ABSTRACT
The host transmembrane protein SERINC5 is incorporated into viral particles and restricts infection by certain retroviruses. However, what motif of SERINC5 mediates this process remains elusive. By conducting mutagenesis analyses, we found that the substitution of phenylalanine with alanine at position 412 (F412A) resulted in a >75-fold reduction in SERINC5’s restriction function. The F412A substitution also resulted in the loss of SERINC5’s function to sensitize HIV-1 neutralization by antibodies recognizing the envelope’s membrane proximal region. A series of biochemical analyses revealed that F412A showed steady-state protein expression, localization at the cellular membrane, and incorporation into secreted virus particles to a greater extent than in the wild type. Furthermore, introduction of several amino acid mutations at this position revealed that the aromatic side chains, including phenylalanine, tyrosine, and tryptophan, were required to maintain SERINC5 functions to impair the virus-cell fusion process and virion infectivity. Moreover, the wild-type SERINC5 restricted infection of lentiviruses pseudotyped with envelopes of murine leukemia viruses, simian immunodeficiency virus, and HIV-2, and F412A abrogated this function. Taken together, our results highlight the importance of the aromatic side chain at SERINC5 position 412 to maintain its restriction function against diverse retrovirus envelopes.
IMPORTANCE The host protein SERINC5 is incorporated into progeny virions of certain retroviruses and restricts the infectivity of these viruses or sensitizes the envelope glycoprotein to a class of neutralizing antibodies. However, how and which part of SERINC5 engages with the diverse array of retroviral envelopes and exerts its antiretroviral functions remain elusive. During mutagenesis analyses, we eventually found that the single substitution of phenylalanine with alanine, but not with tyrosine or tryptophan, at position 412 (F412A) resulted in the loss of SERINC5’s functions toward diverse retroviruses, whereas F412A showed steady-state protein expression, localization at the cellular membrane, and incorporation into progeny virions to a greater extent than the wild type. Results suggest that the aromatic side chain at position 412 of SERINC5 plays a critical role in mediating antiviral functions toward various retroviruses, thus providing additional important information regarding host and retrovirus interaction.
KEYWORDS: SERINC5, host restriction factor, infectivity, HIV-1, retroviruses
INTRODUCTION
Recent reports demonstrate that a family of serine incorporator (SERINC) proteins (1) acts as a host restriction protein against HIV-1 (2, 3). Among them, SERINC5, and to a lesser extent SERINC3, most potently inhibits viral infectivity toward target cells. Being a 40- to 55-kDa transmembrane protein, SERINC5 is incorporated into nascent viral particles and may compromise the formation of the virus-cell fusion pore for viral entry into target cells (2–4). Several models have been proposed to explain how SERINC5 inhibits HIV-1 infectivity; e.g., in one model, SERINC5 induces conformational remodeling of viral envelopes, and in another, clustering of SERINC5 on the viral membrane results in increased rigidity of the viral membrane (5–7). The former model could be supported at least to some extent by data showing that envelopes of certain HIV-1 strains are less susceptible to SERINC5-mediated inhibition of infectivity (2, 8) and that, in the presence of SERINC5, some neutralization-resistant envelopes could become susceptible to neutralization by certain monoclonal antibodies targeting the membrane-proximal external region (MPER) and the gp120-gp41 interface region of the HIV-1 envelope (4, 9, 10). More recently, cryo-electron microscopy (EM) structural analyses of human SERINC5 and its orthologue in Drosophila melanogaster revealed that potential functional sites for HIV-1 restriction are composed of surface-exposed regions of SERINC5 formed by clusters of amino acid residues conserved across biological species (11). However, how specific amino acid residues of SERINC5 are involved in HIV-1 restriction activity remains enigmatic. In addition to HIV-1, human SERINC5 can also effectively restrict the infectivity of other retroviruses, including murine leukemia virus (MLV) (2, 12), simian immunodeficiency virus (SIV) (13) and equine infectious anemia virus (EIAV) (14). However, how SERINC5 can interact with such highly diverse envelope sequences to restrict viral infectivity remains elusive.
In this study, we sought to investigate detailed roles of the surface-exposed regions (extracellular loops) of SERINC5 in restriction functions against HIV-1 as well as other retroviruses. By introducing a series of amino acid insertions and substitutions, we narrowed down potential roles of individual amino acid residues in the extracellular loops and revealed that the aromatic side chains at position 412 play an important role in SERINC5’s restriction function against not only HIV-1 but also other lentiviruses and retroviruses.
RESULTS
Important role of extracellular loop 5 of SERINC5 in HIV-1 restriction.
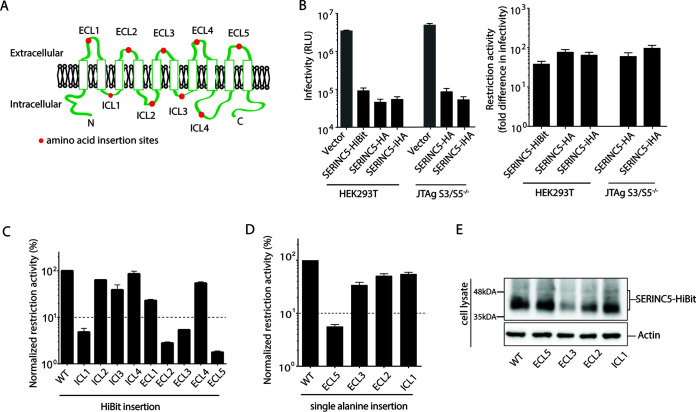
Human SERINC5 harbors 10 transmembrane domains connected by 4 intracellular loops (ICL) and 5 extracellular loops (ECL) (10, 11) (Fig. 1A) and exhibited a 40- to 100-fold reduction in HIV-1 infectivity in the absence of Nef expression when producer cells were cotransfected with an HIV-1ΔNef proviral vector and various types of SERINC5-expressing vectors (Fig. 1B, left). The restriction function was calculated based on the fold difference in infectivity between viruses produced in the absence and presence of SERINC5 (Fig. 1B, right). First, to look for a loop structure important for the restriction function, we inserted a 11-mer peptide, HiBit (VSGWRLFKKIS) (15) into each loop of SERINC5 and examined its functional effects. HiBit insertions at ICL1, ECL2, ECL3, and ECL5 resulted in a >20-fold impairment of the SERINC5 function, whereas those at other loops resulted in <5-fold impairment (Fig. 1C). Next, we tested the functional effects by inserting an alanine residue (instead of HiBit to reduce the size of the inserted peptide) at the same positions, designated ICL1Ala115-116, ECL2Ala147-148, ECL3Ala223-224, and ECL5Ala417-418. ECL5Ala417-418 impaired the SERINC5 restriction function by >20-fold, whereas the others impaired it by <3-fold (Fig. 1D). Steady-state expression levels of ECL5Ala417-418 and ICL1Ala115-116 in the cell lysates were comparable to those in the wild type (WT), whereas those of ECL3Ala223-224 and ECL2Ala147-148 were impaired (Fig. 1E). These findings suggested that ECL5 of SERINC5 contained an important determinant for the restriction of HIV-1 infectivity.
FIG 1.
Important role of extracellular loop 5 of SERINC5 in HIV-1 restriction. (A) Schematic illustration of SERINC5 based on TOPCON’s consensus prediction of transmembrane protein topology (39). Red dots represent the location of HiBit peptide or alanine insertions. (B) Infectivity of HIV-1 NL43ΔEnvΔNef pseudotyped with EnvNL produced in HEK293T and JTAg-SERINC−/− cells in the presence of vector only or various constructs of SERINC5 as indicated (left). SERINC5-HiBit and SERINC5-HA contained HiBit and HA peptide tags, respectively, at the C terminus of SERINC5. SERINC5-iHA contained an HA peptide tag located between residues 290 and 291 of SERINC5. Fold difference in infectivity in the absence and presence of SERINC5 constructs (as a measure of restriction activity of SERINC5 molecules) is shown (right). (C and D) Restriction activity of WT and a series of HiBit insertion mutants of SERINC5 (C) and a series of alanine insertion mutants of SERINC5 (D) against HIV-1 NL43ΔEnvΔNef pseudotyped with EnvNL that had been produced in HEK293T cells. The single-round infectivity was measured as β-galactosidase activity in TZM-bl target cells. The restriction activity was calculated as the fold difference of infectivity relative to the empty vector control, and the resultant values were normalized to SERINC5 WT, which was set to 100%. Mutants with normalized restriction activity of <10% (dotted lines) were considered to be significantly affected. (E) Steady-state cellular expression levels of the WT and indicated alanine-inserted mutants of SERINC5-HiBit that had been visualized with a Nano-Glo HiBit blotting system (Promega). Data are means and SD from 3 independent experiments.
Important role of F412 of SERINC5 in HIV-1 restriction.
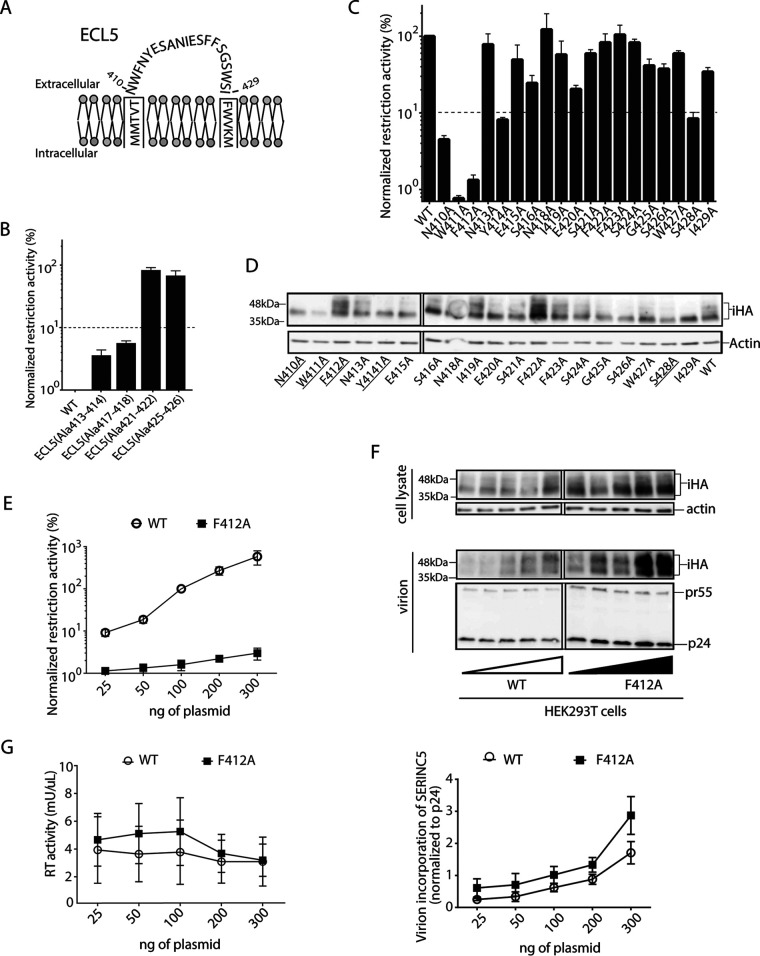
ECL5 comprises a total of 20 amino acid residues spanning positions 410 to 429 (Fig. 2A). We next introduced alanine insertions at 3 additional positions of ECL5, namely, at ECL5Ala413-414, ECL5Ala421-422, and ECL5Ala425-426. ECL5Ala413-414 also exhibited a >20-fold impairment of the restriction function with a level similar to that of ECL5Ala417-418, whereas ECL5Ala421-422 and ECL5Ala425-426 displayed no substantial effect (Fig. 2B), indicating the functional importance of the N-terminal part of the loop. Next, we sought to specify the functionally important residue(s) in ECL5 by introducing the alanine substitution at each residue. In this experiment, we used a SERINC5 variant harboring an internal HA tag inserted at positions 290 and 291 within ECL4 (designated SERINC5-iHA) (3), as we observed no functional effect by the insertion of the peptide tag at this site (Fig. 1B). The restriction function of SERINC5-iHA was significantly impaired by >10-fold when the alanine substitutions were introduced at position 410, 411, 412, 414, or 428, whereas the substitutions at other locations were less impaired (Fig. 2C). Immunoblot analysis of the lysate of HEK293T cells transfected with the gene encoding WT SERINC5 revealed multiple species, including a major 40-kDa form and a 55-kDa glycosylated form (Fig. 2D), as reported previously (16). Among the 5 alanine substitutions leading to impaired function, N410A, W411A, Y414A, and S428A showed a reduction in the steady-state protein expression level compared with that of the WT, whereas, interestingly, only the F412A mutant showed an expression level even greater than that of the WT (Fig. 2D). The restriction function of SERINC5 increased in direct proportion to the amount of plasmid DNA used for transfection (Fig. 2E). In contrast, the extent of the restriction function of the F412A mutant did not reach the SERINC5 WT level (Fig. 2E), even though the steady-state expression level of F412A increased proportionally to the amount of the plasmid (Fig. 2F). The level of the virion production was unchanged regardless of the amount of SERINC5 WT and F412A plasmid DNAs used for transfection (Fig. 2G).
FIG 2.
Important role of F412 residue of SERINC5 in HIV-1 restriction. (A) Schematic illustration of ECL5 depicting residues N410 to W429 of SERINC5. (B and C) Restriction activity of WT and a series of the indicated ECL5 alanine insertion mutants of SERINC5-iHA (B) and of a series of alanine substitution mutants of ECL5 of SERINC5-iHA (C) against HIV-1 NL43ΔEnvΔNef pseudotyped with EnvNL that had been produced in HEK293T cells. The single-round infectivity was measured as β-galactosidase activity in TZM-bl target cells. The restriction activity was calculated as the fold difference of infectivity relative to the empty vector control, and the resultant values were normalized to SERINC5 WT, which was set to 100%. Mutants with normalized restriction activity of <10% (dotted lines) were considered to be significantly affected. (D) Immunoblots showing the steady-state cellular expression levels of the indicated WT and alanine substitution mutants of SERINC5-iHA that had been visualized with anti-HA tag antibody. Mutants with significantly reduced restriction activity are underlined. (E) Restriction activity of WT and F412A mutant of SERINC5-iHA proportional to the indicated amount of DNA at transfection. (F) Immunoblots depicting the virion incorporation levels of WT and F412A mutant with increasing transfection dosage (25 to 300 ng) (top) and virion incorporation levels of WT and the F412A mutant with increasing transfection dosage (25 to 300 ng) normalized to p24 Gag levels (bottom). (G) Virion production of HIV-1 NL43ΔEnvΔNef pseudotyped with EnvNL in the presence of increasing transfection dosage (25 to 300 ng) of the genes encoding WT and F412A SERINC5 in HEK293T cells. Culture supernatant containing viral particles was harvested and assessed for reverse transcriptase (RT) activity. The immunoblot shown is representative of 3 independent replicates. Data are means and SD from 3 independent experiments.
Localization, cell surface expression, and virion incorporation of the F412A mutant.
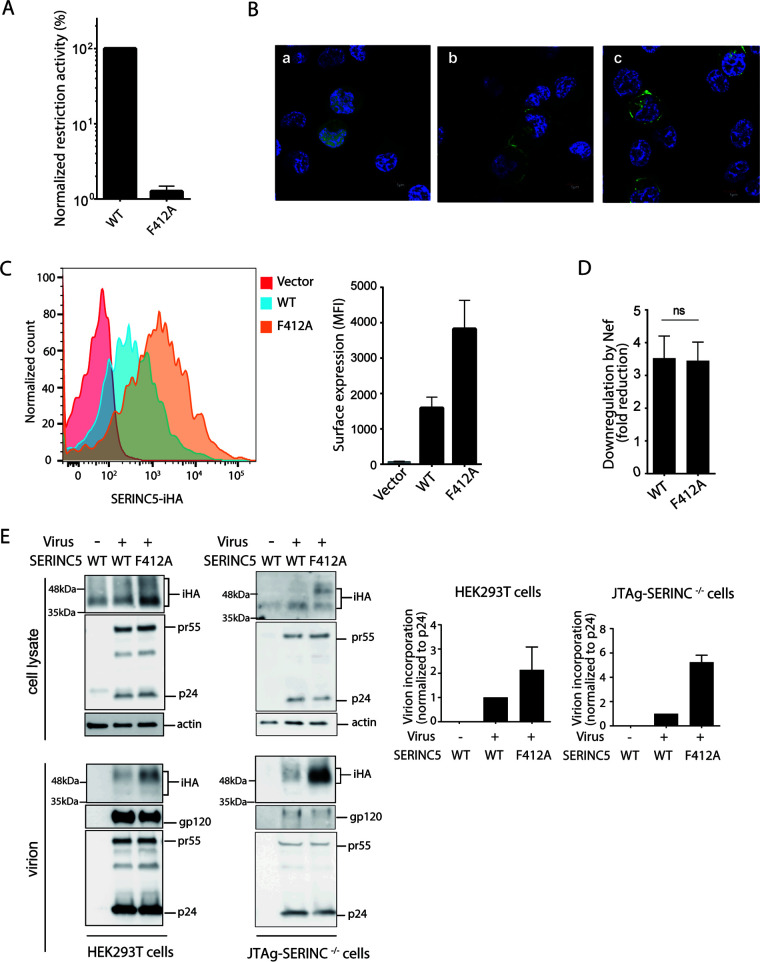
Virus-producing capacity may differ depending on the cellular lineage examined (2, 3); in particular, HEK293T cells are known to express, albeit to a lesser extent, endogenous SERINC5 (3). We therefore repeated the experiments using another virus producer cell line, Jurkat derivative JTAg cells, in which SERINC3/5 had been knocked out (designated JTAg-SERINC−/−). The SERINC5 restriction function appeared to be consistent regardless of the virus-producing cell line, i.e., HEK293T and JTAg-SERINC−/− (Fig. 1B). In addition, F412A SERINC5 exhibited a >75-fold impairment of restriction function when the virus was produced in JTAg-SERINC−/− cells (Fig. 3A), in good agreement with the data obtained from HEK293T cells as a virus producer cell line (Fig. 2C).
FIG 3.
Cell-surface localization of SERINC5 F412A and its incorporation into HIV-1 virions. (A) Restriction activity of WT and F412A mutant of SERINC5-iHA against HIV-1 NL43ΔEnvΔNef pseudotyped with EnvNL that had been produced in JTAg-SERINC−/− cells. (B) JTAg-SERINC−/− cells transfected with the genes encoding GFP alone (a), WT SERINC5-GFP (green) (b), or the F412A mutant (c) of SERINC5-GFP (green) were visualized under a fluorescence microscope. DAPI was used to stain cellular nuclei (in blue). (C) JTAg-SERINC−/− cells transfected with empty vector or genes encoding WT and F412A mutant of SERINC5-iHA were stained with anti-HA antibody and analyzed for cell surface expression by flow cytometry. (D) The cell surface expression of WT and F412A SERINC5-iHA was analyzed in the presence of GFP only or NefSF2-GFP. Fold reduction of MFI of SERINC5-iHA in GFP-gated JTAg-SERINC−/− cells was shown. (E) Immunoblots showing total cellular expression and virion incorporation of the WT and the F412A mutant of SERINC5-iHA in HEK293T and JTAg-SERINC−/− producer cells. A mock-infected control was included. The virion incorporation level is depicted in the graphs (right). Data are means ± SD from 3 independent experiments.
We then assessed whether the F412A mutation could alter the subcellular localization of SERINC5. JTAg-SERINC−/− cells were transfected with genes encoding green fluorescent protein (GFP)-tagged SERINC5 WT and the F412A, and the subcellular localization was observed by confocal microscopy. Consistent with previous reports (2, 17), SERINC5 WT was largely located around the plasma membrane, as was the F412A mutant (Fig. 3B). In addition, we assessed cell surface expression levels of SERINC5-iHA WT and F412A by flow cytometry. The mean fluorescence intensity was substantially increased when the JTAg-SERINC−/− cells were transfected with the gene encoding SERINC5-iHA and stained with anti-HA antibody (Fig. 3C). In contrast, the F412A mutant showed even greater mean fluorescence intensity than SERINC5-iHA WT, in good agreement with the data showing greater steady-state expression level of F412A than that of WT in the cells (Fig. 2D and F). HIV-1 Nef downregulates SERINC5 from the cell surface by interacting at ICL4 in conjunction with host adaptor protein 2 (18, 19). We asked whether F412A affects Nef’s sensitivity to downregulate SERINC5. The data showed that fold reduction of the cell surface expression level of SERINC5-iHA by Nef was unchanged after the introduction of F412A (Fig. 3D).
We further assessed whether F412A could impair SERINC5 incorporation into nascent virions. Both fractions of cell lysates and culture supernatant containing virion particles were prepared from HEK293T and JTAg-SERINC−/− cells transfected with the genes encoding WT and F412A SERINC5-iHA, and the steady-state expression level of SERINC5-iHA as well as that of gp120 and Gag proteins (as components of HIV-1 virions) was quantified by immunoblot analysis (Fig. 3E). Consistent with the data obtained with HEK293T cells (Fig. 2D), immunoblot analysis of the lysate of JTAg-SERINC−/− cells expressing WT and F412A SERINC5-iHA showed multiple species, including a glycosylated form of SERINC5-iHA (Fig. 3E). The steady-state expression level of the F412A mutant in the cell lysate was greater than that of the WT regardless of the cell line used, whereas the expression levels of viral antigens were comparable (Fig. 3E). Of note, F412A was again incorporated into the nascent virions to a greater extent than was the WT, whereas the expression levels of viral antigens were comparable (Fig. 3E). Overall, these results demonstrated that F412A impaired the restriction function of SERINC5 but was efficiently incorporated into virions.
Importance of aromatic side chains at position 412 for SERINC5 restriction function.
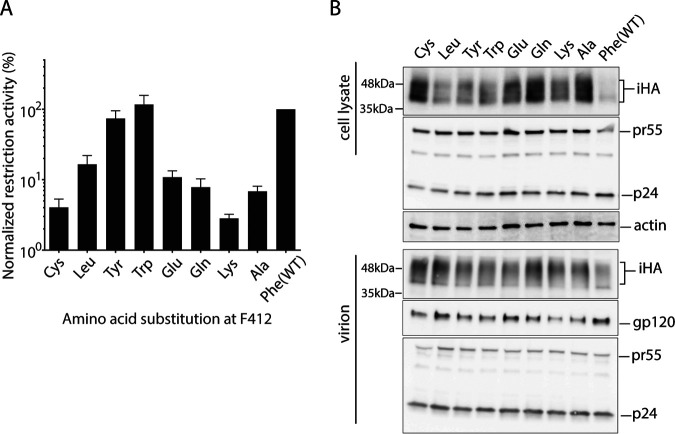
Next, we replaced the F412 residue with several other amino acids, including cysteine, leucine, tyrosine, tryptophan, glutamine, glutamic acid, and lysine, and examined their effects on the restriction function and the intracellular steady-state expression level of SERINC5 in HEK293T cells. The SERINC5-iHA restriction function was preserved when F412Y and F412W mutations were introduced, whereas the function was substantially impaired by the other mutations (Fig. 4A). Notably, all mutants tested here showed steady-state cellular expression and virion incorporation to a greater extent than the WT (Fig. 4B).
FIG 4.
Effects of a panel of F412 mutations on the restriction function and steady-state expression level of SERINC5. (A and B) Restriction activity (A) and immunoblot analysis (B) of WT and variants of SERINC5-iHA having various amino acid substitutions at F412 against EnvNL pseudotyped HIV-1 NL43ΔEnvΔNef produced in HEK293T cells. Data are means and SD from 3 independent experiments.
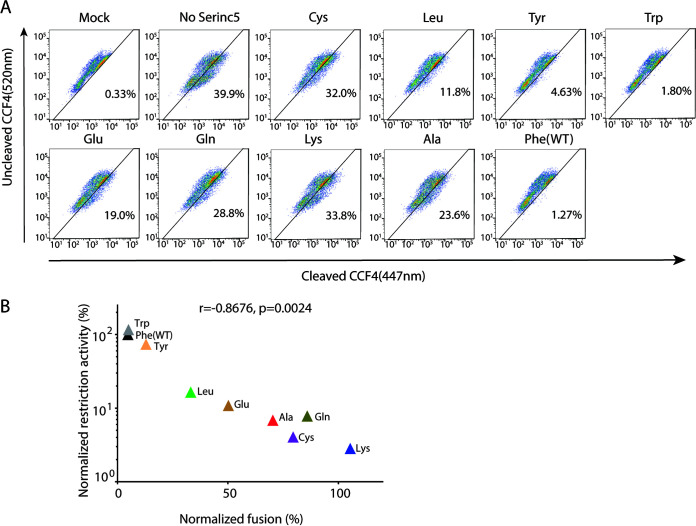
SERINC5 is known to abrogate HIV-1 entry, at least partly, by impairing the virus-cell fusion process (2–4), although additional factors may be involved in exerting its restriction function (20). We tested the panel of SERINC5 mutants at position 412 for the virus-cell fusion process by using the BlaM-Vpr fusion assay (21). As expected, the virus-cell fusion was rarely observed in the absence of the envelope (mock infection; 0.33%), whereas it was clearly observed in the presence of the envelope and the absence of SERINC5 (39.9%) (Fig. 5A). WT (Phe) SERINC5 inhibited this fusion process to 1.27%, whereas the virus-cell fusion remained at 23.6% in the presence of the SERINC5 F412A (Ala) mutant, suggesting that F412A impaired the SERINC5-mediated inhibition of the fusion process. In contrast, the substitution of phenylalanine within the aromatic side chain groups (i.e., tyrosine and tryptophan) of SERINC5 exhibited comparable inhibitory activity toward the fusion process (Fig. 5A). The substitution with leucine and glutamic acid affected to some extent the SERINC5-mediated inhibition of the fusion process. Overall, the restriction function (Fig. 4A) and the fusion efficiency (Fig. 5A) by SERINC5 were significantly inversely correlated (r = −0.8676; P < 0.01) (Fig. 5B), suggesting that the SERINC5 function to restrict HIV-1 infectivity was mediated dominantly through inhibition of the virus-cell fusion process. The data indicated again the importance of the aromatic side chains at SERINC5 position 412 for the restriction function.
FIG 5.
Effects of the panel of F412 mutations of SERINC5 on the virus-cell fusion process. (A) HIV-1 NL43ΔEnvΔNef viruses pseudotyped without (mock) or with EnvNL supplemented with genes encoding BlaM-Vpr and SERINC5-iHA WT or a panel of F412 mutants were produced in HEK293T cells. Fusion formation between the resultant viral particles and the TZM-bl target cells, as assessed by fluorescence signals from the cleaved form of CCF4 (excitation of 447 nm), was analyzed by flow cytometry. The percentage of cells that successfully achieved virus-cell fusion is indicated in the dot plots. Data are representative of 3 independent experiments. (B) Correlation between the restriction function and fusion efficiency in the context of WT and the panel of F412 mutants of SERINC5. Fusion efficiency was normalized to the absence of SERINC5-iHA, which was set to 100%. Data are means from 3 independent experiments. The statistical significance was analyzed by using Pearson’s correlation coefficient test.
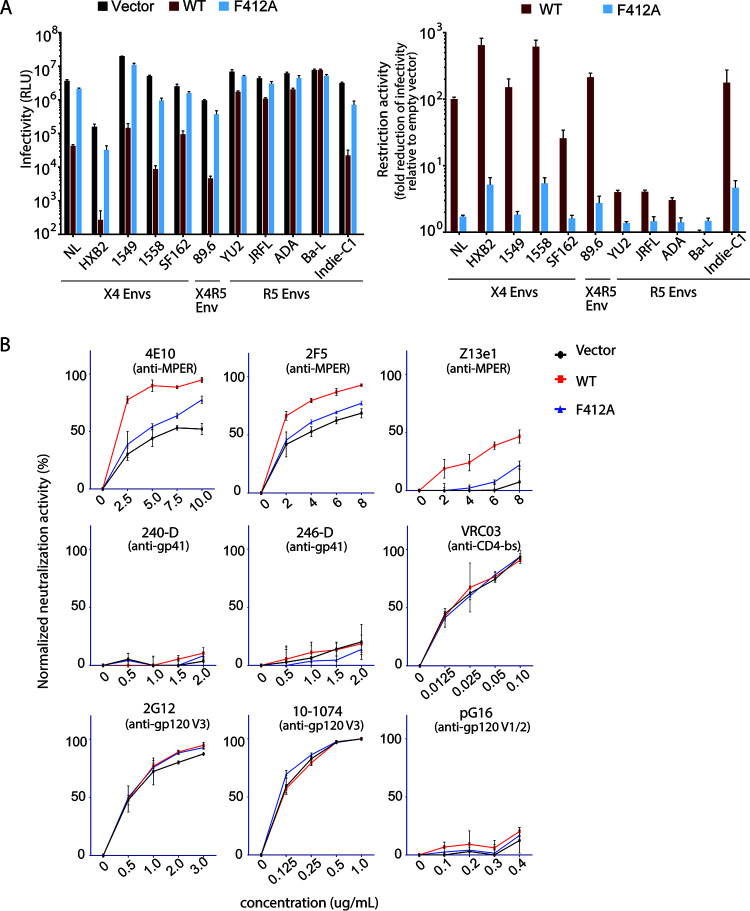
Effects of the F412A mutation on SERINC5 function toward various HIV-1 envelopes.
The restriction function of SERINC5 is known to vary among various strains of HIV-1 envelopes (2–4, 8, 9). We therefore sought to test WT and F412A SERINC5 restriction function for virions pseudotyped with various strains of HIV-1 envelopes, including NL4-3, HXB2, YU2, JRFL, ADA, and Ba-L. We also tested envelopes of primary isolates, including CXCR4-tropic QH1549 and QH1558, derived from 2 individuals with late-stage AIDS (22), and CCR5-tropic Indie-C1, isolated in India (23). As expected, WT SERINC5 exhibited the restriction function toward all envelopes except for Ba-L, and the extent of the restriction function was only <4-fold for envelopes of YU2, JRFL, and ADA compared with those of the others (Fig. 6A). In contrast, the envelopes of HXB2, 1558, Indie-C1, 89.6, and 1549 showed higher sensitivity to SERINC5 restriction than the NL4-3 envelope. Also, F412A SERINC5 showed decreased restriction function compared with the WT toward all envelopes tested except for the case of Ba-L (Fig. 6A).
FIG 6.
Effects of F412A on SERINC5-mediated restriction of various HIV-1 envelopes. (A) Infectivity values of HIV-1 NL43ΔEnvΔNef pseudotyped with various HIV-1 envelopes produced in the presence of WT or F412A mutant of SERINC5-HA in JTAg-SERINC−/− cells. The single-round infectivity was measured as β-galactosidase activity in TZM-bl target cells, and the background levels of luminescence (cells only) were deducted as relative light units. The restriction activity is shown as fold difference in infectivity in the absence and presence of WT or mutant SERINC5. (B) HIV-1 NL43ΔEnvΔNef pseudotyped with EnvJRFL was produced in JTAg-SERINC−/− cells in the presence of empty vector or genes encoding WT or F412A of SERINC5-HA. The pseudoviruses were preincubated with increasing concentrations of the indicated neutralizing antibodies for 1 h at 37°C, and then TZM-bl indicator cells were exposed to them. Neutralization activity was calculated as the percentage reduction in infectivity relative to infectivity in the absence of antibody. Data are means and SD from 3 independent experiments.
It was reported that SERINC5 has an additional function to render certain envelopes (including neutralization-resistant JRFL) susceptible to neutralization by antibodies targeting epitopes at the membrane-proximal external region (MPER) of gp41 (4, 9, 11) and at the gp41-gp120 interface (9). We wanted to see whether the F412A mutation could affect this additional SERINC5 function. Therefore, HIV-1 virions pseudotyped with JRFL were prepared in the absence and presence of WT and F412A SERINC5. The sensitivity of these viruses to neutralization was tested with a panel of monoclonal antibodies, including the anti-gp41 MPER antibodies 2F5, 4E10, and Z13; anti-gp41 antibodies 240-D and 246-D; anti-gp120 CD4 binding site antibody VRC03; anti-gp120 V1/V2 antibody PG16; and anti-gp120 V3 antibodies 2G12 and 10-1074. As expected, JRFL-pseudotyped viruses produced in the presence of WT SERINC5 showed enhanced neutralization by anti-gp41 MPER antibodies 2F5, 4E10, and Z13, whereas the viruses remained resistant to neutralization by the other classes of antibodies (Fig. 6B). Notably, F412A SERINC5 failed to sensitize the susceptibility of JRFL-pseudotyped viruses to anti-MPER neutralizing antibodies (Fig. 6B).
Effects of the F412A mutation on SERINC5 function toward retroviral envelopes.
Although HIV-2 Nef is known to counteract SERINC5 and enhance infectivity of HIV-1ΔNef (13), it remains unknown whether HIV-2 infectivity is restricted by SERINC5. Therefore, we prepared HIV-2 pseudoviruses by transfecting HEK293T cell with the nef- and env-deficient HIV-2 proviral vector pGL-ANΔEnvΔNef and the plasmid encoding the envelope of HIV-2 GH123 or vesicular stomatitis virus glycoprotein (VSV-G). As expected, SERINC5 did not inhibit the infectivity of HIV-2 pseudotyped with VSV-G. In contrast, infectivity of HIV-2 pseudotyped with EnvGH123 showed a 4-fold reduction in the presence of SERINC5 (Fig. 7A), indicating the ability of SERINC5 to restrict HIV-2 infectivity. We then tested the effect of the F412A mutation on SERINC5 restriction function against HIV-2. The results clearly showed that the F412A mutation abrogated the restriction function of SERINC5 against HIV-2 (Fig. 7A).
FIG 7.
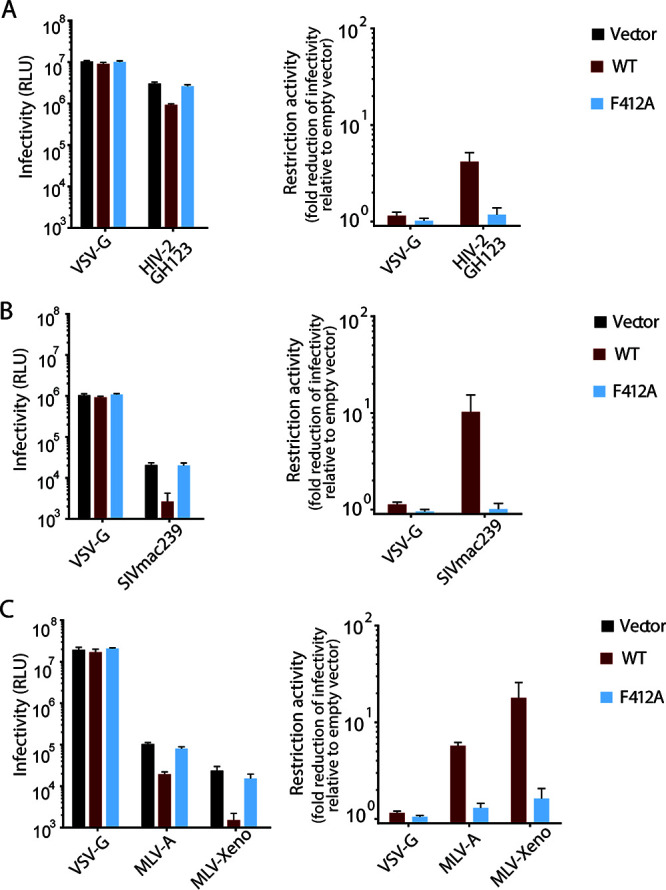
Effects of F412A on SERINC5-mediated restriction of various retroviral envelopes. Infectivity values (left) of HIV-2 GL-ANΔEnvΔNef pseudotyped with HIV-2 EnvGH123 (A), SIVmacΔEnvΔNef pseudotyped with SIVmac239 Env (B), and HIV-1 NL43ΔEnvΔNef pseudotyped with MLV Envs (C) in the absence and presence of the indicated SERINC5. Fold difference in infectivity (as restriction activity; right) of the same pseudoviruses (A to C) between the absence and the presence of WT or F412A mutant of SERINC5-HA. The pseudoviruses were produced in the absence and the presence of WT or F412A mutant of SERINC5-HA in HEK293T cells and tested for the single-round infectivity as assessed in terms of firefly luciferase activity in MAGIC5 target cells; and the background levels were deducted. Data are means and SD from 3 independent experiments.
Infectivity of simian immunodeficiency viruses has been shown to be restricted by human SERINC5 (13). We prepared SIVmac pseudoviruses by transfecting HEK293T cells with the nef- and env-deficient SIVmac proviral vector pSIVmacΔEnvΔNef and the plasmid encoding the envelope of SIVmac239 or VSV-G. As expected, infectivity of SIVmac239 pseudovirus showed over a 10-fold reduction in the presence of SERINC5, whereas infectivity of the VSV-G pseudovirus remained comparable (Fig. 7B), confirming the ability of human SERINC5 to restrict SIVmac infectivity. We then tested the effect of the F412A mutation on SERINC5 restriction function against SIVmac. The results clearly showed that F412A mutation abrogated the restriction function of human SERINC5 against SIVmac (Fig. 7B).
In addition, certain retroviruses, including murine leukemia viruses have been shown to be restricted by human SERINC5 (2). We prepared HIV-1 pseudotyped with the envelopes of amphotropic (MLV-A) and xenotropic (MLV-Xeno) MLV strains or VSV-G. As expected, infectivity of MLV-A and MLV-Xeno pseudoviruses showed 6- and 18-fold reductions, respectively, in the presence of SERINC5, whereas the infectivity of the VSV-G-pseudovirus remained comparable (Fig. 7C), confirming the ability of human SERINC5 to restrict MLV-A and MLV-Xeno infectivity. We then tested the effect of the F412A mutation on SERINC5 restriction function against MLVs. The results clearly showed that F412A mutation abrogated the restriction function of human SERINC5 against MLVs (Fig. 7C).
DISCUSSION
We demonstrated here that the aromatic side chains at position 412 located at the base of ECL5 of SERINC5 played an essential role in the restriction function against various strains of HIV-1 as well as HIV-2, SIVmac, and lentiviruses pseudotyped with amphotropic and xenotropic envelopes of Moloney murine leukemia virus (M-MLV), thus suggesting a common antiviral mechanistic pathway of SERINC5 against these viruses. The observation that the F412A mutant failed to restrict viral infectivity while increasing the levels of intracellular expression and virion incorporation compared to those of the WT indicates that the SERINC5 restriction function took place at the surface of these diverse lentiviral particles. In addition, F412A failed to sensitize HIV-1 envelopes to certain neutralizing antibodies, suggesting fundamental roles of the F412 residue for potential interaction between SERINC5 and viral envelopes on the virion surface. These results are a substantial extension of the recently reported cryo-EM structures of SERINC5 and its orthologue in Drosophila melanogaster showing that the antiviral function of SERINC5 can be mediated by interaction of ECL5, in conjunction with ECL3 to some extent, of SERINC5, resulting in the conformational remodeling of surface-exposed regions of the HIV-1 Env (11). The results suggest that SERINC5 mediated its antiviral function by interacting with a diverse array of viral envelopes through amino acids in ECL5, including the F412 residue of SERINC5.
Aromatic side chains of amino acid residues (i.e., phenylalanine, tryptophan, and tyrosine) play a critical role in intra- and intermolecular interactions of proteins (24, 25). In our study, substitution of F412 of SERINC5 with tryptophan or tyrosine resulted in no substantial change in anti-HIV-1 restriction function, whereas substitution with other amino acids (e.g., alanine, cysteine, and lysine) substantially impaired this function. The aromatic side chain at position 412 located at the base of ECL5 may be important for maintaining conformation of the loop, or alternatively, this amino acid residue may participate in interaction with or be located in close proximity to other molecules, including HIV-1 envelopes (8, 26). In fact, a recent study demonstrates the possibility of a SERINC5-induced conformational change in SERINC5-sensitive envelopes, but not SERINC5-resistant ones, of viral particles as assessed by an antibody-dependent virus capture assay (7). Moreover, the finding in this study, obtained with a panel of SERINC5 mutants with mutations at position 412, demonstrated that the restriction function of SERINC5 significantly inversely correlated with the efficiency of the virus-cell fusion formation, which is in line with previous reports (2–4, 20) indicating that the SERINC5-mediated restriction of viral infectivity occurs at least partly at the point of the virus-cell fusion process. Further investigation is needed to reveal how aromatic side chains of the amino acid at SERINC5 position 412 are involved in inducing a conformational change in viral envelopes that may subsequently lead to restricted infectivity of diverse retroviruses.
Our present report, at least to our knowledge, is the first to show that the infectivity of an HIV-2 reporter virus pseudotyped with the HIV-2 envelope (as a model of HIV-2 infection) is inhibited by human SERINC5. Because infectivity of SIVmac239 reporter virus pseudotyped with SIVmac239 envelope was also shown to be inhibited by human SERINC5, in good agreement with a previous report (13), these results suggest that SERINC5 has the capacity to restrict the infectivity of diverse lentiviruses. In contrast, an HIV-1 reporter virus pseudotyped with certain strains of HIV-1 envelopes demonstrated here and in several previous studies (2, 3, 27) was less sensitive to restriction by human SERINC5, whereas SERINC5 could instead sensitize these envelopes (which are resistant to SERINC5-mediated inhibition of viral infectivity) to certain neutralizing antibodies, suggesting that SERINC5 maintains the ability to interact with all these envelopes. Thus, a quite interesting question remains to be answered as to how such different phenotypic effects arise in the context of SERINC5 and retroviral envelopes. The observation demonstrated here that a single F412A mutation could impair both SERINC5 functions may provide a key to investigate this issue in future studies.
Some limitations of our study merit mention. Because the mutational screening in this study focused on the SERINC5 loop structures between the transmembrane domains (ECL5 in particular), this approach could not capture all residues or motifs responsible for restricting the diverse array of retroviral envelopes and may overlook roles of the transmembrane domains. We used a total of 4 different cell lines as virus-producing and target cells for the analysis of SERINC5 function and obtained consistent results across the cell lines used. However, primary cell lineages responsible for virus-producing and virus-targeting cells in vivo might have differential expression levels and localization of SERINC5. The role of SERINC5 in cell-to-cell HIV-1 infection in vivo also remains unclear. Nevertheless, we clearly demonstrated herein that a single amino acid change (F412A) in SERINC5 did not affect the ability of this protein to be incorporated into progeny virions but did cause it to almost entirely lose the restriction function against a panel of retroviruses and that, in addition, the amino acids with aromatic side chains at this position of SERINC5 were required to exert inhibitory activity against HIV-1 infection, at least in part, at the point of the virus-cell fusion process. These results highlight a common antiretroviral mechanism by a host protein at the virion surface, revealing a novel aspect of establishing viral entry to target cells.
MATERIALS AND METHODS
Cell lines.
HEK293T cell (American Type Culture Collection), TZM-bl indicator cell (NIH AIDS Reagent Program), and MAGIC5 cells expressing CD4, CXCR4, and CCR5 receptors (23) were cultured in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (Sigma). A derivative of the Jurkat T cell line expressing simian virus 40 large T antigen (JTAg) and JTAg cells that had been engineered to knock out both SERINC3 and SERINC5 (JTAg-SERINC−/−) (2) were provided by M. Pizzato and maintained in RPMI supplemented with 10% fetal bovine serum.
Plasmid construction.
The plasmids pBJ5-SERINC5-HA and pBJ5-SERINC5-iHA (3), harboring a hemagglutinin (HA) tag (YPYDVPDYA) at the C terminus and ECL4 (inserted between positions 290 and 291) of SERINC5, respectively, were provided by H. Göttlinger. The C-terminal HA tag was replaced with an HiBit tag (VSGWRLFKKIS), giving rise to pBJ5-SERINC5-HiBit. A proviral plasmid of HIV-1, pNL43 (28), was engineered to introduce 2 stop codons in the env and the nef regions, giving rise to pNLΔEnvΔNef. Also, a pNL43 derivative harboring the luciferase gene as a marker, pNLΔEnvΔNef-LUC2/InHibit (29), was used as needed. An env-deficient proviral plasmid of SIVmac239, pSIVmacΔEnv-LUC2 (30) was engineered to introduce a stop codon in the nef region, giving rise to pSIVmacΔEnvΔNef-LUC2. Plasmid vectors encoding HIV-1 envelopes 89.6, JRFL, and YU2 (31) were provided by Y. Maeda, and SF162 and HXB2 were provided by NIH AIDS Reagent Program; NL4-3, 1549, 1558, ADA, BaL, and IndieC1 were previously described (22, 23, 30, 32). A nef- and env- deficient HIV-2 proviral vector, pGL-ANΔEnvΔNef-LUC (33), was provided by A. Adachi, and plasmid vectors encoding envelopes of MLV-A (2), MLV-X (2), SIVmac239 (30), and HIV-2 GH123 (30) were previously described. The plasmid pMM310, encoding BlaM-Vpr (34), was provided by the NIH AIDS Reagent Program. The plasmid pcDNA3.1-NefSF2-GFP, which allowed expression of the NefSF2-GFP fusion protein, was previously described (35, 36).
Virus production and infectivity measurement.
HIV-1 pseudoviruses were produced in HEK293T cells and JTAg-SERINC−/− (36). In brief, HEK293T cells were transfected with 500 ng of pNL43ΔEnvΔNef, 20 ng of an Env-encoding plasmid (such as pCAGGS-EnvNL43), and 50 ng of pBJ5-SERINC5 or empty vector that had been premixed with Lipofectamine 2000 (Thermo Fisher). JTAg cells were electroporated with 5 μg of pNL43ΔEnvΔNef, 1 μg of Env-encoding plasmid (such as pCAGGS-EnvNL43), and 900 ng pBJ5-SERINC5 or empty vector by using a Gene Pulser Xcell system (Bio-Rad) operating at 250 V and 950 μF. In both cases, 48 h later, the secreted viral particles were harvested from the culture supernatant, clarified by centrifugation at 3,000 × g for 3 min at 4°C, and quantified in terms of reverse transcriptase activity, as described previously (37). TZM-bl indicator cells (1 × 104 cells per well in 96-well plate) were exposed to equal amounts of viral inoculum (normalized by reverse transcription activity); and the single-round infectivity was determined 48 h later by measuring the chemiluminescence intensity with the Tropix Galacto-Star system (Applied Biosystems). For viruses pseudotyped with MLV, HIV-2 GH123, and SIVmac239 envelopes, the infectivity was determined by measuring firefly luciferase expression with the One-Glo luciferase assay system (Promega) in MAGIC5 target cells. Fold restriction activity of each SERINC5 mutant was calculated by dividing the infectivity of pseudovirus produced in the absence of SERINC5 (empty vector control) by that obtained in the presence of the SERINC5 mutant. The resultant value was then expressed relative to that for SERINC5 WT, which was set as 100% restriction.
Western blotting.
For analysis of cell lysates, transfected cells were lysed on ice for 15 min in a buffer {100 mM NaCl, 1 mM TCEP [Tris(2-carboxyethyl)phosphine hydrochloride], 2× protease inhibitor, and 10 mM HEPES; pH 7.5} containing 1% n-dodecyl-β-d-maltoside (DDM; Thermo Scientific). For analysis of virion particles, the culture supernatant was clarified by filtration (0.45-μm pore size), pelleted on a 20% sucrose cushion by ultracentrifugation at 50,000 rpm for 15 min at 4°C (Beckman Coulter Optima-TLX), and then lysed in 1% DDM buffer. The resultant samples were separated in SDS-PAGE gels and transferred to nitrocellulose membranes (Wako). The membranes were soaked in a blocking buffer (Nacalai Tesque) for 1 h at room temperature and then mixed with primary antibodies, including mouse anti-HA (1:1,000; BioLegend), mouse anti-HIV gp120 0.5β (1:5,000; provided by T. Kuwata) (38), rabbit anti-HIV Gag p24 (1:5,000; Bioacademia), and mouse anti-β-actin (1:5,000; Wako), followed by staining with the appropriate secondary antibodies, horseradish peroxidase (HRP)-conjugated anti-mouse (1:50,000; Wako) or anti-rabbit (1:50,000; Wako) Ig antibodies. The membrane was developed with the enhanced chemiluminescence reagents (Wako) and visualized by using ImageQuant LAS 400 (GE Healthcare). For detection of SERINC5-HiBit, the membranes were washed in Tris-buffered saline containing Tween (TBS-T) for 4 h before detection of bioluminescence by using the Nano-Glo HiBit blotting system based on the manufacturer’s recommendation (Promega).
Viral fusion assay.
HIV-1 particles incorporating BlaM-Vpr were produced by cotransfection of 293T cells with pNLΔEnvΔNef., pCA-NL43 Env, pMM310 encoding BlaM-Vpr (34), and either a pBJ5-based empty vector or mutant or WT SERINC5. Resultant viruses were collected and quantified by using p24 antigen capture enzyme-linked immunosorbent assay (ELISA) (ZeptoMetrix). TZM-bl cells (1 × 105) seeded overnight on a 12-well plate were spin-inoculated with 100 ng of the viruses at 1,500 × g and 4°C for 1 h. The remaining inocula were carefully removed and replaced with fresh medium, followed by incubation at 37°C for 3 h to allow viral entry. After having been washed 3 times with phosphate-buffered saline (PBS), cells were loaded with a 2 μM concentration of the cell-penetrating ester form of β-lactamase negatively charged fluorescent substrates (CCF4-AM) in a LiveBLAzer FRET-B/G loading kit (Invitrogen) supplemented with 0.5% solution D, an anion transport inhibitor (Invitrogen), in Opti-MEM–10% fetal bovine serum (FBS) and allowed to develop for 12 h at room temperature. Cells were washed 3 times with PBS and fixed in 2% paraformaldehyde, followed by flow cytometry analysis to quantify uncleaved and cleaved CCF4 at fluorescence intensities of 520 nm and 447 nm, respectively.
Flow cytometry.
JTAg-SERINC−/− cells (3 × 106) were electroporated at 250 V and 950 μF with 5 μg of DNA encoding WT SERINC5 or a series of mutant SERINC5-iHA proteins supplemented with GFP-encoding DNA as an internal control by using a Gene Pulser Xcell system (Bio-Rad). A day later, the resultant cells were collected and stained for 30 min at 4°C with Zombie Aqua fixable viability dye and Alexa Fluor-647-conjugated anti-HA (BioLegend) in PBS containing 2% fetal bovine serum. A live GFP+ subset was gated and then analyzed for SERINC5-iHA surface expression (as an HA tag) by flow cytometry (FACSVerse; Becton Dickinson).
Confocal microscopy.
JTAg-SERINC−/− cells (3 × 106) were electroporated as described above with 8 μg of DNA encoding WT or mutant SERINC5-GFP fusion protein. A day later, the resultant cells (2 × 105) were seeded onto 0.01% poly-l-lysine-coated slides (Nunc Lab-Tek II chamber) and fixed with 4% paraformaldehyde (Nacalai Tesque). The cells were then stained with 0.5 μg/ml of 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI; Invitrogen) for 10 min and mounted on NEO micro cover glasses (Matsunami Glass) layered with SlowFade Diamond antifade mountant (Invitrogen). A confocal microscope (Olympus FV1200-IX-KU) equipped with a 60× oil lens objective was used to examine a minimum of 100 random cells per slide under the same exposure settings.
Statistical analysis.
Statistical tests employed are described in the figure legends and were performed by using GraphPad Prism. Standard deviations (SD) were calculated to estimate variance. A P value of <0.05 was considered significant.
ACKNOWLEDGMENTS
We thank Massimo Pizzato (University of Trento) for providing the JTAg and its derivative cell lines; Heinrich G. Göttlinger (University of Massachusetts) for providing the pBJ5-SERINC5-HA, pBJ5-SERINC5-HA, and pBJ5-SERINC5-iHA plasmids; Takeo Kuwata and Shuzo Matsushita (Kumamoto University) for providing the anti-HIV gp120 0.5β antibody; Yosuke Maeda (Kumamoto University) for providing the 89.6, JRFL, and YU2 Env plasmids; Akio Adachi (Kansai Medical University) for providing HIV-2 proviral vector pGH123ΔEnvΔNef-LUC2; and the NIH AIDS Reagent Program for providing several neutralizing antibodies used in this study.
This study was supported by grants from Japan Agency for Medical Research and Development, AMED (Research Program on HIV/AIDS), and JSPS KAKENHI Grants-in-Aid for Scientific Research. T.S.T. is a recipient of a Japanese Government scholarship.
Conceptualization, T.S.T. and T.U.; reagents and specimens, M.T. and K.T.; data collection, T.S.T. and M.T.; manuscript writing, T.S.T., M.T., K.T., and T.U.; supervision, T.U.; funding acquisition, M.T. and T.U.
We declare no competing interests.
Contributor Information
Takamasa Ueno, Email: uenotaka@kumamoto-u.ac.jp.
Viviana Simon, Icahn School of Medicine at Mount Sinai.
REFERENCES
- 1.Inuzuka M, Hayakawa M, Ingi T. 2005. Serinc, an activity-regulated protein family, incorporates serine into membrane lipid synthesis. J Biol Chem 280:35776–35783. 10.1074/jbc.M505712200. [DOI] [PubMed] [Google Scholar]
- 2.Rosa A, Chande A, Ziglio S, De Sanctis V, Bertorelli R, Goh SL, McCauley SM, Nowosielska A, Antonarakis SE, Luban J, Santoni FA, Pizzato M. 2015. HIV-1 Nef promotes infection by excluding SERINC5 from virion incorporation. Nature 526:212–217. 10.1038/nature15399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Usami Y, Wu Y, Gottlinger HG. 2015. SERINC3 and SERINC5 restrict HIV-1 infectivity and are counteracted by Nef. Nature 526:218–223. 10.1038/nature15400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sood C, Marin M, Chande A, Pizzato M, Melikyan GB. 2017. SERINC5 protein inhibits HIV-1 fusion pore formation by promoting functional inactivation of envelope glycoproteins. J Biol Chem 292:6014–6026. 10.1074/jbc.M117.777714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beitari S, Wang Y, Liu SL, Liang C. 2019. HIV-1 envelope glycoprotein at the interface of host restriction and virus evasion. Viruses 11:311. 10.3390/v11040311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Firrito C, Bertelli C, Vanzo T, Chande A, Pizzato M. 2018. SERINC5 as a new restriction factor for human immunodeficiency virus and murine leukemia virus. Annu Rev Virol 5:323–340. 10.1146/annurev-virology-092917-043308. [DOI] [PubMed] [Google Scholar]
- 7.Featherstone A, Aiken C. 2020. SERINC5 inhibits HIV-1 infectivity by altering the conformation of gp120 on HIV-1 particles. J Virol 94:e00594-20. 10.1128/JVI.00594-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang X, Shi J, Qiu X, Chai Q, Frabutt DA, Schwartz RC, Zheng Y-H. 2019. CD4 expression and Env conformation are critical for HIV-1 restriction by SERINC5. J Virol 93:e00544-19. 10.1128/JVI.00544-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beitari S, Ding S, Pan Q, Finzi A, Liang C. 2017. Effect of HIV-1 Env on SERINC5 antagonism. J Virol 91:e02214-16. 10.1128/JVI.02214-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schulte B, Selyutina A, Opp S, Herschhorn A, Sodroski JG, Pizzato M, Diaz-Griffero F. 2018. Localization to detergent-resistant membranes and HIV-1 core entry inhibition correlate with HIV-1 restriction by SERINC5. Virology 515:52–65. 10.1016/j.virol.2017.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pye VE, Rosa A, Bertelli C, Struwe WB, Maslen SL, Corey R, Liko I, Hassall M, Mattiuzzo G, Ballandras-Colas A, Nans A, Takeuchi Y, Stansfeld PJ, Skehel JM, Robinson CV, Pizzato M, Cherepanov P. 2020. A bipartite structural organization defines the SERINC family of HIV-1 restriction factors. Nat Struct Mol Biol 27:78–83. 10.1038/s41594-019-0357-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Timilsina U, Umthong S, Lynch B, Stablewski A, Stavrou S. 2020. SERINC5 potently restricts retrovirus infection in vivo. mBio 11:e00588-20. 10.1128/mBio.00588-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heigele A, Kmiec D, Regensburger K, Langer S, Peiffer L, Stürzel CM, Sauter D, Peeters M, Pizzato M, Learn GH, Hahn BH, Kirchhoff F. 2016. The potency of Nef-mediated SERINC5 antagonism correlates with the prevalence of primate lentiviruses in the wild. Cell Host Microbe 20:381–391. 10.1016/j.chom.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chande A, Cuccurullo EC, Rosa A, Ziglio S, Carpenter S, Pizzato M. 2016. S2 from equine infectious anemia virus is an infectivity factor which counteracts the retroviral inhibitors SERINC5 and SERINC3. Proc Natl Acad Sci U S A 113:13197–13202. 10.1073/pnas.1612044113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwinn MK, Machleidt T, Zimmerman K, Eggers CT, Dixon AS, Hurst R, Hall MP, Encell LP, Binkowski BF, Wood KV. 2018. CRISPR-mediated tagging of endogenous proteins with a luminescent peptide. ACS Chem Biol 13:467–474. 10.1021/acschembio.7b00549. [DOI] [PubMed] [Google Scholar]
- 16.Sharma S, Lewinski MK, Guatelli J. 2018. An N-glycosylated form of SERINC5 is specifically incorporated into HIV-1 virions. J Virol 92:e00753-18. 10.1128/JVI.00753-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang X, Zhou T, Yang J, Lin Y, Shi J, Zhang X, Frabutt DA, Zeng X, Li S, Venta PJ, Zheng Y-H. 2017. Identification of SERINC5-001 as the predominant spliced isoform for HIV-1 restriction. J Virol 91:e00137-17. 10.1128/JVI.00137-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dai W, Usami Y, Wu Y, Gottlinger H. 2018. A long cytoplasmic loop governs the sensitivity of the anti-viral host protein SERINC5 to HIV-1 Nef. Cell Rep 22:869–875. 10.1016/j.celrep.2017.12.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stoneham CA, Ramirez PW, Singh R, Suarez M, Debray A, Lim C, Jia X, Xiong Y, Guatelli J. 2020. A conserved acidic-cluster motif in SERINC5 confers partial resistance to antagonism by HIV-1 Nef. J Virol 94:e01554-19. 10.1128/JVI.01554-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Passos V, Zillinger T, Casartelli N, Wachs AS, Xu S, Malassa A, Steppich K, Schilling H, Franz S, Todt D, Steinmann E, Sutter K, Dittmer U, Bohne J, Schwartz O, Barchet W, Goffinet C. 2019. Characterization of endogenous SERINC5 protein as anti-HIV-1 factor. J Virol 93:e01221-19. 10.1128/JVI.01221-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cavrois M, De Noronha C, Greene WC. 2002. A sensitive and specific enzyme-based assay detecting HIV-1 virion fusion in primary T lymphocytes. Nat Biotechnol 20:1151–1154. 10.1038/nbt745. [DOI] [PubMed] [Google Scholar]
- 22.Tokunaga K, Greenberg ML, Morse MA, Cumming RI, Lyerly HK, Cullen BR. 2001. Molecular basis for cell tropism of CXCR4-dependent human immunodeficiency virus type 1 isolates. J Virol 75:6776–6785. 10.1128/JVI.75.15.6776-6785.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mochizuki N, Otsuka N, Matsuo K, Shiino T, Kojima A, Kurata T, Sakai K, Yamamoto N, Isomura S, Dhole TN, Takebe Y, Matsuda M, Tatsumi M. 1999. An infectious DNA clone of HIV type 1 subtype C. AIDS Res Hum Retroviruses 15:1321–1324. 10.1089/088922299310223. [DOI] [PubMed] [Google Scholar]
- 24.Anjana R, Vaishnavi MK, Sherlin D, Kumar SP, Naveen K, Kanth PS, Sekar K. 2012. Aromatic-aromatic interactions in structures of proteins and protein-DNA complexes: a study based on orientation and distance. Bioinformation 8:1220–1224. 10.6026/97320630081220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lanzarotti E, Biekofsky RR, Estrin DA, Marti MA, Turjanski AG. 2011. Aromatic–aromatic interactions in proteins: beyond the dimer. J Chem Inf Model 51:1623–1633. 10.1021/ci200062e. [DOI] [PubMed] [Google Scholar]
- 26.Staropoli I, Dufloo J, Ducher A, Commere PH, Sartori-Rupp A, Novault S, Bruel T, Lorin V, Mouquet H, Schwartz O, Casartelli N. 2020. Flow cytometry analysis of HIV-1 Env conformations at the surface of infected cells and virions: role of Nef, CD4, and SERINC5. J Virol 94:e01783-19. 10.1128/JVI.01783-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu Y, Olety B, Weiss ER, Popova E, Yamanaka H, Göttlinger H. 2019. Potent enhancement of HIV-1 replication by Nef in the absence of SERINC3 and SERINC5. mBio 10:e01071-19. 10.1128/mBio.01071-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol 59:284–291. 10.1128/JVI.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ozono S, Zhang Y, Tobiume M, Kishigami S, Tokunaga K. 2020. Super-rapid quantitation of the production of HIV-1 harboring a luminescent peptide tag. J Biol Chem 295:13023–13030. 10.1074/jbc.RA120.013887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tada T, Zhang Y, Koyama T, Tobiume M, Tsunetsugu-Yokota Y, Yamaoka S, Fujita H, Tokunaga K. 2015. MARCH8 inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins. Nat Med 21:1502–1507. 10.1038/nm.3956. [DOI] [PubMed] [Google Scholar]
- 31.Shimura K, Nameki D, Kajiwara K, Watanabe K, Sakagami Y, Oishi S, Fujii N, Matsuoka M, Sarafianos SG, Kodama EN. 2010. Resistance profiles of novel electrostatically constrained HIV-1 fusion inhibitors. J Biol Chem 285:39471–39480. 10.1074/jbc.M110.145789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu QX, Barry AP, Wang ZX, Connolly SM, Peiper SC, Greenberg ML. 2000. Evolution of the human immunodeficiency virus type 1 envelope during infection reveals molecular corollaries of specificity for coreceptor utilization and AIDS pathogenesis. J Virol 74:11858–11872. 10.1128/jvi.74.24.11858-11872.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nomaguchi M, Doi N, Adachi A. 2014. Virological characterization of HIV-2 vpx gene mutants in various cell systems. Microbes Infect 16:695–701. 10.1016/j.micinf.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 34.Tobiume M, Lineberger JE, Lundquist CA, Miller MD, Aiken C. 2003. Nef does not affect the efficiency of human immunodeficiency virus type 1 fusion with target cells. J Virol 77:10645–10650. 10.1128/jvi.77.19.10645-10650.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ueno T, Idegami Y, Motozono C, Oka S, Takiguchi M. 2007. Altering effects of antigenic variations in HIV-1 on antiviral effectiveness of HIV-specific CTLs. J Immunol 178:5513–5523. 10.4049/jimmunol.178.9.5513. [DOI] [PubMed] [Google Scholar]
- 36.Toyoda M, Kamori D, Tan TS, Goebuchi K, Ohashi J, Carlson J, Kawana-Tachikawa A, Gatanaga H, Oka S, Pizzato M, Ueno T. 2020. Impaired ability of Nef to counteract SERINC5 is associated with reduced plasma viremia in HIV-infected individuals. Sci Rep 10:19416–19416. 10.1038/s41598-020-76375-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pizzato M, Erlwein O, Bonsall D, Kaye S, Muir D, McClure MO. 2009. A one-step SYBR green I-based product-enhanced reverse transcriptase assay for the quantitation of retroviruses in cell culture supernatants. J Virol Methods 156:1–7. 10.1016/j.jviromet.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 38.Maeda H, Matsushita S, Eda Y, Kimachi K, Tokiyoshi S, Bendig MM. 1991. Construction of reshaped human antibodies with HIV-neutralizing activity. Hum Antibodies Hybridomas 2:124–134. 10.3233/HAB-1991-2302. [DOI] [PubMed] [Google Scholar]
- 39.Tsirigos KD, Peters C, Shu N, Käll L, Elofsson A. 2015. The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic Acids Res 43:W401–W407. 10.1093/nar/gkv485. [DOI] [PMC free article] [PubMed] [Google Scholar]