Abstract
Recent studies have identified the metazoan ER-resident proteins, TMEM41B and VMP1, and so structurally related VTT-domain proteins, as glycerolipid scramblases.
Keywords: scramblase, TMEM41B, VMP1, VTT-domain
Most cellular phospholipids are synthesized in the cytosolic leaflet of the ER, then equilibrated to the luminal leaflet by lipid scramblases. Lipids from the ER are then redistributed to other organelles either via the secretory pathway or else via protein-mediated lipid transport at membrane contact sites. With the arguable exception of vesicular trafficking, the molecular mechanisms underlying these lipid redistribution steps remain murky. In particular, even the identity of ER-resident scramblases is still being discovered, as are their roles in subsequent lipid trafficking steps.
A number of recent studies have highlighted the role of two metazoan ER-resident proteins, TMEM41B and VMP1, in processes broadly related to lipid dynamics. Thus, the proteins are implicated in lipid droplet maturation, in apolipoprotein maturation and secretion, and in the formation of the autophagosome (Tabara and Escalante, 2016; Morita et al., 2018; Morishita et al., 2019; Shoemaker et al., 2019). They were also reported to be critical in flavi- and coronavirus replication (Hoffmann et al., 2021; Schneider et al., 2021) potentially via a role in replication compartment formation. Both TMEM41B and VMP1 are integral membrane proteins in the DedA family, which also includes bacterial homologs involved in membrane integrity, all of whose members comprise signature 3-helix “VTT domains” (Okawa et al., 2021). Bioinformatics analyses suggested that the members of the family might be half-transporters that might oligomerize to form transporters (Okawa et al., 2021) and, indeed, TMEM41B and VMP1 are reported to form a 1:1 complex (Morita et al., 2018). Still, the function of DedA proteins was unknown until now, with several groups (including ours) reporting that TMEM41B and VMP1, and so likely all members of the DedA family, are glycerolipid scramblases (Ghanbarpour et al., 2021; Li et al., 2021).
TMEM41B and VMP1 as ER-Resident Lipid Scramblases
In a pair of recent publications, our groups as well as Yang and colleagues established TMEM41B and VMP1 as lipid scramblases using a series of in vitro reconstitution assays (Ghanbarpour et al., 2021; Li et al., 2021). Either protein alone, or in combination, was able to redistribute a variety of fluorescently-labeled glycerophospholipids from the luminal to the exposed face of proteoliposomes as assessed by both dithionite sensitivity and BSA-mediated lipid removal. The lipid redistribution is constitutive, ATP-independent and increases with increasing concentrations of reconstituted protein, but critically occurs without exposure of other soluble luminal contents as measured in a parallel leakage assay. Thus, by each of these measures, TMEM41B and VMP1 are lipid scramblases.
The Yang group also demonstrated that these proteins contribute to lipid distribution throughout the cell (Li et al., 2021). They expressed GFP-tagged sensors of different lipid populations in wildtype cells and cells depleted for TMEM41B and/or VMP1 and discovered that depletion of either scramblase leads to a mis-sorting of both phosphatidylserine (PS) and cholesterol. PS normally accumulates on the cytoplasmic face of the plasma membrane and endolysosomal compartments, but can be found on the lumenal face of the ER and early Golgi (e.g. Fairn et al., 2011; Holthuis and Menon, 2014; Hankins et al., 2015; note there is some cell-type dependence to this distribution: Tsuji et al., 2019). However, in cells depleted for TMEM41B and/or VMP1, they find that the luminal PS pool is missing, and instead these cells accumulate about twice as much cytoplasm-facing PS. In addition, these cells redistribute cholesterol from internal organelles to the plasma membrane which, as the authors note, is consistent with other work connecting PS levels to cholesterol trafficking (Maekawa and Fairn, 2015; Ercan et al., 2021). Likewise, Huang et al. (2021) introduced click-chemistry reactive choline headgroups into phosphatidylcholine (PC) to track PC on each leaflet. They discovered a similar loss of PC from the lumenal but not cytoplasmic face of the ER when TMEM41B is depleted. These results demonstrate that the VTT family scramblases play a major role in balancing the two leaflets in the biogenic ER membrane.
The identity of the scramblases moving newly synthesized lipids off the cytoplasmic face of the ER has been a long-standing question (Holthuis and Menon, 2014). Recently, several groups have shown that TMEM16K is a calcium-dependent ER-scramblase (Bushell et al., 2019; Tsuji et al., 2019), and there may still be other scramblases necessary to move specific subpopulations of lipid (Verchere et al., 2021). We suggest below that scramblase activity is required locally, and thus it may be advantageous to recruit specific scrambling proteins to different ER membrane events. However, the apparent complete loss of PS from luminal membranes when TMEM41B and VMP1 are missing suggests the intriguing possibility that these two scramblases may also play a fundamental and constitutive role in ER lipid homeostasis. This idea is consistent with new discoveries from the Chen lab, showing that the absence of these proteins promotes a new type of stress signal originated from imbalanced ER membrane leaflets, which are interpreted as a trigger of a new lipid-homeostasis sensing pathway, connecting in part to the SREBP response (Huang et al., 2021).
TMEM41B and VMP1 in Lipid Droplet and Apolipoprotein Production on the ER
Biogenesis of both lipid droplets (on the cytoplasmic face of the ER) and apolipoproteins (on the luminal face of the ER) depends upon ER-derived phospholipids to coat their growing neutral lipid cores. Genetic depletion of either TMEM41B or VMP1 drives the accumulation of oversized lipid droplets (Tabara and Escalante, 2016; Moretti et al., 2018; Morita et al., 2018) and incompletely formed or missing apolipoproteins with a concomitant loss of apolipoprotein-dependent lipid secretion (Morishita et al., 2019; Huang et al., 2021). The mechanism(s) through which TMEM41B or VMP1 regulate each of these events has not been understood. One possibility is that these events are a result of the global changes in lipid homeostasis detected when each is depleted in the cell (Huang et al., 2021; Li et al., 2021). For example, in vitro reconstitution of LD biogenesis suggests that both the levels of local cholesterol and the structure of local lipid asymmetry across the bilayer are key regulators in LD budding (Chorlay et al., 2019), each of which is now established to be disrupted in the TMEM41B/VMP1 depleted cells.
Alternatively, there are indications that TMEM41B and VMP1 in fact act locally, and perhaps directly engage the machinery responsible for membrane remodeling events on the ER. For example, in the case of LD biogenesis several groups have shown a colocalization of VMP1 with LD budding sites (Tabara and Escalante, 2016; Zhao et al., 2017). How it is localized is not yet known, although Yang and colleagues ruled out direct interactions between VMP1 or TMEM41B with the LD biogenesis protein Seipin (Li et al., 2021). A similar local role in lipoprotein production is now suggested by the work from Huang et al. (2021). The biogenesis of lipoproteins starts with the co-translational insertion of the major structural protein apolipoprotein B (ApoB) into the ER lumen, where ApoB is stabilized by the loading of lipids via the microsomal triglyceride transfer proteins (MTP). Through combinations of proteomics, co-immunoprecipitations and localization of endogenous tagged proteins, Huang et al. (2021) discovered that TMEM41B directly interacts with both ApoB and the apolipoprotein carrier protein SURF4, concentrated at the so-called ER exit sites. These data imply a possible protein complex-mediated coordination of local lipid availability, apolipoprotein production and lipoprotein trafficking to secretion sites on the ER. They also noted that the depletion of TMEM41B presents a more severe phenotype than depletion of VMP1, consistent with other recent reports that these two proteins are functionally similar but not perfectly redundant (e.g. Morita et al., 2018; Shoemaker et al., 2019) and perhaps indicative of the specific need for TMEM41B-apolipoprotein interactions during biogenesis.
TMEM41B and VMP1 as Pieces of a Cohesive Lipid-Transport System Functioning at Contact Sites
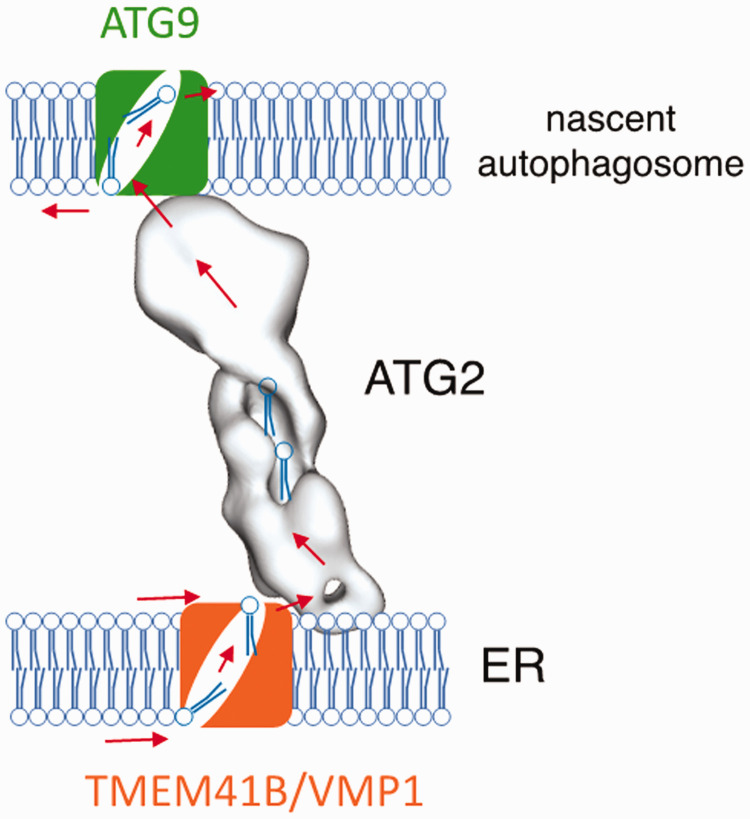
New discoveries by the Melia and Reinisch labs (Ghanbarpour et al., 2021) indicate that TMEM41B and VMP1 also act locally in autophagosome biogenesis, as part of a complex lipid transport system at ER-autophagosome contact sites that moves lipids from the ER to expand the autophagosome. They found a direct interaction between TMEM41B or VMP1 and ATG2, a club-shaped structure proposed to function as bridge that allows for bulk lipid flow between compartments (Valverde et al., 2019). While this interaction requires the N-terminal end of the ATG2 “club” (Ghanbarpour et al., 2021), another portion of ATG2 interacts directly with a second scramblase, ATG9, in Golgi derived vesicles that initiate autophagosome formation (Maeda et al., 2020; Matoba et al., 2020; Ghanbarpour et al., 2021). Thus, they proposed that TMEM41B/VMP1, ATG2, and ATG9 form a lipid transfer complex (Ghanbarpour et al., 2021) analogous to those observed in prokaryotes, such as the lipopolysaccharide transport system that moves lipids between the inner and outer membranes of Gram-negative bacteria (Okuda et al., 2016) (Figure 1). Bacterial lipopolysaccharide transport couples a lipid transport bridge with integral membrane proteins in both the lipid donor and acceptor compartments. These proteins re-equilibrate the membrane leaflets as lipids are removed or added and perhaps help in partitioning lipids between the transport protein and membranes, steps that are energetically costly and rate limiting (Reinisch and Prinz, 2021). During autophagosome formation, TMEM41B/VMP1 likely channel lipids onto ATG2 while also re-equilibrating the leaflets of the ER as lipids are extracted, then the lipids flow along ATG2 to ATG9 in the nascent autophagosome. ATG9 could help to insert arriving lipids into the autophagosome membrane and likely re-equilibrates the cytosolic and luminal leaflets to allow for membrane expansion (Maeda et al., 2020; Matoba et al., 2020; Ghanbarpour et al., 2021).
Figure 1.
TMEM41B and VMP1 Are Glycerolipid Scramblases (Ghanbarpour et al., 2021; Li et al., 2021) Important for Cellular Lipid Distribution From the ER. They can also act locally as components of lipid transport machinery, as shown here cooperating with the lipid transport protein ATG2 and a scramblase in the nascent autophagosome, ATG9, in supplying lipids for autophagosome biogenesis at ER-autophagosome membrane contact sites. They are also components of a protein assembly that facilitates maturation of lipoproteins in the ER (Huang et al., 2021).
Autophagosome biogenesis has evolved into a prominent model system in which to study de novo growth of organelles, especially those not connected to vesicular trafficking pathways. Importantly, ATG2 is structurally and functionally similar to VPS13-proteins, which are involved in various membrane biogenesis events, including prospore formation in yeast (Park and Neiman, 2012), and mitochondrial maintenance (Baldwin et al., 2021; Guillen-Samander et al., 2021) and peroxisome (Baldwin et al., 2021) and acrosome (Da Costa et al., 2020) formation in metazoa. Thus, we propose that VPS13-proteins are like ATG2 in cooperating with scramblases in bulk lipid transport. Indeed, human VPS13A has been reported to interact directly with a putative scramblase XK (Park and Neiman, 2020), and yeast Vps13 interacts directly with an integral membrane protein in the outer mitochondrial membrane ( Mcp1: John Peter et al., 2017; Bean et al., 2018) which has scrambling activity (Reinisch lab, unpublished). In humans, VPS13’s may transport lipids from the ER as VPS13A, -C, and -D all localize to contact sites with the ER (Kumar et al., 2018; Baldwin et al., 2021; Guillen-Samander et al., 2021), but no ER scramblase has yet been identified as an interaction partner. Because VPS13’s and ATG2 share sequences at their N-terminal ends (chorein-N motif, Munoz-Braceras et al., 2015) one intriguing possibility is that the human VPS13’s might also interact with TMEM41B or VMP1, which are already known to have such broad roles in membrane dynamics, in coordinating bulk lipid transfer.
Footnotes
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: KMR and TJM are funded by the NIH (GM131715 to K.M.R. and GM135290 to T.J.M.). X-WC is funded by National Key R&D Program grant 2018YFA0506900, National Science Foundation of China (NSFC) grants 91754000, 91957119, 31571213, 31521062 and 31771307.
ORCID iD: Karin M. Reinisch https://orcid.org/0000-0001-9140-6150
References
- Baldwin HA, Wang C, Kanfer G, Shah HV, Velayos-Baeza A, Dulovic-Mahlow M, Bruggemann N, Anding A, Baehrecke EH, Maric D, et al. (2021). VPS13D promotes peroxisome biogenesis. J Cell Biol 220, e202001188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean BDM, Dziurdzik SK, Kolehmainen KL, Fowler CMS, Kwong WK, Grad LI, Davey M, Schluter C, Conibear E. (2018). Competitive organelle-specific adaptors recruit Vps13 to membrane contact sites. J Cell Biol 217, 3593–3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushell SR, Pike ACW, Falzone ME, Rorsman NJG, Ta CM, Corey RA, Newport TD, Christianson JC, Scofano LF, Shintre CA, et al. (2019). The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K. Nat Commun 10, 3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chorlay A, Monticelli L, Verissimo Ferreira J, Ben M’barek K, Ajjaji D, Wang S, Johnson E, Beck R, Omrane M, Beller M, et al. (2019). Membrane asymmetry imposes directionality on lipid droplet emergence from the ER. Dev Cell 50, 25–42.e7. [DOI] [PubMed] [Google Scholar]
- Da Costa R, Bordessoules M, Guilleman M, Carmignac V, Lhussiez V, Courot H, Bataille A, Chlemaire A, Bruno C, Fauque P, et al. (2020). Vps13b is required for acrosome biogenesis through functions in golgi dynamic and membrane trafficking. Cell Mol Life Sci 77, 511–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ercan B, Naito T, Koh DHZ, Dharmawan D, Saheki Y. (2021). Molecular basis of accessible plasma membrane cholesterol recognition by the GRAM domain of GRAMD1b. Embo J 40, e106524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairn GD, Schieber NL, Ariotti N, Murphy S, Kuerschner L, Webb RI, Grinstein S, Parton RG. (2011). High-resolution mapping reveals topologically distinct cellular pools of phosphatidylserine. J Cell Biol 194, 257–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanbarpour A, Valverde DP, Melia TJ, Reinisch KM. (2021). A model for a partnership of lipid transfer proteins and scramblases in membrane expansion and organelle biogenesis. Proc Natl Acad Sci U S A 118, 4–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillen-Samander A, Leonzino M, Hanna MG, Tang N, Shen H, De Camilli P. (2021). VPS13D bridges the ER to mitochondria and peroxisomes via miro. J Cell Biol 220, e202010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hankins HM, Baldridge RD, Xu P, Graham TR. (2015). Role of flippases, scramblases and transfer proteins in phosphatidylserine subcellular distribution. Traffic 16, 35–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann HH, Schneider WM, Rozen-Gagnon K, Miles LA, Schuster F, Razooky B, Jacobson E, Wu X, Yi S, Rudin CM, MacDonald MR, et al. (2021). TMEM41B is a pan-flavivirus host factor. Cell 184, 133–148.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holthuis JC, Menon AK. (2014). Lipid landscapes and pipelines in membrane homeostasis. Nature 510, 48–57. [DOI] [PubMed] [Google Scholar]
- Huang D, B X, Liu L, Wu L, Zhu Y, Ghanbarpour A, Wang Y, Chen F-J, Lyu J, Hu Y, et al. (2021). TMEM41B acts as an ER scramblase required for lipoprotein biogenesis and lipid homeostasis. Cell Metab 33, 1–6. [DOI] [PubMed] [Google Scholar]
- John Peter AT, Herrmann B, Antunes D, Rapaport D, Dimmer KS, Kornmann B. (2017). Vps13-Mcp1 interact at vacuole-mitochondria interfaces and bypass ER-mitochondria contact sites. J Cell Biol 216, 3219–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N, Leonzino M, Hancock-Cerutti W, Horenkamp FA, Li P, Lees JA, Wheeler H, Reinisch KM, De Camilli P. (2018). VPS13A and VPS13C are lipid transport proteins differentially localized at ER contact sites. J Cell Biol 217, 3625–3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YE, Wang Y, Du X, Zhang T, Mak HY, Hancock SE, McEwen H, Pandzic E, Whan RM, Aw YC, et al. (2021). TMEM41B and VMP1 are scramblases and regulate the distribution of cholesterol and phosphatidylserine. J Cell Biol 220, e202103105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda S, Yamamoto H, Kinch LN, Garza CM, Takahashi S, Otomo C, Grishin NV, Forli S, Mizushima N, Otomo T. (2020). Structure, lipid scrambling activity and role in autophagosome formation of ATG9A. Nat Struct Mol Biol 27, 1194–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa M, Fairn GD. (2015). Complementary probes reveal that phosphatidylserine is required for the proper transbilayer distribution of cholesterol. J Cell Sci 128, 1422–1433. [DOI] [PubMed] [Google Scholar]
- Matoba K, Kotani T, Tsutsumi A, Tsuji T, Mori T, Noshiro D, Sugita Y, Nomura N, Iwata S, Ohsumi Y, et al. (2020). Author correction: Atg9 is a lipid scramblase that mediates autophagosomal membrane expansion. Nat Struct Mol Biol 27, 1209. [DOI] [PubMed] [Google Scholar]
- Moretti F, Bergman P, Dodgson S, Marcellin D, Claerr I, Goodwin JM, DeJesus R, Kang Z, Antczak C, Begue D, et al. (2018). TMEM41B is a novel regulator of autophagy and lipid mobilization. EMBO Rep 19, e45889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita H, Zhao YG, Tamura N, Nishimura T, Kanda Y, Sakamaki Y, Okazaki M, Li D, Mizushima N. (2019). A critical role of VMP1 in lipoprotein secretion. Elife 8, e48834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita K, Hama Y, Izume T, Tamura N, Ueno T, Yamashita Y, Sakamaki Y, Mimura K, Morishita H, Shihoya W, et al. (2018). Genome-wide CRISPR screen identifies TMEM41B as a gene required for autophagosome formation. J Cell Biol 217, 3817–3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Braceras S, Calvo R, Escalante R. (2015). TipC and the chorea-acanthocytosis protein VPS13A regulate autophagy in dictyostelium and human HeLa cells. Autophagy 11, 918–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okawa F, Hama Y, Zhang S, Morishita H, Yamamoto H, Levine TP, Mizushima N. (2021). Evolution and insights into the structure and function of the DedA superfamily containing TMEM41B and VMP1. J Cell Sci 134. doi: 10.1242/jcs.255877 [DOI] [PubMed] [Google Scholar]
- Okuda S, Sherman DJ, Silhavy TJ, Ruiz N, Kahne D. (2016). Lipopolysaccharide transport and assembly at the outer membrane: the PEZ model. Nat Rev Microbiol 14, 337–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JS, Neiman AM. (2012). VPS13 regulates membrane morphogenesis during sporulation in Saccharomyces cerevisiae. J Cell Sci 125, 3004–3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JS, Neiman AM. (2020). XK is a partner for VPS13A: a molecular link between Chorea-Acanthocytosis and McLeod syndrome. Mol Biol Cell 31, 2425–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinisch KM, Prinz WA. (2021). Mechanisms of nonvesicular lipid transport. J Cell Biol 220. doi: 10.1083/jcb.202012058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider WM, Luna JM, Hoffmann HH, Sanchez-Rivera FJ, Leal AA, Ashbrook AW, Le Pen J, Ricardo-Lax I, Michailidis E, Peace A, et al. (2021). Genome-scale identification of SARS-CoV-2 and pan-coronavirus host factor networks. Cell 184, 120–132.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker CJ, Huang TQ, Weir NR, Polyakov NJ, Schultz SW, Denic V. (2019). CRISPR screening using an expanded toolkit of autophagy reporters identifies TMEM41B as a novel autophagy factor. PLoS Biol 17, e2007044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabara LC, Escalante R. (2016). VMP1 establishes ER-Microdomains that regulate membrane contact sites and autophagy. PLoS One 11, e0166499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji T, Cheng J, Tatematsu T, Ebata A, Kamikawa H, Fujita A, Gyobu S, Segawa K, Arai H, Taguchi T, et al. (2019). Predominant localization of phosphatidylserine at the cytoplasmic leaflet of the ER, and its TMEM16K-dependent redistribution. Proc Natl Acad Sci U S A 116, 13368–13373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde DP, Yu S, Boggavarapu V, Kumar N, Lees JA, Walz T, Reinisch KM, Melia TJ. (2019). Transports lipids to promote autophagosome biogenesis. J Cell Biol 218, 1787–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verchere A, Cowton A, Jenni A, Rauch M, Haner R, Graumann J, Butikofer P, Menon AK. (2021). Complexity of the eukaryotic dolichol-linked oligosaccharide scramblase suggested by activity correlation profiling mass spectrometry. Sci Rep 11, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao YG, Chen Y, Miao G, Zhao H, Qu W, Li D, Wang Z, Liu N, Li L, Chen S, et al. (2017). The ER-Localized transmembrane protein EPG-3/VMP1 regulates SERCA activity to control ER-Isolation membrane contacts for autophagosome formation. Mol Cell 67, 974–989.e6. [DOI] [PubMed] [Google Scholar]