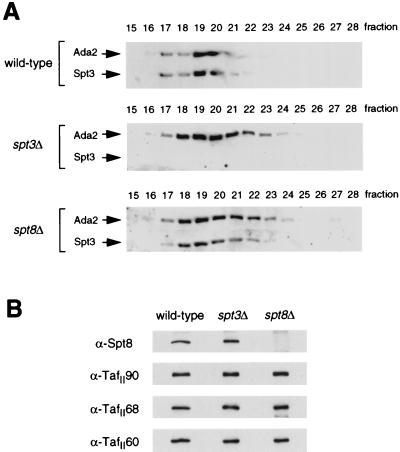
First, extracts were prepared from both SPT3 and SPT8 disruption strains to determine whether the SAGA complex exists in the absence of these subunits. The extracts were chromatographed through Ni2+-agarose and Mono Q resins to separate four previously identified nucleosomal HAT complexes (24). Western blotting using Ada2 and Gcn5 antibodies demonstrated that the mutant SAGA complexes were chromatographically distinct from wild-type SAGA: spt3Δ and spt8Δ SAGA complexes elute from Mono Q approximately two and seven fractions earlier, respectively (data not shown).
To investigate these altered complexes further, larger-scale preparations of SAGA from spt3Δ and spt8Δ mutants were fractionated with an additional Superose 6 column step (24). Size determination via anti-Ada2 Western blots of the Superose fractions indicated that the SAGA complexes derived from both mutants were only slightly smaller and eluted in a broader peak than wild-type SAGA (Fig. 1A), suggesting that few, if any, additional subunits were lost along with either Spt3 or Spt8. In addition, Spt3 is present in spt8Δ SAGA (Fig. 1A), and Spt8 is present in spt3Δ SAGA (Fig. 1B). Furthermore, deletion of Spt3 or Spt8 has no effect on TafII90, TafII68, or TafII60 interaction with SAGA (Fig. 1B).
FIG. 1.
Western blot characterization of Superose 6-purified spt3Δ and spt8Δ SAGA complexes. (A) SAGA complexes derived from wild-type (FY631), spt3Δ (FY294), and spt8Δ (FY462) strains were pooled, concentrated, and run on a Superose 6 column for separation based on molecular weight. Western blots of these fractions were performed to compare the sizes of the complexes. Blots were visualized with dilutions of antisera raised against the Ada2 or Spt3 proteins as indicated. (B) Western blots of wild-type (fraction 20), spt3Δ (fraction 21), and spt8Δ (fraction 21) SAGA, visualized with antisera specific to Spt8, TafII90, TafII68, or TafII60.
Ada1 is integral to SAGA structure.
In contrast to the modest effects on structure from loss of either Gcn5 (24), Spt3, or Spt8, our previous biochemical analyses of Spt20 and Spt7 suggested that these proteins are critical to the structural integrity of the SAGA complex (24). The large size of SAGA (1.8 MDa) indicates that there may be additional proteins having a similar role in holding the complex together. The observation that a strain bearing an ADA1 disruption exhibits severe phenotypes similar to those of SPT20 mutants, along with the indication that Ada1 may exist in a large complex with Ada2, Ada3, Gcn5, and Spt20 (5, 37), suggested that Ada1 might be present in SAGA and required for its integrity.
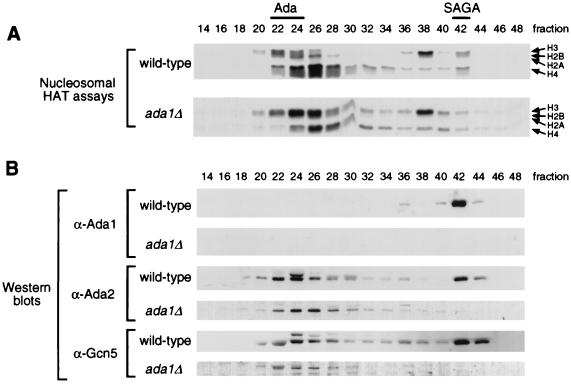
To test whether Ada1 has a role in SAGA, cell extracts were prepared from wild-type and ada1Δ strains and tested for the presence and activity of SAGA. Nucleosomal HAT assays were performed on Mono Q column fractions, following Ni2+ chelate affinity chromatography (24). Extract from ada1Δ cells specifically lacked the nucleosomal HAT activity of SAGA (around fraction 42), while the activity of the Ada complex (peak in fractions 24 to 26) was intact (Fig. 2A). This profile is identical to that previously observed for both SPT20 and SPT7 disruptions.
FIG. 2.
Presence of Ada1 in the SAGA complex. HAT complexes were derived from wild-type (PSY316) and ada1Δ (SB11) yeast strains through the Mono Q column step of the previously described purification procedure. (A) Fluorographs of nucleosome acetylation assays of the even-numbered column fractions. The arrows denote the relative positions of the four histone proteins as separated on SDS–18% polyacrylamide gels; visualized species contain acetyl groups labeled with 3H. Wild-type fractions displaying the H3/H2B-acetylating activities of the Ada and SAGA complexes are indicated at the top. (B) Western blots of the fractions, visualized with dilutions of antisera raised against Ada1, Ada2, or Gcn5 protein.
To confirm that Ada1 is directly involved in the SAGA complex, Western blot analysis was performed. An anti-Ada1 blot revealed that wild-type SAGA, but not the Ada complex, does contain Ada1 (Fig. 2B). Furthermore, immunoblotting analyses using anti-Ada2 and anti-Gcn5 antibodies indicate that SAGA itself is lost in the ada1Δ mutant preparation, while the Ada complex was unaffected (Fig. 2B). Ada1 is therefore similar to Spt20 and Spt7 in that it is required for the overall structural integrity of SAGA.
Three phenotypic classes of SAGA components.
Thus, structural analysis of SAGA provides a biochemical correlation with previous observations that disruptions of SAGA components cause phenotypes with a range of severity. We further tested the hypothesis that loss of components that have little detectable effect on SAGA complex integrity result in modest phenotypes, and in contrast, loss of components that are completely disruptive to the complex result in severe phenotypes.
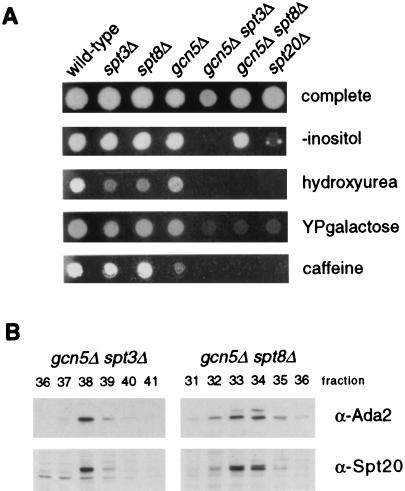
Phenotypic effects of disruptions of representatives of each putative phenotypic class are presented in Fig. 3. Growth was tested under various conditions known to cause growth defects in some yeast mutant strains (31). As shown previously and here, Ada2/Gcn5 and Spt3/Spt8 each have modest phenotypic effects. In contrast, ada1Δ causes genetic phenotypes similar in severity to those caused by spt7Δ and spt20Δ mutations and more severe than those of the others (spt3Δ, spt8Δ, gcn5Δ, ada2Δ, and ada3Δ). In particular, as reported previously (37), ada1Δ is like spt20Δ in that it is an inositol auxotroph and displays an Spt− phenotype; we show here that both mutants also have the additional phenotype of failure to grow on media containing caffeine or the DNA synthesis inhibitor hydroxyurea.
FIG. 3.
Phenotypic comparison of ada1Δ and other SAGA deletion mutants. Strains FY630, FY1106, FY1559, FY297, FY463, FY1599, and FY1553 were grown overnight in liquid YPD medium. For each strain, approximately 5 × 103 cells were spotted on the indicated plates and grown for 2 days at 30°C. All strains contain the his4-917δ insertion mutation. Otherwise, wild-type strains containing this allele are His−, while strains containing a mutation able to suppress this allele are His+ (Spt− phenotype).
We tested another phenotype that differed for the subgroups of SAGA mutants: double-mutant lethality in combination with mutations in either SNF/SWI genes or Srb/mediator genes. These results (Table 2) also demonstrate that ada1Δ mutations cause the same phenotypes as spt20Δ and spt7Δ mutations (59): ada1Δ is lethal in combination with every snf/swi or Srb/mediator mutation tested. In contrast, ada2Δ and ada3Δ mutations did not cause inviability in similar double mutants (Table 2), as was also previously seen for spt3Δ, spt8Δ, and gcn5Δ mutations (59), indicating a less critical role for these subunits in cellular growth. These results contrast with those of Pollard and Peterson (58), who observed lethality in double mutants of gcn5Δ, ada2Δ, or ada3Δ combined with swi1Δ, albeit in a different yeast strain background.
Thus, both biochemical and genetic evidence places Ada1 in the Spt20/Spt7 class; i.e., it is required for SAGA complex integrity, and loss of it is strongly debilitating to function in vivo. In contrast, either Gcn5, Spt3, or Spt8 has modest effects on SAGA structure and function in vivo. In summary, we detect three phenotypic and structural classes of SAGA components: Gcn5/Ada2/Ada3, Spt3/Spt8, and Ada1/Spt7/Spt20.
Double mutants gcn5Δ spt3Δ and gcn5Δ spt8Δ are similar in phenotype to spt20Δ and spt7Δ mutants.
Based on the above data, it appears that each of the two moderate phenotypic classes contributes distinct functions to SAGA. Because moderate phenotypes result from deletions of genes of either class, we postulated that in the absence of one class, the function of the second class remains intact. However, in the cases of ada1Δ, spt7Δ, and spt20Δ mutations, where the complex is completely disrupted, both functions are lost and severe phenotypic defects result. Based on these observations and ideas, we hypothesized that double mutants that disrupt the function of each moderate phenotypic class simultaneously would cause severe phenotypes similar to those of ada1Δ, spt7Δ, and spt20Δ mutations.
To test this idea, gcn5Δ spt3Δ and gcn5Δ spt8Δ mutants were constructed. As had been seen previously in other phenotypic assays (reference 59 and Fig. 3), neither spt3Δ, spt8Δ, nor gcn5Δ cells were severely defective (Fig. 4). In fact, spt3Δ and spt8Δ strains grew well in the absence of inositol, in the presence of hydroxyurea or caffeine, or with galactose as a sole carbon source; gcn5Δ cells also grew well under all conditions except caffeine. In contrast, the double mutants gcn5Δ spt3Δ and gcn5Δ spt8Δ were significantly more defective, failing to grow under the latter three conditions, and gcn5Δ spt8Δ grew less vigorously than the single mutants in media lacking inositol (Fig. 4A). These levels of impairment are similar to what is seen in an spt20Δ mutant tested under the same conditions (Fig. 4A). These results show that the double mutants have severe phenotypes similar to those of single mutations that disrupt SAGA altogether. Importantly, SAGA is intact in the double mutants and can be purified by standard methods, as shown by immunoblot analysis of SAGA from these strains (Fig. 4B). Overall, the results support the idea that the two phenotypically moderate subgroups of components contribute distinct biochemical functions to SAGA.
FIG. 4.
Comparison of gcn5Δ spt3Δ and gcn5Δ spt8Δ double mutants with an spt20Δ mutant. (A) Strains FY2, FY293, FY1299, FY1286, FY1440, FY1719, and FY1098 were grown overnight in liquid YPD medium. For each strain, approximately 2 × 103 cells were spotted on the indicated plates. Growth is shown after 2 days of incubation at 30°C, except in the case of the caffeine plates, which were photographed after 3 days. (B) Double-mutant SAGA complexes are intact. Mono Q fractions from gcn5Δ spt3Δ (FY1441) and gcn5Δ spt8Δ (FY1720) HAT complex preparations were analyzed by Western blotting. SAGA subunits were visualized with dilutions of antisera raised against Ada2 or Spt20 protein as indicated.
Gcn5 bromodomain is important for normal levels of nucleosome acetylation by SAGA.
We wished to characterize the biochemical functions of the Gcn5/Ada2/Ada3 and Spt3/Spt8 subgroups in SAGA. Each member of the Gcn5/Ada2/Ada3 subgroup previously was shown to be required for histone acetylation, and Gcn5 was identified as the HAT catalytic subunit. Recombinant Gcn5 acetylates free core histones and not nucleosomes; however, both the Ada and SAGA complexes acetylate nucleosomes (24). Thus, an important question is the determinant(s) of physical interaction with nucleosomes.
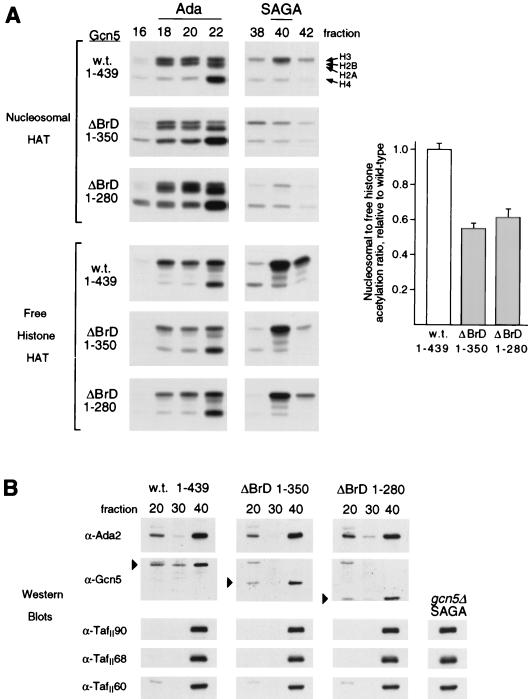
One clue to a possible determinant of nucleosome access is within the Gcn5 protein itself. The bromodomain at the carboxyl terminus of Gcn5 (aa 349 to 422 [22]) is a conserved sequence that is found in a variety of transcription-related proteins but has no clear function (33, 41, 72). In particular, the bromodomain is found in several transcription cofactors and coactivators that possess HAT activity or other nucleosome remodeling activity, thus making the bromodomain a candidate domain to mediate interaction with nucleosomes. It was shown previously that the Gcn5 bromodomain is not required for acetylation of free histones by recombinant Gcn5 in vitro or for transcriptional activation by strong activators, but its deletion does result in minor growth defects and reduced activation by weak activators (13, 21, 48). To investigate the role of the bromodomain in nucleosome acetylation, we prepared Ada and SAGA complexes from extracts of gcn5Δ cells containing integrated copies of wild-type or bromodomain-deleted (ΔBrD) GCN5. To control for potential construct-specific differences in stability or expression, two independent bromodomain mutants (13), containing the sequences for either the first 350 or 280 aa of the 439-aa full-length Gcn5, were used for these experiments.
Wild-type Ada and SAGA complexes (Mono Q peak fractions 20 and 40, respectively) showed comparable levels of histone H3-acetylating activity on nucleosomal substrates (Fig. 5A). Ada complex fractions derived from the bromodomain mutants had nucleosome-acetylating activities similar to that of the wild-type Ada complex (fraction 20), but both mutants displayed significantly reduced activity by SAGA (fraction 40). A different situation was observed when free histones were used as a substrate for acetylation: the levels of acetylation by the ΔBrD SAGA fractions were only moderately lower than that of wild-type SAGA. Quantitation of the H3/H2B species in the HAT assays shown for fraction 40 revealed that the nucleosomal substrates were acetylated approximately half as well as free histones by ΔBrD SAGA compared to wild-type SAGA (Fig. 5A, bar graph). In addition, in each case acetylation of free histones by SAGA was higher than free histone acetylation by Ada and nucleosome acetylation by either SAGA or Ada. Anti-Ada2 and anti-Gcn5 Western blots (Fig. 5B) confirmed that these free histone acetylation results accurately reflected the amount of complex in the fractions, since SAGA contained more protein than Ada. The immunoblots also show that the bromodomain mutants had nearly wild-type amounts of both Ada and SAGA.
FIG. 5.
Effect of Gcn5 bromodomain deletion on nucleosomal HAT activity. Cells containing wild-type (w.t.; residues 1 to 439) or ΔBrD (residues 1 to 350 or 1 to 280) GCN5 were used to prepare Mono Q fractions by the standard purification method for the HAT complexes. (A) HAT assays with nucleosomal or free histone substrates were performed with even-numbered fractions surrounding the Ada and SAGA peaks; fluorographs of the resulting gels are presented. The arrows denote the relative positions of the four histone proteins as separated on SDS–18% polyacrylamide gels; visualized species contain acetyl groups labeled with 3H. The bar graph at right presents the ratio of nucleosomal to free histone H3/H2B acetylation for SAGA fraction 40, normalized to the wild-type ratio. (B) Samples of fractions 20 (Ada peak), 30 (negative control; no HAT complex), and 40 (SAGA peak) were also analyzed by Western blotting. Five microliters of each was run on SDS-polyacrylamide gels and electroblotted to nitrocellulose, which underwent immunodetection with dilutions of anti-Ada2, anti-Gcn5, or anti-TafII antibodies. Markers on the Gcn5 panels indicate the Gcn5 species visualized on equivalent portions of the blots, and their positions of migration agreed with their predicted sizes (51, 40, and 32 kDa for the 1-439, 1-350, and 1-280 constructs, respectively). The peak SAGA fraction from a preparation of gcn5Δ (FY1370) cells was also tested for TafIIs (right).
Overall, the results show that despite the fact that SAGA fractions derived from wild-type and Gcn5 ΔBrD strains have copious amounts of Gcn5 and innate HAT activity, they acetylate nucleosomes less effectively than the less concentrated Ada complex. Furthermore, the observed defects may be due to absence of the bromodomain and not due to loss of other associated protein factors, since no major differences in size were observed in Superose 6 fractionations of wild-type and bromodomain mutant SAGA complexes (data not shown). In addition, Western analysis showed that TafII60, TafII68, and TafII90 were present in the ΔBrD SAGA complexes, as well as in gcn5Δ SAGA (Fig. 5B).
Since Spt7 is another SAGA component harboring a bromodomain, we also analyzed a similar deletion within this protein. Ada and SAGA complexes prepared from SPT7 ΔBrD cells (20) or from a double mutant with GCN5 ΔBrD were indistinguishable from their wild-type SPT7 counterparts in terms of nucleosome acetylation or growth phenotypes under various conditions (data not shown). Thus, the Spt7 bromodomain has no discernible functional effect or synthetic interaction with the Gcn5 bromodomain in these assays.
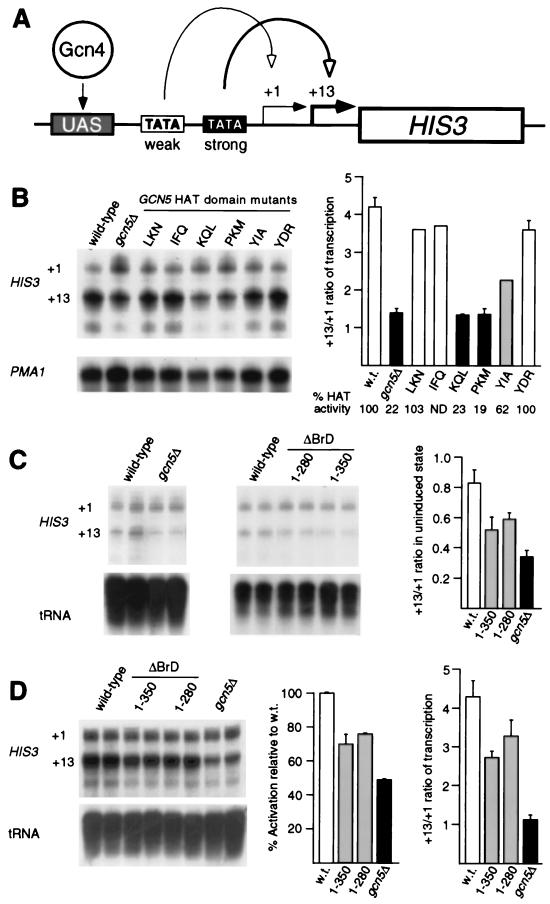
Gcn5 bromodomain and HAT domain mutants display similar HIS3 transcriptional defects.
In vitro, nucleosomal acetylation by Gcn5 was specifically lowered by deletion of the bromodomain. The effect of Gcn5 bromodomain deletion in vivo was investigated by S1 analysis of RNA produced from the HIS3 gene, which has been shown to be regulated by Gcn5’s nucleosome acetylation activity (44). There are two RNA start sites in HIS3 (Fig. 6A), and in wild-type cells under basal conditions these sites are used equally (Fig. 6C and reference 70). In strains lacking Gcn5 or bearing Gcn5 ΔBrD, usage of the +13 start site was reduced (Fig. 6C). Also similar to the absence of Gcn5, the Gcn5 ΔBrD strains show diminished activation potential (Fig. 6D).
FIG. 6.
HIS3 transcriptional defects observed in GCN5 null, HAT domain substitution, and ΔBrD mutants. (A) Diagram of HIS3 promoter function. Weak and strong TBP-binding sequences (TATA boxes) direct transcription from the +1 and +13 start sites, respectively. The activator Gcn4 binds to multiple upstream sites (UAS). (B) Start site usage for activated HIS3 transcription in wild-type (w.t.), gcn5Δ, and GCN5 HAT domain substitution mutant strains. Activating conditions were achieved with 3-aminotriazole. PhosphorImager quantitation of the S1 assays is presented as the ratio of +13 to +1. Shown at the bottom are in vitro HAT activities (as a percentage of wild-type activity) of native SAGA complexes purified from the indicated strains (27). (C) Start site preference for wild-type, gcn5Δ, and ΔBrD strains under noninducing conditions (no 3-aminotriazole). (D) Activated HIS3 transcription in wild-type, gcn5Δ, and ΔBrD strains. PMA1 RNA and tRNA were included as internal controls for sample loading.
Under activated conditions in wild-type cells, there is a strong preference for the +13 start site over the +1 site (Fig. 6B and D and reference 70). In gcn5Δ cells, +13 and +1 start sites were used equally in induced conditions (Fig. 6B and D). To examine the potential role of acetylation in start site usage, we tested the effects of HAT domain and bromodomain mutations. HAT domain substitution mutations, which exhibit a correlation between HAT competence and transcriptional activation at a model promoter (78), also show a clear correlation with start site usage (Fig. 6B). The +13 start site was preferred in yeast strains bearing the LKN, IFQ, or YDR mutation (HAT-competent mutants), and hence these were similar to the wild type. KQL and PKM (HAT-defective mutants) had much reduced start site preference for +13 and were similar to the gcn5Δ strain. Finally, YIA (HAT-intermediate mutant) showed an intermediate preference for start sites between GCN5+ and gcn5Δ. Thus, the results demonstrate a strong correlation between HAT activity and start site preference, indicating that HAT activity of Gcn5 is critical for normal start site selection at the HIS3 promoter.
We then tested the effects of the two ΔBrD mutations on the +13/+1 start site ratio under activated conditions. Interestingly, these mutants also showed a change in start site usage (Fig. 6D), and the effect was intermediate between those of the wild type and HAT domain mutants that result in complete loss of HAT activity (compare Fig. 6D with Fig. 6B). Thus, several aspects of transcriptional control of HIS3 are altered in the bromodomain mutants in a manner resembling a defect specifically in Gcn5 HAT catalytic function.
TBP binding by SAGA in vitro requires Spt8 but not Spt3.
We then characterized SAGA prepared from strains bearing deletions of the other moderate class, Spt3 or Spt8, to determine whether the biochemical functions of the complexes were altered. SAGA complexes purified from spt3Δ and spt8Δ cells were tested for HAT activity, and no major differences in acetylation were observed (data not shown). Thus, in contrast to Gcn5, neither Spt3 nor Spt8 is crucial for the HAT activity of SAGA.
SAGA complexes prepared from SPT3 or SPT8 mutant strains were therefore intact and possessed HAT activity, but as described above, genetic evidence had indicated that these Spt proteins are important for normal function. Previous results suggested that Spt3 and Spt8 interact functionally, and possibly physically, with TBP. Although TBP does not cofractionate with SAGA (23), pull-downs of GST-Spt20 from yeast extracts recovered TBP along with the other Spt proteins (59). Therefore, we tested whether SAGA interacts with TBP, and if so, whether Spt3 and/or Spt8 was required for this interaction.
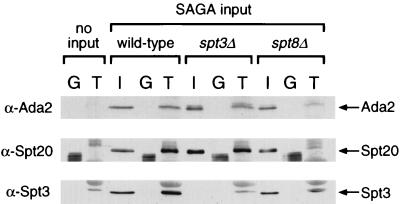
GST-TBP was used for pull-down experiments with the Superose-fractionated SAGA complexes derived from wild-type, spt3Δ, and spt8Δ strains, and the binding of complexes was monitored by Western analysis. Wild-type SAGA interacted with GST-TBP but not with the GST negative control protein, as visualized by using antibodies to Ada2, Spt3, and Spt20 (Fig. 7). Interestingly, while SAGA lacking Spt3 also bound to GST-TBP, SAGA lacking Spt8 showed a very low level of binding (Fig. 7). The finding that Spt3 was not a critical determinant of SAGA interaction with TBP was supported by the observation that Spt3 was present in SAGA prepared from the Spt8 disruption, which bound poorly to TBP (Fig. 7). Similarly, the potential participation of Spt8 in TBP binding is bolstered by the finding that Spt8 is present in the spt3Δ SAGA sample (Fig. 1B). Altogether, these data indicate that the Spt3/Spt8 subgroup is not critical for SAGA structure, but at least Spt8 is required for SAGA interaction with TBP in vitro.
FIG. 7.
In vitro binding of wild-type, spt3Δ, and spt8Δ SAGA complexes to TBP. Glutathione-Sepharose beads containing bacterially expressed GST or GST-TBP were incubated alone or with similar amounts of wild-type, spt3Δ, or spt8Δ SAGA complexes at 4°C under conditions of approximately 60 mM NaCl. After washing, Western blotting was performed with these beads to analyze the proteins that had bound to GST (G lanes) or GST-TBP (T lanes). The input (I) lanes contained half of the amount of SAGA used for the binding experiments. The position of the antibody-visualized Ada2, Spt20, and Spt3 bands on the Western blots are indicated by arrows. An unidentified species in the GST-TBP preparation (T lanes) had a mobility similar to but slightly slower than that of Spt3 and cross-reacted with anti-Spt3 antibodies; the band seen in the spt8Δ T lane is a combination of this species and a small amount of Spt3.
DISCUSSION
A number of interconnected processes are involved in the activation of transcription, and the recently discovered SAGA complex apparently combines several of them: activator interaction (through Ada2, Gcn5, and Ada3), histone acetylation (by Gcn5), and TBP interaction (through Spt8 and Spt3). We have sought to characterize the latter two of these functions in vivo and in vitro and to analyze the structure of SAGA by studying mutant complexes and individual subunits in vitro. These studies indicate that while neither the Gcn5/Ada2/Ada3 nor Spt3/Spt8 subgroup is critical for SAGA structure, each confers a distinct and important biochemical function. Either may be partially dispensable, but loss of both is highly detrimental to overall SAGA function.
Components necessary for SAGA structure.
In this study, we have identified Ada1 as a component of the SAGA complex, and we have further shown that its loss leads to the physical disruption of SAGA and severe phenotypic defects. This places it in a class with two other previously described SAGA components with similar qualities, Spt20 and Spt7. These three subunits each appear to be necessary for the structure of the complex, with the critical role of holding it together. The precise interactions among these proteins and with other SAGA components remain to be determined, but their function could be to link the Spt and Ada subunits within SAGA.
We have also characterized spt3Δ and spt8Δ mutants and find that in contrast to loss of Spt20/Spt7/Ada1, they maintain overall SAGA integrity, as was seen with gcn5Δ. Moreover, SAGA integrity is maintained in the double disruption of Gcn5 and either Spt3 or Spt8. Based on these data, we conclude that the Gcn5/Ada2/Ada3 and Spt3/Spt8 groups of proteins are likely to be peripheral within the complex, i.e., exposed and in a position to form protein contacts with other factors, including histones and TBP. Overall, the biochemical characterization of structure nicely mirrors the genetic analysis of function: disruptions of the SAGA components that are integral for structure are extremely debilitating in vivo, while disruptions of the more structurally peripheral components are less severe.
Nucleosomal acetylation by the SAGA complex.
The most thoroughly characterized function within the SAGA complex has been the HAT activity of Gcn5, and recent studies indicate that the HAT domain is required for Gcn5’s role in transcription (13, 44, 78). The experiments presented here suggest that another region of Gcn5, the bromodomain, may have significant effects on the function of the SAGA complex in acetylation. Specifically, removal of the bromodomain reduces the HAT activity of SAGA on nucleosomal substrates, even though the complex and its activity on free histones are otherwise largely intact. One interpretation of these data is that the bromodomain is required for physical interaction either with histones or with other components in SAGA that bind to histones (and other proteins would fulfill such a function in the Ada complex). Relevant to this is the observation that the size of SAGA prepared from the ΔBrD strain was not grossly altered. However, small changes in size or shape of SAGA would not be apparent due to the low sensitivity of the Superose 6 sizing column near the void volume (where SAGA elutes). In this view, Gcn5 possesses separate domains for catalytic HAT function and for interaction with nucleosomal substrates. This is consistent with two previous observations. First, recombinant Gcn5 acetylates free core histones but not nucleosome-assembled histones (24); second, mutations in TafII68 similarly reduce nucleosomal histone acetylation without reducing free histone acetylation (25). However, the SAGA complexes bearing Gcn5 deletions of the bromodomain are not altered for TafII60, TafII68, or TafII90, indicating that there may be multiple determinants for nucleosome interaction.
A second interpretation of the data is that the bromodomain defect could be related to the fact that nucleosome acetylation by wild-type SAGA is already somewhat inhibited relative to that of the 0.8-MDa Ada complex, as shown in Fig. 5. This unidentified inhibitory activity, not present in the Ada complex, may reside with the Spt, TafII, or unknown subunits of SAGA and may act by interfering with Gcn5’s interaction with nucleosomes. Thus, the function of the Gcn5 bromodomain in SAGA could be to counteract this inhibition partially, so that deletion of the domain leads to a more complete inhibition. Such a process would be relevant only in the context of SAGA, since ΔBrD Ada complex acetylates nucleosomes very well. Further study will be required to determine which of these two models is correct and how regulation of acetylation activity may be achieved through the bromodomain.
In either case, it is important to note that the deletion of the bromodomain affected transcription of the HIS3 gene in vivo, causing both a slight reduction in activation potency and an alteration in start sites. These effects were also seen in a total disruption of Gcn5 or in HAT-defective substitution mutants of Gcn5. Thus, the effect of the bromodomain deletion in vivo clearly ties this domain to acetylation function of Gcn5.
Bromodomains occur in numerous proteins having a role in chromatin remodeling, including the acetyltransferases CBP and TAFII250, and Snf2/Swi2, a component of the ATP-dependent remodeling complex Swi-Snf (41). Our data do not necessarily indicate that in general, bromodomains are required for nucleosome association. First, the deletion of the bromodomain in Spt7 did not affect acetylation; second, the bromodomain of human Gcn5 interacts with a nonhistone DNA binding protein complex, called Ku70/80 (4). The bromodomain-Ku physical interaction results in phosphorylation of human GCN5 and repression of its HAT activity, apparently via a Ku-dependent recruitment of DNA-dependent protein kinase. Thus, we believe that there may not be a unified mechanism for bromodomains, but rather, there may be a multiplicity of roles involved in modulation of activity of the resident complex.
TBP interaction by the SAGA complex.
TBP interaction is another function of SAGA that had been suggested by previous studies but has been directly demonstrated here. In vitro binding experiments show specific interaction between purified SAGA and GST-TBP. Thus, these data agree with previous evidence that TBP interacts with Ada2 (3), Ada3 (64), or Spt20 (59) in the context of yeast whole-cell extract.
GST-TBP interaction analysis of two mutant complexes indicated that Spt8 is required for SAGA-TBP interaction in vitro, but Spt3 is not. We do not yet know whether Spt8 makes direct contact with TBP. While the mutant SAGA is not obviously smaller than wild-type SAGA, the sensitivity of Superose 6 sizing is low, as discussed above. The three largest TafIIs in SAGA are not sufficient for TBP interaction, because they are present in complex prepared from the spt8Δ mutant strain, which is impaired in TBP binding. In addition, SAGA prepared from the tafII68 mutant strain maintains binding to TBP but has lost Spt3 (25), which underscores that Spt3 is not required for TBP interaction in vitro.
The requirement of Spt8, as opposed to Spt3, for TBP binding was unexpected in light of previous in vivo results, which suggested a direct interaction between Spt3 and TBP and an auxiliary role for Spt8 (17, 18). It is possible that Spt3 contributes significantly to the interaction under physiological conditions within the cell. Thus, there may be multiple ways that SAGA recruits TBP, including through TafIIs, and our in vitro binding assay detects only a subset of them.
Model for multiple functionality of SAGA.
The spt3Δ gcn5Δ and spt8Δ gcn5Δ double mutants (Fig. 4) apparently disrupt two in vivo functions, namely, TBP binding and nucleosome acetylation, leading to major phenotypic defects. The combined losses cause defects which approximate those of a mutant which disrupts SAGA altogether (spt20Δ). This provides further evidence that multiple functions are incorporated in the SAGA complex in vivo, and that the two discussed above are needed for its full functionality but are partially redundant. Thus, one hypothesis that emerges from these data is that two functions of SAGA are directed at the same objective: regulation of TBP binding to the TATA box. Activation domain interaction is a third presumptive function of the SAGA complex. This was indicated in earlier studies of Ada2, Ada3, and Gcn5 function and has been shown directly by recent studies with SAGA itself (75).
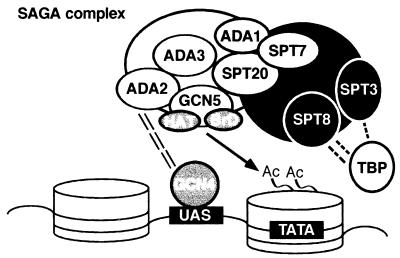
These results, along with additional findings presented in this and previous studies, suggest an overall model for the structure and function of the SAGA complex, shown in Fig. 8. In this model, SAGA consists of two major parts with discrete functions: adaptor related and Spt related. A knockout of any of the identified adaptor-related components (Gcn5, Ada2, or Ada3) leads to moderate growth defects and resistance to the toxicity of overexpressed GAL4-VP16, while knockouts of the shown Spt proteins result in defective phenotypes of varying severity and in suppression of Ty element insertion in a promoter. At the interface between these two parts would be Ada1, Spt20, and Spt7 proteins, identified as being important to SAGA structural integrity; knockouts of any of these have combined Ada− and Spt− phenotypes. A number of unknown proteins, as well as the several TafIIs mentioned above, are also present in SAGA.
FIG. 8.
Model for SAGA structure and function in transcription. Depicted is a hypothetical gene with an upstream activation sequence (UAS) and TATA box; the DNA is wound around nucleosomes (cylinders). The SAGA complex, composed of adaptor (white) and Spt (black) functional regions and held together by several structurally important proteins (grey), is proposed to interact with the an activator (such as Gcn4) at the UAS through Ada2. This allows the HAT activity of Gcn5, in cooperation with its bromodomain (BrD), to acetylate (Ac = acetyl group) the amino-terminal tails of nucleosomal histones, providing more effective TATA binding of TBP. Further regulation is provided by SAGA-TBP interaction, principally through Spt8, in conjunction with Spt3 and/or other factors. The large white and black circles represent unidentified members of SAGA; several TafIIs are also present in SAGA (25), but their exact role in the complex has yet to be defined.
As the complex approaches a promoter, the Ada2 subunit would interact with the activation domain of an activator bound to an upstream activating sequence, bringing Gcn5 and the Spt components in proximity with the promoter. We propose that after this recruitment, the Gcn5 moiety of SAGA functions to acetylate histones within the nucleosome(s) of the promoter region, with the assistance of the bromodomain, thereby remodeling the chromatin structure in a localized region. This makes the TATA box available to TBP, and its DNA binding and/or interaction with general transcription factors may be regulated through contact with Spt8, Spt3, TafIIs, or undetermined SAGA subunits. In this model, loss of either acetylation of the nucleosomes at the TATA box (e.g., gcn5Δ) or loss of TBP regulation (e.g., spt8Δ) is tolerated, but loss of both functions (e.g., gcn5Δ spt3Δ and ada1Δ) is highly detrimental.
This model assumes that SAGA is an independent complex with multiple functions, but the exact relationship between Ada and SAGA has not yet been established. The two HAT complexes are known to have Gcn5, Ada2, and Ada3 in common, but other subunit identities await the direct characterization and comparison of purified samples—Ada and SAGA may well be functionally distinct complexes that only share a few subunits. Further characterization of SAGA will shed light on this issue.
The present functional analyses of SAGA illustrate an important concept, i.e., in large complexes, individual components have distinct biochemical functions. The combined biochemical and genetic analysis of SAGA yields unique insights into function. In general, these studies are a paradigm for further studies of this multicomponent complex, as well as other large acetylation and deacetylation complexes now under investigation.
ACKNOWLEDGMENTS
S. M. Roberts and P. A. Grant contributed equally to this work.
We thank R. Candau for preparation of the ADA1 disruption strain SB11, Z. Yang for technical assistance in the preparation of Spt7ΔBrD HAT complexes, and N. Barlev for advice on binding assays. We thank R. Reeder for the gift of GST-TBP plasmid, J. Reese and M. Green for TafII antibodies, and A. Navas, Z. Zhou, and S. Elledge for informing us that spt20 mutants have an HUS phenotype. We thank G. Moore for helpful discussions and critical comments on the manuscript.
This research was supported by grants from the National Institutes of General Medical Sciences to S.L.B., F.W., and J.L.W. and from the National Science Foundation and the Council for Tobacco Research to S.L.B. P.A.G. was supported by a postdoctoral fellowship from the American Cancer Society. An NIH Cancer Core training grant to the Wistar Institute supported D.E.S. and L.J.D.; D.E.S. was also supported by an NIH postdoctoral fellowship.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Wiley & Sons, Inc.; 1994. [Google Scholar]
- 2.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 3.Barlev N A, Candau R, Wang L, Darpino P, Silverman N, Berger S L. Characterization of physical interactions of the putative transcriptional adaptor, ADA2, with acidic activation domains and TATA-binding protein. J Biol Chem. 1995;270:19337–19344. doi: 10.1074/jbc.270.33.19337. [DOI] [PubMed] [Google Scholar]
- 4.Barlev N A, Poltoratsky V, Owen-Hughes T, Ying C, Liu L, Workman J L, Berger S L. Repression of GCN5 histone acetyltransferase activity via bromodomain-mediated binding and phosphorylation by the Ku/DNA-dependent protein kinase complex. Mol Cell Biol. 1998;18:1349–1358. doi: 10.1128/mcb.18.3.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berger S L, Piña B, Silverman N, Marcus G A, Agapite J, Regier J L, Triezenberg S J, Guarente L. Genetic isolation of ADA2: a potential transcriptional adaptor required for function of certain acidic activation domains. Cell. 1992;70:251–265. doi: 10.1016/0092-8674(92)90100-q. [DOI] [PubMed] [Google Scholar]
- 6.Boeke J D, LaCroute F, Fink G R. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 7.Bradbury E M. Reversible histone modifications and the chromosome cell cycle. Bioessays. 1992;14:9–16. doi: 10.1002/bies.950140103. [DOI] [PubMed] [Google Scholar]
- 8.Braunstein M, Rose A B, Holmes S G, Allis C D, Broach J R. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- 9.Brownell J E, Zhou J, Ranalli T, Kobayashi R, Edmondson D G, Roth S Y, Allis C D. Tetrahymena histone acetyltransferase A: a transcriptional co-activator linking gene expression to histone acetylation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 10.Cairns B R, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg R D. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- 11.Candau R, Berger S L. Structural and functional analysis of yeast putative adaptors: evidence for an adaptor complex in vivo. J Biol Chem. 1996;271:5237–5345. doi: 10.1074/jbc.271.9.5237. [DOI] [PubMed] [Google Scholar]
- 12.Candau R, Moore P A, Wang L, Barlev N, Ying C Y, Rosen C A, Berger S L. Identification of functionally conserved human homologues of the yeast adaptors ADA2 and GCN5. Mol Cell Biol. 1996;16:593–602. doi: 10.1128/mcb.16.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Candau R, Zhou J, Allis C D, Berger S L. Histone acetyltransferase activity and interaction with ADA2 are critical for GCN5 function in vivo. EMBO J. 1997;16:555–565. doi: 10.1093/emboj/16.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiang Y C, Komarnitsky P, Chase D, Denis C L. ADR1 activation domains contact the histone acetyltransferase GCN5 and the core transcriptional factor TFIIB. J Biol Chem. 1996;271:32359–32365. doi: 10.1074/jbc.271.50.32359. [DOI] [PubMed] [Google Scholar]
- 15.Côté J, Quinn J, Workman J L, Peterson C L. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- 16.Côté J, Utley R T, Workman J L. Analysis of transcription factor binding to nucleosomes. Methods Mol Genet. 1995;6:108–128. doi: 10.1016/s0076-6879(96)74024-7. [DOI] [PubMed] [Google Scholar]
- 17.Eisenmann D M, Arndt K M, Ricupero S L, Rooney J W, Winston F. SPT3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes Dev. 1992;6:1319–1331. doi: 10.1101/gad.6.7.1319. [DOI] [PubMed] [Google Scholar]
- 18.Eisenmann D M, Chapon C, Roberts S M, Dollard C, Winston F. The Saccharomyces cerevisiae SPT8 gene encodes a very acidic protein that is functionally related to SPT3 and TATA-binding protein. Genetics. 1994;137:647–657. doi: 10.1093/genetics/137.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eisenmann D M, Dollard C, Winston F. SPT15, the gene encoding the yeast TATA binding factor TFIID, is required for normal transcription initiation in vivo. Cell. 1989;58:1183–1191. doi: 10.1016/0092-8674(89)90516-3. [DOI] [PubMed] [Google Scholar]
- 20.Gansheroff L J, Dollard C, Tan P, Winston F. The Saccharomyces cerevisiae SPT7 gene encodes a very acidic protein important for transcription in vivo. Genetics. 1995;139:523–536. doi: 10.1093/genetics/139.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Georgakopoulos T, Gounalaki N, Thireos G. Genetic evidence for the interaction of the yeast transcriptional co-activator proteins GCN5 and ADA2. Mol Gen Genet. 1995;246:723–728. doi: 10.1007/BF00290718. [DOI] [PubMed] [Google Scholar]
- 22.Georgakopoulos T, Thireos G. Two distinct yeast transcriptional activators require the function of the GCN5 protein to promote normal levels of transcription. EMBO J. 1992;11:4145–4152. doi: 10.1002/j.1460-2075.1992.tb05507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grant, P. A. Unpublished data.
- 24.Grant P A, Duggan L, Côté J, Roberts S M, Brownell J E, Candau R, Ohba R, Owen-Hughes T, Allis C D, Winston F, Berger S L, Workman J L. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 25.Grant P A, Schieltz D, Pray-Grant M G, Steger D J, Reese J C, Yates III J R, Workman J L. A subset of TAFIIs are integral components of the SAGA complex required for nucleosome acetylation and transcription stimulation. Cell. 1998;94:45–53. doi: 10.1016/s0092-8674(00)81220-9. [DOI] [PubMed] [Google Scholar]
- 26.Grant P A, Sterner D E, Duggan L J, Workman J L, Berger S L. The SAGA unfolds: convergence of transcription regulators in chromatin-modifying complexes. Trends Cell Biol. 1998;8:193–197. doi: 10.1016/s0962-8924(98)01263-x. [DOI] [PubMed] [Google Scholar]
- 27.Gregory P D, Schmid A, Zavari M, Liu L, Berger S L, Hörz W. Absence of Gcn5 HAT activity defines a novel state in the opening of chromatin at the PHO5 promoter in yeast. Mol Cell. 1998;1:495–505. doi: 10.1016/s1097-2765(00)80050-7. [DOI] [PubMed] [Google Scholar]
- 28.Guarente L. Transcriptional coactivators in yeast and beyond. Trends Biochem Sci. 1995;20:517–521. doi: 10.1016/s0968-0004(00)89120-3. [DOI] [PubMed] [Google Scholar]
- 29.Hager G, Smith C, Svaren J, Hörz W. Initiation of expression: remodelling genes. In: Elgin S C R, editor. Chromatin structure and gene expression. Vol. 9. Oxford, England: IRL Press; 1995. pp. 89–103. [Google Scholar]
- 30.Hahn S, Buratowski S, Sharp P A, Guarente L. Isolation of the gene encoding the yeast TATA binding protein TFIID: a gene identical to the SPT15 suppressor of Ty element insertions. Cell. 1989;58:1173–1181. doi: 10.1016/0092-8674(89)90515-1. [DOI] [PubMed] [Google Scholar]
- 31.Hampsey M. A review of phenotypes in Saccharomyces cerevisiae. Yeast. 1997;13:1099–1133. doi: 10.1002/(SICI)1097-0061(19970930)13:12<1099::AID-YEA177>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 32.Harlow E, Lane I. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 33.Haynes S R, Dollard C, Winston F, Beck S, Trowsdale J, Dawid I B. The bromodomain: a conserved sequence found in human, Drosophila and yeast proteins. Nucleic Acids Res. 1992;20:2603. doi: 10.1093/nar/20.10.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hebbes T R, Clayton A L, Thorne A W, Crane-Robinson C. Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken beta-globin chromosomal domain. EMBO J. 1994;13:1823–1830. doi: 10.1002/j.1460-2075.1994.tb06451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hirschhorn J N, Brown S A, Clark C D, Winston F. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev. 1992;6:2288–2298. doi: 10.1101/gad.6.12a.2288. [DOI] [PubMed] [Google Scholar]
- 36.Horiuchi J, Silverman N, Marcus G A, Guarente L. ADA3, a putative transcriptional adaptor, consists of two separable domains and interacts with ADA2 and GCN5 in a trimeric complex. Mol Cell Biol. 1995;15:1203–1209. doi: 10.1128/mcb.15.3.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horiuchi J, Silverman N, Piña B, Marcus G A, Guarente L. ADA1, a novel component of the ADA/GCN5 complex, has broader effects than GCN5, ADA2, or ADA3. Mol Cell Biol. 1997;17:3220–3228. doi: 10.1128/mcb.17.6.3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Imbalzano A N, Kwon H, Green M R, Kingston R E. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature. 1994;370:481–485. doi: 10.1038/370481a0. [DOI] [PubMed] [Google Scholar]
- 39.Ito T, Bulger M, Pazin M J, Kobayashi R, Kadonaga J T. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- 40.Iyer V, Struhl K. Absolute mRNA levels and transcriptional initiation rates in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:5208–5212. doi: 10.1073/pnas.93.11.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jeanmougin F, Wurtz J-M, Le Douarin B, Chambon P, Losson R. The bromodomain revisited. Trends Biochem Sci. 1997;22:151–153. doi: 10.1016/s0968-0004(97)01042-6. [DOI] [PubMed] [Google Scholar]
- 42.Jeppesen P, Turner B M. The inactive X chromosome in female mammals is distinguished by a lack of histone H4 acetylation, a cytogenetic marker for gene expression. Cell. 1993;74:281–289. doi: 10.1016/0092-8674(93)90419-q. [DOI] [PubMed] [Google Scholar]
- 43.Kingston R E, Bunker C A, Imbalzano A N. Repression and activation by multiprotein complexes that alter chromatin structure. Genes Dev. 1996;10:905–920. doi: 10.1101/gad.10.8.905. [DOI] [PubMed] [Google Scholar]
- 44.Kuo M-H, Zhou J, Jambeck P, Churchill M E A, Allis C D. Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes Dev. 1998;12:627–639. doi: 10.1101/gad.12.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kwon H, Imbalzano A N, Khavari P A, Kingston R E, Green M R. Nucleosome disruption and enhancement of activator binding by a human SWI/SNF complex. Nature. 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- 46.Laurent B C, Treitel M A, Carlson M. The SNF5 protein of Saccharomyces cerevisiae is a glutamine- and proline-rich transcriptional activator that affects expression of a broad spectrum of genes. Mol Cell Biol. 1990;10:5616–5625. doi: 10.1128/mcb.10.11.5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marcus G A, Horiuchi J, Silverman N, Guarente L. ADA5/SPT20 links the ADA and SPT genes, which are involved in yeast transcription. Mol Cell Biol. 1996;16:3197–3205. doi: 10.1128/mcb.16.6.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marcus G A, Silverman N, Berger S L, Horiuchi J, Guarente L. Functional similarity and physical association between GCN5 and ADA2: putative transcriptional adaptors. EMBO J. 1994;13:4807–4815. doi: 10.1002/j.1460-2075.1994.tb06806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mizzen C A, Yang X-J, Kokubo T, Brownell J E, Bannister A J, Owen-Hughes T, Workman J, Wang L, Berger S L, Kouzarides T, Nakatani Y, Allis C D. The TAFII250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 50.O’Neill L P, Turner B M. Histone H4 acetylation distinguishes coding regions of the human genome from heterochromatin in a differentiation-dependent but transcription-independent manner. EMBO J. 1995;14:3946–3957. doi: 10.1002/j.1460-2075.1995.tb00066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ogryzko V V, Schlitz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 52.Owen-Hughes T, Utley R T, Côté J, Peterson C L, Workman J L. Persistent site-specific remodeling of a nucleosome array by transient action of the SWI/SNF complex. Science. 1996;273:513–516. doi: 10.1126/science.273.5274.513. [DOI] [PubMed] [Google Scholar]
- 53.Owen-Hughes T, Workman J L. Experimental analysis of chromatin function in transcription control. Crit Rev Eukaryotic Gene Expr. 1994;4:403–441. [PubMed] [Google Scholar]
- 54.Ozer J, Lezina L E, Ewing J, Audi S, Lieberman P M. Association of transcription factor IIA with TATA binding protein is required for transcriptional activation of a subset of promoters and cell cycle progression in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:2559–2570. doi: 10.1128/mcb.18.5.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Paranjape S M, Kamakaka R T, Kadonaga J T. Role of chromatin structure in the regulation of transcription by RNA polymerase II. Annu Rev Biochem. 1994;63:265–297. doi: 10.1146/annurev.bi.63.070194.001405. [DOI] [PubMed] [Google Scholar]
- 56.Peterson C L, Herskowitz I. Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell. 1992;68:573–583. doi: 10.1016/0092-8674(92)90192-f. [DOI] [PubMed] [Google Scholar]
- 57.Piña B, Berger S, Marcus G A, Silverman N, Agapite J, Guarente L. ADA3: a gene, identified by resistance to GAL4-VP16, with properties similar to and different from those of ADA2. Mol Cell Biol. 1993;13:5981–5989. doi: 10.1128/mcb.13.10.5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pollard K J, Peterson C L. Role for ADA/GCN5 products in antagonizing chromatin-mediated transcriptional repression. Mol Cell Biol. 1997;17:6212–6222. doi: 10.1128/mcb.17.11.6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roberts S M, Winston F. Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/mediator complexes. Genetics. 1997;147:451–465. doi: 10.1093/genetics/147.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roberts S M, Winston F. SPT20/ADA5 encodes a novel protein functionally related to the TATA-binding protein and important for transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:3206–3213. doi: 10.1128/mcb.16.6.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rose M D, Winston F, Hieter P. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 62.Rothstein R J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 63.Ruiz-García A B, Sendra R, Pamblanco M, Tordera V. Gcn5p is involved in the acetylation of histone H3 in nucleosomes. FEBS Lett. 1997;403:186–190. doi: 10.1016/s0014-5793(97)00049-5. [DOI] [PubMed] [Google Scholar]
- 64.Saleh A, Lang V, Cook R, Brandl C J. Identification of native complexes containing the yeast coactivator/repressor proteins NGG1/ADA3 and ADA2. J Biol Chem. 1997;272:5571–5578. doi: 10.1074/jbc.272.9.5571. [DOI] [PubMed] [Google Scholar]
- 65.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 66.Scherer S, Davis R W. Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc Natl Acad Sci USA. 1979;76:4951–4955. doi: 10.1073/pnas.76.10.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Silverman N, Agapite J, Guarente L. Yeast ADA2 protein binds to the VP16 protein activation domain and activates transcription. Proc Natl Acad Sci USA. 1994;91:11665–11668. doi: 10.1073/pnas.91.24.11665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Steger D J, Workman J L. Remodeling chromatin structures for transcription: What happens to the histones? Bioessays. 1996;18:875–884. doi: 10.1002/bies.950181106. [DOI] [PubMed] [Google Scholar]
- 70.Struhl K. Constitutive and inducible Saccharomyces cerevisiae promoters: evidence for two distinct molecular mechanisms. Mol Cell Biol. 1986;6:3847–3853. doi: 10.1128/mcb.6.11.3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Svaren J, Hörz W. Regulation of gene expression by nucleosomes. Curr Opin Genet Dev. 1996;6:164–170. doi: 10.1016/s0959-437x(96)80046-3. [DOI] [PubMed] [Google Scholar]
- 72.Tamkun J W, Deuring R, Scott M P, Kissinger M, Pattatucci A M, Kaufman T C, Kennison J A. brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell. 1992;68:561–572. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- 73.Tsukiyama T, Wu C. Purification and properties of an ATP dependent nucleosome remodeling factor. Cell. 1995;83:1011–1020. doi: 10.1016/0092-8674(95)90216-3. [DOI] [PubMed] [Google Scholar]
- 74.Turner B M, Birley A J, Lavender J. Histone H4 isoforms acetylated at specific lysine residues define individual chromosomes and chromatin domains in Drosophila polytene nuclei. Cell. 1992;69:375–384. doi: 10.1016/0092-8674(92)90417-b. [DOI] [PubMed] [Google Scholar]
- 75.Utley R T, Ikeda K, Grant P A, Côté J, Steger D J, Eberharter A, John S, Workman J L. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature. 1998;394:498–502. doi: 10.1038/28886. [DOI] [PubMed] [Google Scholar]
- 76.Varga-Weisz P D, Wilm M, Bonte E, Dumas K, Mann M, Becker P B. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature. 1997;388:598–602. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]
- 77.Vettese-Dadey M, Grant P A, Hebbes T R, Crane-Robinson C, Allis C D, Workman J L. Acetylation of histone H4 plays a primary role in enhancing transcription factor binding to nucleosomal DNA in vitro. EMBO J. 1996;15:2508–2518. [PMC free article] [PubMed] [Google Scholar]
- 78.Wang L, Liu L, Berger S L. Critical residues for histone acetylation by GCN5, functioning in Ada and SAGA complexes, are also required for transcriptional function in vivo. Genes Dev. 1998;12:640–653. doi: 10.1101/gad.12.5.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Winston F. Analysis of SPT genes: a genetic approach toward analysis of TFIID, histones, and other transcription factors of yeast. In: McKnight S L, Yamamoto K R, editors. Transcriptional regulation. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 1271–1293. [Google Scholar]
- 80.Winston F, Dollard C, Ricupero-Hovasse S L. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- 81.Wolffe A P. Chromatin: structure and function. London, England: Academic Press; 1992. [Google Scholar]
- 82.Yang X-J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral E1A oncoprotein. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]