ABSTRACT
Feline leukemia virus (FeLV) is associated with a range of clinical signs in felid species. Differences in disease processes are closely related to genetic variation in the envelope (env) region of the genome of six defined subgroups. The primary hosts of FeLV are domestic cats of the Felis genus that also harbor endogenous FeLV (enFeLV) elements stably integrated in their genomes. EnFeLV elements display 86% nucleotide identity to exogenous, horizontally transmitted FeLV (FeLV-A). Variation between enFeLV and FeLV-A is primarily in the long terminal repeat (LTR) and env regions, which potentiates generation of the FeLV-B recombinant subgroup during natural infection. The aim of this study was to examine recombination behavior of exogenous FeLV (exFeLV) and enFeLV in a natural FeLV epizootic. We previously described that of 65 individuals in a closed colony, 32 had productive FeLV-A infection, and 22 of these individuals had detectable circulating FeLV-B. We cloned and sequenced the env gene of FeLV-B, FeLV-A, and enFeLV spanning known recombination breakpoints and examined between 1 and 13 clones in 22 animals with FeLV-B to assess sequence diversity and recombination breakpoints. Our analysis revealed that FeLV-A sequences circulating in the population, as well as enFeLV env sequences, are highly conserved. We documented many recombination breakpoints resulting in the production of unique FeLV-B genotypes. More than half of the cats harbored more than one FeLV-B variant, suggesting multiple recombination events between enFeLV and FeLV-A. We concluded that FeLV-B was predominantly generated de novo within each host, although we could not definitively rule out horizontal transmission, as nearly all cats harbored FeLV-B sequences that were genetically highly similar to those identified in other individuals. This work represents a comprehensive analysis of endogenous-exogenous retroviral interactions with important insights into host-virus interactions that underlie disease pathogenesis in a natural setting.
IMPORTANCE Feline leukemia virus (FeLV) is a felid retrovirus with a variety of disease outcomes. Exogenous FeLV-A is the virus subgroup almost exclusively transmitted between cats. Recombination between FeLV-A and endogenous FeLV analogues in the cat genome may result in emergence of largely replication-defective but highly virulent subgroups. FeLV-B is formed when the 3′ envelope (env) region of endogenous FeLV (enFeLV) recombines with that of the exogenous FeLV (exFeLV) during viral reverse transcription and integration. Both domestic cats and wild relatives of the Felis genus harbor enFeLV, which has been shown to limit FeLV-A disease outcome. However, enFeLV also contributes genetic material to the recombinant FeLV-B subgroup. This study evaluates endogenous-exogenous recombination outcomes in a naturally infected closed colony of cats to determine mechanisms and risk of endogenous retroviral recombination during exogenous virus exposure that leads to enhanced virulence. While FeLV-A and enFeLV env regions were highly conserved from cat to cat, nearly all individuals with emergent FeLV-B had unique combinations of genotypes, representative of a wide range of recombination sites within env. The findings provide insight into unique recombination patterns for emergence of new pathogens and can be related to similar viruses across species.
KEYWORDS: feline leukemia virus, genetic recombination, retroviruses
INTRODUCTION
Endogenous retroviruses influence pathogenesis of exogenous viral infections by a wide variety of mechanisms (1, 2). An endogenous virus forms when proviral elements integrate into host germ line cells and become heritable components of the host genome. Over time, these endogenous viruses often acquire mutations that render them incapable of generating infectious virus (3–8). A prime example of this is feline leukemia virus (FeLV), a gammaretrovirus that infects domestic and wild felid species and is known to cause a range of diseases, including lymphadenopathy, anemia, bone marrow suppression, immune suppression, lymphoma, leukemia, and, ultimately, death (9, 10). FeLV has the highest case fatality rate in domestic cats of all the major feline viruses, which include feline immunodeficiency virus and feline coronaviruses (11). Although multiple species of felids are susceptible to FeLV infection, only domestic and wild cats of the Felis genus harbor endogenous FeLV (enFeLV) in their genome (7). Endogenous FeLV has been correlated with resistance against exFeLV infections and is also a precursor to the FeLV-B recombinant subgroup (12, 13).
Recombination events between exogenous and endogenous FeLV is one factor in the formation of subgroups of exogenous FeLV, which include FeLV-A, -B, -C, -D, -E, and -T (3–5, 9, 14–16). The most epidemiologically relevant subgroup, FeLV-A, is an exclusively exogenous virus and is horizontally transmitted through social behaviors, via contact with blood, feces, and during suckling (13, 17, 18). The remainder have arisen via mutations, deletions, and recombination between enFeLV (or endogenous retrovirus-domestic cat [ERV-DC]) and exogenous FeLV-A (exFeLV) in the envelope gene (env) (7, 9, 15, 17, 19, 20); FeLV-B is, by far, the most common recombinant variant described. Recombination in env allows for a change in cellular tropism and results in a different disease progression. Although these recombination events and mutations in env typically have been associated with increased pathogenicity, these FeLV variants are associated with decreased ability for horizontal transmission and require FeLV-A as a helper virus (9, 16, 17, 20–22). For example, horizontal transmission of FeLV-B has been documented in three different cases among many putative emergent infections (23, 24). Therefore, it is thought that, with rare exception, each enFeLV–FeLV-A recombination event occurs de novo and is unique to each infected individual (9, 20, 21).
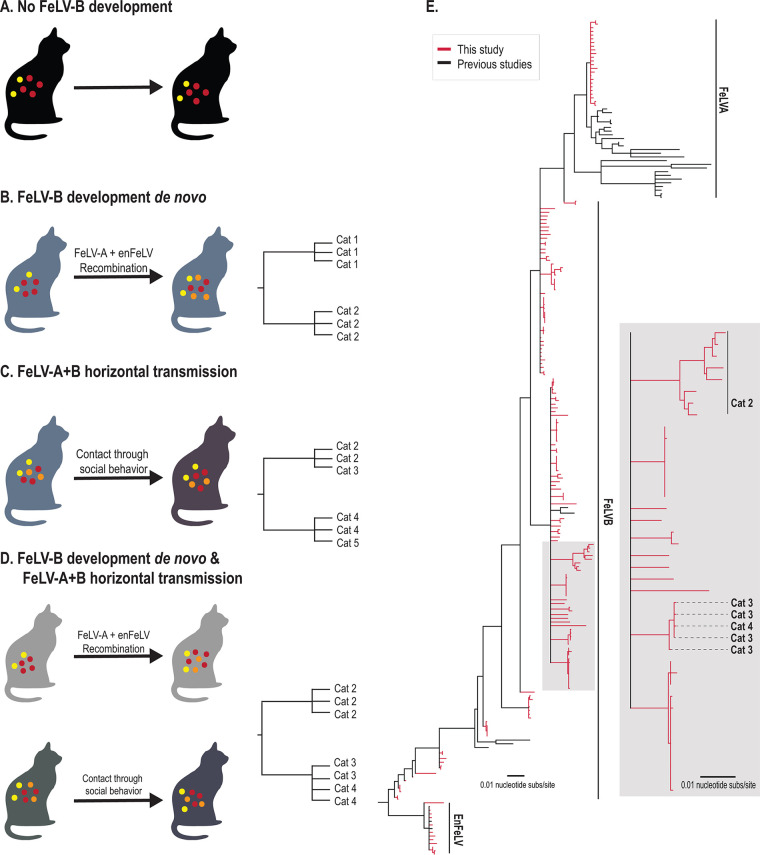
While the presence of FeLV-B has been well documented, little work has been done to understand recombination events that predispose to FeLV-B emergence during natural infection. Defining the molecular underpinnings of this process provides a better understanding of the molecular basis for FeLV-B disease and provides important mechanistic details about endogenous-exogenous retroviral interactions (25). Therefore, we genotyped naturally emergent FeLV-B isolates from a closed, privately owned colony of domestic cats with epizootic FeLV (13) to investigate how FeLV-B infection arises and spreads within a population. We hypothesized four scenarios that could be tested by our analysis (Fig. 1A to D): (i) no recombination events (i.e., no FeLV-B detected), (ii) FeLV-B formation resulting exclusively via de novo recombination within individuals, (iii) FeLV-A+B arising in a minority of individuals and subsequently transmitted horizontally, or (iv) FeLV-B de novo emergence occurring simultaneously with horizontal transmission. We analyzed FeLV-A, FeLV-B, and enFeLV env sequences from natural FeLV-A and FeLV-B infection to assess phylogenetic relatedness and recombination breakpoints from 22 individuals to determine relationships among these agents.
FIG 1.
FeLV-B phylogeny reveals a high degree of genetic dissimilarity within the closed cat colony and supports de novo recombination as a primary source of FeLV-B emergence. (A to D) Four potential scenarios for FeLV-B emergence were considered, which could be distinguished by phylogenetic analysis. All members of the Felis genus harbor enFeLV (yellow circles as shown for each scenario). (A) No recombination. Active FeLV-A infection (red circles) does not develop into FeLV-B, despite the cat harboring enFeLV (yellow circles). Ten of 32 cats evaluated were in this category. (B) A cat has an active FeLV-A infection (red circles) that recombines with enFeLV (yellow circles) to develop an FeLV-B (orange) infection. If all FeLV-B infections developed de novo and horizontal transmission was not possible or rare, genomic analysis would cluster viral sequences from the same individual together or show recombination events that are not identical between individuals. (C) A cat horizontally transmits both FeLV-A (red circles) and FeLV-B (orange circles) to another cat via social contact. In this case, phylogenies indicate branches with shared FeLV-B ancestry between infected individuals. (D) FeLV-B infection (orange circles) emerges via both de novo recombination and horizontal transmission. Resulting phylogeny would demonstrate both independent FeLV-B branches related to each cat as well as mixed branches indicating FeLV-B infections shared between individuals. It is important to note that different FeLV-B recombination break points that occur within an individual could be shared across individuals and would generate a phylogenic structure similar to that of direct transmission of FeLV-B and could confound interpretations. (E) Neighbor-joining phylogenetic tree illustrates the relationship between FeLV sequences (FeLV-A and -B and enFeLV) recovered in this study (red) and sequences reported previously (black). Two primary clades and several minor clades were identified. Several FeLV-B clades are seen here, which are indicative of the differing recombination patterns between FeLV-A and enFeLV. This results in FeLV-B isolates that are more closely related to FeLV-A, isolates that are more enFeLV-like, or variants that are intermediate between these two identities. Our data suggest scenario D, although the majority of FeLV-B variants arise de novo.
RESULTS
Prevalence of FeLV within the colony.
We previously reported that 32 cats from a colony with 65 individuals were diagnosed with FeLV antigenemia by a commercially available point-of-care test (49.2%). Results were verified using a FeLV-A specific PCR. FeLV-B-specific PCR determined that 22 of 32 FeLV-A positive individuals had detectable FeLV-B infection (68.8% FeLV-A+/B+). No individuals were FeLV-B positive without a concurrent FeLV-A infection (FeLV-A−/B+) (13).
Genetic analyses of FeLV subgroups. (i) FeLV-A and enFeLV sequences collected are nearly identical in all cats.
One hundred ninety-two FeLV sequences were analyzed from the env region of FeLV-A (22 sequences from 16 individuals), FeLV-B (156 sequences from 22 individuals), and enFeLV (14 sequences from 14 individuals) and were sequenced from domestic cats (Table 1). Phylogenetic analysis of this data set indicated that all FeLV-A sequences collected from the colony are highly similar and share 99 to 100% identity (Fig. 1E; pairwise data not shown). As anticipated, enFeLV sequences, which are fixed in the domestic cat genome, were 98 to 100% similar (Fig. 1E; pairwise data not shown). FeLV-B sequences were more variable, with 88 to 100% sequence identity (Fig. 1E; pairwise data not shown). FeLV-B phylogeny documented emergent viruses in this colony form multiple clades. Specifically, FeLV-B variants could be grouped into clades that were FeLV-A-like, enFeLV-like, or intermediate (Fig. 1).
TABLE 1.
Sample information, including cat ID, demographics, FeLV group, and accession numbers
(ii) FeLV-B recombination sites were variable.
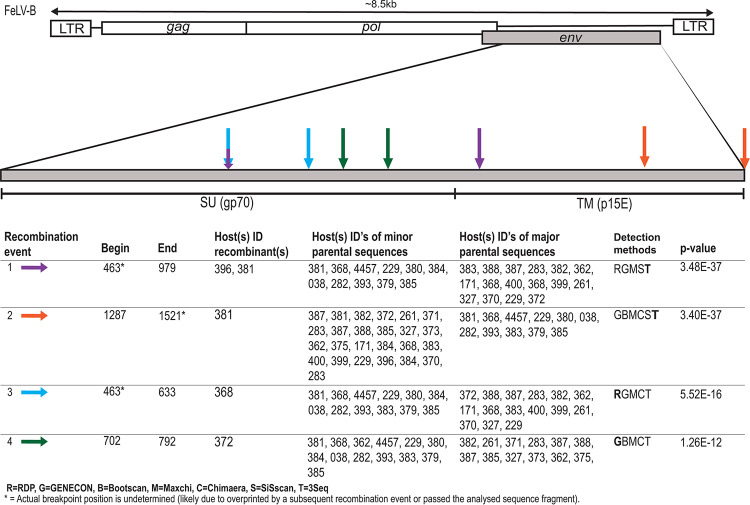
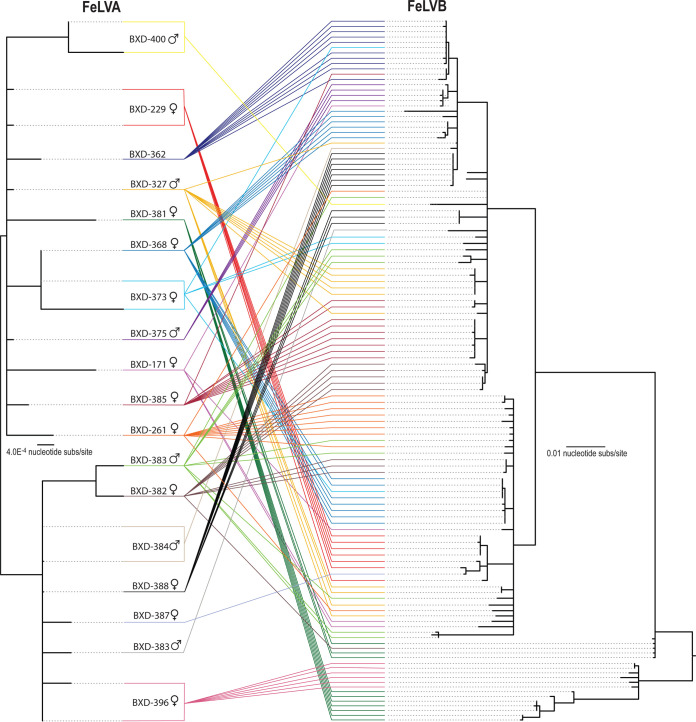
Recombination Detection Program 4 (RDP4) analysis identified numerous recombination breakpoint sites. A large number of recombination events were identified. Due to the similarities in sequence identity, it should be noted that many recombination events detected likely contribute to the genetic diversity of FeLV-B; however, for the purpose of this study, we only highlighted those events that specifically were detected to have occurred between enFeLV, FeLV-A, and FeLV-B. Our analyses detected 4 recombination sites within p15E and one site at the 3′ end of gp70 (Fig. 2). The majority of sites were located in the 5′ end of gp70. We were unable to detect all potential 5′ recombination sites due to the location of our forward primer and because of the complexity of recombination patterns in this region. FeLV-A and enFeLV recombination sites are surprisingly numerous, resulting in a large number of FeLV-B variants. A tanglegram analysis of FeLV-A and FeLV-B variants from single hosts determined that variants are not unique to a single host (Fig. 3). Of 18 individuals where both FeLV-A/B sequences were recovered, nine (50%) had multiple FeLV-B variants (Fig. 3). While several FeLV-B sequences from this study cluster with those sequenced previously, we were able to identify multiple unique clades within this single colony (Fig. 1).
FIG 2.
Several recombination sites in env identified among FeLV-A, enFeLV, and FeLV-B isolates. The schematic illustrates recombination events in FeLV-B inferred from parental FeLV-A and enFeLV sequences. The majority of sites were concentrated in the 5′ end of gp70. Recombination sites for each event are represented by arrows of different colors. Information below schematic the shows recombination event information, including event, cat ID harboring recombinant sequence, cat ID harboring parental sequence, detection methods, with best method (boldface), and the corresponding highest E value. For cat IDs, see Table 1.
FIG 3.
Comparison of FeLV-A and FeLV-B sequences from individual cats illustrates both de novo recombination and horizontal transmission. Neighbor-joining phylogeny on the left illustrates FeLV-A env recovered from a single individual. (Right) The tanglegram illustrates the relationship with FeLV-B env recovered from the same individual. Colored lines highlight env of two subtypes recovered from the same individual. This figure demonstrates that env variants from an individual are usually closely related; however, in half the cats with multiple variants detected, FeLV-B variants were distinct enough to group into separate clades.
DISCUSSION
This study of FeLV-B variants emerging during natural FeLV-A infection in a closed colony of domestic cats has important implications not only for pathogenesis of FeLV disease in cats but also in furthering our general understanding of endogenous-exogenous virus interactions (26). The most thoroughly characterized of these interactions have been described in mice (mouse mammary tumor virus and murine leukemia virus), sheep (jaagsiekte sheep retrovirus), chickens (Rous sarcoma virus and avian sarcoma leukosis virus), koalas (koala retrovirus), and cats with FeLV (27). Few studies have been performed in times of modern genomics analysis, particularly in infections occurring in a natural setting that have been well-characterized clinically and virologically; thus, our findings provide novel insights that lay the groundwork for future studies.
Although enFeLV is defective and incapable of forming infectious virions, open reading frames in several genomic segments, most notably env, result in full-length transcripts in certain tissues (19). FeLV-A recombination with enFeLV occurs during retroviral transcriptase-directed DNA synthesis following copackaging of endogenous and exogenous viral genomes (28). Our findings of common but varied sites of recombination concentrated in the N-terminal portion of env support previous results (12, 15). These hot spots likely represent locations within env where template switching between FeLV-A and enFeLV is favorable. The resultant recombinant FeLV-B strains are more virulent but are replication defective and require an FeLV-A helper virus for successful replication. The variation in recombination sites often results in changes in pathology or infectivity of FeLV-B associated with the ability to utilize new receptors. For example, certain FeLV-B variable region A (VRA) sequences within the receptor binding domain facilitate binding to both Pit 1 and Pit 2 receptors (29). Conversely, some variants may also result in a nonfunctional recombinant. The FeLV surface glycoprotein (SU gp70) is often under strong selective pressure from host immune response, as it is responsible for binding to host cell receptors and a common target of neutralizing antibodies (26, 30, 31). This selection has resulted in extensive genetic variation in FeLV SU coding regions (26), which is consistent with our findings of multiple recombination sites at a variety of locations with the SU gp70 region. Our sequences exclude an approximately 400-bp portion of the 5′ regions of env; therefore, phylogenetic analysis in Fig. 1 is not representative of the entire env. Although this region lies outside the FeLV-A–enFeLV recombination zone (Fig. 1E and 2), omission of this region does not alter our conclusions about FeLV-B composition. The frequency and variation of recombination events in this region suggest selective pressures have resulted in mechanisms for greater viral variation in this region. One event was identified in the transmembrane (TMp15E) region and another that spans much of the SU and the 5′ region of the transmembrane. The functional changes associated with these recombination sites are unknown, but previous work using chimeric viruses has established that differences in the amount of enFeLV in SU gp70 alters viral replication efficiency, cell tropism, and tendency to induce aggregation of lymphoid cells in tissue culture (12). This study investigates these variant patterns in individual cats living in a closed cat colony, validating previous in vitro results (12) and illustrating the extent to which within- and between-individual recombination between FeLV-A and enFeLV occurs, resulting in high diversity in key coding regions of FeLV-B. It is notable that the recombination patterns in the SU gp70 region correlate with those predicted by in vitro experiments nearly 30 years ago by Pandey et al. (12).
Understanding the emergence of FeLV-B has important implications for feline health, as the emergence of this recombinant virus is associated with poorer health outcomes and higher viral loads (13, 14, 22). For example, should FeLV-B be readily transmitted among cats, strain-specific testing may be important prior to housing infected cats together. Alternatively, if FeLV-B only forms within individual cats from recombination between enFeLV and FeLV-A, this may be a frequent occurrence that could increase variation of FeLV should recombination occur at different locations. Our findings suggest that the FeLV-B infections arise de novo within individual cats, as no cats were singularly infected with FeLV-B and the majority of FeLV-B infections from a single cat clustered together phylogenetically. A tanglegram comparing FeLV-A with multiple clones of FeLV-B from the same cat further supports that the variation in FeLV-B is primarily a function of FeLV-A variation. However, we identified 9 cats with more than one distinct FeLV-B variant, indicating that FeLV-B at times is copackaged and transmitted with FeLV-A and/or that more than one recombination event can emerge in an individual cat. While variation in recombination sites complicates phylogenic analysis of FeLV-B and, thus, assessment of cat-to-cat transmission of FeLV-B is inconclusive, we have documented some shared variants between cats suggesting that horizontal transmission cannot be completely excluded. Further, we have previously reported concurrent FeLV-A/FeLV-B cross-species transmissions in Florida panthers lacking enFeLV, supporting the potential for horizontal FeLV-A/FeLV-B cotransmission events (23). However, our data strongly support that FeLV-B predominately arises de novo, and variation in FeLV-B is largely driven by FeLV-A recombination site promiscuity.
Finally, while the majority of cats in this colony with progressive FeLV-A infection were FeLV-B positive, one-third of the cats were FeLV-B negative, consistent with other reports that FeLV-B does not emerge in every case of FeLV-A infection (14). Examining cellular mechanisms that relate to resistance to FeLV-B emergence and, concurrently, better prognosis, is an area for future studies to understand factors restricting ERV-XRV recombination.
MATERIALS AND METHODS
Animals and sampling.
Blood samples were collected in EDTA from 65 individual cats (leopard cat × domestic cat cross) in a privately owned breeding colony at a single time point and shipped overnight on ice. The colony consists of domestic cat-Asian leopard cat (Prionailurus bengalensis) hybrids that have been backcrossed with domestic cats for several generations. At the time of blood collection, animals ranged in age from 8 weeks to 9 years, with 30 males, 32 females, and 3 animals of unknown sex. All procedures and handling of samples were done by a licensed veterinarian (13).
Sample processing.
DNA was isolated from peripheral blood mononuclear cells (PBMCs) using a DNeasy blood and tissue extraction kit (Qiagen, Inc., Valencia, CA). DNA concentration was determined using a NanoDrop-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Blood samples were stored at −80°C and working samples stored at −20°C.
FeLV status.
During a previous study, commercially available enzyme-linked immunosorbent assay (ELISA) SNAP tests (IDEXX Laboratories, Westbrook, ME, USA) were used to determine FeLV status using plasma and serum samples (13). FeLV-A and FeLV-B status was established using an FeLV-specific PCR (described below).
Amplification of FeLV-A, -B, and enFeLV sequences.
We adapted previously published FeLV-A and enFeLV PCR protocols to compare enFeLV and FeLV-A among cats in this colony. We developed a protocol that would specifically amplify FeLV-B sequences. To accomplish this, we designed a forward primer specific for enFeLV in a known recombination site in the env gene and reverse primer specific for exFeLV in the long terminal repeat (LTR) region. Amplicons bridged recombination sites between enFeLV and FeLV-A in 3′ env. Primers used are described in Table 2.
TABLE 2.
Primers for amplification of enFeLV, FeLV-A, and FeLV-B env segments
| FeLV type | Orientation | Primer sequence | Primer location | Amplicon size (kb) |
|---|---|---|---|---|
| FeLV-A | Forward | 5′-ACCCAAGCTAATGCCACCTC-3′ | FeLV-A env | ∼1.9 |
| Reverse | 5′-CCTCTAACTTCCTTGTATCTCATGG-3′ | FeLV-A 3′-LTR | ||
| enFeLV | Forward | 5′-CTGACAGACGCCTTCCCTAC-3′ | enFeLV env | ∼2 |
| Reverse | 5′-CTAGGCTCATCTCTAGGGTCTATC-3′ | enFeLV 3′LTR | ||
| FeLV-B | Forward | 5′-CAGATCAGGAACCATTCCCAGG-3′ | enFeLV env and FeLV-B | ∼1.8 |
| Reverse | 5-CCTCTATCTTCCTTGTATCTCATGG-3′ | FeLV-A and FeLV-B 3′-LTR |
Primers were used in a PCR with the following specifications. Each reaction mix contained 1 μl of 10 μM concentration of forward and reverse primer, 10 μl of Kapa HiFi master mix (Kapa Biosystems), 7 μl water, and 2 μl DNA template in a 20-μl reaction volume. Thermal cycling conditions were 95°C for 3 min, followed by 30 cycles of 95°C for 30 s, 58°C for 30 s, and 72°C for 2 min, ending with 72°C for 3 min. Each set of PCRs included negative controls to ensure positive reactions were FeLV-B specific, namely, specific-pathogen-free (SPF) cat DNA containing enFeLV sequences (to ensure that enFeLV was not the amplification product) and a no-template control (DNA-free water). A confirmed FeLV-B sample from the colony was included as a positive control. Positive PCR samples were run on a 0.7% agarose gel at 80 V, 400 mA for 30 to 45 min. Bands of interest were excised, and DNA was extracted using a MEGAquick-spin plus fragment DNA purification kit (iNtRON Biotechnology, South Korea). Purified DNA was ligated into a Pjet1.2 plasmid, and plasmids were then isolated using a DNA-spin plasmid DNA purification kit (iNtRON Biotechnology, South Korea), which were then sequenced by a commercial laboratory using Sanger protocols (Macrogen Inc., Rockville, MD). One to 13 clones from each host were sent, depending upon sample availability, and the number of colonies was obtained.
Sequence analysis.
Sequences were trimmed and aligned in Geneious (Biomatters Inc., Newark, NJ) with representative FeLV subtypes downloaded from GenBank. Initial analysis confirmed grouping with FeLV-B/FeLV-B-like, FeLV-A, or enFeLV-like sequences. Data sets were compiled of those sequences recovered in this study and representatives available in GenBank from the three FeLV groups using the software Molecular Evolutionary Genetic Analysis (MEGA) (32), and sequences were aligned using MUSCLE (33). This data set was used to construct a neighbor-joining phylogenetic tree using a Jukes-Cantor model and 1,000 bootstrap replicates in MEGA (32). Completed trees were then uploaded to iTOL (interactive tree of life) (34), which allowed for the trees to be further modified. This comparison was used to inform the spread of FeLV-B infection versus FeLV-A infection within a single individual as well as between infected individuals in the study group.
Recombination analysis.
Recombination Detection Program 4 (RDP4) was used to detect recombination sites within FeLV-B (35). A recombination event was accepted if detected by three or more methods with associated P values of <103 and strong phylogenetic support. Analysis methods included RDP (37), GENECONV (38), Bootscan (39), Maxchi (40), Chimera (41), Siscan (42), and 3Seq (43). Sequences also underwent analysis by the Sequence Demarcation Tool, version 1.2 (SDTv1.2) (36). Tanglegrams consist of neighbor-joining phylogenetic trees of the FeLV-A and FeLV-B data sets from those cats where sequencing of both variants was recovered, constructed using the parameters described above.
ACKNOWLEDGMENTS
This work was supported by NSF-EID award 1413925 (S.V.W.) and by the Office of the Director, National Institutes Of Health, under award number F30OD023386 (E.C.).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Sue VandeWoude, Email: sue.vandewoude@colostate.edu.
Viviana Simon, Icahn School of Medicine at Mount Sinai.
REFERENCES
- 1.Chiu ES. 2019. Role of endogenous retrovirus in control of feline leukemia virus infection and implications for cross species transmission. Colorado State University Libraries, Fort Collins, CO. [Google Scholar]
- 2.Chiu ES, VandeWoude S. 2020. Presence of endogenous viral elements negatively correlates with FeLV susceptibility in puma and domestic cat cells. J Virol 94:e01274-20. 10.1128/JVI.01274-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kawasaki J, Nishigaki K. 2018. Tracking the continuous evolutionary processes of an endogenous retrovirus of the domestic cat: ERV-DC. Viruses 10:179. 10.3390/v10040179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiu ES, Hoover EA, VandeWoude S. 2018. A retrospective examination of feline leukemia subgroup characterization: viral interference assays to deep sequencing. Viruses 10:29. 10.3390/v10010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anai Y, Ochi H, Watanabe S, Nakagawa S, Kawamura M, Gojobori T, Nishigaki K. 2012. Infectious endogenous retroviruses in cats and emergence of recombinant viruses. J Virol 86:8634–8644. 10.1128/JVI.00280-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roca AL, Pecon-Slattery J, O'Brien SJ. 2004. Genomically intact endogenous feline leukemia viruses of recent origin. J Virol 78:4370–4375. 10.1128/jvi.78.8.4370-4375.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Polani S, Roca AL, Rosensteel BB, Kolokotronis S-O, Bar-Gal GK. 2010. Evolutionary dynamics of endogenous feline leukemia virus proliferation among species of the domestic cat lineage. Virology 405:397–407. 10.1016/j.virol.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 8.Rasmussen HB. 1997. Interactions between exogenous and endogenous retroviruses. J Biomed Sci 4:1–8. 10.1007/BF02255587. [DOI] [PubMed] [Google Scholar]
- 9.Levy LS. 2008. Advances in understanding molecular determinants in FeLV pathology. Vet Immunol Immunopathol 123:14–22. 10.1016/j.vetimm.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartmann K. 2011. Clinical aspects of feline immunodeficiency and feline leukemia virus infection. Vet Immunol Immunopathol 143:190–201. 10.1016/j.vetimm.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Addie D, Toth S, Reid S, Jarrett O, Dennis J, Callanan J. 2000. Long-term impact on a closed household of pet cats of natural infection with feline coronavirus, feline leukaemia virus and feline immunodeficiency virus. Vet Rec 146:419–424. 10.1136/vr.146.15.419. [DOI] [PubMed] [Google Scholar]
- 12.Pandey R, Ghosh A, Kumar D, Bachman B, Shibata D, Roy-Burman P. 1991. Recombination between feline leukemia virus subgroup B or C and endogenous env elements alters the in vitro biological activities of the viruses. J Virol 65:6495–6508. 10.1128/JVI.65.12.6495-6508.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Powers JA, Chiu ES, Kraberger SJ, Roelke-Parker M, Lowery I, Erbeck K, Troyer R, Carver S, VandeWoude S. 2018. Feline leukemia virus (FELV) disease outcomes in a domestic cat breeding colony: relationship to endogenous felv and other chronic viral infections. J Virol 92:e00649-18. 10.1128/JVI.00649-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miyazawa T. 2002. Infections of feline leukemia virus and feline immunodeficiency virus. Front Biosci 7:d504–d518. 10.2741/A791. [DOI] [PubMed] [Google Scholar]
- 15.Watanabe S, Kawamura M, Odahara Y, Anai Y, Ochi H, Nakagawa S, Endo Y, Tsujimoto H, Nishigaki K. 2013. Phylogenetic and structural diversity in the feline leukemia virus env gene. PLoS One 8:e61009. 10.1371/journal.pone.0061009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elder JH, Mullins JI. 1983. Nucleotide sequence of the envelope gene of Gardner-Arnstein feline leukemia virus B reveals unique sequence homologies with a murine mink cell focus-forming virus. J Virol 46:871–880. 10.1128/JVI.46.3.871-880.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stewart MA, Warnock M, Wheeler A, Wilkie N, Mullins J, Onions D, Neil J. 1986. Nucleotide sequences of a feline leukemia virus subgroup A envelope gene and long terminal repeat and evidence for the recombinational origin of subgroup B viruses. J Virol 58:825–834. 10.1128/JVI.58.3.825-834.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.HardyWD, Jr, McClelland A, MacEwen EG, Hess PW, Hayes AA, Zuckerman EE. 1977. The epidemiology of the feline leukemia virus (FeLV). Cancer 39:1850–1855. . [DOI] [PubMed] [Google Scholar]
- 19.Kumar DV, Berry BT, Roy-Burman P. 1989. Nucleotide sequence and distinctive characteristics of the env gene of endogenous feline leukemia provirus. J Virol 63:2379–2384. 10.1128/JVI.63.5.2379-2384.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boomer S, Gasper P, Whalen L, Overbaugh J. 1994. Isolation of a novel subgroup B feline leukemia virus from a cat infected with FeLV-A. Virology 204:805–810. 10.1006/viro.1994.1597. [DOI] [PubMed] [Google Scholar]
- 21.Ahmad S, Levy LS. 2010. The frequency of occurrence and nature of recombinant feline leukemia viruses in the induction of multicentric lymphoma by infection of the domestic cat with FeLV-945. Virology 403:103–110. 10.1016/j.virol.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jarrett O, Russell PH. 1978. Differential growth and transmission in cats of feline leukaemia viruses of subgroups A and B. Int J Cancer 21:466–472. 10.1002/ijc.2910210411. [DOI] [PubMed] [Google Scholar]
- 23.Chiu ES, Kraberger S, Cunningham M, Cusack L, Roelke M, VandeWoude S. 2019. Multiple introductions of domestic cat feline leukemia virus in endangered Florida panthers. Emerg Infect Dis 25:92–101. 10.3201/eid2501.181347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stewart H, Jarrett O, Hosie M, Willett B. 2013. Complete genome sequences of two feline leukemia virus subgroup B isolates with novel recombination sites. Genome Announc 1:e00036-12. 10.1128/genomeA.00036-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen H, Bechtel MK, Shi Y, Phipps A, Mathes LE, Hayes KA, Roy-Burman P. 1998. Pathogenicity induced by feline leukemia virus, Rickard strain, subgroup A plasmid DNA (pFRA). J Virol 72:7048–7056. 10.1128/JVI.72.9.7048-7056.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sheets R, Pandey R, Jen W, Roy-Burman P. 1993. Recombinant feline leukemia virus genes detected in naturally occurring feline lymphosarcomas. J Virol 67:3118–3125. 10.1128/JVI.67.6.3118-3125.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chiu ES, VandeWoude S. 2021. Endogenous retroviruses drive resistance and promotion of exogenous retroviral homologs. Annu Rev Anim Biosci 9:225–248. 10.1146/annurev-animal-050620-101416. [DOI] [PubMed] [Google Scholar]
- 28.Luo G, Taylor J. 1990. Template switching by reverse transcriptase during DNA synthesis. J Virol 64:4321–4328. 10.1128/JVI.64.9.4321-4328.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anderson MM, Lauring AS, Robertson S, Dirks C, Overbaugh J. 2001. Feline Pit2 functions as a receptor for subgroup B feline leukemia viruses. J Virol 75:10563–10572. 10.1128/JVI.75.22.10563-10572.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jarrett O, Laird HM, Hay D. 1972. Restricted host range of a feline leukaemia virus. Nature 238:220–221. 10.1038/238220a0. [DOI] [PubMed] [Google Scholar]
- 31.Ramsey I, Spibey N, Jarrett O. 1998. The receptor binding site of feline leukemia virus surface glycoprotein is distinct from the site involved in virus neutralization. J Virol 72:3268–3277. 10.1128/JVI.72.4.3268-3277.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Letunic I, Bork P. 2019. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47:W256–W259. 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin DP, Murrell B, Golden M, Khoosal A, Muhire B. 2015. RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol 1:vev003. 10.1093/ve/vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muhire BM, Varsani A, Martin DP. 2014. SDT: a virus classification tool based on pairwise sequence alignment and identity calculation. PLoS One 9:e108277. 10.1371/journal.pone.0108277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin D, Rybicki E. 2000. RDP: detection of recombination amongst aligned sequences. Bioinformatics 16:562–563. [DOI] [PubMed]
- 38.Padidam M, Sawyer S, Fauquet CM. 1999. Possible emergence of new geminiviruses by frequent recombination. Virology 265:218–225. [DOI] [PubMed]
- 39.Martin DP, Posada D, Crandall KA, Williamson C. 2005. A modified bootscan algorithm for automated identification of recombinant sequences and recombination breakpoints. AIDS Res Hum Retrovir 21:98–102. [DOI] [PubMed]
- 40.Smith JM. 1992. Analyzing the mosaic structure of genes. J Mol Evol 34:126–129. [DOI] [PubMed]
- 41.Posada D, Crandall KA. 2001. Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc Natl Acad Sci U S A 98:13757–13762. [DOI] [PMC free article] [PubMed]
- 42.Gibbs MJ, Armstrong JS, Gibbs AJ. 2000. Sister-scanning: a Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics 16:573–582. [DOI] [PubMed]
- 43.Boni MF, Posada D, Feldman MW. 2007. An exact nonparametric method for inferring mosaic structure in sequence triplets. Genetics 176:1035–1047. [DOI] [PMC free article] [PubMed]