Abstract
CCAAT binding factors (CBFs) positively regulating the expression of the amdS gene (encoding acetamidase) and two penicillin biosynthesis genes (ipnA and aatA) have been previously found in Aspergillus nidulans. The factors were called AnCF and PENR1, respectively. Deletion of the hapC gene, encoding a protein with significant similarity to Hap3p of Saccharomyces cerevisiae, eliminated both AnCF and PENR1 binding activities. We now report the isolation of the genes hapB and hapE, which encode proteins with central regions of high similarity to Hap2p and Hap5p of S. cerevisiae and to the CBF-B and CBF-C proteins of mammals. An additional fungus-specific domain present in HapE was revealed by comparisons with the homologs from S. cerevisiae, Neurospora crassa, and Schizosaccharomyces pombe. The HapB, HapC, and HapE proteins have been shown to be necessary and sufficient for the formation of a CCAAT binding complex in vitro. Strains with deletions of each of the hapB, hapC, and hapE genes have identical phenotypes of slow growth, poor conidiation, and reduced expression of amdS. Furthermore, induction of amdS by omega amino acids, which is mediated by the AmdR pathway-specific activator, is abolished in the hap deletion mutants, as is growth on γ-aminobutyric acid as a sole nitrogen or carbon source. AmdR and AnCF bind to overlapping sites in the promoters of the amdS and gatA genes. It is known that AnCF can bind independently of AmdR. We suggest that AnCF binding is required for AmdR binding in vivo.
The sequence CCAAT is found in the 5′ region of approximately 30% of eukaryotic genes between 50 and 200 bp from the start point of transcription and can be present in either orientation (8). There is evidence for a conserved multimeric protein activator that recognizes this sequence in a wide variety of organisms (reviewed in references 31 and 32). The first complex identified was the Hap complex of Saccharomyces cerevisiae, which consists of four subunits, Hap2p, Hap3p, Hap4p, and Hap5p (33, 39, 42). Hap2p, Hap3p, and Hap5p are essential for the formation of a DNA binding complex, while Hap4p is not directly involved in DNA binding but carries a functional activation domain (18, 53). The Hap complex is required for the expression of genes involved in oxidative phosphorylation in response to growth on nonfermentable carbon sources (42). In addition, genes not involved in respiration have been found to be subject to regulation by the Hap complex (13, 14). Conserved homologs of HAP2, HAP3, and HAP5 have been found in a variety of organisms (reviewed in reference 32). The mammalian CCAAT binding factor (CBF; also known as NF-Y) consists of three subunits, CBF-A, -B, and -C; in this case, CBF-B and CBF-C have been found to carry transcriptional activation domains. Evidence for a fourth subunit equivalent to Hap4p is lacking (11; reviewed in reference 31).
In the filamentous fungus Aspergillus nidulans, CCAAT sequences in the promoters of the amdS gene (encoding the catabolic enzyme acetamidase) and the acvA, ipnA, and aatA genes (encoding enzymes for biosynthesis of the secondary metabolite penicillin) have been shown to affect the expression of these genes (27, 29, 49). Proteins binding to these sequences have been detected—AnCF in the case of amdS (51) and PENR1 in the case of the penicillin biosynthesis genes (29, 49). Similarly, a protein designated AnCP from A. nidulans has been found to bind to a CCAAT sequence present in the Taka-amylase gene promoter of Aspergillus oryzae (24, 36).
The hapC gene of A. nidulans was identified as a homolog of the HAP3 gene of S. cerevisiae (40). Deletion of this gene resulted in low levels of expression of amdS, aatA, and ipnA reporter gene fusions (28, 40). In addition, deletion of hapC resulted in loss of binding of AnCF and PENR1 to CCAAT sequences derived from amdS, the penicillin biosynthesis genes, and the Taka-amylase gene, indicating that these complexes contain a hapC-encoded component (25, 28, 40). This was confirmed directly in assays using anti-HapC antiserum (25, 28). A CCAAT binding complex was reconstituted by using recombinant HapC together with expressed Hap2p and Hap5p from S. cerevisiae (25).
We have now cloned genes encoding two additional components of the AnCF complex. These genes, hapB and hapE, encode proteins with a central core with a high similarity to Hap2p and Hap5p of S. cerevisiae as well as to the corresponding mammalian proteins. We have shown that the HapB, HapC, and HapE subunits are necessary and sufficient for DNA binding by reconstitution of the complex in vitro. Furthermore, deletions of the hapB and hapE genes result in loss of AnCF binding activity and in phenotypes similar to those observed with the hapC deletion. However, resolution of the binding complex into two bands by using electrophoretic mobility shift assays together with the observation that HapE shares a fungus-specific domain with the S. cerevisiae and Schizosaccharomyces pombe homologs has raised the possibility that one or more additional components may associate with the core HapB/C/D complex and may carry activation domains.
We also have found for the first time evidence that AnCF also affects a pathway-specific regulatory circuit. The amdR gene encodes a Zn(II)2Cys6 binuclear cluster DNA binding protein that is required for omega-amino acid induction of the amdS gene and the genes for omega-amino acid utilization (3). Deletion of the hapB and hapE genes results in loss of omega-amino acid induction of amdS expression. In addition, the hapB and hapE deletion strains are unable to use γ-aminobutyric acid (GABA) as a sole nitrogen or carbon source.
MATERIALS AND METHODS
Strains, media, genetic techniques, and transformation.
A. nidulans strains used in this study are described in Table 1. Standard A. nidulans growth media and conditions were as described by Cove (12). Nitrogen sources were added to a final concentration of 10 mM. In plate growth tests, sodium acetate (50 mM), ethanol (1%, vol/vol), glycerol (1%, vol/vol), GABA (10 mM), and proline (10 mM) were used as carbon sources. Genetic manipulation of haploid A. nidulans strains was carried out as described by Clutterbuck (10). Protoplast preparation and DNA transformation were performed as described by Andrianopoulos and Hynes (2). Escherichia coli NM522 (New England Biolabs) was used for propagation of plasmids and DNA manipulation; strain BL21(DE3) (Novagen) was used for expression of His6-HapBct encoded by pT7-hapBct (see below). Molecular cloning techniques were performed as specified by Sambrook et al. (45).
TABLE 1.
A. nidulans strains used in this study
| Strain | Genotypea |
|---|---|
| MH54 | biA1; niiA4 |
| MH3018 | yA1, pabaA1; amdS368 argB2; amdA7 |
| MH3408 | biA1; amdS::lacZ; niiA4 |
| MH9092 | yA1, pabaA1; amdS368 argB2; amdA7; hapEΔ::argB |
| MH9099 | yA1, pabaA1; amdS368 argB2; amdA7; hapBΔ::argB |
| MH9094 | MH9092 transformed with pPTR2 |
| MH9210 | MH9099 transformed with pBShapB4.0 |
| MH9206 | yA1, pabaA1; amdS::lacZ; hapEΔ::argB |
| MH9207 | yA1, pabaA; amdS::lacZ; hapBΔ::argB |
| MH8194 | hapCΔ::riboB, amdS::lacZ; pyroA4, niiA4 |
Isolation of hapB and hapE genes.
For amplification of hapB cDNA fragments, degenerate oligonucleotides HAPBA1 (5′ CARCCITTYTAYGTIAAYGC 3′) and HAPBB1 (5′ GCRTTIACRTARAAIGGYTG 3′) were used together with the T7 or T3 oligonucleotide or primer HAPB5 (5′ CAGACATCATACCAGCATAAC 3′) and an A. nidulans cDNA library in λ ZAPII (constructed by R. Aramayo; available from the Fungal Genetics Stock Center, Kansas City, Kans.) previously amplified by using the universal and reverse primers (52) as the template for PCR. With the primer combination HAPB1A and T7, a single 700-bp fragment was generated and upon sequencing found to have a high level of similarity to Hap2p of S. cerevisiae. The primer combination HAPBB1 and T3 yielded a fragment of 800 bp with no similarity to Hap2p but which subsequently proved to contain the 5′ region of hapB. To isolate a genomic clone, a 520-bp PCR fragment, amplified with the primers HAPBA1 and HAPB5, was used to screen a genomic A. nidulans cosmid library (7) by colony hybridization. The clone pCOSHAPB was isolated and a 4.0-kb BamHI fragment that hybridized to the probe in Southern blot analysis was subcloned into pBluescriptII SK+. To obtain the corresponding cDNA, a T3 or T7 primer together with internal primers derived from this open reading frame (ORF) were used to amplify PCR fragments from the A. nidulans cDNA library.
Plasmid pCAAB1-1.5 (kindly provided by J. A. Kinsey, Kansas City, Kans.) contains a 1.5-kb EcoRI aab-1 cDNA clone (9). An A. nidulans cDNA library was probed with the 1.5-kb EcoRI fragment derived from this clone. A hybridizing clone (pHAPE-9) containing a cDNA insert of 1.2 kb was sequenced, and the insert showed 63% sequence identity to aab-1 at the DNA level. The gene was designated hapE. The 1.2-kb insert of pHAPE-9 was found to hybridize to a 6.0-kb BglII-PstI genomic fragment in Southern blot analysis. A plasmid partial library was created by cloning genomic BglII-PstI fragments of approximately 6.0 kb into BamHI-PstI-digested pBluescriptII SK+ and a hapE-containing clone was identified by colony hybridization with the pHAPE-9 cDNA insert. Southern blot analysis showed that the hapE gene was contained within a 2.8-kb BamHI-PstI fragment, which was subcloned into pBluescriptII SK+ and sequenced.
Oligonucleotides and plasmids.
Plasmid pBRhapB carries a genomic 4.0-kb BamHI hapB fragment in the BamHI site of pBR322. pKOhapB was constructed by excising a 1.6-kb SmaI-XhoI hapB fragment from pBRhapB and replacing it with a 1.8-kb SmaI-XhoI argB fragment derived from plasmid pDC1 (4). pPTR18 carries a genomic 2.5-kb BamHI-SalI hapE fragment in pBluescriptII SK+. pKOhapE was constructed by replacing an 0.8-kb EcoRV-SphI fragment of pPTR18 with a SmaI-SphI argB fragment derived from plasmid pDC1, thereby eliminating the evolutionary conserved domain of hapE. pMalE-HapC and pMalE-HapE, were kindly provided by M. Kato and N. Tsukagoshi (Nagoya, Japan).
pT7-hapBct, used for expression of a N-terminal truncated HapB protein, was generated as follows. A full-length hapB PCR product was cloned in frame into the XmnI/BamHI sites of pMal-c2 (New England Biolabs) to give pMalE-hapB. The integrity of the cloned PCR product was verified by DNA sequencing. A SacI-HindIII digest was performed to transfer the insert of pMalE-hapB into pQE31 (Qiagen), resulting in pQE31-hapB. An EcoRI-HindIII fragment carrying the full-length hapB, an E. coli ribosome binding site, and a His6 tag was then cloned into pT7-5 (48) to give pT7-hapB. The expression constructs pMalE-hapB, pQE31-hapB, and pT7-hapB did not allow high-level expression of full-length HapB in E. coli. However, an N-terminal part of HapB (amino acid [aa] 166 to 349) could be highly expressed. After an inverse PCR using primers Bct1 (TGGATAGGTACCGGTCATCCTTCC) and Bct2 (CCATGTCGGTACCCCGCACG), with pT7-hapB as the template, a PCR fragment missing the hapB N-terminal part coding for aa 3 to 165 was created. Digestion with KpnI and ligation of the PCR fragment resulted in plasmid pT7-hapBct, which allows expression of the His6-tagged C-terminal 183 aa of HapB (His6-HapBct). The hapB sequence was checked by DNA sequencing.
DNA sequencing.
Double-stranded plasmids were sequenced by the dideoxy-chain termination method of Sanger et al. (46) on an ABI Prism (Perkin-Elmer) automated sequencer.
Expression and purification of recombinant proteins.
MalE-HapC and MalE-HapE fusion proteins were prepared as described by Kato et al. (25). Cultures (200 ml) of E. coli NM522 harboring plasmid pMalE-HapC or pMalE-HapE were grown to an optical density at 600 nm of ∼0.5, and induction was achieved by the addition of 1 mM isopropyl-β-d-thiogalactopyranoside. After induction for 3 h at 37°C, cells were harvested, taken up in 15 ml of buffer A (20 mM Tris-Cl, 1 mM EDTA, 1 mM dithiothreitol [DTT], 200 mM NaCl [pH 7.5]), lysed by sonication, and centrifuged at 12,100 × g for 20 min. The crude cellular extracts were applied to amylose columns (New England Biolabs) and washed with buffer A until the flowthrough optical density at 280 nm was <0.01. The fusion proteins were eluted with buffer A containing 10 mM maltose. His6-HapBct-containing crude cellular E. coli extracts were obtained by the same protocol. The crude cellular extracts were applied to a nitrilotriacetic acid-agarose column (Qiagen). Washing steps were performed according to the supplier. The His6-tagged protein was eluted by a 50 to 500 mM gradient of imidazole in washing buffer. The purity of all recombinant proteins was examined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Coomassie blue staining.
Preparation of A. nidulans nuclear extracts and electrophoretic mobility shift assays.
A. nidulans nuclear extracts were prepared by the method of Papagiannopoulos et al. (40). Oligonucleotides MH10 (GATCGCCAGCCAATCACCAGCTAGGCACCAGCTAAACCC) and MH11 (GATCGGGTTTAGCTGGTGCCTAGCTGGTGATTGGCTGGC) (referred to as oligonucleotides 10 and 11) are complementary oligonucleotides derived from the CCAAT box region of the amdS promoter which have 5′ GATC overhangs after annealing (51). Labeling of the probe and electrophoretic mobility shift assays were performed as described by Papagiannopoulos et al. (40) except that 20 μg of nuclear protein extract was incubated at 25°C for 30 min together with the probe before the mixtures were loaded on 6% polyacrylamide gels. For reconstitution experiments, the DNA binding reaction mixture contained DTT at a concentration of 5 mM and 10 ng of each recombinant purified protein.
β-Galactosidase assays.
Reporter gene studies used strains containing a single copy of the amdS::lacZ fusion lacking vector sequences in place of the native amdS gene (16). β-Galactosidase assays were carried out by the method of Davis et al. (16).
Nucleotide sequence accession numbers.
The complete hapB DNA and protein sequences have been submitted to EMBL/GenBank databases under accession no. Y13768; hapE DNA and protein sequences are available under accession no. U96847.
RESULTS
Cloning and sequencing of hapB.
The DNA binding and subunit interaction domains of Hap2p and CBF-B reside in an essential conserved 65-aa core (30, 39). Degenerate oligonucleotides were derived from this conserved region and used to generate probes for screening a genomic cosmid library (see Materials and Methods). A 4.0-kb BamHI fragment derived from the resulting hybridizing cosmid was subcloned in pBluescriptII SK+. The resulting plasmid, pBShapB4.0, was sequenced and found to contain a 1,047-bp ORF encoding a polypeptide of 349 aa with a calculated molecular mass of 37.2 kDa. Sequencing of cDNA derived by PCR from an A. nidulans cDNA library showed that a putative initiation codon lay 39 bp downstream from the cDNA start and that no introns were present. The gene was designated hapB, and the derived HapB protein was found to show high conservation in the DNA binding and subunit interaction domains with the S. cerevisiae (HAP2) and rat (CBF-B) gene products (Fig. 1A); no significant blocks of similarity were found in any other region (not shown).
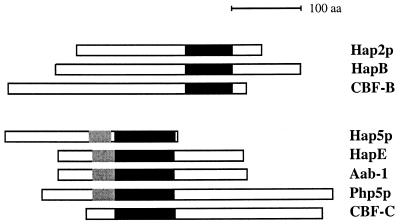
FIG. 1.
Comparison of the evolutionary conserved domains of HapB and HapE with their homologs. (A) Alignment of the DNA binding and subunit interaction domains from Hap2p, HapB, and CBF-B. (B) Alignment of the conserved domain derived from Hap5p, HapE, Aab-1, Php5p, and CBF-C. Invariant amino acids that are present in at least two of the three sequences (A) or three out of five sequences (B) are boxed in black. The numbers indicate the first and last residues of the domains depicted. Abbreviations for organisms: S. c., S. cerevisiae; A. n., A. nidulans; N. c., N. crassa; Sc. p., S. pombe; R. n., R. norvegicus. The asterisks in panel B indicate amino acids that are identical in at least three of the fungal proteins but are not found in the mammalian homolog CBF-C. Alignments were performed with the PILEUP program from the Wisconsin package (version 8; Genetics Computer Group, Madison, Wis.), made available via the Australian National Genomics Information Service.
Cloning and sequencing of hapE.
The aab-1 gene from Neurospora crassa has been found to be a homolog of the HAP5 gene of S. cerevisiae (9). A fragment of this gene was used to clone the hapE homolog from A. nidulans (see Materials and Methods). Comparison of the cDNA sequence with the genomic sequence revealed the presence of four introns within an ORF predicted to encode a polypeptide of 265 aa with a molecular mass of 29.2 kDa. Three of the introns (1, 2, and 4) are at positions identical to those of the N. crassa homolog (9). HapE has significant similarity to a conserved block of approximately 85 aa present in the Hap5p (S. cerevisiae), Aab-1 (N. crassa), Php5p (S. pombe), and CBF-C (rat [Rattus norvegicus]) homologs (Fig. 1B). Furthermore, HapE shows similarity to a Hap5p domain that is located directly N terminal of the conserved core (Fig. 1B). Significantly, this domain is conserved in all four fungal homologs but not in the mammalian homolog.
Deletion of hapB and hapE.
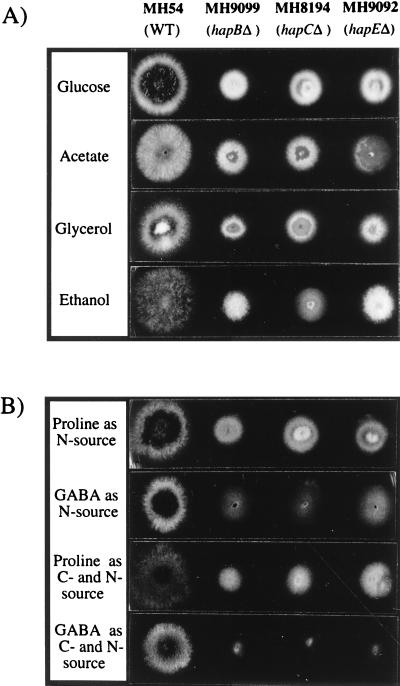
Strains deleted for hapC show a viable but slow-growth phenotype (40). If hapB and hapE encode protein subunits that form a CCAAT binding complex with HapC, then the deletion of each of these genes should result in the same phenotype as seen in the hapC deletion strain. The deletion plasmids pKOhapB and pKOhapE were constructed by replacing internal regions of the genes hapB (aa 13 to 349) and hapE (aa 1 to 172) with the selectable marker argB from plasmid pDC1 (Fig. 2B and Materials and Methods). pKOhapB was linearized by EcoRI digestion, pKOhapE was digested with XbaI, and then the argB mutant strain MH3018 was transformed with each of the linear deletion constructs. Among the transformants with an Arg+ phenotype, one pKOhapB transformant and three pKOhapE transformants showed the phenotype of the hapC deletion strain previously observed (40). Southern blot analysis showed that strain MH9099 had a single integration of the deletion construct (pKOhapB) at the hapB locus, whereas strain MH9092 carried a single integration of the hapE deletion construct (pKOhapE) at the hapE locus (Fig. 2B). Both strains grew more slowly than a wild-type strain on all media tested but showed a phenotype similar to that of the hapC deletion strain MH8194 (Fig. 3). Conidiation in all three hap deletion strains was reduced but normal, as described previously for the hapC deletion (40). Strains containing hap deletions have been found to be extremely difficult to cross to other strains, but it cannot be determined whether this is a general effect of poor growth or is due to a specific defect in sexual reproduction.
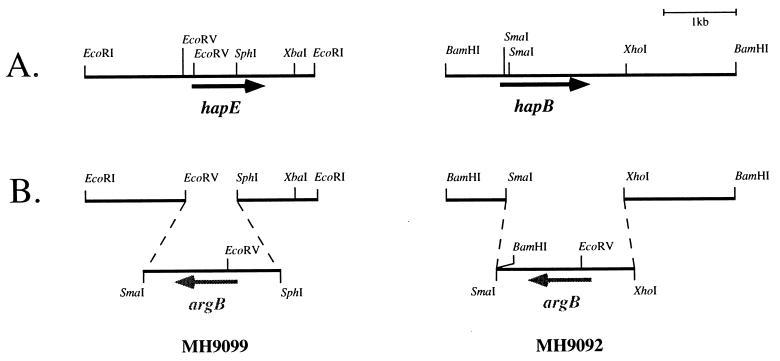
FIG. 2.
Deletion strategy for hapB and hapE. (A) Partial restriction maps of the genomic regions of hapE and hapB. ORFs and direction of transcription are indicated by rightward arrows. (B) Schematic representation of the deletions generated by gene replacement of hapB (strain MH9099) and hapE (strain MH9092) by the selectable marker argB after a homologous double-recombination event with the linearized deletion constructs pKOhapB and pKOhapE, respectively. The direction of argB transcription is shown (leftward arrows).
FIG. 3.
Effects of hap deletions on growth and morphology. Strains MH9099 (hapBΔ), MH8194 (hapCΔ), and MH9092 (hapEΔ) are compared to a wild-type (WT) strain (MH54). (A) Growth on complete medium and sodium acetate (50 mM), ethanol (1%, vol/vol) and glycerol (1%, vol/vol) as sole carbon sources in the presence of 10 mM NH4Cl. (B) Growth on proline (10 mM) or GABA (10 mM) as the sole nitrogen source in the presence of 1% glucose or as the sole carbon and nitrogen source. The strains were incubated at 37°C for 48 h, except for the growth tests on ethanol and on proline or GABA as the sole carbon and nitrogen source, in which cases incubation was for 78 h at 37°C.
The mutant phenotypes of deletion strain MH9099 could be complemented by cotransforming with plasmid pAmPh520, encoding bleomycin resistance, and pBShapB4.0, carrying a 4.0-kb BamHI fragment containing the hapB gene. In the case of MH9092, complementation was achieved by cotransformation with pAmPh520 and pPTR2, which contains a 2.2-kb KpnI-PstI hapE fragment. Complemented strains which show normal growth and morphology could be used for crossing to other strains, allowing the isolation of recombinant progeny containing hap deletion mutations. This analysis showed that ascospores containing the deletions developed normally and could germinate to produce colonies with the typical hap deletion phenotype.
Loss of AnCF binding activity in hapBΔ and hapEΔ strains.
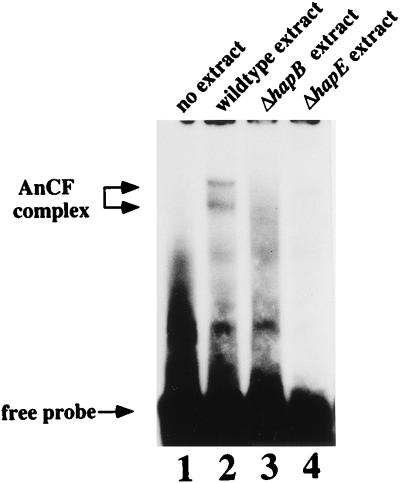
The oligonucleotide pair 10-11 (51) spanning the CCAAT box derived from the amdS promoter was end labeled and used in an electrophoretic mobility shift assay with nuclear extracts prepared from strains MH9092 and MH9099. Nuclear extracts from a wild-type strain resulted in two complexes with different mobilities (Fig. 4, lane 2), whereas the nuclear extracts prepared from the hapB and hapE deletion strains showed loss of both complexes (Fig. 4, lanes 3 and 4), clearly indicating that HapB and HapE are also required for AnCF binding, as has been shown for HapC (40). In contrast to previous results where the resolution of two complexes was not clearly seen (40), two CCAAT binding complexes were consistently resolved with different preparations of wild-type nuclear extracts and different CCAAT-containing probes (not shown), although the intensity of the two complexes showed some variation. Two complexes binding to a CCAAT-containing sequence have also been observed in N. crassa (9).
FIG. 4.
Loss of CCAAT binding activity in hapBΔ and hapEΔ strains. For mobility shift analysis, the end-labeled oligonucleotide pair 10-11 was incubated with 20 μg of crude nuclear extract prepared from a hap+ strain (lane 2), a hapBΔ strain (MH9099; lane 3), or a hapEΔ strain (MH9092; lane 4), and the mixtures were fractionated on a 6% polyacrylamide gel. Free probe and AnCF complexes are marked by arrows.
Expression of amdS is reduced in hapBΔ and hapEΔ backgrounds.
To assess amdS expression in hapBΔ and hapEΔ backgrounds, strains MH9094 and MH9210 were crossed with strain MH3408, carrying a single-copy amdS::lacZ reporter gene at the amdS locus (16). β-Galactosidase expression in the resulting hapBΔ and hapEΔ backgrounds was compared to that in the parent strain MH3408. The two deletions reduced amdS expression in similar manners (Table 2). As noted for a hapC deletion strain (40), the reduction of amdS expression was more significant under carbon-limiting conditions (0.1% glucose) than with 1% glucose. However, the amdS::lacZ fusion was sensitive to nitrogen metabolite repression in the deletion strains, as shown by derepression by growth on the limiting nitrogen source l-alanine compared with ammonium-grown mycelia (Tables 2 and 3).
TABLE 2.
Expression of amdS::lacZ fusions in the hapBΔ and hapEΔ backgrounds
| Strain | Relevant genotype | β-Galactosidase activitya (U)
|
|
|---|---|---|---|
| 1% Glucose + 10 mM NH4-tartrate | 0.1% Glucose + 10 mM NH4-tartrate | ||
| MH3408 | amdS::lacZ | 1.94 (0.12) | 20.18 (2.49) |
| MH9207 | amdS::lacZ; hapBΔ | 0.38 (0.12) | 1.07 (0.24) |
| MH9206 | amdS::lacZ; hapEΔ | 0.31 (0.11) | 0.42 (0.06) |
Mycelia were grown in 100 ml of medium at 37°C for 16 h (1% glucose) and 20 h (0.1% glucose). Data represent the results of four separate experiments; standard errors are given in parentheses. Assays of endogenous β-galactosidase in a wild-type strain and in hapBΔ and hapEΔ strains lacking the inserted amdS::lacZ fusion gene gave activities of 0.1 to 0.4 and were not affected by the growth conditions used for the experiments reported here or in Table 3.
TABLE 3.
Expression of amdS::lacZ fusions in the hapBΔ and hapEΔ backgrounds under induction with omega amino acids
| Strain | Relevant genotype | β-Galactosidase activitya (U)
|
||
|---|---|---|---|---|
| 10 mM l-alanine | 10 mM l-alanine + 10 mM GABA | 10 mM l-alanine + 10 mM β-alanine | ||
| MH3408 | amdS::lacZ | 19.14 (2.02) | 94.24 (7.16) | 148.70 (3.56) |
| MH9207 | amdS::lacZ; hapBΔ | 1.95 (0.34) | 2.26 (0.55) | 1.10 (0.07) |
| MH9206 | amdS::lacZ; hapEΔ | 1.14 (0.08) | 1.21 (0.07) | 0.83 (0.07) |
Mycelia were grown in 100 ml of medium at 37°C for 16 h with 1% glucose. Nitrogen sources were added as indicated. Data represent the results of four separate experiments; standard errors are given in parentheses.
In vitro reconstitution of a CCAAT binding complex.
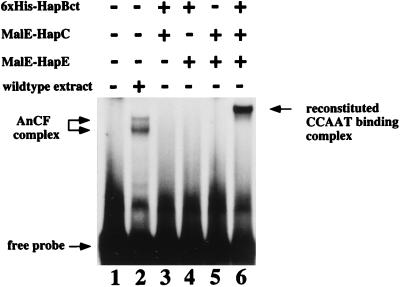
HapB, HapC, and HapE were expressed in E. coli as fusion proteins and purified by affinity chromatography (Materials and Methods). HapC (aa 1 to 186) and HapE (aa 1 to 265) were purified as MalE fusion proteins. HapB was purified as a His6-tagged truncated protein (HapBct) containing the C-terminal 183 aa of HapB (aa 168 to 349), which includes the predicted conserved DNA binding and subunit interaction domains (Fig. 1A) characterized in S. cerevisiae Hap2p (39) and rat CBF-B (30). When combinations of two of the A. nidulans Hap proteins were mixed and incubated with the DNA probe 10-11, no DNA binding was detected in mobility gel shifts (Fig. 5, lanes 3 to 5). Only when all three proteins were combined was binding observed (Fig. 5, lane 6). The subunits HapC and HapE were each fused to the 42.7-kDa MalE protein; therefore, the newly formed DNA binding protein complex had a mobility lower than that of the wild-type Hap complex(es) (lane 2). The MalE-HapE fusion protein was cleaved into the two components MalE and HapE, using the protease factor Xa cleavage site in the linker region. When this HapE-MalE mixture was used in a mobility shift together with His6-HapBct and MalE-HapC, a complex with increased mobility was observed, indicating that the presence of HapE instead of the larger MalE-HapE fusion protein reduced the complex size. DNA binding could be reconstituted only in the presence of 5 mM DTT. Subunit association and DNA binding of the mammalian CBF have previously been shown to be dependent on reduction of the CBF subunits (37).
FIG. 5.
In vitro reconstitution of a CCAAT binding complex. The recombinant proteins His6-HapBct, MalE-HapC, and MalE-HapE were expressed in E. coli and purified by affinity chromatography (Materials and Methods). The recombinant proteins were tested for DNA binding in electrophoretic mobility shift analysis using the oligonucleotide 10-11 probe. Binding reaction mixtures contained 10 ng of each recombinant protein, indicated by + (lanes 3 to 6), or 20 μg of crude nuclear extract prepared from a wild-type A. nidulans strain (lane 2). Free probe, AnCF complexes, and the reconstituted CCAAT binding complex are marked by arrows.
The A. nidulans Hap complex is required for AmdR-dependent induction.
The binding sites for the transcriptional activator AmdR and the AnCF complex are contained within a 19-bp sequence in the amdS promoter (51). Furthermore, other functional AmdR binding sites are found in the lamA/lamB and gatA promoters, directly flanked by a GCAAT or a CCAAT sequence within the 19-bp element (44). The hap deletion mutants allowed us to examine whether a functional AnCF complex is a prerequisite for AmdR-mediated activation of amdS. Induction by omega amino acid GABA or β-alanine (five- or eightfold, respectively, in a wild-type strain) mediated by AmdR was eliminated in hapB and hapE deletion backgrounds (Table 3). This result suggested that AmdR is dependent on a functional AnCF complex for activation of amdS expression.
Utilization of GABA as carbon or nitrogen source requires the expression of gabA (GABA permease) and gatA (GABA transaminase), which are positively regulated by AmdR (6, 43). The three hap deletion strains show greatly reduced levels of growth on GABA as a nitrogen source compared to the wild-type strain MH54 and do not form any aerial mycelia (Fig. 1B). Similarly, the hap deletion strains grew extremely poorly on GABA as a sole carbon and nitrogen source or as a sole carbon source in the presence of ammonium. Strain MH8194 (hapCΔ) carries an additional deletion of amdR (40), whereas strains MH9092 (hapEΔ) and MH9099 (hapBΔ) are amdR+. Although amdR is intact in these two strains, growth levels were comparable to those observed for the hapCΔ amdRΔ double mutant MH8194. This finding indicated that AmdR- mediated activation of GABA utilization does not occur in the absence of the AnCF complex. Proline and GABA are metabolized via independent pathways leading to glutamate. Growth of the hap deletion mutants on proline either as a sole nitrogen source or as a sole carbon and nitrogen source was comparable to growth on glucose and ammonium medium (Fig. 3). In addition, growth on l-proline, l-alanine, and l-glutamate as sole carbon sources in the presence of ammonium was not affected by the hap deletion mutations (results not shown). Therefore, the observed weak growth on GABA reflects a pathway-specific effect on the genes responsible for the utilization of GABA.
DISCUSSION
The finding that deletion of each of the three genes hapB, hapC, and hapE results in identical slowly growing and weakly conidiating phenotypes is consistent with all three gene products interacting to form a functional complex in which each subunit is essential. Loss of AnCF binding in each of the three hap deletion strains together with in vitro reconstitution of a binding complex with expressed subunits provides further compelling evidence for this.
The in vitro reconstitution of a binding complex shows that the absence of the N-terminal 182 HapB amino acids in His6-HapBct does not impair subunit interaction or DNA binding. These residues are therefore not essential for functional DNA binding complex formation in vitro. Furthermore, this experiment demonstrates that a heteromeric complex consisting only of the subunits HapB, HapC, and HapE creates a protein surface able to bind to DNA containing CCAAT sequences, as has been found for the equivalent three subunits in S. cerevisiae (33) and mammals (47).
Although the DNA binding and subunit association domains of HapB and HapE have been highly conserved among fungi and mammals (Fig. 1), the proteins have diverged significantly in overall structure. The functional domains are not conserved with respect to their positions within the proteins (Fig. 6), and when regions apart from the conserved domains are compared, no significant homology between yeast, mammals, and A. nidulans can be found. Therefore, while the ability of the subunits of the HAP-CBF complexes to interact with each other and with DNA has been maintained, the specific functions of the complexes have been altered by the divergence of the flanking sequences during evolution. HapE and the N. crassa Aab-1 protein show considerable similarity within their central conserved domains and also in the flanking sequences. Although these species are the closest relatives in this group, they represent distinct taxa within the fungi, suggesting that the Hap complexes in these two filamentous fungi may be involved in the regulation of similar genes and processes.
FIG. 6.
Positions of the functional domains of the HapB and HapE homologs are not conserved. Each protein is depicted with the N terminus on the left and aligned with respect to the conserved domains. Highlighted boxes indicate the evolutionarily conserved domains that are depicted in Fig. 1. Black boxes represent the conserved domains found in all of the HapB or HapE homologs, whereas grey boxes represent the homology in the Hap4p interaction domain characterized for Hap5p (33, 34) present in all of the fungal homologs.
McNabb et al. (34) identified a second domain in S. pombe that is conserved in S. cerevisiae and showed that this domain is involved in recruiting the activator subunit Hap4p to the Hap complex in S. cerevisiae. This domain is also present in HapE and in Aab-1 but not in CBF-C (Fig. 1B and 6) and therefore may be specific to the fungi. As suggested by McNabb et al. (34), this is a strong indication that fungal CCAAT binding complexes may have a fourth Hap4p-like subunit. Our results provide evidence that this may be the case for A. nidulans. When wild-type extracts are used in electrophoretic mobility shifts, two distinct DNA-protein complexes are formed, but both are lost when nuclear extracts derived from hapΔ mutants are used (Fig. 4). The complex with higher mobility may be composed of the DNA binding core complex HapB/C/E, whereas the complex with lower mobility contains a fourth, unknown component. The predicted molecular mass of a HapB/C/E complex is 87 kDa. Gel filtration experiments determining the molecular mass of the AnCF and PENR1 complexes have given estimates of 120 to 130 kDa (25, 28). These results provide further support for the idea that one (or more) additional subunit is associated with the HapB/C/E core. It is possible that different Hap4p-like activator subunits can be associated with the core complex and provide specificity for particular sets of genes. In S. cerevisiae, there is evidence for the regulation of genes by the Hap complex that is not absolutely dependent on Hap4p (13, 14).
In S. cerevisiae, the Hap complex plays a major role in the control of genes involved in respiration in response to the carbon source, and the activator subunit Hap4p is strongly up-regulated by a shift from glucose to nonfermentable carbon sources (18). The A. nidulans Hap complex, in contrast, appears to be not absolutely required for activation of genes involved in oxidative phosphorylation. The ability of hapB/C/E deletion strains to grow on carbon sources such as acetate and glycerol (Fig. 3) clearly distinguishes A. nidulans, an obligate aerobe, from S. cerevisiae hap mutants, which are not viable on nonfermentable carbon sources (33, 42).
The CBF complex in mammals has a wide range of target genes (32). In filamentous fungi, the complex has been implicated in the regulation of genes involved in the catabolism of sole carbon sources (e.g., Taka-amylase [24, 36]), sole nitrogen sources (formamide hydrolysis [19]), and sources of both carbon and nitrogen (acetamide [reference 40 and references therein]). In addition, the complex affects the expression of genes involved in secondary metabolism (penicillin biosynthesis [28, 29, 49]) and in ammonium assimilation (NADP-dependent glutamate dehydrogenase [9, 15]). A protein binding to a CCAAT sequence in the 5′ region of the conidiation-specific yA gene of A. nidulans has been detected, but it has not been established that the protein is AnCF (1, 5).
The viability of the A. nidulans hapB, hapC, and hapE deletion strains indicates that the hap complex is not absolutely required for the transcription of any essential genes. It is likely that the phenotype of slow growth results from suboptimal expression of many genes which are subject to additional regulatory controls. This is exemplified by the amdS gene: although the CCAAT sequence and the hap complex have a major effect on expression, the gene can still respond to other control mechanisms. In addition, penicillin production is reduced but not abolished in a hapC deletion strain (28).
The amdS gene is subject to multiple wide domain- and pathway-specific regulatory controls, one of which is mediated by the amdR gene, which is necessary for omega-amino acid induction (reviewed in reference 23). The amdI93 deletion, which removes both the CCAAT site and the binding site for AmdR, does not affect regulation by the amdA, areA, facB, or creA gene (21, 22, 50). Mutation of the amdS 5′ CCAATCA 3′ sequence to 5′ CAAGTCT 3′ reduces amdS expression to levels seen in the hap deletion strains and eliminates amdR-mediated induction (20). However, the effects of the other regulatory circuits, including responses to the gain-of-function mutations amdA7 and areA102 are still observed (20). We have shown here that deletion of the hapB and hapE genes eliminates omega-amino acid induction of the amdS::lacZ reporter gene and also specifically affects GABA utilization, which is dependent on AmdR activity. Therefore, we conclude that activation of gene expression by the pathway-specific regulator AmdR is dependent on AnCF function.
AmdR contains an N-terminal Zn(II)2Cys6 binuclear cluster DNA binding domain which has been shown to confer DNA binding specificity in vivo (3, 41). Studies on transformants overexpressing AmdR due to multiple copies (2, 3) together with domain swapping experiments with the FacB activator and the analysis of amdRc constitutive mutations (41) lead to the suggestion that omega-amino acid inducers do not affect AmdR DNA binding but rather increase the ability of bound protein to activate transcription. Both AmdR and AnCF have been found to bind in vitro to conserved sequences found within a 19-bp element in the amdS, gatA, and lamA/B 5′ regions (44, 51). Therefore, in each gene controlled by AmdR, there is a close association with AnCF binding sites.
The dependence of AmdR function on the AnCF complex could result from effects on DNA binding and/or activation. AnCF activity is not dependent on AmdR, since all genes regulated by AnCF are not also regulated by AmdR and the basal levels of amdS expression can be restored by the insertion of the CCAAT-containing oligonucleotide pair 10-11 without restoring AmdR control. Fusion of the activation domains of AmdR to the DNA binding domain of the acetate-specific regulator FacB results in activation of acetate utilization genes and in omega-amino acid induction of amdS when tested in amdR and facB mutant strains (41). Since AnCF is not required for acetate utilization (Fig. 3) and does not affect FacB-mediated regulation of amdS, this suggests that it is less likely that AmdR dependence on AnCF results from cooperative activation of transcription. It is unlikely that AnCF is required for AmdR expression, since no CCAAT sites are present in the 5′ region of amdR and the amdR transcript is present at a low constitutive level (2, 3). It is therefore more likely that AnCF facilitates AmdR binding to DNA in vivo. Previous studies of in vitro binding of nuclear extracts showed that AmdR could bind to an oligonucleotide pair derived from the amdS 5′ region in which the CCAAT motif was changed to CCTTT, while AnCF could not bind to this sequence. This demonstrates the independent nature of the binding sites but does not necessarily reflect the in vivo binding capacity of AmdR. We suggest that AnCF binding to its target sequence is a prerequisite for a change in chromatin structure necessary for AmdR binding. Support for this view comes from preliminary data from this laboratory which indicate that a CCAAT sequence and the integrity of the AnCF complex are necessary for a change in DNase I hypersensitivity (38).
Interactions of HAP-like CCAAT binding complexes with other transcription factors have been observed in both mammals and fungi. The Kluyveromyces lactis Hap complex stabilizes the binding of Abf1p, and activation of transcription is achieved synergistically (35). Mutations in the CCAAT boxes of the invariant chain promoter have been shown to abolish the binding of two other transcription factors (Sp1 and RFX) to their target sites in in vivo footprinting studies (26). In the 3-hydroxy-3-methylglutaryl coenzyme A synthase gene, a binding site for the sterol regulatory element binding proteins is separated by 17 bp from an inverted CCAAT box. It has been shown that binding of the CBF complex to the CCAAT box is required for sterol-mediated regulation of 3-hydroxy-3-methylglutaryl coenzyme A synthase. Furthermore, a direct interaction between CBF and the basic helix-loop-helix sterol regulatory element binding proteins occurs (17). Other examples of CBF interactions with regulatory circuits are reviewed by Maity and de Crombrugghe (31).
Complex protein-protein interactions between transcription factors have been discovered with increasing frequency in recent years. Characterization of interactions between the HapB/C/E complex and Hap4p homologs as well as gene-specific regulatory proteins such as AmdR are required to determine how the specificity of Hap complex-mediated regulation of gene expression in filamentous fungi is modulated.
ACKNOWLEDGMENTS
We gratefully acknowledge Norihiro Tsukagoshi and Masashi Kato for providing the expression plasmids pMalE-HapC and pMalE-HapE and Jack Kinsey for plasmid pCAAB1-1.5.
This work was supported by the Australian Research Council, by the Deutsche Forschungsgemeinschaft (Sonderforschungsbereich 369), and by a DAAD Doktorandenstipendium im Rahmen des gemeinsamen Hochschulsonderprogramms III von Bund und Laendern granted to S.S.
REFERENCES
- 1.Andrianopoulos, A. Unpublished data.
- 2.Andrianopoulos A, Hynes M J. Cloning and analysis of the positively acting regulatory gene amdR from Aspergillus nidulans. Mol Cell Biol. 1988;8:3532–3541. doi: 10.1128/mcb.8.8.3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrianopoulos A, Hynes M J. Sequence and functional analysis of the positively acting regulatory gene amdR from Aspergillus nidulans. Mol Cell Biol. 1990;10:3194–3203. doi: 10.1128/mcb.10.6.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aramayo R, Adams T H, Timberlake W E. A large cluster of highly expressed genes is dispensable for growth and development in Aspergillus nidulans. Genetics. 1989;122:65–71. doi: 10.1093/genetics/122.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aramayo R, Timberlake W E. The Aspergillus nidulans yA gene is regulated by abaA. EMBO J. 1993;12:1039–2048. doi: 10.1002/j.1460-2075.1993.tb05853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arst H N. Integrator gene in Aspergillus nidulans. Nature. 1976;262:231–234. doi: 10.1038/262231a0. [DOI] [PubMed] [Google Scholar]
- 7.Borges-Walmsley M I, Turner G, Bailey A M, Brown J, Lehmbeck J, Clausen I G. Isolation and characterisation of genes for sulphate activation and reduction in Aspergillus nidulans: implications for evolution of an allosteric control region by gene duplication. Mol Gen Genet. 1995;247:423–429. doi: 10.1007/BF00293143. [DOI] [PubMed] [Google Scholar]
- 8.Bucher P. Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. J Mol Biol. 1990;212:563–578. doi: 10.1016/0022-2836(90)90223-9. [DOI] [PubMed] [Google Scholar]
- 9.Chen H, Crabb J W, Kinsey J A. The Neurospora aab-1 gene encodes a CCAAT binding protein homologous to yeast HAP5. Genetics. 1998;148:123–130. doi: 10.1093/genetics/148.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clutterbuck A J. Loci and linkage map of the filamentous fungus Aspergillus nidulans. In: O’Brien S J, editor. Genetic maps. Vol. 3. New York, N.Y: Cold Spring Harbor Laboratory Press; 1984. pp. 265–273. [Google Scholar]
- 11.Coustry F, Sinha S, Maity S N, de Crombrugghe B. The two activation domains of the CCAAT-binding factor CBF interact with the dTAFII110 component of the Drosophila TFIID complex. Biochem J. 1998;331:291–297. doi: 10.1042/bj3310291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cove D J. The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim Biophys Acta. 1966;113:51–56. doi: 10.1016/s0926-6593(66)80120-0. [DOI] [PubMed] [Google Scholar]
- 13.Dang V D, Bohn C, Bolotin-Fukuhara M, Daignan-Fornier B. The CCAAT box-binding factor stimulates ammonium assimilation in Saccharomyces cerevisiae, defining a new cross-pathway regulation between nitrogen and carbon metabolisms. J Bacteriol. 1996;178:1842–1849. doi: 10.1128/jb.178.7.1842-1849.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dang V D, Valens M, Bolotin-Fukuhara M, Daignan-Fornier B. Cloning of the ASN1 and ASN2 genes encoding asparagine synthetases in Saccharomyces cerevisiae: differential regulation by the CCAAT-box-binding factor. Mol Microbiol. 1996;22:681–692. doi: 10.1046/j.1365-2958.1996.d01-1715.x. [DOI] [PubMed] [Google Scholar]
- 15.Davis, M. A. Unpublished data.
- 16.Davis M A, Cobbet C S, Hynes M J. An amdS-lacZ fusion for studying gene regulation in Aspergillus. Gene. 1988;63:199–212. doi: 10.1016/0378-1119(88)90525-2. [DOI] [PubMed] [Google Scholar]
- 17.Dooley K A, Millinder S, Osborne T F. Sterol regulation of 3-hydroxy-3-methylglutaryl-coenzyme A synthase gene through a direct interaction between sterol regulatory element binding protein and the trimeric CCAAT-binding factor/nuclear factor Y. J Biol Chem. 1998;273:1349–1356. doi: 10.1074/jbc.273.3.1349. [DOI] [PubMed] [Google Scholar]
- 18.Forsburg S L, Guarente L. Identification and characterization of HAP4: a third component of the CCAAT-bound HAP2/HAP3 heteromer. Genes Dev. 1989;3:1166–1178. doi: 10.1101/gad.3.8.1166. [DOI] [PubMed] [Google Scholar]
- 19.Fraser, J. A. Unpublished data.
- 20.Hynes, M. J. Unpublished data.
- 21.Hynes M J. A mutation, adjacent to the gene amdS, defining the site of action of positive-control gene amdR in Aspergillus nidulans. J Bacteriol. 1980;142:400–406. doi: 10.1128/jb.142.2.400-406.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hynes M J, Corrick C M, Kelly J M, Littlejohn T G. Identification of the sites of action for regulatory genes controlling the amdS gene of Aspergillus nidulans. Mol Cell Biol. 1988;8:2589–2596. doi: 10.1128/mcb.8.6.2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hynes M J, Davis M A. Regulation of acetamide catabolism. In: Bramble R, Marzluf G A, editors. The Mycota, biochemistry and molecular biology. III. Heidelberg, Germany: Springer-Verlag; 1996. pp. 381–393. [Google Scholar]
- 24.Kato M, Aoyama A, Naruse F, Kobayashi T, Tsukagoshi N. An Aspergillus nidulans nuclear protein, AnCP, involved in enhancement of Taka-amylase A gene expression, binds to the CCAAT-containing taaG2, amdS, and gatA promoters. Mol Gen Genet. 1997;254:119–126. doi: 10.1007/s004380050399. [DOI] [PubMed] [Google Scholar]
- 25.Kato M, Aoyama A, Naruse F, Tateyama Y, Hayashi K, Miyazaki M, Papagiannopoulos P, Davis M A, Hynes M J, Kobayashi T, Tsukagoshi N. The Aspergillus nidulans CCAAT-binding factor AnCP/AnCF is a heteromeric protein analogous to the HAP complex of Saccharomyces cerevisiae. Mol Gen Genet. 1998;257:404–411. doi: 10.1007/s004380050664. [DOI] [PubMed] [Google Scholar]
- 26.Linhoff M W, Wright K L, Ting J P. CCAAT-binding factor NF-Y and RFX are required for in vivo assembly of a nucleoprotein complex that spans 250 base pairs: the invariant chain promoter as a model. Mol Cell Biol. 1997;17:4589–4596. doi: 10.1128/mcb.17.8.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Littlejohn T G, Hynes M J. Analysis of the site of action of the amdR product for the regulation of the amdS gene of Aspergillus nidulans. Mol Gen Genet. 1992;235:81–88. doi: 10.1007/BF00286184. [DOI] [PubMed] [Google Scholar]
- 28.Litzka O, Papagiannopolous P, Davis M A, Hynes M J, Brakhage A A. The penicillin regulator PENR1 of Aspergillus nidulans is a HAP-like transcriptional complex. Eur J Biochem. 1998;251:758–767. doi: 10.1046/j.1432-1327.1998.2510758.x. [DOI] [PubMed] [Google Scholar]
- 29.Litzka O, Then Bergh K, Brakhage A A. The Aspergillus nidulans penicillin-biosynthesis gene aat(penDE) is controlled by a CCAAT-containing DNA element. Eur J Biochem. 1996;238:675–682. doi: 10.1111/j.1432-1033.1996.0675w.x. [DOI] [PubMed] [Google Scholar]
- 30.Maity S N, de Crombrugghe B. Biochemical analysis of the B subunit of the heteromeric CCAAT-binding factor. A DNA-binding domain and a subunit interaction domain are specified by two separate segments. J Biol Chem. 1992;267:8286–8292. [PubMed] [Google Scholar]
- 31.Maity S N, de Crombrugghe B. Role of the CCAAT-binding protein CBF/NF-Y in transcription. Trends Biochem Sci. 1998;23:174–178. doi: 10.1016/s0968-0004(98)01201-8. [DOI] [PubMed] [Google Scholar]
- 32.Mantovani R. A survey of 178 NF-Y binding CCAAT boxes. Nucleic Acids Res. 1998;26:1135–1143. doi: 10.1093/nar/26.5.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McNabb D S, Xing Y, Guarente L. Cloning of yeast HAP5: a novel subunit of a heteromeric complex required for CCAAT binding. Genes Dev. 1995;9:47–58. doi: 10.1101/gad.9.1.47. [DOI] [PubMed] [Google Scholar]
- 34.McNabb D S, Tseng K A, Guarente L. The Saccharomyces cerevisiae Hap5p homolog from fission yeast reveals two conserved domains that are essential for assembly of heterotetrameric CCAAT-binding factor. Mol Cell Biol. 1997;17:7008–7018. doi: 10.1128/mcb.17.12.7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mulder W, Scholten I H, Grivell L A. Distinct transcriptional regulation of a gene coding for a mitochondrial protein in the yeasts Saccharomyces cerevisiae and Kluyveromyces lactis despite similar promoter structures. Mol Microbiol. 1995;17:813–824. doi: 10.1111/j.1365-2958.1995.mmi_17050813.x. [DOI] [PubMed] [Google Scholar]
- 36.Nagata O, Takashima T, Tanaka M, Tsukagoshi N. Aspergillus nidulans nuclear proteins bind to a CCAAT element and the adjacent upstream sequence in the promoter region of the starch-inducible Taka-amylase A gene. Mol Gen Genet. 1993;237:251–260. doi: 10.1007/BF00282807. [DOI] [PubMed] [Google Scholar]
- 37.Nakshatri H, Bhat-Nakshatri P, Currie R A. Subunit association and DNA binding activity of the heterotrimeric transcription factor NF-Y is regulated by cellular redox. J Biol Chem. 1996;271:28784–28791. doi: 10.1074/jbc.271.46.28784. [DOI] [PubMed] [Google Scholar]
- 38.Narendja, F. Unpublished data.
- 39.Olesen J T, Guarente L. The HAP2 subunit of yeast CCAAT transcriptional activator contains adjacent domains for subunit association and DNA recognition: model for the HAP2/3/4 complex. Genes Dev. 1990;4:1714–1729. doi: 10.1101/gad.4.10.1714. [DOI] [PubMed] [Google Scholar]
- 40.Papagiannopoulos P, Andrianopoulos A, Sharp J A, Davis, M. A. M A, Hynes M J. The hapC gene of Aspergillus nidulans is involved in the expression of CCAAT-containing promoters. Mol Gen Genet. 1996;251:412–421. doi: 10.1007/BF02172369. [DOI] [PubMed] [Google Scholar]
- 41.Parsons L M, Davis M A, Hynes M J. Identification of functional regions of the positively acting regulatory gene amdR from Aspergillus nidulans. Mol Microbiol. 1992;6:2999–3007. doi: 10.1111/j.1365-2958.1992.tb01758.x. [DOI] [PubMed] [Google Scholar]
- 42.Pinkham J, Guarente L. Cloning and molecular analysis of the HAP2 locus: a global regulator of respiratory genes in Saccharomyces cerevisiae. Mol Cell Biol. 1985;5:3410–3416. doi: 10.1128/mcb.5.12.3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Richardson I B, Hurley S K, Hynes M J. Cloning and molecular characterisation of the amdR controlled gatA gene of Aspergillus nidulans. Mol Gen Genet. 1989;217:118–125. doi: 10.1007/BF00330950. [DOI] [PubMed] [Google Scholar]
- 44.Richardson I B, Katz M E, Hynes M J. Molecular characterisation of the lam locus and sequences involved in regulation by the AmdR protein of Aspergillus nidulans. Mol Cell Biol. 1992;12:337–346. doi: 10.1128/mcb.12.1.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1989. [Google Scholar]
- 46.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain- terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sinha S, Maity S N, Lu J, de Crombrugghe B. Recombinant rat CBF-C, the third subunit of CBF/NFY, allows formation of a protein-DNA complex with CBF-A and CBF-B and with yeast HAP2 and HAP3. Proc Natl Acad Sci USA. 1995;92:1624–1628. doi: 10.1073/pnas.92.5.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tabor S, Richardson C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Then Bergh K, Litzka O, Brakhage A A. Identification of a major cis-acting DNA element controlling the bidirectionally transcribed penicillin biosynthesis genes acvA (pcbAB) and ipnA (pcbC) of Aspergillus nidulans. J Bacteriol. 1996;178:3908–3916. doi: 10.1128/jb.178.13.3908-3916.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Todd R B, Andrianopoulos A, Davis M A, Hynes M J. FacB, the Aspergillus nidulans activator of acetate utilization genes, binds dissimilar DNA sequences. EMBO J. 1998;17:2042–2054. doi: 10.1093/emboj/17.7.2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Heeswijck R, Hynes M J. The amdR product and a CCAAT-binding factor bind to adjacent, possibly overlapping DNA sequences in the promoter region of the Aspergillus nidulans amdS gene. Nucleic Acids Res. 1991;19:2655–2660. doi: 10.1093/nar/19.10.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weidner G, Steffan B, Brakhage A A. The Aspergillus nidulans lysF gene encodes homoaconitase, an enzyme involved in the fungus-specific lysine biosynthesis pathway. Mol Gen Genet. 1997;255:237–247. doi: 10.1007/s004380050494. [DOI] [PubMed] [Google Scholar]
- 53.Xing Y, Fikes J D, Guarente L. Mutations in yeast HAP2/HAP3 define a hybrid CCAAT box binding domain. EMBO J. 1993;12:4647–4655. doi: 10.1002/j.1460-2075.1993.tb06153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]